Abstract
Protein misfolding is monitored by a variety of cellular “quality control” systems. Endoplasmic reticulum (ER) quality control handles misfolded secretory and membrane proteins and is well characterized. However, less is known about the quality control of misfolded cytosolic proteins (CytoQC). To study CytoQC, we have employed a genetic system in Saccharomyces cerevisiae using a transplantable degron, CL1 (1). Attachment of CL1 to the cytosolic protein Ura3p destabilizes Ura3p, targeting it for rapid proteasomal degradation. We have performed a comprehensive analysis of Ura3p-CL1 degradation requirements. As shown previously, we observe that the ER-localized ubiquitin E2 (Ubc6p, Ubc7p, and Cue1p) and E3 (Doa10p) machinery involved in ER-associated degradation (ERAD) are also responsible for the degradation of the cytosolic substrate Ura3p-CL1. Importantly, we find that the cytosol/ER membrane-localized chaperones Ydj1p and Ssa1p, known to be necessary for the ERAD of membrane proteins with misfolded cytosolic domains, are also required for the ubiquitination and degradation of Ura3p-CL1. In addition, we show a role for the Cdc48p-Npl4p-Ufd1p complex in the degradation of Ura3p-CL1. When ubiquitination is blocked, a portion of Ura3p-CL1 is ER membrane-localized. Furthermore, access to the cytosolic face of the ER is required for the degradation of CL1 degron-containing proteins. The ER is distributed throughout the cytosol, and our data, together with previous studies, suggest that the cytosolic face of the ER membrane serves as a “platform” for the degradation of Ura3p-CL1, which may also be the case for other CytoQC substrates.
Mutation, errors in transcription or translation, and cellular stress can cause alterations in amino acids that may prevent proteins from attaining their properly folded, native conformations. Protein “quality control” is an essential process monitoring protein folding, ultimately targeting misfolded proteins for degradation via the ubiquitin-proteasome system. The importance of protein quality control is best exemplified by the numerous human diseases that can result from protein misfolding due to mutational or physiological causes and include cystic fibrosis, Parkinson disease, and α1-antitrypsin deficiency (2, 3).
Distinct protein quality control systems appear to exist in various cellular compartments, including the nucleus, mitochondria, and endoplasmic reticulum (ER),2 with the best-characterized system being ER quality control (4–7). Studies of ER quality control and, in particular, ER-associated degradation (ERAD) have revealed discrete chaperone and ubiquitination machinery required for the recognition and ubiquitination of different classes of misfolded secretory or membrane proteins. Much of this work has been greatly aided by the use of “model” ER quality control substrates, such as CPY* or Ste6p*, in the yeast Saccharomyces cerevisiae (8–12). For example, it has become clear that model membrane proteins with misfolded cytosolic domains (called ERAD-C substrates), such as Ste6p*, require specific cytosol/ER-localized Hsp70 (Ssa1p, Ssa2p, Ssa3p, Ssa4p) and Hsp40 (Ydj1p and Hlj1p) chaperones for recognition and targeting to E3 ubiquitin ligases (8, 13–15). The ubiquitination of ERAD-C substrates is mediated primarily through Doa10p in conjunction with the E2 ubiquitin-conjugating enzymes, Ubc6p and Ubc7p/Cue1p. Ubiquitinated substrates are removed to the cytosol via the action of the Cdc48p-Ufd1p-Npl4p AAA-ATPase complex, and Cdc48p complex co-factors target substrates to the cytosolic 26S proteasome for degradation (8, 15–20).
Another quality control system, only recently examined with the model substrates von Hippel Lindau protein (VHL) and ΔssCPY* (21, 22), is cytosolic quality control (CytoQC). CytoQC studies to date have used a candidate gene approach to identify the machinery required for the degradation of cytosolic proteins and have begun to implicate various molecular chaperones and E2 enzymes (21, 22). It remains to be seen whether distinct degradation pathways exist for different substrates and what overlap, if any, there may be with the ERAD-C pathway.
The identification of many ubiquitin-proteasome components in yeast was pioneered using the Deg1 degron of the MATα2 transcriptional repressor. Deg1 is a transplantable, 67-amino acid sequence within MATα2 that targets the repressor for ubiquitination and degradation by the proteasome (23–25). Selection for mutants that stabilized Deg1 revealed, among others, the ER-localized, E2 ubiquitin-conjugating enzymes, Ubc6p and Ubc7p, and the E3 ubiquitin ligase, Doa10p, found to also be involved in ERAD-C (26, 27). Further studies demonstrated that Deg1 has a nuclear localization signal and needs to be localized to the nucleus for efficient degradation and is in fact ubiquitinated by a specific nuclear subpopulation of Doa10p (28, 29).
Deg1 is predicted to form an amphipathic helix, and the hydrophobic residues of the helix are crucial for its instability (30). In the context of MATα2, Deg1 is masked by interactions with the MATα2 binding partner, MATa1, and MATα2 is stable. In the absence of MATa1, Deg1 is exposed, and MATα2is targeted for degradation. Similarly, hydrophobic residues or other degradation signals in a wide variety of proteins, normally buried in the context of native protein conformation, may be exposed by protein misfolding and act as degrons to target the misfolded or binding partner-deficient protein for degradation by the ubiquitin-proteasome system. In this way, the Deg1 degron may resemble a misfolded protein when it is exposed.
The hypothesis that degrons display characteristics similar to misfolded protein domains, in light of the insight gained through characterization of the degradation requirements of Deg1, has led us to make use of another less well characterized degron, the CL1 degron (1), to study CytoQC. The CL1 degron was identified in a powerful screen to identify genomic sequences that destabilized the cytosolic yeast protein, Ura3p, by targeting it for degradation via Ubc6p/Ubc7p (1). The degradation of Ura3p-CL1 has subsequently been shown to be dependent on Doa10p and the proteasome (31, 32). CL1 is a 16-amino acid sequence (ACKNWFSSLSHFVIHL) predicted to form an amphipathic helix that, like Deg1, depends on hydrophobic residues to function as a degron (31). Additionally, our sequence analysis suggests that CL1 is an out of frame region of the yeast PMD1 gene.3 Thus, studies of CL1 may also reveal insight into the fate of improper translation products.
Here, we have extended the analysis of Ura3p-CL1 by analyzing its ubiquitination status and determining its additional degradation requirements. Interestingly, we find several parallels (in addition to the involvement of Ubc6p/Ubc7p, Cue1p, and Doa10p) between the degradation requirements of Ura3p-CL1 and ERAD-C substrates, such as Ste6p*, with misfolded cytosolic domains. In particular, we find a requirement for the cytosol/ER-localized chaperones Ydj1p and Ssa1p for the ubiquitination and degradation of Ura3p-CL1. We also find a role for the Cdc48p-Npl4p-Ufd1p AAA-ATPase complex in the degradation of Ura3p-CL1. Interestingly, when ubiquitination is blocked, a portion of Ura3p-CL1 localizes to the ER membrane, and access of CL1 to the cytosolic face of the ER is required for the degradation of CL1 degron-containing proteins. Taken together, these results suggest that the cytosolic face of the ER serves as a “platform” for the degradation of Ura3p-CL1, and, as the ER forms a reticular network throughout the cytosol, this may also be the case for many CytoQC substrates.
MATERIALS AND METHODS
Yeast Strains, Media, and Growth Conditions—The S. cerevisiae strains used in this study are listed in Table 1. Solid and liquid drop-out media were prepared as described previously (33). 5-Fluoroorotic acid (5-FOA) was present in solid media at a final concentration of 0.045%. Yeast strains and cultures were grown at 30 °C, except for temperature-sensitive strains, which were grown at room temperature (25 °C) or 37 °C, as indicated. Yeast transformations were performed by the lithium acetate method (34).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference/Source |
|---|---|---|
| SM3417 (RSY155) | MATaleu2-3,112 ura3-52 ade2-1 pep4-3 sec63-1 | R. Scheckman |
| SM3419 (RSY156) | MATaleu2-3,112 ura3-52 ade2-1 pep4-3 SEC63 | R. Scheckman |
| SM3865 (MS10) | MATa ura3-52, leu2-3,112 ade2-101 KAR2 | 45 |
| SM3867 (MS193) | MATa ura3-52, leu2-3,112 ade2-101 kar2-133 | 45 |
| SM4177 (JN516) | MATahis3-11,15 leu2-3,112 ura3-52 trp1-Δ1 lys2 ssa1-45 ssa2-1::LEU2 ssa3-1::TRP1 ssa4-2::LYS2 | 46 |
| SM4247 (JB67) | MATahis3-11,15 leu2-3,112 ura3-52 trp1-Δ1 lys2 SSA1 ssa2-1::LEU2 ssa3-1::TRP1 ssa4-2::LYS2 | 46 |
| SM4552 (XII-22 p82a) | MATaade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 hsc82::LEU2 hsp82::LEU2 [CEN TRP1 HSP82] | 74 |
| SM4553 (XII-23 G313) | MATaade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 hsc82::LEU2 hsp82::LEU2 [CEN TRP1 hsp82-G313N] | 74 |
| SM4779 (BWGI-7A) | MATahis4-519 ura3-52 ade1-100 leu2-3,112 UFD1 | 20 |
| SM4780 (PM373) | MATahis4-519 ura3-52 ade1-100 leu2-3,112 ufd1-1 | 20 |
| SM4784 (PSY580) | MATaura3-52 leu2Δ1 trp1Δ63 NPL4 | 20 |
| SM4785 (PSY2340) | MATaura3-52 leu2Δ1 trp1Δ63 npl4-1 | 20 |
| SM4333 (WCG4a) | MATaura3-52 leu2-3,112 his3-11,15 PRE1 PRE2 | 75 |
| SM4334 (WCG4-11/21a) | MATaura3-52 leu2-3,112 his3-11,15 pre1-1 pre2-1 | 75 |
| SM4460 (BY4741) | MATahis3 leu2 met15 ura3 | Open Biosystems |
| SM4783 | MATaura3-52 leu2-3,112 ade2-1 trp1-1 his3 cdc48-3 | T. Rapoport |
| SM4820 | MATahis3 leu2 met15 ura3 doa10::KanMX | Open Biosystems |
