Abstract
The p38 MAPK is a family of serine/threonine protein kinases that play important roles in cellular responses to external stress signals, e.g. UV irradiation. To assess the role of p38 MAPK pathway in nucleotide excision repair (NER), the most versatile DNA repair pathway, we determined the efficiency of NER in cells treated with p38 MAPK inhibitor SB203580 and found that p38 MAPK is required for the prompt repair of UV-induced DNA damage CPD. We further investigated the possible mechanism through which p38 MAPK regulates NER and found that p38 MAPK mediates UV-induced histone H3 acetylation and chromatin relaxation. Moreover, p38 MAPK also regulates UV-induced DDB2 ubiquitylation and degradation via phosphorylation of the target protein. Finally, our results showed that p38 MAPK is required for the recruitment of NER factors XPC and TFIIH to UV-induced DNA damage sites. We conclude that p38 MAPK regulates chromatin remodeling as well as DDB2 degradation for facilitating NER factor assembly.
Living organisms are incessantly exposed to a variety of DNA-damaging agents with potentially devastating consequences. To overcome the deleterious effects of such exposures, cells respond with a variety of defensive strategies to eliminate damage and maintain the integrity of their genome. Among them, nucleotide excision repair (NER)3 is the most important repair system that removes UV-induced photolesions, including cyclobutane pyrimidine dimers (CPD) and pyrimidine-pyrimidone photoproducts (6-4PP), as well as other bulky DNA adducts caused by different chemical carcinogens (1). There are two subpathways of NER: global genome NER (GG-NER), which removes lesions from the entire genome, and transcription-coupled NER, which eliminates DNA damage from the transcribed strand of the actively transcribed genes (1, 2). The two pathways differ mainly at the initial damage recognition steps. For transcription-coupled NER, the lesion is detected by stalled RNA polymerase during transcription (3, 4), whereas for GG-NER, recognition of DNA lesions depends on specific DNA-binding proteins that have high affinity for damaged DNA, e.g. XPC. Evidence from both in vitro and in vivo studies indicates that XPC comes to the damage site at a very early stage and is a critical GG-NER initiator (5, 6). Although XPC has remarkable binding capacity to damaged DNA, XPC alone is not sufficient to recognize DNA lesions like CPD occurring deep within the chromatin (7, 8). Another equally important factor, UV-damaged DNA binding (UV-DDB) complex, has emerged as a crucial player in recognizing and processing of CPD in the chromatin context (9, 10). DDB has an inherently high binding ability to DNA lesions, especially to 6-4PP (11–15), and its binding to damaged DNA is independent of functional XPC protein, suggesting that DDB arrives at the damage site prior to XPC binding (16, 17). Accumulating evidence has suggested that DDB helps recruit XPC to both CPD and 6-4PP and accelerates the repair of these lesions (17, 18). Thus, DDB can be considered as the initial damage recognition factor for UV-induced photolesions. DDB2, which is a subunit of the UV-DDB complex, is shown to undergo ubiquitin-mediated proteolysis following UV irradiation (19–21), and the timely degradation of DDB2 has an important role in the arrival of XPC to the damage site in vivo (22). Interestingly, despite its critical role in detection and repair of UV-induced DNA lesion in cells, DDB2 is not essential for reconstituted in vitro NER, suggesting that DDB2 is specifically required for the repair of lesions within chromatin (23–27).
The highly condensed nature of the chromatin fiber makes the access to DNA damage difficult for the repair factors and thus inhibitory to damage processing. Therefore, chromatin accessibility is an important feature in the detection and efficient removal of DNA lesions by GG-NER (28, 29). Repair of both CPD and 6-4PP is affected by their location and accessibility because their repair is faster in the nucleosome-free and linker regions than in the nucleosome core (30, 31). Earlier studies have also revealed a relationship of NER to UV-induced chromatin accessibility occurring through histone acetylation (32–34). It is reported that histone acetylation increases the chromatin accessibility and facilitates NER in mammalian cells. Furthermore, DNA repair synthesis is enhanced in hyperacetylated nucleosome (33, 34). Interestingly, the tumor suppressor protein p53, which enhances GG-NER, is also involved in UV-induced histone H3 and H4 acetylation and chromatin relaxation (35). Another family of tumor suppressors, ING1 and ING2, are reported to enhance NER through histone acetylation and increased chromatin accessibility in a p53-dependent manner (36–38). All of these studies suggest that chromatin structure modification is an important parameter in DNA lesion accessibility and consequently efficient NER.
The p38 MAPK, a serine/threonine kinase, is known to play important roles in cellular responses to various external stress signals. It has been shown that p38 MAPK is an important mediator of phosphorylation of histone H3 at serine 10 in response to UV (39). This phosphorylation is involved in the alteration of chromatin condensation during transcription activation and cell division (40). Furthermore, p38 MAPK has been found to mediate H3 phosphorylation at serine 10 induced by cisplatin, which binds to DNA to form a covalent platinum-DNA adduct that is removed by NER (41). Thus far, no report has attributed the UV-induced acetylation and global chromatin relaxation to the action of p38 MAPK, and it is still not known whether p38 MAPK plays any role in facilitating NER in response to UV.
In the present report, we have studied the function of p38 MAPK in NER in cells exposed to UV irradiation. Our studies indicate that p38 MAPK is involved in UV-induced histone H3 acetylation at lysine 14 and chromatin relaxation. Our results also show that p38 MAPK, by regulating DDB2 degradation and consequently the recruitment of XPC and TFIIH, plays a pivotal role in the removal of UV lesion CPD through NER.
EXPERIMENTAL PROCEDURES
Cell Lines and Treatments—Normal human fibroblasts (OSU-2) were established in our laboratory as described previously (42). HeLa cells stably transfected with NH2-terminal FLAG-hemagglutinin-tagged DDB2 (HeLa-DDB2) were a gift from Dr. Yoshihiro Nakatani (Dana-Farber Cancer Institute, Boston, MA). These cell lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C in a humidified atmosphere with 5% CO2. To study the function of p38 MAPK, the cells were pretreated with p38 MAPK inhibitor SB203580 (Calbiochem, San Diego, CA) or DMSO for 30 min before UV irradiation. To measure ubiquitylation of DDB2, HeLa-DDB2 cells were cultured in medium supplemented with MG132 (Calbiochem, San Diego, CA) 30 min before UV treatment. For UV irradiation, the cells were washed twice with phosphate-buffered saline, irradiated at varying UV doses, and incubated in appropriate medium for the desired time period. The irradiation was done with a germicidal lamp at a dose rate of 0.8 J/m2/s as measured by a Kettering model 65 radiometer (Cole Palmer Instrument, Co., Vernon Hill, IL).
In Vivo Ubiquitylation and Phosphorylation Detection—DDB2 ubiquitylation and phosphorylation were detected as described before (43). Briefly, HeLa-DDB2 cells were treated with MG132 (10 μm) 30 min before UV irradiation at 40 J/m2 and kept in the same medium for indicated time periods. Whole cell lysates were prepared by boiling in SDS lysis buffer (2% SDS, 10% glycerol, 10 mm dithiothreitol, 62 mm Tris-HCl, pH 6.8, and protease inhibitor mixture) for 10 min. After centrifugation, the supernatant was diluted with radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5 mm EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor mixture) and subjected to immunoprecipitation with anti-FLAG M2 affinity gel (Sigma). The immunoprecipitates were then subjected to Western blotting and detected with anti-ubiquitin (Santa Cruz Biotechnology, Santa Cruz, CA), anti-phospho-serine, anti-phospho-tyrosine (Cell Signaling Technology, Danvers, MA), and anti-DDB2 antibodies.
