Abstract
The JIL-1 histone H3S10 kinase in Drosophila localizes specifically to euchromatic interband regions of polytene chromosomes and is enriched 2-fold on the male X chromosome. JIL-1 can be divided into four main domains including an NH2-terminal domain, two separate kinase domains, and a COOH-terminal domain. Our results demonstrate that the COOH-terminal domain of JIL-1 is necessary and sufficient for correct chromosome targeting to autosomes but that both COOH- and NH2-terminal sequences are necessary for enrichment on the male X chromosome. We furthermore show that a small 53-amino acid region within the COOH-terminal domain can interact with the tail region of histone H3, suggesting that this interaction is necessary for the correct chromatin targeting of the JIL-1 kinase. Interestingly, our data indicate that the COOH-terminal domain alone is sufficient to rescue JIL-1 null mutant polytene chromosome defects including those of the male X chromosome. Nonetheless, we also found that a truncated JIL-1 protein which was without the COOH-terminal domain but retained histone H3S10 kinase activity was able to rescue autosome as well as partially rescue male X polytene chromosome morphology. Taken together these findings indicate that JIL-1 may participate in regulating chromatin structure by multiple and partially redundant mechanisms.
The JIL-1 tandem kinase is a multidomain protein that localizes specifically to euchromatic chromosome regions, phosphorylates histone H3S10 at interphase, and is enriched almost 2-fold on the transcriptionally hyperactive male X chromosome (1–3). Furthermore, the JIL-1 kinase has been functionally implicated in counteracting heterochromatization and gene silencing and is required for maintaining proper chromosome morphology (4–8). In polytene autosomes loss of JIL-1 leads to misalignment of interband chromatin fibrils and to increased ectopic contacts between nonhomologous regions (7). Furthermore, there is an abnormal coiling of the chromosomes with an intermixing of euchromatic regions and the compacted chromatin characteristic of banded regions. Especially affected by loss of JIL-1 is the male X chromosome where chromatin is dispersed into a diffuse network without any discernable banded regions that leads to a characteristic “puffed” appearance (7).
JIL-1 can be divided into four main domains including an NH2-terminal domain (NTD),2 the first kinase domain (KDI), the second kinase domain (KDII), and a COOH-terminal domain (CTD) (1). Interestingly, mutations resulting in truncations of the COOH-terminal domain of JIL-1 lead to chromatin mislocalization of the protein (5) and give rise to some of strongest suppressor-of-variegation (Su(var)) phenotypes yet described of the wm4 allele (4). Thus, to determine which sequences function to localize JIL-1 to chromatin and to enrich it on the male X chromosome, we have undertaken a domain analysis of the JIL-1 protein by expressing deletion constructs of JIL-1 transgenically in JIL-1 null mutant flies. Our results demonstrate that the CTD of JIL-1 is necessary and sufficient for correct chromosome targeting to autosomes but that both COOH- and NH2-terminal sequences are necessary for enrichment on the male X chromosome. In addition, the CTD has an unusual organization, being highly acidic in its first half (pI < 4) and highly basic in its second half (pI > 11), the latter of which contains a predicted globular tertiary structure (9). We show by in vitro deletion construct analysis that a small 53-amino acid region of the putative CTD globular domain specifically binds to the tail region of histone H3. Thus, our findings indicate that this sequence within the COOH-terminal region of JIL-1 represents a novel histone H3 binding domain that is required for the correct localization of the JIL-1 kinase to chromatin.
MATERIALS AND METHODS
JIL-1 GFP/CFP- and V5-tagged Fusion Constructs—A full-length JIL-1 (1–1207) construct (JIL-1-FL), a NH2-terminal domain construct of JIL-1 from residue 1–260 (NTD), a KDI/KDII construct containing the two kinase domains (255–888), and a NTD/CTD construct containing most of the NH2-terminal domain (1–190) and the COOH-terminal domain (927–1207) in tandem were cloned into the pUAST vector with in-frame V5 tags (V5-pMT vector, Invitrogen) at the COOH termini using standard methods (10). Similarly, a ΔCTD construct from residue 1–926 with an in-frame GFP tag at the NH2 terminus as well as a CTD construct containing sequences from amino acids 927–1207 with an in-frame CFP tag were cloned into the pUAST vector. For those constructs not containing the endogenous JIL-1 nuclear localization sequence (NLS) that is situated in the NH2-terminal domain (1), the NLS from NLS-pECFP vector (Clontech) was added to the NH2 terminus. The fidelity of all constructs was verified by sequencing at the Iowa State University Sequencing facility.
Drosophila melanogaster Stocks—Fly stocks were maintained at 23 °C according to standard protocols (11). Canton-S was used for wild type preparations. The JIL-1z2 null allele is described in Wang et al. (3) as well as in Zhang et al. (12). The GFP-JIL-1 line (P[hsp83-GFP-JIL-1, w+]) has been previously characterized in Jin et al. (1) and in Wang et al. (3). JIL-1 construct pUAST lines were generated by standard P-element transformation (BestGene, Inc.), and expression of the transgenes were driven using the hsp70-GAL4 (P{w[+mC] = GAL4-hsp70.PB}) driver (obtained from the Bloomington Stock Center) introduced by standard genetic crosses. The hsp83 and hsp70 promoters are leaky (1, 13) and were used without heat shock treatment. Expression levels of each of the JIL-1 constructs were monitored by immunoblot analysis as described below. Balancer chromosomes and markers are described in Lindsley and Zimm (14).
