Abstract
We reported that peroxisome proliferator-activated receptor γ (PPARγ) transcriptionally regulates the β-cell differentiation factor pancreatic duodenal homeobox (PDX)-1 based on in vitro RNA interference studies. We have now studied mice depleted of PPARγ within the pancreas (PANC PPARγ-/-) created by a Cre/loxP recombinase system, with Cre driven by the pdx-1 promoter. Male PANC PPARγ-/- mice were hyperglycemic at 8 weeks of age (8.1 ± 0.2 mm versus 6.4 ± 0.3 mm, p = 0.009) with islet cytoarchitecture and pancreatic mass of islet β-cells that were indistinguishable from the controls. Islet PDX-1 mRNA (p = 0.001) and protein levels (p = 0.003) were lowered 60 and 40%, respectively, in tandem with impaired glucose-induced insulin secretion and loss of thiazolidinedione-induced increase in PDX-1 expression. We next identified a putative PPAR-response element (PPRE) in the mouse pdx-1 promoter with substantial homology to the corresponding region of the human PDX-1 promoter. Electrophoretic mobility supershift assays with nuclear extracts from β-cell lines and mouse islets, also in vitro translated PPARγ and retinoid X receptor, and chromatin immunoprecipitation analysis demonstrated specific binding of PPARγ and retinoid X receptor to the human and mouse pdx-1 × PPREs. Transient transfection assays of β-cells with reporter constructs of mutated PPREs showed dramatically reduced pdx-1 promoter activity. In summary, we have presented in vivo and in vitro evidence showing PPARγ regulation of pdx-1 transcription in β-cells, plus our results support an important regulatory role for PPARγ in β-cell physiology and thiazolidinedione pharmacology of type 2 diabetes.
Peroxisome proliferator-activated receptor γ (PPARγ)4 is a member of the nuclear hormone receptor superfamily of ligand-inducible transcription factors (1) and contributes significantly to diverse biological processes such as glucose homeostasis, lipid metabolism, cellular proliferation, and differentiation (2–4). PPARγ regulates transcriptional activity of target genes by forming a heterodimer with retinoid X receptor (RXR), and binding to a specific PPARγ response element sequence (PPRE) within the promoter region (5, 6). The PPRE consists of two hexamer repeats (DR1 and DR2) separated by a single nucleotide, with PPARγ and RXR occupying the 5′- and 3′-half-sites, respectively (7, 8). Thiazolidinediones are PPARγ agonists used for the treatment of type 2 diabetes based on insulin-sensitizing effects in adipose tissue and muscle (9). However, a recent finding is PPARγ-mediated anti-proliferation and pro-survival of islet β-cells (10–12). This is of interest as clinical trials showing glycemic benefits of thiazolidinediones in prediabetes and early type 2 diabetes (13–17) have usually been interpreted that the insulin sensitization effect unloads overstimulated insulin secretion, so-called “β-cell rest.” On the other hand, the possibility of direct PPARγ regulatory effects in islet β-cells is controversial, as a β-cell-specific PPARγ knock-out mouse was normoglycemic basally and after fat feeding (11). Also little is known about PPARγ target genes in β-cells.
PPARγ regulates the function and survival of tissues by acting through a triad of effects as follows: anti-proliferation, prosurvival, and pro-differentiation (18). Because PPARγ-mediated pro-survival and anti-proliferation had been reported for β-cells (10–12), one might also expect pro-differentiation properties. Our group has studied a model of β-cell adaptation to a loss of β-cell mass, 60% pancreatectomy (Px) Sprague-Dawley rats. They are normoglycemic because of partial β-cell regeneration during the 1st week post-Px (19, 20) that is followed by β-cell hyperfunction secondary to increased glucokinase activity (21). We investigated the transition phase, 14 days post-Px, to determine how these events are coordinated, and we found increased nuclear PPARγ expression in isolated islets at mRNA and protein levels (22). We also noted increased nuclear expression of pancreatic duodenal homeobox (PDX)-1 that is an essential regulator of the function and survival of mature β-cells (23–28). We then studied INS-1 β-cells following RNAi-induced 75% knockdown of PPARγ, finding PDX-1 mRNA and protein levels were lowered 80 and 60%, respectively. Based on these collective results, we proposed that pdx-1 is a physiologically regulated target gene for PPARγ in β-cells (22).
We now set out to verify these findings in vivo by studying mice with a pancreas-specific ablation of PPARγ based on a Cre/loxP recombinase system with Cre driven by the pdx-1 promoter (29). Our findings confirmed the expected reduction of PDX-1 expression in isolated islets. Moreover, these mice were characterized by glucose intolerance, impaired glucose-induced insulin secretion, and loss of the pharmacological effect of thiazolidinediones to up-regulate PDX-1 expression. In addition, we have identified and characterized a novel functional PPRE within the mouse and human pdx-1 promoter regions.
EXPERIMENTAL PROCEDURES
Animal Studies—Mice with PPARγ deficiency restricted to pancreatic islets, ducts, and acini (PANC PPARγ-/-) were generated from crossing pdx-1-Cre mice (original source D. A. Melton, described in Ref. 30) and mice with two floxed PPARγ alleles as detailed previously (29). Controls were littermate Cre negative PPARγ floxed mice. All protocols were in accordance with the principles of laboratory animal care and were approved by the Institutional Animal Use and Care Committee of the University of Vermont. At 8 weeks of age, tail vein blood sampling for blood glucose (Freestyle glucose monitor) was performed from nonanesthetized normally fed mice. Some animals underwent an intraperitoneal glucose tolerance test that consisted of an intraperitoneal injection of 2 g/kg glucose with blood glucose measured at 0, 30, 60, 90, and 120 min. Islets were isolated by pancreas duct infiltration with collagenase, Histopaque gradient separation, and hand picking.
Tissue Immunoblots for PPARγ—Mouse tissues (liver, kidney, skeletal muscle, hypothalamus, heart, small intestine, and islets) were homogenized in 1 ml of lysis buffer (50 mm HEPES, pH 7.4, 150 mm sucrose, 2 mm sodium orthovanadate, 80 mm β-glycerophosphate, 10 mm sodium fluoride, 10 mm sodium pyrophosphate, 2 mm sodium EGTA, 2 mm sodium EDTA, 1% Triton X-100, 0.1% SDS, 100 μl of protease inhibitor mixture for mammalian cell culture (Sigma), 1 mm phenylmethylsulfonyl fluoride). Protein aliquots (60 μg) were resolved on 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Bio-Rad) that were incubated overnight at 4 °C with mouse monoclonal PPARγ antibody (Chemicon) and then with goat anti-mouse horseradish peroxidase-conjugated antibody (Jackson ImmunoResearch) for 1 h at room temperature. Detection was by chemiluminescence using HyperFilm-ECL (Amersham Biosciences). Membranes were stripped and reprobed to establish equivalent loading using anti-β-actin (Sigma).
Pancreas Histology and Morphometrics—Pancreata were rapidly excised, cleared of fat and lymph nodes, and blotted before immersion-fixing overnight in 4.0% paraformaldehyde in 0.1 mm phosphate buffer at 4 °C. After washing in several changes of phosphate-buffered saline, tissues were dehydrated and embedded in paraffin. Pancreas sections were incubated overnight at 4 °C in antibody mixtures of sheep anti-amylase (Biogenesis), guinea pig anti-insulin (Linco), and rabbit anti-glucagon (Linco). β-Cell mass was quantified using a computerized planimetric method and β-cell proliferation frequency using Ki-67 as the cell cycle marker, using methods detailed previously (31).
