Abstract
The regulation of mitochondrial degradation through autophagy is expected to be a tightly controlled process, considering the significant role of this organelle in many processes ranging from energy production to cell death. However, very little is known about the specific nature of the degradation process. We developed a new method to detect mitochondrial autophagy (mitophagy) by fusing the green fluorescent protein at the C terminus of two endogenous mitochondrial proteins and monitored vacuolar release of green fluorescent protein. Using this method, we screened several atg mutants and found that ATG11, a gene that is essential only for selective autophagy, is also essential for mitophagy. In addition, we found that mitophagy is blocked even under severe starvation conditions, if the carbon source makes mitochondria essential for metabolism. These findings suggest that the degradation of mitochondria is a tightly regulated process and that these organelles are largely protected from nonspecific autophagic degradation.
The mitochondrion is an organelle that carries out a number of important metabolic processes such as fatty acid oxidation, the Krebs cycle, and oxidative phosphorylation. Mitochondria also have a key role in the regulation of apoptosis (1). Mitochondrial oxidative phosphorylation supplies a large amount of energy that contributes to a range of cellular activities. However, this organelle is also the major source of cellular reactive oxygen species that cause damage to mitochondrial lipid, DNA, and proteins, and the accumulation of this damage is related to aging, cancer, and neurodegenerative diseases (2). Thus, quality control of mitochondria is important to maintain cellular homeostasis. In fact, mitochondria have some of their own quality control systems including a protein degradation system (3), DNA repair enzymes (4, 5), and phospholipid hydroperoxide glutathione peroxidase (6). In addition, it has long been assumed that autophagy is the pathway for mitochondrial recycling, and various theories suggest that a specific targeting of damaged mitochondria to vacuoles or lysosomes occurs by autophagy (7), although there is little direct experimental evidence for selective recognition of mitochondria. Very recently, several studies suggest that selective mitochondrial degradation via autophagy (mitophagy) might play an important role for mitochondrial quality control (8–12).
Macroautophagy is the bulk degradation of cytoplasmic components that allows cells to respond to various types of stress and to adapt to changing nutrient conditions (13, 14). After certain environmental cues such as nutrient deprivation or hormonal stimuli, cells dynamically sequester portions of the cytoplasm within double-membrane cytosolic vesicles, called autophagosomes, and the completed vesicles subsequently fuse with lysosomes/vacuoles (15, 16). There are a number of selective autophagy pathways that appear to target specific cellular components, and the cytoplasm to vacuole targeting (Cvt)2 pathway and pexophagy are well understood examples. The Cvt pathway is an autophagy-like process that encompasses the biosynthetic routes of two known proteins, aminopeptidase I (Ape1) and α-mannosidase, without delivering any additional known cargo to the vacuole (16, 17). Pexophagy is the selective degradation of peroxisomes via autophagy that occurs particularly when cells are shifted from conditions where these organelles are required for metabolism to ones where they are no longer necessary (18). There are several lines of evidence from yeast to mammal that suggest that mitophagy is another type of selective autophagy. In mammalian cells, the mitochondrial permeability transition induces mitochondrial degradation (19, 20), and daughter mitochondria with reduced membrane potential after a fission event are more likely to be targeted by autophagy (11). In yeast, alterations of F0F1-ATPase biogenesis in a conditional mutant triggers autophagy (9). Moreover, Aup1 and Uth1, both mitochondrial proteins, have been shown to be essential in inducing mitophagy (21–23).
The molecular breakthroughs in autophagy including the identification of the molecular components of the Cvt pathway and pexophagy have taken place during only the past decade. The start of the molecular realm of autophagy began with the identification of autophagy-related (ATG) genes by genetic screening in yeast (24, 25). To date, 31 ATG genes have been identified, and the biological properties of some Atg proteins have been characterized. Compared with autophagy, the genetic and molecular mechanisms of mitophagy are still poorly understood. This is in part due to the absence of a sensitive and convenient method for mitophagy induction and detection. For example, Tal et al. (21) culture cells for more than 3 days to induce mitophagy, and Kissova et al., (23) use electron microscopy to detect the presence of mitochondria in the vacuole.
In this study, we established a sensitive and convenient method to monitor mitophagy. Using this method, we screened several atg mutants and found that ATG11, a gene that is essential for only selective autophagy, is also essential for mitophagy. In addition, we found that mitophagy is blocked even under severe starvation conditions, if the carbon source makes mitochondria essential for metabolism. These findings suggest that the degradation of mitochondria is a tightly regulated process and that these organelles may be protected from nonspecific autophagic degradation.