Western Blot Analysis of Proteins—The proteins were quantified and separated by SDS-PAGE, and the immunoblot analysis was done by using chemiluminescent detection. Phosphorylated MK2, phospho-Akt (Ser473), and phospho-H3 (Ser10) were detected by using rabbit polyclonal antibodies purchased from Cell Signaling Technology (Danvers, MA). Phospho-ERK (Santa Cruz Biotechnology) and phospho-JNK (Promega) antibodies were used to detect the activated ERK and JNK, respectively. Acetyl-H3 (K14), acetyl-H3 (K9, 14), acetyl-H3 (K18), and acetyl-H4 were detected using rabbit polyclonal antibodies purchased from Upstate Biotechnology (Billerica, MA). Rabbit anti-DDB2 antibody was generated in our lab as described before (44). To serve as a loading control, total lamin B or actin levels in the cell lysates were detected using anti-lamin B or anti-actin antibody (Santa Cruz Biotechnology).
Micrococcal Nuclease (MNase) Digestion Assay—The cells were trypsinized and resuspended in hypotonic cell lysis buffer (10 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 1 mm dithiothreitol, 25% glycerol, 0.2% Nonidet P-40, 0.5 μm spermidine, 0.15 μm spermine, and protease inhibitor mixture). The cells were incubated on ice for 10 min followed by pipetting 10 times to release nuclei and low speed centrifugation. The nuclei were resuspended in 200 μl of MNase buffer (10 mm Tris-HCl, pH 8.0, 50 mm NaCl, 300 mm sucrose, 3 mm MgCl2, and 1 mm CaCl2) and digested with a concentration range of MNase (Sigma) for 5 min at room temperature. The reaction was stopped by adding 5× stop solution (0.1 m EDTA, 0.01 m EGTA, pH 8.0). DNA was extracted with phenol/chloroform, and 2 μg of DNA was separated on a 2% agarose gel.
Quantification of CPD and 6-4PP by Immuno-Slot Blot (ISB) Analysis—A noncompetitive ISB assay, described earlier (45), was used to quantify the initial formation of UV-induced CPD or 6-4PP and those remaining in cells after desired time of repair. Briefly, after different treatments, the cells were lysed immediately for DNA isolation. The same amount of DNA was loaded on nitrocellulose membranes, and the amounts of CPD or 6-4PP were detected with monoclonal anti-CPD TDM-2 or anti-6-4PP 64 M-2 antibody (MBL International Corporation, Woburn, MA).
Fractionation of Cellular Proteins—OSU-2 cells were pretreated with either SB203580 or DMSO for 30 min before UV irradiation at 20 J/m2. After further incubation for 30 min, the cells were harvested and subjected to cellular protein fractionation procedure as described earlier (46). Briefly, cytoplasm and nuclei were first separated by suspending cells in hypotonic buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, and protease inhibitor mixture) and treated with 0.1% Triton X-100. After centrifugation, the supernatant was saved as S, and the nuclear pellet was then treated with a higher concentration of Triton X-100 (1%) in LS buffer (10 mm Tris-HCl, pH 7.4, 0.2 mm MgCl2, and protease inhibitor mixture) to remove the nuclear envelope and recover the fraction of nucleoplasmic soluble proteins, designated as TW. The nuclear pellet was further extracted consecutively with increasing concentrations (0.3, 0.5, and 2.0 m) of NaCl in LS buffer to result in supernatant fractions, designated as 0.3, 0.5, and 2.0, respectively. Each protein fraction, corresponding to an equivalent cell number, was loaded on SDS-PAGE and analyzed by immunoblotting with anti-DDB2 antibody.
Localized Micropore UV Irradiation and Immunofluorescent Staining—OSU-2 cells growing on glass coverslips were pretreated with either SB203580 or DMSO for 30 min, and the cells were then washed with phosphate-buffered saline and UV-irradiated through a 5-μm isopore polycarbonate filter (Millipore, Bedford, MA) as described previously (47). The cells were then double stained with rabbit anti-XPC and mouse anti-CPD (TDM-2, MBL International Corporation), or rabbit anti-XPB and mouse anti-CPD, or rabbit-anti-phospho-p38 and mouse anti-CPD antibodies. Fluorescein isothiocyanate and Texas Red conjugates of anti-rabbit and anti-mouse IgG were simultaneously used for the detection of primary antibody binding. Fluorescence images were obtained with a Nikon fluorescence microscope E80i (Nikon, Tokyo, Japan) fitted with appropriate filters for fluorescein isothiocyanate and Texas Red. The digital images were then captured with a cooled CCD camera and processed with the help of its SPOT software (Diagnostic Instruments, Sterling Heights, MI).
RESULTS
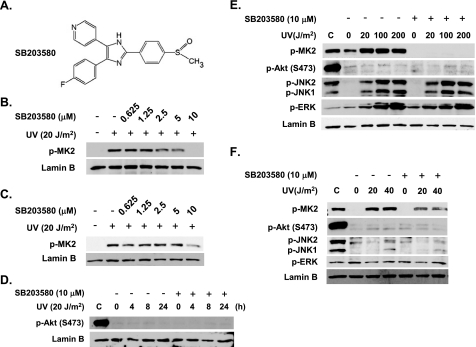
p38 MAPK Is Required for Optimal Repair of UV-induced CPD—To study the role of p38 MAPK in NER, we inhibited p38 MAPK activity with its specific pharmacological inhibitor, SB203580. This pyridinyl imidazole inhibitor (structure shown in Fig. 1A) is now extensively used to understand the role of p38 MAPK. It has been shown to selectively inhibit p38 MAPK activity without significantly affecting the activity of other MAP kinases, e.g. JNK and ERK (48). To determine the inhibition of p38 MAPK activity by SB203580, OSU-2 cells or HeLa-DDB2 cells were pretreated with SB203580 at different concentrations followed by UV irradiation at 20 J/m2. The cell lysates were subjected to Western blot analysis using phospho-MK2-specific antibody. MK2, often referred to as MAPK-activated protein kinase-2, is a physiological target of p38 MAPK (49). Therefore, the phosphorylation of MK2 indicates the activation of p38 MAPK. We found that the phospho-MK2 level was markedly elevated after UV irradiation in control DMSO-treated cells. In contrast, cells treated with 10 μm SB203580 exhibited a dramatic reduction of UV-induced MK2 activation in both OSU-2 (Fig. 1B) and HeLa-DDB2 (Fig. 1C) cell lines. Importantly, however, drug concentrations below 5 μm exhibited a markedly reduced inhibitory effect on p38 MAPK activity (Fig. 1, B and C). Thus, in this study we used 10 μm SB203580 to effectively suppress UV-induced p38 MAPK activity in both OSU and HeLa-DDB2 cells. At this concentration, we did not observe an evident inhibition of ERK and JNK activities in either OSU-2 (Fig. 1E) or HeLa-DDB2 cells (Fig. 1F). It should be noted that higher concentrations (>1 μm) of SB203580 are reported to also inhibit the interleukin-2-stimulated phosphorylation of Akt in T cells independent of p38 MAPK (50). We tested for this in our system and found that in OSU-2 cells UV irradiation at 20 J/m2 failed to exhibit any elevation of phosphorylated Akt (Ser473) within 24 h after UV irradiation (Fig. 1D). Moreover, within a short period of time (30 min) after UV treatment, an even higher dose of UV (100 or 200 J/m2) did not induce any activation of Akt (Fig. 1E). Similarly, in HeLa-DDB2 cells, we did not observe the activation of Akt after UV irradiation at 20 or 40 J/m2 (Fig. 1F). More importantly, 10 μm concentration of SB203580 did not have any inhibitory effect on the basal level of phospho-Akt in both OSU-2 and HeLa-DDB2 cells (Fig. 1, D–F). Therefore, Akt activity is not likely a candidate for the effects observed upon 10 μm SB203580 treatment of cell lines in this study.