| SM4821 | MATahis3 leu2 met15 ura3 ubc7::KanMX | Open Biosystems |
| SM4822 | MATahis3 leu2 met15 ura3 cue1::KanMX | Open Biosystems |
| SM4823 | MATahis3 leu2 met15 ura3 hrd1::KanMX | Open Biosystems |
| SM4947 | MATα ade2 his3 leu2 ura3 trp1 can1-100 YDJ1 | T. Lithgow |
| SM4948 | MATα ade2 his3 leu2 ura3 trp1 can1-100 ydj1-2::HIS3 ydj1-151::LEU2 | T. Lithgow |
| SM5177 | MATahis3 leu2 met15 ura3 hlj1::KanMX | Open Biosystems |
| SM5124 | MATaura3-52 leu2-3,112 ade2-1 trp1-1 his3 CDC48 | T. Rapoport |
| SM5186 | MATahis3 leu2 met15 ura3 dsk2::KanMX rad23::KanMX | 8 |
| SM5362 | MATahis3 leu2 met15 ura3 ubc6::HIS3 ubc7::NatMX | This study |
| SM5377 | MATahis3 leu2 met15 ura3 ubc4::KanMX | Open Biosystems |
| SM5381 | MATahis3 leu2 met15 ura3 ubx2::KanMX | Open Biosystems |
| SM5471 (JN54) | MATahis3-11,3-15 leu2-3,2-112 ura3-52 trp1-1 lys2 SSB1 SSB2 | 76 |
| SM5472 (JN212) | MATahis3-11,3-15 leu2-3,2-112 ura3-52 trp1-1 lys2 ssb1-1::LEU2 ssb2-1::HIS3 | 76 |
| SM5473 (SEY6211) | MATaura3-52 leu2-3,112 his3-Δ200 trp1-Δ902 ade2-101 suc2-Δ9 HSP26 HSP42 | 77 |
| SM5474 | MATaura3-52 leu2-3,112 his3-Δ200 trp1-Δ902 ade2-101 suc2-Δ9 hsp26::HIS3 hsp42::LEU2 | 77 |
| SM5483 | MATahis3 leu2 met15 ura3 sti1::KanMX | Open Biosystems |
| SM5484 | MATahis3 leu2 met15 ura3 sse1::KanMX | Open Biosystems |
| SM5485 | MATahis3 leu2 met15 ura3 hsp104::KanMX | Open Biosystems |
| SM5486 | MATahis3 leu2 met15 ura3 snl1::KanMX | Open Biosystems |
| SM5489 | MATahis3 leu2 met15 ura3 ubc5::KanMX | Open Biosystems |
| SM5492 | MATahis3 leu2 met15 ura3 ubc6::HIS3 | This study |
| SM5494 | MATahis3 leu2 met15 ura3 ufd2::KanMX | Open Biosystems |
| SM5495 | MATahis3 leu2 met15 ura3 ubc6::HIS3 ubc7::NatMX doa10::KanMX | This study |
| SM5547 | MATahis3 leu2 met15 ura3 ubx1::KanMX | Open Biosystems |
| SM5548 | MATahis3 leu2 met15 ura3 ubx3::KanMX | Open Biosystems |
| SM5549 | MATahis3 leu2 met15 ura3 ubx4::KanMX | Open Biosystems |
| SM5550 | MATahis3 leu2 met15 ura3 ubx5::KanMX | Open Biosystems |
| SM5551 | MATahis3 leu2 met15 ura3 ubx6::KanMX | Open Biosystems |
| SM5552 | MATahis3 leu2 met15 ura3 ubx7::KanMX | Open Biosystems |
| SM5553 | MATahis3 leu2 met15 ura3 otu1::KanMX | Open Biosystems |
| SM5554 | MATahis3 leu2 met15 ura3 doa1::KanMX | Open Biosystems |
Strain and Plasmid Constructions—The ubc6:HIS3 mutant strain (SM5492) was constructed in several steps. First, ubc6::HIS3 was PCR-amplified with 500 bp of 5′- and 3′-flanking sequence from MHY552 (27). The PCR product was transformed into BY4742 (Open Biosystems, Huntsville, AL), and recombinants were identified, yielding a MATα ubc6::HIS3 strain (SM5347). This strain was backcrossed to SM4460 (BY4741; Open Biosystems), diploids were sporulated, and tetrads were dissected to yield a MATa ubc6::HIS3 strain (SM5492). The ubc6Δ ubc7Δ double mutant strain (SM5362) was constructed by mating a MATα ubc6::HIS3 strain (SM5347) to a MATa ubc7::NatMX strain, generated by replacing the KanMX cassette of ubc7::KanMX (Open Biosystems) by recombination with the NatMX cassette generated from digestion of pAG25 (35) with NotI. After selection for diploids, cells were sporulated and dissected to recover a MATa ubc6::HIS3 ubc7::NatMX segregant (SM5362). The ubc6::HIS3 ubc7::NatMX doa10::KanMX mutant strain (SM5495) was generated by mating a MATα ubc6::HIS3 ubc7::NatMX strain (SM5361) with a MATa doa10::KanMX (SM4820). Diploids were sporulated, and tetrads were dissected to yield a MATa ubc6::HIS3 ubc7::NatMX doa10::KanMX segregant (SM5495).
Plasmids used in this study are listed in Table 2. Relevant regions of all newly made constructs were verified by DNA sequencing. The plasmid pSM1959 (2μ LEU2 SEC63-RFP) was generated using recombinational cloning to first replace the GFP in pSM1462 (CEN URA3 SEC63-GFP) (36) with mRFP that was PCR-amplified from pRSETB-mRFP (37). The SEC63-RFP insert was subcloned into pRS425 (2μ LEU2) (38) via the NotI and HindIII sites to yield 2μ LEU2 SEC63-RFP (pSM1959).
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant markers | Reference or Source |
|---|---|---|
| pSM1959 | [2μ LEU2 SEC63-RFP] | This study |
| pSM2287 | [CEN LEU2 URA3-HA] | This study |
| pSM2288 | [CEN LEU2 URA3-HA-CL1] | This study |
| pSM2289 | [CEN LEU2 URA3-GFP] | This study |
| pSM2290 | [CEN LEU2 URA3-GFP-CL1] | This study |
| pSM2291 | [CEN HIS3 URA3-HA-CL1] | This study |
| pSM2292 | [CEN TRP1 URA3-HA-CL1] | This study |
| pSM2293 | [CEN LEU2 HIS3-HA-CL1] | This study |
| pSM2294 | [CEN LEU2 VMA12-URA3-HA] | This study |
| pSM2295 | [CEN LEU2 VMA12-URA3-HA-CL1] | This study |
| pSM2296 | [CEN LEU2 VMA12-URA3-GFP] | This study |
| pSM2297 | [CEN LEU2 VMA12-URA3-GFP-CL1] | This study |
| pSM2298 | [CEN LEU2 TOM20-URA3-HA] | This study |
| pSM2299 | [CEN LEU2 TOM20-URA3-HA-CL1] | This study |
| pSM2300 | [CEN LEU2 TOM20-URA3-GFP] | This study |
| pSM2301 | [CEN LEU2 TOM20-URA3-GFP-CL1] | This study |
| pSM2302 | [CEN LEU2 CPY-HA] | This study |
| pSM2303 | [CEN LEU2 CPY-HA-CL1] | This study |
| pSM2318 | [CEN LEU2 HIS3-HA] | This study |
The plasmids pSM2287 (CEN LEU2 URA3-HA), pSM2288 (CEN LEU2 URA3-HA-CL1), pSM2289 (CEN LEU2 URA3-GFP), and pSM2290 (CEN LEU2 URA3-GFP-CL1) were all generated in two steps. First, URA3 (including 5′- and 3′-untranslated regions) was PCR-amplified from pRS316 (38), adding flanking HindIII and SpeI sites to the 5′ and 3′ ends, respectively. Following digestion with HindIII and SpeI, the insert was cloned into the same sites in pRS315 (38) to generate a CEN LEU2 plasmid expressing URA3. HA and GFP with and without the CL1 degron were added by recombination of a PCR product encoding HA, HA-CL1, GFP, or GFP-CL1 into the region 3′ of the URA3 ORF, eliminating its stop codon, to generate CEN LEU2 URA3-HA (pSM2287), CEN LEU2 URA3-HA-CL1 (pSM2288), CEN LEU2 URA3-GFP (pSM2289), and CEN LEU2 URA3-GFP-CL1 (pSM2290).
Plasmids pSM2291 (CEN HIS3 URA3-HA-CL1) and pSM2292 (CEN TRP1 URA3-HA-CL1) were generated by subcloning URA3-HA-CL1 from pSM2288 into pRS313 and pRS314 (38), respectively, using the XhoI and NaeI sites. HIS3 was PCR-amplified from genomic DNA and recombined into pSM2287 and pSM2288 to exactly replace the URA3 ORF with HIS3 to generate pSM2293 (CEN LEU2 HIS3-HA-CL1) or pSM2318 (CEN LEU2 HIS3-HA), respectively.
The plasmids pSM2294 (CEN LEU2 VMA12-URA3-HA), pSM2295 (CEN LEU2 VMA12-URA3-HA-CL1), pSM2296 (CEN LEU2 VMA12-URA3-GFP), and pSM2297 (CEN LEU2 VMA12-URA3-GFP-CL1) were generated by PCR-amplifying the VMA12 ORF (lacking its stop codon) from p414MET25-Deg1-Vma12-URA34 and recombining into pSM2287, pSM2288, pSM2289, pSM2290, respectively, resulting in the introduction of VMA12 upstream of URA3. The plasmids pSM2298 (CEN LEU2 TOM20-URA3-HA), pSM2299 (CEN LEU2 TOM20-URA3-HA-CL1), pSM2300 (CEN LEU2 TOM20-URA3-GFP), and pSM2301 (CEN LEU2 TOM20-URA3-GFP-CL1) were generated by PCR amplifying TOM20 (lacking its stop codon) from genomic DNA and recombining into pSM2287, pSM2288, pSM2289, pSM2290, respectively, resulting in the introduction of the TOM20 ORF upstream of URA3. CPY (PRC1) was PCR-amplified from genomic DNA and recombined into pSM2287 and pSM2288 to exactly replace the URA3 ORF with CPY to generate pSM2302 (CEN LEU2 CPY-HA) and pSM2303 (CEN LEU2 CPY-HA-CL1).
Spot Growth Assay—Log phase cultures growing in selective media were diluted to an A600 of 0.1, and four serial 10-fold dilutions were made in 96-well plates to yield a dilution series in adjacent wells. 10 μl of each dilution was spotted onto the indicated media using a multichannel Pipetman, and plates were incubated for 2 days at 30 °C unless otherwise indicated.
Preparation of Cell Extracts and Immunoblotting—Cell extracts and immunoblotting were prepared as described previously (39). Briefly, 2.5 A600 units of cells were grown logarithmically in synthetic drop-out media and lysed by the addition of β-mercaptoethanol/NaOH. Proteins were precipitated in 5% trichloroacetic acid, and protein pellets were resuspended in trichloroacetic acid sample buffer (3.5% SDS, 0.5 m dithiothreitol, 80 mm Tris, 8 mm EDTA, 15% glycerol, 0.1 mg/ml bromphenol blue). To remove N-linked glycosylations from CPY, 0.2 A600 unit equivalents of protein extracts resuspended in trichloroacetic acid sample buffer were treated with 1,000 units of endoglycosidase Hf (New England Biolabs, Inc., Ipswich, MA) for 1 h at 37 °C.
Protein samples were analyzed by 12% SDS-PAGE, followed by transfer to nitrocellulose and blocking for 1 h at room temperature in 10% blocking reagent (Roche Applied Science). For immunoblotting, all antibodies were diluted in 1× TBST containing 5% blocking reagent for 1 h at room temperature unless otherwise noted. HA epitope-tagged proteins were detected using the 12CA5 mouse anti-HA monoclonal antibody (Roche Applied Science) diluted 1:10,000. GFP-tagged proteins were detected using the mouse anti-GFP monoclonal antibody (Roche Applied Science) diluted 1:2,000. Hexokinase was detected using rabbit anti-hexokinase antibodies (a generous gift of Rob Jensen, The Johns Hopkins University) diluted 1:200,000. Sec61p was detected using rabbit anti-Sec61p antibodies (a generous gift of Colin Stirling, University of Manchester) diluted 1:4,000. Ssa1p was detected using rabbit anti-Ssa1p antibodies (a generous gift of Jeff Brodsky, University of Pittsburgh) diluted 1:2,000.
Ubiquitin-conjugated proteins were detected using rabbit anti-ubiquitin antibodies. Polyclonal antibodies to ubiquitin were generated by Babco (Hazelton, PA) by immunizing rabbits with purified ubiquitin. The antibodies were affinity-purified by C. Pickart against purified ubiquitin. For immunoblotting, these antibodies were diluted 1:4000 in 1× TBST containing 5% blocking reagent and incubated overnight at 4 °C.