EXPERIMENTAL PROCEDURES
Strains and Media—The yeast strains used in this study are listed in Table 1. Yeast cells were grown in rich medium (YPD; 1% yeast extract, 2% peptone, 2% glucose), lactate medium (YPL; 1% yeast extract, 2% peptone, 2% lactate), synthetic minimal medium with glucose (0.67% yeast nitrogen base, 2% glucose, amino acids, and vitamins), synthetic minimal medium with galactose (0.67% yeast nitrogen base, 2% galactose, amino acids, and vitamins), or synthetic minimal medium with lactate (SML; 0.67% yeast nitrogen base, 2% lactate, amino acids, and vitamins). Starvation experiments were performed in synthetic minimal medium lacking nitrogen (SD-N; 0.17% yeast nitrogen base without amino acids, 2% glucose; or SL-N, 0.17% yeast nitrogen base without amino acids, 2% lactate).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Invitrogen |
| CWY239 | SEY6210 atg17Δ::KAN | Ref. 39 |
| D3Y108 | SEY6210 atg24Δ::HIS3 S.k. | Ref. 38 |
| D3Y109 | SEY6210 atg20Δ::HIS3 S.k. | Ref. 38 |
| FRY112 | SEY6210 atg8Δ::HIS5 S.p. | This study |
| HAY456 | SEY6210 atg9Δ::HIS5 S.p. | Ref. 52 |
| HCY109 | SEY6210 atg29Δ::KAN | This study |
| HCY111 | SEY6210 atg31Δ::HIS5 S.p. | This study |
| SEY6210 | MATα his3-Δ200 leu2-3,112 lys2-801 trp1-Δ901 ura3-52 suc2-Δ9 GAL | Ref. 53 |
| SSY31 | SEY6210 atg19Δ:: HIS5 S.p. | Ref. 42 |
| TKYM22 | SEY6210 OM45-GFP::TRP1 | This study |
| TKYM29 | SEY6210 pep4Δ::LEU2, OM45-GFP::TRP1 | This study |
| TKYM50 | SEY6210 IDH1-GFP::KAN | This study |
| TKYM58 | TN124 OM45-GFP::TRP1 | This study |
| TKYM81 | SEY6210 pep4Δ::LEU2, IDH-GFP::TRP1 | This study |
| TN124 | MATa leu2-3,112 ura3-52 trp1 pho8::pho8Δ60 pho13Δ::LEU2 | Ref. 27 |
| WHY1 | SEY6210 atg1Δ::HIS5 S.p. | Ref. 36 |
| YTS147 | SEY6210 atg11Δ::LEU2 | This study |
PCR-based integration of a DNA fragment encoding green fluorescent protein (GFP) at the 3′ end of OM45, IDH1, and PEX14 generated cells expressing chromosomally tagged Om45-GFP, Idh1-GFP, and Pex14-GFP, respectively, for the mitophagy and pexophagy assays (26).
Plasmids and Antibodies—A doxycycline-regulated Mdm38-HA expression strain was generated by PCR amplification of the MDM38 gene with an HA tag from yeast genomic DNA using the following primers: 5-MDM38–1M-BamH1, 5′-ATATGGATCCATGTTGAATTTCGCATCAAGAGCG-3′; 3-MDM38-HAstop-Pst1, 5′-ATATCTGCAGCTAAGCATAGTCAGGCACATCATAGGGGTAATCTTTCTTAATGACAAAAGTCTTCGC-3′. The BamHI-PstI fragment was inserted into the respective site of the vector pCM189, yielding pTet-MDM38HA. The plasmid to express Myc-tagged Atg11 (pMyc-Atg11) has been described previously (17). Monoclonal anti-GFP antibody clone 7.1 and 13.1 mixture (Roche Applied Science), monoclonal anti-YFP antibody clone JL8 (Clontech, Mountain View, CA), and anti-Pgk1 antibody (a generous gift from Dr. Jeremy Thorner (University of California, Berkeley)) were used for immunoblotting.
Fluorescence Microscopy—Cells expressing fusion proteins with fluorescent tags were grown in YPL medium for the indicated times, and then cells were washed in SML medium before observation. Fluorescence signals were visualized on a wide field fluorescence inverted microscope (Olympus IX-70; Mellville, NY) equipped with a 100× oil NA 1.4 objective lens and fluorescein isothiocyanate filters. The images were captured by a Photometrix CoolSnap HQ camera (Photometrics, Tucson, AZ).
Mitophagy Assays—For Assay 1, cells grown in YPD medium to mid-log phase were shifted to YPL medium (starting from A600 = 0.1) for 12 h. For starvation, the cells were washed in water two times and cultured in SD-N. The cells were collected at 0, 4, and 6 h, and the cell lysates equivalent to A600 = 0.1 unit of cells were subjected to immunoblot analysis. For Assay 2, cells grown in YPD medium to mid-log phase were shifted to YPL medium (starting from A600 = 0.1). After 12 ± 2 h (day 1), 36 ± 5 h (day 2), and 60 ± 5 h (day 3), the cells were collected and used for immunoblotting and fluorescence microscopy analysis.
Assays for Autophagy and Pexophagy—For monitoring bulk autophagy, the alkaline phosphatase activity of Pho8Δ60 and processing of GFP-Atg8 were carried out as described previously (27, 28). Pex14-GFP processing to monitor pexophagy has been described previously (29, 30).
RESULTS
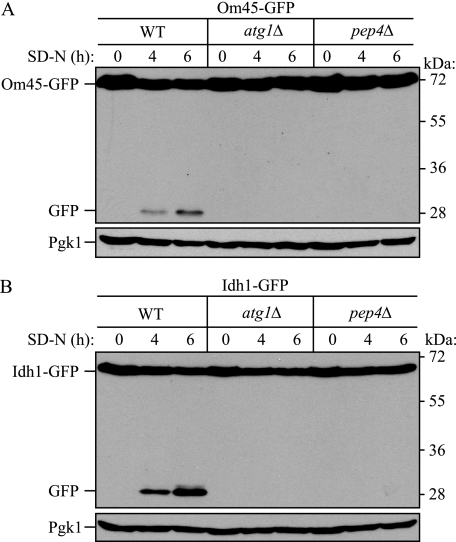
Monitoring Mitophagy Using C-terminal GFP-tagged Mitochondrial Protein Processing and Fluorescence Microscopy—To establish a method to induce and detect mitophagy in yeast, we took advantage of the observation that GFP present in fusion constructs is relatively stable within the vacuole lumen and is often released as an intact protein after delivery to the vacuole (28). Therefore, we tagged the C terminus of several different mitochondrial proteins with GFP and monitored mitophagy by examining the localization and degradation of the chimera, with the concomitant release of free GFP, in the vacuole under several culture conditions. When cells were grown in medium with 2% lactate as the sole carbon source (YPL) for more than 12 h and then shifted to SD-N medium, two of the GFP-tagged mitochondrial proteins, the mitochondrial outer membrane protein Om45 and the matrix protein Idh1, showed GFP processing to an extent that depended on the duration of nitrogen starvation (Fig. 1, WT). This processing was not dependent on mitochondrial endogenous proteinases (such as Lon or Clp1), because both a mitochondrial outer membrane and matrix protein showed the same result. In contrast, strains deleted for ATG1, encoding a protein kinase essential for macroautophagy, or PEP4, one of the main vacuolar hydrolases, did not show GFP processing, suggesting that the appearance of free GFP was autophagy- and vacuole-dependent (Fig. 1).