FIGURE 1.
p38 MAPK inhibitor SB203580 inhibits the activation of p38 MAPK but not JNK, ERK, or Akt in response to UV irradiation. A, chemical structure of SB203580. B and C, inhibition of p38 MAPK kinase activity by different doses of SB203580. OSU-2 cells (B) or HeLa-DDB2 cells (C) were treated with SB203580 for 30 min at indicated concentrations and UV-irradiated at 20 J/m2. The cells were cultured in the same medium for 1 h, and the cell lysates were analyzed by Western blot analysis using anti-phospho-MK2 antibody. Lamin B was detected to serve as an internal control. D–F, inhibition of p38 MAPK, JNK, ERK, and Akt activation by SB203580. OSU-2 cells (D and E) or HeLa-DDB2 cells (F) were treated with DMSO or SB203580 (10 μm) for 30 min and UV-irradiated at indicated doses. The cells were allowed to repair for indicated time periods (D) or 30 min (E) or 2 h (F), and the whole cell lysates were analyzed by Western blotting. Whole cell lysates of CP70 cells (lanes marked C), having overexpressed Akt, were used as a positive control of Akt phosphorylation. Lamin B, from the same gels, was used as a loading control.
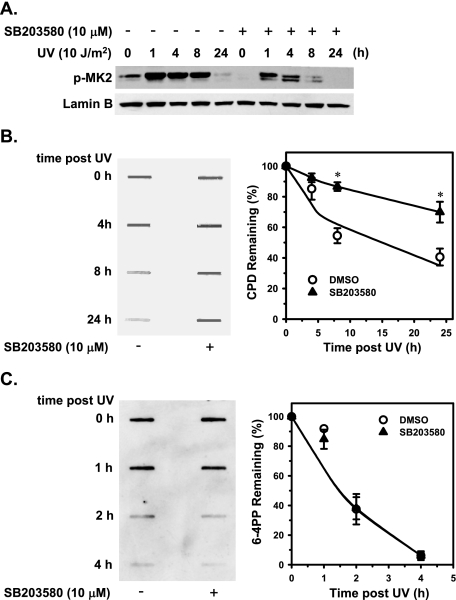
Next, we tested the effect of modulating p38 MAPK activity on the efficiency of NER. OSU-2 cells were treated with SB203580 or DMSO for 30 min and then UV-irradiated at a dose of 10 J/m2. The cells were allowed to repair for indicated time periods and analyzed for the activity of p38 MAPK and the removal of UV-induced CPD using ISB analysis with anti-CPD antibody. As shown in Fig. 2A, p38 MAPK activity was successfully inhibited by SB203580 at each time point. When p38 MAPK was inhibited by SB203580, efficiency of NER was severely compromised as reflected by a decreased removal rate of UV-induced CPD (Fig. 2B). In fact, 8 h after UV treatment, only 55% of CPD remained in control cells in comparison with 87% CPD remaining in cells with suppressed p38 MAPK activity. Similarly, 41% CPD was left in control cells after 24 h of repair in contrast to 70% CPD in cells with curtailed p38 MAPK activity. Notably, however, the repair of another UV-induced DNA lesion, 6-4PP, was not affected by inactivation of p38 MAPK (Fig. 2C). These results indicate that p38 MAPK is required for prompt repair of UV-induced CPD.
FIGURE 2.
p38 MAPK is required for efficient removal of CPD. A, inhibition of p38 MAPK activation by SB203580. OSU-2 cells were pretreated with DMSO or SB203580 (10 μm) for 30 min. The cells were then UV-irradiated at 10 J/m2 and further cultured in the same medium for indicated time periods. The cell lysates were analyzed for phospho-MK2 levels using Western blotting. Lamin B was detected as an internal control. B and C, effect of p38 MAPK on removal of CPD or 6-4PP. OSU-2 cells were cultured in serum-free medium for 48 h before the start of different treatments. The cells were treated with SB203580 (10 μm) or DMSO for 30 min and then UV-irradiated at 10 J/m2 and allowed to repair for indicated times in the same medium. Identical amounts of genomic DNA were subjected to ISB analysis, and the amounts of CPD or 6-4PP at different times were detected with anti-CPD or anti-6-4PP antibody, respectively. The intensity of each band was quantified, and the relative CPD or 6-4PP remaining in different samples were calculated based on the initial CPD or 6-4PP formation. The data presented in the graph on the right indicate the means ± S.E. of the relative CPD or 6-4PP remaining from three independent experiments. *, significant difference between DMSO- and SB203580-treated cells, p < 0.05.
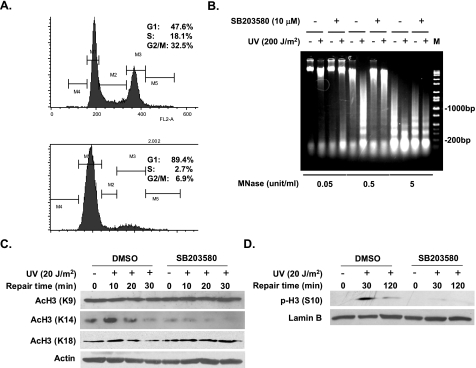
p38 MAPK Affects UV-induced Chromatin Relaxation and Histone Acetylation—Upon UV irradiation of cells, p38 MAPK has been reported to phosphorylate histone H3 at serine 10 (39), indicating its potential role in modulating chromatin remodeling in response to UV treatment. Because repair of DNA damage occurs within highly condensed chromatin fibers, the relaxation of chromatin structure is necessary for the accessibility of damaged DNA to repair proteins (29, 51). Thus, it is reasonable to assume that increased efficiency of NER caused by p38 MAPK function results from enhanced chromatin relaxation. To ascertain this possibility, OSU-2 cells were grown in serum-free medium for 48 h before UV treatment to arrest cells in the G0/G1 phase. Flow cytometric analysis revealed that following serum starvation 89.4% of the cells were arrested in G0/G1 phase (Fig. 3A, upper panel) compared with 47.6% of cells grown in complete medium (Fig. 3A, lower panel). The serum-starved growth-arrested cells were pretreated with SB203580 to inhibit p38 MAPK activity or treated with DMSO as control and then UV-irradiated at 200 J/m2. After a further 30 min of incubation, the cell nuclei were gently isolated and subjected to digestion with appropriately titrated concentrations of MNase. Among the three different concentrations tested, lower and higher MNase concentrations were unable to clearly distinguish the effect of the inhibitor on nuclease digestibility (Fig. 3B). In contrast, intermediate MNase concentration (0.5 units/ml) provided easily discernable digestion patterns between unirradiated versus irradiated as well as in absence and presence of the inhibitor. Chromatin relaxation at 30 min after UV irradiation, observed in control cells, was reflected by dramatically increased DNA sensitivity to MNase. However, when p38 MAPK activity was suppressed, UV-mediated increased DNA sensitivity to MNase was dramatically reversed. (Fig. 3B). This result suggests that UV-induced chromatin relaxation is intimately modulated by the activation of p38 MAPK.
