Abstract
High levels of heparanase are an indicator of poor prognosis in myeloma patients, and up-regulation of the enzyme enhances tumor growth, angiogenesis, and metastasis in animal models. At least part of the impact of heparanase in driving the aggressive tumor phenotype is due to its effect on increasing the expression and shedding of the heparan sulfate proteoglycan syndecan-1, a molecule known to promote myeloma progression. The present work demonstrated that elevation in heparanase expression in myeloma cells stimulates sustained ERK phosphorylation that in turn drives MMP-9 expression. In addition, urokinase-type plasminogen activator (uPA) and uPA receptor expression levels increased, and blocking the proteolytic activation of either MMP-9 or uPA inhibited the heparanase-induced increase in syndecan-1 shedding. Together these data provide a mechanism for heparanase-induced syndecan-1 shedding and, more importantly, demonstrate that heparanase activity in myeloma cells can lead to increased levels of proteases that are known to play important roles in the aggressive behavior of myeloma tumors. This in addition to its other known biological roles, indicates that heparanase acts as a master regulator of the aggressive tumor phenotype by up-regulating protease expression and activity within the tumor microenvironment.
Heparanase is a heparan sulfate degrading endoglycosidase that becomes up-regulated in many human cancers causing significant increases in both the angiogenic and metastatic potential of tumor cells (1, 2). Studies on tissues from human cancer patients strongly support an association between heparanase expression and poor prognosis, and high levels of heparanase are associated with a shorter postoperative survival time compared with cancer patients with low levels of heparanase (2–4). Experimental models have been used to extend these correlative studies in patients and have demonstrated an important role for heparanase in driving aggressive tumor progression. For example, pancreatic cells engineered to express heparanase exhibit increased invasiveness in Boyden chamber assays as compared with controls, and inhibitors of heparanase block tumor growth in an animal model of pancreatic cancer (4, 5). Murine T-lymphoma cells acquire an enhanced metastatic phenotype when engineered to express heparanase (6), and breast cancer cell growth, angiogenesis, and survival in vivo are dramatically increased upon up-regulation of heparanase expression (7). Knockdown of heparanase expression in either lymphoma or melanoma cells using antiheparanase ribozymes decreases metastasis and increases survival in animal models (8). Inhibition of heparanase expression by a heparanase-specific antisense approach inhibits lung and esophageal cancer cells invasion in vitro as well as the pleural dissemination of cells implanted into nude mice (9). The effects of heparanase in cancer may not solely be due to its enzymatic activity because mutated, non-enzymatically active heparanase retains some biological functions. For example, heparanase enhances Akt signaling and stimulates phosphoinositide 3-kinase- and p38-dependent migration and invasion of endothelial cells, and this can occur via mutated heparanase that lacks enzymatic activity (10). In addition, vascular endothelial growth factor expression is up-regulated in several tumor cell lines following their transfection with mutated heparanase and down-regulated in melanoma cells transfected with heparanase-specific small interfering RNA (11). Overall the data from cancer patients coupled with experimental data from animal models strongly point to heparanase as a potent protumorigenic, proangiogenic, and prometastatic enzyme.
Multiple myeloma is a devastating cancer that resides predominantly within the bone marrow microenvironment and is characterized by fatigue, intractable bone pain, renal failure, and recurrent infections. These effects result from widespread tumor dissemination with accompanying high tumor burden, cytokine dysregulation, osteolytic bone disease, and deposition of high levels of immunoglobulin light chain (12). The syndecan-1 heparan sulfate proteoglycan is expressed by almost all myeloma tumors and when present at high levels in the serum of patients is an indicator of poor prognosis (13–15). Moreover the shed form of syndecan-1 promotes tumor growth and metastasis in vivo (16). Because heparanase modulates the structure and function of syndecan-1 by cleaving its heparan sulfate chains, in previous studies we investigated its expression and function in myeloma. We discovered that enzymatically active heparanase is present at high levels in the bone marrow plasma of many myeloma patients, and this correlates with high microvessel density, suggesting that high heparanase is associated with poor prognosis (17, 18). Using in vivo models we also demonstrated that heparanase promotes the growth and spontaneous metastasis of myeloma tumors to bone and that an inhibitor of heparanase potently blocks tumor growth (19, 20). Interestingly heparanase also regulates both the level and location of syndecan-1 within the myeloma microenvironment by enhancing syndecan-1 expression and shedding (18, 21). This suggests an important functional link between heparanase expression, syndecan-1 shedding, and an aggressive tumor phenotype.
The present study was undertaken to examine the mechanism by which heparanase promotes syndecan-1 shedding. We found that elevation of heparanase expression stimulates a prolonged ERK2 signaling by the myeloma cells. This signaling up-regulated expression of MMP-9 and uPA/uPA receptor (uPAR), which appear to work in concert to enhance syndecan-1 shedding. This enhanced shedding of syndecan-1 likely has an impact on tumor behavior, but more importantly, the up-regulated proteases are known to promote tumor growth, angiogenesis, metastasis, and bone disease. Together these data imply that heparanase acts as a master regulator of the aggressive tumor phenotype in myeloma.
EXPERIMENTAL PROCEDURES
Cells and Transfections—CAG cells were established from a bone marrow aspirate of a myeloma patient at the Arkansas Cancer Research Center as described previously (22). ARH-77 cells were obtained from the American Type Culture Collection (Manassas, VA). CAG and ARH-77 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum. As described previously, the cells were transfected with either empty vector or vector containing the human heparanase cDNA to generate heparanase low and heparanase high cells, respectively (17, 19). For construction of vectors carrying mutations in the enzyme active site of heparanase, mutations were generated at Glu-225 or Glu-343 as described previously (21).
Gelatin Zymography—After cells were incubated with serum-free medium for 48 h, supernatants were collected and concentrated in Centriplus columns with a 30-kDa cutoff value (Millipore Corp., Bedford, MA). Protein in the concentrated media was quantified using the BCA protein assay reagent kit (Pierce), and an equal amount of protein (50 μg) was mixed with non-reducing sample buffer (62.5 mm Tris-HCl, pH 6.8, 25% glycerol, 4% SDS, and 0.01% bromphenol blue) and analyzed by SDS-PAGE using 10% polyacrylamide gels co-polymerized with gelatin (Bio-Rad). Electrophoresis was carried out at 10 mA for 2 h. The SDS in the acrylamide gel was extracted by incubation with 2.5% Triton X-100 solution for 2 h at room temperature, and gelatinolytic activities were developed in a buffer containing 50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 5 mm CaCl2, and 0.02% Brij 35 at 37 °C overnight. The gel was then stained with Coomassie Blue. Following destaining, sites of proteolytic activities were visualized as clear bands against the blue background of stained gelatin. For some experiments, cells were treated with recombinant human heparanase (kindly provided by Dr. Israel Vlodavsky and prepared as described previously (23)) or MAPK inhibitor PD98059 (Calbiochem) (50 μm) and incubated at 37 °C in 5% CO2 for 24 h. NIH Image (National Institutes of Health, Bethesda, MD) was used to quantify the bands.