FIGURE 1.
Monitoring mitophagy using C-terminal GFP-tagged mitochondrial protein processing during amino acid starvation. Wild-type (WT), atg1Δ, or pep4Δ strains expressing Om45-GFP (A) or Idh1-GFP (B) were cultured in YPL medium to mid-log growth phase and then shifted to SD-N medium for 0, 4 and 6 h. GFP processing was monitored by immunoblotting with anti-GFP and anti-Pgk1 (loading control) antibody or antiserum, respectively. The positions of molecular mass markers are indicated on the right.
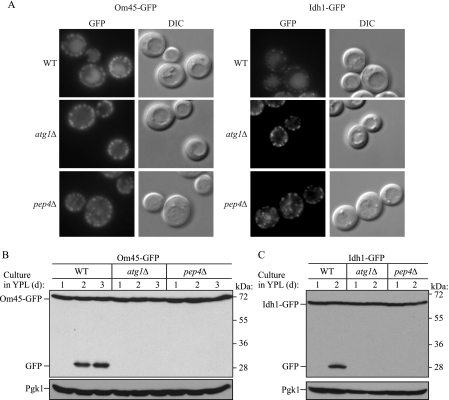
Recently it was reported that a mitochondrially targeted GFP protein accumulates in the vacuole after long term growth in lactate medium (21). We cultured wild-type cells expressing Om45-GFP or Idh1-GFP in YPL medium for 1–3 days and monitored GFP processing and accumulation in the vacuole. After 2 days of growth in YPL, the GFP fluorescence became very clear in the vacuole (Fig. 2A, top panels), and at the same time, GFP was processed (Fig. 2, B and C, WT). The absence of Atg1 blocked both GFP processing and vacuolar GFP accumulation (Fig. 2). In contrast, deletion of PEP4 blocked GFP processing (Fig. 2, B and C) but showed vacuolar dotted fluorescence likely corresponding to autophagic bodies containing mitochondria as a cargo (Fig. 2A, bottom panels). These findings reconfirmed that processing of GFP-tagged mitochondrial proteins is autophagy-dependent and suggests that we are monitoring actual mitophagy. The time course of cell growth and mitophagy in lactate medium revealed that mitophagy was induced when cells reached post-log to stationary phase growth (data not shown).
FIGURE 2.
Monitoring mitophagy using C-terminal GFP-tagged mitochondrial protein processing at post-log phase. Wild-type (WT), atg1Δ, and pep4Δ strains expressing Om45-GFP or Idh1-GFP were cultured in YPL medium for 1–3 days. The localization of GFP was visualized by fluorescence microscopy at day 2 (A), and GFP processing was monitored by immunoblotting with anti-GFP and anti-Pgk1 (loading control) antibody and antiserum, respectively, at the indicated days in the strains expressing Om45-GFP (B) or Idh1-GFP (C). DIC, differential interference contrast.
Previous studies have examined mitochondrial degradation induced by the doxycycline-regulated shut-off of the MDM38 gene (10) or by impairing the bioenergetic status and the biogenesis of mitochondria using mutants defective in assembly or stability of the F0F1-ATPase (9). We used an identical doxycycline-regulated MDM38 strain and separately used the mitochondrial complex V (ATP synthase) inhibitor oligomycin to determine whether our system relying on cleavage of OM45-GFP replicated these results. We found that free GFP was detected in either case, although the amount of processed GFP was extremely low compared with the mitophagy induction shown above (supplemental Fig. S1A and S2). Therefore, we repeated the analysis with oligomycin examining the effect over time and found that there was an increase in mitophagy that corresponded to the exposure time to the drug (supplemental Fig. S1B). These results fit with the previous findings that interference with the function of the F0F1-ATPase induces mitophagy (9).
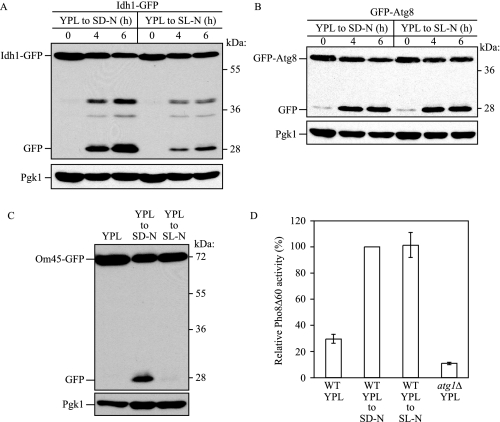
Mitophagy Is Regulated Independent from Macroautophagy—Having developed a system that can be used to monitor mitophagy, we next wanted to address the question of whether this process can occur in a selective manner. When cells are cultured with a nonfermentable carbon source, a situation where cells require mitochondrial oxidative phosphorylation for cell growth, the number of mitochondria and the mass amount of mitochondrial proteins are increased (31–33). If the medium is shifted to a fermentable carbon source, the level of mitochondria is eventually decreased. To determine whether the reduction of mitochondria is caused by mitophagy, wild-type cells expressing Om45-GFP were grown in YPL medium to mid-log phase and shifted to glucose medium (YPD) for 6 h, and mitophagy was monitored by GFP processing. Under these conditions, however, mitophagy was barely detected (data not shown). On the other hand, if we shifted the cells to SD-N medium, we detected a substantial level of free GFP (Fig. 3A). In contrast, when cells cultured in YPL were shifted to nitrogen starvation medium supplemented with lactate (SL-N) instead of glucose, mitophagy occurred at a very low level (Fig. 3A). These results suggest that detectable levels of mitophagy were not induced simply by shifting to a fermentable carbon source (YPD) or by shifting to starvation conditions in the presence of a nonfermentable carbon source (SL-N). To determine whether the lack of GFP processing reflected a block in mitophagy or a general block in macroautophagy, we next examined a marker for the latter process. Atg8 is one of two Atg proteins that remain associated with the completed autophagosome. Therefore, processing of GFP-Atg8 can be used to monitor nonspecific autophagy (34). We expressed GFP-Atg8 in the same cell background and monitored GFP-Atg8 processing after shifting to SD-N or SL-N. In this case, the amount of processed GFP was almost the same in both conditions, indicating that SL-N medium still induced a starvation response (Fig. 3B).
