Abstract
Human glutathione transferase π (GST π) has been crystallized as a homodimer, with a subunit molecular mass of ∼23 kDa; however, in solution the average molecular mass depends on protein concentration, approaching that of monomer at <0.03 mg/ml, concentrations typically used to measure catalytic activity of the enzyme. Electrostatic interaction at the subunit interface greatly influences the dimer-monomer equilibrium of the enzyme and is an important force for holding subunits together. Arg-70, Arg-74, Asp-90, Asp-94, and Thr-67 were selected as target sites for mutagenesis, because they are at the subunit interface. R70Q, R74Q, D90N, D94N, and T67A mutant enzymes were constructed, expressed in Escherichia coli, and purified. The construct of N-terminal His tag enzyme facilitates the purification of GST π, resulting in a high yield of enzyme, but does not alter the kinetic parameters or secondary structure of the enzyme. Our results indicate that these mutant enzymes show no appreciable changes in Km for 1-chloro-2,4-dinitrobenzene and have similar CD spectra to that of wild-type enzyme. However, elimination of the charges of either Arg-70, Arg-74, Asp-90, or Asp-94 shifts the dimer-monomer equilibrium toward monomer. In addition, replacement of Asp-94 or Arg-70 causes a large increase in the KmGSH, whereas substitution for Asp-90 or Arg-74 primarily results in a marked decrease in Vmax. The GST π retains substantial catalytic activity as a monomer probably because the glutathione and electrophilic substrate sites (such as for 1-chloro-2,4-dinitrobenzene) are predominantly located within each subunit.
Cytosolic glutathione S-transferases (GSTs)3 are a family of detoxification enzymes involved in the metabolism of endogenous and xenobiotic electrophilic compounds (1-3). They catalyze the conjugation of glutathione to the electrophilic center of a variety of toxic compounds, resulting in more water-soluble products and facilitating the transport of toxic substances from cells (1, 4). The mammalian GSTs have been grouped into several classes according to their sequence homology, substrate specificities, and physical properties (3-8). GSTs have been implicated as promising therapeutic targets because specific isoenzymes are overexpressed in a variety of tumors, contributing to the development of resistance of tumors toward anticancer drugs (2, 9).
Human GST π has a widespread distribution in most tumor cells and is predominantly found in lung but is absent in liver. This enzyme was crystallized as a homodimer, with a subunit molecular mass of ∼23 kDa. Each subunit contains an active site that consists of a glutathione (GSH) binding site (G site) (7, 8, 10-12) and several xenobiotic substrate binding sites (H site) (7, 8, 13-18) adjacent to the G site.
The crystal structure shows two main areas of interaction between the subunits: a hydrophobic region and an electrostatic region. Unlike the hydrophobic cavity of μ class GST, the xenobiotic binding site of GST π is approximately half hydrophobic and half hydrophilic (7, 8, 11-14). The importance of the hydrophobic region (lock and key interaction) as an influence on the monomer-dimer equilibrium and enzymatic activity has been extensively studied by other groups (3, 7, 14, 19). In contrast, the electrostatic interactions at the subunit interface of GST π have not previously been investigated in depth. Chaotropic salts have been used to disrupt electrostatic interactions in proteins by shielding charges between subunits (20-22). Studies conducted in our laboratory showed that the molecular mass of GST π is decreased by the addition of potassium bromide, implying that electrostatic interactions are important forces holding the subunits together (23).
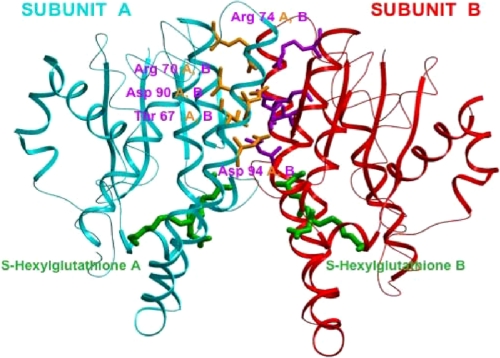
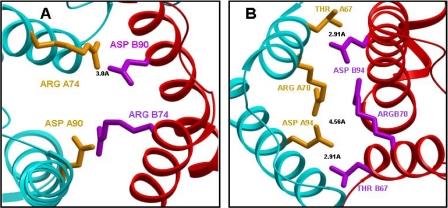
In this report, we focus on the electrostatic interactions at the subunit interface of GST π. The crystal structure of GST π (Fig. 1) shows the charged amino acids at the subunit interface of GST π (7, 8, 11, 13) (Arg-70, Arg-74, Asp-90, and Asp-94), away from the binding site of S-hexylglutathione. These residues are sufficiently close to participate in intra- and intersubunit electrostatic interactions that can influence the monomer-dimer equilibrium of the enzyme. Fig. 2 shows an expanded view of these amino acids. The distance between Arg-74 of subunit A and Asp-90 of the subunit B is only 3.0 Å (Fig. 2A), whereas the closest distance between the two carboxylate oxygens of Asp-94 of subunit A and the guanidinium group of Arg-70 of subunit B is 4.6 Å (Fig. 2B). In addition, Thr-67 of subunit A is only 2.9 Å from the carboxylate oxygens of Asp-94 of subunit B (Fig. 2B), a distance consistent with hydrogen bonding between these amino acids. The two arginines and two aspartates are conserved among GSTs of the π class from other mammals, although Thr-67 is not, as represented by the sequence alignment shown in Fig. 3.
FIGURE 1.
Dimeric structure (side view) of human GST π (PDB 9GSS) crystallized with S-hexylglutathione, which is colored green. This structure shows the electrostatic region between two subunits. Each individual subunit is colored-coded: the backbone of subunit A is shown in cyan, while that of subunit B is red. The residues selected for mutagenesis at the subunit interface are Arg-70, Arg-74, Asp-90, Asp-94, and Thr-67. The side chains of the A subunit amino acid residues are in yellow, whereas those of the B subunit are purple.
FIGURE 2.
An enlargement of the wild-type human GST π interface showing interacting residues between the two subunits. Subunit A is displayed in cyan and subunit B is in red. A, the distance shown is between Arg-74 (A) (yellow) and Asp-90 (B) (purple) (3.0 Å). B, an enlargement of wild-type GST π interface showing Thr-67, Arg-70, and Asp-94 from both subunits. Subunit A is displayed in cyan and subunit B is in red. The distances shown are between the closest oxygen of Asp-94 (A subunit) and the guanido group of Arg-70 (B subunit) (4.56 Å), and between Thr-67A (A subunit) and Asp-94 (B subunit) (2.91 Å).
FIGURE 3.
The ClustalW sequence alignment of three mammalian π class GST isozymes. The mGSTPi, pGSTPi, and hGSTPi sequences are representative and are for mouse, porcine, and human species, respectively. The amino acids selected for mutagenesis in this study are designated by bold letters.
