Abstract
Transforming growth factor β family ligands are neutralized by a number of structurally divergent antagonists. Follistatin-type antagonists, which include splice variants of follistatin (FS288 and FS315) and follistatin-like 3 (FSTL3), have high affinity for activin A but differ in their affinity for other ligands, particularly bone morphogenetic proteins. To understand the structural basis for ligand specificity within FS-type antagonists, we determined the x-ray structure of activin A in complex with FSTL3 to a resolution of 2.5 Å. Similar to the previously resolved FS·activin A structures, the ligand is encircled by two antagonist molecules blocking all ligand receptor-binding sites. Recently, the significance of the FS N-terminal domain interaction at the ligand type I receptor site has been questioned; however, our data show that for FSTL3, the N-terminal domain forms a more intimate contact with activin A, implying that this interaction is stronger than that for FS. Furthermore, binding studies revealed that replacing the FSTL3 N-terminal domain with the corresponding FS domain considerably lowers activin A affinity. Therefore, both structural and biochemical evidence support a significant interaction of the N-terminal domain of FSTL3 with activin A. In addition, structural comparisons with bone morphogenetic proteins suggest that the interface where the N-terminal domain binds may be the key site for determining FS-type antagonist specificity.
The transforming growth factor (TGF)2 β superfamily of ligands controls a wide array of processes that govern stem cell fate, embryonic development, organ and tissue homeostasis, reproduction, response to injury, and immune system function. Thirty-three ligands can be subdivided into three branches: TGFβ, activin/inhibin, and bone morphogenetic protein (BMP) (1). Each ligand is comprised of a disulfide-linked homo- or heterodimer and signals by engaging a pair of type II and type I serine-threonine kinase receptors. Tight control of ligand signaling occurs in the extracellular space, where a number of structurally diverse protein antagonists attenuate or completely inhibit signaling by binding and sequestering ligands. Antagonists include the follistatin-types, noggin, chordin, DAN/Cerberus, gremlin, decorin, and others. Antagonists restrict ligand activity to certain physiological environments and will often selectively inhibit ligands from one of the three branches. Antagonists bind multiple ligands with varying affinities, and for the most part, the structural basis for this specificity remains unresolved (2, 3).
Follistatin-type (FS-type) antagonists, which include the well studied splice variants of follistatin (FS), FS288 and FS315, are potent regulators of activin A signaling (4-7). Also included is a related protein, follistatin-like 3 (FSTL3/FLRG/FSRP), that was originally identified in B-cell leukemia (8). FSTL3 is similar to FS in that it is distributed among many tissues and cell types (9-11). Mice deficient in each form exhibit drastically different phenotypes. FS-deficient mice show retarded growth and death within hours of birth (12), whereas FSTL3-deficient mice are viable and normal-sized but exhibit metabolic phenotypes characterized by changes in glucose and fat homeostasis (13). Although both FSTL3 and FS bind activin A with high affinity (Kd ∼ 30 pm), they exhibit affinity differences with other ligands. FS exhibits broad ligand specificity, interacting at high affinity with numerous ligands including activin B, myostatin, and growth and differentiation factor 11 and at low affinity with several BMPs (11, 14-17). Recently, however, it was determined that FSTL3 is more limited than FS in the number of ligands it antagonizes, and it lacks interaction with BMPs, specifically BMPs 2, 4, 6, and 7 (17).
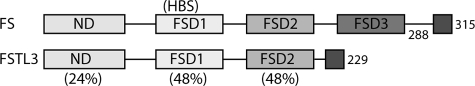
Differences in the domain structures of FSTL3 and FS likely provide a basis for ligand specificity. All of the follistatin antagonists possess an N-terminal domain followed by two or three follistatin domains (FSD) (Fig. 1). Despite similar architecture, there are significant differences between FSTL3 and FS288/FS315: (i) FS288/FS315 both have three FSDs, whereas FSTL3 contains only two; (ii) the sequences of FSD1 and FSD2 are well conserved between FS and FSTL3, but the N-terminal domains are significantly more divergent (24% sequence identity); and (iii) FSTL3 lacks a cell surface heparin-binding motif (located in FSD1 of FS) (7), which makes it the most serum available form (18, 19).
FIGURE 1.
Domain architecture of FSTL3 and FS. FSTL3 lacks FSD3 and a heparin-binding sequence located within FSD1. FS315 and FSTL3 both have an extended acidic C-terminal tail. The percentage of identity is indicated for each domain excluding the conserved cysteines.
The individual contribution of each FS domain to ligand binding and antagonism has been debated, especially with respect to the N-terminal domain. In one case, deletion or disruption of the N-terminal domain of FS prevented both binding to activin A and suppression of activin A activity (20). However, other studies have questioned the importance of the N-terminal domain to binding. For example, a FS derivative only containing FSD1 and FSD2 was shown to form a stable complex with activin A and was able to suppress elongation of Xenopus animal caps (21). However, the Kd for this form was ∼400 nm or 10,000 times higher than for native FS. Similar experiments showed that activin A binding could be achieved through use the same two FSDs of FSTL3 (9, 22). Furthermore, mutations of activin A residues near the N-terminal domain interface did not affect affinity; however, certain activin A residues were difficult to interpret (23).
To understand the structural features that could account for the differences in ligand specificity between FS and FSTL3 and clarify the role of the N-terminal domain in binding, we have solved the x-ray structure at 2.5 Å of FSTL3 in complex with activin A. The FSTL3·activin A structure illustrates that a common binding mechanism exists for FS-type antagonists where FSD1 and FSD2 block the type II receptor surface and the N-terminal domain interacts at the type I receptor interface. Comparison with the previously determined FS·activin A structures (24, 25) indicates that significant conformational differences occur within the N-terminal domain-ligand interface, thus pointing to a structural basis for ligand specificity. Domain exchange experiments were performed to validate our structural observations.