Islet Insulin Secretion—Freshly isolated islets were incubated 1 h in RPMI 1640 media supplemented with 10% fetal calf serum, 10 mm HEPES, 2 mm l-glutamine, 1 mm sodium pyruvate, 100 units/ml penicillin, 100 μg/ml streptomycin, 11.1 mm glucose, at 37 °C in a 5% CO2 incubator. Duplicate batches of 15–25 islets were preincubated in 12-well plates for 2 h in RPMI 1640 medium at 2.8 mm glucose and then 1 h in KRBH plus 0.5% bovine serum albumin and 2.8 mm glucose. Insulin secretion was assessed during a subsequent 1-h incubation in KRBH, 0.5% bovine serum albumin at 2.8 or 16.7 mm glucose at 37 °C, followed by ultrasensitive mouse insulin enzyme-linked immunosorbent assay (Mercodia) of the medium. Islet insulin content was measured after acid-ethanol (0.2 mm HCl in 75% ethanol) extraction, and the insulin secretion results were normalized to the total insulin content.
Islet Culture with Troglitazone—Isolated islets were incubated in culture media supplemented with 10 μm troglitazone or vehicle (DMSO) for 72 h with the media changed each 24 h. After the incubation period, islets were lysed and immunoblots were performed (20 μg) using the technique described above, with rabbit anti-PDX-1 (Chris Wright, Vanderbilt University) followed by reprobing with anti-β-actin (Sigma).
Islet PCR and Immunoblots—Islet total RNA was extracted using the RNeasy Micro kit plus single step on-column DNase digestion (Qiagen). cDNAs were synthesized using 500 ng of extracted RNA with ImProm-II reverse transcriptase (Promega), dNTPs, and random hexamer primers. PCR analyses were carried out in a PTC-200 Peltier Thermal Cycler (MJ Research) using the cDNAs together with TaqDNA polymerase (Promega) and primer combinations (sequences available on request). The thermal cycle program was denaturing step at 95 °C for 2 min followed by 35 cycles for PPARγ, PPARα, and glucagon or 25 cycles for PDX-1, GLUT2, INS-1, and INS-2, at 94 °C for 15 s, 56 °C for 30 s, and 72 °C for 60 s, with extension step of 5 min at 72 °C. α-Tubulin and cyclophilin A were used as internal controls. Gel images were captured using the Gel Doc EQ documentation system (Bio-Rad) and analyzed using ImageJ software (National Institutes of Health), with the results expressed relative to the control genes.
PDX-1 and β-actin immunoblots were performed as detailed previously. Immunoblot and PCR results are expressed as mean ± S.E. from three pairs of littermate floxed control and PANC PPARγ-/- mice. Statistical significance was determined by Student's t test.
Tissue Culture—INS-1 (832/13) cells (gift from Christopher Newgard, Duke University) were maintained in RPMI 1640 medium containing 10% fetal calf serum, 8.3 mm glucose, 10 mm HEPES, 100 mm l-glutamine, 50 mm sodium pyruvate, 100 units/ml penicillin, 100 μg/ml streptomycin, and 50 μm β-mercaptoethanol. βTC6 cells were maintained in Dulbecco's modified Eagle's medium containing 5 mm glucose supplemented with 10% fetal bovine serum, 10 mm HEPES, 200 mm l-glutamine, 1 mm sodium pyruvate, 0.15% sodium bicarbonate, 50 units/ml penicillin, and 50 μg/ml streptomycin.
Electrophoretic Mobility Shift Assay—Nuclear extracts from INS-1 cells, mouse islets, and βTC6 cells were prepared using the NucBuster kit (Novagen). In vitro transcription/translation of PPARγ (plasmid pcDNA-PPARγ, Addgene) and retinoid X receptor-α (RXR-α, plasmid pSVsport-RXR-α, Addgene) was performed using the T7 and SP6 RNA polymerase-specific TnT quick-coupled transcription/translation kits (Promega), respectively. PAGE purified oligos against the mouse pdx-1 × PPRE (FW 5′-AGCTGAGGCAGGGTACCTCCAGTATCA-3′; REV 5′-GATCTGATACTGGAGGTACCCTGCCTC-3′), human PDX-1 × PPRE (FW 5′-AGCTGCCGGCAAGGACCTCCAGTATCA-3′; REV 5′-GATCTGATACTGGAGGTCCTTGCCGGC-3′), and acyl-CoA oxidase PPRE (FW 5′-CCCGAACGTGACCTTTGTCCTGGTCC-3′; REV 5′-AGCTGGACCAGGACAAAGGTCACGTT-3′) were synthesized (IDT), annealed, labeled by end-filling with [α-32P]dCTP (PerkinElmer Life Science), and purified using the QIAquick nucleotide removal kit (Qiagen). DNA binding reaction was performed in 20 μl of reaction mixture containing EMSA buffer (100 mm KCl, 20 mm HEPES, 0.2 mm EDTA, 20% glycerol, 0.5 mm dithiothreitol) and 500 ng of solicited salmon sperm DNA, 0.01 unit of poly(dI-dC), 10 μg of nuclear extract, 60,000 cpm of [α-32P]dCTP-labeled ds probe, and incubated 30 min at room temperature. Overnight cast 6% nondenaturing DNA retardation gels (29:1 acrylamide to bisacrylamide) were pre-run for 30 min in EMSA running buffer in 0.5 m Tris, then loaded with reaction mixture (18 μl of DNA·protein complex mix, 2 μl of 6× DNA loading dye), and samples separated by electrophoresis for 2.5 h at 100 V. For competition studies, DNA binding reaction mixtures were preincubated with unlabeled ds DNA oligos of the mouse wild type pdx-1 × PPRE or mutant sequences (Mut 1, FW 5′-AGCTGAGGCAGGGTACCTAAAATATCA-3′; REV 5′-GATCTGATATTTTAGGTACCCTGCCTC-3′; Mut 2, FW 5′-AGCTGAGGCAAAATAACTCCAGTATCA-3′; REV 5′-GATCTGATACTGGAGTTATTTTGCCTC-3′; Mut 3, FW 5′-AGCTGAGGCAAAATAACTAAAATATCA-3′; REV 5′-GATCTGATATTTTAGTTATTTTGCCTC-3′). Alternatively, unlabeled acyl-CoA oxidase PPRE was used to compete with the [α-32P]dCTP-labeled ds mouse wild type pdx-1 × PPRE probe. For supershift gel retardation assay, binding reactions were followed by addition of PPARγ-specific antibody (Biomol) for another 20 min before resolving the DNA·protein complex on a nondenaturing gel. When in vitro translated PPARγ/RXR-α proteins (2.5 μl) were used, binding reactions were preincubated with PPARγ, pan-RXR, or RXR-α antibodies (Santa Cruz Biotechnology) for 30 min on ice followed by 30 min of room temperature incubation after addition of [α-32P]dCTP-labeled ds PPRE probes for mouse pdx-1, human PDX-1, or acyl-CoA oxidase PPREs.