The aim of this study is to use site-directed mutagenesis and light scattering studies to evaluate the contribution to the stability of the dimer structure of amino acids at the subunit interface, which may be involved in electrostatic interaction. Arg-74, Asp-90, Arg-70, Asp-94, and Thr-67 were selected as target sites for mutagenesis because of their proximity to the subunit interface. In addition, we address the question of whether the dimeric form of GST π is required for activity.
EXPERIMENTAL PROCEDURES
Materials—GSH, 1-chloro-2,4-dinitrobenzene (CDNB), ampicillin, dithiothreitol, and buffer components were purchased from Sigma Co. Ni-NTA resin and the QuikChange-XL kit were obtained from Stratagene (La Jolla, CA). Oligonucleotides were synthesized by Bio-Synthesis, Inc. (Lewisville, TX). The plasmid DNA purification kit was supplied by Qiagen Inc. (Valencia, CA). Human thrombin was purchased from Enzyme Research Lab, Inc. (South Bend, IN). CM-cellulose (CM-52) was supplied by Whatman Inc. (Clifton, NJ).
Plasmid and Mutagenesis—The cDNA encoding human GST P1-1 was inserted into pET-15b vector, which contains a thrombin cleavable hexahistidine N-terminal tag (24). All mutant enzymes were generated using the QuikChange-XL kit. The oligonucleotides used for mutagenesis were as follows: T67A forward primer (5′-C CAG TCC AAT GCC ATC CTG CGT CAC CTG G) and its complement; R70Q forward primer (5′-CC AAT ACC ATC CTG CAG CAC CTG GGC CG) and its complement; R74Q forward primer (5′-G CGT CAC CTG GGC CAG ACC CTT GGG) and its complement; D90N forward primer (5′-GCA GCC CTG GTG AAC ATG GTG AAT GAC G) and its complement; D94N forward primer (5′-G GAC ATG GTG AAT AAC GGC GTG GAC C) and its complement. The underlined codons are for mutated amino acids. DNA extraction and purification were performed using a QIAgen Spin Miniprep Kit. All mutations were confirmed by sequencing performed on an ABI Prism model 377 DNA sequencer (PE Biosystems) at the center for Agricultural Biotechnology, University of Delaware.
Expression and Purification of Wild-type and Mutant Enzyme—All His tag wild-type and mutant enzymes were expressed in BL21 Escherichia coli cells. In each case, the cell cultures were grown at 37 °C in LB medium containing 0.1 mg/ml ampicillin and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 25 °C for 24 h. The cell cultures were then centrifuged and collected, as described previously (25). These His tag enzymes were purified using an Ni-NTA column (25). In this method, the cell pellet of wild-type or mutant enzyme was sonicated, and the supernatant was applied to the Ni-NTA column (12 ml of resin) equilibrated with 20 mm Tris-HCl buffer, pH 7.8, containing 0.2 m NaCl and 0.1 mm dithiothreitol (Buffer A). The column was eluted first with Buffer A, followed by Buffer B (Buffer A containing 20 mm imidazole). The enzyme was eluted at ∼50 mm imidazole using a linear gradient of imidazole (20-200 mm) in buffer A (100 ml of each buffer). The fractions exhibiting activity were pooled, concentrated, and dialyzed into 0.1 m potassium phosphate buffer (pH 6.5) containing 1 mm EDTA.
For some experiments the His tag was cleaved by thrombin and then purified using a CM-52 ion-exchange column. In this procedure, the His tag enzyme was incubated with thrombin (40 units/mg GST protein) in 0.1 m potassium phosphate buffer, pH 7.2, at 25 °C for 2 h. Separation of non-His tag GST and thrombin was achieved by applying the digest to a CM-52 column equilibrated with 0.1 m potassium phosphate buffer (pH 7.2). The isoelectric point of thrombin (7.5) is much higher than that of GST π (5.6); at pH 7.2, the negatively charged cleaved GST is eluted in the column wash, whereas the positively charged thrombin binds to the negatively charged carboxymethyl-cellulose column. The fractions of relatively high GST activity were pooled, concentrated, and dialyzed into 0.1 m phosphate buffer (pH 6.5) containing 1 mm EDTA.
The purity of His tag and non-His tag enzymes was assessed by SDS-PAGE (26) and N-terminal sequencing. N-terminal amino acid sequencing was conducted on an Applied Biosystems instrument (Model Procise) equipped with an on-line microgradient delivery system (Model 140C) and a computer (Model 610 Macintosh). All the purified enzymes were stored in aliquots at -80 °C.
Enzyme Assays—As a standard assay, the enzymatic activity was monitored continuously by the formation of the conjugate of glutathione (GSH) (2.5 mm in assay) and CDNB (3 mm) at 340 nm (Δε = 9.6 mm-1cm-1) in 0.1 m potassium phosphate buffer, pH 6.5, containing 1 mm EDTA at 25 °C (27). All assays contained 2.5% ethanol, which facilitates the solubility of CDNB. The measured enzymatic activity was corrected for the nonenzymatic reaction between GSH and CDNB. To determine the apparent Km for GSH, the concentration of GSH was varied (generally, 0.02 to 20 mm), whereas the concentration of CDNB was held constant at 3 mm in potassium phosphate buffer (pH 6.5) containing 1 mm EDTA. Similarly, to determine the apparent Km for CDNB, a range of CDNB concentration was used (0.05 to 3 mm), while the GSH concentration was maintained at saturating concentrations (2.5-60 mm). The Km and Vmax values were determined from direct plots of velocity versus substrate concentration using SigmaPlot software, and are presented along with their standard errors. The enzyme concentrations used in the assays are given in the tables.
CD Spectroscopy—CD spectra of non-His tag and His tag wild-type and mutant enzymes were recorded using an Aviv CD spectrometer, Model 400. All enzymes (∼0.3 mg/ml) were scanned five times, averaged, and the background from buffer (0.1 m potassium phosphate buffer and 1 mm EDTA, pH 6.5) was subtracted from each spectrum. Ellipticity was measured as a function of wavelength from 250 to 200 nm at 2 nm increments using a quartz cuvette of 0.1 cm path length. The mean molar ellipiticity (θ) (deg cm2dmol-1) was calculated from the equation, (θ) = θ/(10 nCl), where θ is the measured ellipticity in millidegrees, C is the molar concentration of subunit, l is the path length in centimeters, and n is the number of amino acids per subunit (215 and 231 for non-His tag and His tag GST π, respectively). The protein concentration was determined using the Bio-Rad protein assay, based on the Bradford method, with the homogeneous wild-type GST π as the protein standard (28).
Molecular Mass Determination by Light Scattering—Molecular masses of wild-type and mutant enzymes were determined using light scattering (miniDAWN laser photometer from Wyatt Technology). Buffer (0.1 m potassium phosphate and 1 mm EDTA, pH 6.5) and all enzymes (0.02-1.2 mg/ml) were filtered through a 0.2-μm filter before being used. Because of possible changes in the concentration during filtration, the protein concentration (measured by the Bio-Rad assay, as described above) was determined on the sample used for light scattering after removal from the cuvette. Data were collected and analyzed using ASTRA for Windows.