Sheep anti-mouse IgG-horseradish peroxidase and donkey anti-rabbit IgG-horseradish peroxidase secondary antibodies (GE Healthcare, Buckinghamshire, UK) were diluted 1:5000 in 1× TBST containing 5% blocking reagent and incubated with immunoblots for 1 h at room temperature. Proteins were detected using Lumi-Light Western blotting substrate (Roche Diagnostics), and blots were visualized using a VersaDoc quantitative digital imaging system and quantitated using Quantity One software (Bio-Rad).
Immunopurification and Ubiquitin Chain Detection—The ubiquitination status of Ura3p-HA-CL1 in wild-type and mutant strain backgrounds was assessed by immunopurifying Ura3p-HA-CL1 from cell extracts, followed by immunoblotting for ubiquitin chains. Cells were grown logarithmically in synthetic media, 25 A600 units were harvested, and protein extracts were prepared as described above. Extracts were heated at 65 °C for 10 min, followed by the addition of 1 ml of IP dilution buffer (1% Triton X-100, 150 mm NaCl, 5 mm EDTA, 50 mm Tris-HCl, pH 7.5, 2 μg/ml aprotinin, 1 mm phenylmethylsulfonyl fluoride, 10 mm N-ethylmaleimide). The lysate was cleared by centrifugation at 13,000 rpm for 1 min at 4 °C to remove insoluble material, and 50 μl of anti-HA affinity matrix (Roche Applied Science) was added to the cleared lysate and incubated overnight at 4 °C on a rotator wheel to immunopurify Ura3p-HA-CL1. The affinity matrix was pelleted and washed twice in Ub dilution buffer A (0.1% Triton X-100, 0.02% SDS, 150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 5 mm EDTA, 10 mm N-ethylmaleimide), followed by a final wash in Ub dilution buffer B (150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 5 mm EDTA, 10 mm N-ethylmaleimide). Immunopurified Ura3p-HA-CL1 was dissociated from the matrix by the addition of 50 μl of 2× Laemmli sample buffer (20% glycerol, 10% β-mercaptoethanol, 4% SDS, 0.125 m Tris-HCl, pH 6.8, 0.02% bromphenol blue) and incubation at 65 °C for 10 min. Immunopurified proteins were subject to 12% SDS-PAGE, and the entire gel (including the stacking gel) was transferred to nitrocellulose. Ubiquitin blots were boiled in distilled H2O for 20 min between two pieces of Whatman paper prior to blocking, followed by immunoblotting. SDS-PAGE and immunoblotting with anti-HA antibodies was performed first to determine the amount immunopurified from each mutant strain compared with its isogenic wild-type, and then equal amounts of immunopurified Ura3p-HA-CL1 were analyzed by SDS-PAGE and immunoblotting with anti-HA (one-tenth volume loaded) and anti-ubiquitin antibodies.
Cycloheximide Chase Analysis—Cycloheximide chase analysis was done to examine the rates of turnover of degron-containing proteins in wild-type and mutant strain backgrounds. Cells were grown logarithmically in synthetic media, and 6 A600 units were harvested, washed, and resuspended to 3 A600 units/ml in synthetic media. Cells were incubated at 30 °C for 5 min for non-temperature-sensitive strains or at 37 °C for 1 h for temperature-sensitive strains, after which cycloheximide was added to a final concentration of 100 μg/ml to inhibit further protein synthesis. For the zero time point, 500 μl of cells were immediately harvested by addition to an equal volume of 2× azide stop mix (20 mm NaN3, 0.5 mg/ml bovine serum albumin) on ice. At each additional indicated time point after the addition of cycloheximide, 500 μl of cells were harvested as above. Cells were incubated during the chase at 30 °C for non-temperature-sensitive strains or 37 °C for temperature-sensitive strains. Following each time point, cells were pelleted, the supernatant was removed, and pellets were frozen at –80 °C until preparation of cell extracts, SDS-PAGE, and immunoblotting were performed as described above. Blots were visualized using a Versadoc quantitative digital imaging system. Exposures were adjusted to approximate equal levels of signal at the zero time point, and blots were quantitated using Quantity One software (Bio-Rad). Half-lives were calculated from exponential curve fits using Excel (Microsoft). All experiments were repeated at least three times with similar results; a representative experiment is shown in each case.
Fluorescence Microscopy—Cells expressing Ura3p-GFP, Ura3p-GFP-CL1, or Sec63p-RFP were grown to mid-log phase and examined at 100× magnification on concanavalin A-coated slides using a Zeiss Axioskop microscope equipped with fluorescence and Nomarski optics (Zeiss, Thornwood, NY). For GFP, the excitation and emission filters were 470 nm (40-nm bandwidth) and 525 nm (50-nm bandwidth), respectively, and for RFP, the excitation and emission filters were 545 nm (30-nm bandwith) and 610 nm (75-nm bandwith), respectively. Images were captured with a Photometrics Cool Snap EZ CCD camera and IP Lab Spectrum software (Biovision Technologies, Inc., Exton, PA).
Fractionation and Sucrose Flotation Gradients—Fractionation of cell extracts by differential centrifugation at 13,000 × g was used to analyze the membrane association or solubility of Ura3p-HA-CL1 in wild-type and mutant strain backgrounds. The majority of ER membranes, plasma membranes, mitochondria, and protein aggregates fractionate in the pellet under these conditions, whereas soluble proteins, Golgi membranes, and endosomal membranes remain in the supernatant fraction (40).5 Briefly, 25 A600 units of cells growing logarithmically in synthetic media were harvested via centrifugation, and cell pellets were frozen at –80 °C until cell fractionation. Cells pellets were resuspended in 100 μl of fractionation buffer (50 mm HEPES, pH 7.4, 150 mm NaCl, 5 mm EDTA, 1 m DTT, supplemented with phenylmethylsulfonyl fluoride, leupeptin, and pepstatin, according to the manufacturer's instructions). Glass beads were added to the meniscus, and the resuspension was vortexed eight times for 1 min, with 1 min on ice between each agitation. The lysate was removed to a new tube, and the beads were washed with an equal volume of fractionation buffer. The combined extract was brought up to 500 μl with fractionation buffer and centrifuged twice at 300 × g for 2 min to remove unbroken cells. A 25-μl portion of the lysate was removed to a new Eppendorf tube and designated total (T). The remaining lysate (∼400 μl) was centrifuged at 13,000 × g for 20 min at 4 °C. The supernatant (∼300 μl) was transferred to a new tube and designated supernatant (S). The residual supernatant was removed by aspiration, and the pellet was washed in 200 μl of fractionation buffer and resuspended in 400 μl of fractionation buffer. This fraction is designated pellet (P).
For SDS-PAGE and immunoblot, 50 μl of the S and P fractions were reserved. The P fraction was pelleted at 13,000 × g and resuspended in 2× Laemmli sample buffer. The T and S fractions were precipitated in 10% trichloroacetic acid, and protein pellets were resuspended in trichloroacetic acid sample buffer. Equal A600 unit equivalents of each fraction were analyzed by 12% SDS-PAGE and immunoblotting.
Sucrose flotation gradients to determine if proteins are membrane-associated and thus float were done essentially as described (41). Briefly, fractions were loaded from bottom to top in a centrifuge tube (catalog number 349622; Beckman) as follows: 300 μl of 2.3 m sucrose (in fractionation buffer), 100 μl of the S or P fractions from the 13,000 × g spins (above) mixed with 300 μl of 2.3 m sucrose (in fractionation buffer; a final sucrose concentration of ∼1.8 m), 600 μl of 1.5 m sucrose (in fractionation buffer), and 500 μl of 0.25 m sucrose (in fractionation buffer). The tube was centrifuged at 100,000 × g in a Beckman SW55 rotor for 18 h at 4 °C. 150-μl aliquots were removed from the top to the bottom of the gradient and analyzed by 12% SDS-PAGE and immunoblotting.
RESULTS
The CL1 Degron Dramatically Destabilizes Ura3p in a Ubiquitin- and Proteasome-dependent Manner—To study protein degradation in the cytosol, we have utilized a genetically tractable system featuring the cytosolic protein, Ura3p (orotidine-5′-phosphate decarboxylase), required for the biosynthesis of uracil and thus for the growth of yeast cells on media lacking uracil. We have tagged Ura3p with either GFP or HA and the 16-amino acid CL1 degron (1, 31). Whereas previous studies have analyzed the degradation of a Ura3p-HA-CL1 construct (1, 31), here we have also included Ura3p-GFP-CL1 to facilitate localization studies. As expected, cells expressing Ura3p-GFP or Ura3p-HA grow well on media lacking uracil, whereas cells expressing Ura3p-GFP-CL1 or Ura3p-HA-CL1 do not grow on media lacking uracil (Fig. 1A, SC-Leu-Ura), despite showing robust growth on media containing uracil (Fig. 1A, SC-Leu). Thus, the addition of the CL1 degron strongly destabilizes Ura3p-GFP and Ura3p-HA to levels that are insufficient for growth. Additionally, cells expressing Ura3p-GFP-CL1 or Ura3p-HA-CL1 are able to grow on media containing 5-FOA, a drug that counterselects for Ura3p (Fig. 1A, SC-Leu + 5-FOA). As expected, 5-FOA is toxic to cells expressing Ura3p without the CL1 degron.
FIGURE 1.
The CL1 degron dramatically destabilizes the cytosolic protein Ura3p in a ubiquitin- and proteasome-dependent manner. A, serial dilutions of wild-type cells containing plasmids expressing Ura3p-GFP, Ura3p-GFP-CL1, Ura3p-HA, or Ura3p-HA-CL1 (pSM2289, pSM2290, pSM2287, and pSM2288, respectively) were spotted to the indicated media. B, SDS-PAGE of 0.1 A600 units of cells/lane of steady state protein extracts from the strains used in A and immunoblotting with anti-GFP or anti-HA antibodies. Hexokinase (HK) is a loading control. C, differential interference contrast (DIC) (3 and 4) and fluorescence (GFP) (1 and 2) microscopy of wild-type cells expressing Ura3p-GFP or Ura3p-GFP-CL1 (pSM2289 and pSM2290, respectively). D, cycloheximide chase, SDS-PAGE, and immunoblots of the strains in A for the indicated times. Blots were probed with anti-GFP or anti-HA antibodies. Protein extracts from 0.1 A600 units of cells were loaded for strains without the CL1 degron and from 0.3 A600 units of cells for strains with the CL1 degron. E, serial dilutions of PRE1 PRE2 wild-type or pre1-1 pre2-1 mutant cells expressing Ura3p-HA-CL1 (pSM2288) were spotted to the indicated media. Plates were incubated at 25 °C for 4 days. F, cycloheximide chase, SDS-PAGE, and immunoblots of the strains used in E were performed for the indicated times. Blots were probed with anti-HA antibodies to detect Ura3p-HA-CL1. Quantitations of the cycloheximide chases are graphed below. G, anti-HA immunopurification (IP), SDS-PAGE, and ubiquitin immunoblots (WB) in PRE1 PRE2 wild-type or pre1-1 pre2-1 mutant cells expressing either Ura3p-HA-CL1 (pSM2288; lanes 1 and 2) or Ura3p-GFP-CL1 (pSM2290; lanes 3 and 4; negative control). Blots were probed with either anti-ubiquitin (Ub) or anti-HA antibodies (one-tenth volume of sample was loaded for anti-HA immunoblot), as described under “Materials and Methods.” The asterisk indicates a cross-reactive band unrelated to the presence of Ura3p-HA-CL1. The strain used in A–D was SM4460, and the strains used in E–G were SM4333 and SM4334. Cycloheximide chases were repeated at least three times with similar results; a representative experiment is shown for each strain.