FIGURE 3.
p38 MAPK is involved in UV-induced chromatin relaxation. A, cell cycle distribution. OSU-2 cells grown in complete medium (top panel) and serum free medium (bottom panel) were analyzed by flow cytometry for cell cycle distribution. B, MNase sensitivity in UV-irradiated cells. OSU-2 cells were serum-starved for 48 h and then treated with DMSO or SB203580 for 30 min. The cells were UV-irradiated at 200 J/m2 and cultured for additional 30 min before nuclei isolation. MNase digestion and agarose gel electrophoresis was conducted to determine the extent of chromatin relaxation. C, histone acetylation levels in response to UV irradiation. Growth-arrested OSU-2 cells were pretreated with DMSO or SB203580, UV-irradiated at 20 J/m2, and allowed to repair for indicated time periods. The whole cell lysates were subjected to Western blotting, and acetylated histone H3 was detected with anti-AcH3 (K9, 14), anti-AcH3 (K14), or anti-AcH3 (K18). Actin in the same gels was detected as loading control. D, histone H3 phosphorylation upon UV irradiation. Growth-arrested OSU-2 cells were treated as described in C. Anti-p-H3 (Ser10) antibody was used to detect the phosphorylated H3 (Ser10) level in the whole cell lysates.
Histone acetylation is believed to play an important role in chromatin remodeling during the NER process (32–34). To assess whether the effect of p38 MAPK on chromatin relaxation is invoked through histone acetylation, growth-arrested OSU-2 cells were pretreated with SB203580 and then analyzed for the status of histone H3 acetylation immediately after UV irradiation. Fig. 3C shows the H3 acetylation at lysines 9, 14, and 18 by the use of specific anti-AcH3 antibodies. Histone H3 acetylation between p38 MAPK-active and p38 MAPK-inactive cells was essentially unaltered for lysines 9 and 18. On the other hand, acetylation at H3 lysine 14 showed a prompt increase within 10 min of UV treatment and subsequent return to basal levels within 30 min. However, upon p38 MAPK inhibition, histone H3 lysine 14 acetylation levels were negligible under both physiological conditions and after UV treatment. It is clear that specific histone acetylation occurring at specific lysine residues after UV irradiation is impaired in the absence of p38 MAPK, which could be instrumental in decreased chromatin relaxation and NER. We further tested the effect of SB203580 on UV-induced phosphorylation of H3 at serine 10 in growth-arrested OSU-2 cells. UV induced a dramatic increase of phospho-H3 (Ser10) in DMSO-treated cells, whereas in cells deprived of p38 MAPK activity, phospho-H3 (Ser10) level remained the same as nonirradiated cells (Fig. 3D).
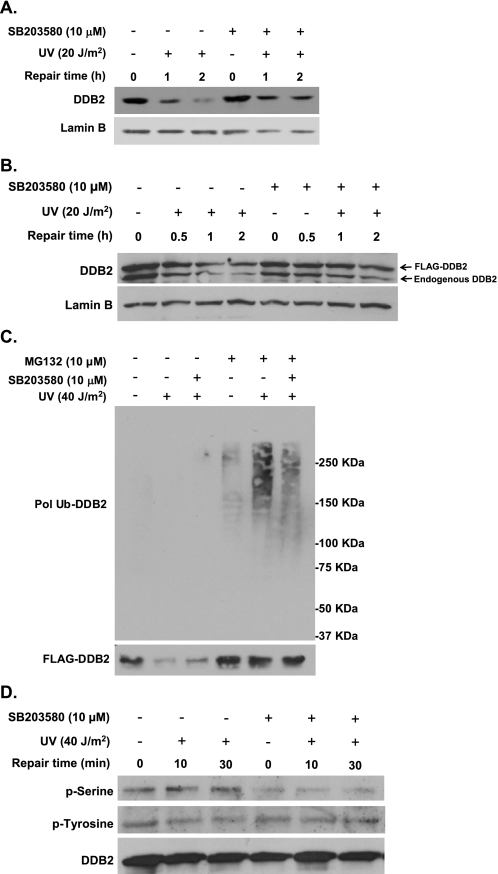
p38 MAPK Participates in UV-induced DDB2 Degradation—p38 MAPK is known to regulate degradation of many cellular proteins through their phosphorylation (52–56). It is already established that the rapid degradation of DDB2 protein after UV irradiation is critical for efficient GG-NER (22, 57). Whether the degradation of DDB2 is also influenced by the p38 MAPK activity was investigated next in OSU-2 cells pretreated with SB203580 before irradiation, and an in-depth analysis of DDB2 was conducted. As expected, DDB2 levels rapidly decreased in cells UV-irradiated at 20 J/m2. For example, in control cells, with normal p38 MAPK activity, only negligible amounts of DDB2 were seen at 2 h post-irradiation (Fig. 4A). However, UV-induced DDB2 degradation was significantly attenuated in cells exposed to p38 MAPK inhibitor SB203580 (Fig. 4A). The degradation of DDB2 was also assessed in HeLa cell line with ectopically overexpressed FLAG-DDB2 (HeLa-DDB2). In DMSO-treated cells, both endogenous and FLAG-tagged DDB2 levels declined as early as 30 min after UV treatment and continued up to 2 h post-irradiation. Once again both endogenous and FLAG-tagged DDB2 degradation was severely compromised by the treatment of cells with SB203580 (Fig. 4B).
FIGURE 4.
p38 MAPK mediates UV-induced DDB2 ubiquitylation and degradation. A and B, OSU-2 (A) or HeLa-DDB2 (B) cells were treated with DMSO or SB203580 for 30 min, UV-irradiated at 20 J/m2, and further incubated for indicated times. The whole cell extracts were generated by boiling of the cells in SDS lysis buffer and then subjected to Western blotting with anti-DDB2 and anti-Lamin B antibodies. C, DDB2 ubiquitylation upon UV treatment. HeLa-DDB2 cells were pretreated with MG132 together with SB203580 for 30 min and UV-irradiated at 40 J/m2. The cells were cultured in the same medium for 2 h, and the cell lysates were immunoprecipitated with anti-FLAG M2 affinity gel. The immunoprecipitates were subjected to Western blot analysis with anti-ubiquitin or anti-DDB2 antibody. D, DDB2 phosphorylation by p38 MAPK. HeLa-DDB2 cells were pretreated with DMSO or SB203580 for 30 min prior to UV irradiation at 20 J/m2. The cells were kept in the same medium for the indicated times, and the whole cell lysates were prepared. FLAG-tagged DDB2 was immunoprecipitated with anti-FLAG M2 affinity gel and analyzed by Western blotting with anti-phospho-serine, anti-phospho-tyrosine, or anti-DDB2 antibody.
