Abstract
Dentin matrix protein-1 (DMP1), bone sialoprotein (BSP), and osteopontin (OPN) are three SIBLINGs (small integrin-binding ligand, N-linked glycoproteins) co-expressed/secreted by skeletal and active ductal epithelial cells. Although etiological mechanisms remain unclear, DMP1 is the only one of these three genes currently known to have mutations resulting in human disease, and yet it remains the least studied. All three contain the highly conserved integrin-binding tripeptide, RGD, and experiments comparing the cell attachment and haptotactic migration-enhancing properties of DMP1 to BSP and OPN were performed using human skeletal (MG63 and primary dental pulp cells) and salivary gland (HSG) cells. Mutation of any SIBLING's RGD destroyed all attachment and migration activity. Using itsαVβ5 integrin, HSG cells attached to BSP but not to DMP1 or OPN. However, HSG cells could not migrate onto BSP in a modified Boyden chamber assay. Expression of αVβ3 integrin enhanced HSG attachment to DMP1 and OPN and promoted haptotactic migration onto all three proteins. Interchanging the first four coding exons or the conserved amino acids adjacent to the RGD of DMP1 with corresponding sequences of BSP did not enhance the ability of DMP1 to bindαVβ5. For αVβ3-expressing cells, intact DMP1, its BMP1-cleaved C-terminal fragment, and exon six lacking all post-translational modifications worked equally well but the proteoglycan isoform of DMP1 had greatly reduced ability for cell attachment and migration. The sequence specificity of the proposed BMP1-cleavage site of DMP1 was verified by mutation analysis. Direct comparison of the three proteins showed that cells discriminate among these SIBLINGs and among DMP1 isoforms.
Dentin matrix protein-1 (DMP1)2 is a member of the small integrin-binding ligand, N-linked glycoprotein (SIBLING) family, which also includes bone sialoprotein (BSP), osteopontin (OPN), dentin sialophosphoprotein (DSPP), and matrix extracellular phosphoglycoprotein (MEPE) (1, 2). SIBLINGs share many common features, such as their tandem gene location on human chromosome 4q21, similar intron/exon organization, multiple phosphorylation sites, disulfide-free flexible hydrophilic structures, and the presence of the integrin-binding tripeptide, arginine-glycine-aspartate (RGD) (1, 2). These proteins have long been thought to play key biological roles during the synthesis of mineralized dentin and bone matrices (3, 4). DMP1 was first found in odontoblasts (5), however, recent studies revealed that, like all other SIBLINGs, it is also expressed in specific soft tissues, including many tumors (2) as well as in normal, metabolically active ductal epithelial cells such as salivary gland, sweat gland, and kidney (6–8). Although DMP1 has been proposed to be involved in regulating tumor cell migration (9), its function in tumors as well as normal ductal cells remains elusive. The importance of DMP1 in the processes of mineralization is supported by observations that the DMP1-knockout mouse has defective dentin development, hypomineralization, and rickets (10, 11) and was associated with severe hypophosphatemia, phosphaturia, and increased levels of the phosphate-regulating hormone, FGF23 (11). Human genetic studies have connected mutations in DMP1 with the autosomal-recessive forms of hypophosphatemic rickets in which blood levels of FGF23 are elevated (11, 12). Thus, DMP1 may be involved directly or indirectly in controlling the production of the bone-derived hormone, FGF23. However, the underlying mechanisms of DMP1 action remain to be explained.
DMP1 has several interesting structural and functional domains, including several acidic domains rich in phosphoserine, Glu, and Asp, that are reported to serve as putative sites for calcium deposition and hydroxyapatite crystal nucleation (3–5). DMP1, like most if not all SIBLINGs (2), is susceptible to proteolytic cleavage resulting in the generation of specific N- and C-terminal fragments (13). In analogy to another SIBLING, DSPP, it has been suggested that the BMP1-cleavage fragments represent the active form(s) of DMP1, whereas the full-length form may be a non-functional precursor (13). The bone morphogenetic protein-1 (BMP1)/Tolloid-like metalloproteinases were proposed to be the enzyme group responsible for cleaving between the conserved Ser212 and Asp213 residues in mouse DMP1 (14). DMP1 exists also as a proteoglycan form with the glycosaminoglycan (GAG) chain being attached at a highly conserved Ser-Gly motif in the N-terminal domain (15). The well known integrin-binding tripeptide, Arg-Gly-Asp (RGD), (5, 16) is one of the most highly conserved motifs for DMP1. Ligand binding of integrins often triggers intracellular signals that control many aspects of cell behavior, including differentiation, formation of focal adhesion complexes, migration, and apoptosis (17, 18). It has been shown that, depending on cell and tissue type, DMP1 is capable of supporting cell attachment via the RGD motif (19). However, the specific integrin(s) involved in this cellular interaction with DMP1 have not been identified. Therefore, we investigated the integrins involved in binding to specific DMP1 isoforms compared with two other more highly studied SIBLING proteins, BSP and OPN, by performing attachment and haptotactic migration assays.
EXPERIMENTAL PROCEDURES
Cell Culture—Human salivary intercalated duct cell line (HSG), established from an irradiated human salivary gland (20), was propagated in DMEM/F-12 medium (Invitrogen) supplemented with 5% fetal bovine serum (FBS). Primary human dental pulp cells (DPC) were obtained from human third molars collected from adults at the Dental Clinic of the NIDCR, National Institutes of Health (NIH) under approved guidelines set by the NIH Office of Human Subjects Research as described previously (21) and were cultured in α-minimal essential medium (Invitrogen) supplemented with 20% FBS (v/v) and ascorbic acid (100 μm). The DPC were a generous gift from Drs. Sayuri Yoshizawa and Pamela Gehron Robey, Craniofacial and Skeletal Diseases Branch/NIDCR/NIH. Human osteosarcoma cells (MG63, ATCC number CRL-1427) and human embryonic kidney 293A (HEK293A) cells were obtained from ATCC (Manassas, VA) and Invitrogen, respectively, and were maintained in DMEM (Invitrogen) containing 10% FBS (v/v). All media were supplemented with l-glutamine (2 mm), penicillin (100 IU/ml), and streptomycin (100 μg/ml). Cells were grown at 37 °C in 5% CO2/air.
Antibodies and Peptides—Monoclonal antibody (mAb) against human α5 integrin subunit (clone 238307) and polyclonal goat anti-human BMP1 antibody (Cat. # AF1927) were from R&D Systems (Minneapolis, MN), mAbs against human integrin subunits αV (clone 272–17E6), β1 (clone 4B7), and β6 (clone 442.5C4) were from EMD Biosciences Calbiochem (La Jolla, CA), mAbs against human αVβ3 (clone LM609), αVβ5 (clone P1F6), and subunit β1 (clone P4C10) were from Chemicon International, Inc. (Temecula, CA), Alexa Flour 488-conjugated F(ab′)2 fragment of goat anti-mouse IgG was from Molecular Probes (Eugene, OR). GRGDS and GRDGS peptides were synthesized and purified at the FDA/NIH Center for Biological Evaluation and Research, Facility for Biotechnology Resources (Bethesda, MD).