In agreement with the growth levels on solid media, the degron-containing proteins Ura3p-GFP-CL1 and Ura3p-HA-CL1 are virtually undetectable by immunoblot (Fig. 1B, top, lanes 2 and 4). This is in contrast to Ura3p-GFP and Ura3p-HA, which are easily visualized when an equal amount of steady state protein extract is analyzed (Fig. 1B, top, lanes 1 and 3). Fluorescence microscopy confirms that, although Ura3p-GFP is diffusely cytosolic and easily detectable, the addition of the CL1 degron renders Ura3p-GFP undetectable (Fig. 1C, compare panels 1 and 2).
Additionally, cycloheximide chase analysis reveals that the addition of the CL1 degron reduces the half-life of Ura3p from ∼31 h for Ura3p-GFP and ∼17 h for Ura3p-HA to less than 5 min for either protein fused to the CL1 degron (Fig. 1D). In order to detect CL1-containing constructs by immunoblot, three times as much protein extract was loaded, and a longer exposure was used. Taken together, these data indicate that the CL1 degron dramatically increases the rate of turnover of Ura3p, resulting in decreased steady-state protein levels and an inability to grow on media lacking uracil, in agreement with and extending the findings of Gilon et al. (1, 31). Although we find that Ura3p tagged with HA or GFP act similarly to one another (Fig. 1, A–D), there is an ∼2-fold greater half-life of Ura3p-GFP compared with Ura3p-HA (31 versus 17 h), suggesting that the addition of a large, folded domain may further stabilize Ura3p. Regardless, we find the CL1 degron destabilizes both dramatically (to a half-life of <5 min) and give similar results for all cases tested. Thus, we have used Ura3p-HA-CL1 and Ura3p-GFP-CL1 interchangeably in these studies.
Previous studies have shown that the CL1 degron promotes degradation of Ura3p by the proteasome (31) but have not examined its ubiquitination status directly. Since several ubiquitin-independent substrates of the proteasome have been identified to date (42, 43), we felt it important to examine the ubiquitination of Ura3p-CL1. First, we confirmed the proteasome dependence of Ura3p-CL1 degradation in our system. The wild-type PRE1 PRE2 stain is unable to grow on media lacking uracil (Fig. 1E, SC-Leu-Ura) and is able to grow on media containing 5-FOA (Fig. 1E, SC-Leu + 5-FOA). In contrast, we find that pre1-1 pre2-1 mutant cells can grow on media lacking uracil (Fig. 1E, SC-Leu-Ura) and are unable to grow on media containing 5-FOA (Fig. 1E, SC-Leu + 5-FOA). There is also a significant stabilization of Ura3p-CL1 by cycloheximide chase in the proteasome mutant strain (Fig. 1F), reaffirming that Ura3p-CL1 is a substrate of the proteasome. It should be noted that the slight difference in the half-life of Ura3p-HA-CL1 observed in the nonisogenic wild-type strains used in Fig. 1 (<5 min in Fig. 1D and 9 min in Fig. 1F) can be attributed to differences in strain background and the 37 °C temperature shift required for the experiment in Fig. 1F.
We also analyzed the polyubiquitination status of Ura3p-CL1. Ura3-HA-CL1 was immunopurified from the pre1-1 pre2-1 mutant and isogenic wild-type strains, followed by immunoblot using either anti-HA antibody to detect Ura3p-HA-CL1 or anti-ubiquitin antibodies to detect ubiquitinated species of Ura3p-HA-CL1. In the wild-type PRE1 PRE2 strain, Ura3p-HA-CL1 is ubiquitinated, as demonstrated by a high molecular weight smear with anti-ubiquitin antibodies (Fig. 1G, lane 1, α-Ub). Despite loading equal amounts of immunopurified Ura3p-HA-CL1 (Fig. 1G, lanes 1 and 2, α-HA), this smear is intensified in the pre1-1 pre2-1 strain, indicating that ubiquitinated forms of Ura3p-CL1 accumulate in the absence of proteasomal degradation (Fig. 1G, lane 2, α-Ub). These smears are specific for ubiquitinated Ura3p-HA-CL1, given that negative control strains not expressing Ura3p-HA-CL1 do not show specific signal with anti-ubiquitin antibodies (Fig. 1G, lanes 3 and 4, α-Ub). Thus, Ura3p-CL1 is ubiquitinated prior to degradation by the proteasome.
Ura3p-CL1 Is Ubiquitinated by ER-localized Ubiquitination Machinery—We wanted to analyze the degradation requirements of Ura3p-CL1. Previous studies have demonstrated that CL1-mediated degradation is dependent on Ubc6p/Ubc7p, Cue1p, and Doa10p (1, 31, 32). However, analysis comparing the growth on media lacking uracil to the degree of stabilization and examination of whether these enzymes are directly responsible for ubiquitination of Ura3p-CL1 has not been reported. It was intriguing to us that ER-localized ubiquitination machinery, rather than cytosolic machinery, would be responsible for the degradation of the cytosolic Ura3p-CL1, and we wished to examine this further.
We assessed the growth phenotype of Ura3p-CL1 in strains deleted for cytosolic and the ER-localized ubiquitination machinery. Notably, only strains deleted for the ERAD-C ubiquitination machinery (ubc6Δ, ubc7Δ, cue1Δ, and doa10Δ) and not cytosolic E2 enzymes (ubc4Δ and ubc5Δ) permit growth on media lacking uracil (Fig. 2A, SC-Leu-Ura), in agreement with Gilon et al. (1, 31). Additionally, we examined mutants in ER-localized ubiquitination machinery known to act on ER lumenal ERAD-L substrates, whose ubiquitination also occurs at the cytosolic face of the ER. However, HRD1 (Fig. 2A, SC-Leu-Ura) (31), HRD3, DER1, YOS9, HTM1, and USA1 mutants had no effect on the turnover rate of Ura3p-CL1; nor did deletion of the vacuolar peptidase, PEP4 (data not shown).
FIGURE 2.
Ura3p-CL1 is ubiquitinated by ER-localized ubiquitination machinery. A, serial dilutions of wild-type and mutant cells expressing Ura3p-HA-CL1 (pSM2288) were spotted to the indicated media. B, immunoblots of steady-state protein levels from wild-type (WT) cells expressing Ura3p-HA (pSM2287) and from the strains used in A. Protein extracts from 0.1 A600 units of cells/lane were probed with anti-HA antibodies, and hexokinase (HK) serves as a loading control. C, quantitation of the data shown in B. Blots were imaged using a VersaDoc digital imaging system and quantitated using Quantity One software. The amount of Ura3p-HA-CL1 in each strain was calculated by normalizing the α-HA signal to the α-hexokinase signal. Fold stabilization was graphed relative to the amount of normalized protein present in the wild-type strain. Data is the average of three independent experiments, and one S.D. is indicated. D, anti-HA immunopurification (IP) and ubiquitin immunoblots (WB) in wild-type and mutant cells expressing either Ura3p-HA-CL1 (pSM2288; lanes 1–3) or Ura3p-GFP-CL1 (pSM2290; lanes 4–6) as a negative control. Immunopurifications were performed as described in the legend to Fig. 1 and under “Materials and Methods.” For lanes 7 and 8, 10 times as much protein sample was loaded as in lanes 2 and 3. The asterisk indicates a cross-reactive band unrelated to Ura3p-HA-CL1. E, cycloheximide chase, SDS-PAGE, and immunoblots of wild-type and mutant cells expressing either Ura3p-HA-CL1 (pSM2288; top), His3p-HA-CL1 (pSM2293; middle), or His3p-HA (pSM2318; bottom) for the indicated time points. Blots were probed with anti-HA antibodies. Cycloheximide chases were repeated at least three times with similar results; a representative experiment is shown for each strain. Strains used in A–E were SM4460, SM5377, SM5489, SM5492, SM4821, SM4822, SM4823, SM4820, and SM5362.
Comparison of the steady state levels of Ura3p-CL1 in these deletion strains (Fig. 2, B and C) reveals that Ubc7p is the major E2 ubiquitin-conjugating enzyme acting on Ura3p-CL1, since there is a significant protein stabilization (>30-fold) in the ubc7Δ or cue1Δ mutant strains (Cue1p is the membrane anchor and activator of Ubc7p) (44), and there is very little stabilization in the ubc6Δ mutant alone (∼5-fold; Fig. 2, B and C). Interestingly, despite the low level of Ura3p-CL1 that accumulates in the ubc6Δ mutant, this strain displays very similar growth on media lacking uracil to ubc7Δ, cue1Δ, or doa10Δ strains (Fig. 2A). Thus, a very small amount of Ura3p is sufficient to support full growth on media lacking uracil, highlighting the sensitivity of this genetic selection.
We also examined the ubiquitination status of Ura3p-CL1 in these mutants. In the wild-type strain, Ura3p-CL1 is ubiquitinated, as evidenced by a high molecular weight smear with anti-ubiquitin antibodies (Fig. 2D, lane 1, α-Ub), not seen in negative control strains (Fig. 2D, lanes 4–6, α-Ub). This smear is absent in the doa10Δ or ubc6Δ/ubc7Δ strains, indicating that ubiquitination of Ura3p-CL1 does not occur in the absence of Doa10p or Ubc6p/Ubc7p (Fig. 2D, lanes 2 and 3, α-Ub). Even when 10 times as much protein was loaded for the doa10Δ and ubc6Δ/ubc7Δ deletion strains, ubiquitination was not detectable (Fig. 2D, lanes 7 and 8). Thus, the ER-localized proteins Ubc6p, Ubc7p, Cue1p, and Doa10p appear to be the sole E2 and E3 ubiquitination components acting on Ura3p-CL1. The requirement for ER-localized ubiquitination machinery is not specific to Ura3p, since the addition of CL1 also destabilizes His3p, and His3p-CL1 is dramatically stabilized in doa10Δ and ubc6Δ/ubc7 deletion strains, similar to Ura3p-CL1 (Fig. 2E).
CL1 Degron-mediated Degradation Is Dependent on the Chaperones Ssa1p and Ydj1p—We wished to analyze whether the degradation of Ura3p-CL1 is chaperone-dependent, as is the case for misfolded proteins. The SSA class of Hsp70 chaperones and the Hsp40 co-chaperones, Ydj1p and Hlj1p, are known to function with Ubc6p/Ubc7p and Doa10p in ERAD-C, as determined by analysis of Ste6p* (8, 15). However, other cytosolic chaperones, such as Sti1p, Sse1p, and Hsp82p, have been shown to be required for the degradation of the CytoQC substrates VHL and VHL-L158P (21). Thus, we decided to determine which set of chaperones act on a cytosolic substrate that is targeted for ubiquitination by ER-localized machinery.
We first analyzed the role of the SSA family of chaperones and its cognate Hsp40, Ydj1p. Because the deletion of all four SSA genes (SSA1, SSA2, SSA3, and SSA4) is inviable and that of YDJ1 grows very poorly and may pick up suppressors, well characterized temperature-sensitive point mutants (ssa1-45 ssa2Δ ssa3Δ ssa4Δ and ydj1-151) that block function (45, 46) were used instead. Even in the presence of uracil, growth of these mutant strains on solid media at the nonpermissive temperature resulted in severely slow growth or lethality, respectively, precluding the use of spot growth assays for this analysis.5 Thus, we used cycloheximide chase analysis to determine the effects that mutation of the chaperones have on the degradation of Ura3p-CL1. Comparison of its fate in the ssa1-45 and ydj1-151 mutants to their isogenic wild-type strains at the nonpermissive temperature (Fig. 3, A and B) demonstrates that Ssa1p and Ydj1p chaperone function is required for degradation of Ura3p-CL1. Ydj1p is ER-localized, at least in part (47, 48). Interestingly, another ER-localized Hsp40, Hlj1p, which has been shown in some cases to function redundantly with Ydj1p (49), does not appear to play a significant role in the degradation of Ura3p-CL1 (Fig. 3D, top), as judged by the lack of growth of the hlj1Δ mutant on media lacking uracil.