Immunohistochemistry—Standard polytene chromosome squash preparations were performed as in Kelley et al. (15) using the 5-min fixation protocol, and acid-free squash preparations were done following the procedure of DiMario et al. (16). Antibody labeling of these preparations was performed as described in Jin et al. (1) and in Wang et al. (3). In most preparations the male X chromosome was identified by double labeling with MSL antibody as previously described (2). Primary antibodies used in this study include rabbit anti-H3S10ph (Cell Signaling), rabbit anti-histone H3 (Cell Signaling), rabbit anti-GFP (Invitrogen), chicken anti-GFP (Aves Labs), anti-Chromator mAb 6H11 (17), rabbit anti-MSL-2 (generous gift of Dr. M. Kuroda, Harvard University, Boston), anti-GST mAb 8C7 (17), anti-V5 mAb (Invitrogen), anti-Myc mAb 9E10 (Developmental Studies Hybridoma Bank, University of Iowa), mouse anti-lamin Dm0 (9), rabbit anti-JIL-1 (1), chicken anti-JIL-1 (2), and anti-JIL-1 mAb 5C9 (2). DNA was visualized by staining with Hoechst 33258 (Molecular Probes) in PBS. The appropriate species- and isotype-specific Texas Red-, TRITC-, and fluorescein isothiocyanate-conjugated secondary antibodies (Cappel/ICN, Southern Biotech) were used (1:200 dilution) to visualize primary antibody labeling. The final preparations were mounted in 90% glycerol containing 0.5% n-propyl gallate. The preparations were examined using epifluorescence optics on a Zeiss Axioskop microscope, and images were captured and digitized using a high resolution Spot CCD camera. Images were imported into Photoshop where they were pseudocolored, image-processed, and merged. In some images nonlinear adjustments were made to the channel with Hoechst labeling for optimal visualization of chromosomes. For quantification of the relative levels of antibody staining of X chromosomes and autosomes, images of polytene chromosome squashes were analyzed using the NIH Image software as previously described (1). In short, the staining intensity of the autosomes from male preparations was averaged and normalized to a value of one and compared with the average pixel intensity for that of the X chromosome.
Immunoblot Analysis—Protein extracts were prepared from whole third instar larvae or in some experiments from dissected salivary glands homogenized in a buffer containing 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 10 mm EDTA, 1 mm EGTA, 0.2% Triton X-100, 0.2% Nonidet P-40, 2 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 1.5 μg/ml aprotinin. Proteins were separated by SDS-PAGE according to standard procedures (10). Electroblot transfer was performed as in Towbin et al. (18) with transfer buffer containing 20% methanol and in most cases including 0.04% SDS. For these experiments we used the Bio-Rad Mini PROTEAN II system, electroblotting to 0.2-μm nitrocellulose, and using anti-mouse or anti-rabbit HRP-conjugated secondary antibody (Bio-Rad) (1:3000) for visualization of primary antibody. Antibody labeling was visualized using chemiluminescent detection methods (SuperSignal West Pico Chemiluminescent substrate, Pierce). The immunoblots were digitized using a flatbed scanner (Epson Expression 1680).
Pulldown Assays—In pulldown assays GST fusion proteins containing JIL-1 sequences were used to pull down endogenous nuclear proteins from S2 cell lysates as previously described (9). In addition, pulldown experiments were performed with histone H3 GST fusion proteins of lysates from an S2 cell line stably transfected with a Myc-tagged CTD domain of JIL-1 (927–1207). This construct was subcloned into the pMT/V5-His vector (Invitrogen) with an NH2-terminal insertion of a 6-Myc tag from the 6-CMYC clone CD3-128 (Arabidopsis Biological Resource Center) as well as of a nuclear localization sequence. Initially the JIL-1 GST fusion proteins CTD-A (886–1033), CTD-B (1033–1207), and CTD-G (1065–1196), which have been previously described (9), and a full-length histone H3 (1–135) construct that was reverse transcription-PCR-amplified from mRNA extractions from S2 cells and cloned into the pGEX4T vector (Amersham Biosciences) were expressed in Escherichia coli using standard techniques (10). For subsequent experiments, GST fusion protein constructs with various truncations of histone H3 and the COOH-terminal domain of JIL-1 were generated by PCR amplification and insertion into the pGEX4T vector. The additional histone H3 GST fusion proteins were GST-H3-T (1–74) and GST-H3-C (56–135), and the additional JIL-1 GST fusion proteins were CTD-G1 (1105–1196), CTD-G2 (1105–1144), and CTD-G3 (1144–1196). All GST fusion protein constructs were verified by sequencing. The GST tag of all constructs was added to the NH2 terminus. For the in vitro protein-protein interaction assays, ∼2 μg of the appropriate GST fusion protein was coupled with glutathione-agarose beads and incubated with 300 μl of S2 cell lysate (from 107 cells/ml) at 4 °C overnight on a rotating wheel. The beads were washed 4 times for 10 min each in 1 ml of PBS with 0.5% Tween 20, and proteins retained on the glutathione-agarose beads were analyzed by SDS-PAGE and immunoblotting as previously described (9).
Overlay Experiments—The truncated GST-JIL-1 fusion proteins GST-NTD (1–211) and GST-CTD (927–1207) have been previously described (2, 9). The JIL-1 GST fusion protein CTD-G3 (1144–1196) is described above. The respective GST fusion proteins were expressed in BL21 cells and purified over a glutathione agarose column (Sigma-Aldrich) according to the pGEX manufacturer's instructions (Amersham Biosciences). For the overlay interaction assays, either bovine histones (Worthington), individually purified bovine histones (Roche Applied Science), or Drosophila histone extractions from S2 cells performed as described below were fractionated by SDS-PAGE and electroblotted to nitrocellulose. The blots were subsequently incubated with ∼2 μg of either GST-NTD, GST-CTD, or CTD-G3 GST fusion protein overnight at 4 °C in PBS with 0.5% Tween 20 and 5% nonfat milk on a rotating wheel. The blots were washed 4 times for 10 min each in PBS with 0.5% Tween 20, and binding was detected by anti-GST mAb 8C7 (17).
Histone Extraction from Drosophila S2 Cells—10 ml of S2 cell culture containing ∼107 cells/ml was pelleted by slow spinning and washed once with 5 ml of ice-cold PBS. The re-pelleted cells were resuspended with 2 ml of lysis buffer (10 mm Tris-HCL, pH 8.0, 1 mm KCl, 1.5 mm MgCl2, 0.5% Nonidet P-40, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1.5 μg/ml aprotinin) and incubated on ice for 30 min. The nuclei were collected by spinning at 5000 rpm for 10 min at 4 °C, resuspended in 0.4 ml 0.4 N H2SO4, and incubated on a rotating wheel at 4 °C for 30 min to overnight. After incubation, the supernatant was collected by spinning at 13,200 rpm for 10 min, and histones were precipitated by adding trichloroacetic acid (100%) to a final concentration of 20% and incubating on ice for 30 min. The precipitated histone proteins were harvested by spinning at 13,200 rpm for 10 min, washed twice with cold acetone (-20 °C), air dried, and resuspended in the appropriate amount of H2O.