Site-directed Mutagenesis—One-kb mouse pdx-1 promoter sequence (-2916 to -1920) containing the putative PPARγ-binding site (-2720 to -2708) was subcloned in pTAL luciferase reporter gene vector (Clontech). Mutation of the wild type pdx-1 fragment was carried out using QuickChange XL site-directed mutagenesis kit (Stratagene). Briefly, using PAGE-purified mutagenic primer pairs: SDM-1 FW 5′-GGA AGA GAG GCA GGG TAC CTA AAA TAT CAG GGA GGA CTA TCA G-3′; SDM-1 REV 5′-CTG ATA GTC CTC CCT GAT ATT TTA GGT ACC CTG CCT CTC TTC C-3′; SDM-2 FW 5′-GGA AGA GAG GCA AAA TAA CTC CAG TAT CAG GGA GGA CTA TCA G-3′; SDM-2 REV 5′-CTG ATA GTC CTC CCT GAT ACT GGA GTT ATT TTG CCT CTC TTC C-3′, and pTAL × PPRE × pdx promoter plasmid as template, PCR was performed with the following cycles: 95 °C for 1 min; 18 cycles of 95 °C for 50 s, 60 °C for 1 min, 68 °C for 6 min. Parental DNA was digested using DpnI (10 units). XL10 gold ultracompetent cells were transformed with the mutagenic PCR product, and after plasmid preparation, incorporation of the mutagenesis was confirmed by sequencing (University of Vermont, DNA core facility).
Chromatin Immunoprecipitation Assay—Chromatin immunoprecipitation assay was performed with mouse-derived βTC6 cells using ChIP-IT kit (Active Motif). Cells were fixed using formaldehyde solution, and the chromatin was sheared by enzymatic digestion according to the instruction manual to DNA fragments that averaged 300–500 bp in length. Mouse monoclonal antibody against PPARγ (E8, Santa Cruz Biotechnology) was added to aliquots of precleared chromatin and incubated overnight, with parallel samples incubated with the negative-control IgG provided with the kit. Protein G-agarose beads were added, and the mixture was incubated for 1.5 h at 4 °C. After reversing the cross-links, DNA was isolated, and PCRs were performed with primers for the mouse pdx-1 promoter region PPRE (GenBank™ accession number AF192495). Primer sequences were as follows: FW 5′-ACACACTCACTCACTCACTCATTGGG-3′; REV 5′-CTGAGATACCCAGCCATTAGGCAAGA-3′ (expected PCR product 312 bp). A second primer pair was as follows: FW 5′-CAATCTAGTCCAAACCAGCCTTTGGC-3′; REV 5′-TGAGATACCCAGCCATTAGGCAAGAG-3′ (expected PCR product 265 bp). As negative control, a primer pair (FW 5′-GCGCTGAGTTCTGCAAGCATTTCT-3′; REV 5′-CGCGAACACCTGCACTTGTTTCAA-3′) was selected that amplified a stretch of 450 bp of DNA 2.56 kb downstream of the mouse pdx-1 × PPRE. PCR conditions were 1 cycle of 94 °C for 3 min, 39 cycles of 94 °C for 30 s, 62 °C for 1 min, 72 °C for 1 min. A mouse-specific positive control for appropriate shearing of DNA and co-immunoprecipitation was performed using a kit (Active Motif) based on binding of transcription factor EFI-α by anti-RNA pol II.
Luciferase Reporter Gene Assay—80–90% confluent INS-1 cells in a 6-well format were incubated overnight in antibiotic-free media. Cells were transfected using Lipofectamine 2000 transfection reagent (Invitrogen) with 2 μg of pTAL empty vector (firefly luciferase vector) or equivalent wild type or mutated pTAL × PPRE pdx-1 promoter vector. Renilla luciferase reporter plasmid (pRL-TK, Promega) was included (0.05 μg) in all transfections as internal control. Twenty four h post-transfection, cells were incubated another 24 h with 10 μm troglitazone or DMSO, and the luciferase assay performed in a TD 20/20 luminometer (Turners Design) using dual luciferase assay kit (Promega). Firefly luciferase activity was normalized with Renilla luciferase and expressed as relative luciferase activity. Each experimental condition was performed in triplicate with the results calculated as relative (%) luciferase value of the wild type PPRE controls.
RESULTS
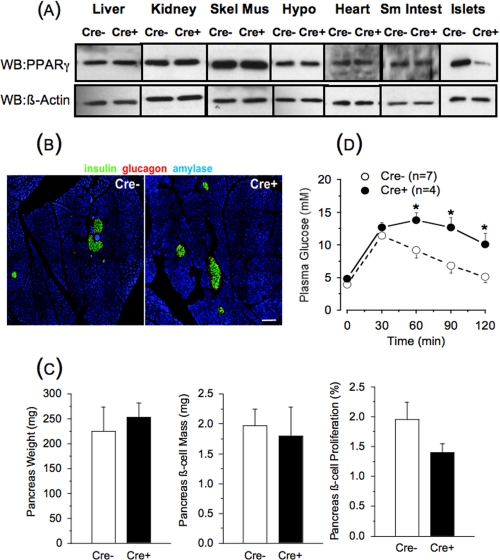
PANC PPARγ-/- Mice—We studied mice with PPARγ deficiency within the pancreas created by a Cre/loxP recombinase system, with Cre driven by the pdx-1 promoter (29). PDX-1 is highly expressed in all pancreatic epithelial cells during embryogenesis, with continued high expression in β-cells postnatally as opposed to a marked reduction in other pancreatic tissues (32). Thus, the pdx-1-driven Cre creates a PPARγ knock-out in β-cells although there remains the possibility of additional effects related to PPARγ deletion in ducts, acini, and other islet cells. The original description of these mice reported increased pancreas weight, with larger islets and normal pancreas histology by gross inspection, compared with control mice (29). We studied 8–10-week-old male animals and confirmed the pancreas specificity of the PPARγ deletion (Fig. 1A). We also found no abnormal pancreas histological features, as islet cytoarchitecture and insulin and glucagon immunostaining intensities were indistinguishable from the controls (Fig. 1B). In contrast, there was no difference in pancreas weight, β-cell mass, or β-cell proliferation rate between the PANC PPARγ-/- and the floxed control mice (Fig. 1C). PANC PPARγ-/- mice had the same body weight (29 ± 1 g versus 28 ± 1 g, n = 7) as the controls, but unexpectedly, nonfasting glucose values were higher (8.1 ± 0.2 mm versus 6.4 ± 0.3 mm, n = 4, p = 0.009) and there was glucose intolerance during an intraperitoneal glucose tolerance test (Fig. 1D).
FIGURE 1.
Tissue panel for PPARγ immunoblot (A), pancreas histology (B), pancreas morphometrics (C), and intraperitoneal glucose tolerance test results (D) in 8-week-old male floxed control (Cre-) and PANC PPARγ-/- mice (Cre+). A, representative PPARγ immunoblot of liver, kidney, skeletal muscle (Skel Mus), hypothalamus (Hypo), heart, small intestine (Sm Intest), and isolated islets. Membranes were stripped and reprobed to establish equivalent loading using anti-β-actin antibody. WB, Western blot. B, representative immunofluorescence staining for insulin, glucagon, and amylase in pancreatic sections of control and PANC PPARγ-/- mice. Scale bars, 50 μm. C, comparison of pancreas weights, pancreas β-cell mass, and β-cell proliferation rates of control and PANC PPARγ-/- mice. None of the parameters statistically differed between the animal groups. D, blood glucose values after intraperitoneal injection of glucose (2 g/kg body weight) in control and PANC PPARγ-/- mice. *, p < 0.05.