UV-induced DDB2 degradation in cells is known to occur through ubiquitin-proteasome pathways (19–21). To determine the influence of p38 MAPK on UV-induced DDB2 degradation through the ubiquitin-proteasome pathway, we compared the UV-induced DDB2 ubiquitylation in cells with or without functional p38 MAPK activity. HeLa-DDB2 cells were either treated with SB203580 to inhibit p38 MAPK activity or treated with DMSO as control. Subsequently, proteasome inhibitor MG132 was added to the cultures 0.5 h before UV irradiation and maintained in cultures during the period of repair to ensure the detection of ubiquitylated proteins. The cell lysates were immunoprecipitated with anti-FLAG M2 affinity gel, and the immunoprecipitates were detected by Western blotting with anti-ubiquitin antibody. As shown in Fig. 4C, UV irradiation enhanced the overall cellular ubiquitylation of DDB2 in the cells with normal p38 MAPK activity. However, upon abolishing p38 MAPK activity by inhibitor treatment, the UV-induced ubiquitylation of DDB2 was significantly decreased. The appearance of polyubiquitylated DDB2 bands in MG132-treated cells coincided with the corresponding accumulation of DDB2 in cells. These results indicate that p38 MAPK influences the repair outcome through its effect on UV-induced DDB2 ubiquitylation, which is required for the clearance of DDB2 from the damaged chromatin and successful commencement of GG-NER of CPD.
Phosphorylation by protein kinases positively or negatively regulates protein ubiquitylation through various pathways (58). Because our data demonstrated an involvement of p38 MAPK in UV-induced DDB2 ubiquitylation, we wanted to explore whether DDB2 is also phosphorylated by p38 MAPK. For this, HeLa-DDB2 cells were UV-irradiated at 40 J/m2 and allowed to repair for indicated time periods before immunoprecipitation of FLAG-tagged DDB2 with anti-FLAG M2 affinity gel. The immunoprecipitates were subjected to Western blot analysis with phosphoserine- or phosphotyrosine-specific antibodies. As shown in Fig. 4D, overall serine phosphorylation of DDB2 was not influenced by the UV treatment but was significantly reduced upon inhibition of p38 MAPK activity. In contrast, tyrosine phosphorylation of DDB2, which has been reported to be regulated by c-Abl tyrosine kinase, did not show any change by irradiation or MAPK inhibition. The data suggest that p38 MAPK is involved in serine phosphorylation of DDB2, which may influence its ubiquitylation following irradiation.
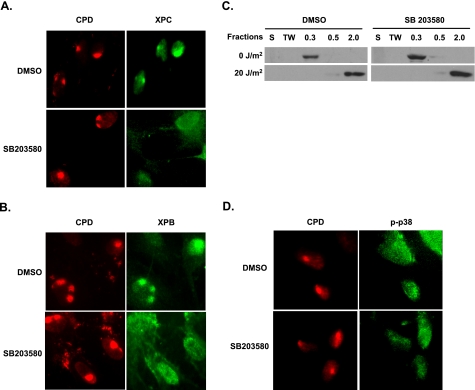
p38 MAPK Regulates in Vivo Recruitment of XPC and TFIIH, but Not DDB2, to DNA Damage Sites—Because p38 MAPK could help open chromatin structure and promote DDB2 degradation following UV irradiation and both of them are essential to the subsequent recruitment of critical damage recognition factor XPC, we speculated that XPC recruitment to DNA damage would be compromised in the absence of p38 MAPK activity. To verify this, we analyzed the XPC recruitment by using a local micropore UV irradiation of cells and immunofluorescent staining of DNA damage and NER factors. OSU-2 cells were pretreated with either SB203580 or DMSO and UV-irradiated through a 5-μm pore filter and allowed to repair the damage induced by UV irradiation. The cells were then fixed and subjected to double staining with anti-XPC and anti-CPD antibodies. As shown in Fig. 5A, recruitment of XPC to the antibody marked UV-induced CPD sites was clearly seen as early as 15 min after irradiation of DMSO treated control cells (upper panels). In contrast, cells lacking the functional p38 MAPK show clear CPD foci but fail to show the arrival of XPC to CPD site (lower panels). We also tested the co-localization of TFIIH, which is essential for the NER of CPD. As seen with XPC recruitment, TFIIH component XPB was unable to localize to CPD sites in p38 MAPK-inactive as compared with p38 MAPK-active cells (Fig. 5B). These results indicate that the effect of p38 MAPK on NER is ultimately through modulating the recruitment of preincision NER factors like XPC and TFIIH.
FIGURE 5.
p38 MAPK affects the recruitment of XPC and XPB to CPD foci. A and B, recruitment of XPC and XPB to CPD. OSU-2 cells were grown on coverslips and pretreated with DMSO or SB203580 for 30 min prior to UV irradiation (100 J/m2) through a 5-μm isopore polycarbonate filter. The cells were incubated in medium for 15 min and then fixed with 2% paraformaldehyde. The cells were double-stained with mouse anti-CPD antibody and rabbit anti-XPC antibody or with mouse anti-CPD antibody and rabbit anti-XPB antibody. C, binding of DDB2 to UV-damaged chromatin. OSU-2 cells were treated with DMSO or SB203580 (10 μm) for 30 min and then UV-irradiated at 20 J/m2. The cells were incubated in the same medium for another 30 min and subjected to cellular protein fractionation. Five fractions as indicated according to the resistance to detergent and salt were obtained. S, cytoplasmic soluble protein; TW, nucleoplasmic soluble proteins; 0.3, proteins binding to chromatin loosely; 0.5, proteins binding to chromatin with intermediate affinity; 2.0, proteins binding to chromatin tightly. Each protein fraction, corresponding to an equivalent cell number, was loaded for SDS-PAGE and analyzed by immunoblotting with anti-DDB2 antibody. D, recruitment of p38 MAPK to the damage site. The cells were treated as described in A and B and double-stained with mouse anti-CPD antibody and rabbit anti-phospho-p38 antibody.
UV-DDB2 has a strong affinity for UV-induced DNA lesions, and it is required for XPC recruitment to CPD during GG-NER (17, 18). Thus, we wanted to determine whether the recruitment of DDB2 to UV-damaged chromatin is also affected by p38 MAPK-mediated chromatin remodeling. Because our DDB2 antibody cannot work in immunofluorescent assay, we applied another technique that has been established in our laboratory to study the binding of DDB2 to UV-damaged chromatin (46). This assay is based on translocation of DDB2 protein into tight association with chromatin upon UV irradiation of cells. OSU-2 cells, with and without p38 MAPK inhibitor, were UV-irradiated at 20 J/m2 and further incubated for 30 min to allow repair of CPD lesions. The cellular proteins were fractionated into five components according to the resistance to extraction in different concentrations of detergent and salt. Proteins in each fraction were analyzed by Western blotting using antibody against DDB2. As shown in Fig. 5C, DDB2 was mainly present in the 0.3 m salt fraction in unirradiated control cells (upper panels). At 30 min after UV irradiation, DDB2 was only detected in the 2 m salt fraction, indicating translocation and a tighter association of DDB2 with chromatin-containing DNA lesions. Interestingly, an identical pattern of DDB2 distribution in different fractions of unirradiated and irradiated cells was observed in cultures exposed to SB203580. These data indicated that p38 MAPK inactivation has no effect on the recognition and binding of DDB2 to UV-induced damage. Because p38 MAPK inactivation inhibited UV-induced chromatin relaxation, it can be concluded that the recruitment and binding of DDB2 to UV-damaged DNA is independent of chromatin remodeling at the damage sites.
Even though p38 MAPK affects the recruitment of NER factors XPC and TFIIH, p38 MAPK itself was not found to colocalize to the damage site (Fig. 5D). Therefore, it is inferred that p38 MAPK does not physically orchestrate the events at or near DNA damage but modulates repair of CPD by affecting downstream targets of its responsive signal transduction cascade.
