Abstract
The genome of the virus that causes salivary gland hypertrophy in Musca domestica (MdSGHV) was sequenced. This non-classified, enveloped, double stranded, circular DNA virus had a 124,279 bp genome. The G+C content was 43.5% with 108 putative methionine-initiated open reading frames (ORFs). Thirty ORFs had homology to database proteins: eleven to proteins coded by both baculoviruses and nudiviruses (p74, pif-1, pif-2, pif-3, odv-e66, rr1, rr2, iap, dUTPase, MMP, and Ac81-like), seven to nudiviruses (mcp, dhfr, ts, tk and three unknown proteins), one to baculovirus (Ac150-like), one to herpesvirus (dna pol), and ten to cellular proteins. Mass spectrum analysis of the viral particles’ protein components identified 29 structural ORFs, with only p74 and odv-e66 previously characterized as baculovirus structural proteins. Although most of the homology observed was to nudiviruses, phylogenetic analysis showed that MdSGHV was not closely related to them or to the baculoviruses.
Keywords: Salivary gland hypertrophy virus, insect viruses, insect salivary gland, Musca domestica, SGHV, genome sequence
INTRODUCTION
Presently, three different dipterans are known to harbor salivary gland hypertrophy viruses (SGHV). These include the house fly Musca domestica (Coler et al., 1993), the narcissus bulb fly Merodon equestris (Amargier et al., 1979), and various tsetse fly species Glossina spp. (Gouteux, 1987; Jaenson, 1978; Minter-Goedbloed and Minter 1989; Otieno et al., 1980; Shaw and Moloo, 1993). Comparisons among different virus-host fly interactions have demonstrated that the different viruses share several morphological and pathological properties (Amargier et al., 1979; Coler et al., 1993; Ellis and Maudlin, 1987; Jaenson, 1978; Kokwaro et al., 1990, 1991; Odindo et al., 1986). Electron microscopy of virus particles in hypertrophic salivary glands showed heterogeneous, elongate (650 by 75 nm), enveloped rod-shaped virions (Geden et al., 2008). In all three fly hosts, the virus replicated in the nuclei of salivary gland cells, resulting in nuclear and salivary gland hypertrophy (SGH) that is easily visualized when the insects are dissected. Infected adults do not exhibit any external disease symptoms, but viral infection is known to inhibit reproduction (Lietze et al., 2007).
The best-studied SGHVs are those associated with the tsetse fly complex that were detected over sixty years ago in feral flies (Burtt, 1945; Whitnall, 1934). SGHV infection in tsetse flies, in addition to causing SGH, results in testicular degeneration and ovarian abnormalities (Feldmann et al., 1992; Jura et al., 1988; Sang et al., 1998, 1999). The tsetse SGHV can infect the female milk gland, resulting in maternal transmission to larvae in this viviparous, hematophagous insect. Venereal transmission between infected and healthy tsetse flies has been reported (Jura et al., 1989). The natural incidence of the tsetse SGHV is low; only 0.4 to 5% of the field-collected flies display SGH (Ellis and Maudlin, 1987; Jaenson, 1978; Jura et al., 1988; Odindo, 1982; Odindo et al., 1981). In colonized G. morsitans, incidence of symptomatic SGH ranges from 1.1% (Jura et al., 1993) to 4.0% (Kokwaro et al., 1990). However, in recent years, the tsetse SGHV has been reported to infect and sometimes decimate the tsetse fly colonies being used for sterile insect release programs. Recently, the development of a diagnostic polymerase chain reaction (PCR) has demonstrated that the tsetse SGHV can exist in an asymptomatic state in host flies (Abd-Alla et al., 2007). Insect colonies displaying low levels (<5%) of SGH symptoms produce a 100% tsetse SGHV PCR-positives.
The SGHV from M. domestica (MdSGHV) is orally transmitted by adult feeding on contaminated substrates and is capable of inducing the symptomatic SGH within days. Like the tsetse SGHV, infection by MdSGHV acts to rapidly and completely inhibit egg production in viremic females (Lietze et al., 2007). Examination of the translational and transcriptional products has demonstrated that the infection blocks hexamerin and yolk protein synthesis. In addition, infection with MdSGHV disrupts house fly mating behavior in that symptomatic females refuse to copulate with healthy males and symptomatic males show reduce avidity to initiate courtship with healthy females. Thus, the MdSGHV-induced mechanisms underlying the inhibition of house fly reproduction appear to function on both physiological and behavioral levels (Lietze et al., 2007).