EXPERIMENTAL PROCEDURES
Production and Purification of FSTL3 and Activin A—Human FSTL3 and activin A were each cloned into the pcDNA3.1/myc-His expression vector (Invitrogen) as described previously for FS (20, 26) and co-expressed in HEK293F cells using the Freestyle™ 293 transient transfection system (Invitrogen). The native stop codon was maintained for activin A and therefore did not include the myc-His tag. Conditioned medium was collected 48 h after transfection. Protein was extracted using a histidine affinity column and eluted with a stepwise imidazole gradient: activin A and FSTL3 eluted together between 150 and 300 mm imidazole. The complex was treated with thrombin protease to remove the C-terminal myc-His tag and passed through a Superdex 75 fast protein liquid chromatography gel filtration column in 100 mm Tris/HCl buffer, pH 7.2. The predominant protein peak was observed at an elution position consistent with a 2:1 complex of FSTL3 and activin A.
Crystal Structure Determination—The FSTL3·activin A complex was concentrated to 7.5 mg/ml and mixed 1:1 in a hanging drop experiment with a well solution containing 25% (w/v) polyethylene glycol 3350, 200 mm ammonium sulfate, and 100 mm Tris, pH 8.5. Thin hexagonal crystals grew to 300 μm within 2-3 weeks. The diffraction experiments were performed at the Argonne National Laboratory Advanced Photon Source 19BM beam line. The data were integrated and scaled to 2.5 Å resolution using HKL2000 (27). The position of two complexes in the asymmetric unit were resolved through molecular replacement with the program PHASER (28), and the FS288·activin A complex (Protein Data Bank identifier 2B0U) with FSD3 removed from the search model (24). The atomic coordinates were refined using CNS (29) and REFMAC (30) along with repeated rounds of model building with COOT (31). Positional displacement of each chain was described by eight translation/libration/screw groups that were identified by the TLSMD server (32, 33). Translation liberation and screw rotation parameters were then further refined against the x-ray data with REFMAC and subsequently held constant while isotropic B-factor contributions and atomic coordinates were refined (34). During refinement with REFMAC, noncrystallographic symmetry restraints with medium weight were utilized. The two complexes of FSTL3·activin A showed a root mean square deviation of 0.40 Å. The final models for activin A monomers are missing various N-terminal residues near the 6-8 region and in the region from 49-53. FSTL3 models are missing residues 1-6 and 218-237. Data collection and refinement statistics are shown in Table 1, and an example of the electron density map of the complex interface is shown in Fig. 5. Coordinates and structure factors have been deposited in the Protein Data Bank with the identifier 3B4V. Analysis of interfaces and buried surface calculations were performed with the PISA server (35).
TABLE 1.
Crystallographic data and refinement statistics
| Data collection | Native (collected at 100 K) |
| Resolution (Å) | 33.02-2.48 (2.59-2.48) |
| Crystallographic cell - P1 | a = 63.6 Å, b = 71.4 Å, c = 100.2 Å, α = 98.5°, β = 90.6°, γ = 90.1° |
| Observations | 255,677 |
| Unique reflections | 61,295 |
| Completeness (%) | 98.6 (98.3) |
| Redundancy | 4.2 (3.6) |
| Rmerge (%) | 8.5 (44.8) |
| <I/σI> | 15.0 (2.3) |
| Wilson plot B factor (Å2) | 48.8 |
| Model refinement | |
| Reflections (total/free) | 61,224/3,144 |
| Rfactor (Rfree), (%) | 22.6 (27.8) |
| Atoms (total/protein) | 9,730/9,508 |
| Root mean square deviation from ideal | |
| Bonds (Å) | 0.010 |
| Angles (°) | 1.26 |
| <B> factors, all atoms (Å2) | 45.3 |
| Complex 1 | |
| Activin A (chain A/chain B) | 33.3/32.8 |
| FSTL3 (chain C/chain D) | 48.1/41.1 |
| Complex 2 | |
| Activin A (chain E/chain F) | 32.6/32.9 |
| ESTL3 (chain G/chain H) | 48.1/41.0 |
| Other molecules (number, <B> factor) | |
| Water | 214 (45.6) |
| Sulfate | 1 (80.0) |
| N-acetylglucosamine | 2 (74.1) |
| Ethylene glycol | 3 (65.4) |
| Ramachandran plot statistics (number) | |
| Most favored | 90.5% (963) |
| Additionally allowed | 8.6% (92) |
| Generously allowed | 0.5% (5) |
| Disallowed | 0.4% (4) (Asn38 in all activin A chains) |
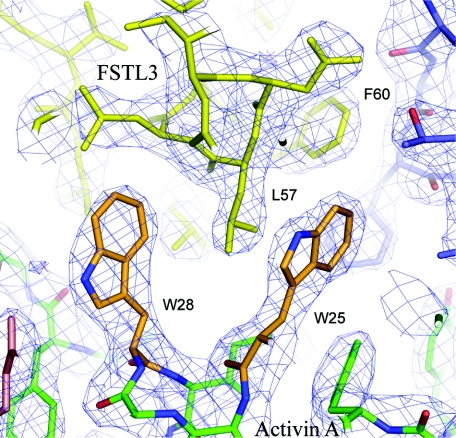
FIGURE 5.
Electron density of FSTL3 N-terminal domain interaction with activin A. Shown is a 2Fo - Fc electron density map contoured at 1.5 σ depicting the activin A tryptophan residues and their interaction with the N-terminal domain helix.
N-terminal Domain Swap—Residues 1-70 of FSTL3 were replaced with the sequence 1-63 of FS288 in the pcDNA3.1/myc-His expression vector as described (26) and designated NDFSFSD1,2FSTL3. A second construct spliced the third FS domain (residues 212-288) into the C terminus of the NDFSFSD1,2FSTL3 vector to generate NDFSFSD1,2FSTL3FSD3FS. The respective myc-His-tagged proteins were expressed in HEK293F cells as described above and isolated by histidine affinity chromatography. Protein was quantified by a C-terminal myc tag solution phase assay (20). Binding of altered FSs to labeled activin A was determined by competition assay as described (20). Relative potencies were calculated by comparison of half-maximal inhibition of labeled activin A binding by domain-exchanged and wild-type FSTL3.