FIGURE 3.
Macroautophagy occurs independent of mitophagy. Wild-type (WT) cells expressing Idh1-GFP (A) or GFP-Atg8 (B) were cultured in YPL medium for 12 h and then starved in SD-N or SL-N for up to 6 h. The cells were collected at the indicated time points, and cell lysates equivalent to A600 = 0.1 unit of cells were subjected to immunoblot analysis with anti-GFP and anti-Pgk1 (loading control) antibody or antiserum, respectively. C, the Pho8Δ60 strain (TN124) expressing Om45-GFP was cultured in YPL medium for 12 h and starved in SD-N or SL-N for 6 h. Cell lysates equivalent to A600 = 0.1 unit of cells were subjected to immunoblot analysis as above. D, cell lysates equivalent to A600 = 2 units of cells were analyzed by the Pho8Δ60 activity assay. The Pho8Δ60 activity of wild-type cells starved in SD-N was set to 100%. The values represent the mean and standard deviation from three independent experiments. The Pho8Δ60 activity of the atg1Δ strain cultured in YPL is shown to indicate the background activity of this assay.
To measure autophagic activity quantitatively, we used an additional assay. Pho8Δ60 is a truncated form of the vacuolar membrane enzyme alkaline phosphatase, which is normally delivered to this organelle through a portion of the secretory pathway. Pho8Δ60 lacks the N-terminal transmembrane domain that serves as an internal uncleaved signal sequence, and the altered protein remains in the cytosol; it is only delivered to the vacuole by autophagy, which results in cleavage of the C-terminal propeptide and enzyme activation. Thus, this assay monitors nonspecific autophagy (27, 35). We expressed Om45-GFP in the Pho8Δ60 strain and then monitored mitophagy and autophagic activity. As before, mitophagy was barely induced after cells were shifted to SL-N compared with SD-N. On the other hand, autophagic activity based on Pho8Δ60 activity was almost the same in both conditions (Fig. 3, C and D).
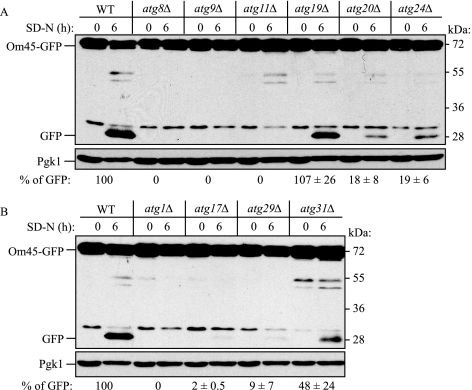
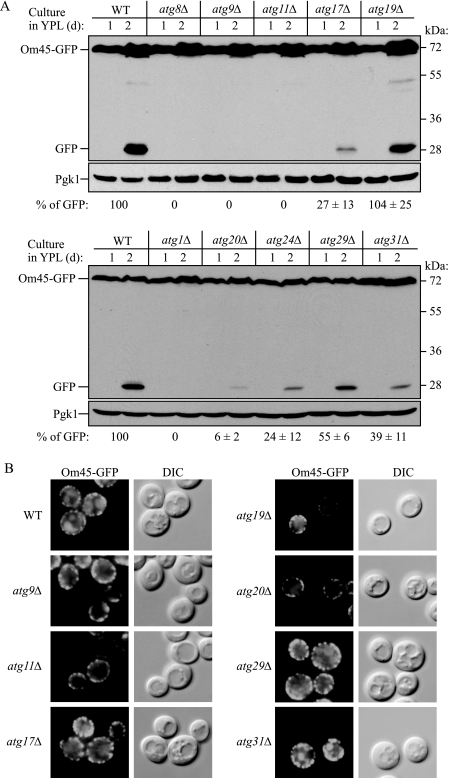
Screening atg Mutants for Potential Mitophagy Defects—Currently, 31 genes are denoted as autophagy-related (ATG). We examined the mitophagy capacity of the atg mutants during starvation. ATG genes that are essential for both specific and nonspecific autophagy such as ATG8 and ATG9 were also essential for mitophagy (Fig. 4A and supplemental Fig. S3), in agreement with previous reports (12, 21, 23). Atg19 functions as a receptor for precursor Ape1 (prApe1) and is specific for the Cvt pathway (36), and the absence of Atg19 did not affect mitophagy. In contrast, the atg11Δ mutant was completely blocked for the degradation of mitochondria (Fig. 4A and supplemental Fig. S3). Atg11 appears to function as an adaptor or scaffold protein and is needed for both the Cvt pathway and pexophagy but not for nonspecific autophagy (37). The block in mitophagy seen in the atg11Δ strain was rescued by expressing Myc-tagged Atg11 (data not shown), indicating that the defect was due to the absence of this protein.
FIGURE 4.