Atomic Force Microscopy—Protein was imaged on a cleaned polylysine-coated glass slide. AFM images were obtained on a BioScope II (Veeco Instruments) in conjunction with an inverted optical microscope (Axiovert, Zeiss, Germany) operated in contact mode in fluid. Protein samples (10 μl of enzyme at 0.25 mg/ml) were incubated with 5 μl of 2.5% glutaraldehyde solution in 0.1 m potassium phosphate buffer, pH 6.5, at room temperature for 15 min on the treated glass slides. Samples were then washed with phosphate buffer (100 μl) before imaging (29). Silicon-nitride probes (MLCT, Veeco Probes) with a nominal spring constant of 0.01 newton/m were used for imaging under phosphate-buffered saline. Samples were flattened using first order flattening and analyzed as described below.
Molecular Mass Determination from AFM Images—Cross-sections (width, w, and height, h) of individual protein molecules were measured using the AFM software (Nanoscope version 7.20). The radius (r) is dependent on the width (w) and is calculated by the equation 1, r = w2/16R, (where r = 30 nm, and is a property of the geometry of the scanning tip). These images were analyzed for molecular volume in accordance with equation 2, Vm = (h/6) (3r2 + h2), which in turn was used to determine the molecular weight of the sample. Briefly, cross-sections were taken of individual proteins and compared with a standard curve of proteins of known molecular weight (29, 30).
Molecular Modeling—Models were generated using the Insight II modeling software from Molecular Simulations, Inc. on an Indigo 2 workstation from Silicon Graphics. The model was based on the crystal structure of human GST π (PDB #9GSS) in complex with S-hexylglutathione. The figures illustrating the models were prepared using ICM software from Molsoft LLC (La Jolla, CA).
RESULTS
Expression and Purification of Wild-type and Mutant GST π—The plasmid encoding human GST π with a thrombin-cleavable hexahistidine N-terminal tag was transformed and expressed in BL21 E. coli cells. The His tag enzymes were purified on an Ni-NTA column, as described under “Experimental Procedures.” Enzymes were purified to homogeneity, yielding a single protein by N-terminal protein sequencing. The N-terminal sequence of GSSHHHHHHSSGLVPRGSHMLEPPYTVVYEFP confirmed the presence of the thrombin cleavable hexahistidine (underlined amino acids) before the N-terminal of GST π (PPYTVV—). The purity and identity of His tag enzymes were also evaluated by SDS-PAGE. A single distinct band, corresponding to the molecular mass of 25.5 kDa, was observed for wild-type and mutant enzymes (data not shown). These results demonstrate that all His tag enzyme preparations contained a single protein.
The non-His tag enzyme was obtained by first digesting the isolated His tag enzyme with thrombin and then purifying it on a CM-52 column at pH 7.2, as described under “Experimental Procedures.” N-terminal sequencing of each purified enzyme indicated the presence of one protein with the correct N-terminal sequence of GSHMLEPPY; the underlined amino acids before the N terminus of GST π (PPY) are the amino acid residues that follow the thrombin cleavage site. The results from SDS-PAGE confirmed the purity and identity of these enzymes: only a single band was observed for each enzyme, corresponding to the correct molecular mass of 23.5 kDa for the non-His tag wild-type and mutant class π GSTs (data not shown). The total yield of GST π purified by these methods was five to ten times higher than that obtained by the earlier procedures (16, 17). The enzyme yield varies with the mutation; typically 100 mg/liter cell culture was obtained for the wild-type enzyme and 50-100 mg/liter cell culture for the mutant enzymes.
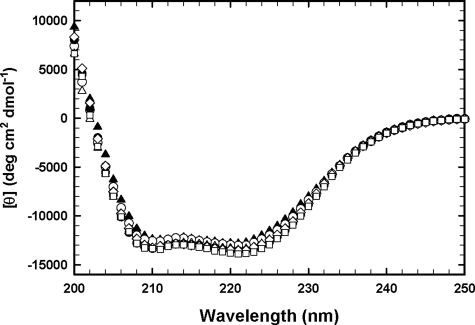
CD Spectra of Wild-type and Mutant Enzymes—CD spectra of all non-His tag and His tag GSTs were measured to evaluate whether the mutations cause changes in the secondary structure. The spectra were determined using protein concentrations of 0.3 mg/ml in potassium phosphate buffer (pH 6.5), as described under “Experimental Procedures.” As shown in Fig. 4, the CD spectra of all non-His tag mutant enzymes were very similar to that of non-His tag wild-type enzyme. The CD spectrum of non-His tag wild-type GST π is superimposable on that of His tag wild-type enzyme, indicating that there is no detectable difference in secondary structure between these two forms of the enzyme. In addition, the CD spectra of all His tag mutant enzymes were the same as that of the His tag wild-type enzyme (data not shown). These results indicate that the mutations at the electrostatic interface do not cause appreciable changes in the enzyme's secondary structure.
FIGURE 4.
CD spectra were determined with the protein (0. 3 mg/ml) in 0.1 m potassium phosphate (pH 6.5) containing 1 mm EDTA. CD spectra for non-His tag wild-type and mutant enzymes: WT (▴), T67A (○), R70Q (▵), R74Q (•), D90N (⋄), D94N (□).
Kinetic Parameters for Enzymes with Subunit Interface Mutations—Table 1 shows the kinetic parameters determined for non-His tag enzymes with replacements for amino acids in the electrostatic portion of the subunit interface. The specific activity of wild-type enzyme, measured under the standard assay conditions in phosphate buffer (pH 6.5), was 75 μmol min-1mg-1. The corresponding values for R70Q, R74Q, D90N, and D94N mutant enzymes were greatly decreased.
TABLE 1.
Kinetic parameters at pH 6.5 for non-His-tag wild type and mutant human GST π enzymes
| Enzyme | Specific activitya | Protein concentration in assay | Vmaxb | Km GSHb | Km CDNBc |
|---|---|---|---|---|---|
| μmol min−1 mg−1 | mg/ml | μmol min−1 mg−1 | mm | mm | |
| WT | 75 ± 3.0 | 0.00028 | 75 ± 1.4 | 0.31 ± 0.03 | 1.00 ± 0.20 |
| T67A | 102 ± 3.4 | 0.00018 | 116 ± 2 | 0.25 ± 0.06 | 1.26 ± 0.19 |
| R70Q | 3.4 ± 0.14 | 0.0038 | 9.62 ± 0.49 | 4.10 ± 0.63 | 1.11 ± 0.38 |
| R74Q | 0.18 ± 0.014 | 0.03-0.09 | 0.20 ± 0.01 | 0.88 ± 0.11 | 0.90 ± 0.03 |
| D90N | 0.019 ± 0.0015d | 0.04 | 0.03 ± 0.001 | 0.73 ± 0.14 | 1.62 ± 0.66 |
| D94N | 0.21 ± 0.014e | 0.04 | 3.00 ± 0.48 | 52 ± 10 | 0.64 ± 0.10 |
The standard assay solution contains 2.5 mm GSH, 3 mm CDNB, and 1 mm EDTA in 0.1 m potassium phosphate buffer (pH 6.5) in a total volume of 1 ml at 25 °C.