Western Blotting—For MMP-9 and uPA detection, serum-free media conditioned for 48 h were concentrated using Centriplus YM30 (Millipore), and proteins (20 μg for MMP-9 blots and 80 μg for uPA blots) were subjected to 10% SDS-PAGE under reducing conditions, transferred to nitrocellulose membrane (Schleicher and Schuell), and probed with either rabbit anti-human MMP-9 antibody (Chemicon, Inc., Temecula, CA) or rabbit anti-human uPA (H-140) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) followed by horseradish peroxidase-conjugated donkey anti-rabbit IgG. Bands were quantified using NIH Image. MMP-9 within tumors was detected by Western blotting following their homogenization in lysis buffer (1:4, w/v) containing 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, and 0.1% SDS. uPAR expression was detected in the cell lysate using a mouse anti-human uPAR antibody (CD87) (R&D System Inc., Minneapolis, MN), and heparanase expression was detected using a rabbit anti-human heparanase polyclonal antibody (kindly provided by Hua-Quan Miao, Imclone, Inc.) (24). Equal loading of protein was confirmed by staining membranes for human β-actin (Sigma).
For analysis of signaling pathways, equal amounts of cell lysate protein from heparanase high or heparanase low cells were subjected to 10% SDS-PAGE, transferred to nitrocellulose membrane, and probed with either of the following antibodies: mouse monoclonal phospho-ERK (Cell Signaling Technology, Inc., Beverly, MA), rabbit polyclonal phospho-p38 (Cell Signaling Technology, Inc.), or rabbit polyclonal phospho-Src (Cell Signaling Technology, Inc.) followed by the corresponding horseradish peroxidase-labeled secondary antibody. Immunoreactive bands were detected using the ECL detection reagent (Amersham Biosciences). After stripping, the same blots were reprobed with antibodies to total ERK, p38, or Src (all from Cell Signaling Technology, Inc.).
Knockdown of Heparanase by shRNA—Using the nucleotide target sequence (423GGAATCAACCTTTGAAGAG441) (6), a double-stranded oligonucleotide was synthesized (Integrated DNA Technologies, Coralville, IA) to knock down heparanase: oligonucleotide 1, CGCGTCCCCGGAATCAACCTTTGAAGAGTTCAAGAGACTCTTCAAAGGTTGATTCCTTTTTTGGAAAT; oligonucleotide 2, CGATTTCCAAAAAAGGAATCAACCTTTGAAGAGTCTCTTGAACTCTTCAAAGGTTGATTCCGGGGA. The control shRNA includes a scrambled sequence that does not match any sequence of human genes: scrambled oligonucleotide 1, CGCGTCCCCGTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGACTTTTTGGAAAT; scrambled oligonucleotide 2, CGATTTCCAAAAAGTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGACGGGGA. The complementary oligonucleotides were annealed, and the double-stranded oligonucleotides were ligated between MluI and ClaI restriction sites on pLVTHM vector (containing the H1 promoter), and lentivirus was packaged by transfection of the pLVTHM vector and plasmids pMD2G and pCMV-dR8.91 (vectors and plasmids kindly provided by Dr. Didier Trono, University of Geneva, Geneva, Switzerland) into 293FT cells (Invitrogen). The conditioned medium of the transfected 293FT cells was filtered (0.45 μm), and the titer of virus was determined by fluorescence-activated cell sorting and calculated by the percentage of green fluorescent protein-positive cells after 48 h postinfection of CAG cells. CAG cells were infected at a multiplicity of infection of 50 followed by cell sorting by green fluorescent protein expression. The reduction of heparanase expression was confirmed by reverse transcription-PCR and immunoblotting. For PCR, the forward and reverse primers were 5′-CGCGTAGTGATGCCATGTAACTGAAT-3′ (forward) and 5′-CGCTTCGATCCCAAGAAGGAATCAAC-3′ (reverse) for heparanase and 5′-ACCACAGTCCATGCCCATCAC-3′ (forward) and 5′-TCCACCACCCTGTTGCTGTA-3′ (reverse) for glyceraldehyde-3-phosphate dehydrogenase. The PCR was performed as an initial denaturation at 95 °C for 2 min followed by 32 cycles of 95 °C for 45 s, 60 °C for 1 min, and 72 °C for 1 min and ended by extension at 72 °C for 10 min. PCR products were separated by 1.0% agarose gel electrophoresis and visualized by ethidium bromide staining.
Immunohistochemistry—Sections from tumors formed from heparanase low and heparanase high cells were deparaffinized with xylene and then rehydrated through graded concentrations of ethanol and distilled water. Epitope retrieval was performed by steaming the slides for 20 min in citrate buffer solution (pH 6.0). Slides were washed and incubated with 2.5% H2O2 for 30 min to quench endogenous peroxide activities and then were blocked with 1% bovine serum albumin in phosphate-buffered saline for 1 h at room temperature. The slides were then stained overnight with rabbit anti-human MMP-9 antibody (10 μg/ml in 1% bovine serum albumin and phosphate-buffered saline) at 4 °C. The sections were washed with phosphate-buffered saline and stained with biotinylated goat anti-rabbit IgG (Vector Laboratories, Burlingame, CA) for 1 h at room temperature followed by Vectastain ABC reagent (Vector Laboratories) for another 1 h at room temperature. Detection was accomplished using a 3,3′-diaminobenzidine substrate kit (Vector Laboratories). Photographs were taken using an Olympus BX60 System Microscope.
Quantification of Syndecan-1—Equal numbers of cells (106 cells/ml) were plated in wells of 12-well plates in complete RPMI 1640 medium. MMP-9 neutralizing antibody 6-6B (Calbiochem) or MMP-9 inhibitor 1 (Calbiochem) was added to some wells. In some experiments the cells were incubated with monoclonal anti-human uPAR antibody, which blocks uPA activation (R&D System, Inc.). After 48 h of incubation at 37 °C in 5% CO2, the cell culture media were collected, and the levels of shed syndecan-1 were assessed by enzyme-linked immunoadsorbent assay using an Eli-pair kit from Diaclone (Cell Sciences Inc., Norwood, MA). The standard curve was linear between 8 and 256 ng/ml, and all samples were diluted to concentrations within that range.
Matrigel Invasion Assay—The invasiveness of myeloma cells was analyzed using Biocoat matrigel invasion chambers (BD Biosciences). The matrigel invasion chamber consists of cell culture inserts containing an 8-μm-pore size polyethylene membrane precoated with a thin layer of matrigel basement membrane matrix. 500 μl of warm RPMI 1640 medium was added to the interior of the inserts and to each well, and the matrigel was allowed to rehydrate for 2 h in a humidified tissue culture incubator at 37 °C and 5% CO2. After rehydration the medium was carefully removed without disturbing the layer of matrigel matrix on the membrane. Cells (2 × 105) suspended in 500 μl of serum-free RPMI 1640 medium were seeded in the upper compartment of chambers. To examine the role of uPA/uPAR in invasion, cells together with mouse anti-human uPAR antibody were loaded into the upper compartment of the matrigel invasion chamber. 750 μl of RPMI 1640 medium with 10% fetal bovine serum, which served as the chemoattractant, was added to the lower compartment of the invasion chamber and incubated for 22 h. Cells that invaded the matrigel-coated filters were recovered from the lower compartment and counted using a Coulter Z1 Particle counter. Each assay was carried out in triplicate.