Screening atg mutants for potential mitophagy defects during starvation. Strains deleted for the indicated ATG genes and expressing Om45-GFP were screened for potential mitophagy activity using the method demonstrated in Fig. 1. The atg mutant strains in A are either defective in all types of autophagy (atg8Δ and atg9Δ) or are defective in the Cvt pathway. The atg mutant strains in B are defective for all types of autophagy (atg1Δ) or are defective in nonspecific macroautophagy, but are normal for the Cvt pathway. The relative amount of processed GFP was calculated. The values represent the means and standard deviation from three independent experiments. WT, wild type.
To determine whether this block was specific to the atg11Δ strain, we next examined other atg mutant strains that are defective for selective autophagy but that have little or no effect on bulk autophagy. The atg20Δ and atg24Δ strains both showed a strong block in mitophagy, although not as severe as that seen with atg11Δ (Fig. 4A). Essentially the same results were obtained at the post-logarithmic phase in lactate medium (Fig. 5). Because the atg11Δ, atg20Δ, and atg24Δ strains displayed substantial blocks in mitophagy but are essentially normal for macroautophagy (37, 38), these findings suggest that mitophagy is a selective process. The atg17Δ, atg29Δ, and atg31Δ strains are normal for the Cvt pathway and display substantial but not complete blocks in autophagy (30, 39, 40). These strains also showed significant blocks in mitophagy that varied depending on the culture conditions (Figs. 4B and 5).
FIGURE 5.
Screening atg mutants for mitophagy defects during post-log phase growth. Strains deleted for the indicated ATG genes and expressing Om45-GFP were cultured in YPL medium for 2 days. A, cells were collected at the indicated days and monitored for GFP processing by immunoblotting. The relative amount of processed GFP was calculated. The values represent the means and standard deviation from three independent experiments. B, the indicated strains cultured in YPL for 2 days were observed by fluorescence microscopy. The phenotype of the atg8Δ strain was essentially identical to that of atg9Δ, and that of atg20Δ was essentially identical to that of atg24Δ. DIC, differential interference contrast. WT, wild type.
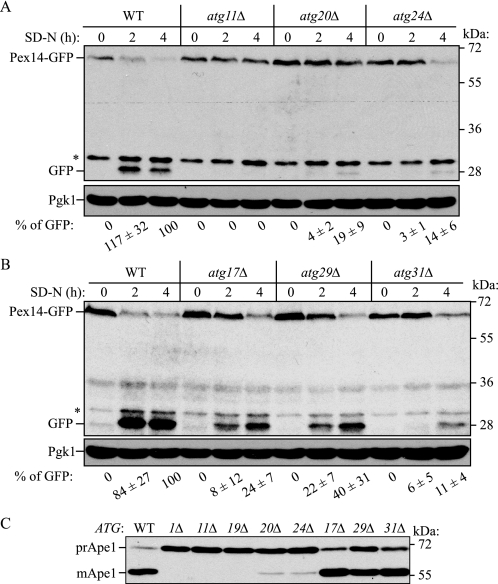
Relationship between Selective Autophagy and ATG Genes—Mitophagy is a selective type of organelle degradation, whereas the Cvt pathway is a biosynthetic route for the delivery of resident vacuolar hydrolases. Even though the Cvt pathway uses most of the same autophagic machinery as nonspecific autophagy, we decided to examine another type of selective organelle degradation, pexophagy. Accordingly, we tagged various atg mutant strains with Pex14-GFP. Pex14 is a peroxisomal membrane protein, and processing of Pex14-GFP can be used to monitor autophagic degradation of peroxisomes (41). Pexophagy has been analyzed in some of the atg mutants (29, 30, 37–39, 42, 43) but not in the atg29Δ or atg31Δ strains. Using wild-type and atg mutant strains, peroxisome biogenesis was induced in medium supplemented with 0.1% oleic acid instead of glucose, and the cells were then shifted to glucose-containing starvation medium (SD-N) to induce pexophagy. After 2 and 4 h, Pex14-GFP processing was monitored by immunoblotting. The wild-type strain displayed clear processing of Pex14-GFP, which reflected pexophagy (Fig. 6). In contrast, the atg11Δ, atg20Δ, and atg24Δ mutants that are defective in selective autophagy (i.e. the Cvt pathway; Fig. 6C) displayed severe blocks in pexophagy, as previously reported (37, 38). The atg17Δ and atg29Δ mutants, which are essentially normal for the Cvt pathway (Fig. 6C) showed partial blocks in pexophagy, whereas atg31Δ cells displayed a more severe block (Fig. 6B).
FIGURE 6.
Analysis of pexophagy and the Cvt pathway. Wild-type (WT) and atg11Δ, atg20Δ, and atg24Δ (A) or atg17Δ, atg29Δ, and atg31Δ (B) strains expressing Pex14-GFP were cultured with oleic acid-containing medium for 19 h, then shifted to SD-N for the indicated times, and monitored for GFP processing by immunoblotting. The asterisks indicate nonspecific bands. The relative amount of processed GFP was calculated. The values represent the means and standard deviation from three independent experiments. C, each mutant was cultured in YPD medium and analyzed for prApe1 maturation by immunoblotting to monitor the Cvt pathway during vegetative growth. The positions of precursor and mature Ape1 are indicated.
Table 2 summarizes the relationship between selective and nonselective autophagy and the ATG genes. ATG1, ATG8, and ATG9 (and most of the ATG genes) are essential for all types of autophagy. Mutants in the corresponding genes display defects in both selective and nonselective autophagic processes. In contrast, ATG11, ATG20, and ATG24 are linked only with selective autophagy and are essentially normal for bulk autophagy; Atg19 is a receptor for prApe1 and is only blocked in the Cvt pathway. On the other hand, ATG17, ATG29, and ATG31 showed a different pattern of autophagy blockage; strains deleted for these genes were defective for macroautophagy, pexophagy, and mitophagy, but not the Cvt pathway. These results are consistent with recent reports showing that these latter three proteins form part of a complex that functions in the early stages of phagophore assembly site (PAS) formation (44, 45). This result also suggests that although the Cvt pathway, pexophagy, and mitophagy are all selective types of autophagy, there are some differences between the Cvt pathway and organelle-specific types of autophagy with regard to the requirement for particular ATG genes.