RESULTS
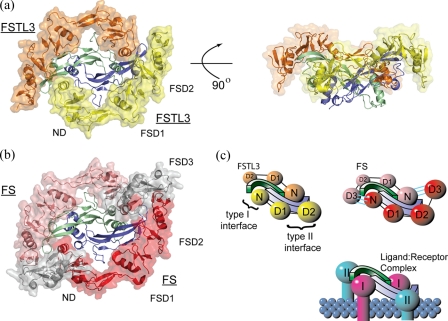
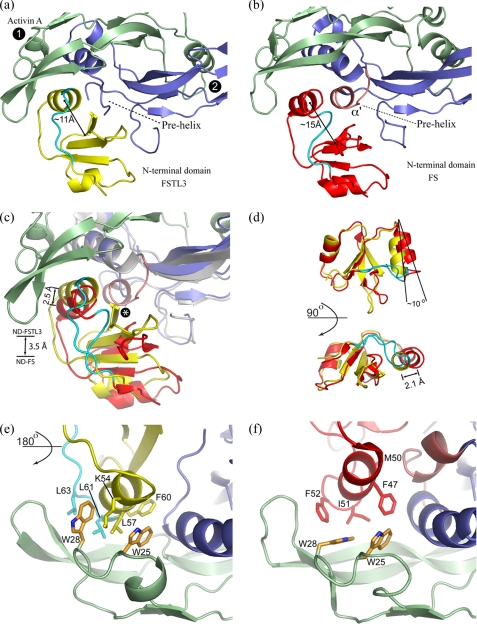
Overview of Structure—The x-ray crystallographic structure of FSTL3·activin A has been determined to 2.5 Å resolution, and an overview of the complex is shown in Fig. 2a. The previously determined structure of FS288·activin A is shown for comparison in Fig. 2b. Similar to FS288 and FS315, two molecules of FSTL3, each forming a C-shape, completely surround the activin A dimer. Consequently, the two FSTL3 molecules bury one-fourth of the activin A dimer surface or a total of 3,403 Å2, which is similar to FS288 (3,074 Å2). As observed with FS, FSTL3 molecules contact activin A at two discontinuous surfaces, interfering with both type I and type II receptor-binding sites (Fig. 2c). One interface is formed when the N-terminal domain of FSTL3 interacts at the activin A dimer interface. Here FSTL3 interacts with the concave portion of the β-strands on one activin A monomer and the long helix or “wrist” of the adjacent activin A monomer (type I interface). The other interface is formed by FSD1 and FSD2, which interact on the convex ligand surface or “knuckle,” and extends toward the tip of one activin A monomer (type II interface). For clarity, the two FS-type contact points will be referred to generically as type I and type II, corresponding to the respective receptor-binding sites that they block.
FIGURE 2.
Overall comparison of the FSTL3·activin A complex with FS288·activin A. Both FSTL3 and FS bind activin A in a similar fashion. a, two molecules of FSTL3 (transparent surface, orange and yellow) bind the central activin A homodimer as compared with the FS288·activin A structure in b. The third FS domain, which is not present in FSTL3, is colored gray. c, schematic depicting the blockade of both type I and type II receptor interfaces (N = ND, D1-3 = FSD1-FSD3). In FS, an interaction is observed between the N-terminal domain of one FS with FSD3 of the adjacent FS (noted by three cyan lines).
In both FS·activin A structures, the N-terminal domain of one FS
interacts with FSD3 of the other FS, hereafter referred to as
FS(ND) FS(FSD3)
(Fig. 2c). Because
FSTL3 does not have a third follistatin domain, this interaction is absent. In
fact, there are no interactions between the two FSTL3 molecules throughout the
whole complex. The significance of the N-terminal domain interaction with
activin A (20) has been
questioned by recent evidence
(21,
23). However, the structure of
FSTL3·activin A (Fig. 2, a
and b) shows that, even in the absence of
FS(ND)
FS(FSD3)
(Fig. 2c). Because
FSTL3 does not have a third follistatin domain, this interaction is absent. In
fact, there are no interactions between the two FSTL3 molecules throughout the
whole complex. The significance of the N-terminal domain interaction with
activin A (20) has been
questioned by recent evidence
(21,
23). However, the structure of
FSTL3·activin A (Fig. 2, a
and b) shows that, even in the absence of
FS(ND) FS(FSD3) contacts, the
N-terminal domain is still positioned at the type I interface.
FS(FSD3) contacts, the
N-terminal domain is still positioned at the type I interface.
Comparison of Other Activin A Structures to the FSTL3·Activin A Complex—Unlike BMP ligands, which exhibit minor fluctuations in the relative position of each monomer, structures of activin A, alone and in complex, have displayed significant flexibility at the dimer interface, resulting in dimers that range from compact or “V-shaped” to fully extended (21, 24, 36, 37) (supplemental Fig. S1). The flexibility of the activin A dimer is a result of numerous glycine and serine residues located in the loop (at positions 45-57) preceding the long α-helix at the dimer interface, referred to as the prehelix loop. This region is disordered in activin A structures (supplemental Fig. S1) that display more compact configurations (21, 36, 37). In all complexes with FS-type antagonists, including FSTL3, the activin A dimer is extended. This is likely a result of the N-terminal domain binding in the type I interface.
Type II Interface Blockade (FSD1 and FSD2)—Similar to the FS·activin A structures, one of the two FSTL3·activin A interfaces is located where the type II receptors bind ligand. Here, FSD1 and FSD2 cover a large portion of the convex surface of activin A. Residues that interact with activin A at the type II interface are well conserved between FSTL3 and FS (19/26 residues conserved). FSTL3 and FS effectively bind matching activin A residues utilized by type II receptors (supplemental Figs. S2 and S3). At this interface, FSTL3/FS cover ∼820 Å2 of activin A as compared with the activin type II receptor (ActRIIB), which has been shown to cover ∼600 and ∼730 Å2 (36, 37) (supplemental Fig. S3). In addition to most of the type II receptor interface, FSTL3/FS extend toward the outer tip of activin A (supplemental Fig. S3). Here FSTL3/FS form additional contacts with Asp27, Asp96, Gln98, and Asn99 of activin A, which are contacts not present in the receptor complex. At this interface, a conserved arginine (Arg192 in FS and Arg199 in FSTL3) bridges the two activin A loops at the ligand fingertips, forming a salt bridge with Asp27 and Gln98 and a hydrophobic interaction with Y94 of activin A. Mutation of Arg192 in a truncated version of FS (FSD1-FSD2) considerably reduces activin A affinity (21). At the other end of the interface, the type II receptors interact with activin A residues that do not contact either FSTL3 or FS (Lys85, Pro88, and Glu111) (supplemental Fig. S3). These differences in activin A binding, which are conserved across both receptors and antagonists, may be attractive sites for modification that would selectively block either antagonist or receptor binding.