Vmax and Km values were determined by extrapolation to infinite concentration of the GSH, while maintaining (CDNB) at 3 mm in 0.1 m potassium phosphate buffer (pH 6.5) containing 1 mm EDTA, using SigmaPlot for data analysis.
Km values were determined by varying the concentration of CDNB (0.05-3 mm), while the GSH concentration was maintained at concentrations (2.5-60 mm), which are 10 to 20 times the Km GSH for that enzyme. These concentrations were close to saturating for all the enzymes except D94N (for which 60 mm GSH was used).
A specific activity of 0.017 ± 0.0004 μmol/min/mg was measured using 0.15 mg of protein/ml in the assay.
A specific activity of 0.17 ± 0.015 μmol/min/mg was measured using 0.06 mg of protein/ml in the assay.
The specific activity of R70Q mutant enzyme was ∼4.5% that of wild-type enzyme, whereas both R74Q and D94N mutant enzymes exhibited only ∼0.2% of the activity of wild-type enzyme. However, when the negatively charged aspartate was replaced by neutral asparagine at position 90, the enzyme lost most of its activity (0.02% of wild-type enzyme). In contrast, substitution by alanine for threonine at position 67 actually resulted in an increase in specific activity (102 μmol min-1 mg-1).
Table 1 also shows the KmGSH, KmCDNB, and Vmax values extrapolated to saturating concentrations of GSH for wild-type and mutant enzymes. The KmGSH value of T67A mutant is similar to that of the wild-type enzyme. The KmGSH values of R74Q and D90N exhibit small increases (∼2- to 3-fold), whereas the KmGSH values of R70Q and D94N increased greatly. Notably, the D94N mutant enzyme had a KmGSH value more than 160 times that of the wild-type enzyme. In contrast, there were no major changes in Km values for CDNB for all mutant enzymes, measured in the presence of a constant concentration of GSH that was high relative to the Km for that enzyme.
Table 2 reports the kinetic parameters of His tag wild-type and His tag mutant enzymes (T67A, R70Q, R74Q, D90N, and D94N) at pH 6.5. The specific activities, KmGSH, KmCDNB, and Vmax values of all His tag enzymes, were similar to those of the corresponding non-His tag enzymes (compare Tables 1 and 2), indicating that the additional 16 amino acids had little effect on the catalytic properties of GST. The kinetic parameters (Tables 1 and 2) suggest that the removal of charge from Arg-70, Asp-94, Arg-74, or Asp-90 results in substantial decreases in Vmax, particularly marked in the cases of Arg-74 and Asp-90. In contrast, replacement of threonine by alanine at position 67 did not diminish the activity. Unchanged values for Km CDNB indicate that these residues are not directly involved in the interaction of the enzyme with CDNB.
TABLE 2.
Kinetic parameters at pH 6.5 for His tag wild-type and mutant human GST π enzymes
| Enzyme | Specific activitya | Protein concentration in assay | Vmaxb | Km GSHb | Km CDNBc |
|---|---|---|---|---|---|
| μmol min−1 mg−1 | mg/ml | μmol min−1 mg−1 | mm | mm | |
| His-WT | 73.5 ± 2.5 | 0.0002 | 71 ± 1 | 0.32 ± 0.03 | 0.83 ± 0.09 |
| His-T67A | 102 ± 0.6 | 0.0004 | 101 ± 3 | 0.22 ± 0.04 | 0.90 ± 0.39 |
| His-R70Q | 6.2 ± 0.2 | 0.0066 | 11 ± 0.23 | 2.11 ± 0.18 | 0.87 ± 0.20 |
| His-R74Q | 0.19 ± 0.014 | 0.04 | 0.21 ± 0.006 | 0.41 ± 0.06 | 0.51 ± 0.08 |
| His-D90N | 0.02 ± 0.006d | 0.04 | 0.02 ± 0.001 | 1.18 ± 0.26 | 0.85 ± 0.24 |
| His-D94N | 0.20 ± 0.018e | 0.04 | 3.40 ± 0.83 | 61 ± 19 | 0.70 ± 0.17 |
The standard assay solution contains 2.5 mm GSH, 3 mm CDNB, and 1 mm EDTA in 0.1 M potassium phosphate buffer (pH 6.5) in a total volume of 1 ml at 25 °C.
Vmax and Km values were determined by extrapolation to infinite concentration of the GSH, while maintaining (CDNB) at 3 mm in 0.1 M potassium phosphate buffer (pH 6.5) containing 1 mm EDTA using SigmaPlot for data analysis.
Km values were determined by varying the concentration of CDNB (0.05-3 mm), while the GSH concentration was maintained at high concentrations (2.5-60 mm), which are close to saturating for all the enzymes except D94N.
A specific activity of 0.018 ± 0.0017 μmol/min/mg was measured using 0.37 mg of protein/ml in the assay.
A specific activity of 0.20 ± 0.008 μmol/min/mg was measured using 0.08 mg of protein/ml in the assay.
Determination of Molecular Masses by Light Scattering—Molecular masses for non-His tag and His tag wild-type enzymes were measured using light scattering at pH 6.5 (Table 3). Each enzyme was measured over a range of protein concentrations (usually 0.03-1.0 mg/ml). The average molecular masses of non-His tag wild-type enzyme were 32,000-34,000 at a protein concentration of 0.06-0.4 mg/ml. As shown in Table 3 (compare columns 6 and 4), the addition of glutathione exerted little effect on the average molecular mass of the non-His tag enzyme at low protein concentrations; however, at protein concentrations above 0.2 mg/ml, GSH caused a small increase in the weight average molecular mass. The substrate CDNB also has only a small influence on the molecular mass (compare columns 8 and 4). Similarly, the addition of a reaction product S-hexylglutathione, which occupies both the glutathione and electrophilic substrate sites and is also an inhibitor of the GST-catalyzed reaction of glutathione and CDNB (1), does not change the enzyme's molecular weight more than the addition of each substrate individually (compare columns 10, 8, 6, and 4). Because each subunit has a mass of 23,500, these results suggest that the non-His tag enzyme exists as an equilibrium mixture of monomer-dimer, with the monomer dominating below 0.3 mg/ml. In contrast, the average molecular mass of wild-type His tag GST (40,000-47,000) was substantially higher than that of non-His tag enzymes over the same protein concentration range (compare columns 4 and 2). Above 0.1 mg/ml, the average molecular mass of the His tag enzyme was close to that of the dimer. When the protein concentration was higher than 1 mg/ml, both the His tag and the non-His tag enzymes existed as dimers, and when the protein concentration was lower than 0.03 mg/ml, the enzymes were predominantly monomers, even in the presence of glutathione, CDNB, or S-hexylglutathione. These results indicate that both forms of GST π exist in an equilibrium mixture of monomer and dimer, but that the additional 16 amino acids of the His tag enzyme stabilize the dimeric form.
