Abstract
Transforming growth factor β (TGF-β) signals through two distinct pathways to regulate endothelial cell proliferation, migration, and angiogenesis, the ALK-1/Smad 1/5/8 and ALK-5/Smad2/3 pathways. Endoglin is a co-receptor predominantly expressed in endothelial cells that participates in TGFβ-mediated signaling with ALK-1 and ALK-5 and regulates critical aspects of cellular and biological responses. The embryonic lethal phenotype of knock-out mice because of defects in angiogenesis and disease-causing mutations resulting in human vascular diseases both support essential roles for endoglin, ALK-1, and ALK-5 in the vasculature. However, the mechanism by which endoglin mediates TGF-β signaling through ALK-1 and ALK-5 has remained elusive. Here we describe a novel interaction between endoglin and GIPC, a scaffolding protein known to regulate cell surface receptor expression and trafficking. Co-immunoprecipitation and immunofluorescence confocal studies both demonstrate a specific interaction between endoglin and GIPC in endothelial cells, mediated by a class I PDZ binding motif in the cytoplasmic domain of endoglin. Subcellular distribution studies demonstrate that endoglin recruits GIPC to the plasma membrane and co-localizes with GIPC in a TGFβ-independent manner, with GIPC-promoting cell surface retention of endoglin. Endoglin specifically enhanced TGF-β1-induced phosphorylation of Smad 1/5/8, increased a Smad 1/5/8 responsive promoter, and inhibited endothelial cell migration in a manner dependent on the ability of endoglin to interact with GIPC. These studies define a novel mechanism for the regulation of endoglin signaling and function in endothelial cells and demonstrate a new role for GIPC in TGF-β signaling.
Transforming growth factor (TGF)2-β signaling in endothelial cells requires a heteromeric receptor complex comprising the type II TGF-β receptor (TβRII) and two type I TGF-β superfamily receptors, activin-like kinase-1 (ALK1), expressed in the endothelium, and ALK5 (also referred to as the type I TGF-β receptor, TβRI), which is ubiquitously expressed (1). In addition, endothelial cells express a TGF-β superfamily co-receptor, endoglin, which is involved in ALK1 and ALK5 signaling (2–8). Importantly, germline mutations in endoglin and ALK-1 result in the human vascular disease, hereditary hemorrhagic telangiectasia type I (HHT1) and HHT2, respectively, and ALK1, ALK5, TβRII and endoglin-null mice are all embryonic lethal due to vascular defects, supporting essential roles for these signaling pathways in the vasculature (9–13).
TGF-β superfamily receptors signal by phosphorylating intracellular effectors, termed Smads, which upon phosphorylation, form a complex with Smad 4, accumulate in the nucleus and regulate the expression of specific target genes (5, 14–17). In the case of ALK1, its catalytic engagement leads to the phosphorylation of Smads1/5/8, while ALK5 targets Smads2/3 for phosphorylation (5, 6). In addition to binding and signaling downstream of TGF-β ligands, ALK-1 has also been demonstrated to bind and respond to BMP-9 and BMP-10, suggesting that these might also be physiological ligands for ALK-1 (18, 19). BMP-9-induced activation of ALK-1 and constitutively active ALK-1 have been reported to inhibit endothelial cell proliferation, migration, and angiogenesis (18–20). In contrast, TGF-β-induced activation of ALK-1 has been reported to stimulate endothelial cell proliferation and migration, while TGF-β-induced activation of ALK-5 has been reported to inhibit endothelial cell proliferation and migration (6, 21). In addition, cross-talk between these pathways has been reported, with a role for ALK5 in TGF-β-induced ALK-1 signaling, and TGF-β-induced ALK-1 signaling inhibiting ALK-5 signaling (21). However, a recent report in murine and zebrafish models suggests that ALK-1 has a ALK-5/TβRII-independent role in HHT1, lending further support for a role of BMP-9/10 as physiological ALK-1 ligands (22).
Endoglin, while unable to bind TGF-β superfamily ligands besides BMP-9 directly, associates with a number of TGF-β superfamily receptors, including TβRII, ALK1, and ALK5, and binds TGF-β1 and -β3, Activin A, BMP-2, and BMP-7 in the context of their respective ligand-binding receptors (4, 23). Although the mutation of endoglin in HHT1 and the embryonic lethal phenotype of endoglin-null mice both support an essential role for endoglin in the vasculature, the mechanisms by which endoglin exert its influence on TGF-β superfamily signaling, or its role in cardiovascular development is poorly understood. In particular, the capacity for TGF-β to propagate two distinct signaling pathways in endothelial cells has been the subject of intense investigation. While endoglin has been characterized as a mediator of TGF-β-dependent activation of two opposing pathways in endothelial cells, there have been conflicting views on the nature of its actions. For instance, Lebrin et al. demonstrated that small interfering RNA-mediated knockdown of endoglin resulted in the reduction of TGF-β-induced Smad1 activation, suggesting a role of endoglin in enhancing ALK-1/Smad1 signaling, whereas Pece-Barbara et al. demonstrated an enhancement of ALK-1/Smad1 signaling in endoglin-null cell lines, suggesting a role of endoglin in inhibiting ALK-1/Smad1 signaling (24–26). Determining how endoglin participates in TGF-β superfamily signaling at the molecular level may help reconcile these disparate reports.
In our effort to characterize endoglin function we have investigated how the association between endoglin and specific intracellular scaffolding proteins influence TGF-β superfamily signaling in endothelial cells. Recently, we reported a novel interaction between endoglin and β-arrestin2, an association that resulted in their internalization in endocytic vesicles (27–29). While the endoglin-β-arrestin2 complex had little impact on either Smad1/5/8 or Smad 2/3 activation, the endoglin-β-arrestin2 complex regulated ERK activation, localization, and endothelial cell migration in a TGF-β1-dependent manner. Here we report a novel interaction between endoglin and the scaffolding protein GAIP-interacting protein C terminus (GIPC), and the impact on TGF-β-induced Smad signaling and endothelial cell migration.