DISCUSSION
MAP kinase pathways, including ERKs, JNKs, and p38 MAPK, have been reported to be involved in NER in a number of studies. Activation of JNK upon UV enhances both the mRNA and protein levels of XPF and stimulates NER in mouse fibroblast (59, 60). Moreover, blocking JNK activity sensitizes tumor cells to cisplatin through modulating NER (61, 62). ERK has been shown to participate in NER through mediating induction of an essential NER incision factor ERCC1 by epidermal growth factor in human hepatoma cells (63). Although p38 MAPK is known to be involved in DNA damage response and negatively modulates tumorigenesis through apoptosis and cell cycle regulation, very little is known about its role in NER. In this report, we have demonstrated that p38 MAPK is a critical regulator of NER in response to cellular UV irradiation. In contradiction to a recent report that failed to observe an effect on NER by all three MAP kinase pathways (64), our data provide multifaceted evidence for the integral role of p38 MAPK in the prompt repair of UV-induced photolesion CPD. We conclude that p38 MAPK mechanistically operates through chromatin relaxation and DDB2 degradation, which are essential for initial access and recognition factor recruitment during the preincision phase of NER.
p38 MAPK and Chromatin Remodeling—In its native state damage inflicted upon DNA of living cells occurs primarily in the context of chromatin. Chromatin structure presents a demanding obstacle to the function of various DNA templated processes including DNA repair. Remodeling of chromatin through an elaborate and highly regulated multistep process allows access to protein nanomachines involved in DNA replication, transcription, and repair. Localized modification of histone proteins is known to play an important role in chromatin accessibility to NER. In yeast cells, UV-induced global hyperacetylation of histone H3 through histone acetyltransferase Gcn5p was associated with the relaxation of specific repressed loci and enhanced efficiency of NER. Moreover, these histone acetylations and chromatin relaxation are independent of actual damage processing by NER (65), suggesting that UV-induced histone acetylation and chromatin relaxation occur before NER and are associated with chromatin accessibility to repair complex. Similar findings were also reported in mammalian cells. A number of tumor suppressor proteins, including p53, ING1, and ING2, were found to enhance chromatin relaxation and NER through histone modification (35–38). Our data ascribe the chromatin accessibility function in NER to the activity of p38 MAPK. We have shown that chromatin relaxation in response to UV irradiation treatment depends on normal p38 MAPK activity. Furthermore, p38 MAPK is indispensible for UV-induced acetylation of histone H3 at lysine 14 but does not have an effect on acetylation at lysine 9 and 18 (Fig. 3, B and C). Nevertheless, the mechanism through which p38 MAPK mediates UV-induced H3 acetylation is not yet established. It is known that p38 MAPK mediates UV-induced histone H3 phosphorylation at serine 10 (39) (Fig. 3D). However, the function of this histone modification in NER is not elucidated. A number of studies in vitro have shown that several histone acetyltransferases have strong preference for H3 with phosphorylated serine 10 over unmodified H3 (66, 67). In vivo studies also reveal dynamic interplay between H3 acetylation and H3 serine 10 phosphorylation (66, 68). Based on the data presented here, we reason that p38 MAPK regulates H3 acetylation through phosphorylation of H3 at serine 10 in response to UV irradiation to evoke chromatin relaxation.
XPC and DDB2 are essential DNA damage recognition factors for GG-NER, and the recruitment of these two factors to the damage sites is a prerequisite for the subsequent events of repair. Because XPC has been shown to have a lower affinity for nucleosomes than for naked DNA (69), relaxation of the chromatin may facilitate the recruitment of XPC to the damage site. Thus, it is plausible that the impaired localization of XPC to the UV-induced damage site is partially due to rigid chromatin conformation induced by the suppression of p38 MAPK (Fig. 5A). Interestingly, however, the binding of DDB2 to the chromatin was tighter after UV irradiation, and it was not affected by p38 MAPK-regulated chromatin remodeling (Fig. 5C), indicating that the recruitment of DDB2 to the damaged chromatin may not require chromatin relaxation. However, because the repair of 6-4PP was not affected by p38 MAPK activity, we cannot rule out the possibility that in cells treated with p38 MAPK inhibitor, DDB2 was recruited to 6-4PP lesion sites instead of CPD damage sites.
p38 MAPK and DDB2 Degradation—An interesting and novel finding of this study is that p38 MAPK is intimately involved in UV-induced DDB2 ubiquitylation and degradation (Fig. 4, A–C). What could be the mechanism through which p38 MAPK controls DDB2 ubiquitylation? We found that in addition to its role in DDB2 ubiquitylation, p38 MAPK activity is also required for serine phosphorylation of DDB2 independent of UV irradiation (Fig. 4D). Cross-talk between phosphorylation and ubiquitylation has been widely reported (see review by Hunter (58)). Phosphorylation can either promote or inhibit ubiquitylation, which can lead to proteasomal degradation. As for p38 MAPK, phosphorylation of ATF2 by p38 MAPK renders ATF2 resistant to ubiquitylation and degradation (56). In contrast, p38 MAPK has been shown to phosphorylate cyclin D1 and cyclin D2 and trigger the ubiquitin/proteasome-dependent degradation of these two proteins in response to osmotic stress (54, 55). Based on our observation, we speculate that p38 MAPK facilitates DDB2 ubiquitylation and degradation by phosphorylating DDB2 at serine moiety. However, additional investigations are needed to clarify the specific site of serine phosphorylation by p38 MAPK and demonstrate whether this phosphorylation is indispensable for UV-induced DDB2 ubiquitylation.
Both in vitro and in vivo studies have shown that DDB2 breakdown through ubiquitin-proteasome pathway following UV irradiation is crucial for subsequent XPC recruitment and NER efficiency. Sugasawa et al. (57) found that UV-DDB breakdown increases the binding of XPC to the damaged DNA using reconstituted NER component in vitro. Moreover, interference with DDB2 proteolysis compromises the removal of UV-induced CPD in vivo (22). Therefore, the impaired recruitment of XPC and TFIIH to the UV induced damage site may be partly due to the reduced DDB2 degradation caused by suppression of p38 MAPK activity.
p38 MAPK and 6-4PP—Despite its role in the repair of CPD, p38 MAPK does not seem to have an effect on the removal of another UV-induced DNA lesion 6-4PP (Fig. 2C). As reported before, even though DDB2 has very high binding affinity for 6-4PP, it does not have a significant effect on the recruitment of repair complex to 6-4PP and repair of this UV lesion. In DDB2-deficient XP-E cells, XPC was able to bind 6-4PP but not CPD (17), and the repair of CPD is profoundly reduced in XP-E cells, whereas 6-4PP removal is only moderately affected (10, 70). Moreover, whereas DDB2 degradation is required for efficient repair of CPD, it does not affect the repair of 6-4PP (22). Therefore, it is not surprising that p38 MAPK, which is critical for UV-induced DDB2 degradation, is not required for the repair of 6-4PP. Moreover, p38 MAPK-regulated chromatin remodeling also does not affect the removal of 6-4PP, perhaps because 6-4PP itself induces a significant DNA helix distortion and may be more accessible to NER factors without any accompanying chromatin relaxation.