TABLE 3.
Weight average molecular masses, as determined by light scattering, of the His tag and non-His tag WT enzymes
In all cases, the enzyme was in 0.1 M potassium phosphate buffer (pH 6.5), containing 1 mm EDTA.
|
His-tag WT enzyme
|
Non-His tag WT enzyme
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
No additions
|
No additions
|
+2.5 mm glutathione
|
+3 mm CDNB
|
+2.5 mmS-hexylglutathione
|
|||||
| Conc. | Molecular mass | Conc. | Molecular mass | Conc. | Molecular mass | Conc. | Molecular mass | Conc. | Molecular mass |
| mg/ml | Da | mg/ml | Da | mg/ml | Da | mg/ml | Da | mg/ml | Da |
| 0.03 | 32,900 ± 1,000 | 0.03 | 29,600 ± 3,900 | 0.03 | 28,720 ± 1880 | 0.03 | 29,500 ± 1,600 | 0.03 | 24,630 ± 1,160 |
| 0.04 | 25,760 ± 1,790 | ||||||||
| 0.06 | 38,100 ± 300 | 0.06 | 34,100 ± 2,300 | 0.07 | 31,520 ± 370 | 0.06 | 27,290 ± 3,600 | ||
| 0.08 | 28,910 ± 840 | ||||||||
| 0.09 | 40,500 ± 3,000 | 0.10 | 33,200 ± 1,800 | 0.09 | 33,500 ± 1,600 | 0.09 | 31,200 ± 1,150 | ||
| 0.15 | 46,400 ± 5,100 | 0.12 | 33,200 ± 1,300 | 0.14 | 33,590 ± 360 | 0.13 | 31,750 ± 460 | ||
| 0.18 | 47,300 ± 2,800 | 0.19 | 34,600 ± 2,300 | 0.19 | 33,520 ± 1,450 | ||||
| 0.29 | 46,200 ± 2,300 | 0.25 | 32,800 ± 4,100 | 0.22 | 34,960 ± 300 | ||||
| 0.39 | 46,800 ± 2,200 | 0.33 | 33,100 ± 2,800 | 0.32 | 38,200 ± 1,100 | 0.37 | 37,860 ± 570 | ||
| 0.44 | 47,000 ± 1,300 | 0.40 | 33,400 ± 1,400 | 0.40 | 37,680 ± 720 | 0.47 | 41,780 ± 1,460 | ||
| 0.71 | 45,300 ± 2,400 | 0.79 | 41,800 ± 2,500 | 0.82 | 46,520 ± 1700 | 0.76 | 46,020 ± 1,300 | ||
| 1.07 | 44,800 ± 2,600 | 1.20 | 47,500 ± 2,700 | ||||||
Table 4 records the effect of interface mutations on the average molecular masses of His tag GST enzymes as assessed by light scattering under the same conditions, including protein concentration, used for wild-type enzyme (Table 3). His-tagged T67A mutant enzyme is similar in average molecular mass to that of wild-type His tag enzyme (44,000-50,000). In contrast, His-tagged R70Q, R74Q, D90N, and D94N mutant enzymes are systematically lower than wild-type enzyme in molecular mass; these mutants give average molecular masses of 23,000-27,000, measured at low protein concentration (<0.1 mg/ml). These results indicate that only the His tag T67A mutant enzyme exhibits the dimer molecular mass, whereas, the other four His tag mutant enzymes are predominantly monomers. The protein concentrations used here were the same or higher than those used to assay these enzymes (Table 2).
TABLE 4.
Weight average molecular masses, as determined by light scattering experiments, for His-tag WT and His tag mutant enzymes
In all cases, the His tag enzyme was in 0.1 m potassium phosphate buffer (pH 6.5), containing 1 mm EDTA. At higher protein concentrations, these His tag mutant enzymes were unstable and tended to aggregate.
| Enzyme | Concentration | Molecular mass |
|---|---|---|
| mg/ml | Da | |
| His-WT | 0.03 | 32,900 ± 1,000 |
| 0.06 | 38,100 ± 300 | |
| 0.09 | 40,500 ± 3,000 | |
| 0.15 | 46,400 ± 5,100 | |
| His-T67A | 0.05 | 43,500 ± 3,000 |
| 0.13 | 47,500 ± 2,500 | |
| 0.26 | 50,000 ± 2,500 | |
| His-R70Q | 0.02 | 26,900 ± 2,000 |
| 0.07 | 25,900 ± 1,700 | |
| His-R74Q | 0.01 | 24,700 ± 5,800 |
| 0.04 | 24,500 ± 2,500 | |
| His-D90N | 0.01 | 23,300 ± 500 |
| 0.04 | 24,300 ± 9,100 | |
| His-D94N | 0.04 | 25,400 ± 8,900 |
| 0.07 | 25,200 ± 1,200 |
Non-His tag T67A mutant GST π is similar to the wild-type enzyme: it behaves as a mixture of monomer and dimer, with a molecular mass of 33,000 ± 1500, measured at protein concentrations ranging from 0.05 to 0.3 mg/ml. The average molecular masses of Non-His tag R70Q, R74Q, D90N, and D94N mutant enzymes could not be obtained, because these mutant enzymes are unstable over the time period needed for the light scattering measurements (data not shown).
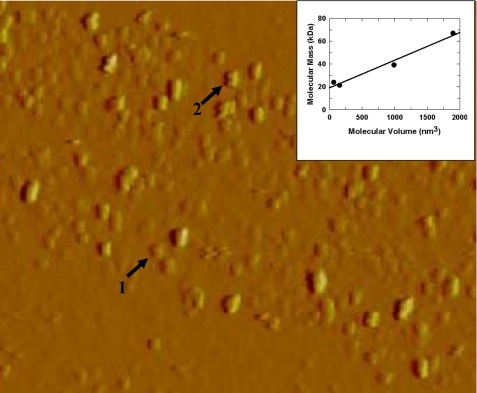
Molecular Mass Determination from Atomic Force Microscopy—Non-His tag wild-type enzyme was examined and scanned in phosphate buffer (pH 6.5) under an AFM, as described under “Experimental Procedures.” As shown in Fig. 5, the wild-type GST π exhibited a mixture of molecules of two different sizes. The inset of Fig. 5 shows the molecular mass of protein standards plotted against the molecular volumes measured by AFM, demonstrating that molecular mass increased with molecular volume. The average molecular mass of these molecules of two sizes found in solutions of wild-type enzyme can be estimated from Fig. 5 as 28.2 ± 2.3 kDa and 58.2 ± 5.5 kDa, respectively. Non-His tag T67A mutant enzyme exhibited an AFM similar to that of wild-type enzyme. This result is consistent with our findings from light scattering that wild-type and T67A enzymes exist as a mixture of monomer and dimer.
FIGURE 5.