FIGURE 3.
The chaperones Ssa1p and Ydj1p are required for the degradation of Ura3p-CL1. A–C, cycloheximide chase, SDS-PAGE, and immunoblots of SSA1 and ssa1-45 cells (SM4247 and SM4177; A), YDJ1 and ydj1-151 cells (SM4947 and SM4948; B), or KAR2 and kar2-133 cells (SM3865 and SM3867; C), expressing Ura3p-HA-CL1 (pSM2291, pSM2292, and pSM2288, respectively) for the indicated times. Blots were probed using anti-HA antibodies and quantitations of the cycloheximide chases are graphed at the right. Cycloheximide chases were repeated at least three times with similar results; a representative experiment is shown for each strain. D, serial dilutions of wild type and mutant cells expressing Ura3p-HA-CL1 (pSM2288) were spotted to the indicated media. Wild-type cells expressing Ura3p-HA (pSM2287) was included as a positive control. Strains examined in D are SM4460, SM5483, SM5486, SM5485, SM5177, SM5484, SM4552, SM4553, SM571, SM5472, SM5473, and SM5474.
Not surprisingly, the ER-lumenal Hsp70 Kar2p does not play a role in the degradation of Ura3p-CL1, since there is no stabilization in a kar2-133 mutant (Fig. 3C). Further, other cytosolic chaperones, including other Hsp70s (Ssb1p/Ssb2p), Hsp90 (Hsp82p), Hsp100 (Hsp104p), the small heat shock proteins (Hsp26p/Hsp42p), or other co-chaperones (Sti1p, Snl1p, and Sse1p) are not required for degradation of Ura3p-CL1, as demonstrated by the lack of growth on media lacking uracil (Fig. 3D, SC-Leu-Ura). Taken together, it is quite notable that ER-bound chaperones and not other cytosolic chaperones are required for the degradation of Ura3p-CL1.
CL1 Degron-mediated Degradation Is Impaired in Mutants in the Cdc48p AAA-ATPase Complex—Since the degradation requirements of Ura3p-CL1 are similar to those of membrane proteins with misfolded cytosolic domains (ERAD-C substrates), we decided to analyze the contribution of other cellular components known to act in ERAD to the degradation of Ura3p-CL1. A partial stabilization of Ura3p-CL1 is apparent in the cdc48-3 (Fig. 4A), npl4-1 (Fig. 4B), and ufd1-1 (Fig. 4C) mutants, as compared with their isogenic wild-type strains. This is similar to the partial stabilization of the ERAD-C substrate, Ste6p*, also seen in these mutants (8). During ERAD, the Cdc48p-Npl4p-Ufd1p AAA-ATPase complex is thought to couple mechanical force to ubiquitin binding, probably for membrane extraction or proteasomal targeting of ubiquitinated substrates (18–20, 50, 51). Our findings with Ura3p-CL1 suggest that the Cdc48p complex may also be playing such a role in the degradation of Ura3p-CL1. However, since the stabilization of Ura3p-CL1 in the Cdc48p complex mutants is incomplete, some degradation also probably occurs without the aid of the Cdc48p complex.
FIGURE 4.
The Cdc48p AAA-ATPase complex is required for efficient degradation of Ura3p-CL1. A–C, cycloheximide chase, SDS-PAGE, and immunoblots of CDC48 and cdc48-3 cells (SM5124 and SM4783; A), NPL4 and npl4-1 cells (SM4784 and SM4785; B), or UFD1 and ufd1-1 cells (SM4779 and SM4780; C), expressing Ura3p-HA-CL1 (pSM2288) for the indicated time points. Blots were probed using anti-HA antibodies, and quantitations of the cycloheximide chases are graphed below. Cycloheximide chases were repeated at least three times with similar results; a representative experiment is shown for each strain. D, immunoblots of steady-state protein levels from wild-type (WT) and mutant cells expressing Ura3p-HA-CL1 (pSM2288). The strains used are SM5186, SM5381, SM5494, SM5547, SM5548, SM5549, SM5550, SM5551, SM5552, SM5553, and SM5554. E, quantitation of the data shown in D, calculated as in Fig. 2C. Data are the average of three independent experiments with one S.D. indicated.
We also examined the contribution of Cdc48p complex co-factors to the degradation of Ura3p-CL1 and find that there is a slight stabilization of Ura3p-CL1 in the ubx2Δ mutant and significant stabilization in the doa1Δ mutant compared with wild-type cells (Fig. 4, D and E). However, because the doa1Δ mutant is depleted for ubiquitin, this effect could be indirect (52, 53).
Blocking Ubiquitination Reveals That Ura3p-CL1 Is Distributed between the Cytosol and ER Membrane—We analyzed the localization properties of Ura3p-GFP-CL1 by fluorescence microscopy. In contrast to wild-type cells, where Ura3p-GFP-CL1 is undetectable (Fig. 5A, panel 5), Ura3p-GFP-CL1 is readily apparent in ubiquitination machinery mutants (doa10Δ, ubc7Δ, and cue1Δ; Fig. 5A, panels 11, 17, and 23). In these mutants, much of the protein shows a diffuse cytosolic localization, but interestingly a portion of Ura3p-GFP-CL1 is localized to the ER, as demonstrated by co-localization with the integral ER membrane protein Sec63p-RFP (Fig. 5A, panels 12, 18, and 24). In addition to its cytosolic and ER localization, there is also a punctate localization of Ura3p-GFP-CL1 in these mutant strains. The localization pattern of Ura3p-GFP-CL1 is in contrast to the exclusively cytosolic localization of Ura3p-GFP in these strains (Fig. 5A, panels 8, 14, and 20).
FIGURE 5.
Ura3p-CL1 is localized to the cytosol and ER. A, differential interference contrast (DIC) and fluorescence microscopy of wild-type (WT) and mutant cells co-expressing either Ura3p-GFP (pSM2289; left three columns) or Ura3p-GFP-CL1 (pSM2290; right three columns) and Sec63p-RFP (pSM1959). B, fractionation of mutant cells expressing Ura3p-HA-CL1 (pSM2288) by 13,000 × gspin to yield total (T), supernatant (S), and pellet (P) fractions, which were analyzed by SDS-PAGE and immunoblotting using anti-HA antibodies. Hexokinase (HK) and Sec61p were used as controls for soluble and membrane proteins, respectively. C, the P and S fractions from the doa10Δ strain in B were further separated using sucrose flotation gradient ultracentrifugation, as described under “Materials and Methods,” and subject to SDS-PAGE and immunoblotting as in B, except blots were also probed for Ssa1p. Fractions are labeled from the top (fraction 1) to bottom (fraction 12) of the gradient, and load indicates where samples were loaded onto the gradient (1.8 m sucrose). Sucrose concentrations of the gradient are shown at the bottom. Strains used are SM4460, SM4820, SM4821, SM4822, SM5362, and SM5495.
To further examine the localization pattern of Ura3p-CL1 when its ubiquitination is blocked, we performed fractionation using 13,000 × g centrifugation for 20 min, a condition where greater than 90% of ER and plasma membrane but almost no Golgi or endosomes pellet (40).5 In the doa10Δ and ubc6Δ/ubc7Δ strains, the majority of Ura3p-CL1 is in the supernatant (s) fraction after a 13,000 × g spin, as is the control cytosolic protein, hexokinase (HK in Fig. 5B), suggesting that the majority of Ura3p-CL1 is in a soluble and probably cytosolic form. However, it is notable that a small portion of Ura3p-CL1 (∼10%) is found in the 13,000 × g pellet (p) fraction in these mutant strains (Fig. 5B, Ura3p-HA-CL1), as is the ER membrane protein, Sec61p (Fig. 5B, Sec61p). This could be an ER-associated portion of Ura3p-CL1 or, alternatively, aggregates. To distinguish between ER membrane association and aggregation, we analyzed the 13,000 × g pellet and supernatant fractions by sucrose flotation gradient centrifugation. Samples were loaded near the bottom of a discontinuous sucrose gradient and centrifuged at 100,000 × g until equilibrium. Membranes and membrane-associated proteins float up in the gradient to lower sucrose concentrations, whereas aggregated proteins remain near the load in the bottom of the gradient (41, 54, 55). If the portion of Ura3p-CL1 found in the 13,000 × g pellet fraction were ER membrane-associated, it would float up in the gradient. If, instead, this portion of Ura3p-CL1 represents protein aggregates or other insoluble forms of the protein, it would remain near where it was loaded onto the gradient.
Fig. 5C shows a representative experiment using the doa10Δ mutant, and similar results were obtained using a ubc6Δ/ubc7Δ strain (data not shown). Ura3p-CL1 in the P fraction is membrane-associated rather than aggregated, since it floats up from the 1.8 m load fractions into the 1.5 m fractions (Fig. 5C, top panels, lanes 5–7), as does the ER membrane protein, Sec61p. Combined with the fluorescence microscopy (Fig. 5A), we conclude that a portion of Ura3p-CL1 is ER membrane-associated. Interestingly, no Ura3p-CL1 is found at the bottom of the gradient, suggesting the punctate structures seen by fluorescence microscopy are not aggregates and instead may represent a specific subdomain of the ER membrane, where some of Ura3p-CL1 is concentrated, perhaps similar to the ER-associated subcompartment induced by misfolded Ste6p alleles (36). Analysis of the 13,000 × g supernatant (S in Fig. 5C; bottom panels) reveals that Ura3p-CL1 and hexokinase co-fractionate (lanes 7–9), confirming that the Ura3p-CL1 in the soluble fraction behaves identically to a soluble, cytosolic protein. Taken together, our findings suggest that at steady state, much of Ura3p-CL1 is cytosolic, whereas some is associated with the ER membrane.
Importantly, analysis of a ubc6Δ/ubc7Δ/doa10Δ triple mutant strain expressing Ura3p-CL1 examined by fractionation at 13,000 × g, fluorescence microscopy, and sucrose gradient flotation analysis reveals an identical localization and fractionation pattern to the doa10Δ or ubc6Δ/ubc7Δ strains (Fig. 5B) (data not shown). Since a portion of Ura3p-CL1 is still present in an ER membrane-associated form in this triple mutant strain, it is not likely that components of the ubiquitination machinery are solely responsible for the ER membrane localization of Ura3p-CL1.
Ssa1p and Ydj1p Are Required for the Ubiquitination of Ura3p-CL1—We wished to further address the role of the chaperones Ssa1p and Ydj1p in the degradation of Ura3p-CL1. It has been shown that chaperones are required for the ubiquitination of ERAD substrates, such as Ste6p*, and provide a “bridge” between the misfolded protein in the ER membrane and ER-localized ubiquitination machinery (15, 56). Ydj1p is ER membrane-localized via its prenylated CAAX motif (47, 48), but, although implicated in functions at the ER (15, 46, 57), ER localization has not been documented for Ssa1p. To fulfill a bridging function between Ura3p-CL1 and the ER-localized ubiquitination machinery, some of Ssa1p would need to be membrane-localized. In the ubiquitination mutants, we find a portion of Ssa1p in the pellet fraction, all of which co-migrates with Ura3p-CL1 and Sec61p (Fig. 5C, top panels, Ssa1p), suggesting that indeed some of Ssa1p is ER membrane-associated. Also, like Ura3p-CL1, a large portion Ssa1p is soluble (Fig. 5C, bottom panels, Ssa1p).