Protein Production and Purification—Recombinant human BSP, BSP-KAE, OPN, and OPN-KAE, as well as bovine DMP1 and DMP1-KAE were purified from conditioned medium of human bone marrow stromal cells (BMSCs) (gift of Drs. Pamela Gehron Robey and Serger Kuznetsov) infected with the corresponding adenoviruses as described previously (22, 23). Full-length human DMP1 was made by reverse transcription-PCR of human kidney poly(A)+ RNA (Clontech/BD Biosciences, Palo Alto, CA) with oligonucleotides, including Asp718 (5′ to the natural start codon) and BamHI (3′ to the stop codon) restriction sites for eventual ligation into the prepared adenovirus expression plasmid. An adenoviral expression plasmid containing the DMP1MQΔIH cDNA (Ad-DMP1MQΔIH) was generated by site-directed mutagenesis of the Met215-Gln216 residues into IIe-His of the human DMP1 cDNA. An adenoviral expression plasmid containing the DMP1-BSP-likeRGD sequence (Ad-DMP1-BSP-likeRGD) was similarly produced by site-directed mutagenesis of Ser364 and Pro369 residues into Pro and Tyr, respectively, to match BSP's EPRGDNY conserved sequence. The mutated cDNA forms were generated by using oligonucleotide pairs containing the new sequences and the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). An adenoviral expression plasmid containing the hBSP/bDMP1 hybrid cDNA was generated by first making the human BSP exons 2–5 from the B6-5g cDNA clone (24) by PCR using an oligonucleotide pair for introducing the necessary restriction sites. The BSP portion of the PCR construct begins with the Asp718 restriction site (5′ to the natural start codon) and ends with the 3′-end being an in-frame BsrBI blunt end restriction site that, upon digestion, makes the last codon of exon 5 the end of the construct. The bDMP1 exon 6 was made with a blunt end restriction site of FspI added to the 5′-end (which after digestion leaves the first codon of the last exon as the first bases) and a BamHI after the stop codon on the 3′-end by PCR of the bovine DMP1 cDNA (a gift from Dr. M. J. Dixon, University of Manchester, UK). The two amplicons were gel-purified, digested with Asp718 plus BsrBI (BSP) or BamHI site plus FspI (DMP1), purified, and ligated together. A second PCR was performed on the ligation product using the 5′-BSP oligonucleotide and the 3′-DMP1 oligonucleotide to amplify the ligated, full-length product. The Asp718 and BamHI-treated amplicon was ligated into the prepared adenoviral expression plasmid. All adenovirus expression plasmids were sequenced to confirm the entire sequence. Each adenovirus expression plasmid was recombined with the replication-deficient adenovirus type 5 (Ad5) plasmid in appropriate HEK293A cells, selected, amplified, and purified for use. In each case, the mRNA transcription was under the control of the strong cytomegalovirus (CMV) promoter (except for the normal BSP construct, which was under the control of the EF-1 promoter). The recombinant proteins were produced by infecting midpassage, subconfluent normal human BMSCs. Proteins (usually representing ∼50% of the total protein in the serum-free media) were purified from the conditioned serum-free media to >95% purity by means of anion exchange chromatography, as described previously (22, 23).
The bacterial recombinant human DMP1/exon 6 was generated by PCR of human genomic DNA with oligonucleotides containing in-frame NdeI (5′) and BamHI (3′) for ligation into pET-15B (Novagen/EMD Biosciences, La Jolla, CA). Competent BL21(DE3) Escherichia coli cells were transformed with the plasmid and grown to mid log phase, and the fusion protein was stimulated with isopropyl β-d-thiogalactopyranoside (Novagen) for 1 h. DMP1exon6 protein was purified on His-Bind (Novagen) resin using standard denaturing (urea) conditions followed by dialysis against water and freeze drying. The purity of all proteins was analyzed by electrophoresing in SDS on NuPAGE 4–12% Bis-Tris gels, followed by staining with SimplyBlue SafeStain (Invitrogen) and StainsAll (Eastman Kodak Co., Rochester, NY).
Adenoviral Constructs for Integrin and BMP1 Expression—The coding domain of human β3 integrin was PCR-amplified from a pGEM-1/β3 construct containing human β3 integrin cDNA (a gift from Dr. Kenneth Yamada, NIDCR, NIH, Bethesda, MD). The coding sequence of human αV integrin was PCR-amplified from human αV plasmid (IOH38619, Invitrogen). Each cDNA was subcloned into the pVQ-CMV-K-NpA Ad5 vector (ViraQuest, North Liberty, IA) under control of the CMV promoter, and the resulting plasmids were converted into the adenovirus form, amplified, and purified by ViraQuest according to their protocols. Adenoviruses bearing full-length cDNA for human integrin β1 (IOH21479) and β5 (IOH57077) were made using the ViraPower™ Adenoviral Expression System (all from Invitrogen) according to the manufacturer's protocol. Briefly, the coding domain of integrin β1 and β5 flanked by attL sites in a Gateway entry vector (Invitrogen) were recombined into the ViraPower™ pAd/CMV/V5-Dest™ vector using Gateway LR Clonase. The adenoviruses were produced and amplified in HEK293A cells according to the manufacturer's protocol. An adenovirus expressing human BMP1 (Ad-BMP1) was generated using the splice variant 5 of the human BMP1 cDNA in a Gateway Entry vector (IOH36655, Invitrogen) as described above. The expression of the recombinant BMP1 protein was analyzed in conditioned media from BMSCs infected with the Ad-BMP1 by means of both Western blot using the goat anti-human BMP1 antibody and by noting the processing of fibroblast procollagen on SDS-PAGE with Coomassie Blue staining.
Adenoviral Expression of Integrins and FACS Analysis—3 × 105 cells were seeded in 6-well plates in DMEM/F-12 medium and grown until subconfluent. Viruses were than added in 1.5 ml of DMEM/F-12 medium containing 2% FBS. After 24 h, medium was changed to complete medium. 48 h post infection, cells were harvested by means of Enzyme-Free Cell Dissociation Buffer (Invitrogen), and the expression of integrins was analyzed by flow cytometry. Control, “Empty” Aden5 viruses were purchased from ViraQuest (VQ-013). Harvested cells were washed with PBS and resuspended in cold PBS containing 1% FBS. Cells were then incubated with primary monoclonal antibodies for 30 min on ice followed by a cold wash and incubation with Alexa Flour 488-conjugated F(ab′)2 fragment of goat anti-mouse IgG for 30 min on ice. Fluorescence was measured using FACScan flow cytometer (BD Biosciences, Mountain View, CA) and analyzed using CellQuest software.
Attachment Assay—Wells of 96-well non-tissue culture polystyrene microtiter plates (Cat. #655061, Greiner Bio-One Inc., Longwood, FL) were coated with 100 μl of 100 nm solutions of SIBLINGs or their mutant constructs in PBS overnight at 4 °C. Wells were rinsed with PBS and blocked with 10 mg/ml BSA in RPMI 1640 medium for 1 h at 37 °C. Control wells were coated with only BSA. For in situ chondroitinase ABC digestion, 4 milliunits of the enzyme (Seikagaku America Inc., Ijamsville, MD) in 100 μl of reaction buffer (40 mm Tris-HCl, 40 mm sodium acetate, pH 8.0, and 1 mg/ml BSA) were added to wells previously coated with DMP1MQΔIH-PG, incubated for 2 h at 37 °C, and then washed. Control wells were coated with DMP1MQΔIH-PG and incubated with the enzyme reaction buffer only. Cells were harvested using Enzyme-Free Dissociation Buffer and added to the coated wells at 5 × 104 cells per 50 μl of serum-free RPMI 1640 containing 1 mg/ml BSA in the presence or absence of 0.250–1 mm MnCl2 as noted. For blocking experiments, cells were preincubated with 1 mm GRGDS, 1 mm GRDGS peptide for 15 min at room temperature, or with blocking antibodies as indicated for 30 min at 4 °C. After 1-h incubation at 37 °C, non-adherent cells were removed by washing with PBS. The attached cells were fixed for 10 min with 2% paraformaldehyde, stained for 10 min with 0.5% crystal violet in 20% ethanol, rinsed with water, solubilized in 100 μl of 2% SDS, and quantified by measuring the absorbance at 560 nm in a microtiter plate reader.
Migration Assay—Haptotactic migration assays were performed in 24-well Transwell plates with 8-μm pore size (Cat. #353097, Falcon System Inc., Columbia, MD). The lower surface of the polyethylene terephthalate membranes were pre-coated with 50 μl of 1 μm SIBLING solutions overnight at room temperature. After rinsing with PBS, both sides of the membrane were blocked with 10 mg/ml BSA in PBS for 1 h at 37 °C. 5 × 104 cells in 250 μl of serum-free culture medium were added to the upper chambers while 750 μl of the same medium was added to the bottom chamber and incubated at 37 °C for 19 h (HSG cells) or 5 h (MG63 cells). For blocking experiments, cells were preincubated with 1 mm GRGDS, 1 mm GRDGS, or function-blocking antibodies as described above prior to adding to the Transwells. Cells remaining in the top chamber (non-migrated) were removed from the upper membranes surfaces with cotton swabs, whereas cells that had migrated through the pores were fixed with 2% paraformaldehyde, stained with 100 nm blue-fluorescent 4′,6-diamidino-2-phenylindole nucleic acid stain (Molecular Probes) and quantified by performing cell counts on the underside of the filter on 10 randomly selected fields at 400× magnification.
In Vitro BMP1 Enzyme Digestion—10 μg of recombinant human DMP1 and DMP1MQΔIH mutant protein were separately incubated with 0.5 μg of recombinant BMP1 (R&D Systems) in a 50-μl total volume of reaction buffer (25 mm HEPES, 0.01% Brij, pH 7.5) or reaction buffer alone for 19 h at 37 °C. Reactions were stopped by adding 4× NuPAGE LDS sample buffer (SB, Invitrogen) containing 10 mm dithiothreitol and heated for 10 min at 70 °C. Samples were electrophoresed in SDS on NuPAGE 4–12% Bis-Tris gels, and proteins were visualized with StainsAll as previously described (25).