AFM images of human wild-type GST π (from a solution of 0. 25 mg/ml). Image size 25 nm × 4.5 μm. Arrow 1: monomer; arrow 2: dimer. Inset: the calculated volume of proteins imaged by AFM plotted against the molecular mass. The proteins used are xylanase from T. longibraciatum (21.6 kDa), polygalacturonase from A. nigar (24.0 kDa), xylanase from C. japonicus (39.2 kDa), and bovine serum albumin (67.0 kDa).
DISCUSSION
GST π is crystallized as a dimer (7, 31), as illustrated in Fig. 1. However, when the non-His tag wild-type GST π is in solution in the protein concentration range of 0.03-1 mg/ml, it exists as an equilibrium mixture of monomer and dimer. The average molecular mass is considerably lower than the 47,000 Da expected for a dimer, and the average mass increases as the total protein concentration increases (Table 3). The percentage of dimer and monomer can be calculated from the measured average molecular mass at a known protein concentration. For example, at a protein concentration of 0.12 mg/ml, the average molecular mass is 33,200, which indicates that the enzyme is only ∼40% dimeric in solution. In contrast, the α class GST is ∼88% dimeric under similar conditions (25), and the μ class GST is ∼82% dimeric (22). Using the average molecular mass measured for the non-His tag wild-type GST π over the protein concentration range 0.03-0.4 mg/ml, we calculated an average Kd of 5.1 ± 2.6 μm for dissociation of the dimer to the monomer. This value is ∼10 times higher than that measured for the α class GST (25). Although it is likely that all GSTs undergo a reversible equilibrium between dimer and monomer, GST π is the only one of the three major mammalian GST isozymes in which the monomer predominates under most conditions (22, 25). Indeed, the more extensive dissociation of GST π may account for the distinctive ability of GST π (as compared with GST α or μ) to form heterodimers with other proteins such as 1-cysteine peroxireductase (23, 32).
GST π was assayed at protein concentrations (typically 0.0002-0.0005 mg/ml for wild-type enzyme), which were much lower than those used for the molecular mass determinations (Tables 1 and 2). Based on the data for the protein concentration dependence of molecular mass shown in Table 3, GST π certainly exists as a monomer in the assay solution, even in the presence of glutathione, S-hexylglutathione, or CDNB. Because the measured specific activity of wild-type enzyme was ∼75 μmol/min/mg (Table 1), we conclude that the monomer of GST π is catalytically highly active. We earlier concluded that a monomer of the μ class of GST can be almost as active as the dimer form (22), whereas the monomer of the α class of GST retains only partial activity as compared with the dimer (25).
Previous results from studies in this laboratory indicate that electrostatic interactions between the two subunits of GST π can be disrupted by the addition of KBr, which shields the electrostatic interactions at the subunit interface and shifts the dimer-monomer equilibrium toward a monomer (23). In that case, we determined quantitatively the effect of changing ionic strength, by varying the concentration of KBr (0-4 m), on the catalytic properties and molecular mass of GST π (23). We found that, under these conditions, appreciable activity is retained by the monomeric form of the enzyme and estimated that, in the presence of KBr, the monomer is 38% as active as the dimer (23). In our initial report on isolation of a stable heterodimer complex of GST π and 1-cysteine peroxiredoxin, we showed that the GST activity of the complex (expressed per milligram of GST π present) was 28% of that of GST π alone (32). Because the catalytic activity of the GST π monomer within the complex may be influenced by the 1-cysteine peroxiredoxin subunit, this gives only an estimate of the activity of the GST π monomer. Nevertheless, it is clear that a dimer of GST π is not required for enzymatic activity.
An earlier study described the construction of a stable monomeric form of GST π by the introduction of ten specific mutations in the hydrophobic regions of the subunit interface (33). This stable monomer had lost catalytic activity, which is not surprising given the drastic changes in its amino acid sequence (33); that study did not answer the question of whether the normal monomer of GST π is catalytically active.
In the present study, we constructed single mutations for amino acids in the electrostatic region at the subunit interface of cytosolic GST π and studied the effect of these mutations on the active site, kinetic parameters, and molecular mass. Arg-74, Asp-90, Arg-70, Asp-94, and Thr-67 (Figs. 1, 2, 3) were selected as target sites for mutagenesis, because they are at the subunit interface. (None of these target sites for mutagenesis is among the ten sites mutated earlier by Abdalla et al. (33).) The amino acid residues involved in homodimer interaction of GST π at the dimer interface are highly specific and are predominantly distinct from the amino acid residues involved in heterodimer interaction of GST π with other proteins (23). The oppositely charged residues at the subunit's interface are important for the subunit-subunit recognition and the stability of the dimer.
Although it has been suggested that the hydrophobic lock and key interaction is the major interaction between subunit interfaces in all GSTs (7, 14, 15, 25, 34, 35), the electrostatic attractions definitely enhance the interaction between subunits and play an important role at the dimer interface (22, 23, 25, 36). Electrostatic interactions have also been shown to be involved in both the catalytic reaction and the subunit interactions of the δ class GST of Anopheles dirus (37, 38) and of the cephalopod σ class GST (39).
For human GST π, elimination of the charges of Arg-70, Arg-74, Asp-90, and Asp-94 caused a marked decrease in enzymatic activity (Tables 1 and 2) and shifts the equilibrium toward lower average masses (Table 4). These results suggest that the subunit interface can be perturbed by a single mutation. Because GST π can retain substantial catalytic activity as a monomer (23, 32), the loss of activity in these new mutants is not simply due to dissociation of the dimer.
The molecular masses of the wild-type and mutant GSTs are compared in Table 4 as His tag preparations. Data over the protein concentration range of 0.03-0.29 mg/ml can be used to calculate an average Kd of 0.61 μm for dissociation of dimer to monomer for the His tag wild-type GST π. Because the molecular masses of most of the His tag mutant enzymes shown in Table 4 are markedly lower than that of the corresponding wild-type GST π, it is apparent that the dissociation constants of the dimers of these mutant enzymes must be considerably higher. For the His tag R70Q, R74Q, D90N, and D94N, approximate Kd values of 21 μm, 35 μm, 45 μm, and 33 μm, respectively, can be estimated. However, because the measured molecular masses of these mutant enzymes are very close to that of the monomer, the Kd values should be taken only as rough approximations.
The Asp-90 of subunit A is only 3.0 Å away from Arg-74 of subunit B and can contribute both a very strong electrostatic interaction, as well as hydrogen bonding (Fig. 2A). Removal of charge at either position greatly interferes with the interaction and diminishes dimer formation (Table 4). Mutagenesis at these positions also results in the greatest decreases in Vmax (Tables 1 and 2). Asp-90 is conserved in GST π from all mammals (Fig. 3) and seems to be important for maintaining the proper conformation in that region of the enzyme. Asp-90 and Arg-74 are far from the active site, as indicated by the location of S-hexylglutathione (Fig. 1). Replacing the negatively charged aspartate with neutral asparagine is thus likely to have an indirect effect of perturbing the active site, perhaps mediated through a hydrogen bonding and electrostatic network in the vicinity of the active site leading to local conformational changes, as in the case of mμ class GST (22). The loss in activity of D90N and R74Q cannot be attributed solely to monomer formation, because other mutant enzymes that are also monomers exhibit higher Vmax values.