EXPERIMENTAL PROCEDURES
Cell Culture and Antibodies—HMEC-1s were grown in MCDB-131 medium (Invitrogen), supplemented with 10% fetal bovine serum (FBS), 1 μg/ml hydrocortisone (Sigma), 10 ng/ml EGF (Sigma) and 2 mm l-glutamine (Invitrogen). Endoglin-null (endo-/-) and control (endo+/+) MEECs were grown in MCDB-131 supplemented with 15% FBS, 2 mm l-glutamine, 1 mm sodium pyruvate (Invitrogen), 100 μg of heparin (Sigma), and 50 μg/ml endothelial cell growth supplement (ECGS) (Sigma). Both HMEC-1s and MEECs were cultured in flasks coated with 0.05% gelatin. Endoglin antibody, P3D1, which recognizes the extracellular domain, was obtained from Chemicon. Phosphospecific Smad1/5/8 and Smad2/3, as well as their respective total Smad antibodies, were all purchased from Cell Signaling. Sulfo-NHS-LC-Biotin was obtained from Pierce. GIPC antibody (H-55) and the Dual-Luciferase Reporter assay system were purchased from Santa Cruz Biotechnology and Promega, respectively.
Protein Expression and Knockdown—Endo-/- and endo+/+ MEECs were nucleofected with cDNA constructs using Amaxa nucleofection system as described previously. HMEC-1s were infected with either a non-targeting vector control or shRNA endoglin-expressing adenovirus (targeting intracellular sequence) at 50 MOI and incubated for 72 h before harvest. All constructs used in nucleofection were PCR-amplified and inserted into pDispay vector. Adenoviral expression required the PCR cloning of cDNA into pDNR vector supplied by Clonetech BD Bioscience. Subsequent generation and amplification of adenoviruses were as directed by the manufacturer's protocols. shRNA-mediated GIPC knockdown was achieved by nucleofection of MEECs with a plasmid (generous gift from Dr. Jeffrey Rathmell) encorporating the shRNAi mouse sequence: GCATCAAGGAAGGCAGTGTGATTCCTCGAGCAATCAACTGCCTTCCTTGATGC.
Immunofluorescence—MEECs and HMEC-1s were serum-starved for 2 h, washed with PBS, fixed with 4% paraformaldehyde, permeabilized in 0.1% Triton X-100/PBS for 5 min and then blocked with 5% bovine serum albumin in PBS containing 0.05% Triton X-100 for 1 h. Endoglin-L and endoglin-DEL, and GIPC expressions were detected using P3D1 and GIPC antibodies, respectively, for 1 h at room temperature. Cells were washed with PBS and incubated with appropriate secondary fluorophore-conjugated antibodies for 1 h at room temperature, washed, then mounted with Prolong Anti-fade (Sigma).
Co-immunoprecipitation—MEECs expressing GIPC with either endoglin-L or endoglin-DEL were washed with PBS, then lysed on ice with lysis buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, 0.5% Nonidet P-40, 2 mm EDTA, 10 mm NaF, 10% w/v glycerol) supplemented with protease and phosphatase inhibitors (Sigma protease and phosphatase inhibitor cocktails). The lysates were precleared by centrifugation and incubated with either P3D1 or GIPC antibody and protein G-agarose beads for 4 h at 4 °C. The immunoprecipitates were collected by centrifugation, pellets washed with lysis buffer, and stored in 2× sample buffer prior to Western blot analyses.
Cell Surface Biotinylation—MEECs expressing endoglin-L, endoglin-Del, shRNA against GIPC or a non-targeting vector control were washed with PBS prior to treatment with 0.5 mg/ml Biotin-LC (Pierce) on ice for 30 min. Upon extensive washes with PBS, cells were lysed with lysis buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, 0.5% Nonidet P-40, 2 mm EDTA, 10 mm NaF, 10% w/v glycerol) supplemented with protease and phosphatase inhibitors (Sigma protease and phosphatase inhibitor cocktails), lysates normalized for total protein content via Bradford, then immunoprecipitated with P3D1 as described. HMEC-1s expressing GIPC via adenovirus infection were subjected to biotinylation as described previously. Lysates were normalized for protein levels by Bradford assay prior to immunoprecipitation of endoglin using P3D1 antibody. All biotinylated proteins were resolved by SDS-PAGE and probed for biotinylated endoglin with streptavidin-HRP.
Luciferase Reporter Assay—Dual reporter luciferase assays were performed as described by manufacturer (Promega). TGFβ-responsive vectors used as reporters for ALK5/Smad2 and ALK1/Smad1/5 signaling were PE2.1 and 3GC2, respectively. MEECs were nucleofected with GIPC and either endoglin-L or endoglin-DEL, along with either PE2.1 or 3GC2. The expression plasmid containing the Renilla luciferase gene served as an internal control to correct for transfection efficiency. Briefly, 106 cells were subjected to nucleofection with ∼2 μg of endoglin-L, or endoglin-Del, with 2 μg of either PE2.1 or 3GC2 vectors and 50 ng of Renilla vector. 24 h postnucleofection, cells were subjected to serum starvation or treatment with 100 pm TGFβ1 in serum-free media for 12–16 h prior to cell lysis and subsequent luciferase activity analyses.
Transwell Migration Assay—Transwells (Costar Corning Inc. 8 μm polycarbonate membrane 6.5 mm insert, 24-well plate) were coated with 0.02% gelatin prior to plating. Approximately 30,000 cells were plated onto the top of each of the 24-well membrane and allowed to migrate for 12 h prior to cell fixation and staining of the nuclei of the migrated cells on the bottom side of the membrane. The transwell membrane containing the stained cells were digitally imaged, then counted using image J software to tabulate cells as they were manually identified.