BMP1-mediated DMP1 Processing in Vivo—HSG cells (4 × 104) were seeded in 96-well cell culture plates (Falcon System Inc.), and the next day infected with a single fixed dose of viruses expressing DMP1 or DMP1MQΔIH mutant protein, some in combination with increasing doses of the BMP1 adenovirus described above. After 3 days of incubation, samples of serum-free conditioned media (DMEM/F-12) were electrophoresed in SDS on NuPAGE 4–12% Bis-Tris gels and stained with StainsAll.
Verification of BMP1 Cleavage Site by Amino Acid Sequence Analysis—100 μg of purified recombinant DMP1 was incubated with or without 5 μg of recombinant BMP1 at 37 °C for 19 h in a 100-μl total volume of reaction buffer. Enzyme was removed by applying the samples to ∼0.2 ml minicolumns of Q-Sepharose Fast Flow beads (Amersham Biosciences) and washed with PBS. DMP1 and fragments were eluted with 0.4 m NaCl in PBS. Products were resolved on a SDS NuPAGE 4–12% Bis-Tis gel and stained with SimplyBlue SafeStain until faint bands were just detected. Intact and C-terminal DMP1 protein bands were excised from the gels, transferred to a D-Tube Dialyzer (Novagen/EMD Chemicals, Inc., Gibbstown, NJ), and electroeluted according to the manufacturer's instructions. The N-terminal amino acid sequences were determined by automated Edman degradation at the NIH Facility for Biotechnology Resources (FDA) using the Procise ABI Model 494A Protein Sequencing System.
Statistical Analysis—All statistical analyses were performed using GraphPad software (San Diego, CA). Multiple comparisons were performed with a one-way analysis of variance followed by Newman-Keuls test, which compares all pairs of columns. Pairwise comparisons were carried out by using the non-parametric two-sided t test for unpaired observations. Results are expressed as mean ± S.E. and were considered statistically different at p < 0.05.
RESULTS
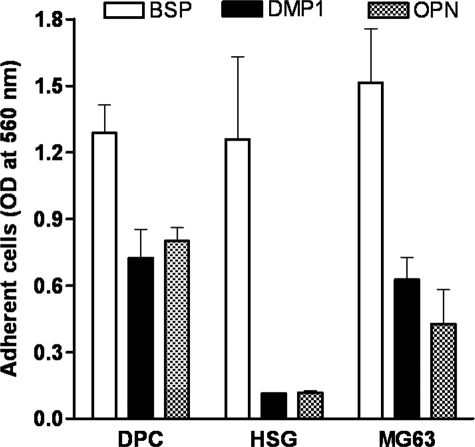
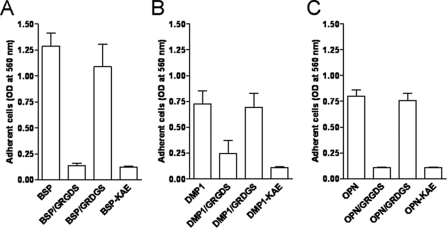
Differential Attachment of DPC, HSG, and MG63 Cells to DMP1, BSP, and OPN—The ability to attach to DMP1-coated plastic has been shown to be cell-type-specific and RGD-dependent (19), but the one or more integrin receptors involved are not known. A panel of cells (bone, tooth, and salivary gland-related cells) was used to test their ability to attach to DMP1 compared with two more frequently studied SIBLINGs, BSP and OPN. DPC, HSG, and MG63 cells were plated on wells coated with purified recombinant DMP1, BSP, OPN, or their respective RGD-inactivated control proteins, DMP1-KAE, BSP-KAE, or OPN-KAE. The SIBLINGs used in these experiments were recombinant proteins purified from human BMSC-conditioned media after infection with the appropriate adenoviral constructs (22, 23). These cells have the ability to add each of the types of post-translational modification reported to be found on the various SIBLING members, although the exact combination of such modifications on the BMSC-synthesized recombinant proteins have not been verified to match any one natural tissue source completely (supplemental Fig. S1 shows evidence for the presence of CS/DS glycosaminoglycan chains, N-linked and O-linked oligosaccharide chains, as well as sialic acid, phosphate, and tyrosine sulfate on various SIBLING members). The purified recombinant DMP1 protein used in this experiment was a mixture of the full-length protein as well as some of its cleavage products of ∼52 kDa and ∼33 kDa, as determined by gel electrophoresis. The sizes of these cleavage products were similar to the previously described 57- and 37-kDa fragments found in rat dentin (13) and corresponded to the C- and N-terminal fragments of DMP1, respectively. All three cell types showed a similar robust attachment to BSP with the majority of the plated cells attaching to the substratum within 1 h of incubation at 37 °C (Fig. 1). Fewer DPC and MG63 cells attached to DMP1- or OPN-coated wells, whereas HSG cells failed to attach to either DMP1 or OPN (Fig. 1). Attachment by all cells was found to be RGD-dependent, because both a soluble GRGDS peptide as well as each SIBLING's respective RGD-inactivated mutants (KAE) totally abolished cell attachment. Representative results are shown for the DPC cells in Fig. 2.
FIGURE 1.
Differential attachment of human dental pulp cells (DPC), salivary gland cells (HSG), and osteoblastic cells (MG63) to bone sialoprotein (BSP)-, dentin matrix protein-1 (DMP1)-, or osteopontin (OPN)-coated wells. Single-cell suspensions of DPC, HSG, and MG63 cells were incubated for 1 h on non-tissue culture wells precoated with BSP, DMP1, or OPN. Note that neither DMP1 nor OPN can support attachment of HSG cells, whereas all three cell types attach well to BSP. No significant number of cells attached to BSA-coated wells (not shown). The adherent cells were fixed, stained with crystal violet, and solubilized with SDS before reading at 560 nm. Bars show the mean ± S.E. from at least three independent experiments, each performed in triplicate.
FIGURE 2.
Cell attachment to BSP, DMP1, and OPN is RGD-dependent. DPCs were plated on wells coated with BSP (A), DMP1 (B), and OPN (C) or their respective KAE mutants. Noted cells were preincubated with 1 mm GRGDS competing peptide or the scrambled control peptide GRDGS for 15 min at room temperature prior to adding to wells. The attachment assays were performed and evaluated as described above. Bars show the mean ± S.E. from six independent experiments, each performed in triplicate.