RESULTS
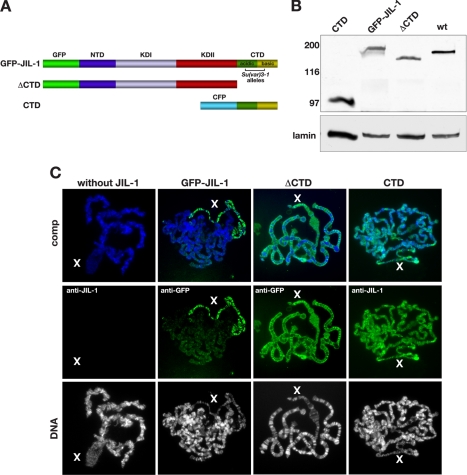
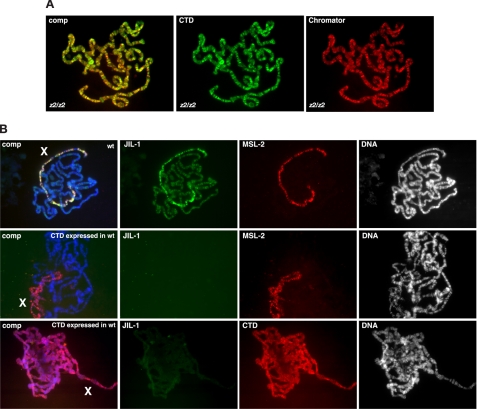
The CTD Domain Is Required for Proper Targeting of JIL-1 to Chromatin—Recently, Ebert et al. (4) mapped six JIL-1Su(var)3-1 alleles to a small region in the JIL-1 COOH-terminal domain (Fig. 1A). The mutations in these alleles lead to COOH-terminal-truncated proteins that mislocalize to ectopic chromatin sites and are not enriched on the male X chromosome (4, 5) implying an important function for the CTD domain in proper chromatin localization of JIL-1. To further explore the role of the different JIL-1 domains in chromosome targeting, we expressed three GFP- or CFP-tagged JIL-1 pUAST constructs transgenically in JIL-1 null mutant animals; a full-length construct (GFP-JIL-1), a construct without the COOH-terminal domain (ΔCTD), and a construct containing only the COOH-terminal region (CTD) (Fig. 1A). In these studies a hsp70-GAL4 driver line was used without heat shock. The hsp70 promoter is leaky (13) and promoted sufficient construct expression at or near wild type levels (Fig. 1B). As previously reported (3) expression of a full-length GFP-JIL-1 construct in a JIL-1 null mutant background rescued all aspects of the mutant polytene chromosome phenotype, and like endogenous JIL-1, GFP-JIL-1 was enriched on the male X chromosome (Fig. 1C, Table 1). Interestingly, as illustrated in Fig. 1C, we found that expression of the CTD domain alone was sufficient for localization to chromatin and for restoration of polytene chromosome morphology to or near that of wild type preparations. That the CTD localized to the same interband chromatin sites normally occupied by full-length JIL-1 was further indicated by double labeling studies with antibody to Chromator, a chromosomal protein that is specific to interband polytene regions in a pattern identical to that of endogenous JIL-1 (17, 19). Fig. 2A shows such a double-labeled polytene squash preparation where CTD localization is indicated in green, Chromator localization in red, and where co-localization is indicated by the predominantly yellow regions in the composite image. At some interband locations in the composite image the color is more green or orange than pure yellow as a result of slight differences in the relative staining intensity of the two antibodies, which can vary from preparation to preparation. We also expressed the CTD domain in a wild type background. As illustrated in Fig. 2B, under these conditions detection of endogenous JIL-1 on the polytene chromosomes was significantly reduced, and enrichment was no longer observed on the male X chromosome. However, localization of the MSL complex as detected by MSL-2 antibody labeling was not affected (Fig. 2B, middle panel). These findings suggest that the CTD domain competed with endogenous JIL-1 for the same chromatin binding sites.
FIGURE 1.
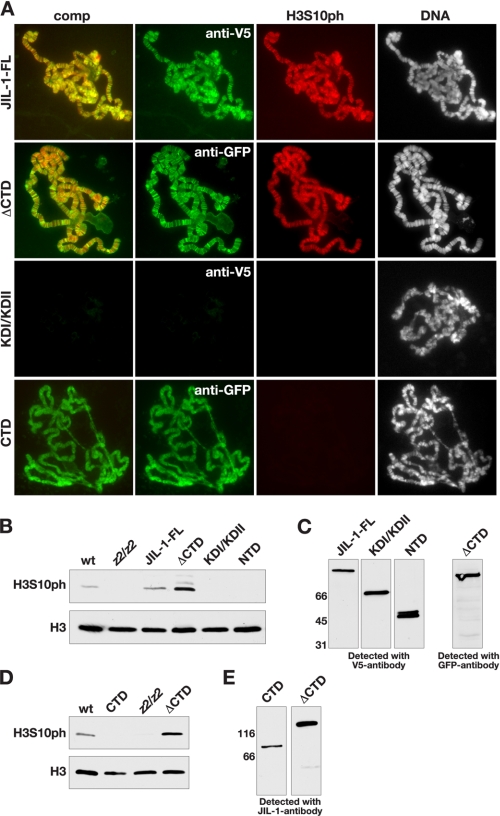
Expression of JIL-1 deletion constructs transgenically in a JIL-1 null mutant background. A, diagrams of the JIL-1 GFP- or CFP-tagged constructs analyzed. The region in the CTD where JIL-1Su(var)3-1 alleles, resulting in COOH-terminally truncated proteins have been mapped (4), is indicated by a bracket. B, immunoblot labeled with JIL-1 antibody of protein extracts from wild type (wt) and from JIL-1 null larvae expressing GFP-JIL-1, CTD, and ΔCTD. Labeling with lamin antibody was used as a loading control. The relative migration of molecular size markers is indicated to the left of the immunoblot in kDa. C, polytene chromosome squash preparations from male JIL-1z2/JIL-1z2 third instar larval salivary glands expressing GFP-JIL-1, ΔCTD, or CTD. The left panel shows a squash preparation from a JIL-1 null mutant larvae for comparison. Protein localization (in green) was identified using either JIL-1 (anti-JIL-1) or GFP (anti-GFP) antibodies, whereas DNA (in blue or gray) was labeled by Hoechst. The male X chromosome is indicated by an X.
TABLE 1.