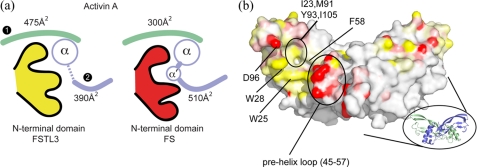
Type I Interface Blockade (N-terminal Domain)—Consistent with the FS·activin A structures, the N-terminal domain of FSTL3 binds at the activin A dimer interface, or the type I receptor site (Fig. 2, a and c). This interface is formed by the curved β-strand portion of one monomer (colored green and referred to as monomer 1 in Figs. 3 and 4) and the central helix and surrounding loops (“wrist”) of the other monomer (colored blue and referred to as monomer 2). Largely, the N-terminal domains of both FSTL3 and FS bind in a similar location and bury a comparable amount of activin A surface (865 and 805 Å2, respectively). But unlike their similarity in binding at the type II interface, we observed significant structural differences between the FSTL3 and FS N-terminal domains. The two N-terminal domains have a Cα root mean square deviation of 2.2 Å, which mirrors the divergence seen in sequence identity (Fig. 1) and is significantly greater than differences in FSD1 (1.3 Å) and FSD2 (1.0 Å) (38). Moreover, marked differences were observed in how each N-terminal domain interacts with activin A. FSTL3 interacts to a greater extent with monomer 1, whereas FS has additional interactions with the prehelix loop of monomer 2 (Fig. 3a). The variations in buried surface area of the two complexes have been plotted on the ligand surface in Fig. 3b. FSTL3 interacts to a greater degree with the hydrophobic surface on monomer 1, whereas FS interacts substantially more with the prehelix loop of monomer 2.
FIGURE 3.
Buried surface area differences on activin A at the type I interface. a, schematic depicting the distribution of buried surface area on each activin A subunit by interactions with the N-terminal domain of FSTL3 (yellow) and FS (red). The type I interface consists of both monomers (monomer 1, green; monomer 2, blue). FSTL3 buries more surface on monomer 1, whereas FS buries more surface on monomer 2 through interactions with the prehelix loop, which adopts a novel helix conformation termed α′. b, surface of activin A representing the difference in buried surface area for individual activin A atoms from interaction with FSTL3 and FS (ΔBSA = FSTL3BSA - FSBSA). The surfaces are colored with a three-step (yellow/white/red) gradient from 25 to -25 Å2. The yellow surfaces depict where FSTL3 buries more activin surface than FS, and the red surfaces depict where FS buries more than FSTL3. The white surfaces indicate that either no interaction occurs with antagonists, or the difference in buried surface area upon binding FSTL3 versus FS is minimal.
FIGURE 4.
Comparison of FSTL3 and FS N-terminal domain interactions with activin A. a and b, the N-terminal domain of FSTL3 is oriented more closely than that of FS to activin A monomer 1 (green). The prehelix loop region on activin A is disordered when bound to FSTL3, whereas in FS it forms the α′ helix (pink). There are also significant structural differences in the C-terminal loop segment on FSTL3/FS following the helix (cyan). c, superposition of only activin A monomer 1 in both complexes, which depicts the position of the each N-terminal domain relative to activin A. The N-terminal domain of FSTL3, including the helix, is closer to activin A by 3.5 and 2.9 Å, respectively. The distances were calculated from the center of mass (COM) of activin A backbone residues 25-28 to the COM of each N-terminal domain or the COM of each helix (FSTL3 residues 51-63 and FS residues 42-53). The asterisk indicates where FSTL3 would clash with the α′ of activin A from the FS structure. d, superposition of only the N-terminal domains. The FSTL3 domain is much more compact with reference to the main helix. The distance between the helices was measured from the COM of FSTL3 (residues 51-63) to the COM of FS (residues 42-53). All of the COM calculations were performed with the program CALCOM (50). In e and f, the structure is rotated 180° from a-c and shows differences in how the N-terminal helices interact with the two conserved tryptophan residues (at positions 25 and 28) of activin A. A significant difference in the orientation of Trp28 is observed in the two structures; this permits Leu57 of FSTL3 to wedge between Trp25 and Trp28 of activin A.
A helix in the N-terminal domain (residues 53-61 in FSTL3 and residues 42-53 in FS; Fig. 4, a and b) comprises a major portion of the interface with the activin A dimer. Differences in surface residues allow the helix of FSTL3 to pack closer to the activin A dimer crevice, especially toward the concave surface of monomer 1 (Fig. 4, a and b). In fact, this helix in FSTL3 buries 200 Å2 more surface area than the corresponding FS helix (for residue details see supplemental Fig. S2). Superimposing activin A monomer 1 of each complex reveals that the N-terminal domain of FSTL3, including its helix, is wedged closer to the activin A dimer interface (Fig. 4c).