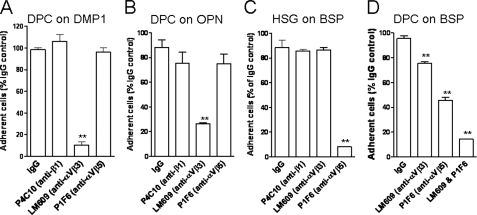
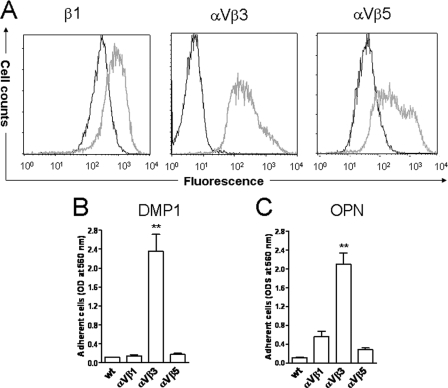
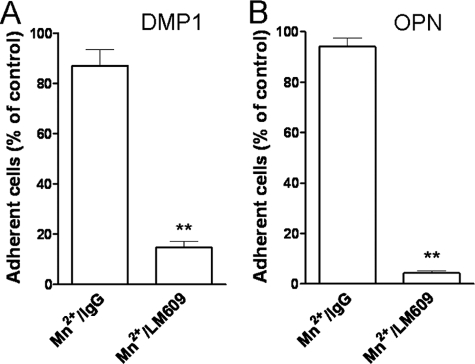
Cell Attachment to DMP1 and OPN Is Mediated through αVβ3 Integrin, whereas Attachment to BSP Can Be Promoted by Either αVβ3 or αVβ5 Integrin—The analysis of cell surface expression of various RGD-dependent integrins in DPC, HSG, and MG63 cells by means of flow cytometry revealed: 1) all three cell types had a strong cell surface expression of αV and β1 integrin subunits; 2) all showed moderate expression of αVβ5 integrin as well as α5 subunit; 3) all three were weakly positive for β6 integrin subunit; whereas 4) αVβ3 integrin receptor was found in MG63 and DPC but not in HSG cells (Table 1). Due to the observation that HSG cells attached to BSP but not to DMP1 or OPN, the latter two were hypothesized to use αVβ3 while BSP may have been able to utilize one or more additional integrins for attachment. To investigate this hypothesis, attachment assays were performed in the presence of function-blocking antibodies. The attachment of cells to DMP1 as well as OPN were found to be αVβ3-dependent, because the anti-αVβ3 monoclonal antibody (mAb), LM609, inhibited most cell attachment to DMP1 (10% of the IgG control, p < 0.001) and OPN (26% of the IgG control, p < 0.001) (Fig. 3, A and B). The anti-β1 mAb (P4C10) and the anti-αVβ5 (P1F6) mAb had no significant effects on cell interaction with DMP1 or OPN (Fig. 3, A and B). In contrast, attachment of HSG cells to BSP was efficiently blocked by anti-αVβ5 antibody alone (Fig. 3C). To effectively block DPC cell attachment to BSP, a combination of both anti-αVβ3 and anti-αVβ5 antibodies was required (Fig. 3D). Based on these results, we hypothesized that cells can use αVβ3 or αVβ5 integrins to attach to BSP while only αVβ3 integrin is effective for attachment to DMP1 or OPN. To verify that αVβ3 integrin is the major receptor mediating cell attachment to DMP1, HSG cells were infected with two adenoviral constructs expressing cDNAs for human αV and β3. As controls, some cells were infected with adenoviral constructs to overexpress αVβ1 and αVβ5 integrins. 48 h after infections, corresponding strong de novo cell surface expressions of αVβ3 integrin and overexpression of αVβ5 as well as the β1 integrin subunit were observed (Fig. 4A). To determine whether the newly acquired integrin expression changed cell attachment to DMP1 or OPN, the parental cells (wt), αVβ3-, αVβ1-, and αVβ5-expressing HSG cells were allowed to attach to immobilized DMP1 or OPN. As shown in Fig. 4 (B and C), HSG cells were able to attach to DMP1- or OPN-coated wells only when expressing αVβ3 integrin. The αVβ1- and αVβ5-infected cells failed to attach to DMP1 and showed only an insignificant increase in attachment to OPN. This confirmed that neither αVβ1 nor αVβ5 integrins are relevant for in vitro cell attachment to DMP1 or OPN under our conditions. The attachment of αVβ3-expressing HSG cells to DMP1- or OPN-coated wells was blocked by soluble GRGDS peptide and was missing entirely in wells coated with the DMP1-KAE or OPN-KAE mutant proteins (supplemental Fig. S2A). Finally, attachment of αVβ3-infected HSG cells to DMP1- or OPN-coated wells was reduced to 24% or 8%, respectively, compared with IgG control (supplemental Fig. S2, B and C), strongly supporting the hypothesis that both DMP1 and OPN share the same integrin receptor-binding selectivity for αVβ3 during cell attachment in vitro.
TABLE 1.
Summary of cell surface expression of selected RGD-dependent integrins in DPC, HSG, and MG63 cells as assessed by flow cytometry Monoclonal antibodies were used against the following integrin complexes or integrin subunits: β1 (4B7), αVβ3 (LM609), αVβ5 (P1F6), β6 (442.5C4), αV (AV1), and α5 (MAB1864) and detected with Alexa Flour 488-conjugated F(ab′)2 fragment of goat anti-mouse IgG. The results were scored semiquantitatively on a scale of: –, negative; +, weak; ++, moderate; and +++, strong positive.
| DPC | HSG | MG63 | |
|---|---|---|---|
| β1 | +++ | +++ | +++ |
| αVβ3 | ++ | – | ++ |
| αVβ5 | ++ | ++ | ++ |
| β6 | + | + | + |
| αV | +++ | +++ | +++ |
| α5 | ++ | ++ | ++ |
FIGURE 3.
Cell attachment to DMP1 and OPN requires αVβ3, whereas attachment to BSP can be promoted by either αVβ3 or αVβ5 integrins. Cells, as indicated, were plated on BSP-, DMP1-, or OPN-coated wells. Some cells were pretreated with 10 μg/ml anti-β1(P4C10), anti-αVβ3(LM609), and/or anti-αVβ5(P1F6) function-blocking antibodies prior to and during the incubation. The attachment assays were performed and evaluated as described above. Cell attachment in the presence of IgG control was assigned a value of 100%. Bars show the mean ± S.E. from six independent experiments, each performed in triplicate. **, p < 0.001, as compared with the corresponding IgG control value.
FIGURE 4.
De novo expression ofαVβ3 integrin causes HSG cells to attach to DMP1-coated surfaces. A, cell surface expression of β1 subunit, αVβ3, or αVβ5 integrins on HSG cells before (black profiles) and after transduction with viral construct expressing αV cDNA in combination with viruses expressing either β1(left), or β3 (middle), or β5(right) cDNA (gray profiles) as assessed by flow cytometry using monoclonal antibodies (4B7 for β1, LM609 for αVβ3, and P1F6 for αVβ5) and detected with Alexa Flour 488-labeled goat anti-mouse IgG. The expression level of each integrin on the cell surfaces is indicated by log fluorescence intensity (x-axis). Note that, among parental and adenoviral-infected HSG cells, only those expressing αVβ3 significantly attached to either DMP1 (B)- or OPN (C)-coated wells. Cells infected with an adenovirus encoding no recombinant protein exhibited a profile (not shown) identical to parental cells (wt). The attachment assays were performed and evaluated as described above. Bars show the mean ± S.E. from seven independent experiments, each performed in triplicate. **, p < 0.001, as compared with the control (wt) cells by means of t test.
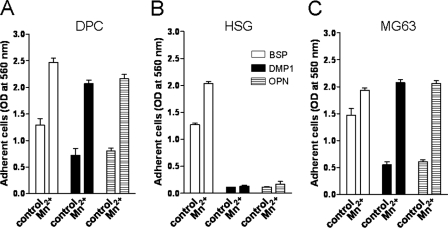
Mn2+-induced Integrin Activation Enhances Cell Attachment—Previous studies have shown that interaction of some integrins with their ligands may be regulated by changes in their activation state, and this may account for some cell type-specific differential responses among cells otherwise expressing the same integrins (26). The physiological stimuli that regulate integrin activity are not well understood, but MnCl2 is widely used to activate many integrins (26–28). Manganese cations appear to bind to the extracellular domain of the integrin and induce a “switchblade-like” opening of the stalks that stabilizes the receptor in a high affinity “ligand occupied-like” conformation (29). To address the question whether integrin activation may regulate cell attachment to DMP1 (as well as to OPN and BSP), DPC, HSG, and MG63 cells were treated with 0.25–1.0 mm MnCl2 during their incubation on SIBLING-coated wells. As was previously the case, BSP was the best at supporting cell attachment of all three cell types, and Mn2+ treatment resulted in only modest increases of 1.9-, 1.5-, and 1.3-fold over the untreated control in DPC, HSG, and MG63 cells, respectively (Fig. 5). The Mn2+-mediated cell attachment to DMP1 was more dramatic with 3- and 3.5-fold increases for DPC and MG63 cells, respectively (Fig. 5, A and C). The Mn2+-stimulated attachment on OPN was comparable with that of DMP1 showing 2.8- and 3.5-fold induction in DPC and MG63 cells, respectively (Fig. 5, A and C). Fig. 6 shows that the blocking antibodies for αVβ3 integrin continue to be effective in stopping the attachment of the Mn2+-activated MG63 cells to DMP1 and OPN reaffirming the integrin specificity of this activation event in these cells. In contrast, Mn2+ did not have any effect on HSG cell attachment to either DMP1 or OPN (Fig. 5B). This shows that even in the presence of an activating reagent, αVβ5 integrin (or any other integrins that HSG cell possess) cannot promote attachment to either DMP1 or OPN in vitro.
FIGURE 5.
Integrin-activation by treatment with Mn2+ enhances cell attachment on all three SIBLINGs. Cells, as indicated, were plated on wells precoated with BSP, DMP1, or OPN with or without 1 mm MnCl2 (DPC and HSG cells) or 0.25 mm MnCl2 (MG63 cells). The attachment assays were performed and evaluated as described above. The bars show the mean ± S.E. of three experiments, each performed in triplicate.
FIGURE 6.
Mn2+-mediated induction of cell attachment to DMP1 and OPN is αVβ3-dependent. Single-cell suspension of MG63 cells was incubated with control IgG or the function blocking anti-αVβ3 antibody (LM609) in the presence of 0.25 mm MnCl2 before plating on wells coated with either DMP1 (A) or OPN (B). The attachment assays were performed and evaluated as described above. Attachment of cells in the presence of IgG control was assigned a value of 100%. The bars show the mean ± S.E. of four experiments each performed in triplicate. **, p < 0.001, as compared with the corresponding IgG control value by means of t test.