Properties of JIL-1 constructs expressed in a JIL-1z2/JIL-1z2 null background
| Construct | Localization to chromatin | Enrichment on male X chromosome | H3S10 phosphorylation | Rescue of autosome morphology | Rescue of male X chromosome morphology |
|---|---|---|---|---|---|
| GFP-JIL-1 | Yes | Yes | Yes | Yes | Yes |
| JIL-1-FL | Yes | Yes | Yes | Yes | Yes |
| CTD | Yes | No | No | Yes | Yes |
| ΔCTD | Yesa | No | Yes | Yes | Partial |
| KDI/KDII | No | No | No | Partial | No |
| NTD | Yesa | No | No | No | No |
| NTD/CTD | Yes | Yes | No | Partial | Partial |
Ectopic chromatin localization
FIGURE 2.
Expression of the CTD construct in JIL-1 null and in wild type larvae. A, double labeling of a female polytene squash preparation from a JIL-1z2/JIL-1z2 third instar larvae expressing the CTD construct. The CTD (in green) was labeled with GFP antibody, and the interband specific protein Chromator (in red) was labeled with mAb 6H11. The composite image (comp) shows the co-localization between the CTD, and Chromator as indicated by the predominantly yellow color. B, expression of the CTD in wild type (wt) larvae dramatically reduced chromosomal levels of endogenous JIL-1 and prevented enrichment on the male X chromosome (X). The upper panel shows the distribution of JIL-1 and the enrichment of JIL-1 on the male X chromosome (X) in a control wild type polytene squash preparation. The labeling of endogenous JIL-1 was with a JIL-1 NH2-terminal domain-specific antibody that does not recognize the CTD domain. The polytene chromosome squash preparations in the upper and middle panels were double-labeled with MSL-2 antibody to show MSL complex localization and with GFP antibody in the lower panel to indicate the distribution of CTD-CFP. The middle and lower panels are from independent experiments.
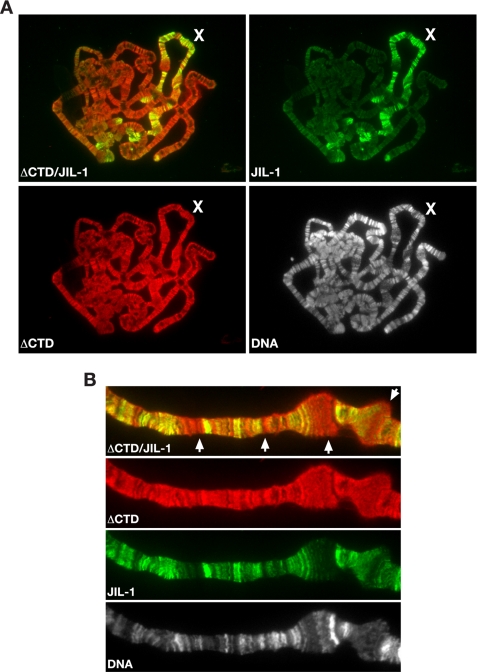
For comparison, we next expressed the ΔCTD construct, which is without the CTD domain in JIL-1 null mutant larvae. As illustrated in Fig. 1C, the ΔCTD was also able to fully rescue autosome morphology in polytene squash preparations. However, in most cases the morphology of the male X chromosome was only partially restored with remnants of puffed regions (Fig. 1C). In addition, it should be noted that autosome chromatin morphology in the polytene squash preparations was “better” than normal when the ΔCTD construct was expressed in JIL-1 mutant (Fig. 1C) as well as in wild type (Fig. 3) backgrounds. In such preparations, the chromosomes spread very easily and were wider with a more crisp and well resolved banding pattern as compared with typical wild type preparations. Furthermore, although the antibody labeling demonstrated that the ΔCTD was localized to chromatin (Fig. 1A), its distribution was broader and present at numerous ectopic locations (Fig. 3B) as compared with the CTD. This was especially evident in polytene squash preparations from third instar larvae where the ΔCTD construct was expressed in a wild type background (Fig. 3). In these preparations the distribution of the ΔCTD was determined by labeling with antibody to its GFP tag, and endogenous JIL-1 was determined using JIL-1 antibody (2) to a COOH-terminal epitope not present in the ΔCTD (Fig. 3). Furthermore, the antibody labeling indicated that the ΔCTD, similar to the CTD, does not show enrichment on the male X chromosome (Figs. 1C and 3A). However, unlike the CTD, ΔCTD did not appear to have any effect on the endogenous JIL-1 chromatin distribution including on the male X chromosome (Fig. 3A). This indicates that the binding sites and/or mechanisms for chromatin association of the CTD and ΔCTD are different.
FIGURE 3.
Expression of the ΔCTD construct in wild type larvae. A, polytene squash preparation from a wild type larvae expressing the ΔCTD construct. The ΔCTD (in red) was labeled with GFP antibody, endogenous JIL-1 (in green) was labeled with JIL-1 antibody to a COOH-terminal epitope not present in the ΔCTD, and the DNA (in gray) was labeled with Hoechst. The male X chromosome is indicated by an X. B, higher magnification image of a region of a male X chromosome from a preparation similar to that in A showing the extensive ectopic localization of ΔCTD (arrows).