RESULTS
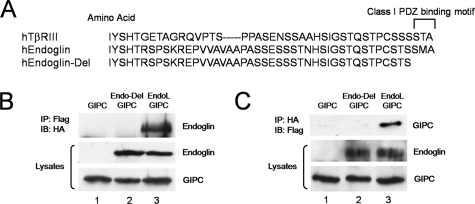
Interaction Between Endoglin and GIPC—The endoglin cytoplasmic domain contains several potential binding sites for signal transducing proteins, including zyxin, zyxin related protein-1, and the recently reported recognition motif for β-arrestin2 (27–29). In addition, the extreme C terminus contains a conserved type I PDZ binding domain with a consensus sequence of X(S/T)X(V/L), which is shared by the closely related TGF-β superfamily co-receptor, the type III TGF-β receptor (TβRIII) (30, 31) (Fig. 1A), and several other transmembrane receptors including the co-receptors neuropilin-1 and syndecan-4. In these receptors this motif has been demonstrated to mediate the interaction with the PDZ domain-containing protein, GIPC, which functions to regulate, at least in part, their cell surface expression. To investigate whether endoglin might interact with GIPC in endothelial cells, we performed co-immunoprecipitation studies in endoglin-null mouse embryonic endothelial cell line (MEECs, denoted endo-/-). HA epitope-tagged wild-type endoglin (endoglin-L) and an HA epitope-tagged endoglin mutant in which the last three C-terminal residues are truncated (ΔSMA, denoted endoglin-Del, Fig. 1A) were tested for their ability to bind GIPC. While immunoprecipitation of GIPC resulted in the co-immunoprecipitation of endoglin-L, endoglin-Del did not co-immunoprecipitate with of GIPC (Fig. 1B, compare lane 3 versus lane 2). In a reciprocal experiment, immunoprecipitation of endoglin-L, but not endoglin-Del, resulted in the co-immunoprecipitation of GIPC (Fig. 1C, compare lane 3 versus lane 2). Taken together, these studies support an interaction between endoglin and GIPC mediated by the class I PDZ binding motif in the cytoplasmic domain of endoglin.
FIGURE 1.
Endoglin interacts with GIPC. A, schematic representation of the cytoplasmic domains of human TβRIII, endoglin wild type, and endoglin-Del. B, anti-Flag immunoprecipitates were prepared from endo-/- MEECs expressing Flag-tagged GIPC with empty vector (lane 1), endoglin-Del (lane 2), and endoglin-L (lane 3). Upper panel, immunoblot of HA-tagged endoglin constructs co-precipitated with Flag-tagged GIPC. Middle and lower panels, immunoblots of endoglin and GIPC in total cell lysates, respectively. C, anti-HA immunoprecipitates were prepared from endo-/- MEECs expressing Flag-tagged GIPC with empty vector (lane 1), endoglin-Del (lane 2), and endoglin-L (lane 3). Upper panel, immunoblot of Flag-tagged GIPC co-precipitated with HA-tagged endoglin constructs. Middle and lower panels, immunoblots of endoglin and GIPC in total cell lysates, respectively. Data are representative of three independent experiments.
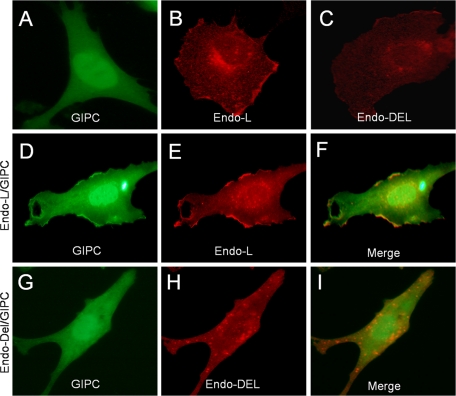
Subcellular Distribution of Endoglin and GIPC—The interaction between endoglin and GIPC was examined next in the context of their subcellular distribution. In endo-/- MEECs, a GFP fusion protein of GIPC was diffusely localized in the nucleus and cytoplasm (Fig. 2A), while endoglin-L had more prominent steady-state membrane localization (Fig. 2B) than endoglin-Del (Fig. 2C). When GIPC was co-expressed with endoglin-L, some of the GIPC localized to distinct clusters at or near the plasma membrane (Fig. 2D, compared with panels A or G), where it co-localized with endoglin-L (Fig. 2, E and F) irrespective of TGFβ1 stimulation (data not shown). In contrast, when GIPC was co-expressed with endoglin-Del, there was no alteration in GIPC localization (Fig. 2G, compared with 2A) and no co-localization of GIPC with endoglin-Del (Fig. 2, H and I, merged image). These studies suggest that endoglin recruits GIPC to distinct parts of the plasma membrane through its PDZ binding motif in its cytoplasmic domain in a TGFβ-independent manner.
FIGURE 2.
Recruitment and co-localization of GIPC with endoglin. Endo-/- MEECs transiently expressing GIPC-GFP alone (A), HA-tagged endoglin-L alone (B), HA-tagged endoglin-Del alone (C), GIPC-GFP with endoglin-L (D–F) or GIPC-GFP with endoglin-Del (G–I) were fixed, permeabilized, and immunostained for endoglin with endoglin antibody (P3D1) to visualize endoglin and GIPC localization using confocal imaging (60×). GIPC-GFP co-localizes with endoglin-L at distinct parts at or near the plasma membrane (D, E, and merged image in F), but not with endoglin-Del (G, H, and merged image I). Data are representative of three independent experiments.