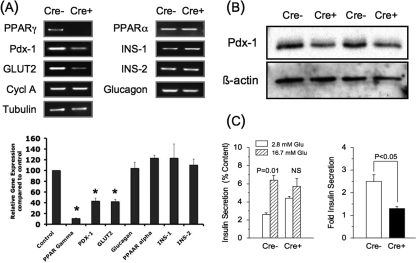
Isolated Islet Studies—Analysis of mRNA levels from isolated islets confirmed the ablation of PPARγ expression (p < 0.001) as opposed to PPARα that was unaffected, demonstrating the specificity of the gene inactivation (Fig. 2A). As predicted, islet PDX-1 mRNA (43 ± 6% of control islets, n = 3, p = 0.001) and protein levels (58 ± 2% of control islets, n = 3, p = 0.003) were markedly lowered in the PANC PPARγ-/- mice compared with the controls (Fig. 2, A and B, respectively). GLUT2 mRNA also was lowered (p = 0.006) as opposed to no change in glucagon, insulin 1, or insulin 2 mRNA levels.
FIGURE 2.
Quantitative PCR analysis (A), PDX-1 immunoblot (B), and glucose-stimulated insulin secretion (C), in isolated islets from 8-week-old male floxed control (Cre-) and PANC PPARγ-/- mice (Cre+). A, quantitative PCR analysis of various genes from total RNA preparations of isolated islets. α-Tubulin and cyclophilin A (Cycl A) were used as internal controls. A representative gel is shown, and the graph contains the mean ± S.E. band intensities from the three separate experiments, with the Cre+ islets expressed as % intensity compared with the Cre- islets. *, p < 0.05. B, representative immunoblot for PDX-1 and β-actin from islet lysates of two control and two PANC PPARγ-/- mice. C, insulin secretion from freshly isolated Cre- (n = 4) and Cre+ (n = 3) islets stimulated for 1 h with 2.8 or 16.7 mm glucose expressed as percentage of total insulin content. Also shown is the fold increase of the insulin response at 16.7 mm glucose versus 2.8 mm glucose.
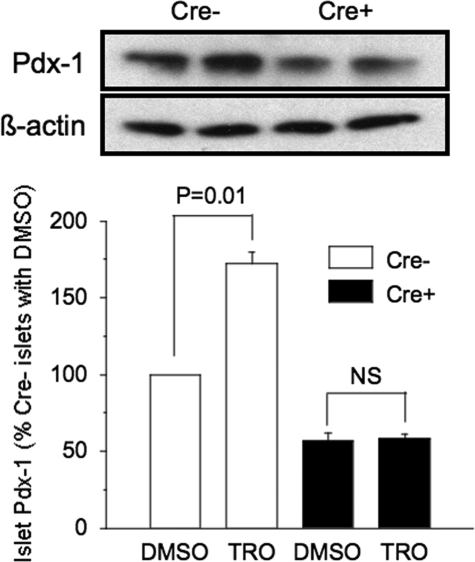
Functional assessment showed a near-total absence of glucose-induced insulin secretion in the PPARγ-/- islets, based on no statistical difference in the in vitro insulin response to 16.7 mm glucose versus 2.8 mm glucose (1.3 ± 0.1-fold), compared with a 2.5 ± 0.3-fold increase in the control islets (Fig. 2C). Also, control islets cultured for 3 days with troglitazone had a near-doubling of Pdx-1 levels. In contrast, troglitazone induced no change of the lowered PDX-1 level in PPARγ-/- islets (Fig. 3).
FIGURE 3.
PDX-1 immunoblot post 72 h of incubation with troglitazone or the diluent DMSO in isolated islets from 8-week-old male floxed control (Cre-) and PANC PPARγ-/- mice (Cre+). Isolated islets were cultured 72 h in medium with 10 μm troglitazone (Tro) or vehicle (DMSO) and then islet lysates underwent PDX-1 immunoblotting. Membranes were stripped and reprobed with β-actin antibody. A representative gel is shown, and the graph contains the mean ± S.E. band intensities from the three separate experiments expressed as % intensity of each experimental condition compared with Cre- islets incubated with DMSO. NS = not significant.
PPRE Sequence on Mouse and Human pdx-1 Promoters—We next investigated the molecular basis for PPARγ regulation of pdx-1 transcription. A PPRE consists of hexamer repeats (DR1 and DR2) separated by one nucleotide, with the consensus sequence being RGGTCA-A-AGGTCA. However, a characteristic feature is a high degree of homology for the 3′-half-site but substantial variance for the 5′-half-site (33), reflecting highly efficient RXR binding to the 3′ DR2 half-site as part of the PPARγ/RXR heterodimer as compared with PPARγ binding to the 5′ DR1 half-site (34). As such, we searched for a PPRE on the mouse pdx-1 promoter using the DR2 consensus sequence to probe for a homologous area within the 4.531-kb mouse pdx-1 promoter region (GenBank™ accession number AF192495). A putative PPRE consisting of GGGTAC-C-TCCAGT was identified at positions -2716/-2698 relative to the transcription initiation site in area 1 of the mouse pdx-1 promoter (Table 1A) as characterized by Gerrish et al. (35). The identified DR2 element was fully homologous to the consensus sequence. In contrast, we compiled a data base of known PPREs (data not shown), and we noted the DR1 sequence was unique from reported PPREs. Comparison of the putative mouse pdx-1 × PPRE with the analogous human PDX-1 promoter sequence revealed partial homology (a three-nucleotide mismatch) for the DR-1 element and full homology for the DR-2 (Table 1, part B).
TABLE 1.
PPRE within the mouse and human pdx-1 promoters
partial sequence of the mouse pdx-1 promoter region (GenBank™ accession number AF192495). The underlined region represents the putative PPRE and complementary sequence
comparison of PPRE sequence of mouse pdx-1 promoter to the analogous sequence stretch of area 1 within the human PDX-1 promoter (35)
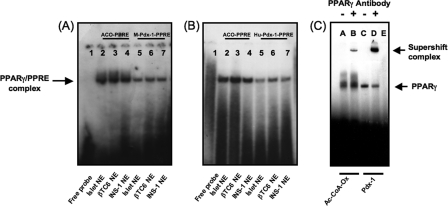
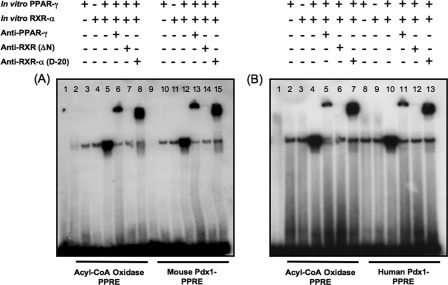
Electrophoretic Mobility Shift and Mutational Studies—We determined whether the putative mouse and human pdx-1 × PPREs bind PPARγ using nuclear extracts of isolated mouse islets, mouse-derived βTC6 cells, and rat-derived INS-1 cells, incubated with 32P-labeled double-stranded oligos of the mouse (Fig. 4A) and human (Fig. 4B) pdx-1 × PPRE sequences. An oligo for the PPRE of acyl-CoA oxidase (36) was included as internal control. In all tissues, complexes were apparent with the pdx-1 × PPREs that migrated at an identical position as the acyl-CoA oxidase PPRE complex. The specificity of the PPARγ·PPRE complex was confirmed using PPARγ antibody that supershifted the mouse pdx-1 × PPRE band (Fig. 4C, lane D) in the same fashion as the PPRE in acyl-CoA oxidase (Fig. 4C, lane B). We also determined the ability of the mouse pdx-1, human PDX-1, and acyl-CoA oxidase PPRE probes to bind in vitro translated PPARγ and RXR-α, alone and in combination (Fig. 5, A and B). The translated PPARγ and RXR-α proteins individually and in combination resulted in a complex with identical mobility using all three probes (Fig. 5A, lanes 3–5 and 10–12; Fig. 5B, lanes 2–4 and 8–10). Analysis by immunoblot demonstrated the reticulocyte lysate used for programming the in vitro translated proteins contained residual amounts of PPARγ and RXR-α accounting for the complexes observed in Fig. 5. Addition of PPARγ antibody or N terminus RXR-α antibody supershifted the complex. In contrast, pan-RXR antibody to the DNA and ligand binding domain of RXR protein, but lacking the N terminus, prevented formation of the complex, indicating that it interfered with PPARγ and RXR-α heterodimerization and DNA binding.