We next asked whether Ssa1p or Ydj1p chaperone function is required for the ubiquitination of Ura3p-CL1. In the SSA1 or YDJ1 parental wild-type strains, Ura3p-HA-CL1 is ubiquitinated, as demonstrated by a high molecular weight smear with anti-ubiquitin antibodies (Fig. 6, A and B, lane 1, α-Ub, respectively). Despite loading approximately equal amounts of immunopurified Ura3p-HA-CL1 (Fig. 6, A and B, lanes 1 and 2, α-HA, respectively), this smear is absent in the ssa1-45 and ydj1-151 strains at nonpermissive temperature, indicating that ubiquitination of Ura3p-CL1 does not occur in either of these mutants (Fig. 6, A and B, lane 2, α-Ub, respectively). These data suggest that the activity of each of these chaperones is absolutely required for the ubiquitination of Ura3p-CL1, probably because the chaperones provide a bridge between Ura3p-CL1 and the ER-localized ubiquitination machinery.
FIGURE 6.
Ssa1p and Ydj1p are required for ubiquitination and to maintain solubility of Ura3p-CL1. A, anti-HA immunopurification (IP) and ubiquitin immunoblots (WB) in SSA1 (SM4247) or ssa1-45 (SM4177) cells expressing either Ura3p-HA-CL1 (pSM2291; lanes 1 and 2) or Ura3p-GFP-CL1 (pSM2290; lanes 3 and 4) as a negative control. Immunopurifications were performed as described in the legend to Fig. 1 and under “Materials and Methods.” The asterisk indicates a cross-reacting band. B, anti-HA immunopurification and ubiquitin immunoblots to detect ubiquitinated Ura3p-HA-CL1 in YDJ1 (SM4947) or ydj1–151 (SM4948) cells expressing either Ura3p-HA-CL1 (pSM2291; lanes 1 and 2) or Ura3p-GFP-CL1 (pSM2290; lanes 3 and 4) as a negative control, done as in A. C, fractionation of the strains in A and B expressing Ura3p-HA-CL1 by 13,000 × g spins as described under “Materials and Methods” and in the legend to Fig. 5B. D, the pellets (P) from C were further separated using sucrose flotation gradient ultracentrifugation as described under “Materials and Methods” and in the legend to Fig. 5C.
Ura3p-CL1 Is Exclusively Membrane-associated When Chaperone Function Is Blocked—We next determined the localization of Ura3p-CL1 when chaperone function is blocked. Despite being dramatically stabilized in the ssa1-45 or ydj1-151 mutant strains by cycloheximide chase (Fig. 3, A and B), we were unable to visualize Ura3p-GFP-CL1 by fluorescence microscopy, possibly due to an inability of GFP to properly fluoresce in these chaperone mutant strains at the nonpermissive temperature. To circumvent this, we analyzed the localization of Ura3p-CL1 by fractionation at 13,000 × g. Strikingly, at the nonpermissive temperature in the ssa1-45 or ydj1-151 mutants, Ura3p-CL1 is found almost exclusively in the 13,000 × g pellet fraction, with very little in the 13,000 × g supernatant fraction (Fig. 6C, Ura3p-HA-CL1). This is in contrast to the fractionation pattern of Ura3p-CL1 in ubiquitination mutants, where it is mostly in the supernatant fraction (Fig. 5B). Further analysis of the pellet fraction by flotation gradient reveals that most of the Ura3p-CL1 in each these mutants is ER membrane-associated, since it floats in the gradient and co-migrates with Sec61p (Fig. 6D, lanes 5–7). A small portion of Ura3p-CL1 clearly remains in the load fractions or additionally migrates into fractions at the bottom of the tube (Fig. 6D, lanes 8–12). This nonfloating fraction probably represents protein aggregates, since a similar flotation pattern has been shown previously for aggregated proteins (55).
Taken together, these data indicate that the chaperones Ssa1p and Ydj1p are required for the ubiquitination of Ura3p-CL1. When Ssa1p and Ydj1p are present but when either one is impaired due to mutation, the majority of Ura3p-CL1 appears to be irreversibly “stuck” at the ER membrane. Presumably, this is because Ura3p-CL1 is bound to one or both of these chaperones but is unable to be properly “passed off” to the ubiquitination machinery in the absence of functional chaperone activity. Apparently, under these conditions, Ura3p-CL1 cannot be released back into the cytosol either, accounting for the predominant ER membrane fractionation pattern of Ura3p-CL1 in the ssa1-45 or ydj1-151 mutant strains. In addition, a small portion of Ura3p-CL1 appears to aggregate in these mutants.
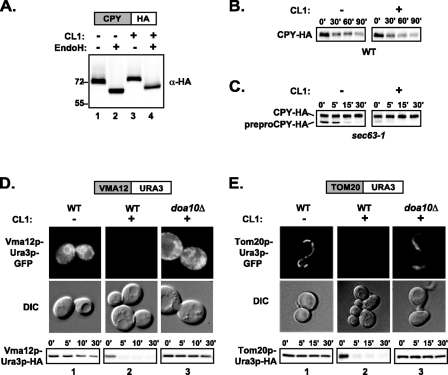
Access to the Cytosolic Face of the ER Is Required for Recognition and Degradation of CL1 Degron-containing Proteins—To determine whether access to the cytosolic face of the ER is absolutely required for recognition and degradation of CL1 degron-containing proteins, we prevented this access by fusing CL1 to CPY-HA. Both CPY-HA and CPY-HA-CL1 are glycosylated, as judged by sensitivity to endoglycosidase H treatment (Fig. 7A, compare lanes 1 and 3 with lanes 2 and 4), indicating that they are efficiently translocated into the ER. Interestingly, the addition of the CL1 degron does not affect the stability of CPY-HA (Fig. 7B), suggesting that the CL1 degron is not recognized or degraded inside the lumenal compartment of the secretory pathway. The half-life of the misfolded allele of CPY, CPY*-HA, which is an ERAD substrate and does not exit the ER, is also unchanged by the addition of the CL1 degron.5 On the other hand, when CPY-HA-CL1 is expressed in the ER import-defective strain, sec63-1 (58, 59), cells accumulate a cytosolic precursor form (prepro-CPY-HA; Fig. 7C). Whereas prepro-CPY-HA has a relatively short half-life itself, presumably due to its misfolding in the cytosol (as has been seen for CPY lacking a signal sequence) (22), the addition of the CL1 degron dramatically destabilizes it even further (Fig. 7C). Thus, access to the cytosolic face of the ER appears to be critical for the recognition and degradation of CL1 degron-containing proteins.
FIGURE 7.
Access to the cytosolic face of the ER is required for recognition of the CL1 degron. A, SDS-PAGE and immunoblotting of wild-type cells expressing CPY-HA (pSM2302; lanes 1 and 2) or CPY-HA-CL1 (pSM2303; lanes 3 and 4) either untreated (lanes 1 and 3) or treated with endoglycosidase Hf (EndoH; lanes 2 and 4). Immunoblots were probed with anti-HA antibodies. B, cycloheximide chase analysis, SDS-PAGE, and immunoblots of wild-type cells expressing CPY-HA (pSM2302) or CPY-HA-CL1 (pSM2303) for the indicated time points. Blots were probed with anti-HA antibodies. C, cycloheximide chase analysis, SDS-PAGE, and immunoblots of sec63-1 cells expressing CPY-HA (pSM2302) or CPY-HA-CL1 (pSM2303) as in B. D, differential interference contrast (DIC) and fluorescence microscopy and cycloheximide chase, SDS-PAGE, and immunoblots of wild type (WT; columns 1 and 2) or doa10Δ (column 3) cells expressing Vma12p-Ura3p-GFP (pSM2296), Vma12p-Ura3p-GFP-CL1 (pSM2297), Vma12p-Ura3p-HA (pSM2294), or Vma12p-Ura3p-HA-CL1 (pSM2295), as indicated. Cycloheximide chase was performed using the indicated time points, and immunoblots were probed using anti-HA antibodies. E, as in D, except expressing Tom20p-Ura3p-GFP (pSM2300), Tom20p-Ura3p-GFP-CL1 (pSM2301), Tom20p-Ura3p-HA (pSM2298), or Tom2p-Ura3p-HA-CL1 (pSM2299), as indicated. All cycloheximide chases were repeated at least three times with similar results; a representative experiment is shown for each strain. Strains used in A–E are SM4460, SM3417, and SM4820.
To further analyze the significance of the cytosolic face of the ER in the degradation of the CL1 degron, we tethered CL1 to the cytosolic face of the ER by generating a protein fusion between the ER membrane protein, Vma12p, and Ura3p-CL1. Vma12p-Ura3p is stable at the protein level and ER-localized by fluorescence microscopy (Fig. 7D, column 1). The presence of the CL1 degron significantly destabilizes the protein (Fig. 7D, column 2). The same ER-localized ubiquitination machinery that acts on Ura3p-CL1 (Doa10p, Ubc6p, Ubc7p, and Cue1p) also stabilizes Vma12p-Ura3p-CL1 (Fig. 7D, column 3) (data not shown).
We also queried the fate of Ura3p and Ura3p-CL1 fused to the C terminus of the mitochondrial outer membrane protein, Tom20p. Tom20p is a type I transmembrane protein with an N-terminal transmembrane domain and its C terminus in the cytosol (60). Tom20p-Ura3p without the CL1 degron is a stable protein and localized to mitochondrial tubular structures by fluorescence microscopy (Fig. 7E, column 1). Interestingly, attachment of the CL1 degron to Tom20-Ura3p dramatically destabilizes the protein (Fig. 7E, column 2), very similar to the cytosolic Ura3p-CL1 or the ER membrane-localized Vma12p-Ura3p-CL1. Quite surprisingly, Tom20p-Ura3p-CL1 is also stabilized by doa10Δ and ubc6Δ/ubc7Δ, and when examined by fluorescence microscopy in the doa10Δ strain, Tom20p-Ura3p-GFP-CL1 is still found to be localized to mitochondria (Fig. 7E, column 3). There is strong evidence for interorganelle contact sites between mitochondrial and ER membranes, which are thought to be specialized subdomains of the ER that function for lipid or calcium transfer to the mitochondria (61–63). Our results suggest that these ER-mitochondrial contact sites can also function for mitochondrial protein ubiquitination by the ER-localized machinery and degradation from the mitochondrial outer membrane. Taken together, these data suggest that access to the cytosolic face of the ER, either from the cytosol or ER membrane or through ER-mitochondrial contact sites, is required for the degradation of the CL1 degron.
DISCUSSION
The quality control of misfolded cytosolic proteins is not well understood. The best characterized CytoQC substrates in yeast are alleles of the mammalian VHL, ΔssCPY, and ubiquitin fusion proteins (21, 22, 52). Thus far, there is not a consensus of the machinery involved in CytoQC. To study CytoQC, we have examined the degradation requirements of Ura3p-CL1. Previous studies and our results here indicate that the degradation of Ura3p-CL1 is proteasome-dependent and requires ER-localized ubiquitination machinery (Ubc6p, Ubc7p, Cue1p, and Doa10p) (1, 31). Although Ura3p-CL1 is not an ER membrane protein, we find that it requires cytosol/ER-localized chaperones (Ydj1p and Ssa1p) for its ubiquitination and degradation. These requirements are similar to those seen for membrane proteins with misfolded cytosolic domains subject to ERAD-C, such as Ste6p*, KSS, or Pma1p-D378S (13, 56). However, there are several interesting differences. For the degradation of Ura3p-CL1, Ydj1p is not functionally redundant with Hlj1p, as is the case for the ERAD-C substrates, Ste6p* and CFTR, where mutation of either Ydj1p or Hlj1p individually has no effect on turnover (8, 49). The ubiquitination and degradation of Ura3p-CL1 is severely impaired in the ydj1-151 mutant alone and is independent of Hlj1p (Fig. 3). Hlj1p is an ER integral membrane protein (49), so this difference may reflect a role for Hlj1p exclusively in the degradation of misfolded membrane proteins.