RESULTS
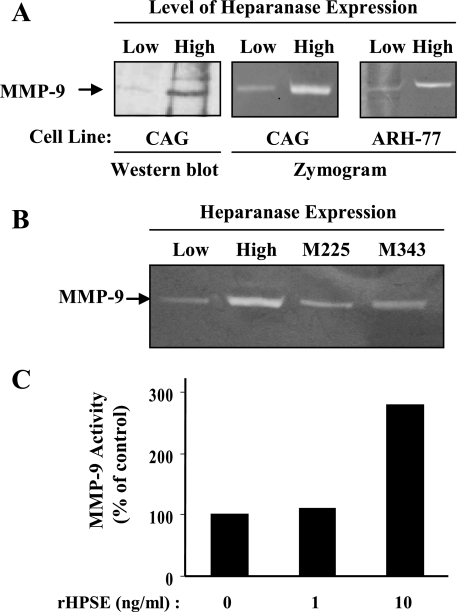
Heparanase Enhances the Expression of MMP-9 in Myeloma Cells—Previously we demonstrated that up-regulated expression of heparanase enhances the synthesis and shedding of syndecan-1 from the surface of myeloma cells, thereby contributing to tumor progression by elevating levels of syndecan-1 in the tumor microenvironment (16, 21). Work by others has shown that several matrix metalloproteinases can mediate syndecan-1 shedding (25–29), suggesting that heparanase may enhance syndecan-1 shedding by mediating the up-regulation of one or more of these proteases. Because MMP-9 is known to be present in myeloma tumors we examined its level of expression by CAG myeloma cells expressing high or low levels of the enzyme. Western blotting revealed a significant increase in MMP-9 protein levels in medium conditioned by the cells having high heparanase expression as compared with those having low expression of the enzyme (Fig. 1A). Reverse transcription-PCR analysis demonstrated a 3-fold increase in MMP-9 mRNA expressed in the heparanase high versus low cells,3 thus confirming the Western blot data. As we have shown previously, it is important to note that the level of heparanase expression and enzymatic activity in the heparanase high CAG cells is similar to that found in some myeloma patient tumors (17, 19, 21). Thus, the increase in MMP-9 expression by these cells is not due to an enhancement of heparanase expression beyond that likely to be found in the human myeloma microenvironment.
FIGURE 1.
Heparanase enhances MMP-9 expression and activity in myeloma cells. A, Western blot of MMP-9 in serum-free conditioned media from CAG cells demonstrates that pro-MMP-9 protein levels are up-regulated in CAG cells expressing high levels of heparanase as compared with cells expressing low levels of heparanase. (Heparanase high cells were transfected with vector containing the cDNA for human heparanase; heparanase low cells were transfected with empty vector (21).) Gelatin zymography of conditioned medium from either CAG or ARH-77 cells confirms Western blotting data by showing an elevation in levels of MMP-9 activity in cells expressing high levels of heparanase. (Note that the MMP-9 seen in both the Western blot and zymogram is the 92-kDa inactive proform of the enzyme rather than the cleaved 82-kDa active form. The gelatinolytic activity of MMP-9 routinely seen in zymograms is thought to be due to activation of the uncleaved enzyme resulting from the gel electrophoresis procedure.) B, CAG cells expressing heparanase that is mutated at amino acids 225 or 343 (M225 or M343) and thus lacking heparan sulfate-degrading activity do not show enhanced MMP-9 levels in zymograms as compared with cells expressing enzymatically active heparanase. C, addition of recombinant heparanase to CAG cells enhances levels of MMP-9. Recombinant heparanase (rHPSE) was added to wild-type CAG cells at the indicated concentrations, and conditioned medium was collected after 24 h and subjected to gelatin zymography. Quantification of MMP-9 gelatinolytic activity in these zymograms revealed an almost 3-fold higher level in cells treated with 10 ng/ml recombinant heparanase versus untreated cells. Shown are results of densitometric analysis of a single, representative gel.
To further explore MMP-9 expression in these cells, serum-free conditioned medium from myeloma cells was subjected to zymography. Heparanase high cells exhibited high gelatinolytic activity corresponding to pro-MMP-9 (92-kDa gelatinase) as compared with heparanase low cells (Fig. 1A). Analysis of heparanase-transfected ARH-77 human lymphoblastoid (myeloma-like) cells also demonstrated a similar increase in MMP-9 enzyme activity as compared with heparanase low cells, suggesting that the up-regulation of MMP-9 is not cell-line specific. However, in contrast to what was observed in myeloma cells, zymogram analysis of heparanase-transfected MDA-MET breast cancer cells (a highly metastatic variant of MDA-MB-231 cells) (30) demonstrated a decrease in MMP-9 expression as compared with controls.3 Thus, the effect of heparanase on MMP-9 expression may vary depending on the tumor type.
To determine whether the enhanced expression of MMP-9 by heparanase high myeloma cells requires the enzymatic (heparan sulfate-degrading) activity of heparanase, we examined MMP-9 activity levels in media from CAG cells expressing heparanase mutated at the active site of the enzyme (mutated at amino acid 225 (M225) or 343 (M343)) (31). In previous work we demonstrated that these cells express high levels of heparanase protein but exhibit low heparan sulfate-degrading activity as compared with cells expressing high levels of wild-type heparanase (21). When the conditioned medium from cells expressing these heparanase mutants was analyzed by zymogram, the gelatinase activity corresponding to MMP-9 was not elevated as compared with cells expressing the active enzyme (heparanase high cells) (Fig. 1B). This indicates that heparanase enzymatic activity is required to enhance MMP-9 expression in these cells.
We previously found that addition of recombinant heparanase to cells would enhance their shedding of syndecan-1 (21). To determine whether recombinant heparanase would affect MMP-9 expression, recombinant heparanase was added to wild-type CAG cells growing in culture. Over a 24-h time period, the activity of MMP-9 in the medium of cells treated with 10 ng/ml recombinant heparanase was almost 3-fold higher than that from cells not exposed to exogenous heparanase (Fig. 1C). This indicates that the up-regulation of MMP-9 in the heparanase high CAG myeloma cells is not simply an artifact related to their transfection with the cDNA for heparanase and that heparanase can stimulate a relatively rapid increase in MMP-9 levels. Moreover this result provides further evidence that extracellular heparanase can influence behavior of cells that are not actually expressing the enzyme (32, 33).
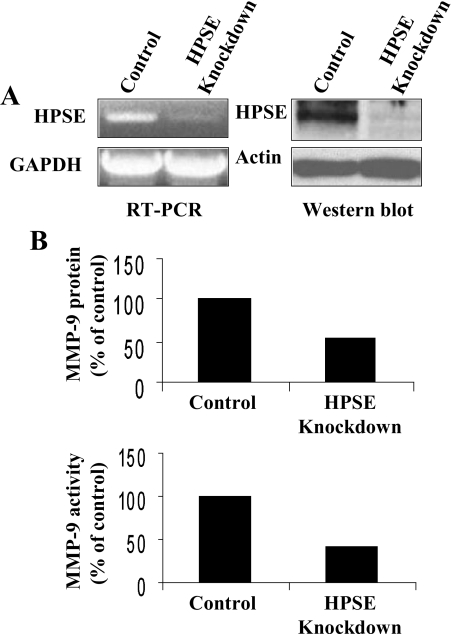
Heparanase Down-regulation Correlates with Reduced MMP-9 Expression Levels—As a final confirmation that heparanase regulates MMP-9 expression in myeloma cells, we used lentiviral vectors coding for an shRNA designed to block heparanase expression. We hypothesized that a reduction in heparanase expression in CAG wild-type cells would reduce their relatively low endogenous level of MMP-9 expression. PCR and Western blotting results confirmed that cells infected with the heparanase shRNA have dramatically reduced expression of heparanase as compared with cells infected with control shRNA (Fig. 2A). This reduction in heparanase expression correlated with a decrease in MMP-9 protein and activity levels (Fig. 2B) and message level as determined by reverse transcription-PCR (not shown). These results support the findings in Fig. 1 and the conclusion that heparanase regulates expression of MMP-9 in these myeloma cells.
FIGURE 2.