TABLE 2.
Defects in selective versus nonselective autophagy
The number of plus signs is meant to indicate the approximate severity of the defect. —, no defect.
| Gene | Macroautophagy defect | Cvt pathway defect | Pexophagy defect | Mitophagy defect | Source | |
|---|---|---|---|---|---|---|
| Mutants blocked in all pathways | ATG1 | +++ | +++ | +++ | +++ | Refs. 27, 29, and 54 |
| ATG8 | +++ | +++ | +++ | +++ | Refs. 29, 55, and 56 | |
| ATG9 | +++ | +++ | +++ | +++ | Refs. 43 and 57 | |
| Mutants blocked in the Cvt pathway and pexophagy | ATG11 | - | +++ | +++ | +++ | Ref. 37 |
| ATG20 | - | +++ | ++ | ++ | Ref. 38 | |
| ATG24 | - | +++ | ++ | ++ | Ref. 38 | |
| Mutants blocked in nonselective autophagy | ATG17 | ++ | - | ++ | +++ | Ref. 39 |
| ATG29 | +++ | - | ++ | +++ | Ref. 40 | |
| ATG31 | +++ | - | +++ | ++ | Ref. 44 | |
| Mutants blocked in the Cvt pathway only | ATG19 | - | +++ | - | - | Ref. 42 |
DISCUSSION
The Case for Selective Mitophagy—The presence of mitochondria within an autophagosome was first reported by Clark (46) in mammalian cells and by Takeshige et al. (47) in yeast. Since then, it is thought that mitochondria are nonselectively engulfed by autophagosomes as a part of the cytoplasm during macroautopahgy. Recently, however, it has been suggested that engulfment of mitochondria also occurs through a selective process (22, 23, 48, 49). This idea of selective degradation is supported by the following: 1) the mitochondrial permeability transition or reduced mitochondrial membrane potential induces mitochondrial degradation in mammalian cells (11, 19, 20); 2) in yeast, alterations of F0F1-ATPase biogenesis in a conditional mutant trigger mitophagy (9); and 3) Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity and induces mitophagy (10).
In this study, we established a sensitive and convenient method to monitor mitophagy (Figs. 1 and 2). The screening of several atg mutants revealed that the ATG11, ATG20, and ATG24 genes, which are essential only for selective-autophagy, are also essential for mitophagy (Figs. 4 and 5). In addition, we demonstrated that mitophagy is induced when the carbon source makes mitochondria nonessential but blocked when the function of these organelles is needed for metabolism of a particular carbon source, suggesting that mitophagy is controlled separately from bulk macroautophagy. These findings suggest that mitophagy is one type of selective autophagy, controlled by the same Atg components as other autophagic processes, but that it may be regulated by a unique set of conditions.
Mitophagy requires certain ATG genes such as ATG1, ATG8, and ATG9, similar to other types of nonselective and selective autophagy (12, 21, 23) (Table 2). In addition, as noted above, ATG11, ATG20, and ATG24, which are essential for the Cvt pathway and pexophagy but not macroautophagy, are also essential for mitophagy (Figs. 4, 5, 6). These data strongly support the idea that mitophagy is a selective form of autophagy. The function of Atg11 has been well characterized in the Cvt pathway and pexophagy. In the Cvt pathway, Atg19 forms a complex with the Cvt pathway cargo prApe1 and α-mannosidase. The prApe1-Atg19-α-mannosidase complex is then targeted to the PAS by Atg11 via the Atg19-Atg11 interaction (17, 36, 42). In the same way, Atg30 makes a complex with the peroxisomal proteins Pex3 and Pex14, and then the peroxisome is targeted to the PAS via the Atg30-Atg11 interaction (50). In both cases, Atg19 or Atg30 work as a receptor to recognize specific cargo, and Atg11 functions to recruit the selected cargo to the PAS, the place of cargo engulfment by the autophagosome. From this point of view, Atg11 may recruit mitochondria to the PAS via binding with an unidentified receptor protein that can recognize mitochondria, although the recognition system may be more complicated compared with other selective types of autophagy because it may be able to distinguish damaged and intact mitochondria (11, 48).
ATG17 is essential for pexophagy but is not required for the Cvt pathway (39). Thus, the importance of ATG17 for selective autophagy had been unclear. Three ATG genes have been reported that are not required for the Cvt pathway but are essential for macroautophagy (ATG17, ATG29, and ATG31). Our analysis indicated that ATG29 and ATG31 are essential for pexophagy similar to ATG17 (Fig. 6). In addition, we found that these three proteins are required for mitophagy. These findings allow us to speculate that among selective types of autophagy, there are mutants that fall into two categories, those that affect the biosynthetic Cvt pathway and organelle degradation but not bulk macroautophagy, and those that are normal for the Cvt pathway, but block organelle degradation and nonspecific autophagy. In addition, Atg19 and Atg30 comprise a third class of proteins that encompass specific cargo receptors.
Mitophagy Is Regulated by Nutrient Conditions—Mitophagy was induced when cells were cultured with lactate as a sole carbon source (YPL) and then shifted to amino acid starvation medium supplemented with glucose; however, mitochondrial degradation was blocked when lactate was the sole carbon source (Fig. 3, A and C). Under both conditions, macroautophagy was strongly activated (Fig. 3, B and D). This finding further supports the idea that mitochondria are selectively degraded.