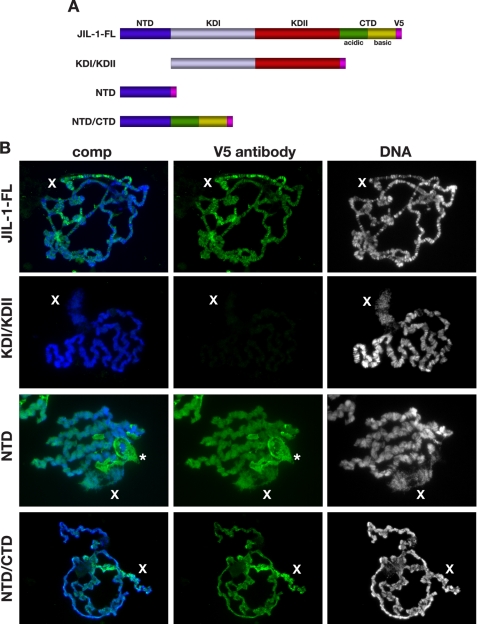
Both NTD and CTD Domain Sequences Are Required for JIL-1 Enrichment on the Male X Chromosome—The findings that neither CTD nor ΔCTD was enriched on the male X chromosome suggested that contributions from more than one JIL-1 domain are needed for this enrichment to occur. To explore this possibility, we generated a series of V5-tagged JIL-1 pUAST constructs and expressed them transgenically in JIL-1 mutant backgrounds. These constructs are diagrammed in Fig. 4A and included a full-length JIL-1 construct (JIL-1-FL), a construct containing the two kinase domains (KDI/KDII), a construct containing only the NH2-terminal domain (NTD), as well as a construct where the NH2- and COOH-terminal domains were fused together (NTD/CTD). The criteria for assessment of chromosome morphology rescue were based on the degree to which autosomes were uncoiled, on the degree to which “puffing” of the male X chromosome was reduced, as well as on the extent of restoration of discrete band/interband regions of the chromosome arms. As illustrated in Fig. 4B, JIL-1-FL had the same properties as GFP-JIL-1 and rescued all aspects of the JIL-1 null polytene chromosome phenotype (Table 1). However, KDI/KDII had no detectable localization to chromosomes although partial rescue of autosome coiling and restoration of some band/interband regions were observed (Fig. 4B). Interestingly, in contrast to the autosomes, there was no improvement in male X chromosome morphology (Fig. 4B). Furthermore, we found that like the CTD, the NTD could associate with chromatin; however, the localization was relatively evenly distributed on the chromosome arms, and there was no discernable rescue of either autosome or male X chromosome morphology (Fig. 4B). Nonetheless, we found that the construct bringing together the NTD and CTD domains (NTD/CTD) was sufficient both for chromosome localization to interband regions and for enrichment on the male X chromosome (Fig. 4B). To quantify the level of this enrichment, we determined the average pixel intensity of NTD/CTD immunostaining on the male X chromosome and compared it to the autosomal staining intensity, which was normalized to a value of 1.0. In five polytene chromosome squashes the difference in X chromosome NTD/CTD staining intensity compared with that of autosomes was 1.8 ± 0.3, identical to the value of 1.8 previously determined for male wild type polytene chromosomes by Jin et al. (1). Interestingly, chromosome morphology was only partially restored by the NTD/CTD in contrast to the full rescue by the CTD domain alone (Fig. 4B). This suggests that the addition of the NTD domain without the kinase domains in some way interfered with the rescue of chromosome morphology provided by the CTD domain alone.
FIGURE 4.
Expression of JIL-1 deletion constructs transgenically in a JIL-1 null mutant background. A, diagrams of the JIL-1 V5-tagged constructs analyzed. B, polytene chromosome squash preparations from male JIL-1z2/JIL-1z2 third instar larval salivary glands expressing JIL-1-FL, KDI/KDII, NTD, and NTD/CTD. Protein localization (in green) was identified using V5 antibody whereas DNA (in blue or gray) was labeled by Hoechst. The male X chromosome is indicated by an X. The asterisk indicates labeling of NTD in the nucleolus. comp, composite image.
Histone H3S10 Phosphorylation by JIL-1 Deletion Proteins—To test whether the histone H3S10 residue could be phosphorylated when the three JIL-1 deletion proteins ΔCTD, CTD, and KDI/KDII were expressed, we labeled acid-free polytene squash preparations (16, 20) with H3S10ph antibody. As illustrated in Fig. 5A, H3S10 phosphorylation was detected by the H3S10ph antibody in an overlapping pattern with that of ΔCTD or JIL-1-FL but not with KDI/KDII or CTD. These results were further corroborated by immunoblot analysis, which showed that JIL-1-FL and ΔCTD were able to phosphorylate H3S10, whereas KDI/KDII, CTD, and NTD were not (Fig. 5, B and D). Fig. 5, C and E, show immunoblots of the JIL-1-FL, ΔCTD, KDI/KDII, CTD, and NTD proteins assayed for H3S10 phosphorylation activity in Fig. 5, B and D. Although H3S10 phosphorylation by JIL-1-FL was slightly above wild type levels, phosphorylation of H3S10 by the ΔCTD construct was considerably more robust, possibly reflecting additional phosphorylation activity at the ectopic sites.
FIGURE 5.
Immunocytochemical and immunoblot analysis of histone H3S10 phosphorylation in JIL-1 null larvae expressing JIL-1 transgenes. A, acid-free polytene chromosome squash preparations from female JIL-1z2/JIL-1z2 third instar larval salivary glands expressing JIL-1-FL, ΔCTD, KΔI/KΔII, and CTD. Protein localization (in green) was identified using V5 (anti-V5) or GFP (anti-GFP) antibody, histone H3 phosphorylation (in red) was identified using H3S10ph antibody, and DNA (in blue or gray) was labeled by Hoechst. B, immunoblots labeled with H3S10ph antibody of protein extracts from salivary glands from wild type (wt) and JIL-1z2/JIL-1z2 (z2/z2) larvae as well as from JIL-1 null larvae expressing JIL-1-FL, ΔCTD, KDI/KDII, and NTD. Labeling with histone H3 (H3) antibody was used as a loading control. C, immunoblots of the JIL-1-FL, ΔCTD, KDI/KDII, and NTD proteins assayed for H3S10 phosphorylation activity in B as detected by V5 and GFP antibody. The relative migration of molecular size markers is indicated to the left of the immunoblots in kDa. D, immunoblots labeled with H3S10ph antibody of protein extracts from salivary glands from wild type and JIL-1z2/JIL-1z2 (z2/z2) larvae as well as from JIL-1 null larvae expressing CTD and ΔCTD, respectively. Labeling with histone H3 (H3) antibody was used as a loading control. E, immunoblots of the CTD and ΔCTD proteins assayed for H3S10 phosphorylation activity in D as detected by JIL-1 antibody. The relative migration of molecular size markers is indicated to the left of the immunoblots in kDa.