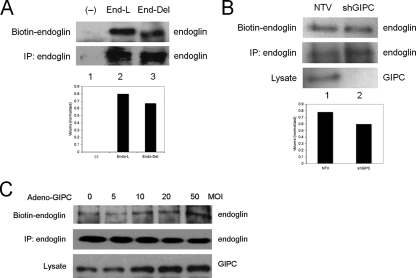
Effect of GIPC on Endoglin Expression—GIPC has been proposed to mediate the cell surface stabilization and/or subcellular trafficking of interacting receptors (30, 32). To determine whether GIPC stabilizes endoglin at the cell surface, endo-/- MEECs expressing either endoglin-L or endoglin-Del were cell surface-biotinylated to assess their steady-state expression at the cell surface. While endoglin-L and endoglin-Del were expressed at approximately equal levels (Fig. 3A, lane 2 versus lane 3, bottom panel), more endoglin-L was expressed at the cell surface compared with endoglin-Del (Fig. 3A, lane 2 versus lane 3, top panel), suggesting that GIPC might mediate cell surface retention of endoglin. To gain further support for GIPC in regulating endoglin cell surface expression, the expression of endogenous GIPC was knocked down via shRNA and levels of endogenous endoglin were assessed. When GIPC expression was decreased (Fig. 3B, lane 1 versus lane 2, lower panel), cell surface endoglin expression decreased (Fig. 3B, lane 1 versus lane 2, top panel), while total cellular levels, if anything, increased (Fig. 3B, lane 1 versus lane 2, middle panel). We also investigated the ability for GIPC to promote cell surface retention of endoglin in another endothelial cell line, human microvascular endothelial cells (HMEC-1s). Here, GIPC expression was increased by infection with adenovirus (Fig. 3C, bottom panel), with increasing GIPC expression correlating with increased cell surface endoglin expression (Fig. 3C, top panel), without altering total cellular endoglin levels (Fig. 3C, middle panel). Taken together, these results support a role for GIPC in stabilizing endoglin on the cell surface through interactions mediated by the type I PDZ binding domain in the cytoplasmic domain of endoglin.
FIGURE 3.
GIPC stabilizes endoglin on the cell surface. A, an empty vector control (lane 1), endoglin-L (lane 2), or endoglin-Del (lane 3) were expressed in Endo-/- MEECs for 40 h prior to cell surface biotinylation. Cells were subsequently immunoprecipitated with P3D1 endoglin antibody, and then resolved on SDS-PAGE before being observed with streptavidin-conjugated HRP (upper panel, biotinylated cell surface endoglin, lower panel, immunoprecipitated total cellular endoglin). Cell surface biotinylated endoglin and endoglin-DEL levels were quantified by densitometry scan, normalized to total endoglin expression and represented in arbitrary units (lower panel). B, non-targeting vector (lane 1) or shRNA against GIPC (lane 2) were nucleofected into endo+/+ MEECs for 48 h prior to cell surface biotinylation followed by immunoprecipitation of endogenous endoglin (upper panel, biotinylated cell surface endoglin; middle panel, immunoprecipitated endogenous total cellular endoglin; lower panel, endogenous GIPC in cell lysate). Cell surface biotinylated endoglin levels were quantified by densitometry scan, normalized to total endoglin expression, and represented in arbitrary units (lower panel). C, after infection with adenovirus encoding GIPC at the indicated MOIs, HMEC-1 cells were cell surface biotinylated. Cells were subsequently immunoprecipitated with P3D1 endoglin antibody, then resolved on SDS-PAGE before being observed with streptavidin-conjugated HRP (upper panel, biotinylated cell surface endoglin; middle panel, immunoprecipitated total cellular endoglin). Expression levels of GIPC were confirmed in total cell lysate (lower panel). Data are representative of two to three independent experiments.
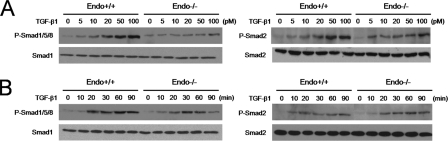
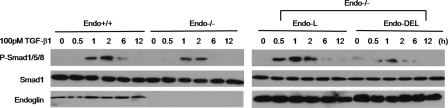
Effect of the Endoglin GIPC Interaction on Smad Signaling—The precise role of endoglin in regulating TGF-β signaling to the Smad pathways in endothelial cells has been elusive. To gain insight into how the endoglin-GIPC interaction might contribute to regulating Smad signaling the levels of Smad activation were assessed upon TGF-β1 treatment in the presence and absence of endoglin competent to bind GIPC (endoglin-L) and endoglin unable to bind GIPC (endoglin-Del) in endo-/- MEECs. We initially assessed the effects of TGF-β1 on endo-/- MEECs relative to endo+/+ MEECs. In both endo+/+ and endo-/- MEECs, TGF-β1 increased the phosphorylation of Smad1/5/8 and Smad2/3 in a dose-dependent manner (Fig. 4A). In endo+/+ MEECs, TGF-β1 more potently activated Smad1/5/8 relative to endo-/- MEECs, suggesting that endoglin might be necessary for full activation of the Smad 1/5/8 pathway. A similar pattern was observed in time course experiments, where TGF-β1 promoted greater and more sustained Smad1/5/8 phosphorylation in endo+/+ MEECs than in endo-/- MEECs (Fig. 4B). In contrast, while TGF-β1 stimulated Smad2/3 phosphorylation was slightly greater in endo+/+ MEECs than in endo-/- MEECs, these effects were modest when compared with the effects on Smad1/5/8.
FIGURE 4.