The amount of mitochondria within the cell is regulated by its biosynthesis, segregation during cell division, and degradation. Mitophagy may contribute not only to eliminate damaged mitochondria (9–11) but also to regulate the amount of mitochondria. During the mid-log growth phase, even if cells are shifted from YPL to glucose medium (YPD), mitophagy is barely detected. This may be because the amount of mitochondria is decreased by segregation into daughter cells. In contrast, under nitrogen starvation conditions in the presence of glucose, we could detect mitophagy. In this case, mitochondrial segregation is probably blocked by cell cycle arrest, and some amount of mitochondria that are not needed for fermentative growth may be degraded by mitophagy. This idea can explain the induction of mitophagy that occurs at the post-log to stationary phase in lactate medium. In the stationary phase, the cell energy requirement is reduced, and accordingly the requirement for mitochondria is decreased. Because mitochondrial segregation is also stopped at this growth phase, mitophagy can be induced to adapt the cell to these conditions and reduce the amount of mitochondria.
Autophagy is the process by which cells recycle cytoplasm and dispose of excess or defective organelles. Thus, autophagy is related with many cellular phenomena and also many diseases (51). Mitophagy, a part of selective autophagy, may play an important role in maintaining mitochondrial function and integrity. Recent data suggest that a deficiency in mitophagy causes the accumulation of damaged mitochondria and an increase in oxidative damage (12). Thus, mitophagy may be important in preventing certain diseases. Clearly there needs to be a greater understanding of the molecular mechanisms and regulatory pathways that control this process.
Supplementary Material
Acknowledgments
We thank Dr. Fulvio Reggiori (University Medical Centre Utrecht) for providing strain FRY112.
This work was supported, in whole or in part, by National Institutes of Health Grant GM53396 (to D. J. K.). This work was also supported by Japan Society for the Promotion of Science Postdoctoral Fellowships for Research Abroad (to T. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: Cvt, cytoplasm to vacuole targeting; GFP, green fluorescent protein; HA, hemagglutinin; PAS, phagophore assembly site.
References
- 1.Keeble, J. A., and Gilmore, A. P. (2007) Cell Res. 17 976–984 [DOI] [PubMed] [Google Scholar]
- 2.Wallace, D. C. (2005) Annu. Rev. Genet. 39 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rep, M., and Grivell, L. A. (1996) Curr. Genet. 30 367–380 [DOI] [PubMed] [Google Scholar]
- 4.Larsson, N. G., and Clayton, D. A. (1995) Annu. Rev. Genet. 29 151–178 [DOI] [PubMed] [Google Scholar]
- 5.Bogenhagen, D. F. (1999) Am. J. Hum. Genet. 64 1276–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arai, M., Imai, H., Koumura, T., Yoshida, M., Emoto, K., Umeda, M., Chiba, N., and Nakagawa, Y. (1999) J. Biol. Chem. 274 4924–4933 [DOI] [PubMed] [Google Scholar]
- 7.Abeliovich, H., and Klionsky, D. J. (2001) Microbiol. Mol. Biol. Rev. 65 463–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mijaljica, D., Prescott, M., and Devenish, R. J. (2007) Autophagy 3 4–9 [DOI] [PubMed] [Google Scholar]
- 9.Priault, M., Salin, B., Schaeffer, J., Vallette, F. M., di Rago, J. P., and Martinou, J. C. (2005) Cell Death Differ. 12 1613–1621 [DOI] [PubMed] [Google Scholar]
- 10.Nowikovsky, K., Reipert, S., Devenish, R. J., and Schweyen, R. J. (2007) Cell Death Differ. 14 1647–1656 [DOI] [PubMed] [Google Scholar]
- 11.Twig, G., Elorza, A., Molina, A. J., Mohamed, H., Wikstrom, J. D., Walzer, G., Stiles, L., Haigh, S. E., Katz, S., Las, G., Alroy, J., Wu, M., Py, B. F., Yuan, J., Deeney, J. T., Corkey, B. E., and Shirihai, O. S. (2008) EMBO J. 27 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, Y., Qi, H., Taylor, R., Xu, W., Liu, L. F., and Jin, S. (2007) Autophagy 3 337–346 [DOI] [PubMed] [Google Scholar]
- 13.Klionsky, D. J. (2005) J. Cell Sci. 118 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yorimitsu, T., and Klionsky, D. J. (2007) Trends Cell Biol. 17 279–285 [DOI] [PubMed] [Google Scholar]
- 15.Klionsky, D. J., and Ohsumi, Y. (1999) Annu. Rev. Cell Dev. Biol. 15 1–32 [DOI] [PubMed] [Google Scholar]
- 16.Klionsky, D. J., and Emr, S. D. (2000) Science. 290 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yorimitsu, T., and Klionsky, D. J. (2005) Mol. Biol. Cell 16 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn, W. A., Jr., Cregg, J. M., Kiel, J. A. K. W., van der Klei, I. J., Oku, M., Sakai, Y., Sibirny, A. A., Stasyk, O. V., and Veenhuis, M. (2005) Autophagy. 1 75–83 [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Enriquez, S., He, L., and Lemasters, J. J. (2004) Int. J. Biochem. Cell Biol. 36 2463–2472 [DOI] [PubMed] [Google Scholar]
- 20.Elmore, S. P., Qian, T., Grissom, S. F., and Lemasters, J. J. (2001) FASEB J. 15 2286–2287 [DOI] [PubMed] [Google Scholar]