Overall the N-terminal domain of FSTL3 is more compact than that of FS, and this contributes to differential activin A binding, especially with the prehelix loop of activin (Fig. 4, a, b, and d). In FS, a crevice is created between the N-terminal domain helix and the central sheets (15 Å line in Fig. 4b). Residues in the prehelix loop of activin A pack into this crevice and stabilize the novel α′ helix. Conversely, the more compact N-terminal domain of FSTL3 lacks this crevice (11 Å line in Fig. 4a). Thus, FSTL3 does not have an equivalent interaction that stabilizes a helical conformation of the prehelix loop in activin A. Superposition of FSTL3 onto activin A in the FS structure shows that if activin A adopted the α′ helix conformation, a steric clash would occur with FSTL3 (asterisk in Fig. 4c). The structural difference in compactness of the N-terminal domain may partially arise from variation in the loop leading out of the N-terminal domain helix (residues 62-66 for FSTL3 and residues 54-59 for FS; cyan in Fig. 4). FS has a longer loop that inserts into the crevice to maintain a more open configuration, whereas the same loop in FSTL3 travels away from the domain, allowing it to collapse (Fig. 4, a and b). In fact, the loop in FSTL3 forms at least one new contact with activin A not observed in FS as described below. A superposition of the two domains in Fig. 4d further demonstrates that FSTL3 is more compact than FS. The N-terminal domain helix of FSTL3 is tilted by 10°, bringing the helix 2.1 Å closer toward the center of the domain than the corresponding FS helix.
Interface Residues Imply a Stronger Type I Interaction for FSTL3—Although a similar amount of surface is buried by both N-terminal domains of FSTL3 and FS, FSTL3 interacts more substantially with the rigid portion of activin A, implying that its interaction is stronger and more favorable. A significant portion of the FSTL3 N-terminal domain interaction occurs at the more static, hydrophobic-rich surface of monomer 1, whereas the N-terminal domain of FS interacts mainly with small flexible amino acids in the α′ helix of monomer 2 (Fig. 3a and supplemental Fig. S2). At the center of the monomer 1 hydrophobic surface is a pair of tryptophan residues (Trp25 and Trp28) that are highly conserved across all TGFβ family ligands. The tryptophan side chain conformations and their corresponding interaction with antagonist are markedly different in the two structures (Fig. 4, e and f). When FSTL3 binds, Trp28 adopts a side chain rotamer that creates a large gap between the tryptophan residues. Leu57 on the N-terminal domain packs into the gap between the tryptophan residues, bringing the whole N-terminal domain closer to activin A. Fig. 5 shows the electron density of residues at this interface. On the other hand, in the FS complex, Trp25 and Trp28 are in a closed arrangement, similar to structures of activin A alone or in complex with receptors (21, 24, 36, 37). This restricts how closely the N-terminal domain contacts activin A.
Additional side chain interactions suggest a stronger interaction between activin A and the N-terminal domain of FSTL3 over FS. The close proximity to activin A monomer 1 enables a charged interaction between Lys54 of FSTL3 and Asp27 of activin A, and another charged interaction between His65 of FSTL3 and Asp96 of activin A. His65 lies on the loop extending distally from the N-terminal domain helix, and its interaction with Asp96 may influence the position of this loop and contribute further to the compactness of the N-terminal domain. These interactions are not seen for homologous residues in FS.
Domain Exchanges Support a Stronger Interaction for the FSTL3
N-terminal Domain—The structure of FSTL3 in complex with activin A
has implied that the interaction between the N-terminal domain and the type I
interface is stronger than that observed for the N-terminal domain of FS. To
validate our structural observation, we exchanged the N-terminal domain of
FSTL3 with FS (NDFSFSD1,2FSTL3). This reduced activin A
binding ∼60-fold as compared with native FS
(Table 2), demonstrating that
the N-terminal domains are not interchangeable. We next tested whether
attaching the third follistatin domain to the
NDFSFSD1,2FSTL3 chimera would reconstitute strong
activin A binding. The expectation was that FSD3 would contact the N-terminal
domain on the adjacent FS chimera and secure it in the type I receptor slot as
observed in the FS288·activin A structure
(24). Indeed, when the third
follistatin domain is introduced to make
NDFSFSD1,2FSTL3FSD3FS, strong activin A
binding is restored (Table 2),
indicating that the
FS(ND) FS(FSD3) contacts
enhance the ability of the N-terminal domain of FS to effectively bind activin
A. This differs markedly from native FS, where removal of FSD3 causes only a
40% loss in activin A binding
(26). Taken together with the
structural data, we conclude that the N-terminal domain of FSTL3 interacts
more strongly with activin A, thus obviating the need for a third follistatin
domain.
FS(FSD3) contacts
enhance the ability of the N-terminal domain of FS to effectively bind activin
A. This differs markedly from native FS, where removal of FSD3 causes only a
40% loss in activin A binding
(26). Taken together with the
structural data, we conclude that the N-terminal domain of FSTL3 interacts
more strongly with activin A, thus obviating the need for a third follistatin
domain.
TABLE 2.
Comparative activin A binding activity of FS/FSTL3 domain swap mutants in competitive binding assay using radiolabeled activin A as described in Ref. 20
| FS/FSTL3 construct | IC50 | Relative potency |
|---|---|---|
| nm | ||
| NDFS-FSD1FS-FSD2FS-FSD3FS (native FS) | 0.30 | 1.0 |
| NDFSTL3-FSD1FSTL3-FSD2FSTL3 (native FSTL3) | 0.12 | 2.5 |
| NDFS-FSD1FSTL3-FSD2FSTL3 | 7.50 | 0.04 |
| NDFS-FSD1FSTL3-FSD2FSTL3-FSD3FS | 0.05 | 6.0 |
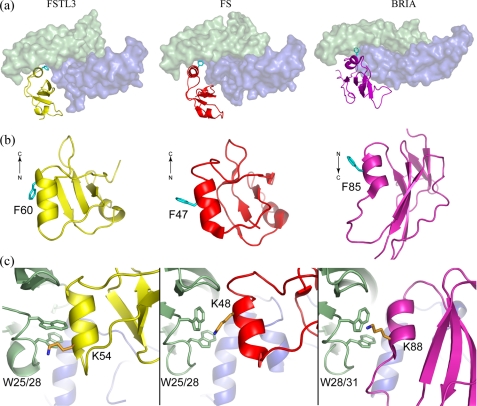
Type I Receptor Antagonism through Mimicry—Multiple x-ray structures have demonstrated that type I receptor binding occurs at or near the dimer interface of the ligand (39-42). With resolution of the FS structure, we identified a striking similarity between type I receptor (BRIA) binding and N-terminal domain binding. Our current structure extends this similarity to include the N-terminal domain of FSTL3. Despite different folds, there are structural similarities between the receptor and the N-terminal domains that further imply a similar strategy for ligand binding. For example, both utilize a single domain of similar size that consists of mainly β-strands with a single helix at the interface. This is in stark contrast to the antagonist noggin, which utilizes an extended loop and not a domain to interact at the type I interface (43).