Endoglin promotes Smad1/5/8 activation. A, Endo+/+ and endo-/- MEECs were serum-starved for 6 h prior to treatment with TGF-β1 for 20 min at the indicated doses. Cells were harvested, the lysates normalized for protein levels via Bradford assay, and levels of Smad 1/5/8 and Smad 2 phosphorylation were assessed with phosphospecific Smad 1/5/8 and Smad 2 antibodies. Upper panels, immunoblot of phospho-Smad1/5/8 (left) and phospho-Smad 2 (right) levels. Lower panels, immunoblot of total Smad 1 (left) and Smad 2(right) levels. B, Endo+/+ and endo-/- MEECs were treated with 50 pm TGF-β1 for the indicated times and analyzed for Smad 1/5/8 and Smad 2 phosphorylation (upper panels, left and right, respectively). Lower panels, immunoblots of total Smad 1 (left) and Smad 2 (right) levels. Data are representative of three independent experiments.
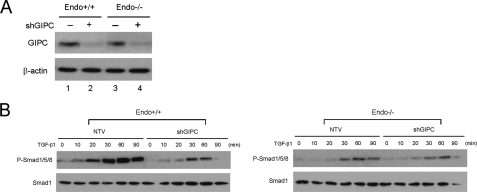
To investigate whether these differences were endoglin-mediated and the effect of the endoglin-GIPC interaction, endoglin-L and endoglin-Del were expressed in endo-/- MEECs. In endo-/- MEECs expression of endoglin-L increased TGF-β1-stimulated Smad1/5/8 and restored sustained Smad1/5/8 phosphorylation, while endoglin-Del expression had no effect (Fig. 5). In a reciprocal experiment, we used shRNA against GIPC to markedly decrease GIPC expression in endo+/+ and endo-/- MEECs (Fig. 6A, upper panel), and examine its effect on Smad signaling. GIPC knockdown sharply reduced Smad 1/5/8 activation in endo+/+ MEECs, whereas minimal changes were observed in endo-/- MEECs (Fig. 6B).
FIGURE 5.
Endoglin interacts with GIPC to promote Smad 1/5/8 activation. Endo+/+, endo-/- MEECs or endo-/- MEECs nucleofected with endoglin-L and endoglin-Del were serum-starved for 6 h prior to 100 pm TGF-β1 stimulation for the indicated times (in hours (h)). Cells were harvested and normalized for protein levels via Bradford, and levels of Smad 1/5/8 were assessed by use of phosphospecific Smad 1/5/8 antibodies. Upper panels, immunoblot of phospho-Smad1/5/8 levels. Middle panels, immunoblot of total Smad 1 levels. Lower panels, immunoblot of endoglin expression. Data are representative of three independent experiments.
FIGURE 6.
GIPC knockdown attenuates Smad 1/5/8 activation. A, both endo+/+ and endo-/- MEECs were nucleofected with either non-targeting vector control (lanes 1 and 3) or shRNA against GIPC (lanes 2 and 4), and the expression of GIPC assessed by Western blot (upper panel) with β-actin assessed as a loading control (lower panel). B, 48 h post-nucleofection with either the non-targeting vector (NTV) or GIPC shRNA, cells were serum-starved for 6 h prior to 100 pm TGF-β1 treatment for the indicated time (hours (h)). Upper panels, immunoblot of phospho-Smad1/5/8 levels. Lower panels, immunoblot of total Smad 1 levels. Data are representative of two independent experiments.
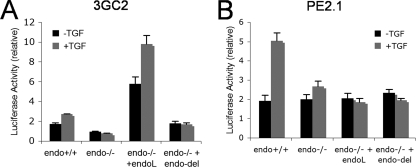
As the main intracellular mediators of TGF-β signaling, the Smad effectors accumulate in the nucleus upon their phosphorylation to engage in the transcriptional responses of target genes. To further examine the effect of the endoglin-GIPC interaction we next examined Smad-dependent transcriptional responses, using two promoter-reporter constructs, 3GC2, which requires Smad1/5/8 signaling, and PE2.1, which requires Smad3 signaling. Consistent with the results with Smad1/5/8 phosphorylation, the relative luciferase activity for 3GC2 in endo+/+ MEECs cells was higher than endo-/- MEECs both in the absence and presence of TGF-β treatment (Fig. 7A). Ectopic expression of endoglin-L in endo-/- MEEC yielded a significant enhancement of basal and TGF-β1-stimulated 3GC2 luciferase activity, while ectopic expression of endoglin-Del had minimal impact (Fig. 7A). In the case of Smad2/3-mediated transcriptional activity, the basal luciferase activity for PE2.1 in endo+/+ MEECs and endo-/- MEECs was equivalent; however, endo-/- MEECs failed to respond to TGF-β1 relative to endo+/+ MEECs (Fig. 7B). In addition, in contrast to the results with the 3GC2 reporter, ectopic expression of endoglin-L of endoglin-DEL in endo-/- MEEC failed to increase basal or TGF-β1-stimulated PE2.1 luciferase activity (Fig. 7B). Taken together, these results indicate that endoglin specifically promotes ALK1/Smad1/5/8 signaling, in part through its interaction with GIPC.
FIGURE 7.
Endoglin up-regulates Smad1 reporter gene activation. Endo+/+ and endo-/- MEECs were nucleofected with either (A) 3GC2 (a Smad 1 reporter) or (B) PE2.1 (a Smad 2 reporter) along with an empty vector, endoglin-L, or endoglin-Del construct. SV40 plasmid nucleofection was included in all the samples for Renilla activity (SV40) and used to control for transfection efficiency. The indicated error bars represent the S.E. from triplicates for each of the represented experimental conditions. Data shown are representative of three independent experiments.