- 21.Tal, R., Winter, G., Ecker, N., Klionsky, D. J., and Abeliovich, H. (2007) J. Biol. Chem. 282 5617–5624 [DOI] [PubMed] [Google Scholar]
- 22.Kissova, I., Deffieu, M., Manon, S., and Camougrand, N. (2004) J. Biol. Chem. 279 39068–39074 [DOI] [PubMed] [Google Scholar]
- 23.Kissova, I., Salin, B., Schaeffer, J., Bhatia, S., Manon, S., and Camougrand, N. (2007) Autophagy 3 329–336 [DOI] [PubMed] [Google Scholar]
- 24.Xie, Z., and Klionsky, D. J. (2007) Nat. Cell Biol. 9 1102–1109 [DOI] [PubMed] [Google Scholar]
- 25.Klionsky, D. J. (2007) Nat. Rev. Mol. Cell. Biol. 8 931–937 [DOI] [PubMed] [Google Scholar]
- 26.Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998) Yeast. 14 953–961 [DOI] [PubMed] [Google Scholar]
- 27.Noda, T., Matsuura, A., Wada, Y., and Ohsumi, Y. (1995) Biochem. Biophys. Res. Commun. 210 126–132 [DOI] [PubMed] [Google Scholar]
- 28.Shintani, T., and Klionsky, D. J. (2004) J. Biol. Chem. 279 29889–29894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutchins, M. U., Veenhuis, M., and Klionsky, D. J. (1999) J. Cell Sci. 112 4079–4087 [DOI] [PubMed] [Google Scholar]
- 30.Yen, W.-L., Legakis, J. E., Nair, U., and Klionsky, D. J. (2007) Mol. Biol. Cell 18 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ibrahim, N. G., Stuchell, R. N., and Beattie, D. S. (1973) Eur. J. Biochem. 36 519–527 [DOI] [PubMed] [Google Scholar]
- 32.Visser, W., van Spronsen, E. A., Nanninga, N., Pronk, J. T., Gijs Kuenen, J., and van Dijken, J. P. (1995) Antonie Van Leeuwenhoek 67 243–253 [DOI] [PubMed] [Google Scholar]
- 33.Damsky, C. H. (1976) J. Cell Biol. 71 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klionsky, D. J., Cuervo, A. M., and Seglen, P. O. (2007) Autophagy 3 181–206 [DOI] [PubMed] [Google Scholar]
- 35.Klionsky, D. J. (2007) Methods Mol. Biol. 390 363–372 [DOI] [PubMed] [Google Scholar]
- 36.Shintani, T., Huang, W.-P., Stromhaug, P. E., and Klionsky, D. J. (2002) Dev. Cell 3 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, J., Kamada, Y., Stromhaug, P. E., Guan, J., Hefner-Gravink, A., Baba, M., Scott, S. V., Ohsumi, Y., Dunn, W. A., Jr., and Klionsky, D. J. (2001) J. Cell Biol. 153 381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nice, D. C., Sato, T. K., Stromhaug, P. E., Emr, S. D., and Klionsky, D. J. (2002) J. Biol. Chem. 277 30198–30207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheong, H., Yorimitsu, T., Reggiori, F., Legakis, J. E., Wang, C.-W., and Klionsky, D. J. (2005) Mol. Biol. Cell 16 3438–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawamata, T., Kamada, Y., Suzuki, K., Kuboshima, N., Akimatsu, H., Ota, S., Ohsumi, M., and Ohsumi, Y. (2005) Biochem. Biophys. Res. Commun. 338 1884–1889 [DOI] [PubMed] [Google Scholar]
- 41.Reggiori, F., Monastyrska, I., Shintani, T., and Klionsky, D. J. (2005) Mol. Biol. Cell 16 5843–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott, S. V., Guan, J., Hutchins, M. U., Kim, J., and Klionsky, D. J. (2001) Mol. Cell 7 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tucker, K. A., Reggiori, F., Dunn, W. A., Jr., and Klionsky, D. J. (2003) J. Biol. Chem. 278 48445–48452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabeya, Y., Kawamata, T., Suzuki, K., and Ohsumi, Y. (2007) Biochem. Biophys. Res. Commun. 356 405–410 [DOI] [PubMed] [Google Scholar]
- 45.Kawamata, T., Kamada, Y., Kabeya, Y., Sekito, T., and Ohsumi, Y. (2008) Mol. Biol. Cell 20, 19 2039–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clark, S. L., Jr. (1957) J. Biophys. Biochem. Cytol. 3 349–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takeshige, K., Baba, M., Tsuboi, S., Noda, T., and Ohsumi, Y. (1992) J. Cell Biol. 119 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemasters, J. J. (2005) Rejuvenation Res. 8 3–5 [DOI] [PubMed] [Google Scholar]
- 49.Abeliovich, H. (2007) Autophagy 3 275–277 [DOI] [PubMed] [Google Scholar]
- 50.Farre, J. C., Manjithaya, R., Mathewson, R. D., and Subramani, S. (2008) Dev. Cell 14 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shintani, T., and Klionsky, D. J. (2004) Science 306 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reggiori, F., Wang, C.-W., Nair, U., Shintani, T., Abeliovich, H., and Klionsky, D. J. (2004) Mol. Biol. Cell 15 2189–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson, J. S., Klionsky, D. J., Banta, L. M., and Emr, S. D. (1988) Mol. Cell. Biol. 8 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamada, Y., Funakoshi, T., Shintani, T., Nagano, K., Ohsumi, M., and Ohsumi, Y. (2000) J. Cell Biol. 150 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lang, T., Schaeffeler, E., Bernreuther, D., Bredschneider, M., Wolf, D. H., and Thumm, M. (1998) EMBO J. 17 3597–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang, W.-P., Scott, S. V., Kim, J., and Klionsky, D. J. (2000) J. Biol. Chem. 275 5845–5851 [DOI] [PubMed] [Google Scholar]
- 57.Noda, T., Kim, J., Huang, W.-P., Baba, M., Tokunaga, C., Ohsumi, Y., and Klionsky, D. J. (2000) J. Cell Biol. 148 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.