The N-terminal domains and the receptor BRIA contain a phenylalanine on the helix (Phr60-FSTL3, Phe47-FS, and Phe85-BRIA) that projects into the hydrophobic ligand dimer crevice (Fig. 6a). Despite both FSTL3 and FS sharing this feature, the two antagonists use a nonhomologous phenylalanine with different side chain orientations (Fig. 6b). In FSTL3, Phe60 is located at the distal end of the N-terminal domain helix and is bent sharply to fit into the hydrophobic pocket rather than lying in a central position in the helix and adopting an extended conformation as is seen for Phe47 in FS (Fig. 6b). The bent conformation of Phe60 (FSTL3) versus Phe47 (FS) may also help position the N-terminal domain helix closer to activin A. An additional resemblance among the three interactions is found in a nonhomologous lysine that borders the conserved tryptophan residues (Fig. 6c). These similarities are preserved even though the helix from the antagonist is antiparallel to the helix from the receptor.
FIGURE 6.
Similarities between N-terminal domain and type I receptor ligand binding strategies. a, overall ligand binding position and domain size are similar among FSTL3/FS N-terminal domains and the BRIA type I receptor with a significant contribution to the binding interface coming from a single helix. b, side view of each domain with the common phenylalanine that is found at the ligand interface highlighted. The direction of the helix is indicated and runs in the reverse direction in BRIA. c, in each case a lysine, located in different regions of the helix, partially buries the conserved tryptophan residues.
DISCUSSION
Neutralization of TGFβ family ligands by means of binding extracellular antagonists is a vital mode of signal regulation. Antagonists fall into structurally distinct classes that bind certain ligands with different affinities (44). The structure of FSTL3 in complex with activin A and its comparison with previously determined FS structures provide a system to examine the similarities and differences in specificity and affinity within a class of structurally related antagonists. Our discussion will largely center on the significance of the N-terminal domain and structural differences between FSTL3 and FS.
The Role of the N-terminal Domain in Antagonism—An
unexpected result from the FS·activin A structure was that in addition
to the type II receptor site, FS also blocks the type I receptor site through
interactions of its N-terminal domain
(24). This observation is now
supported through resolution of the structure of the FSTL3·activin A
complex. In fact, the interaction of the FSTL3 N-terminal domain with activin
A appears stronger. In the FS·activin A complex, the N-terminal domain
interacts with FSD3 of an adjacent FS molecule, which may hold the N-terminal
domain at the type I interface. No such interaction is observed in the
FSTL3·activin A structure because there is no FSD3; yet the N-terminal
domain of FSTL3 still binds activin A. To better interact with the hydrophobic
surface of activin A and possibly compensate for not having the
FS(ND) FS(FSD3) interaction,
the N-terminal domain of FSTL3 adopts a more compact conformation. In
particular, Leu57 of FSTL3 is sandwiched by the two tryptophan
residues conserved in all TGFβ family ligands. This observation is
supported by our domain exchange experiments that demonstrate that the
N-terminal domain of FS cannot replace the N-terminal domain of FSTL3 for full
activity. The implication is that the N-terminal domains in both FS and FSTL3
form a unique but significant interaction at the ligand interface.
FS(FSD3) interaction,
the N-terminal domain of FSTL3 adopts a more compact conformation. In
particular, Leu57 of FSTL3 is sandwiched by the two tryptophan
residues conserved in all TGFβ family ligands. This observation is
supported by our domain exchange experiments that demonstrate that the
N-terminal domain of FS cannot replace the N-terminal domain of FSTL3 for full
activity. The implication is that the N-terminal domains in both FS and FSTL3
form a unique but significant interaction at the ligand interface.
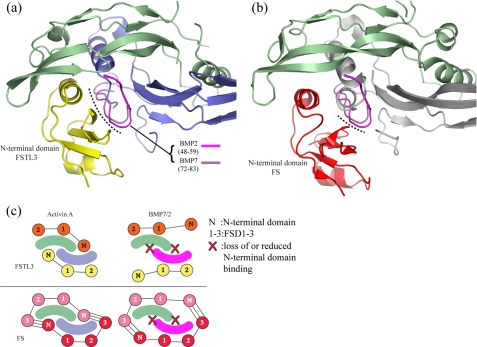
Implications for Ligand Specificity—The molecular basis for why FS and FSTL3 both bind activin A tightly but have either reduced or no affinity for BMP ligands has not been resolved (14, 17, 45). To investigate this we superimposed BMPs 2 and 7 onto activin A in the structures of the FS288 and FSTL3 complexes. In both cases, the N-terminal domains would clash with the prehelix loop of the BMP ligands (Fig. 7, a and b). The molecular overlap (Fig. 7, a and b, dotted lines) appears more extensive for the N-terminal domain of FSTL3 than FS. In contrast to the flexibility of activin A, the rigid nature of the BMP ligands in the prehelix loop suggests that N-terminal domain binding would be significantly, if not completely, disrupted (39, 40, 43, 46, 47).
FIGURE 7.
Structural differences between activin A and BMP may explain antagonist
specificity. a and b, superposition of BMP2 and BMP7
onto the FSTL3·activin A complex, aligning only monomer 1 of activin A.
The ribbon of activin A is shown, but for clarity, only the prehelix loop
residues for each BMP (BMP2 (pink) and BMP7 (purple)) are
depicted. The dotted lines indicate regions on BMP close enough to
clash with the N-terminal domains. These appear more extensive for FSTL3.
c, overall scheme for FSTL3 and FS binding to activin A and BMPs. We
propose that the N-terminal domain of FS-type antagonists does not interact
favorably with BMPs, thus accounting for the decreased affinity. We propose
that the low affinity still observed for FS may be a result of the
FS(ND) FS(FSD3) interactions
and/or structural variation in the antagonist N-terminal domains.