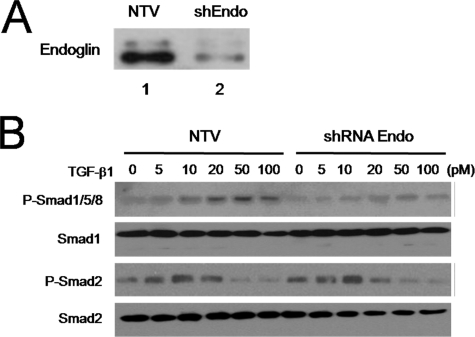
To determine whether the observed effect of endoglin on Smad1/5/8 signaling was cell-type specific, we examined Smad activation in another endothelial cell line, HMEC-1. In addition as the endo-/- MEECs could have adapted to the chronic loss of endoglin expression, we used adenoviral vector-based shRNA to effectively and acutely knockdown endogenous endoglin expression (Fig. 8A). Compared with the non-targeting vector control (NTV), the endoglin-knockdown cells displayed reduced levels of TGF-β1-induced Smad1/5/8 phosphorylation (Fig. 8B), while Smad2/3 activation remained comparable irrespective of endoglin expression. Taken together, these results support a specific role for endoglin in TGF-β1-mediated activation of the Smad1/5/8 pathway, and a role for endoglin interacting with GIPC to mediate this effect.
FIGURE 8.
Endoglin promotes Smad 1/5/8 activation in HMEC-1 cells. A, endoglin expression was assessed in HMEC-1s 72 h after infection with an adenovirus containing a non-targeting vector (NTV, lane 1) or shEndoglin (lane 2). B, phosphorylation levels of Smad 1/5/8 and Smad 2 were assessed via phosphospecific Smad 1/5/8 and Smad 2 antibodies 20 min after TGF-β1 treatment. The first and third panels indicate the phosphorylation of Smad 1/5/8 and Smad 2, respectively. The second and fourth panels indicate the total Smad 1 and Smad 2, respectively. All cells were harvested and normalized for protein levels via Bradford. Data shown are representative of three independent experiments.
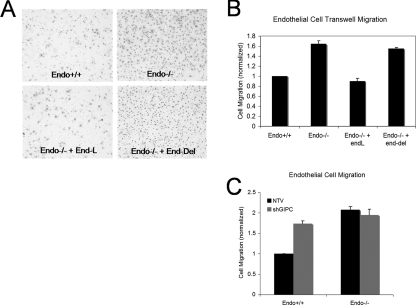
Effect of the Endoglin GIPC Interaction on Endothelial Cell Migration—Endoglin has an established role in inhibiting endothelial cell migration and this function has been reported to require the entire cytoplasmic domain of endoglin (28). Accordingly, we investigated whether endothelial cell migration might be regulated by the interaction between endoglin and GIPC. Consistent with previous reports endo+/+ MEECs migrated slower than endo-/- MEECs in transwell assays (Fig. 9A). When endoglin-L was transiently expressed in endo-/- MEECs, their migration was inhibited to a level comparable to endo+/+ MEECs, confirming a negative role for endoglin in endothelial cell migration (Fig. 9B). In contrast, transient expression of endoglin-Del in endo-/- MEECs failed to inhibit the migration of endo-/- MEECs (Fig. 9B), supporting an important role for the Class I PDZ binding motif of endoglin in interacting with GIPC to mediate this endoglin function. To further test this hypothesis, GIPC expression was knocked down in endo+/+ and endo-/- MEECs and monitored for their cell motility. As expected, silencing GIPC expression up-regulated the cell motility of endo+/+ MEECs while exerting little impact on endo-/- MEECs (Fig. 9C). These results support a role for endoglin in regulating endothelial cell migration through its ability to interact with GIPC.
FIGURE 9.
Endoglin interacts with GIPC to regulate migration. Endo+/+ and endo-/- MEECs were plated on transwells coated with 0.02% gelatin and assessed for migration through the transwells 12 h later. A, migrated cells found on the bottom side of the membrane were fixed, stained for their nuclei, and imaged for endo+/+ (upper left panel), endo-/- (upper right), endo-/- with endoglin-L expression (lower left), and endo-/- with endoglin-Del (lower right). B, number of migrated cells on the membrane bottom (shown as stained nuclei) were counted using Image J software. Cell migration is represented as a percentage, normalized to endo+/+ MEECs, from triplicates for each of the four independent experiments. The error bars indicate the standard error of the mean of the percentage of the migrated cells. C, transwell migration analyses upon shRNA-mediated GIPC knockdown. 48 h post-nucleofection with either non-targeting vector (NTV) or shRNA (shGIPC) endo+/+ and endo-/- MEECs were plated onto transwells for 12 h. Cells were fixed, stained, and quantitated as above.
DISCUSSION
While the mutation of endoglin in the human vascular disease HHT1 and the embryonic lethal phenotype of endoglin-null mice due to defects in angiogenesis both support an essential role for endoglin in endothelial cell biology, how endoglin regulates endothelial cell function is poorly understood. Genetic evidence support a role for TGF-β-superfamily signaling in the pathogenesis of HHT, as mutations in the TGF-β superfamily type I receptor ALK-1 result in HHT2, and mutation in the common TGF-β superfamily transcription factor Smad4, have also been implicated in HHT. We have been investigating how intracellular signal transducing proteins might impact TGF-β signaling and endoglin-mediated endothelial cell biology upon their interaction with endoglin. Indeed, we recently reported a novel interaction of endoglin with β-arrestin2, an association that resulted in their co-internalization, attenuation of ERK activation and inhibition of endothelial cell migration. In the present study, we examined the ability of endoglin to interact with GIPC, and its impact on the Smad signaling pathways. An interaction between endoglin and GIPC is supported by the ability of endoglin-L but not endoglin-Del to co-immunoprecipitate GIPC and for GIPC to co-immunoprecipitate endoglin but not endoglin-Del. Moreover, GIPC was shown to co-localize with endoglin-L but not endoglin-Del near the plasma membrane. Functionally, GIPC stabilized endoglin on the cell surface, as evidenced by cell surface biotinylation assays comparing endoglin-L with endoglin-Del, as well as GIPC knockdown studies. GIPC-interacting motif of endoglin, in particular, was essential for endoglin-mediated stimulation of sustained Smad1/5/8 activation and endoglin-mediated inhibition of endothelial cell migration.