Down-regulation of heparanase reduces MMP-9 expression levels. A, wild-type CAG myeloma cells were infected with lentiviral vectors coding for control or heparanase knockdown shRNAs. Analysis by both reverse transcription (RT)-PCR and Western blotting from extracts of stably infected cells demonstrates effective knockdown of heparanase (HPSE) expression. B, densitometric quantification of Western blots and gelatin zymograms of MMP-9 levels in conditioned medium of control or heparanase knockdown cells indicates that MMP-9 protein and activity levels are reduced when heparanase is knocked down as compared with controls. Shown are results from single, representative gels. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
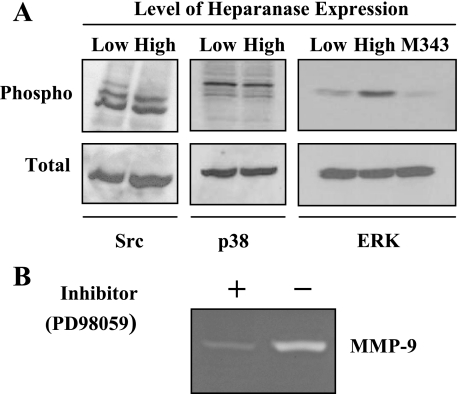
Elevation of MMP-9 Is Regulated by ERK Phosphorylation—To identify the intracellular signaling pathway underlying MMP-9 up-regulation in heparanase-transfected cells, we examined the activation status of several signaling mediators known to be involved in regulating MMP-9 expression including Src, p38 MAPK, and ERK1/2 (34, 35). Lysates from cells expressing low or high levels of heparanase were subjected to immunoblotting with antibodies directed against phosphorylated Src, p38, and ERK. Levels of phospho-Src and phospho-p38 were not affected by an elevation in heparanase expression (Fig. 3A). In contrast, ERK activation was significantly enhanced in the heparanase high cells as compared with the heparanase low cells. Interestingly levels of ERK phosphorylation were not elevated in cells expressing the mutated form of heparanase that lacks enzymatic activity (M343), suggesting that the elevation in ERK signaling is dependent on heparanase-mediated degradation of heparan sulfate chains. We next examined the involvement of these signaling molecules in MMP-9 expression by using inhibitors that block the activation of ERK, Src, and p38. By zymogram analysis of conditioned medium from heparanase high cells, we noted a significant inhibition in the levels of MMP-9 in cells treated with the MAPK/ERK inhibitor (PD98059) (Fig. 3B) but not by a Src inhibitor (PP2) or by a p38 MAPK inhibitor (SB203580).3 This indicates that activation of the ERK signaling is crucial for enhancement of MMP-9 expression.
FIGURE 3.
Expression of enzymatically active heparanase enhances ERK phosphorylation and up-regulation of MMP-9. A, cell lysates from CAG cells expressing low or high levels of heparanase were subjected to immunoblotting with antibodies against the phosphorylated forms of Src, p38, or ERK. Cell lysates from cells expressing the mutated, enzymatically inactive form of heparanase (M343) were also probed for ERK. Blots were subsequently stripped and probed for total Src, p38, or ERK. B, CAG cells expressing high levels of heparanase were treated with MAPK/ERK pathway inhibitor PD98059 (50 μm), and MMP-9 activity in the serum-free conditioned medium was assessed by gelatin zymography.
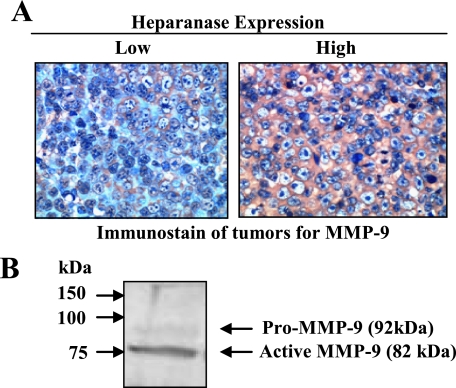
Heparanase Enhances MMP-9 Expression in Vivo—Previously we demonstrated that elevation of heparanase expression in CAG myeloma cells enhances their growth and metastasis in vivo as compared with control cells (19). Because MMP-9 plays an important role in tumor growth, angiogenesis, and metastasis, we investigated whether the heparanase-mediated up-regulation of MMP-9 expression that we saw in vitro was also present within the tumor microenvironment when these cells were injected into severe combined immunodeficient mice. Immunostaining of tumors formed from heparanase high CAG cells revealed that they have high levels of MMP-9 (Fig. 4A). In contrast, tumors formed from heparanase low CAG cells contained very low levels of the enzyme. Importantly Western blotting of tumor lysates revealed that most of the MMP-9 present is in the enzymatically active 82-kDa form (Fig. 4B). This dramatic increase in active MMP-9 correlates with the aggressive phenotype seen in these tumors formed from cells expressing high levels of heparanase (19).
FIGURE 4.
Heparanase induces expression of MMP-9 that becomes activated in vivo. A, CAG myeloma cells expressing low or high levels of heparanase were injected subcutaneously into severe combined immunodeficient mice as described previously (19). After tumors formed, they were removed and immunostained for MMP-9 (original magnification, ×200). B, Western blot of an extract from a tumor formed by heparanase high cells demonstrates that MMP-9 is present predominantly in its enzymatically active (82-kDa) form.
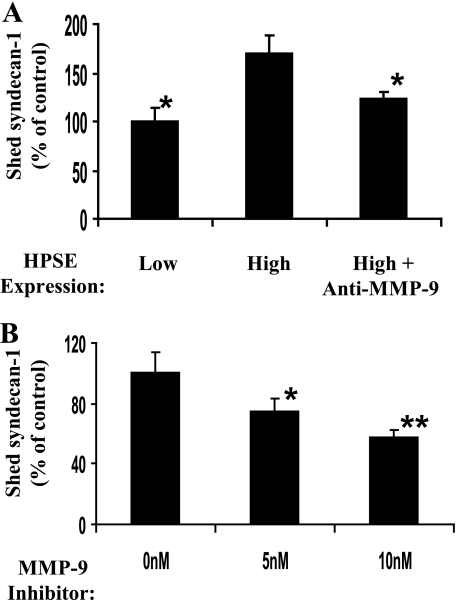
MMP-9 Mediates Enhanced Shedding of Syndecan-1 in Heparanase-expressing Cells—To determine whether MMP-9 mediates the shedding of syndecan-1, we measured the accumulation of the syndecan-1 ectodomain in cell culture media in the presence or absence of 6-6B, an antibody that blocks activation of MMP-9 (36). As expected based on our previous work (21), analysis of conditioned media 48 h after plating cells revealed that the level of shed syndecan-1 is significantly higher in heparanase high as compared with heparanase low cells (Fig. 5A). Antibody 6-6B significantly blocked shedding of syndecan-1 by the heparanase high cells indicating that elevation of MMP-9 is the mechanism by which heparanase expression enhances syndecan-1 shedding. Addition of the antibody to heparanase low-expressing cells had no significant effect on levels of syndecan-1 shedding,3 indicating that MMP-9 is not responsible for constitutive low level shedding that occurs when heparanase expression is low. As another confirmation of the role of MMP-9 in syndecan-1 shedding, MMP-9 inhibitor 1, a selective inhibitor of MMP-9, was introduced into cells growing in vitro. This also significantly and in a dose-dependent manner inhibited the shedding of syndecan-1 by cells expressing high levels of heparanase (Fig. 5B).
FIGURE 5.