Also, although Ura3p-CL1 requires the AAA-ATPase Cdc48p-Npl4p-Ufd1p complex for its degradation, several Cdc48p complex co-factors involved in ERAD (Dsk2p/Rad23p and Ufd2p) (15–18, 64, 65) are dispensable for the degradation of Ura3p-CL1 (Fig. 4). There are seven UBX domain proteins in yeast and various co-factors that interact with Cdc48p and may aid in substrate delivery to the proteasome (18, 53, 66, 67). We find that mutation of Ubx2p causes a very mild defect in the turnover of Ura3p-CL1, whereas mutation of Doa1p (Ufd3p) causes a more severe defect (Fig. 4, D and E). A decrease in ubiquitin levels has been observed in a doa1Δ mutant strain (52, 53), and this may contribute significantly to the stabilization of Ura3p-CL1 in the doa1Δ mutant.
It is notable that the Cdc48p complex is involved in Ura3p-CL1 degradation, since several cytosolic proteins examined to date (Deg1-Ura3p and ΔssCPY*) do not require it for their degradation, whereas others do (Ub-Pro-β-gal) (22, 32, 52). This difference may suggest a unique role for the Cdc48p complex in the degradation of some cytosolic proteins, including Ura3p-CL1, such as separating ubiquitinated Ura3p-CL1 from the ubiquitination machinery or the ER membrane for proteasomal degradation. Future work will be directed toward determining the exact role of the Cdc48p complex and its co-factors in the degradation of cytosolic proteins.
Not only is ER-localized machinery required for the degradation of Ura3p-CL1, but access to the cytosolic face of the ER is essential for the degradation of CL1-containing proteins. CL1 exposed to the cytosol, either by fusion to Ura3p, to an ER membrane protein, or to a mitochondrial outer membrane protein (where ER-mitochondrial contact sites are probably accessed), promotes rapid degradation, whereas CPY-CL1 is metabolically stable and not degraded inside the ER (Fig. 7). These data suggest that the cytosolic face of the ER may serve as a “platform” or “organization center” for a large portion of cellular quality control, including many misfolded cytosolic proteins. Visualization of the ER reveals that this organelle is present throughout the cytosol and makes contact with virtually all organelles (61, 68). Additionally, a significant portion of proteasomes in yeast may be localized to the ER/nuclear envelope membrane (69, 70). Thus, the ER is properly positioned to fulfill a generalized role in cellular quality control, in contrast to the previous view of its role being strictly confined to ERAD.
Taken together, our data here regarding the degradation pathway of Ura3p-CL1, combined with previous studies (1, 31, 32), suggest the following working model (Fig. 8A). First, Ura3p-CL1 is proposed to be recognized and targeted to the cytosolic face of the ER by the HSP70 (Ssa1p) and its HSP40 co-factor (Ydj1p) (Fig. 8A, step 1). The chaperones facilitate interaction between Ura3p-CL1 and the ER-localized ubiquitination machinery (Fig. 8A, step 2) and are required for ubiquitination of Ura3p-CL1 by this machinery. Next, Ura3p-CL1 is ubiquitinated by the E2s (mainly Ubc7p/Cue1p but also Ubc6p) and the E3 (Doa10p; Fig. 8A, step 3). Ubiquitinated Ura3p-CL1 is acted on by the Cdc48p-Npl4p-Ufd1p complex (Fig. 8A, step 4), perhaps to separate it from the ER, and ultimately direct it to the proteasome, where it is degraded (Fig. 8A, step 5). Due to the short half-life of Ura3p-CL1, this process occurs very rapidly and efficiently in wild-type cells.
FIGURE 8.
Model for the degradation of Ura3p-CL1 at the cytosolic face of the ER. A, based on the present study and previous work, in a wild-type strain, the ER-localized portions of the Hsp70 (Ssa1p) and Hsp40 (Ydj1p) chaperones are proposed to be responsible for recognizing Ura3p-CL1 and targeting it to the ER membrane for degradation (step 1). These chaperones are further proposed to direct Ura3p-CL1 to the E2 and E3 ubiquitination machinery (Ubc6p, Ubc7p, Cue1p, and Doa10p) at the cytosolic face of the ER (step 2), where Ura3p-CL1 undergoes ubiquitination (step 3; represented by white triangles). The Cdc48p-Npl4p-Udf1p complex is proposed to be involved in separating ubiquitinated Ura3p-CL1 from the ubiquitination machinery or the ER membrane (step 4) and targeting it to the 26 S proteasome for complete degradation (step 5). B, in the ubc6Δ/ubc7Δ/doa10Δ mutant strain, the Ssa1p and Ydj1p chaperones would target Ura3p-CL1 to the ER membrane (step 1), but in the absence of the ubiquitination machinery, Ura3p-CL1 cannot be ubiquitinated (step 2). Under these conditions, much of Ura3p-CL1 is cytosolic, presumably because it is released from the chaperones (step 3); a small portion is at the ER membrane, perhaps in dynamic equilibrium with the cytosolic population of Ura3p-CL1. C, in the temperature-sensitive chaperone mutants at the nonpermissive temperature (ydj1-151 is shown here), the chaperone is present but nonfunctional, and Ura3p-CL1 is still targeted to the ER membrane (step 1), but in the absence of functional chaperone activity, Ura3p-CL1 remains “stuck” at the ER membrane (step 2).
How, then, are cytosolic proteins targeted to the ER for degradation? As mentioned above and shown in the model in Fig. 8A, in the case of Ura3p-CL1, the chaperones Ssa1p and Ydj1p, rather than the ubiquitination machinery, appear to be involved. The absence of all of the ER-localized E2 and E3 ubiquitination machinery (Doa10p/Ubc6p/Ubc7p) does not alter the amount of Ura3p-CL1 at the ER (Fig. 5B), indicating that the ubiquitination machinery is not responsible for the ER localization. Indeed, only a small portion of Ura3p-CL1 in the doa10Δ/ubc6Δ/ubc7Δ triple mutant is ER-localized, whereas the majority is soluble and cytosolic. In this case, modeled in Fig. 8B, the ER localization may represent Ura3p-CL1 at the ER interacting with Ssa1p and Ydj1p (Fig. 8B, step 1). Normally, the chaperones would rapidly hand off Ura3p-CL1 to the E2/E3 machinery for its ubiquitination and degradation. When this ubiquitination machinery is missing (Fig. 8B, step 2), Ura3p-CL1 may instead be released from the chaperones at the ER membrane into the cytosol (Fig. 8B, step 3) and continue to cycle on and off of the chaperone complex.
On the other hand, in temperature-sensitive mutant strains of Ssa1p (ssa1-45) or Ydj1p (ydj1-151), Ura3p-CL1 is irreversibly “stuck” at the ER membrane, manifested as a dramatic increase in the amount present at the ER (Fig. 6, C and D). In this case, since Ssa1p and Ydj1p can both bind substrate (71–73), either chaperone may independently target Ura3p-CL1 to the ER membrane (Fig. 8C, step 1). However, as Ssa1p and Ydj1p collaborate to carry out ATPase-dependent chaperone function, Ura3p-CL1 cannot be further targeted for ubiquitination and degradation without both chaperones functioning (Fig. 8C, step 2), and Ura3p-CL1 is stuck at the ER membrane, unable to undergo ubiquitination but also unable to cycle on and off of the chaperones to the cytosol, as occurs in Fig. 8B.
In support of Ssa1p and Ydj1p fulfilling their chaperone function at the ER membrane, a portion of Ssa1p co-migrates with ER membranes by flotation gradient (Fig. 5C), and recent in vitro studies analyzing the ubiquitination of Ste6p* demonstrated that Ssa1p and Ydj1p had to be provided in the microsome fraction, and not the cytosolic fraction, to facilitate ERAD (15). Additionally, Ydj1p assists Ssa1p in protein translocation into the ER (46).
We cannot exclude the possibility that CL1 serves as a novel type of ER “targeting signal” and interacts with ER membranes or proteins in a passive manner, but this seems unlikely, since Tom20p-Ura3p-CL1 is post-translationally localized to mitochondria and not aberrantly targeted to the ER. Additionally, aside from signal sequences that direct proteins to translocation machinery, targeting signals that direct proteins specifically to the cytosolic face of the ER have not been identified. We also cannot exclude the possibility that unidentified proteins target Ura3p-CL1 to the ER, and we are currently performing selections to identify additional machinery required for the degradation of Ura3p-CL1.
The degradation requirements for other cytosolic proteins show some similarity to those of Ura3p-CL1. For the limited number of proteins examined thus far (VHL, ΔssCPY*, and their variants), a requirement for Ssa1p appears to be universal, whereas only some substrates require Ydj1p, and others require additional cytosolic chaperones (Hsp90 and Sti1p/Sse1p) (21, 22). The differences in the co-chaperones may reflect the degree of “misfoldedness” that the proteins display and/or may determine whether additional folding attempts should be initiated. The ubiquitination machinery that acts on specific misfolded cytosolic proteins is still being identified, and it will be interesting to see whether these, like Doa10p, Ubc6p, Ubc7p, and Cue1p, are ER-localized.
The source of CL1 was previously reported to be ambiguous, since its amino acid sequence does not match known yeast proteins (1). Our sequence analysis, however, suggests that the CL1 amino acid sequence represents a frameshifted region of the yeast PMD1 gene.3 Additionally, we find that the SL17 degron and the other CL degrons identified by Gilon and co-workers (1) also represent out of frame sequences from yeast ORFs. Frameshift mutations often eliminate gene function, and study of CL1 and other degrons in yeast may thus reveal mechanisms for the removal of improper translation products. Further work using novel degrons isolated from the yeast proteome and this sensitive Ura3p-based genetic selection are expected to reveal new degradation machinery and commonalties and differences among degrons.
Acknowledgments
We thank Jeff Brodsky, Mark Hochstrasser, Stefan Kreft, Rob Jensen, and Colin Stirling for generously supplying plasmids, strains, and antibodies. We also thank Jeff Brodsky, Kunio Nakatsukasa, and Jim Mullally for critical reading of the manuscript and members of the Michaelis laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM51508 (to S. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; 5-FOA, 5-fluoroorotic acid; CPY, carboxypeptidase Y; CytoQC, cytosolic quality control; ERAD, endoplasmic reticulum-associated degradation; GFP, green fluorescent protein; HA, hemagglutinin; RFP, red fluorescent protein; Ub, ubiquitin; VHL, von Hippel Lindau protein; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; ORF, open reading frame; AAA-ATPase, ATPase associated with various cellular activities.
M. Maurer and S. Michaelis, unpublished data.
Kreft and Hochstrasser, unpublished data.