In contrast with the effects on Smad1/5/8 activation, endoglin-L expression did not result in sustained Smad2 phosphorylation or Smad2 responsive reporter activation despite the modestly higher Smad2 phosphorylation observed for endo+/+ than endo-/- MEECs. Moreover, endoglin knockdown in HMEC-1 cells did not significantly alter Smad2/3 activation. These observations suggest a specific role for endoglin and its interaction with GIPC in mediating signaling through the Smad1/5/8 pathway. There are several possible underlying mechanisms for this effect. One the one hand, GIPC could facilitate cell surface retention of endoglin to promote its interaction with ALK-1, thereby enhancing TGF-β1-induced recruitment and activation of Smad 1/5/8 at the plasma membrane. Endoglin, and perhaps its interaction with GIPC, might also modulate the intermediate mechanisms that regulate Smad phosphorylation and subsequent nuclear translocation. While our studies do not address these issues directly, we have observed more potent nuclear translocation of phospho-Smad 1/5/8 upon expression of endoglin-L than endoglin-Del in endo-/- MEECs (data not shown), argue against this latter case.
We examined endothelial cell migration as a biological response. Our results indicate an inhibitory role for endoglin, in part facilitated by its interaction with GIPC. An inhibitory role for endoglin on endothelial cell migration is consistent with most reports in the literature. However, the role of the ALK-1/Smad1/5/8 on endothelial cell migration has been controversial, as TGF-β-induced activation of ALK-1 has been reported to stimulate endothelial cell proliferation and migration (6, 21), while BMP-9-induced activation of ALK-1 and constitutively active ALK-1 have been reported to inhibit endothelial cell proliferation, migration, and angiogenesis (18–20).. Here the ability of endoglin to inhibit endothelial cell migration while activating the Smad1/5/8 pathway suggests a role the ALK-1/Smad1/5/8 in inhibiting endothelial cell migration. On the other hand, the role of endoglin and ALK-1 may depend on the ligand utilized to stimulate these pathways. Furthermore, while the Smad signaling pathways have been characterized to play integral roles in endothelial cell migration, other non-Smad pathways influencing these processes have been reported. Given our findings, it is an attractive possibility that the endoglin-GIPC interaction orchestrates one or several Smad-dependent and Smad-independent signaling pathways to influence TGF-β and BMP-mediated cellular responses. Precisely how endoglin, through interacting with GIPC, responds to TGF-β superfamily ligands to regulate cell motility is likely to be complex, involving distinct signaling receptor complexes, and remains an area of active investigation.
The novel interaction between endoglin and GIPC reported here further expands the role of endoglin in TGF-β-mediated signaling in endothelial cells. Thus far, several proteins have been identified to target the intracellular domain of endoglin: zyxin, zyxin-related protein, and the recently reported scaffolding protein, β-arrestin2. While the precise structural determinants mediating the interaction between endoglin and zyxin or ZRP-1 remains to be defined, β-arrestin2 has been demonstrated to bind endoglin near the C terminus that incorporates the PDZ binding motif. These findings raise an interesting possibility that endoglin is subjected to regulation by opposing scaffolding mediators: GIPC that anchors endoglin and promotes its cell surface retention and promotes Smad 1/5/8 activation, while β-arrestin2 internalizes the co-receptor and attenuates ERK signaling. As the critical structural determinants for their binding to endoglin reside within a few residues apart, whether β-arrestin2 and GIPC compete for binding to endoglin, and what mechanisms, if any, determine their interaction remain to be defined. Efforts are underway to address these questions and to characterize how endoglin modulates downstream Smad and non-Smad-dependent signaling as result of its interaction with β-arrestin2, GIPC.
In summary, we have demonstrated a novel association between endoglin and GIPC. Our findings indicate that the endoglin-GIPC interaction is necessary promote Smad1/5/8 activation and ALK-1-mediated TGF-β1 signaling downstream at the transcriptional level in endothelial cells, as well as for endoglin to mediate its inhibitory effect on migration in endothelial cells.
Acknowledgments
Endoglin+/+ and endoglin-/- MEECs were generous gifts from Dr. Dejana (University of Milan, Italy). The shRNA construct against GIPC was a generous gift from Dr. Jeffrey Rathmell.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-CA105255 from the NCI (to G. C. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TGF, transforming growth factor; PBS, phosphate-buffered saline; HA, hemagglutinin; GFP, green fluorescent protein; GIPC, GAIP-interacting protein C terminus; ALK, activin-like kinase; MOI, multiplicity of infection.