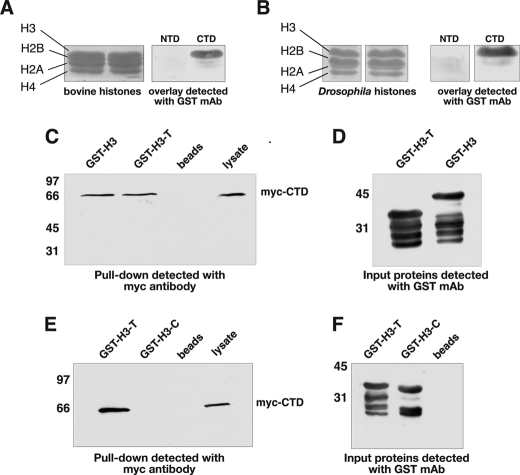
The CTD Domain of JIL-1 Interacts with the Tail Region of Histone H3—The polytene chromosome localization studies of the various JIL-1 domains indicated that both the CTD and the NTD may have the ability to bind to chromatin. Major constituents of chromatin include the four histones H2A, H2B, H3, and H4 that together with DNA form nucleosomes (for review, see Ref. 21). We, therefore, used overlay assays to test for interactions between these chromatin components and the NTD and CTD domains. For the initial screening we used two GST fusion proteins with the NTD (GST-NTD) and CTD (GST-CTD) domains of JIL-1. In the overlay assays purified bovine histones and Drosophila histone extracts from S2 cells were fractionated by SDS-PAGE, transferred to nitrocellulose paper, and incubated with glutathione agarose bead-purified GST-NTD and GST-CTD. Protein interactions were detected with a mAb to GST. As illustrated in Fig. 6, A and B, we found that the CTD domain, but not the NTD domain, could specifically interact with both bovine and Drosophila histone H3 in these assays. To further verify this interaction and to determine whether tail or core regions of histone H3 were involved, we performed additional pulldown assays. For these experiments we generated a Myc-tagged CTD construct (Myc-CTD) that was stably expressed in Drosophila S2 cells. In addition, we generated GST fusion proteins of full-length histone H3 (GST-H3), the tail region of histone H3 (GST-H3-T), and the core region (GST-H3-C). In the pulldown experiments the three histone H3 GST fusion proteins were coupled to glutathione-agarose beads, incubated with lysate from Myc-CTD-expressing S2 cells, washed, fractionated by SDS-PAGE, and analyzed by immunoblot analysis using a Myc-specific antibody. Whereas the beads-only controls and GST-H3-C showed no pulldown activity, both GST-H3 and GST-H3-T were able to pull down Myc-CTD as detected by Myc antibody (Fig. 6, C and E). Immunoblot blot analysis of the GST proteins purified in these experiments and detected with GST antibody showed that approximately equivalent levels of the three fusion proteins were present in these assays (Fig. 6, D and F). Taken together these experiments indicate that the CTD domain of JIL-1 can directly interact with the tail region of histone H3.
FIGURE 6.
The CTD domain of JIL-1 interacts with the tail region of histone H3. In overlay experiments purified bovine histones (A) and Drosophila histone extractions from S2 cells (B) were fractionated by SDS-PAGE, immunoblotted, and incubated with JIL-1 NTD or CTD GST fusion protein, and interactions were detected with a GST mAb (right panels). Ponceau S labeling of the fractionated histone proteins is shown in the left panels. In pulldown experiments (C and E) lysate from S2 cells stably expressing an Myc-tagged CTD domain of JIL-1 (Myc-CTD) and incubated with GST-histone H3 fusion constructs (GST-H3, GST-H3-T, and GST-H3-C) or with beads-only was pelleted with glutathione-agarose beads, and the interacting proteins were fractionated by SDS-PAGE, immunoblotted, and probed with Myc antibody. Unincubated S2 cell lysate was included as a control (right lanes). In these experiments interactions between JIL-1 Myc-CTD and GST-H3 as well as GST-H3-T were detected but not with GST-H3-C. D and F, immunoblots of the GST fusion proteins used for the pulldown experiments in C and E were detected with GST antibody. The relative migration of molecular size markers is indicated to the left of the immunoblots in kDa.
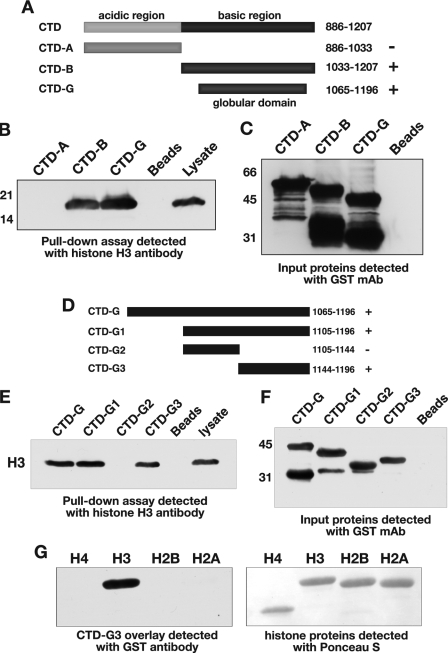
The region of JIL-1 that was found to interact with histone H3, the JIL-1 CTD domain, can be further divided into two distinct regions; that is, an acidic region from residue 887–1033 and a basic region from residue 1034–1207 (Fig. 7A). Furthermore, Bao et al. (9) identified a putative globular domain present within the basic region that spans residues 1065–1196. Thus, to better define the sequences of JIL-1 responsible for the molecular interaction between JIL-1 and histone H3, we generated GST fusion proteins comprising these three regions, CTD-A, CTD-B, and CTD-G (Fig. 7A) and performed pull-down experiments of proteins from S2 cell lysate as described above. As shown in Fig. 7B, both the CTD-B and CTD-G fusion proteins pulled down a 17-kDa protein detected by histone H3 antibody that was also present in the S2 cell lysate. This band was not present in pulldown assays with the CTD-A fusion protein or in beads-only controls (Fig. 7B). Immunoblot analysis of each of the input GST fusion proteins probed with anti-GST antibody showed comparable levels of GST fusion proteins in each of the pulldown assays (Fig. 7C). To further refine the region of the CTD domain responsible for the interaction with histone H3, we generated three additional GST fusion proteins spanning different regions of the globular domain, CTD-G1, CTD-G2, and GST-G3, as diagrammed in Fig. 7D. As shown in Fig. 7E, the CTD-G, CTD-G1, and CTD-G3 fusion proteins all pulled down histone H3 as a 17-kDa protein detected by H3 antibody. This band was not present in pulldown assays with the CTD-G2 fusion protein or in beads-only controls (Fig. 7E). Immunoblot analysis of each of the input GST fusion proteins probed with anti-GST antibody showed comparable levels of GST fusion proteins in each of the pulldown assays (Fig. 7F). We further verified that the interaction of CTD-G3 was specific to histone H3 in overlay assays as described above, only in this case separately purified bovine histone subtypes were used. As illustrated in Fig. 7G, CTD-G3 bound only to histone H3 on immunoblots as detected with GST antibody. These results suggest that a small 53-amino acid region (amino acids 1144–1196) of the prospective globular domain of the COOH-terminal domain of JIL-1 is sufficient for mediating molecular interactions with the tail region of histone H3.