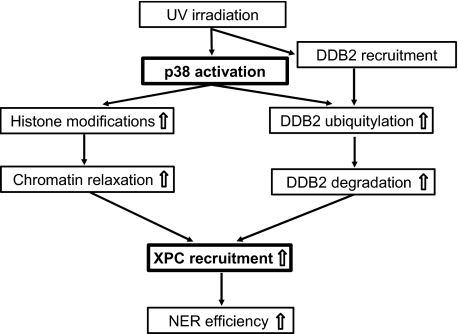
In summary, we have found that p38 MAPK facilitates NER through promoting both UV-induced chromatin relaxation and DDB2 degradation as outlined in a proposed model in Fig. 6. In response to UV irradiation, DDB2 is recruited to the damaged DNA sites independent of p38 MAPK activity. p38 MAPK is promptly activated by UV irradiation, which helps enhance the DDB2 ubiquitylation possibly through phosphorylating DDB2. Consequent degradation of DDB2 invokes the recruitment of XPC needed for continuation of repair process. In tandem, activated p38 MAPK also helps open up the condensed chromatin through increasing histone modifications, e.g. histone H3 acetylation and phosphorylation, and renders UV lesions accessible to NER factor assembly. Thus, the well timed recruitment of XPC engineered by chromatin relaxation as well as DDB2 degradation ensures efficient NER.
FIGURE 6.
Proposed model for positive modulation of NER by p38 MAPK activation following UV irradiation of human cells.
Acknowledgments
We thank Dr. Yoshihiro Nakatani for kindly providing the HeLa-DDB2 cell line.
This work was supported, in whole or in part, by National Institutes of Health Grants ES2388, ES12991, and CA93413. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: NER, nucleotide excision repair; MAPK, mitogen-activated protein kinase; CPD, cyclobutane pyrimidine dimer; 6-4PP, pyrimidine-pyrimidone (6-4) photoproduct; DDB, damaged DNA-binding protein; GG-NER, global genomic NER; ERK, extracellular signal-regulated kinase; JNK, c-Jun NH2-terminal kinase; DMSO, dimethyl sulfoxide; MNase, micrococcal nuclease; ISB, immuno-slot blot.
References
- 1.De Laat, W. L., Jaspers, N. G., and Hoeijmakers, J. H. (1999) Genes Dev. 13 768-785 [DOI] [PubMed] [Google Scholar]
- 2.Hanawalt, P. C. (2002) Oncogene 21 8949-8956 [DOI] [PubMed] [Google Scholar]
- 3.Donahue, B. A., Yin, S., Taylor, J.-S., Reines, D., and Hanawalt, P. C. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 8502-8506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanawalt, P., and Mellon, I. (1994) Curr. Biol. 3 67-69 [DOI] [PubMed] [Google Scholar]
- 5.Sugasawa, K., Ng, J. M. Y., Masutani, C., Iwai, S., Van der Spek, P., Eker, A., Hanoaka, F., Bootsma, D., and Hoeijmakers, J. H. J. (1998) Mol. Cell 2 223-232 [DOI] [PubMed] [Google Scholar]
- 6.Riedl, T., Hanaoka, F., and Egly, J. M. (2003) EMBO J. 22 5293-5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugasawa, K., Okamoto, T., Shimizu, Y., Masutani, C., Iwai, S., and Hanaoka, F. (2001) Genes Dev. 15 507-521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kusumoto, R., Masutani, C., Sugasawa, K., Iwai, S., Araki, M., Uchida, A., Mizukoshi, T., and Hanaoka, F. (2001) Mutat. Res. 485 219-227 [DOI] [PubMed] [Google Scholar]
- 9.Hwang, B. J., Toering, S., Francke, U., and Chu, G. (1998) Mol. Cell. Biol. 18 4391-4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang, J. Y., Hwang, B. J., Ford, J. M., Hanawalt, P. C., and Chu, G. (2000) Mol. Cell 5 737-744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu, G., and Chang, E. (1988) Science 242 564-567 [DOI] [PubMed] [Google Scholar]
- 12.Treiber, D. K., Chen, Z., and Essigmann, J. M. (1992) Nucleic Acids Res. 20 5805-5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reardon, J. T., Nichols, A. F., Keeney, S., Smith, C. A., Taylor, J.-S., Linn, S., and Sancar, A. (1993) J. Biol. Chem. 268 21301-21308 [PubMed] [Google Scholar]
- 14.Keeney, S., Chang, G. J., and Linn, S. (1993) J. Biol. Chem. 268 21293-21300 [PubMed] [Google Scholar]
- 15.Fujiwara, Y., Masutani, C., Mizukoshi, T., Kondo, J., Hanaoka, F., and Iwai, S. (1999) J. Biol. Chem. 274 20027-20033 [DOI] [PubMed] [Google Scholar]
- 16.Wakasugi, M., Shimizu, M., Morioka, H., Linn, S., Nikaido, O., and Matsunaga, T. (2001) J. Biol. Chem. 276 15434-15440 [DOI] [PubMed] [Google Scholar]
- 17.Fitch, M. E., Nakajima, S., Yasui, A., and Ford, J. M. (2003) J. Biol. Chem. 278 46906-46910 [DOI] [PubMed] [Google Scholar]
- 18.Moser, J., Volker, M., Kool, H., Alekseev, S., Vrieling, H., Yasui, A., Van Zeeland, A. A., and Mullenders, L. H. (2005) DNA Repair (Amst.) 4 571-582 [DOI] [PubMed] [Google Scholar]
- 19.Chen, X., Zhang, Y., Douglas, L., and Zhou, P. (2001) J. Biol. Chem. 276 48175-48182 [DOI] [PubMed] [Google Scholar]
- 20.Nag, A., Bondar, T., Shiv, S., and Raychaudhuri, P. (2001) Mol. Cell. Biol. 21 6738-6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rapic-Otrin, V., McLenigan, M. P., Bisi, D. C., Gonzalez, M., and Levine, A. S. (2002) Nucleic Acids Res. 30 2588-2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Mahdy, M. A., Zhu, Q., Wang, Q. E., Wani, G., Praetorius-Ibba, M., and Wani, A. A. (2006) J. Biol. Chem. 281 13404-13411 [DOI] [PubMed] [Google Scholar]
- 23.Aboussekhra, A., Biggerstaff, M., Shivji, M. K. K., Vilpo, J. A., Moncollin, V., Podust, V. N., Protic, M., Hubscher, U., Egly, J.-M., and Wood, R. D. (1995) Cell 80 859-868 [DOI] [PubMed] [Google Scholar]
- 24.Mu, D., Park, C.-H., Matsunaga, T., Hsu, D. S., Reardon, J. T., and Sancar, A. (1995) J. Biol. Chem. 270 2415-2418 [DOI] [PubMed] [Google Scholar]
- 25.Bessho, T., Sancar, A., Thompson, L. H., and Thelen, M. P. (1997) J. Biol. Chem. 272 3833-3837 [DOI] [PubMed] [Google Scholar]
- 26.Araujo, S. J., Tirode, F., Coin, F., Pospiech, H., Syvaoja, J. E., Stucki, M., Hubscher, U., Egly, J. M., and Wood, R. D. (2000) Genes Dev. 14 349-359 [PMC free article] [PubMed] [Google Scholar]
- 27.Kulaksiz, G., Reardon, J. T., and Sancar, A. (2005) Mol. Cell. Biol. 25 9784-9792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, Z., Wu, X., and Friedberg, E. C. (1991) J. Biol. Chem. 269 19034-19040 [Google Scholar]