MMP-9 mediates enhanced syndecan-1 shedding by heparanase high cells. A, CAG cells expressing low or high levels of heparanase were plated at equal density, and 0.5 μg/ml MMP-9 function blocking antibody 6-6B was added. After 48 h, conditioned media were harvested, and the level of shed syndecan-1 was determined by enzyme-linked immunoadsorbent assay (values represents means of triplicate determination ±S.D.). *, p < 0.001 versus heparanase (HPSE) high cells. B, cells expressing high levels of heparanase were grown in the presence of varying levels of MMP-9 inhibitor 1, and syndecan-1 shedding was measured by enzyme-linked immunoadsorbent assay. Values represent means ± S.D. of triplicate determinations. *, p < 0.05 versus 0 nm; **, p < 0.01 versus 0 nm.
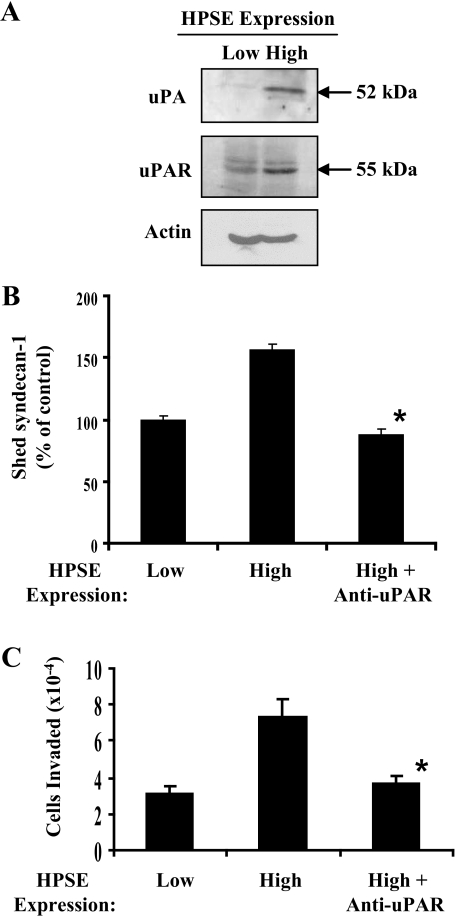
Heparanase Up-regulates the Expression of uPA and uPAR—It has been demonstrated that pro-MMP-9 is activated by a protease cascade initiated by activated uPA (37). Importantly this functional coupling between the two proteases is required for intravasation of tumor cells into the vasculature, a rate-limiting step for metastasis of cancer cells (38). Thus, we sought to determine whether the up-regulation of MMP-9 by heparanase is coupled with the up-regulation of uPA/uPAR. CAG cells expressing low or high levels of heparanase were incubated in serum-free medium for 24 h, and the conditioned media were examined for levels of uPA. Although uPA was absent from the medium of cells expressing low levels of heparanase, it was clearly present in significant amounts in medium from cells expressing high levels of heparanase (Fig. 6A). In addition, Western blots of cell extracts showed that uPAR is also up-regulated. Because uPA/uPAR are known to be involved in proteolysis and cell invasion (39), we explored their effects on syndecan-1 shedding and myeloma cell invasion. Addition to heparanase high cells of an antibody that blocks the activation of uPA significantly inhibited syndecan-1 accumulation in the culture medium (Fig. 6B). This implies that uPA participates in regulating syndecan-1 shedding. The blocking antibody had no significant effect on the levels of syndecan-1 shedding by the heparanase low cells.3 To further test uPA/uPAR function, cells were plated on the surface of matrigel-coated chambers, and the invasion of tumor cells was quantified. The number of invasive cells was almost 3 times higher when cells expressed high levels of heparanase as compared with those expressing low levels of the enzyme (Fig. 6C). Moreover the antibody that blocks uPA activation inhibited the aggressive invasive behavior of the heparanase high cells indicating that activation of uPA is important for enhancing the invasive phenotype of these cells.
FIGURE 6.
Enhanced expression of heparanase up-regulates uPA and uPAR. A, immunoblotting of serum-free conditioned media with an antibody to uPA or cell lysates with an antibody to uPAR or actin reveals up-regulation of uPA/uPAR by cells expressing high levels of heparanase. B, heparanase-mediated up-regulation of syndecan-1 shedding is inhibited by an antibody that blocks activation of uPA. Cells were plated at equal density, and 5 μg/ml antibody was added to cells expressing high levels of heparanase. After 48 h conditioned media was harvested, and the level of syndecan-1 was determined by enzyme-linked immunoadsorbent assay (values represents means of triplicate determination ±S.D.). *, p < 0.001 versus heparanase (HPSE) high cells. C, myeloma invasion is enhanced by expression of heparanase and is blocked by the antibody that inhibits uPA activation. An equal number of CAG cells were seeded on invasion chambers coated with matrigel, and cells were allowed to migrate in the presence or absence of antibody. Cell invasion data represent the mean ± S.D. of three independent experiments. *, p < 0.01 versus heparanase high cells.
DISCUSSION
The present work revealed that heparanase up-regulates the expression of two proteases known to drive an aggressive tumor phenotype. Transfection of heparanase into myeloma cells or addition of recombinant heparanase to wild-type myeloma cells enhanced their expression of pro-MMP-9 in vitro, and knockdown of heparanase expression in wild-type myeloma cells reduced levels of MMP-9 expression. When injected into mice, cells expressing high levels of heparanase retained their high level of MMP-9 expression, and the protease became activated. Up-regulation of heparanase enhanced ERK phosphorylation, and inhibition of ERK phosphorylation by addition of the MAPK/ERK pathway inhibitor PD98059 blocked MMP-9 expression. Moreover we also found that uPA/uPAR, which is known to be functionally coupled to MMP-9, is also up-regulated in response to an increase in heparanase expression. uPA/uPAR participated together with MMP-9 to enhance syndecan-1 shedding and also elevated tumor cell invasion. We also provided evidence that the effects of heparanase on ERK activation and protease expression are dependent on the presence of enzymatically active heparanase. Thus, these biological effects of heparanase occur downstream of heparan sulfate degradation. This is consistent with our previous finding that enzymatically active heparanase is required to enhance shedding of syndecan-1 (21). Together these findings reveal a novel biological pathway in which expression of heparanase increases ERK phosphorylation leading to up-regulation of proteases that act to enhance syndecan-1 shedding. In addition, these proteases when present in the tumor microenvironment are known to be important in promoting tumor growth, angiogenesis, and metastasis.
To our knowledge this is the first report linking heparanase expression with enhanced ERK signaling. Activation of the ERK pathway plays a major role in regulating cell growth, proliferation, differentiation, and angiogenesis and provides a protective effect against apoptosis (40, 41). It has been shown that ERK regulates these processes through modulation of strength or duration of ERK activation (40). Moreover a subtle difference in ERK phosphorylation can result in different biological outcomes (42). Exposure of some melanoma cells to exogenous heparanase enhances fibroblast growth factor-2 binding to cells with a resulting up-regulation of ERK phosphorylation (43). It is possible that by enhancing expression of heparanase in our myeloma cells signaling via growth factors such as hepatocyte growth factor is stimulated. Hepatocyte growth factor is a likely candidate because myeloma cells express hepatocyte growth factor, and hepatocyte growth factor is known to bind to syndecan-1 heparan sulfate and stimulate the ERK signaling pathway via the met receptor (44, 45). ERK signaling appears to be critical for growth of myeloma tumors because inhibitors of the MAPK pathway can inhibit myeloma cell growth and osteoclast differentiation (46, 47). It is interesting that we found that the activation of ERK requires the enzyme activity of heparanase. This suggests that stimulation of signaling occurs as the result of the clipping of heparan sulfate chains by heparanase. We know this clipping occurs in the myeloma cells expressing high levels of heparanase because the syndecan-1 has shorter heparan sulfate chains than those from cells expressing low levels of heparanase (19). Such clipping could “activate” the syndecan-1 by exposing cryptic epitopes within the heparan sulfate chains or the ectodomain core protein to facilitate interactions with ligands (48).