FIGURE 7.
Mapping of the JIL-1 COOH-terminal interaction domain with histone H3. A and D, diagrams of the JIL-1 COOH-terminal domains to which GST fusion proteins were made for mapping. In pulldown experiments (B and E) lysate from S2 cells incubated with the various GST fusion constructs (CTD-A, CTD-B, CTD-G, CTD-G1, CTD-G2, and CTD-G3) or with beads-only was pelleted with glutathione-agarose beads, and the interacting protein(s) was fractionated by SDS-PAGE, immunoblotted, and probed with histone H3 antibody. Un-incubated S2 cell lysate was included as a control (right lanes). C and F, immunoblots of the GST fusion proteins used for the pulldown experiments in B and E were detected with GST antibody. The relative migration of molecular size markers is indicated to the left of the immunoblots in kDa. G, in overlay experiments individually purified bovine histones were fractionated by SDS-PAGE, immunoblotted, and incubated with the JIL-1 CTD-G3 GST fusion protein, and interactions were detected with a GST mAb (left panel). Ponceau S labeling of the fractionated histone proteins is shown in the right panel.
DISCUSSION
In this study we show that the CTD domain of the JIL-1 kinase is required for proper localization to chromatin and that chromosome morphology defects observed in JIL-1 null backgrounds can be fully rescued by expression of this domain. We furthermore provide evidence that a small 53-amino acid region within the CTD domain can interact with the tail-region of histone H3, suggesting that this interaction is necessary for the correct chromatin targeting of the JIL-1 kinase. This is supported by the findings that constructs (ΔCTD, this study) and mutations (JIL-1Su(var)3-1 alleles (4, 5)) that delete this region in the CTD result in mislocalization of the protein to ectopic chromatin sites. Nonetheless, that localization to chromatin still occurs in the absence of the CTD indicates that JIL-1 possesses a second mechanism for chromatin association, and we provide evidence that this association is mediated by sequences residing in the NTD. In contrast, KDI/KDII, which contained only the two kinase domains without the NTD and CTD sequences, did not show any detectable localization to chromatin.
Surprisingly, our data indicate that histone H3S10 phosphorylation is not required for maintaining proper polytene chromosome morphology and band/interband organization and that the CTD alone is sufficient to rescue JIL-1 null mutant polytene chromosome defects including those of the male X chromosome. One possibility is that the CTD may serve as a binding platform for an unidentified protein or protein complex that has the ability to maintain chromosome morphology but which can function only after proper localization through interactions with the CTD of JIL-1. Another possibility is that binding of the CTD to chromatin by itself is sufficient to stabilize chromatin structure through bridging interactions with surrounding molecules. Nonetheless, we also found that a JIL-1 deletion protein (ΔCTD), which was without the CTD domain but retained histone H3S10 kinase activity and which could be localized to chromatin by sequences in the NTD, was able to rescue autosome as well as partially rescue male X polytene chromosome morphology. Taken together these findings indicate that JIL-1 may participate in regulating chromatin structure by multiple and partially redundant mechanisms. That JIL-1 kinase activity is one of these mechanisms is supported by LacI-JIL-1 tethering experiments by Deng et al. (8), which demonstrated that JIL-1-mediated ectopic histone H3S10 phosphorylation is sufficient to induce a change in higher order chromatin structure from a condensed heterochromatin-like state to a more open euchromatic state. This effect was absent when a “kinase dead” LacI-JIL-1 construct with point mutations in each of the two kinase domains of a crucial lysine required for catalytic activity (22) was expressed (8). Interestingly, the expression of KDI/KDII in JIL-1 null mutants was able to partially rescue autosome, but not male X chromosome morphology, despite that it did not localize to chromatin or phosphorylate histone H3S10. We speculate that KDI/KDII either retains or gains the ability to phosphorylate an unknown molecule, which in turn influences chromatin structure.
As indicated above, several of the expressed constructs had differential effects on the rescue in JIL-1 mutants of chromatin structure of autosomes and of the male X polytene chromosome. However, it is well documented that the male X polytene chromosome structure is unique (23, 24), as also reflected in JIL-1 mutant chromosome morphology (7). For example, in JIL-1 mutants the coiling and folding of the autosomes is not observed for the male X chromosome (7). Instead the puffed appearance of the male X chromosome is caused by dispersal of chromatin into a diffuse network of condensed and euchromatic regions (7). One possibility is that the inherent differences in chromosome structure may be linked to the increased transcriptional activity of the male X chromosome, which correlates with a more open chromosome architecture such that despite the fact that it contains half the DNA content, the normal male X chromosome has the same width as the paired female X chromosome and the autosomes (7, 25). This more open chromatin structure is likely to be maintained by the activity of the MSL dosage compensation complex and the MOF (males absent on the first) histone acetyltransferase (23, 24). This leads to hyperacetylation of histone H4, and this particular chromatin modification consequently has the potential to modulate the binding and function of JIL-1 on the male X chromosome. We show that contributions from sequences within both the NTD and CTD are required for enrichment of JIL-1 on the male X chromosome. However, although the CTD showed proper localization to interband regions identical to endogenous JIL-1, the localization of the NTD was relatively evenly distributed on the chromosome arms, which included ectopic regions. This suggests that the CTD may provide high affinity binding to specific chromatin sites, whereas the NTD may mediate a more general and low affinity chromatin association, the nature of which remains undetermined. Thus, the special male X polytene chromatin environment could be envisioned to promote higher affinity interactions with the NTD that only occur in the context of prior CTD-mediated localization and which in turn result in enhanced JIL-1 binding. Alternatively, it has been shown that JIL-1 associates with the MSL complex and that it can physically interact with MSL complex proteins (2), wherefore it is possible that such interactions may govern the JIL-1 enrichment. In this context it should be noted that the in vitro finding of Jin et al. (2) that the MSL complex interaction with JIL-1 may be mediated by the kinase domains is not supported by the present data. In this study we detected no binding of KDI/KDII to the male X chromosome in the JIL-1 null mutant background even though the MSL complex is present on the mutant X chromosome (2, 3).