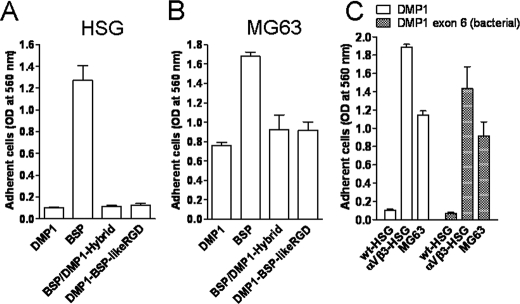
Defining the Biochemical Basis of Integrin Receptor Specificity—As shown previously (27, 30) and verified specifically for the three cell types described above, BSP was capable of utilizing both αVβ3 and αVβ5 integrin receptors for attachment. Both bovine and human DMP1, however, required αVβ3 to support cell attachment and could not use αVβ5. To investigate the underlying elements of the DMP1 protein that may be contributing to its ability to selectively bind αVβ3 and not αVβ5 integrin, two DMP1/BSP hybrid proteins were made and tested for their ability to support attachment through αVβ5 and αVβ3. First, it was noted that specific amino acids flanking the RGD tripeptide of DMP1 were conserved among many different species (e.g. human and opossum). BSP also had conserved amino acids at those same positions (relative to RGD), but they differ from DMP1 (SRGDNP for DMP1 and PRGDNY for BSP). Substituting the Ser363 and the Pro368 residues flanking the DMP1 RGD tripeptide with Pro and Tyr, respectively, resulted in a BSP-like RGD domain within DMP1. HSG cells, however, continued to fail to attach to the DMP1 containing the BSP-like RGD domain (Fig. 7A), suggesting that, although the RGD is necessary for integrin binding, the amino acids immediately flanking the tripeptide motif are not sufficient to endow the DMP1 with ability to bind to αVβ5 integrin. Next, the role of some non-RGD protein components in determining the integrin receptor specificity were investigated by substituting the first four (of five) coding exons of mature DMP1 with those from BSP to generate a BSP/DMP1 protein hybrid. The BSP/DMP1 hybrid protein permitted MG63 cells to attach but failed to promote the BSP-like attachment properties for HSG cells (Fig. 7, A and B) showing that the αVβ5-permissive binding displayed by BSP is not encoded within the first four exons. Finally, to investigate if post-translational modifications within the remaining exon (which encodes 90% of the mature protein, including the RGD motif) are directly involved in this integrin specificity, DMP1's exon 6 (with a short fusion peptide) was expressed in E. coli. This protein, lacking all carbohydrate, phosphate, and other post-translational modifications, is shown in Fig. 7C to retain the ability to support MG63 but not HSG cell attachment (unless the HSG cells are made to express αVβ3) thus proving that the integrin-selective property of DMP1 did not require the eukaryotic modifications. Therefore, further experiments are necessary for more precise, residue-by-residue understanding of the receptor binding specificity of DMP1 and the biochemical basis of the more integrin promiscuous BSP.
FIGURE 7.
Investigating DMP1 sequences involved in integrin specificity. HSG (naturally expressing αVβ5 integrin), HSG-αVβ3 (expressing αVβ5 plus adenovirus-transduced αVβ3 integrins), and MG63 (expressing both αVβ3 and αVβ5 integrins) cells as noted were plated on wells coated with native forms of BSP or DMP1, or one the following DMP1 constructs: BSP/DMP1-Hybrid (DMP1 with the first four coding exons of BSP fused to the last exon of DMP1); DMP1-BSP-likeRGD (DMP1 with its conserved SRGDNP sequence replaced by BSP's conserved PRGDNY); DMP1 exon 6 (bacterial), truncated DMP1 protein without post-translational modification. Note that neither the first four exons nor the RGD domain of BSP replacing DMP1's corresponding regions resulted in αVβ5-attachment activity for HSG (A). Attachment of MG63 cells to BSP/DMP1 hybrid remained DMP1-like (B). Post-translational modifications are not required for DMP1's specificity for αVβ3(C). The attachment assays were carried out and evaluated as described above. The bars show the mean ± S.E. of three experiments each performed in triplicate.
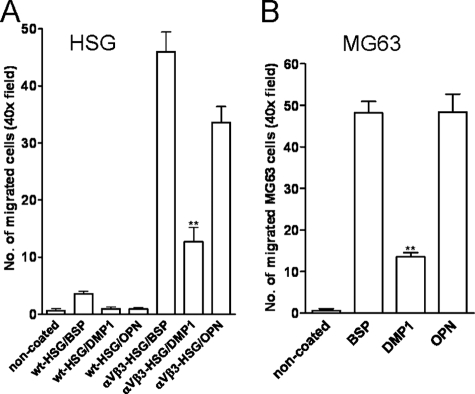
Haptotactic Cell Migration on SIBLINGs Depends on αVβ3 Expression—After attachment, migration can be another vital cellular response involving integrin engagement. Therefore, DMP1 was tested to see if it differed in its support of haptotactic cell migration in comparison to BSP and OPN, and to determine which integrin receptor(s) contributed to this process. To this end, modified Boyden chambers were used in which the bottoms of the 8-μm pore membranes were coated with SIBLINGs (or dilution buffer alone). Cells with the proper complement of receptors can migrate directionally up an adhesion gradient of substrate-bound attractant or, in the simplest case, from an area lacking a ligand to an adjacent region that has been coated with an appropriate protein. This cell movement is referred to as haptotaxis and was judged by counting the number of cells migrating through the pores to the underside of the membranes. HSG cells, which lacked the αVβ3 integrin, failed to migrate onto any of the SIBLING-coated surfaces, including BSP (Fig. 8A). On the other hand, MG63 cells (which endogenously express αVβ3) and αVβ3-transduced HSG cells migrated onto all three SIBLINGs, although to a lower extent onto DMP1 compared with BSP or OPN (Fig. 8, A and B). Migration of cells was significantly reduced by antibodies to αVβ3 confirming the crucial role of αVβ3 for the SIBLING-mediated haptotactic cell migration. For example, the migration of MG63 onto DMP1 was completely blocked by the αVβ3 integrin antibody (data not shown).
FIGURE 8.
αVβ3 integrin is required for haptotactic migration by cells onto any of the three SIBLINGs. Migration permissivity of wt-HSG, αVβ3-HSG (A) or MG63 cells (B) were tested in Transwells for which the bottom of the membranes were coated with the noted SIBLINGs or left uncoated (control) before cells were added to the upper chambers. Cell migration was evaluated by counting the 4′,6-diamidino-2-phenylindole-stained cells on the lower, SIBLINGs-coated surface in random 40× fields after 24 h (HSG) or 5 h (MG63). Native HSG cells (lacking αVβ3) did not migrate onto any SIBLINGs until this integrin was expressed (HSG-αVβ3, A). Note that DMP1 always supported significantly lower levels of migration than BSP or OPN. The bars show the mean from 10 40× fields ± S.E. of a representative experiment. **, p < 0.001 compared with cell migrated on BSP or OPN by using the t test.
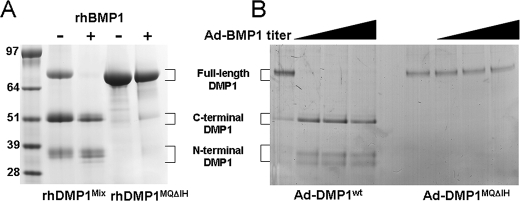
Mutation of Met215-Gln216 to IIe-His Residues in the BMP1-cleavage Site Abolished DMP1 Processing—DMP1 has been hypothesized to be cleaved into its C-terminal active fragment and an N-terminal fragment of unknown (or no) activity (3, 13). BMP1, one of the Tolloid-like proteinases known to remove the C-propeptide of type I collagen during fibril formation, has been shown to also cleave DMP1 into two such fragments with the C-terminal fragment starting with a characteristic DDPES protein sequence (14). It has been reported that fibroblasts, particularly human bone marrow stromal cells, result in good post-translational modifications of DMP1, BSP, and OPN (22, 23). However, like most fibroblastic cells, these cells make a type I collagen matrix and therefore also produce BMP1-like activity, which can result in significant portions of the recombinant DMP1 being cleaved into the ∼52-kDa C-terminal and ∼33-kDa N-terminal fragments (Fig. 9A). These fragments correspond to previously reported 57- and 37-kDa species in rat dentin, respectively (13). Protein microsequencing of gel-purified samples of the ∼52-kDa fragment did result in the expected DDPES sequence, showing that native protease activity in human BMSCs cleave the human DMP1 at the expected, highly conserved site (MQXDDP, where X = G, S, or N in mammals). To both make the full length, well modified recombinant DMP1 and to check the degree of conservation of the BMP1-cleavage motif, a human DMP1 adenovirus was made in which the conserved Met and Gln amino acids of the MQSDDP motif were changed to the chemically similar IHSDDP. Infection of BMSCs with this adenoviral construct (DMP1MQΔIH) resulted in secretion of full-length DMP1MQΔIH only, showing that the MQ portion of the motif is required for processing of DMP1 in normal BMSCs. To verify that the Met215-Gln216 to IIe-His mutation was responsible for the lack of susceptibility of DMP1 to cleavage by authentic BMP1, both native DMP1 and the DMP1MQΔIH were digested with recombinant BMP1 in vitro. Although native DMP1 was cleaved by rBMP1, the DMP1MQΔIH protein remained intact (Fig. 9A). As shown in Fig. 9B, non-mutated DMP1 was also efficiently processed in vivo by overexpressing the BMP1 enzyme in HSG cells. The C-terminal band processed by the co-expressed BMP1 had the expected DDPES amino terminus. The DMP1MQΔIH mutant, however, remained unprocessed in the HSG cells even when increased levels of BMP1 were expressed. These results confirmed that human BMP1 cleaves DMP1 at the predicted site and showed for the first time that one or both of the conserved MQ amino acids at the start of the motif are absolutely required for this activity in vitro and in vivo.