References
- 1.Massague, J. (1998) Annu. Rev. Biochem. 67, 753-791 [DOI] [PubMed] [Google Scholar]
- 2.Cheifetz, S., Andres, J. L. and Massague, J. (1988) J. Biol. Chem. 263 16984-16991 [PubMed] [Google Scholar]
- 3.Cheifetz, S., Bellón, T., Calés, C., Vera, S., Bernabeu, C., Massagué, J., and Letarte, M. (1992) J. Biol. Chem. 267 19027-19030 [PubMed] [Google Scholar]
- 4.Barbara, N. P., Wrana, J. L., and Letarte, M. (1999) J. Biol. Chem. 274 584-594 [DOI] [PubMed] [Google Scholar]
- 5.Ten Dijke, P., Goumans, M. J., Itoh, F., and Itoh, S. (2002) J. Cell. Physiol. 191 1-16 [DOI] [PubMed] [Google Scholar]
- 6.Goumans, M. J., Valdimarsdottir, G., Itoh, S., Rosendahl, A., Sideras, P., and ten Dijke, P. (2002) EMBO J. 21 1743-1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lastres, P., Letamendía, A., Zhang, H., Rius, C., Almendro, N., Raab, U., López, L. A., Langa, C., Fabra, A., Letarte, M., and Bernabéu, C. (1996) J. Cell Biol. 133 1109-1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerrero-Esteo, M., Sanchez-Elsner, T., Letamendia, A., and Bernabeu, C. (2002) J. Biol. Chem. 277 29197-29209 [DOI] [PubMed] [Google Scholar]
- 9.Li, D. Y., Sorensen, L. K., Brooke, B. S., Urness, L. D., Davis, E. C., Taylor, D. G., Boak, B. B., and Wendel, D. P. (1999) Science 284 1534-1537 [DOI] [PubMed] [Google Scholar]
- 10.Bourdeau, A., Dumont, D. J., and Letarte, M. (1999) J. Clin. Investig., 104 1343-1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh, S. P., Seki, T., Goss, K. A., Imamura, T., Yi, Y., Donahoe, P. K., Li, L., Miyazono, K., Dijke, P. t., Kim, S., and Li, E. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 2626-2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang, X., Castilla, L. H., Xu, X., Li, C., Gotay, J., Weinstein, M., Liu, P. P., and Deng, C. X. (1999) Development 126 1571-1580 [DOI] [PubMed] [Google Scholar]
- 13.Fonsatti, E., Altomonte, M., Nicotra, M. R., Natali, P. G., and Maio, M. (2003) Oncogene 22 6557-6563 [DOI] [PubMed] [Google Scholar]
- 14.Blobe, G. C., Schiemann, W. P., and Lodish, H. F. (2000) N. Engl. J. Med. 342 1350-1358 [DOI] [PubMed] [Google Scholar]
- 15.Massague, J., Attisano, L., and Wrana, J. L. Trends Cell Biol. 4 172-178 [DOI] [PubMed]
- 16.Wrana, J. L., Attisano, L., Wieser, R., Ventura, F., and Massagué, J. (1994) Nature 370 341-347 [DOI] [PubMed] [Google Scholar]
- 17.Nakao, A., Imamura, T., Souchelnytskyi, S., Kawabata, M., Ishisaki, A., Oeda, E., Tamaki, K., Hanai, J., Heldin, C. H., Miyazono, K., and ten Dijke, P. (1997) EMBO J. 16, 5353-5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scharpfenecker, M., van Dinther, M., Liu, Z., van Bezooijen, R. L., Zhao, Q., Pukac, L., Löwik, C. W., and ten Dijke, P. (2007) J. Cell Sci. 120 964-972 [DOI] [PubMed] [Google Scholar]
- 19.David, L., Mallet, C., Mazerbourg, S., Feige, J. J., and Bailly, S. (2007) Blood 109 1953-1961 [DOI] [PubMed] [Google Scholar]
- 20.David, L., Mallet, C., Vailhé, B., Lamouille, S., Feige, J. J., and Bailly, S. (2007) J. Cell. Physiol. 213, 484-489 [DOI] [PubMed] [Google Scholar]
- 21.Goumans, M. J., Valdimarsdottir, G., Itoh, S., Lebrin, F., Larsson, J., Mummery, C., Karlsson, S., and ten Dijke, P. (2003) Mol. Cell 12 817-828 [DOI] [PubMed] [Google Scholar]
- 22.Park, S. O., Lee, Y. J., Seki, T., Hong, K. H., Fliess, N., Jiang, Z., Park, A., Wu, X., Kaartinen, V., Roman, B. L., and Oh, S. P. (2008) Blood 111 633-642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koleva, R. I., Conley, B. A., Romero, D., Riley, K. S., Marto, J. A., Lux, A., and Vary, C. P. (2006) J. Biol. Chem. 281 25110-25123 [DOI] [PubMed] [Google Scholar]
- 24.Pece-Barbara, N., Vera, S., Kathirkamathamby, K., Liebner, S., Di Guglielmo, G. M., Dejana, E., Wrana, J. L., and Letarte, M. (2005) J. Biol. Chem. 280 27800-27808 [DOI] [PubMed] [Google Scholar]
- 25.Lebrin, F., Goumans, M. J., Jonker, L., Carvalho, R. L., Valdimarsdottir, G., Thorikay, M., Mummery, C., Arthur, H. M., and ten Dijke, P. (2004) EMBO J. 23 4018-4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanco, F. J., Santibanez, J. F., Guerrero-Esteo, M., Langa, C., Vary, C. P., and Bernabeu, C. (2005) J. Cell. Physiol. 204 574-584 [DOI] [PubMed] [Google Scholar]
- 27.Lee, N. Y., and Blobe, G. C. (2007) J. Biol. Chem. 282 21507-21517 [DOI] [PubMed] [Google Scholar]
- 28.Conley, B. A., Koleva, R., Smith, J. D., Kacer, D., Zhang, D., Bernabéu, C., and Vary, C. P. (2004) J. Biol. Chem. 279 27440-27449 [DOI] [PubMed] [Google Scholar]
- 29.Sanz-Rodriguez, F., Guerrero-Esteo, M., Botella, L. M., Banville, D., Vary, C. P., and Bernabéu, C. (2004) J. Biol. Chem. 279 32858-32868 [DOI] [PubMed] [Google Scholar]
- 30.Blobe, G. C., Liu, X., Fang, S. J., How, T., and Lodish, H. F. (2001) J. Biol. Chem. 276 39608-39617 [DOI] [PubMed] [Google Scholar]
- 31.Letamendia, A., Lastres, P., Botella, L. M., Raab, U., Langa, C., Velasco, B., Attisano, L., and Bernabeu, C. (1998) J. Biol. Chem. 273 33011-33019 [DOI] [PubMed] [Google Scholar]
- 32.Katoh, M., (2002) Int. J. Mol. Med. 9 585-589 [PubMed] [Google Scholar]