FIGURE 4.
Nuclear extract binding to the putative PPRE sequences in the mouse (A) and human pdx-1 promoters (B) along with the PPRE in acyl-CoA oxidase as control, and supershift of the DNA·protein complexes of the acyl-CoA oxidase and mouse pdx-1 × PPREs using PPARγ-specific antibody (C) are shown. 32P-Labeled double-stranded probes for the PPRE in acyl-CoA oxidase and the purported mouse and human pdx-1 × PPREs, underwent DNA binding reaction using nuclear extracts from mouse islets, mouse-derived βTC6 cells, or rat-derived INS-1 cells. A and B, lane 1, free probe; lanes 2–4, nuclear extracts of mouse islets, βTC6 cells, and INS-1 cells with acyl-CoA oxidase PPRE; lanes 5–7, nuclear extracts of mouse islets, βTC6 cells, and INS-1 cells with pdx-1 × PPRE. NE = nuclear extract; ACO = acyl-CoA oxidase. C, INS-1 cell nuclear extract was preincubated with PPARγ-specific antibody or rabbit nonimmune serum on ice for 30 min before adding 32P-labeled acyl-CoA oxidase or mouse pdx-1 × PPRE oligo probes and resolution of the DNA·protein complexes by PAGE. Lanes A and B, acyl-CoA oxidase PPRE with nonimmune serum or PPARγ-specific antibody, respectively; lanes C and D, pdx-1 × PPRE with nonimmune serum or PPARγ-specific antibody, respectively; lane E, free probe.
FIGURE 5.
In vitro translated PPARγ and RXR-α are binding partners for acyl-CoA oxidase, mouse pdx-1, and human pdx-1 × PPRE probes. In vitro translated RXR-α and PPARγ proteins were used in the DNA binding reaction in place of nuclear extracts (alone or in combination) along with 32P-labeled oligo PPRE probes for acyl-CoA oxidase and mouse pdx-1 (A) or human PDX-1 (B). For the supershift assays, binding reactions were preincubated with antibodies for PPARγ, RXR, or RXR-α. A, lanes 2–8, acyl-CoA oxidase PPRE probe; lanes 9–15 mouse pdx-1 × PPRE probe. Lane 1, free probe; lanes 2 and 9, INS-1 nuclear extract; lanes 3 and 10: in vitro translated PPARγ; lanes 4 and 11, in vitro translated RXR-α; lanes 5 and 12, in vitro translated RXR-α and PPARγ together; lanes 6 and 13, in vitro translated RXR-α and PPARγ together showing supershifted complex with PPARγ-specific antibody; lanes 7 and 14, in vitro translated RXR-α and PPARγ showing a lowered band intensity from prevention of the complex formation with the PAN RXR antibody; lanes 8 and 15, in vitro translated RXR-α and PPARγ together showing supershifted complex with RXR-α-specific antibody. B, lanes 2–7, acyl-CoA oxidase PPRE probe; lanes 8–13, human pdx-1 ×PPRE probe. Lane 1, free probe; lanes 2 and 8, in vitro translated PPARγ; lanes 3 and 9, in vitro translated RXR-α; lanes 4 and 10, in vitro translated RXR-α and PPARγ together; lanes 5 and 11, in vitro translated RXR-α and PPARγ together showing supershifted complex with PPARγ-specific antibody; lanes 6 and 12, in vitro translated RXR-α and PPARγ showing a lowered band intensity from prevention of the complex formation with the PAN RXR antibody; lanes 7 and 13, in vitro translated RXR-α and PPARγ together showing supershifted complex with RXR-α-specific antibody.
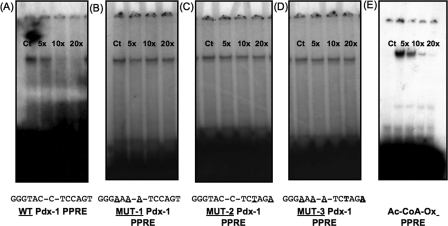
Specificity of binding was shown with mutational studies, in which up to a 20-fold excess of unlabeled oligos that contained mutations in the DR1 half-site of the mouse pdx-1 × PPRE, DR2 half-site, and combined DR1-DR2 mutations failed to elicit competition as reflected in no change in band intensity (Fig. 6, B–D) in contrast to the wild type probe (Fig. 6A). In a reverse competition experiment, increasing concentrations of unlabeled acyl-CoA oxidase PPRE successfully competed with the radiolabeled mouse pdx-1 × PPRE probe (Fig. 6E).
FIGURE 6.
Mutated mouse pdx-1 × PPRE fails to compete with 32P-labeled pdx-1 × PPRE probes. Double-stranded pdx-1 × PPRE oligos were labeled by end filling with [32P], and DNA binding reaction performed using INS-1 cell nuclear extracts as described under “Experimental Procedures.” Competition was assessed by adding 5–20× molar excess of unlabeled wild type or mutated pdx-1 × PPRE oligos (A–D). Mutated sequences are shown at the bottom of each panel. Alternatively, competition was assessed with 5–20× molar excess of unlabeled wild type acyl-CoA oxidase PPRE oligo added to the binding reaction (E). A, wild type unlabeled pdx-1 × PPRE oligo. B, mutated 5′-half-site (DR1) unlabeled pdx-1 × PPRE oligo (MUT 1). C, mutated 3′-half-site (DR2) unlabeled pdx-1 × PPRE oligo (MUT 2). D, combined mutated DR1 and DR2 unlabeled pdx-1 × PPRE oligo (MUT 3). E, wild type unlabeled acyl-CoA oxidase PPRE probe. Ct = control.
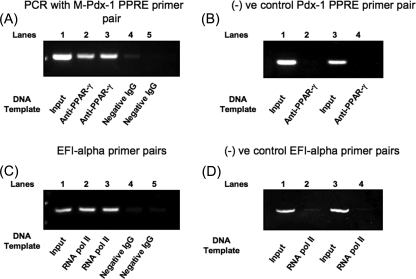
ChIP Assay—Binding of PPARγ to the pdx-1 × PPRE in intact β-cells was confirmed using the ChIP assay. Preparations of 300–500-bp chromatin fragments from mouse-derived βTC6 cells underwent immunoprecipitation with PPARγ monoclonal antibody or negative control IgG, and PCR of the immunoprecipitated DNA and input DNA using mouse pdx-1 × PPRE region flanking primers. The expected 312-bp PCR product was generated with both input DNA and PPARγ antibody-immunoprecipitated DNA from two separate chromatin preparations, whereas only a very faint PCR product was observed from the negative control IgG-immunoprecipitated DNA (Fig. 7A). In contrast, a nonspecific primer pair for a 450-bp PCR product 2.56 kb downstream of the mouse pdx-1 × PPRE showed a PCR product only with input DNA (Fig. 7B). The positive control showed the expected 233-bp PCR product with the EFI primer pair in the RNA pol II antibody-immunoprecipitated DNA but not the negative control IgG-immunoprecipitated DNA (Fig. 7C). Also, the negative control primers generated the expected 245-bp PCR product only with input DNA (Fig. 7D).
FIGURE 7.