FIGURE 9.
Changing of the Met215-Gln216 to the chemically similar IIe-His in DMP1's BMP1-cleavage site completely abolished processing in vitro and in vivo. A, in vitro processing. Recombinant DMP1Mix (containing full-length as well as the BMP1-like cleavage fragments from original production of protein in human bone marrow stromal cells, lane 2) is fully processed upon addition of rBMP1 (lane 3) DMP1MQΔIH (full-length protein with a MQ-to-IH mutation in BMP1-cleavage site, lane 4) was not digested by rBMP1 (lane 5). B, in vivo processing. HSG cells infected with adenoviruses expressing DMP1 (left lanes) shows a small amount of endogenous BMP1-like cleavage activity (first lane) and complete digestion with even lowest dose of BMP1 adenovirus (lane 2). DMP1MQΔIH mutant co-infected without (first of the right lanes) and even the highest dose of BMP1 adenovirus (last lane) showed no processing of the cleavage-site mutant protein. 48 h post-infection conditioned media were electrophoresed with SDS on a 4–12% NuPAGE before detection with StainsAll.
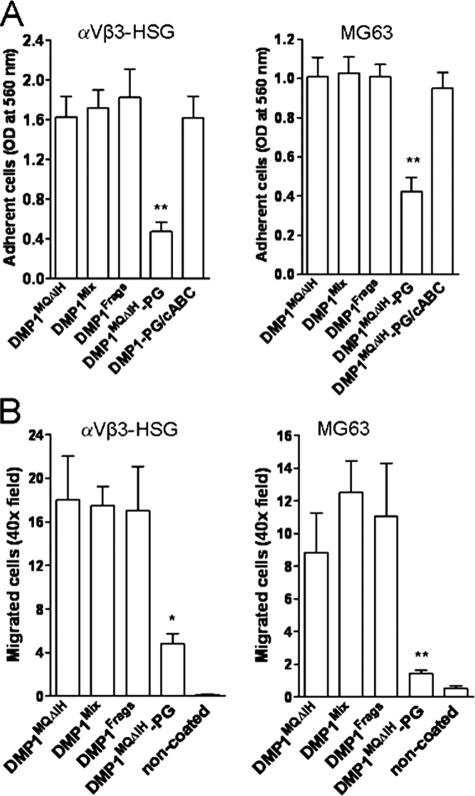
Comparison of Different DMP1 Isoforms in Functional Assays—To determine whether the different DMP1 isoforms differ in attachment/migration functions, the full-length DMP1 (DMP1MQΔIH), the fragments fully processed by recombinant BMP1 in vitro (DMP1Frags), as well as the proteoglycan form of the full-length DMP1 (DMP1MQΔIH-PG) were tested using MG63 and αVβ3-infected HSG cells. The native DMP1 (which, due to endogenous BMP1-like activity in BMSC, contained both full-length and fragments) was used as the positive control. Direct enzyme-linked immunosorbent assays performed with the LF-148 polyclonal antibody against human DMP1 (23), confirmed that very similar amounts of each isoform were present on attachment and haptotactic migration surfaces (data not shown). As shown in Fig. 10, there were no significant differences between the full-length and BMP1-processed DMP1 isoforms in their efficacy in promoting cell attachment or haptotactic migration. However, the proteoglycan form of the full-length DMP1 (DMP1MQΔIH-PG) exhibited a markedly lower ability to support MG63 or αVβ3-expressing HSG cell attachment (Fig. 10A), and those cells that did attach failed to spread effectively (not shown). The migration of both cell types onto DMP1MQΔIH-PG was also significantly inhibited (Fig. 10B). Both attachment and spreading were rescued after the GAG chains were removed by an in situ treatment with chondroitinase ABC directly on the pre-coated plastic/membrane surfaces prior to the addition of the cells.
FIGURE 10.
Proteoglycan form of DMP1 has an impaired ability to support cell adhesion and migration in comparison to full-length and BMP1-processed DMP1 fragments. αVβ3-expressing HSG and MG63 cells were tested for their ability to attach (A) and haptotactically migrate (B) onto different DMP1 isoforms: DMP1MQΔIH (full-length protein only due to a mutation in BMP1-cleavage site); DMP1Mix (full-length plus BMP1-like cleavage fragments); DMP1Frags (fully digested by recombinant BMP1); DMP1MQΔIH-PG (proteoglycan form of full length DMP1); DMP1MQΔIH-PG/chondroitinase ABC (proteoglycan form of DMP1MQΔIH-PG after in situ treatment with chondroitinase ABC (cABC)). The attachment and migration assays were performed and evaluated as described above. Notice that the proteoglycans form significantly inhibited both the attachment (A) and haptotactic migration (B) of both αVβ3-expressing cells and that attachment activity was recovered by the removal of the GAG chain in situ by chondroitinase ABC. The bars show the mean ± S.E. of three experiments each performed in triplicate. **, p < 0.001; *, p < 0.01, as obtained for multiple comparisons by using of analysis of variance and the Newman-Keuls test.
DISCUSSION
One major element that defines all of the SIBLING family members, including DMP1, BSP, and OPN, is the integrin-binding tripeptide motif, RGD. Indeed, the conservation of DMP1's RGD motif (actually the entire hexapeptide, SRGDNP, used in our substitution experiments) over the ∼180 million years of evolution since platypus and human divergence, for example, is significant evidence of the importance of the RGD-related cell-binding properties of these proteins. (The only other DMP1 hexapeptide or slightly larger peptide conserved between humans and platypus are three phosphorylation motifs, the GAG-attachment domain, and the C-terminus.) Because several different mature cells types, including osteoblasts, odontoblasts, and the epithelial cells of metabolically active ducts (e.g. salivary gland, kidney, and sweat gland), express several or sometimes all five of the SIBLINGs at the same time (6–8), it has become increasingly important to ask three questions: 1) which integrins interact with each SIBLING protein; 2) do cells distinguish among the SIBLING members; and 3) do cells distinguish among the different isoforms of a single SIBLING member such as DMP1? Although there is little direct evidence that DMP1, BSP, or OPN are associated to any large degree with insoluble matrices other than mineralized surfaces, in vitro cell attachment and migration assays have long been useful tools to answer some integrin-related aspects of these questions for various matrix and matricellular proteins. This report has directly compared some of the integrin-binding properties of various isoforms and mutations of DMP1 with two of the SIBLING family's more highly studied proteins, OPN and BSP.
First, a survey was taken of several sources of cultured cells that were derived from tissues known to express SIBLINGs. Three different cultured human cells, including MG63 (bone), dental pulp cells (DPCs, tooth), and human salivary gland (HSG), were selected because they were found to have different patterns of attachment and migration onto DMP1, OPN, and BSP-coated surfaces. Under these tissue culture conditions, the cells did not express detectable levels of these three proteins as determined by Western blot analysis of whole cell lysates and conditioned media using specific antibodies (data not shown). Note that any small amounts of SIBLINGs (or other cell attachment protein) that the cells may make would be removed during the cell washing procedures prior to the cells being added to the assay. Furthermore, the assays are all short term experiments that limit the time in which the cells may be producing any cell attachment proteins. The MG63 and the DPCs were found to express αVβ3 integrin, and both could attach well to all three SIBLINGs. Furthermore, the αVβ3 integrin appeared to be in different activation states on DPCs and MG63 cells with conversion into an activated conformation by Mn2+ achieving an optimal engagement of DMP1. This provided the first evidence that DMP1, like BSP and OPN, represents an activation-dependent ligand for αVβ3. In contrast, HSG cells, which expressed αVβ5 but not αVβ3, attached to BSP but not to DMP1 or OPN. These results verified previous reports that BSP can use either αVβ3 or αVβ5 to support attachment in vitro (27, 30). Our results agree with Cheng et al. (31) suggesting that OPN supports cell attachment specifically using αVβ3 and not αVβ5 integrin while contradicting several other reports noting that OPN could apparently use multiple integrins, including αVβ1, αVβ5, and αVβ6, for attachment (32–34). Therefore with respect to attachment, DMP1 appears to be like OPN and to interact effectively with αVβ3, but it is unable to support attachment through αVβ5. It appears likely that DMP1's cell-type attachment specificity is due to both a cell's spectrum of integrins expressed and the activation state of the endogenous integrins such as αVβ3.