M. Metzger and S. Michaelis, unpublished data.
References
- 1.Gilon, T., Chomsky, O., and Kulka, R. G. (1998) EMBO J. 17 2759–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gregersen, N., Bross, P., Vang, S., and Christensen, J. H. (2006) Annu. Rev. Genomics Hum. Genet. 7 103–124 [DOI] [PubMed] [Google Scholar]
- 3.Lin, J. H., Walter, P., and Yen, T. S. (2008) Annu. Rev. Pathol. 3 399–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radke, S., Chander, H., Schafer, P., Meiss, G., Kruger, R., Schulz, J. B., and Germain, D. (2008) J. Biol. Chem. 283 12681–12685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner, R. G., Nelson, Z. W., and Gottschling, D. E. (2005) Cell 120 803–815 [DOI] [PubMed] [Google Scholar]
- 6.Sayeed, A., and Ng, D. T. (2005) Crit. Rev. Biochem. Mol. Biol. 40 75–91 [DOI] [PubMed] [Google Scholar]
- 7.Nakatsukasa, K., and Brodsky, J. L. (2008) Traffic 9 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huyer, G., Piluek, W. F., Fansler, Z., Kreft, S. G., Hochstrasser, M., Brodsky, J. L., and Michaelis, S. (2004) J. Biol. Chem. 279 38369–38378 [DOI] [PubMed] [Google Scholar]
- 9.Taxis, C., Hitt, R., Park, S. H., Deak, P. M., Kostova, Z., and Wolf, D. H. (2003) J. Biol. Chem. 278 35903–35913 [DOI] [PubMed] [Google Scholar]
- 10.Hiller, M. M., Finger, A., Schweiger, M., and Wolf, D. H. (1996) Science 273 1725–1728 [DOI] [PubMed] [Google Scholar]
- 11.Loayza, D., Tam, A., Schmidt, W. K., and Michaelis, S. (1998) Mol. Biol. Cell 9 2767–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bordallo, J., Plemper, R. K., Finger, A., and Wolf, D. H. (1998) Mol. Biol. Cell 9 209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vashist, S., and Ng, D. T. (2004) J. Cell Biol. 165 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvalho, P., Goder, V., and Rapoport, T. A. (2006) Cell 126 361–373 [DOI] [PubMed] [Google Scholar]
- 15.Nakatsukasa, K., Huyer, G., Michaelis, S., and Brodsky, J. L. (2008) Cell 132 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neuber, O., Jarosch, E., Volkwein, C., Walter, J., and Sommer, T. (2005) Nat. Cell Biol. 7 993–998 [DOI] [PubMed] [Google Scholar]
- 17.Medicherla, B., Kostova, Z., Schaefer, A., and Wolf, D. H. (2004) EMBO Rep. 5 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richly, H., Rape, M., Braun, S., Rumpf, S., Hoege, C., and Jentsch, S. (2005) Cell 120 73–84 [DOI] [PubMed] [Google Scholar]
- 19.Jarosch, E., Taxis, C., Volkwein, C., Bordallo, J., Finley, D., Wolf, D. H., and Sommer, T. (2002) Nat. Cell Biol. 4 134–139 [DOI] [PubMed] [Google Scholar]
- 20.Ye, Y., Meyer, H. H., and Rapoport, T. A. (2001) Nature 414 652–656 [DOI] [PubMed] [Google Scholar]
- 21.McClellan, A. J., Scott, M. D., and Frydman, J. (2005) Cell 121 739–748 [DOI] [PubMed] [Google Scholar]
- 22.Park, S. H., Bolender, N., Eisele, F., Kostova, Z., Takeuchi, J., Coffino, P., and Wolf, D. H. (2007) Mol. Biol. Cell 18 153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hochstrasser, M., Ellison, M. J., Chau, V., and Varshavsky, A. (1991) Proc. Natl. Acad. Sci. U. S. A 88 4606–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hochstrasser, M., and Varshavsky, A. (1990) Cell 61 697–708 [DOI] [PubMed] [Google Scholar]
- 25.Chen, P., and Hochstrasser, M. (1995) EMBO J. 14 2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swanson, R., Locher, M., and Hochstrasser, M. (2001) Genes Dev. 15 2660–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen, P., Johnson, P., Sommer, T., Jentsch, S., and Hochstrasser, M. (1993) Cell 74 357–369 [DOI] [PubMed] [Google Scholar]
- 28.Deng, M., and Hochstrasser, M. (2006) Nature 443 827–831 [DOI] [PubMed] [Google Scholar]
- 29.Lenk, U., and Sommer, T. (2000) J. Biol. Chem. 275 39403–39410 [DOI] [PubMed] [Google Scholar]
- 30.Johnson, P. R., Swanson, R., Rakhilina, L., and Hochstrasser, M. (1998) Cell 94 217–227 [DOI] [PubMed] [Google Scholar]
- 31.Gilon, T., Chomsky, O., and Kulka, R. G. (2000) Mol. Cell. Biol. 20 7214–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravid, T., Kreft, S. G., and Hochstrasser, M. (2006) EMBO J. 25 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michaelis, S., and Herskowitz, I. (1988) Mol. Cell. Biol. 8 1309–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983) J Bacteriol 153 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldstein, A. L., and McCusker, J. H. (1999) Yeast 15 1541–1553 [DOI] [PubMed] [Google Scholar]
- 36.Huyer, G., Longsworth, G. L., Mason, D. L., Mallampalli, M. P., McCaffery, J. M., Wright, R. L., and Michaelis, S. (2004) Mol. Biol. Cell 15 908–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A., and Tsien, R. Y. (2002) Proc. Natl. Acad. Sci. U. S. A 99 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sikorski, R. S., and Hieter, P. (1989) Genetics 122 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujimura-Kamada, K., Nouvet, F. J., and Michaelis, S. (1997) J. Cell Biol. 136 271–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcusson, E. G., Horazdovsky, B. F., Cereghino, J. L., Gharakhanian, E., and Emr, S. D. (1994) Cell 77 579–586 [DOI] [PubMed] [Google Scholar]
- 41.Zhang, Y., Nijbroek, G., Sullivan, M. L., McCracken, A. A., Watkins, S. C., Michaelis, S., and Brodsky, J. L. (2001) Mol. Biol. Cell 12 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gandre, S., and Kahana, C. (2002) Biochem. Biophys. Res. Commun. 293 139–144 [DOI] [PubMed] [Google Scholar]
- 43.Jariel-Encontre, I., Bossis, G., and Piechaczyk, M. (2008) Biochim. Biophys. Acta [DOI] [PubMed]
- 44.Bazirgan, O. A., and Hampton, R. Y. (2008) J. Biol. Chem. 283 12797–12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brodsky, J. L., Werner, E. D., Dubas, M. E., Goeckler, J. L., Kruse, K. B., and McCracken, A. A. (1999) J. Biol. Chem. 274 3453–3460 [DOI] [PubMed] [Google Scholar]
- 46.Becker, J., Walter, W., Yan, W., and Craig, E. A. (1996) Mol. Cell. Biol. 16 4378–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cyr, D. M., Lu, X., and Douglas, M. G. (1992) J. Biol. Chem. 267 20927–20931 [PubMed] [Google Scholar]
- 48.Caplan, A. J., Tsai, J., Casey, P. J., and Douglas, M. G. (1992) J. Biol. Chem. 267 18890–18895 [PubMed] [Google Scholar]
- 49.Youker, R. T., Walsh, P., Beilharz, T., Lithgow, T., and Brodsky, J. L. (2004) Mol. Biol. Cell 15 4787–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jentsch, S., and Rumpf, S. (2007) Trends Biochem. Sci. 32 6–11 [DOI] [PubMed] [Google Scholar]
- 51.Raasi, S., and Wolf, D. H. (2007) Semin. Cell Dev. Biol. 18 780–791 [DOI] [PubMed] [Google Scholar]
- 52.Johnson, E. S., Ma, P. C., Ota, I. M., and Varshavsky, A. (1995) J. Biol. Chem. 270 17442–17456 [DOI] [PubMed] [Google Scholar]
- 53.Mullally, J. E., Chernova, T., and Wilkinson, K. D. (2006) Mol. Cell. Biol. 26 822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hrizo, S. L., Gusarova, V., Habiel, D. M., Goeckeler, J. L., Fisher, E. A., and Brodsky, J. L. (2007) J. Biol. Chem. 282 32665–32675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.den Boon, J. A., Chen, J., and Ahlquist, P. (2001) J. Virol. 75 12370–12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han, S., Liu, Y., and Chang, A. (2007) J. Biol. Chem. 282 26140–26149 [DOI] [PubMed] [Google Scholar]
- 57.Verges, E., Colomina, N., Gari, E., Gallego, C., and Aldea, M. (2007) Mol. Cell 26 649–662 [DOI] [PubMed] [Google Scholar]
- 58.Nishikawa, S. I., Fewell, S. W., Kato, Y., Brodsky, J. L., and Endo, T. (2001) J. Cell Biol. 153 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rothblatt, J. A., Deshaies, R. J., Sanders, S. L., Daum, G., and Schekman, R. (1989) J. Cell Biol. 109 2641–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider, H., Sollner, T., Dietmeier, K., Eckerskorn, C., Lottspeich, F., Trulzsch, B., Neupert, W., and Pfanner, N. (1991) Science 254 1659–1662 [DOI] [PubMed] [Google Scholar]
- 61.Levine, T., and Loewen, C. (2006) Curr. Opin. Cell Biol. 18 371–378 [DOI] [PubMed] [Google Scholar]
- 62.Gaigg, B., Simbeni, R., Hrastnik, C., Paltauf, F., and Daum, G. (1995) Biochim. Biophys. Acta 1234 214–220 [DOI] [PubMed] [Google Scholar]
- 63.Filippin, L., Magalhaes, P. J., Di Benedetto, G., Colella, M., and Pozzan, T. (2003) J. Biol. Chem. 278 39224–39234 [DOI] [PubMed] [Google Scholar]
- 64.Wilson, J. D., Liu, Y., Bentivoglio, C. M., and Barlowe, C. (2006) Traffic 7 1213–1223 [DOI] [PubMed] [Google Scholar]
- 65.Schuberth, C., and Buchberger, A. (2005) Nat. Cell Biol. 7 999–1006 [DOI] [PubMed] [Google Scholar]
- 66.Verma, R., Oania, R., Graumann, J., and Deshaies, R. J. (2004) Cell 118 99–110 [DOI] [PubMed] [Google Scholar]
- 67.Schuberth, C., Richly, H., Rumpf, S., and Buchberger, A. (2004) EMBO Rep. 5 818–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Du, Y., Ferro-Novick, S., and Novick, P. (2004) J. Cell Sci. 117 2871–2878 [DOI] [PubMed] [Google Scholar]
- 69.Enenkel, C., Lehmann, A., and Kloetzel, P. M. (1998) EMBO J. 17 6144–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalies, K. U., Allan, S., Sergeyenko, T., Kroger, H., and Romisch, K. (2005) EMBO J. 24 2284–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cyr, D. M. (1995) FEBS Lett. 359 129–132 [DOI] [PubMed] [Google Scholar]
- 72.Lu, Z., and Cyr, D. M. (1998) J. Biol. Chem. 273 27824–27830 [DOI] [PubMed] [Google Scholar]
- 73.Becker, J., and Craig, E. A. (1994) Eur. J. Biochem. 219 11–23 [DOI] [PubMed] [Google Scholar]
- 74.Nathan, D. F., and Lindquist, S. (1995) Mol. Cell. Biol. 15 3917–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heinemeyer, W., Simeon, A., Hirsch, H. H., Schiffer, H. H., Teichert, U., and Wolf, D. H. (1991) Biochem. Soc. Trans. 19 724–725 [DOI] [PubMed] [Google Scholar]
- 76.Nelson, R. J., Ziegelhoffer, T., Nicolet, C., Werner-Washburne, M., and Craig, E. A. (1992) Cell 71 97–105 [DOI] [PubMed] [Google Scholar]
- 77.Ahner, A., Nakatsukasa, K., Zhang, H., Frizzell, R. A., and Brodskv, J. L. (2007) Mol. Biol. Cell. 18 806–814 [DOI] [PMC free article] [PubMed] [Google Scholar]