Chromatin immunoprecipitation assay of BTC6 cells. 500–600-bp chromatin preparations of BTC6 cells were prepared as described under “Experimental Procedures.” A, they were immunoprecipitated using mouse monoclonal PPARγ and negative control IgG, followed by PCR of the immunoprecipitated and nonimmunoprecipitated DNA (input DNA) using flanking primer pairs to mouse pdx-1 × PPRE. The shown bands are 312-bp (expected length) PCR product from two separate experiments, along with absence of PCR product in the control IgG lanes on the right. B, negative control was performed using primer pairs for a 450-bp area 2.7 kilobases downstream of the mouse pdx-1 × PPRE. The expected length band was obtained with input DNA but not the PPARγ-immunoprecipitated DNA. C, as a positive control, in parallel chromatin preparations were precipitated with RNA pol II antibody and underwent PCR of the immunoprecipitated and input DNA using EFI primer pairs. The shown bands are 250-bp (expected length) PCR product from two separate experiments in the RNA pol II antibody and input DNA lanes compared with the expected absence of a PCR product in the control IgG lanes. D, negative control for the RNA pol II immunoprecipitated and input DNA using the negative control primer pairs for RNA pol II.
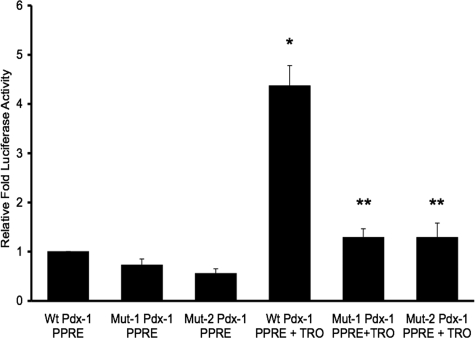
Luciferase Reporter Gene Assay—The 1-kb Pst-Bst (-2916- to -1920-bp) fragment of the mouse pdx-1 promoter region carrying the PPRE was subcloned into the pTAL luciferase reporter vector and used as template to perform site-directed mutagenesis to incorporate the same mutations on the DR1 and DR2 regions as used for the in vitro gel shift competition assay. As shown in Fig. 8, both mutations lowered basal luciferase reporter activity (Mut 1 73 ± 4% of wild type pdx-1 × PPRE, p < 0.001; Mut 2 56 ± 5% of wild type pdx-1 × PPRE, n = 3, p ≤ 0.001). Also, 24 h of stimulation with the PPARγ agonist, troglitazone, of the cells transfected with the wild type pdx-1 × PPRE reporter construct induced a 4-fold increase in luciferase reporter activity (438 ± 15% of nontroglitazone-treated wild type, p < 0.001) that was lowered to less than 2-fold with both mutations (p < 0.001 for troglitazone-treated wild type versus troglitazone-treated Mut 1 and troglitazone-treated Mut 2, respectively). These findings confirmed physiological activity of the identified pdx-1 × PPRE in INS-1 cells basally, and with induction by a PPARγ agonist.
FIGURE 8.
Luciferase reporter activity of wild type and mutated mouse pdx-1 × PPRE transfected into INS-1 cells. INS-1 cells were transfected with wild type (Wt) or mutated pTAL × PPRE × pdx-1 promoter vectors (Mut-1 or Mut-2). Renilla luciferase reporter plasmid was included in all transfections to act as internal control. 24 h post-transfection, cells were treated with 10 μm troglitazone (TRO) or DMSO for 24 h. Firefly luciferase activity was measured by luminometer, normalized with Renilla luciferase, and expressed as relative luciferase activity. Mut 1 contains the mutation of the 5′ DR1 half-site of pdx-1 × PPRE from Fig. 5B. Mut 2 contains the mutation of the 3′ DR2 half-site of pdx-1 × PPRE from Fig. 5C. There were three wells for each experimental condition per experiment. Data are expressed as the mean ± S.E. relative luciferase activity of three separate experiments compared with the wild type pdx-1 × PPRE without troglitazone in lane 1.*, p < 0.001 wild type plus TRO versus wild type plus DMSO. **, p < 0.001 wild type plus troglitazone versus Mut-1 or Mut-2 plus troglitazone.
DISCUSSION
This study stems from our work in the 60% pancreatectomy rat model of β-cell adaptive compensation, in which an initial period of partial β-cell regeneration is followed by enhanced β-cell function, so normoglycemia is preserved (19–21). We investigated the transition between these phases of adaptation, and we identified heightened β-cell nuclear expression of PPARγ and the β-cell differentiation factors PDX-1 and NKX6.1 (22). The known effects of these factors in β-cells (anti-proliferation and pro-survival for PPARγ (10–12), pro-survival and enhanced insulin secretion for PDX-1 (23–28), and suppressed glucagon expression plus enhanced glucose-induced insulin secretion with NKX6.1 (37)) could explain the transition. On the other hand, PPARγ acts in tissues through a triad of effects, anti-proliferation, pro-survival, and pro-differentiation (18), and PPARγ-mediated proliferation arrest and pro-survival effects were described for β-cells (10–12). As such, another possibility was PPARγ-mediated regulation of these β-cell differentiation factors. We focused on PDX-1, as it is the most studied transcription factor in mature β-cells and is a crucial regulator of β-cell function (23–25), viability (26), and compensatory capacity (38, 39). Support for PPARγ-mediated regulation of PDX-1 expression was obtained in INS-1 cells by showing a tripling of PDX-1 expression with the PPARγ agonist, troglitazone, and a lowering of pdx-1 transcription following RNAi-induced reduction of PPARγ expression (22). The current results have confirmed in vivo PPARγ regulation of pdx-1 transcription in mouse β-cells, with 40% of PDX-1 expression being PPARγ-dependent. They also have shown an absolute requirement for pancreas PPARγ expression in terms of normal β-cell function (glucose-induced insulin secretion) and whole animal glucose tolerance. Of potential clinical importance, we have confirmed in primary mouse β-cells the action of thiazolidinediones to augment β-cell PDX-1 expression that we had previously seen in INS-1 cells (22). Collectively, this panoply of findings supports a key role of PPARγ to regulate PDX-1 expression in β-cells physiologically and pharmacologically, and thus to have pro-differentiation effects on β-cell function and survival.