FS(FSD3) interactions
and/or structural variation in the antagonist N-terminal domains.
Therefore, we propose a model where a significant reduction in affinity for
BMPs is a result of impaired N-terminal domain binding
(Fig. 7c). The low
affinity interactions of FS for BMPs may be a consequence of the additional
FS(ND) FS(FSD3) interaction
not observed for FSTL3 or may result from structural differences in the
N-terminal domains. In support of this model, ligands discriminate type I
receptors effectively through structural variation in the prehelix loop
(40,
48,
49). This is consistent with
our current results showing that N-terminal domains, in several respects,
mimic type I receptors. Furthermore, activin A can be converted to a BMP-like
ligand by swapping the major α-helix and flanking loops, including the
prehelix loop (48,
49). Therefore, we expect
FS-type antagonists to also utilize the type I interface for ligand
specificity, much like type I receptors.
FS(FSD3) interaction
not observed for FSTL3 or may result from structural differences in the
N-terminal domains. In support of this model, ligands discriminate type I
receptors effectively through structural variation in the prehelix loop
(40,
48,
49). This is consistent with
our current results showing that N-terminal domains, in several respects,
mimic type I receptors. Furthermore, activin A can be converted to a BMP-like
ligand by swapping the major α-helix and flanking loops, including the
prehelix loop (48,
49). Therefore, we expect
FS-type antagonists to also utilize the type I interface for ligand
specificity, much like type I receptors.
The physiological basis for multiple FS-type molecules with varying affinities for heparin/cell surfaces has not been resolved. Evidence supports the trend that antagonist-ligand specificity tightens as the serum availability of the FS-type antagonist increases. This is logical, because releasing a broad activin/myostatin/BMP antagonist into the serum would likely have dramatic effects on multiple systems. Our x-ray structure of FSTL3·activin A complex and its comparison with the previously determined FS·activin A structures provides an explanation for how this ligand discrimination occurs. The insights gained through our structural studies will aid in the modification and adaptation of antagonists for potential therapeutic applications, because ligand specificity is an important factor in targeting.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants GM084186 (to T. B. T.) and DK053828 (to H. T. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3.
Footnotes
The abbreviations used are: TGF, transforming growth factor; FS, follistatin; FSTL3, follistatin-like 3; BMP, bone morphogenetic protein; FSD, follistatin domain; ND, N-terminal domain; BRIA, BMP type IA receptor; COM, center of mass.
References
- 1.Shi, Y., and Massague, J. (2003) Cell 113 685-700 [DOI] [PubMed] [Google Scholar]
- 2.Shimmi, O., Umulis, D., Othmer, H., and O'Connor, M. B. (2005) Cell 120 873-886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amthor, H., Christ, B., Rashid-Doubell, F., Kemp, C. F., Lang, E., and Patel, K. (2002) Dev. Biol. 243 115-127 [DOI] [PubMed] [Google Scholar]
- 4.Nakamura, T., Takio, K., Eto, Y., Shibai, H., Titani, K., and Sugino, H. (1990) Science 247 836-838 [DOI] [PubMed] [Google Scholar]
- 5.Inouye, S., Guo, Y., DePaolo, L., Shimonaka, M., Ling, N., and Shimasaki, S. (1991) Endocrinology 129 815-822 [DOI] [PubMed] [Google Scholar]
- 6.Michel, U., Farnworth, P., and Findlay, J. K. (1993) Mol. Cell Endocrinol. 91 1-11 [DOI] [PubMed] [Google Scholar]
- 7.Sugino, K., Kurosawa, N., Nakamura, T., Takio, K., Shimasaki, S., Ling, N., Titani, K., and Sugino, H. (1993) J. Biol. Chem. 268 15579-15587 [PubMed] [Google Scholar]
- 8.Hayette, S., Gadoux, M., Martel, S., Bertrand, S., Tigaud, I., Magaud, J. P., and Rimokh, R. (1998) Oncogene 16 2949-2954 [DOI] [PubMed] [Google Scholar]
- 9.Tsuchida, K., Arai, K. Y., Kuramoto, Y., Yamakawa, N., Hasegawa, Y., and Sugino, H. (2000) J. Biol. Chem. 275 40788-40796 [DOI] [PubMed] [Google Scholar]
- 10.Schneyer, A., Tortoriello, D., Sidis, Y., Keutmann, H., Matsuzaki, T., and Holmes, W. (2001) Mol. Cell Endocrinol. 180 33-38 [DOI] [PubMed] [Google Scholar]
- 11.Tsuchida, K., Nakatani, M., Matsuzaki, T., Yamakawa, N., Liu, Z., Bao, Y., Arai, K. Y., Murakami, T., Takehara, Y., Kurisaki, A., and Sugino, H. (2004) Mol. Cell Endocrinol. 225 1-8 [DOI] [PubMed] [Google Scholar]
- 12.Matzuk, M. M., Lu, N., Vogel, H., Sellheyer, K., Roop, D. R., and Bradley, A. (1995) Nature 374 360-363 [DOI] [PubMed] [Google Scholar]
- 13.Mukherjee, A., Sidis, Y., Mahan, A., Raher, M. J., Xia, Y., Rosen, E. D., Bloch, K. D., Thomas, M. K., and Schneyer, A. L. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1348-1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iemura, S., Yamamoto, T. S., Takagi, C., Uchiyama, H., Natsume, T., Shimasaki, S., Sugino, H., and Ueno, N. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 9337-9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welt, C., Sidis, Y., Keutmann, H., and Schneyer, A. (2002) Exp. Biol. Med. (Maywood) 227 724-752 [DOI] [PubMed] [Google Scholar]