MMP-9 and uPA/uPAR are potent promoters of tumor growth and angiogenesis (39, 49–51) and can work in concert to enhance tumor cell intravasation, a rate-limiting step for metastatic diffusion of cancer cells (38). In myeloma, there is evidence that MMP-9 and uPA/uPAR contribute to disease severity. High MMP-9 levels in myeloma are associated with disease recurrence and poor patient survival (52, 53), and expression of uPA/uPAR is an independent factor predicting poor prognosis (54, 55). Evidence suggests that MMP-9 and uPA/uPAR help promote the widespread dissemination of myeloma, a hallmark of this cancer. For example, MMP-9 produced by myeloma cells promotes their invasion across basement membranes in vitro (56, 57), and there is evidence to suggest that inhibition of MMPs including MMP-9 may have antimyeloma effects (58). Inhibition of uPA inhibits invasion of myeloma cells (59), and uPA and MMP-9 have been shown to mediate invasion of the bone marrow extravascular compartment once cells have exited the marrow endothelium (60).
Enhanced MMP-9 and uPA/uPAR expression by myeloma cells may also facilitate the osteolytic phenotype that is responsible for much of the morbidity of this cancer. MMP-9 participates in the recruitment of osteoclasts to sites of bone resorption (61), and high levels of MMP-9 correlate well with bone turnover rate and are a useful prognostic index of bone disease (52, 53). High levels of soluble uPAR are associated with poor prognosis and bone disease in myeloma patients (55), and interactions between osteoclasts and myeloma cells stimulate expression of MMP-9 and uPA possibly creating a microenvironment conducive to bone degradation (62).
In addition to the multiple effects that MMP-9 and uPA/uPAR have in myeloma tumor progression, the present work revealed that the mechanism for the heparanase-induced shedding of syndecan-1 is via up-regulation of proteases. Shedding of syndecan-1 in myeloma is an important disease feature because the shed proteoglycan promotes tumor growth and metastasis (16, 21). Similarly like shed syndecan-1, heparanase also promotes tumor growth and metastasis, and at least part of its protumor effect may be due to its positive effect on syndecan-1 shedding. Also consistent with these findings is the fact that high levels of shed syndecan-1 and heparanase in myeloma patients are indicators of poor prognosis (15, 18). Shedding of syndecan-1 has been attributed to a number of metalloproteinase enzymes including MMP-1, MMP-7, MMP-9, MMP-14 (MT1-MMP), and MMP-16 (MT3-MMP) (25–29). A role for MMP-9 as a sheddase has been demonstrated in several cancer types where it can participate in shedding of various cell surface proteins such as E-cadherin and heparin-binding epidermal growth factor-like growth factor as well as syndecan-1 (24, 34, 35). However, to our knowledge, our data are the first to link uPA/uPAR to shedding of syndecan-1. The blocking antibody we used for these studies blocks uPA binding to uPAR and thus blocks the proteolytic activation of uPA by uPAR. Activation of uPA initiates a cascade of proteolytic events where uPA activates plasmin, plasmin activates MMP-3, and then MMP-3 activates MMP-9 (37). Thus, it is likely that in the myeloma cells uPA does not directly cleave the syndecan-1 ectodomain at the cell surface; rather it activates the cascade that results in MMP-9 activation and subsequent syndecan-1 shedding. This scenario is consistent with our finding that when both MMP-9 and uPAR blocking antibodies were added together to cells the level of inhibition of shedding of syndecan-1 was the same as when antibodies were added singly (i.e. there is no additive effect when both antibodies are present).3
Also it is interesting that addition of MMP-9- or uPA/uPAR-inhibiting antibodies does not alter the constitutive level of shedding of syndecan-1 in cells that express low levels of heparanase. This suggests that up-regulation of heparanase activates a shedding mechanism that is distinct from that mediating constitutive shedding. A number of factors (e.g. chemokines and epidermal growth factor family growth factors) have been shown to enhance or activate shedding of syndecans via action on specific intracellular signaling pathways (63). For example, epidermal growth factor-accelerated shedding of syndecans can be inhibited by the ERK inhibitor PD98059 (28). Similarly we found that this inhibitor blocks the heparanase-mediated increase in MMP-9 levels in the myeloma cells, thus linking receptor activation to the downstream shedding that occurs in these cells.
Our data support a model in which up-regulation of heparanase stimulates enhanced ERK signaling, thereby up-regulating expression of MMP-9 and uPA/uPAR, which then catalyze shedding of syndecan-1 from the cell surface. But perhaps more importantly, the increase within the tumor microenvironment of these proteases can dramatically stimulate aggressive behavior of the tumors. This effect on protease expression adds to a growing list of functions of heparanase that includes the release of tumor-promoting growth factors (64, 65), promotion of signaling via the Akt pathway (10), stimulation of expression of vascular endothelial growth factor and tissue factor, and regulation of cell adhesion (2). Based on these data and the overwhelming correlation between heparanase expression and poor prognosis in many cancers, we propose that heparanase is a master regulator of the aggressive tumor phenotype, and the onset of its expression marks a key defining event in tumor progression.
Acknowledgments
We thank Dr. Hua-Quan Miao (Imclone Systems, Inc., New York, NY) for providing antibody to heparanase, Dr. Israel Vlodavsky (The Bruce Rappaport Faculty of Medicine, Technion, Haifa, Israel) for providing recombinant heparanase, and Dr. Didier Trono (University of Geneva, Geneva, Switzerland) for providing viral vectors for expression of shRNA.
This work was supported, in whole or in part, by National Institutes of Health Grants CA103054, CA055819, and CA013148 (to R. D. S.). This work was also supported by a VISN7 Research Career Development Award from the Department of Veterans Affairs (to R. D. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; MMP, matrix metalloproteinase; shRNA, small hairpin RNA; uPA, urokinase-type plasminogen activator; uPAR, urokinase-type plasminogen activator receptor.