In addition to the interaction with histone H3 demonstrated here, previous studies have identified lamin Dm0 (9) as well as the chromodomain-containing protein, Chromator (19), as direct interaction partners with the CTD domain of JIL-1. Although lamin Dm0-like histone H3 binds to the predicted globular domain of the basic region of the CTD, Chromator binds to sequences in the acidic region. Furthermore, Zhang et al. (26) showed that JIL-1 physically interacts via its first kinase domain with the zf5 splice variant of the complex lola locus, the expression of which is restricted to early embryogenesis. Thus, JIL-1 is a multidomain protein that functions in a number of developmental contexts in addition to its general role in regulating chromatin structure. In future experiments it will be of interest to determine the structural basis for these interactions and specifically how JIL-1, through its COOH-terminal domain, is integrated into nucleosome and chromatin organization.
Acknowledgments
We thank members of the laboratory for discussion, advice, and critical reading of the manuscript. We also acknowledge V. Lephart for maintenance of fly stocks and Laurence Woodruff for technical assistance. We especially thank Dr. M. Kuroda for providing the MSL antibody and Dr. Y. Wang for the Drosophila histone extraction protocol.
This work was supported, in whole or in part, by National Institutes of Health Grant GM62916 (to K. M. J.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: NTD, NH2-terminal domain; CTD, COOH-terminal domain; KDI, kinase domain I; KDII, kinase domain II; Su(var), suppressor-of-variegation; MSL, male-specific lethal; FL, full length; GFP, green fluorescent protein; GST, glutathione S-transferase; mAb, monoclonal antibody; PBS, phosphate-buffered saline; TRITC, tetramethylrhodamine isothiocyanate.
References
- 1.Jin, Y., Wang, Y., Walker, D. L., Dong, H., Conley, C., Johansen, J., and Johansen, K. M. (1999) Mol. Cell 4 129-135 [DOI] [PubMed] [Google Scholar]
- 2.Jin, Y., Wang, Y., Johansen, J., and Johansen, K. M. (2000) J. Cell Biol. 149 1005-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang, Y., Zhang, W., Jin, Y., Johansen, J., and Johansen, K. M. (2001) Cell 105 433-443 [DOI] [PubMed] [Google Scholar]
- 4.Ebert, A., Schotta, G., Lein, S., Kubicek, S., Krauss, V., Jenuwein, T., and Reuter, G. (2004) Genes Dev. 18 2973-2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang, W., Deng, H., Bao, X., Lerach, S., Girton, J., Johansen, J., and Johansen, K. M. (2006) Development 133 229-235 [DOI] [PubMed] [Google Scholar]
- 6.Bao, X., Deng, H., Johansen, J., Girton, J., and Johansen, K. M. (2007) Genetics 176 1355-1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, H., Zhang, W., Bao, X., Martin, J. N., Girton, J., Johansen, J., and Johansen, K. M. (2005) Chromosoma 114 173-182 [DOI] [PubMed] [Google Scholar]
- 8.Deng, H., Bao, X., Cai, W., Blacketer, M. J., Belmont, A. S., Girton, J., Johansen, J., and Johansen, K. M. (2008) Development 135 699-705 [DOI] [PubMed] [Google Scholar]
- 9.Bao, X., Zhang, W., Krencik, R., Deng, H., Wang, Y., Girton, J., Johansen, J., and Johansen, K. M. (2005) J. Cell Sci. 118 5079-5087 [DOI] [PubMed] [Google Scholar]
- 10.Sambrook, J., and Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual, pp. 1.1-18.136, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 11.Roberts, D. B. (1998) Drosophila: A Practical Approach, pp. 1-389, IRL Press at Oxford University Press, Oxford
- 12.Zhang, W., Jin, Y., Ji, Y., Girton, J., Johansen, J., and Johansen, K. M. (2003) Genetics 165 1341-1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallrath, L. L., and Elgin, S. C. R. (1995) Genes Dev. 9 1263-1277 [DOI] [PubMed] [Google Scholar]
- 14.Lindsley, D. L., and Zimm, G. G. (1992) The Genome of Drosophila melanogaster, pp. 1071-1076, Academic Press, Inc., New York
- 15.Kelley, R. L., Meller, V. H., Gordadze, P. R., Roman, G., Davis, R. L., and Kuroda, M. I. (1999) Cell 98 513-522 [DOI] [PubMed] [Google Scholar]
- 16.DiMario, P., Rosby, R., and Cui, Z. (2006) Drosophila Information Service 89 115-118 [Google Scholar]
- 17.Rath, U., Wang, D., Ding, Y., Xu, Y.-Z., Qi, H., Blacketer, M. J., Girton, J., Johansen, J., and Johansen, K. M. (2004) J. Cell. Biochem. 93 1033-1047 [DOI] [PubMed] [Google Scholar]
- 18.Towbin, H., Staehelin, T., and Gordon, J. (1979) Proc. Natl. Acad. Sci. U. S. A. 9 4350-4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rath, U., Ding, Y., Deng, Y., Qi, H., Bao, X., Zhang, W., Girton, J., Johansen, J., and Johansen, K. M. (2006) J. Cell Sci. 119 2332-2341 [DOI] [PubMed] [Google Scholar]
- 20.Cai, W., Bao, X., Deng, H., Jin, Y., Girton, J., Johansen, J., and Johansen, K. M. (2008) Development 135 2917-2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khorasanizadeh, S. (2004) Cell 116 259-272 [DOI] [PubMed] [Google Scholar]
- 22.Bjørbæk, C., Zhao, Y., and Moller, D. E. (1995) J. Biol. Chem. 270 18848-18852 [DOI] [PubMed] [Google Scholar]
- 23.Bone, J. R., Lavender, J., Richman, R., Palmer, M. J., Turner, B. M., and Kuroda, M. I. (1994) Genes Dev. 8 96-104 [DOI] [PubMed] [Google Scholar]
- 24.Hilfiker, A., Hilfiker-Kleiner, D., Pannuti, A., and Lucchesi, J. C. (1997) EMBO J. 16 2054-2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorman, M., Franke, A., and Baker, B. S. (1995) Development 121 463-475 [DOI] [PubMed] [Google Scholar]
- 26.Zhang, W., Wang, Y., Long, J., Girton, J., Johansen, J., and Johansen, K. M. (2003) J. Biol. Chem. 278 11696-11704 [DOI] [PubMed] [Google Scholar]