An unexpected observation was the different phenotype of the PANC PPARγ-/- mice versus that reported for mice with a β-cell-specific knock-out of PPARγ using a Cre/loxP recombinase system based on the rat insulin promoter (RIP) (11). The β-cell phenotype of that model consisted of twice normal pancreatic β-cell mass because of loss of the PPARγ anti-proliferation effect, and blunting of the thiazolidinedione-induced augmentation of glucose-induced insulin secretion in isolated islets, confirming PPARγ regulation of β-cell mass and function (11). Regardless, the normoglycemia of these mice basally and after fat feeding led the investigators to conclude those observations were inconsequential. This study investigated mice with Cre expression driven by the pdx-1 promoter that creates PPARγ deletion in β-cells, but also with the possibility of impacting other pancreatic epithelial cells such as non-β islet cells, ducts, and acini (29). The potential for this additional effect is most pronounced during fetal development when PDX-1 is expressed in all pancreatic epithelial cells, whereas post-birth, PDX-1 is expressed at high levels only in β-cells (32). In contrast to the RIP-Cre PPARγ knock-out mice, male pdx-1-Cre PPARγ knock-out mice were hyperglycemic on a normal chow diet at 8 weeks of age. The basis for this disparity is not fully understood, although concern over the rat insulin II promoter for Cre expression has been raised because of modest hypothalamic expression of the transgene (32) and glucose intolerance of RIP-Cre mice with no loxP gene deletion (40). In contrast, this line of pdx-1-Cre mice is known to have no defect in glucose tolerance (41). Indeed there is an ongoing debate over Cre/loxP technology for β-cell gene deletions as reflected in a recent discussion of the different phenotypes for mice with leptin deletions using RIP and pdx-1 Cre promoters (42). We considered the possibility that the whole pancreas epithelial deletion of PPARγ had caused developmental abnormalities that were responsible for the hyperglycemia. However, in this current study and a previous analysis of this model (29), no abnormal pancreatic histology was noted, plus we measured β-cell mass and found no reduction compared with the control mice. Alternatively, we had concern over the potential for enhanced glucagon expression in the PANC PPARγ-/- mice, as PPARγ is highly expressed in islet α-cells (43, 44) and acts to repress glucagon transcription (37). However, there was no change in islet glucagon expression by mRNA analysis or immunohistochemistry. Furthermore, PDX-1 expression is lacking in adult α-cells. Rather, the results in the PANC PPARγ+/- mice agree with our prior in vitro results in terms of PPARγ regulation of β-cell PDX-1 expression (22). Unanswered is why the PANC PPARγ+/- mice failed to show the β-cell hyperproliferation and larger β-cell mass of the RIP-Cre PPARγ null mice (11), although those measurements were performed in PANC PPARγ+/- mice that already were hyperglycemic. Another issue for consideration is the phenotype of the PANC PPARγ+/- mice is more severe than that reported for PDX-1 haploinsufficient mice, and they also have glucose intolerance, but glucose-induced insulin secretion is unimpaired in isolated islets (25, 26), leading us to speculate there are additional PPARγ regulatory effects in β-cells that are PDX-1-independent.
We next investigated the molecular basis for PPARγ regulation of pdx-1 transcription. A typical PPRE consists of two hexamer repeats, DR1 and DR2, separated by a nucleotide, with a consensus sequence of RGGTCA-A-AGGTCA. However, there is considerable divergence in the DR1 sequence, with a comparison of 73 reported functional PPREs showing only 2 with the consensus sequence, and 8% having five or more mismatches (33). In contrast, 20 of 73 matched the ideal DR2, and another 30 had only 1 nucleotide mismatch. These observations are consistent with the greater importance of RXR binding to the DR2 half-site as part of the PPARγ/RXR heterodimer in terms of PPRE functionality, compared with PPARγ binding to the DR1 half-site, leading to the suggestion that the PPRE consensus sequence may need modification (34). We searched the mouse pdx-1 promoter region for homology to the DR2 consensus sequence, and found a site (CCCATG-G-AGGTCA, -2720 to -2708) that matched the DR2 consensus sequence. In contrast, comparison of the associated DR1 hexamer to reported PPREs showed its sequence was unique. Notably, the so-called area I encompassing the DNase I-hypersensitive site I within the mouse pdx-1 promoter is a regulatory site for β-cell-specific expression of PDX-1 (35), and our identified sequence was located in this area. Also, the human sequence of this area is 89% homologous with the mouse sequence and plays a similar regulatory role for PDX-1 expression (35). Investigation of the analogous human PDX-1 sequence showed partial homology for the DR1 element and 100% homology for DR2 compared with the putative mouse pdx-1 × PPRE.
We went on to show binding of radiolabeled oligos for the mouse and human pdx-1 × PPREs with nuclear extracts from mouse islets, mouse-derived βTC6 cells, and rat-derived INS-1 cells. Supershift analysis with PPARγ antibody confirmed formation of a PPARγ·PPRE complex. Binding studies with in vitro translated PPARγ and RXR-α showed a heterodimerized complex binds to the mouse and human pdx-1 × PPREs. Binding specificity of PPARγ to the mouse PPRE was shown in vitro and in intact cells by competition assay and chromatin immunoprecipitation, followed by showing its capacity to regulate pdx-1 transcription. The latter entailed making DR1 and DR2 mutations for a luciferase reporter gene assay, in the absence and presence of 24 h of troglitazone treatment. As expected, troglitazone enhanced reporter activity of the wild type pdx-1 × PPRE 4-fold, and this increase was lowered more than half with the mutated PPREs. Thus, we have identified and characterized a functional PPRE within the mouse pdx-1 promoter that has a unique sequence from previously reported PPREs and is highly conserved in humans.
In summary, we have provided in vivo and in vitro evidence showing PPARγ regulation of pdx-1 transcription, and thus indirectly β-cell function and mass. Also, we have shown in primary β-cells that thiazoledionediones increase PDX-1 expression through a PPARγ-mediated mechanism. Given the crucial role of PDX-1 in the function (23–25), survival (26), and compensatory ability (38, 39) of mature β-cells, along with the β-cell benefits attributed to thiazoledinediones in prediabetes and type 2 diabetes (13–17), our results strongly support an important regulatory role for PPARγ in β-cell physiology and disease pharmacology.
Acknowledgments
We thank Dr. Roland Stein (Vanderbilt University) for the gift of the mouse pdx-1 promoter construct. We also thank Navjot Monga and Alexander Gokin for expert technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK56818 and DK66635 (to J. L. L.) and DK59851 (to T. L. J.). This work was also supported by American Diabetes Association (to T. L. J. and M. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PPARγ, peroxisome proliferator-activated receptor γ; RXR, retinoid X receptor; PPRE, PPAR-response element sequence; Px, partial pancreatectomy; PDX-1, pancreatic duodenal homeobox-1; PANC PPARγ-/-, pancreas-specific PPARγ null mice; ChIP, chromatin immunoprecipitation; RIP, rat insulin promoter; pol, polymerase; RNAi, RNA interference; oligos, oligonucleotides; FW, forward; REV, reverse; ds, double strand; Mut, mutant.