Next, the possible differences among the three SIBLINGs with respect to haptotactic migration were investigated. Cells attached to a membrane surface can elect to migrate through the pores of a modified Boyden chamber and onto the protein-coated under surface of the membrane. MG63 cells (naturally expressing the αVβ3 integrin) and αVβ3-expressing HSG cells migrated from the commercially prepared tissue culture surface onto all three SIBLING-coated surfaces, although the DMP1 coating was always statistically less efficient at attracting/supporting the migrating cells than either BSP or OPN. Parental HSG cells (which lacked αVβ3) remained on the original surface suggesting that, irrespective of initial attachment status, the αVβ3 integrin was required for haptotactic migration onto these SIBLINGs. Our data are in line with previous reports demonstrating that αVβ3 is necessary for migration on BSP and OPN (31, 33) and extend this observation to DMP1. However, DMP1's poorer showing in attracting and/or supporting the migrating cells illustrates an interesting if unexplained difference from OPN, which it had otherwise so closely mimicked throughout these assays.
The three major isoforms of DMP1, fully intact, the BMP1-cleaved C-terminal fragment, and the proteoglycan forms, were also tested for their relative abilities to support cell attachment and haptotactic migration. The alternatively spliced isoform of DMP1, that we have observed dominating the DMP1 mRNA in human kidney, lacking the 16-amino acid-encoding exon 5 (data not shown), was not tested. The intact DMP1 was indistinguishable from the C-terminal fragment suggesting that at least as far as these in vitro assays are concerned, this SIBLING did not need to be “activated” by BMP1-like cleavage to function. In contrast, the proteoglycan form of the intact DMP1 protein greatly reduced cell attachment and haptotactic migration events. Removal of the glycosaminoglycan chain by chondroitinase ABC after the protein was already bound to the surface restored the attachment activity. Because the GAG chain attachment site is ∼250 amino acids away from the RGD motif, its effect on the DMP1-αVβ3 integrin interaction would not appear to due to direct steric interference. Interestingly, previous studies revealed that the GAG chains on other proteins did not directly interfere with focal contacts, but did so indirectly via interactions with relevant cell-surface receptor(s) supporting the clustering of actin-linked integrins to form focal adhesion and stress fibers as a strong cell-matrix adhesion junction (35, 36). More recent work has suggested annexin 6 as one possible candidate receptor for some of these GAG chains (37). Because the GAG chain is attached to the N-terminal fragment domain of DMP1, BMP1 will uncouple this inhibitory GAG chain from the RGD domain. If this event was to occur in solution, the GAG chain domain could diffuse away from the C-terminal, RGD-containing fragment and thus “activate” it. Additional studies are necessary to quantify the relative abundance of the standard and proteoglycan forms of DMP1 in normal and tumor tissues to determine the possible importance of this observation.
The RGD motif remained necessary for attachment and migration events as shown by the loss of these activities upon the addition of the competitive peptide, GRGDS, and when the surfaces were coated with SIBLINGs in which their RGD had been replaced by the chemically similar but inactive amino acids (KAE). Because all of these proteins contain the same basic RGD motif and yet the αVβ5 integrin distinguished DMP1 from the more promiscuous BSP, an investigation of several possible domains and post-translational modifications of DMP1 and BSP was undertaken. Replacing the conserved amino acids of the DMP1 RGD domain (SRGDNP) with those found to be highly conserved for BSP (PRGDNY) did not change the DMP1 into a BSP-like attachment protein. It has been previously shown that fibronectin's RGD-mediated binding to α5β1 integrin is dramatically enhanced by a so-called synergy site within fibronectin III domain 9 (38). Apparently αIIbβ3 integrin uses a similar synergy site suggesting that this phenomenon may have a more general significance (39). Therefore, the first four coding exons of DMP1 (including exon 5 that we and others have seen deleted in alternative splicing events (40)) were substituted with those found in BSP, but this also did not confer on DMP1 the BSP-like ability to support attachment via αVβ5 integrin suggesting that the synergy site resides within the last and largest coding exon. When the last coding exon of DMP1 was produced in bacteria it retained this SIBLING's ability to support attachment of the tested cells only through αVβ3 suggesting that none of the many post-translational modifications of DMP1 were necessary for the specificity of this interaction. Future experiments exchanging the many short, conserved amino acid domains in the remaining, untested C-terminal portions between BSP and DMP1 (and OPN) will be necessary to precisely identify the synergy-like domain of BSP that enables it to functionally interact with αVβ5 integrins.
Steiglitz et al. (14) recently showed that BMP1 and up to three other related Tolloid-like proteases that are involved in the processing of many different matrix proteins are also capable of cleaving DMP1 and probably another SIBLING member, DSPP. Because BMSCs naturally make a type I collagen matrix that is processed by BMP1, our recombinant DMP1 was also often at least partially processed into the C- and N-terminal fragments. To make solely intact DMP1 in the presence of the cell's native BMP1-like enzymes and to test the necessity of the Met and Gln in the highly conserved cleavage domain, the MQSDDP motif was changed into IHSDDP in specific DMP1-expressing adenovirus constructs. These conservative changes completely stopped the cleavage of the DMP1 by not only the endogenous Tolloid-like proteases in the BMSCs but also by both commercial recombinant BMP1 (in vitro) and by BMP1 overexpressed in HSG cells co-infected with the DMP1MQΔIH construct. The unprocessed procollagen observed in the media of BMSCs was clearly seen on Coomassie-stained gels to be processed when the BMP1 adenovirus was added to these cells thereby providing additional evidence of the functionality of this construct in tissue culture settings. Although we do not know which of the complement of Tolloid-like proteases are expressed in normal BMSCs, it is clear that the one or both of the Met and Gln amino acids in the cleavage motif are critical to at least BMP1 cleavage of DMP1 and, by inference, DSPP. This rigid requirement for sequence conservation in DMP1 is a curious observation given that BMP1 is thought to proteolytically process several other matrix and matricellular proteins whose cleavage sites differ substantially from DMP1 and DSPP. However, the observation previously made that, with the exception of mouse chordin (and by sequence analogy, the human protein also), an aromatic amino acid or a methionine is required at the -3 amino acid of the cleavage site (14) is supported by our loss of activity upon substituting the Met with the chemically conserved (i.e. hydrophobic) but non-aromatic Ile. Curiously OPN, the SIBLING we found to have attachment/migration properties most similar to DMP1, is known to be cleaved by thrombin-like proteases rather than BMP1 (41).
In conclusion, DMP1 shares many of its attachment and haptotactic properties with OPN except that DMP1 appears to be somewhat less effective in supporting the migration process. Like OPN, DMP1 binds and supports attachment through αVβ3 but, unlike BSP, neither can use αVβ5 integrins. Most isoforms of DMP1 behave similarly in these in vitro assays except for the proteoglycans form that cannot be efficiently used without removal of the inhibitory glycosaminoglycan chain. Although none of these properties directly explain the etiology of the various DMP1 human genetic diseases, the differences among the three SIBLINGs may lead to explanations as to why neither OPN nor BSP may be able to compensate for the loss of DMP1 activity in the disorders.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Division of Intramural Research, NIDCR, and the Intramural Research Program. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: DMP1, dentin matrix protein-1; SIBLING, small integrin-binding ligand, N-linked glycoprotein; BSP, bone sialoprotein; OPN, osteopontin; DSPP, dentin sialophosphoprotein; BMP1, bone morphogenetic protein-1; DMEM, Dulbecco's modified Eagle's medium; GAG, glycosaminoglycan; FBS, fetal bovine serum; DPC, dental pulp cell; mAb, monoclonal antibody; PBS, phosphate-buffered saline; BSA, bovine serum albumin; BMSC, bone marrow stromal cell; CMV, cytomegalovirus; Bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; FACS, fluorescence-activated cell sorting.