The highly specific G-site for binding GSH is formed by Tyr-7, Arg-13, Trp-38, Lys-44, Gln-51, Leu-52, and Ser-65 from one subunit and Asp-98 from the other subunit (7, 10, 11, 40, 41). Tyr-7, which is conserved in all cytosolic GSTs, is an essential residue for catalytic activity (41, 42). The structure of mouse GST π confirms that the role of Tyr-7 is to stabilize the thiolate by hydrogen bonding and to position it in the right orientation (40). Asp-98, an evolutionally conserved aspartyl residue across the dimer interface, participates in GSH recognition (7, 11, 43). Crystal structures show that the negatively charged carboxylate of Asp-98 interacts with the α-amino group of the γ-glutamyl moiety of GSH bound to the opposite subunit. Thus, any mutation that promotes monomerization of GST π will indirectly diminish the interactions between Asp-98 and glutathione, thereby weakening the affinity of GSH for the enzyme. Eliminating the negative charge at Asp-94 causes disruption of its electrostatic interaction with Arg-70 of the opposite subunit, and shifts the dimer-monomer equilibrium toward the monomer (Table 4). Although the carboxylate of Asp-94 is ∼7 Å away from that of Asp-98, removing the charge at position 94 may also indirectly affect the interaction of Asp-98 with the substrate by perturbing the hydrogen bonding network. These cumulative effects can account for the marked increase in Km for GSH (to ∼200 times that of WT enzyme).
Arg-70 of subunit A is only 4.6-5 Å away from Asp-94 of subunit B (Fig. 2B). Removing the charge from Arg-70 affects the electrostatic interaction with Asp-94 of the opposite subunit and subsequently affects both the binding of GSH and the dimer stability. The Km value for GSH for R70Q mutant enzyme increases substantially when compared with that of wild-type enzyme (Tables 1 and 2). In addition, removing the charge from either Asp-94 or Arg-70 lowers the enzymatic activity (Vmax values of D94N and R70Q mutant enzymes are 4.8% and 15.5%, respectively, that of the wild-type enzyme), although the effects on Vmax are not as large as those caused by replacing Asp-90 or Arg-74. The mutagenesis results suggest that substitution for Asp-94 and Arg-70 predominantly affect dimer stability and affinity for GSH. In contrast, Asp-90 and Arg-74 are not determinants of GSH affinity: the Km values for GSH exhibit only small increases in D90N and R74Q. These observations are consistent with the crystal structure of GST π, because Asp-90 and Arg-74 do not participate in GSH binding and are located far from the GSH binding site (7).
R70Q, R74Q, D90Q, D94Q, and T67A mutant enzymes show no appreciable change in Km for CDNB, as compared with that of the wild-type enzyme (Tables 1 and 2). This agrees with the crystal structure in which the electrophilic substrate binding site is near the C-terminal domain of one subunit and away from the subunit interface (3, 7). Therefore, it is not surprising that CDNB has little effect on the average molecular weight of the wild-type enzyme measured at various protein concentrations (Table 3).
In contrast to the other four mutants, the Thr-67 mutant enzyme is similar to the wild-type enzyme. It exists as a monomer-dimer mixture (Table 4) and has a similar Km for GSH (Tables 1 and 2). However, the enzymatic activity is higher than that of the wild-type enzyme. This may be because of the proximity of Thr-67 of subunit A (only 2.9 Å away) to Asp-94 of subunit B (Fig. 2B); these two residues can form a hydrogen bond. The crystal structure of GST π (Fig. 1) shows that there is a large cleft in the center of the dimer interface, which is open to the active sites and bulk solvent (7). The subunit interactions are, in part, mediated by the hydrogen bond between Thr-67 of human GST π of subunit A and Asp-94 of subunit B. Replacement of threonine with alanine at position 67 removes the hydrogen bond and increases the flexibility of the intersubunit cleft, perhaps making possible interactions within the protein that are more favorable to catalysis.
Recombinant proteins, which were expressed in Escherichia coli with an N-terminal His tag containing a thrombin cleavable site, were used in this study. This construct was used to facilitate the purification of the GST π and to obtain a high yield of enzyme. However, the results from our studies indicate that the average molecular mass of the His tag enzyme is considerably higher than that of the non-His tag enzyme when measured over the protein concentration range 0.03-0.8 mg/ml (Table 3, columns 2 and 4). This observation is likely due to the extra 16 amino acids at the N-terminal of each subunit, which may interact with the other subunit; thus, the dimerization of this His tag GST π may be an artifact. However, this non-physiological dimerization does not seem to alter the kinetic parameters or the secondary structure of the enzyme (Table 2 and Fig. 5). In our studies, all GST π in solution undergoes reversible association and dissociation; the extent of association depends on the protein concentration and the equilibrium constant (44). The His tag and the non-His tag wild-type enzymes were assayed at a very low protein concentration (∼0.0002 mg/ml) at which they are expected to exist mainly as monomers (Table 3). Thus, it is not surprising that these two wild-type enzymes exhibit similar specific activities and kinetic parameters (Tables 1 and 2).
The results of this study demonstrate that the His tag R70Q, R74Q, D90N, and D94N mutant enzymes exist as monomers when measured at protein concentrations at which the wild-type enzyme is predominantly dimeric. We conclude that the loss in catalytic activity upon mutation at these sites is due to the diminution in affinity for GSH and to an indirect effect on the active site, rather than directly to the dissociation of dimer. The dimerization of GST π may improve the stability of the enzyme, but it is not required for catalysis.
Acknowledgments
We thank Sharmila Sivendran for her helpful discussions and Mayura Dange for her assistance in making the figures.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-CA66561 (to R. F. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GST, GST; CDNB, 1-chloro-2,4-dinitrobenzene; WT, wild-type; AFM, atomic force microscope; Ni-NTA, nickel-nitriloacetic acid; p, porcine; m, mouse; h, human.