A. Purushothaman and R. Sanderson, unpublished observation.
References
- 1.Vlodavsky, I., Goldshmidt, O., Zcharia, E., Atzmon, R., Rangini-Guatta, Z., Elkin, M., Peretz, T., and Friedmann, Y. (2002) Semin. Cancer Biol. 12 121-129 [DOI] [PubMed] [Google Scholar]
- 2.Ilan, N., Elkin, M., and Vlodavsky, I. (2006) Int. J. Biochem. Cell Biol. 38 2018-2039 [DOI] [PubMed] [Google Scholar]
- 3.Gohji, K., Hirano, H., Okamoto, M., Kitazawa, S., Toyoshima, M., Dong, J., Katsuoka, Y., and Nakajima, M. (2001) Int. J. Cancer 95 295-301 [DOI] [PubMed] [Google Scholar]
- 4.Koliopanos, A., Friess, H., Kleeff, J., Shi, X., Liao, Q., Pecker, I., Vlodavsky, I., Zimmermann, A., and Buchler, M. W. (2001) Cancer Res. 61 4655-4659 [PubMed] [Google Scholar]
- 5.Joyce, J. A., Freeman, C., Meyer-Morse, N., Parish, C. R., and Hanahan, D. (2005) Oncogene 24 4037-4051 [DOI] [PubMed] [Google Scholar]
- 6.Vlodavsky, I., Friedmann, Y., Elkin, M., Aingorn, H., Atzmon, R., Ishai-Michaeli, R., Bitan, M., Pappo, O., Peretz, T., Michal, I., Spector, L., and Pecker, I. (1999) Nat. Med. 5 793-802 [DOI] [PubMed] [Google Scholar]
- 7.Cohen, I., Pappo, O., Elkin, M., San, T., Bar-Shavit, R., Hazan, R., Peretz, T., Vlodavsky, I., and Abramovitch, R. (2005) Int. J. Cancer 118 1609-1617 [DOI] [PubMed] [Google Scholar]
- 8.Edovitsky, E., Elkin, M., Zcharia, E., Peretz, T., and Vlodavsky, I. (2004) J. Natl. Cancer Inst. 96 1219-1230 [DOI] [PubMed] [Google Scholar]
- 9.Uno, F., Fujiwara, T., Takata, Y., Ohtani, S., Katsuda, K., Takaoka, M., Ohkawa, T., Naomoto, Y., Nakajima, M., and Tanaka, N. (2001) Cancer Res. 61 7855-7860 [PubMed] [Google Scholar]
- 10.Gingis-Velitski, S., Zetser, A., Flugelman, M. Y., Vlodavsky, I., and Ilan, N. (2004) J. Biol. Chem. 279 23536-23541 [DOI] [PubMed] [Google Scholar]
- 11.Zetser, A., Bashenko, Y., Edovitsky, E., Levy-Adam, F., Vlodavsky, I., and Ilan, N. (2006) Cancer Res. 66 1455-1463 [DOI] [PubMed] [Google Scholar]
- 12.Barlogie, B., Shaughnessy, J., Epstein, J., Sanderson, R., Anaissie, E., Walker, R., and Tricot, G. (2006) in Williams Hematology (Lichtman, M. A., Beutler, E., Kipps, T. J., Seligsohn, U., Kaushansky, K., and Prchal, J. T., eds) 7th Ed., pp. 1501-1534, McGraw-Hill, New York
- 13.Ridley, R. C., Xiao, H. Q., Hata, H., Woodliff, J., Epstein, J., and Sanderson, R. D. (1993) Blood 81 767-774 [PubMed] [Google Scholar]
- 14.Dhodapkar, M. V., Kelly, T., Theus, A., Athota, A. B., Barlogie, B., and Sanderson, R. D. (1997) Br. J. Haematol. 99 368-371 [DOI] [PubMed] [Google Scholar]
- 15.Seidel, C., Sundan, A., Hjorth, M., Turesson, I., Dahl, I. M., Abildgaard, N., Waage, A., and Borset, M. (2000) Blood 95 388-392 [PubMed] [Google Scholar]
- 16.Yang, Y., Yaccoby, S., Liu, W., Langford, J. K., Pumphrey, C. Y., Theus, A., Epstein, J., and Sanderson, R. D. (2002) Blood 100 610-617 [DOI] [PubMed] [Google Scholar]
- 17.Kelly, T., Miao, H. Q., Yang, Y., Navarro, E., Kussie, P., Huang, Y., MacLeod, V., Casciano, J., Joseph, L., Zhan, F., Zangari, M., Barlogie, B., Shaughnessy, J., and Sanderson, R. D. (2003) Cancer Res. 63 8749-8756 [PubMed] [Google Scholar]
- 18.Mahtouk, K., Hose, D., Raynaud, P., Hundemer, M., Jourdan, M., Jourdan, E., Pantesco, V., Baudard, M., De Vos, J., Larroque, M., Moehler, T., Rossi, J. F., Reme, T., Goldschmidt, H., and Klein, B. (2007) Blood 109 4914-4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, Y., Macleod, V., Bendre, M., Huang, Y., Theus, A. M., Miao, H. Q., Kussie, P., Yaccoby, S., Epstein, J., Suva, L. J., Kelly, T., and Sanderson, R. D. (2005) Blood 105 1303-1309 [DOI] [PubMed] [Google Scholar]
- 20.Yang, Y., MacLeod, V., Dai, Y., Khotskaya-Sample, Y., Shriver, Z., Venkataraman, G., Sasisekharan, R., Naggi, A., Torri, G., Casu, B., Vlodavsky, I., Suva, L. J., Epstein, J., Yaccoby, S., Shaughnessy, J. D., Jr., Barlogie, B., and Sanderson, R. D. (2007) Blood 110 2041-2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang, Y., Macleod, V., Miao, H. Q., Theus, A., Zhan, F., Shaughnessy, J. D., Jr., Sawyer, J., Li, J. P., Zcharia, E., Vlodavsky, I., and Sanderson, R. D. (2007) J. Biol. Chem. 282 13326-13333 [DOI] [PubMed] [Google Scholar]
- 22.Borset, M., Hjertner, O., Yaccoby, S., Epstein, J., and Sanderson, R. D. (2000) Blood 96 2528-2536 [PubMed] [Google Scholar]
- 23.Nardella, C., Lahm, A., Pallaoro, M., Brunetti, M., Vannini, A., and Steinkuhler, C. (2004) Biochemistry 43 1862-1873 [DOI] [PubMed] [Google Scholar]
- 24.Miao, H. Q., Navarro, E., Patel, S., Sargent, D., Koo, H., Wan, H., Plata, A., Zhou, Q., Ludwig, D., Bohlen, P., and Kussie, P. (2002) Protein Expr. Purif. 26 425-431 [DOI] [PubMed] [Google Scholar]
- 25.Li, Q., Park, P. W., Wilson, C. L., and Parks, W. C. (2002) Cell 111 635-646 [DOI] [PubMed] [Google Scholar]
- 26.Brule, S., Charnaux, N., Sutton, A., Ledoux, D., Chaigneau, T., Saffar, L., and Gattegno, L. (2006) Glycobiology 16 488-501 [DOI] [PubMed] [Google Scholar]
- 27.Endo, K., Takino, T., Miyamori, H., Kinsen, H., Yoshizaki, T., Furukawa, M., and Sato, H. (2003) J. Biol. Chem. 278 40764-40770 [DOI] [PubMed] [Google Scholar]
- 28.Fitzgerald, M. L., Wang, Z., Park, P. W., Murphy, G., and Bernfield, M. (2000) J. Cell Biol. 148 811-824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding, K., Lopez-Burks, M., Sanchez-Duran, J. A., Korc, M., and Lander, A. D. (2005) J. Cell Biol. 171 729-738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bendre, M. S., Gaddy-Kurten, D., Mon-Foote, T., Akel, N. S., Skinner, R. A., Nicholas, R. W., and Suva, L. J. (2002) Cancer Res. 62 5571-5579 [PubMed] [Google Scholar]
- 31.Hulett, M. D., Hornby, J. R., Ohms, S. J., Zuegg, J., Freeman, C., Gready, J. E., and Parish, C. R. (2000) Biochemistry 39 15659-15667 [DOI] [PubMed] [Google Scholar]