References
- 1.Fisher, L. W., Torchia, D. A., Fohr, B., Young, M. F., and Fedarko, N. S. (2001) Biochem. Biophys. Res. Commun. 280 460-465 [DOI] [PubMed] [Google Scholar]
- 2.Bellahcene, A., Castronovo, V., Ogbureke, K. U., Fisher, L. W., and Fedarko, N. S. (2008) Nat. Rev. Cancer 8 212-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin, C., Baba, O., and Butler, W. T. (2004) Crit. Rev. Oral Biol. Med. 15 126-136 [DOI] [PubMed] [Google Scholar]
- 4.Huq, N. L., Cross, K. J., Ung, M., and Reynolds, E. C. (2005) Arch. Oral Biol. 50 599-609 [DOI] [PubMed] [Google Scholar]
- 5.George, A., Sabsay, B., Simonian, P. A., and Veis, A. (1993) J. Biol. Chem. 268 12624-12630 [PubMed] [Google Scholar]
- 6.Ogbureke, K. U., and Fisher, L. W. (2004) J. Dent. Res. 83 664-670 [DOI] [PubMed] [Google Scholar]
- 7.Ogbureke, K. U., and Fisher, L. W. (2005) Kidney Int. 68 155-166 [DOI] [PubMed] [Google Scholar]
- 8.Ogbureke, K. U., and Fisher, L. W. (2007) J. Histochem. Cytochem. 55 403-409 [DOI] [PubMed] [Google Scholar]
- 9.Bucciarelli, E., Sidoni, A., Bellezza, G., Cavaliere, A., Brachelente, G., Costa, G., Chaplet, M., Castronovo, V., and Bellahcene, A. (2007) Breast Cancer Res. Treat. 105 95-104 [DOI] [PubMed] [Google Scholar]
- 10.Ling, Y., Rios, H. F., Myers, E. R., Lu, Y., Feng, J. Q., and Boskey, A. L. (2005) J. Bone Miner. Res. 20 2169-2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, J. Q., Ward, L. M., Liu, S., Lu, Y., Xie, Y., Yuan, B., Yu, X., Rauch, F., Davis, S. I., Zhang, S., Rios, H., Drezner, M. K., Quarles, L. D., Bonewald, L. F., and White, K. E. (2006) Nat. Genet. 38 1310-1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorenz-Depiereux, B., Bastepe, M., Benet-Pages, A., Amyere, M., Wagenstaller, J., Muller-Barth, U., Badenhoop, K., Kaiser, S. M., Rittmaster, R. S., Shlossberg, A. H., Olivares, J. L., Loris, C., Ramos, F. J., Glorieux, F., Vikkula, M., Juppner, H., and Strom, T. M. (2006) Nat. Genet. 38 1248-1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin, C., Brunn, J. C., Cook, R. G., Orkiszewski, R. S., Malone, J. P., Veis, A., and Butler, W. T. (2003) J. Biol. Chem. 278 34700-34708 [DOI] [PubMed] [Google Scholar]
- 14.Steiglitz, B. M., Ayala, M., Narayanan, K., George, A., and Greenspan, D. S. (2004) J. Biol. Chem. 279 980-986 [DOI] [PubMed] [Google Scholar]
- 15.Qin, C., Huang, B., Wygant, J. N., McIntyre, B. W., McDonald, C. H., Cook, R. G., and Butler, W. T. (2006) J. Biol. Chem. 281 8034-8040 [DOI] [PubMed] [Google Scholar]
- 16.Takagi, J. (2004) Biochem. Soc. Trans. 32 403-406 [DOI] [PubMed] [Google Scholar]
- 17.van der Flier, A., and Sonnenberg, A. (2001) Cell Tissue Res. 305 285-298 [DOI] [PubMed] [Google Scholar]
- 18.Takada, Y., Ye, X., and Simon, S. (2007) Genome Biology http://genomebiology.com/2007/8/5/215
- 19.Kulkarni, G. V., Chen, B., Malone, J. P., Narayanan, A. S., and George, A. (2000) Arch. Oral Biol. 45 475-484 [DOI] [PubMed] [Google Scholar]
- 20.Shirasuna, K., Sato, M., and Miyazaki, T. (1981) Cancer 48 745-752 [DOI] [PubMed] [Google Scholar]
- 21.Gronthos, S., Mankani, M., Brahim, J., Robey, P. G., and Shi, S. (2000) Proc. Natl. Acad. Sci. of U. S. A. 97 13625-13630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fedarko, N. S., Fohr, B., Robey, P. G., Young, M. F., and Fisher, L. W. (2000) J. Biol. Chem. 275 16666-16672 [DOI] [PubMed] [Google Scholar]
- 23.Jain, A., Karadag, A., Fohr, B., Fisher, L. W., and Fedarko, N. S. (2002) J. Biol. Chem. 277 13700-13708 [DOI] [PubMed] [Google Scholar]
- 24.Fisher, L. W., McBride, O. W., Termine, J. D., and Young, M. F. (1990) J. Biol. Chem. 265 2347-2351 [PubMed] [Google Scholar]
- 25.Green, M. R., Pastewka, J. V., and Peacock, A. C. (1973) Anal. Biochem. 56 43-51 [DOI] [PubMed] [Google Scholar]
- 26.Lampugnani, M. G., Bernasconi, S., Neri, P., Lozzi, L., Gavazzi, I., Marchisio, P. C., and Dejana, E. (1991) Lab. Invest. 65 96-103 [PubMed] [Google Scholar]
- 27.Byzova, T. V., Kim, W., Midura, R. J., and Plow, E. F. (2000) Exp. Cell Res. 254 299-308 [DOI] [PubMed] [Google Scholar]
- 28.Helluin, O., Chan, C., Vilaire, G., Mousa, S., DeGrado, W. F., and Bennett, J. S. (2000) J. Biol. Chem. 275 18337-18343 [DOI] [PubMed] [Google Scholar]
- 29.Shimaoka, M., Takagi, J., and Springer, T. A. (2002) Annu. Rev. Biophys. Biomol. Struct. 31 485-516 [DOI] [PubMed] [Google Scholar]
- 30.Sung, V., Stubbs, J. T., 3rd, Fisher, L., Aaron, A. D., and Thompson, E. W. (1998) J. Cell. Physiol. 176 482-494 [DOI] [PubMed] [Google Scholar]
- 31.Cheng, S. L., Lai, C. F., Fausto, A., Chellaiah, M., Feng, X., McHugh, K. P., Teitelbaum, S. L., Civitelli, R., Hruska, K. A., Ross, F. P., and Avioli, L. V. (2000) J. Cell. Biochem. 77 265-276 [DOI] [PubMed] [Google Scholar]
- 32.Hu, D. D., Lin, E. C., Kovach, N. L., Hoyer, J. R., and Smith, J. W. (1995) J. Biol. Chem. 270 26232-26238 [DOI] [PubMed] [Google Scholar]
- 33.Liaw, L., Skinner, M. P., Raines, E. W., Ross, R., Cheresh, D. A., Schwartz, S. M., and Giachelli, C. M. (1995) J. Clin. Investig. 95 713-724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokosaki, Y., Tanaka, K., Higashikawa, F., Yamashita, K., and Eboshida, A. (2005) Matrix Biol. 24 418-427 [DOI] [PubMed] [Google Scholar]
- 35.Yamagata, M., and Kimata, K. (1994) J. Cell Sci. 107 2581-2590 [DOI] [PubMed] [Google Scholar]
- 36.Yamagata, M., Saga, S., Kato, M., Bernfield, M., and Kimata, K. (1993) J. Cell Sci. 106 55-65 [DOI] [PubMed] [Google Scholar]
- 37.Takagi, H., Asano, Y., Yamakawa, N., Matsumoto, I., and Kimata, K. (2002) J. Cell Sci. 115 3309-3318 [DOI] [PubMed] [Google Scholar]
- 38.Aota, S., Nomizu, M., and Yamada, K. M. (1994) J. Biol. Chem. 269 24756-24761 [PubMed] [Google Scholar]
- 39.Bowditch, R. D., Hariharan, M., Tominna, E. F., Smith, J. W., Yamada, K. M., Getzoff, E. D., and Ginsberg, M. H. (1994) J. Biol. Chem. 269 10856-10863 [PubMed] [Google Scholar]
- 40.Qin, C., D'Souza, R., and Feng, J. Q. (2007) J. Dent. Res. 86 1134-1141 [DOI] [PubMed] [Google Scholar]
- 41.Smith, L. L., Cheung, H. K., Ling, L. E., Chen, J., Sheppard, D., Pytela, R., and Giachelli, C. M. (1996) J. Biol. Chem. 271 28485-28491 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.