- 29.Ura, K., Araki, M., Saeki, H., Masutani, C., Ito, T., Iwai, S., Mizukoshi, T., Kaneda, Y., and Hanaoka, F. (2001) EMBO J. 20 2004-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smerdon, M. J., and Thoma, F. (1990) Cell 61 675-684 [DOI] [PubMed] [Google Scholar]
- 31.Wellinger, R. E., and Thoma, F. (1997) EMBO J. 16 5046-5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramanathan, B., and Smerdon, M. J. (1986) Carcinogenesis 7 1087-1094 [DOI] [PubMed] [Google Scholar]
- 33.Ramanathan, B., and Smerdon, M. J. (1989) J. Biol. Chem. 264 11026-11034 [PubMed] [Google Scholar]
- 34.Smerdon, M. J., Lan, S. Y., Calza, R. E., and Reeves, R. (1982) J. Biol. Chem. 257 13441-13447 [PubMed] [Google Scholar]
- 35.Rubbi, C. P., and Milner, J. (2003) EMBO J. 22 975-986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheung, K. J., Jr., Mitchell, D., Lin, P., and Li, G. (2001) Cancer Res. 61 4974-4977 [PubMed] [Google Scholar]
- 37.Wang, J., Chin, M. Y., and Li, G. (2006) Cancer Res. 66 1906-1911 [DOI] [PubMed] [Google Scholar]
- 38.Kuo, W. H., Wang, Y., Wong, R. P., Campos, E. I., and Li, G. (2007) Exp. Cell Res. 313 1628-1638 [DOI] [PubMed] [Google Scholar]
- 39.Zhong, S. P., Ma, W. Y., and Dong, Z. (2000) J. Biol. Chem. 275 20980-20984 [DOI] [PubMed] [Google Scholar]
- 40.Cheung, P., Allis, C. D., and Sassone-Corsi, P. (2000) Cell 103 263-271 [DOI] [PubMed] [Google Scholar]
- 41.Wang, D., and Lippard, S. J. (2004) J. Biol. Chem. 279 20622-20625 [DOI] [PubMed] [Google Scholar]
- 42.Venkatachalam, S., Denissenko, M., and Wani, A. A. (1997) Oncogene 14 801-809 [DOI] [PubMed] [Google Scholar]
- 43.Li, J., Wang, Q. E., Zhu, Q., El-Mahdy, M. A., Wani, G., Praetorius-Ibba, M., and Wani, A. A. (2006) Cancer Res. 66 8590-8597 [DOI] [PubMed] [Google Scholar]
- 44.Wang, Q. E., Zhu, Q., Wani, G., El-Mahdy, M. A., Li, J., and Wani, A. A. (2005) Nucleic Acids Res. 33 4023-4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wani, A. A., D'Ambrosio, S. M., and Alvi, N. K. (1987) Photochem. Photobiol. 46 477-482 [DOI] [PubMed] [Google Scholar]
- 46.Wang, Q. E., Zhu, Q., Wani, G., Chen, J., and Wani, A. A. (2004) Carcinogenesis 25 1033-1043 [DOI] [PubMed] [Google Scholar]
- 47.Wang, Q. E., Zhu, Q., Wani, M. A., Wani, G., Chen, J., and Wani, A. A. (2003) DNA Repair 2 483-499 [DOI] [PubMed] [Google Scholar]
- 48.Davies, S. P., Reddy, H., Caivano, M., and Cohen, P. (2000) Biochem. J. 351 95-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rouse, J., Cohen, P., Trigon, S., Morange, M., onso-Llamazares, A., Zamanillo, D., Hunt, T., and Nebreda, A. R. (1994) Cell 78 1027-1037 [DOI] [PubMed] [Google Scholar]
- 50.Lali, F. V., Hunt, A. E., Turner, S. J., and Foxwell, B. M. (2000) J. Biol. Chem. 275 7395-7402 [DOI] [PubMed] [Google Scholar]
- 51.Wang, Z. G., Wu, X. H., and Friedberg, E. C. (1997) J. Biol. Chem. 272 24064-24071 [DOI] [PubMed] [Google Scholar]
- 52.Poizat, C., Puri, P. L., Bai, Y., and Kedes, L. (2005) Mol. Cell. Biol. 25 2673-2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li, J. P., and Yang, J. L. (2007) J. Cell. Physiol. 212 481-488 [DOI] [PubMed] [Google Scholar]
- 54.Kida, A., Kakihana, K., Kotani, S., Kurosu, T., and Miura, O. (2007) Oncogene 26 6630-6640 [DOI] [PubMed] [Google Scholar]
- 55.Casanovas, O., Miro, F., Estanyol, J. M., Itarte, E., Agell, N., and Bachs, O. (2000) J. Biol. Chem. 275 35091-35097 [DOI] [PubMed] [Google Scholar]
- 56.Fuchs, S. Y., Tappin, I., and Ronai, Z. (2000) J. Biol. Chem. 275 12560-12564 [DOI] [PubMed] [Google Scholar]
- 57.Sugasawa, K., Okuda, Y., Saijo, M., Nishi, R., Matsuda, N., Chu, G., Mori, T., Iwai, S., Tanaka, K., Tanaka, K., and Hanaoka, F. (2005) Cell 121 387-400 [DOI] [PubMed] [Google Scholar]
- 58.Hunter, T. (2007) Mol. Cell 28 730-738 [DOI] [PubMed] [Google Scholar]
- 59.Christmann, M., Tomicic, M. T., Aasland, D., and Kaina, B. (2007) Carcinogenesis 28 183-190 [DOI] [PubMed] [Google Scholar]
- 60.Christmann, M., Tomicic, M. T., Origer, J., Aasland, D., and Kaina, B. (2006) Nucleic Acids Res. 34 6530-6539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayakawa, J., Depatie, C., Ohmichi, M., and Mercola, D. (2003) J. Biol. Chem. 278 20582-20592 [DOI] [PubMed] [Google Scholar]
- 62.Potapova, O., Haghighi, A., Bost, F., Liu, C., Birrer, M. J., Gjerset, R., and Mercola, D. (1997) J. Biol. Chem. 272 14041-14044 [DOI] [PubMed] [Google Scholar]
- 63.Andrieux, L. O., Fautrel, A., Bessard, A., Guillouzo, A., Baffet, G., and Langouet, S. (2007) Cancer Res. 67 2114-2123 [DOI] [PubMed] [Google Scholar]
- 64.Rouget, R., Auclair, Y., Loignon, M., Affar, e. B., and Drobetsky, E. A. (2008) J. Biol. Chem. 283 5533-5541 [DOI] [PubMed] [Google Scholar]
- 65.Yu, Y., Teng, Y., Liu, H., Reed, S. H., and Waters, R. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8650-8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheung, P., Tanner, K. G., Cheung, W. L., Sassone-Corsi, P., Denu, J. M., and Allis, C. D. (2000) Mol. Cell 5 905-915 [DOI] [PubMed] [Google Scholar]
- 67.Lo, W. S., Trievel, R. C., Rojas, J. R., Duggan, L., Hsu, J. Y., Allis, C. D., Marmorstein, R., and Berger, S. L. (2000) Mol. Cell 5 917-926 [DOI] [PubMed] [Google Scholar]
- 68.Clayton, A. L., Rose, S., Barratt, M. J., and Mahadevan, L. C. (2000) EMBO J. 19 3714-3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hara, R., Mo, J., and Sancar, A. (2000) Mol. Cell. Biol. 20 9173-9181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hwang, B. J., Ford, J. M., Hanawalt, P. C., and Chu, G. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 424-428 [DOI] [PMC free article] [PubMed] [Google Scholar]