- 16.Lin, S. Y., Morrison, J. R., Phillips, D. J., and de Kretser, D. M. (2003) Reproduction 126 133-148 [DOI] [PubMed] [Google Scholar]
- 17.Sidis, Y., Mukherjee, A., Keutmann, H., Delbaere, A., Sadatsuki, M., and Schneyer, A. (2006) Endocrinology 147 3586-3597 [DOI] [PubMed] [Google Scholar]
- 18.Hill, J. J., Davies, M. V., Pearson, A. A., Wang, J. H., Hewick, R. M., Wolfman, N. M., and Qiu, Y. (2002) J. Biol. Chem. 277 40735-40741 [DOI] [PubMed] [Google Scholar]
- 19.Schneyer, A. L., Wang, Q., Sidis, Y., and Sluss, P. M. (2004) J. Clin. Endocrinol. Metab. 89 5067-5075 [DOI] [PubMed] [Google Scholar]
- 20.Sidis, Y., Schneyer, A. L., Sluss, P. M., Johnson, L. N., and Keutmann, H. T. (2001) J. Biol. Chem. 276 17718-17726 [DOI] [PubMed] [Google Scholar]
- 21.Harrington, A. E., Morris-Triggs, S. A., Ruotolo, B. T., Robinson, C. V., Ohnuma, S., and Hyvonen, M. (2006) EMBO J. 25 1035-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arai, K. Y., Tsuchida, K., Li, C., Watanabe, G., Sugino, H., Taya, K., and Nishiyama, T. (2006) Protein Expression Purif. 49 78-82 [DOI] [PubMed] [Google Scholar]
- 23.Harrison, C. A., Chan, K. L., and Robertson, D. M. (2006) Endocrinology 147 2744-2753 [DOI] [PubMed] [Google Scholar]
- 24.Thompson, T. B., Lerch, T. F., Cook, R. W., Woodruff, T. K., and Jardetzky, T. S. (2005) Dev. Cell 9 535-543 [DOI] [PubMed] [Google Scholar]
- 25.Lerch, T. F., Shimasaki, S., Woodruff, T. K., and Jardetzky, T. S. (2007) J. Biol. Chem. 282 15930-15939 [DOI] [PubMed] [Google Scholar]
- 26.Keutmann, H. T., Schneyer, A. L., and Sidis, Y. (2004) Mol. Endocrinol. 18 228-240 [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 28.McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C., and Read, R. J. (2005) Acta Crystallogr. Sect. D Biol. Crystallogr. 61 458-464 [DOI] [PubMed] [Google Scholar]
- 29.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 30.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 31.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 32.Painter, J., and Merritt, E. A. (2006) Acta Crystallogr. Sect. D Biol. Crystallogr. 62 439-450 [DOI] [PubMed] [Google Scholar]
- 33.Painter, J., and Merritt, E. A. (2006) J. Appl. Crystallogr. 39 109-111 [Google Scholar]
- 34.Winn, M. D., Isupov, M. N., and Murshudov, G. N. (2001) Acta Crystallogr. Sect. D Biol. Crystallogr. 57 122-133 [DOI] [PubMed] [Google Scholar]
- 35.Krissinel, E., and Henrick, K. (2007) J. Mol. Biol. 372 774-797 [DOI] [PubMed] [Google Scholar]
- 36.Greenwald, J., Vega, M. E., Allendorph, G. P., Fischer, W. H., Vale, W., and Choe, S. (2004) Mol. Cell 15 485-489 [DOI] [PubMed] [Google Scholar]
- 37.Thompson, T. B., Woodruff, T. K., and Jardetzky, T. S. (2003) EMBO J. 22 1555-1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holm, L., and Park, J. (2000) Bioinformatics 16 566-567 [DOI] [PubMed] [Google Scholar]
- 39.Allendorph, G. P., Vale, W. W., and Choe, S. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 7643-7648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirsch, T., Sebald, W., and Dreyer, M. K. (2000) Nat. Struct. Biol. 7 492-496 [DOI] [PubMed] [Google Scholar]
- 41.Weber, D., Kotzsch, A., Nickel, J., Harth, S., Seher, A., Mueller, U., Sebald, W., and Mueller, T. D. (2007) BMC Struct. Biol. 7 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Groppe, J., Hinck, C. S., Samavarchi-Tehrani, P., Zubieta, C., Schuermann, J. P., Taylor, A. B., Schwarz, P. M., Wrana, J. L., and Hinck, A. P. (2008) Mol. Cell 29 157-168 [DOI] [PubMed] [Google Scholar]
- 43.Groppe, J., Greenwald, J., Wiater, E., Rodriguez-Leon, J., Economides, A. N., Kwiatkowski, W., Affolter, M., Vale, W. W., Belmonte, J. C., and Choe, S. (2002) Nature 420 636-642 [DOI] [PubMed] [Google Scholar]
- 44.Zhang, J. L., Huang, Y., Qiu, L. Y., Nickel, J., and Sebald, W. (2007) J. Biol. Chem. 282 20002-20014 [DOI] [PubMed] [Google Scholar]
- 45.Glister, C., Kemp, C. F., and Knight, P. G. (2004) Reproduction 127 239-254 [DOI] [PubMed] [Google Scholar]
- 46.Greenwald, J., Groppe, J., Gray, P., Wiater, E., Kwiatkowski, W., Vale, W., and Choe, S. (2003) Mol. Cell 11 605-617 [DOI] [PubMed] [Google Scholar]
- 47.Keller, S., Nickel, J., Zhang, J. L., Sebald, W., and Mueller, T. D. (2004) Nat. Struct. Mol. Biol. 11 481-488 [DOI] [PubMed] [Google Scholar]
- 48.Nickel, J., Kotzsch, A., Sebald, W., and Mueller, T. D. (2005) J. Mol. Biol. 349 933-947 [DOI] [PubMed] [Google Scholar]
- 49.Korupolu, R. V., Muenster, U., Read, J. D., Vale, W., and Fischer, W. H. (2008) J. Biol. Chem. 283 3782-3790 [DOI] [PubMed] [Google Scholar]
- 50.Costantini, S., Paladino, A., and Facchiano, A. M. (2008) Bioinformation 2 271-272 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.