References
- 1.Mannervik, B., and Danielson, U. H. (1988) CRC Crit. Rev. Biochem. 23 283-337 [DOI] [PubMed] [Google Scholar]
- 2.Pickett, C. B., and Lu, A. Y. H. (1989) Annu. Rev. Biochem. 58 743-764 [DOI] [PubMed] [Google Scholar]
- 3.Wilce, M. C., and Parker, M. W. (1994) Biochim. Biophys. Acta 1205 1-18 [DOI] [PubMed] [Google Scholar]
- 4.Armstrong, R. N. (1997) Chem. Res. Toxicol. 10 2-18 [DOI] [PubMed] [Google Scholar]
- 5.Mannervik, B. (1985) Adv. Enzymol. 57 357-415 [DOI] [PubMed] [Google Scholar]
- 6.Mannervik, B., Board, P. G., Hayes, J. D., Listowsky, I., and Pearson, W. R. (2005) Methods Enzymol. 401 1-8 [DOI] [PubMed] [Google Scholar]
- 7.Reinemer, P., Dirr, H. W., Ladenstein, R., Huber, R., Lo Bello, M., Federici, G., and Parker, M. W. (1992) J. Mol. Biol. 227 214-226 [DOI] [PubMed] [Google Scholar]
- 8.Sinning, I., Kleywegt, G. J., Cowan, S. W., Reinemer, P., Dirr, H. W., Huber, R., Gilliland, G. L., Armstrong, R. N. Ji, X., Board, P. G., Olin, B., Mannervik, B., and Jones, T. A. (1993) J. Mol. Biol. 232 192-212 [DOI] [PubMed] [Google Scholar]
- 9.Townsend, D. M., and Tew, K. D. (2003) Oncogene 22 7369-7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manoharan, T. H., Gulick, A. M., Puchalski, R. B., Servais, A. L., and Fahl, W. E. (1992) J. Biol. Chem. 26 18940-18945 [PubMed] [Google Scholar]
- 11.Dirr, H., Reinemer, P., and Huber, R. (1994) J. Mol. Biol. 243 72-92 [DOI] [PubMed] [Google Scholar]
- 12.Erhardt, J., and Dirr, H. W. (1996) FEBS Lett. 391 313-316 [DOI] [PubMed] [Google Scholar]
- 13.Oakley, A. J., Lo Bello, M., Battistoni, A., Ricci, G., Rossjohn, J., Villar, H. O., and Parker, M. W. (1997) J. Mol. Biol. 274 84-100 [DOI] [PubMed] [Google Scholar]
- 14.Ji, X., Tordova, M., O'Donnell, R., Parsons, J. F., Hayden, J. B., Gilliland, G. L., and Zimniak, P. (1997) Biochemistry 36 9690-9702 [DOI] [PubMed] [Google Scholar]
- 15.Ji, X., Blaszczyk, J., Xiao, B., O'Donnell, R., Hu, X., Herzog, C., Singh, S. V., and Zimniak, P. (1999) Biochemistry 38 10231-10238 [DOI] [PubMed] [Google Scholar]
- 16.Ralat, L. A., and Colman, R. F. (2006) Biochemistry 45 12491-12499 [DOI] [PubMed] [Google Scholar]
- 17.Ralat, L. A., and Colman, R. F. (2004) J. Biol. Chem. 279 50204-50213 [DOI] [PubMed] [Google Scholar]
- 18.Ralat, L. A., and Colman, R. F. (2003) Protein Sci. 12 2575-2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenberg, D., Addalla, A. M., and Mannervik, B. (2000) Biochem. Biophys. Res. Commun. 271 59-63 [DOI] [PubMed] [Google Scholar]
- 20.Ramos, C. H., and Baldwin, R. L. (2002) Protein Sci. 11 1771-1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baldwin, R. L. (1996) Biophys. J. 71 2056-2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hearne, J. L., and Colman, R. F. (2006) Biochemistry 45 5974-5984 [DOI] [PubMed] [Google Scholar]
- 23.Ralat, L. A., Misquitta, S. A., Manevich, Y., Fisher, A. B., and Colman, R. F. (2008) Arch. Biochem. Biophys. 474 109-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang, M., Bolton, J. L., and Blond, S. Y. (1999) Protein Expr. Purif. 17 443-448 [DOI] [PubMed] [Google Scholar]
- 25.Vargo, M. A., Nguyen, L., and Colman, R. F. (2004) Biochemistry 43 3327-3335 [DOI] [PubMed] [Google Scholar]
- 26.Soundar, S., and Colman, R. F. (1993) J. Biol. Chem. 268 5264-5271 [PubMed] [Google Scholar]
- 27.Habig, W. H., Pabst, M. J., and Jakoby, W. B. (1974) J. Biol. Chem. 249 7130-7139 [PubMed] [Google Scholar]
- 28.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 29.Adams, E. L., Kroon, P. A., Williamson, G., Gilbert, H. J., and Morris, V. (2004) Carbohydrate Res. 339 579-590 [DOI] [PubMed] [Google Scholar]
- 30.Schneider, S. W., Larmer, J., Henderson, R. M., and Oberleithner, H. (1998) Eur. J. Physiol. 435 362-367 [DOI] [PubMed] [Google Scholar]
- 31.Parker, M. W., Lo Bello, M., and Federici, G. (1990) J. Mol. Biol. 213 221-222 [DOI] [PubMed] [Google Scholar]
- 32.Ralat, L. A., Manevich, Y., Fisher, A. B., and Colman, R. F. (2006) Biochemistry 45 360-372 [DOI] [PubMed] [Google Scholar]
- 33.Abdalla, A.-M., Bruns, C. M., Tainer, J. A., Mannervik, B., and Stenberg, G. (2002) Protein Eng. 15 827-834 [DOI] [PubMed] [Google Scholar]
- 34.Hornby, J. A., Codreanu, S. G., Armstrong, R. N., and Dirr, H. W. (2002) Biochemistry 41 14238-14247 [DOI] [PubMed] [Google Scholar]
- 35.Hagazy, U. M., Mannervik, B., and Stenberg, G. (2004) J. Biol. Chem. 279 9586-9596 [DOI] [PubMed] [Google Scholar]
- 36.Thompson, L. C., Walters, J., Burke, J., Parsons, J. F., Armstrong, R. N., and Dirr, H. W. (2006) Biochemistry 45 2267-2273 [DOI] [PubMed] [Google Scholar]
- 37.Vararattanavech, A., Prommeenate, P., and Ketterman, A. J., (2006) Biochem. J. 393 89-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winayanuwattikun, P., and Ketterman, A. J. (2007) Biochem. J. 402 339-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevens, J. M., Armstrong, R. N., and Dirr, H. W. (2000) Biochem. J. 347 193-197 [PMC free article] [PubMed] [Google Scholar]
- 40.Párraga, A., García-Sáez, I., Walsh, S. B., Mantle, T. J., and Coll, M. (1998) Biochem. J. 333 811-816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong, K. H., Takasu, K., Inoue, H., and Takahashi, K. (1992) Biochem. Biophys. Res. Commun. 184 194-197 [DOI] [PubMed] [Google Scholar]
- 42.Kong, K. H., Nishida, M., Inoue, H., and Takahashi, K. (1992) Biochem. Biophys. Res. Commun. 182 1122-1129 [DOI] [PubMed] [Google Scholar]
- 43.Kong, K. H., Inoue, H., and Takahashi, K. (1993) Protein Eng. 6 93-99 [DOI] [PubMed] [Google Scholar]
- 44.Carey, J., Lindman, S., Bauer, M., and Linse, S. (2007) Protein Sci. 16 2317-2333 [DOI] [PMC free article] [PubMed] [Google Scholar]