- 32.Gingis-Velitski, S., Zetser, A., Kaplan, V., Ben-Zaken, O., Cohen, E., Levy-Adam, F., Bashenko, Y., Flugelman, M. Y., Vlodavsky, I., and Ilan, N. (2004) J. Biol. Chem. 279 44084-44092 [DOI] [PubMed] [Google Scholar]
- 33.Vreys, V., Delande, N., Zhang, Z., Coomans, C., Roebroek, A., Durr, J., and David, G. (2005) J. Biol. Chem. 280 33141-33148 [DOI] [PubMed] [Google Scholar]
- 34.Yao, J., Xiong, S., Klos, K., Nguyen, N., Grijalva, R., Li, P., and Yu, D. (2001) Oncogene 20 8066-8074 [DOI] [PubMed] [Google Scholar]
- 35.Cortes-Reynosa, P., Robledo, T., Macias-Silva, M., Wu, S. V., and Salazar, E. P. (2008) Matrix Biol. 27 220-231 [DOI] [PubMed] [Google Scholar]
- 36.Ramos-DeSimone, N., Moll, U. M., Quigley, J. P., and French, D. L. (1993) Hybridoma 12 349-363 [DOI] [PubMed] [Google Scholar]
- 37.Ramos-DeSimone, N., Hahn-Dantona, E., Sipley, J., Nagase, H., French, D. L., and Quigley, J. P. (1999) J. Biol. Chem. 274 13066-13076 [DOI] [PubMed] [Google Scholar]
- 38.Kim, J., Yu, W., Kovalski, K., and Ossowski, L. (1998) Cell 94 353-362 [DOI] [PubMed] [Google Scholar]
- 39.Dass, K., Ahmad, A., Azmi, A. S., Sarkar, S. H., and Sarkar, F. H. (2008) Cancer Treat. Rev. 34 122-136 [DOI] [PubMed] [Google Scholar]
- 40.Marshall, C. J. (1995) Cell 80 179-185 [DOI] [PubMed] [Google Scholar]
- 41.Schaeffer, H. J., and Weber, M. J. (1999) Mol. Cell. Biol. 19 2435-2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hulit, J., Suyama, K., Chung, S., Keren, R., Agiostratidou, G., Shan, W., Dong, X., Williams, T. M., Lisanti, M. P., Knudsen, K., and Hazan, R. B. (2007) Cancer Res. 67 3106-3116 [DOI] [PubMed] [Google Scholar]
- 43.Reiland, J., Kempf, D., Roy, M., Denkins, Y., and Marchetti, D. (2006) Neoplasia 8 596-606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derksen, P. W., Keehnen, R. M., Evers, L. M., van Oers, M. H., Spaargaren, M., and Pals, S. T. (2002) Blood 99 1405-1410 [DOI] [PubMed] [Google Scholar]
- 45.Holt, R. U., Fagerli, U. M., Baykov, V., Ro, T. B., Hov, H., Waage, A., Sundan, A., and Borset, M. (2008) Haematologica 93 619-622 [DOI] [PubMed] [Google Scholar]
- 46.Hideshima, T., Akiyama, M., Hayashi, T., Richardson, P., Schlossman, R., Chauhan, D., and Anderson, K. C. (2003) Blood 101 703-705 [DOI] [PubMed] [Google Scholar]
- 47.Breitkreutz, I., Raab, M. S., Vallet, S., Hideshima, T., Raje, N., Chauhan, D., Munshi, N. C., Richardson, P. G., and Anderson, K. C. (2007) Br. J. Haematol. 139 55-63 [DOI] [PubMed] [Google Scholar]
- 48.Ma, P., Beck, S. L., Raab, R. W., McKown, R. L., Coffman, G. L., Utani, A., Chirico, W. J., Rapraeger, A. C., and Laurie, G. W. (2006) J. Cell Biol. 174 1097-1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duffy, M. J. (2004) Curr. Pharm. Des. 10 39-49 [DOI] [PubMed] [Google Scholar]
- 50.Kunigal, S., Lakka, S. S., Gondi, C. S., Estes, N., and Rao, J. S. (2007) Int. J. Cancer 121 2307-2316 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Rao, J. S., Gondi, C., Chetty, C., Chittivelu, S., Joseph, P. A., and Lakka, S. S. (2005) Mol. Cancer Ther. 4 1399-1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sfiridaki, A., Miyakis, S., Tsirakis, G., Alegakis, A., Passam, A. M., Kandidaki, E., Margioris, A. N., and Alexandrakis, M. G. (2005) Clin. Chem. Lab. Med. 43 934-938 [DOI] [PubMed] [Google Scholar]
- 53.Van Valckenborgh, E., Bakkus, M., Munaut, C., Noel, A., St Pierre, Y., Asosingh, K., Van Riet, I., Van Camp, B., and Vanderkerken, K. (2002) Int. J. Cancer 101 512-518 [DOI] [PubMed] [Google Scholar]
- 54.Hjertner, O., Qvigstad, G., Hjorth-Hansen, H., Seidel, C., Woodliff, J., Epstein, J., Waage, A., Sundan, A., and Borset, M. (2000) Br. J. Haematol. 109 815-822 [DOI] [PubMed] [Google Scholar]
- 55.Rigolin, G. M., Tieghi, A., Ciccone, M., Bragotti, L. Z., Cavazzini, F., Della Porta, M., Castagnari, B., Carroccia, R., Guerra, G., Cuneo, A., and Castoldi, G. (2003) Br. J. Haematol. 120 953-959 [DOI] [PubMed] [Google Scholar]
- 56.Parmo-Cabanas, M., Molina-Ortiz, I., Matias-Roman, S., Garcia-Bernal, D., Carvajal-Vergara, X., Valle, I., Pandiella, A., Arroyo, A. G., and Teixido, J. (2006) J. Pathol. 208 108-118 [DOI] [PubMed] [Google Scholar]
- 57.Vande Broek, I., Asosingh, K., Allegaert, V., Leleu, X., Facon, T., Vanderkerken, K., Van Camp, B., and Van Riet, I. (2004) Leukemia 18 976-982 [DOI] [PubMed] [Google Scholar]
- 58.Van Valckenborgh, E., Croucher, P. I., De Raeve, H., Carron, C., De Leenheer, E., Blacher, S., Devy, L., Noel, A., De Bruyne, E., Asosingh, K., Van Riet, I., Van Camp, B., and Vanderkerken, K. (2004) Am. J. Pathol. 165 869-878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hecht, M., Heider, U., Kaiser, M., von Metzler, I., Sterz, J., and Sezer, O. (2007) Br. J. Haematol. 138 446-458 [DOI] [PubMed] [Google Scholar]
- 60.Menu, E., Asosingh, K., Van Riet, I., Croucher, P., Van Camp, B., and Vanderkerken, K. (2004) Blood Cells Mol. Dis. 33 111-119 [DOI] [PubMed] [Google Scholar]
- 61.Engsig, M. T., Chen, Q. J., Vu, T. H., Pedersen, A. C., Therkidsen, B., Lund, L. R., Henriksen, K., Lenhard, T., Foged, N. T., Werb, Z., and Delaisse, J. M. (2000) J. Cell Biol. 151 879-889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hecht, M., von Metzler, I., Sack, K., Kaiser, M., and Sezer, O. (2008) Exp. Cell Res. 314 1082-1093 [DOI] [PubMed] [Google Scholar]
- 63.Bartlett, A. H., Hayashida, K., and Park, P. W. (2007) Mol. Cells 24 153-166 [PubMed] [Google Scholar]
- 64.Elkin, M., Ilan, N., Ishai-Michaeli, R., Friedmann, Y., Papo, O., Pecker, I., and Vlodavsky, I. (2001) FASEB J. 15 1661-1663 [DOI] [PubMed] [Google Scholar]
- 65.Vlodavsky, I., Miao, H. Q., Medalion, B., Danagher, P., and Ron, D. (1996) Cancer Metastasis Rev. 15 177-186 [DOI] [PubMed] [Google Scholar]