References
- 1.Francis, G. A., Fayard, E., Picard, F., and Auwerx, J. (2003) Annu. Rev. Physiol. 65 261-311 [DOI] [PubMed] [Google Scholar]
- 2.Rosen, E. D., and Spiegelman, B. M. (2001) J. Biol. Chem. 276 37731-37734 [DOI] [PubMed] [Google Scholar]
- 3.Picard, F., and Auwerx, J. (2002) Annu. Rev. Nutr. 22 167-197 [DOI] [PubMed] [Google Scholar]
- 4.Lehrke, M., and Lazar, M. A. (2005) Cell 123 992-999 [DOI] [PubMed] [Google Scholar]
- 5.Kliewer, S. A., Umesono, K., Noonan, D. J., Heyman, R. A., and Evans, R. M. (1992) Nature 358 771-774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gearing, K. L., Gottlicher, M., Teboul, M., Widmark, E., and Gustafsson, J. A. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 1440-1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ijpenberg, A., Jeannin, E., Wahli, W., and Desvergne, B. (1997) J. Biol. Chem. 272 20108-20117 [DOI] [PubMed] [Google Scholar]
- 8.Hsu, M. H., Palmer, C. N., Song, W., Griffin, K. J., and Johnson, E. F. (1998) J. Biol. Chem. 273 27988-27997 [DOI] [PubMed] [Google Scholar]
- 9.Yki-Jarvinen, H. (2004) N. Engl. J. Med. 351 1106-1118 [DOI] [PubMed] [Google Scholar]
- 10.Ohtani, K. I., Shimizu, H., Sato, N., and Mori, M. (1998) Endocrinology 139 172-178 [DOI] [PubMed] [Google Scholar]
- 11.Rosen, E. D., Kulkarni, R. N., Sarraf, P., Ozcan, U., Okada, T., Hsu, G. H., Eisenman, D., Magnuson, M. A., Gonzalez, F. J., Kahn, C. R., and Spiegelman, B. M. (2003) Mol. Cell. Biol. 23 7222-7229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin, C. Y., Gurlo, T., Haataja, L. O., Hsueh, W. A., and Butler, P. C. (2005) J. Clin. Endocrinol. Metab. 90 6678-6686 [DOI] [PubMed] [Google Scholar]
- 13.Buchanan, T. A., Xiang, A. H., Peters, R. K., Kjos, S. L., Marroquin, A., Goico, J., Ochoa, C., Tan, S., Berkowitz, K., Hodis, H. N., and Azen, S. P. (2002) Diabetes 51 2796-2803 [DOI] [PubMed] [Google Scholar]
- 14.Knowler, W. C., Hamman, R. F., Edelstein, S. L., Barrett-Connor, E., Ehrmann, D. A., Walker, E. A., Fowler, S. E., Nathan, D. M., Kahn, S. E., and Diabetes Prevention Program Research Group (2005) Diabetes 54 1150-1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang, A. H., Peters, R. K., Kjos, S. L., Marroquin, A., Goico, J., Ochoa, C., Kawakubo, M., and Buchanan, T. A. (2006) Diabetes 55 517-522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DREAM (Diabetes REduction Assessment with Ramipril and Rosiglitazone Medication) Trial Investigators, Gerstein, H. C., Yusuf, S., Bosch, J., Pogue, J., Sheridan, P., Dinccag, N., Hanefeld, M., Hoogwerf, B., Laakso, M., Mohan, V., Shaw, J., Zinman, B., and Holman, R. R. (2006) Lancet 368 1096-1105 [DOI] [PubMed] [Google Scholar]
- 17.Kahn, S. E., Haffner, S. M., Heise, M. A., Herman, W. H., Holman, R. R., Jones, N. P., Kravitz, B. G., Lachin, J. M., O'Neill, M. C., Zinman, B., Viberti, G., and ADOPT Study Group (2006) N. Engl. J. Med. 355 2427-2443 [DOI] [PubMed] [Google Scholar]
- 18.Feige, J. N., Gelman, L., Michalik, L., Desvergne, B., and Wahli, W. (2006) Prog. Lipid Res. 45 120-159 [DOI] [PubMed] [Google Scholar]
- 19.Liu, Y. Q., Montanya, E., and Leahy, J. L. (2001) Diabetologia 44 1026-1033 [DOI] [PubMed] [Google Scholar]
- 20.Jetton, T. L., Liu, Y. Q., Trotman, W. E., Nevin, P. W., Sun, X.-J., and Leahy, J. L. (2001) Diabetologia 44 2056-2065 [DOI] [PubMed] [Google Scholar]
- 21.Liu, Y. Q., Nevin, P. W., and Leahy, J. L. (2000) Am. J. Physiol. 279 E68-E73 [DOI] [PubMed] [Google Scholar]
- 22.Moibi, J. A., Gupta, D., Jetton, T. L., Peshavaria, M., Desai, R., and Leahy, J. L. (2007) Diabetes 56 88-95 [DOI] [PubMed] [Google Scholar]
- 23.Ahlgren, U., Jonsson, J., Jonsson, L., Simu, K., and Edlund, H. (1998) Genes Dev. 12 1763-1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holland, A. M., Hale, M. A., Kagami, H., Hammer, R. E., and MacDonald, R. J. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 12236-12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brissova, M., Shiota, M., Nicholson, W. E., Gannon, M., Knobel, S. M., Piston, D. W., Wright, C. V., and Powers, A. C. (2002) J. Biol. Chem. 277 11225-11232 [DOI] [PubMed] [Google Scholar]
- 26.Johnson, J. D., Ahmed, N. T., Luciani, D. S., Han, Z., Tran, H., Fujita, J., Misler, S., Edlund, H., and Polonsky, K. S. (2003) J. Clin. Investig. 111 1147-1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Servitja, J. M., and Ferrer, J. (2004) Diabetologia 47 597-613 [DOI] [PubMed] [Google Scholar]
- 28.Wang, H., Iezzi, M., Theander, S., Antinozzi, P. A., Gauthier, B. R., Halban, P. A., and Wollheim, C. B. (2005) Diabetologia 48 720-731 [DOI] [PubMed] [Google Scholar]
- 29.Ivashchenko, C. Y., Duan, S. Z., Usher, M. G., and Mortensen, R. M. (2007) Am. J. Physiol. 293 G319-G326 [DOI] [PubMed] [Google Scholar]
- 30.Gu, G., Dubauskaite, J., and Melton, D. A. (2002) Development (Camb.) 129 2447-2457 [DOI] [PubMed] [Google Scholar]
- 31.Jetton, T. L., Lausier, J., LaRock, K., Trotman, W. E., Larmie, B., Habibovic, A., Peshavaria, M., and Leahy, J. L. (2005) Diabetes 54 2294-2304 [DOI] [PubMed] [Google Scholar]
- 32.Gannon, M., Herrera, P.-L., and Wright, C. V. E. (2000) Genesis 26 143-144 [DOI] [PubMed] [Google Scholar]
- 33.Lemay, D. G., and Hwang, D. H. (2006) J. Lipid Res. 47 1583-1587 [DOI] [PubMed] [Google Scholar]
- 34.Temple, K. A., Cohen, R. N., Wondisford, S. R., Yu, C., Deplewski, D., and Wondisford, F. E. (2005) J. Biol. Chem. 280 3529-3540 [DOI] [PubMed] [Google Scholar]
- 35.Gerrish, K., Gannon, M., Shih, D., Henderson, E., Stoffel, M., Wright, C. V., and Stein, R. (2000) J. Biol. Chem. 275 3485-3492 [DOI] [PubMed] [Google Scholar]
- 36.Tugwood, J. D., Issemann, I., Anderson, R. G., Bundell, K. R., McPheat, W. L., and Green, S. (1992) EMBO J. 11 433-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schisler, J. C., Jensen, P. B., Taylor, D. G., Becker, T. C., Knop, F. K., Takekawa, S., German, M., Weir, G. C., Lu, D., Mirmira, R. G., and Newgard, C. B. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 7297-7302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulkarni, R. N., Jhala, U. S., Winnay, J. N., Krajewski, S., Montminy, M., and Kahn, C. R. (2004) J. Clin. Investig. 114 828-836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brissova, M., Blaha, M., Spear, C., Nicholson, W., Radhika, A., Shiota, M., Charron, M. J., Wright, C. V., and Powers, A. C. (2005) Am. J. Physiol. 288 E707-E714 [DOI] [PubMed] [Google Scholar]
- 40.Lee, J.-Y., Ristow, M., Lin, X., White, M. F., Magnuson, M. A., and Hennighausen, L. (2006) J. Biol. Chem. 281 2649-2653 [DOI] [PubMed] [Google Scholar]
- 41.Lee, J.-Y., and Hennighausen, L. (2005) Biochem. Biophys. Res. Commun. 334 764-768 [DOI] [PubMed] [Google Scholar]
- 42.Niswender, K. D., and Magnuson, M. A. (2007) J. Clin. Investig. 117 2753-2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braissant, O., Foufelle, F., Scotto, C., Dauca, M., and Wahli, W. (1996) Endocrinology 137 354-366 [DOI] [PubMed] [Google Scholar]
- 44.Dubois, M., Pattou, F., Kerr-Conte, J., Gmyr, V., Vandewalle, B., Desreumaux, P., Auwerx, J., Schoonjans, K., and Lefebvre, J. (2000) Diabetologia 43 1165-1169 [DOI] [PubMed] [Google Scholar]