Comparisons reveal that MdSGHV possesses biological properties that are distinct from the tsetse SGHV. The house fly is highly susceptible to the virus. Injection of an estimated 100 virions results in 100% SGH of test flies (Lietze et al., 2007), whereas injection with millions of virions results in ~20% SGH in colony-reared adult tsetse flies (Abd-Alla, personal communication). The SGHVs in tsetse flies have been reported to be transmitted vertically from mother to offspring, whereas the MdSGHV has been shown to be only orally transmitted (Lietze et al., 2007). The incidence of symptomatic SGH, reported to be as high as 34% in feral house fly populations (Geden et al., 2008), is much greater than the 0.4 to 5.0% in tsetse fly populations (Jaenson, 1978; Otieno et al., 1980; Odindo et al., 1981; Odindo, 1982; Ellis and Maudlin, 1987; Jura et al., 1988).
The MdSGHV, as well as the tsetse SGHV, share general characteristics with the non-occluded insect nudiviruses, such as being insect-pathogenic, having an enveloped, rod-shaped morphology, and possessing a circular dsDNA genome (Wang et al., 2007a). Members of this group were once placed within subgroup C of the Baculoviridae but have since been placed in the unassigned nudivirus group and classified as sedis incertae. Analyses of complete genome sequences of over 40 baculoviruses have shown the presence of a core of 29 conserved genes (Garcia-Maruniak et al., 2004; Lauzon et al., 2004). The Heliothis zea nudiviruses 1 and 2 (HzNV-1, HzNV-2), the Gryllus bimaculatus nudivirus (GbNV), and the Oryctes rhinoceros nudivirus (OrNV) have been shown to contain 15 core baculovirus genes (Cheng et al., 2002; Wang et al., 2007a, 2007b, 2007c). Analysis of selected nudivirus sequences has indicated that they form a monophyletic sister group that apparently diverged from a common ancestor of the baculoviruses (Wang et al., 2007b). Whether the SGHV fits within this monophyletic nudivirus group is unknown. Preliminary studies on both the tsetse fly and housefly SGHVs have demonstrated that they possess structural and biological properties that are distinct from the insect nudiviruses (Coler et al., 1993; Abd-Alla et al., 2007). During submission of this manuscript, the complete genome sequence of the related GpSGHV became available (Abd-Alla et al., 2008). In order to determine the genetic basis for the SGHV group classification and to further understand the MdSGHV, its genome has been sequenced. The results of this effort are presented in this manuscript.
RESULTS AND DISCUSSION
Virus morphology and SDS-PAGE analysis
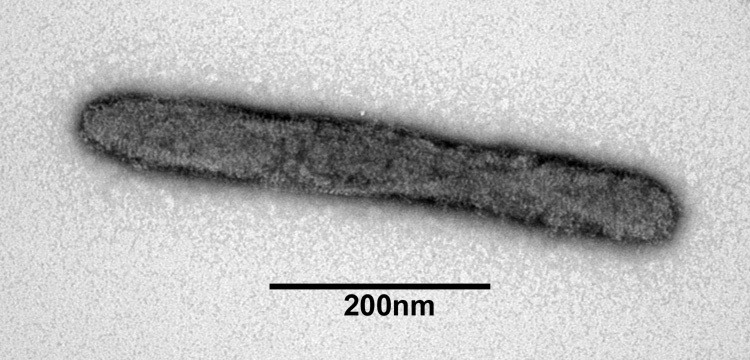
Injection of the filtered homogenate from MdSGHV infected salivary glands into healthy houseflies resulted in 100% infection. Gradient purification on osmotically stabililized Nycodenz gradients produced a uniform population of enveloped rod-shaped virus particles measuring 65 by 550 nm (Fig. 1). Previous attempts involving sucrose gradient purification resulted in preparations consisting of partly enveloped nucleocapsids (Coler et al., 1993). Negative staining of the Nycodenz-purified virus revealed enveloped particles possessing a unique braided, bead-like surface topography not observed with other rod-shaped insect viruses (Fig. 1). Furthermore, the ends of the virus particle were rounded and not blunt-ended as observed with the nudiviruses. The length of the enveloped MdSGHV, although shorter than the Glossina pallipedes GpSGHV (1.0 µm in length) (Odindo et al., 1986), was significantly longer than the enveloped forms of GbNV and OrNV (100 by 200 nm, Wang et al., 2007a).
Figure 1.
Transmission electron micrograph of MdSGHV particle purified on Nycodenz gradients and stained with uranyl acetate.
SDS-PAGE of purified MdSGHV produced a complex of Coomassie R-250 blue-stained bands ranging from 10 to >150 KDa (Fig. 2). The presence and distribution of the two major-stained (bands b and h) and moderately-stained bands agreed with the protein profile previously reported (Coler et al., 1993) and those results obtained with various other (putative) nudiviruses (Boucias et al., 1989; Payne, 1974). However, the low molecular bands (<20 kDa) were not resolved in a previous analysis (Coler et al., 1993) that utilized a non-gradient 12% separating gel. In addition to the 16 bands, multiple light-stained bands were detected throughout the gel, suggesting proteolysis had occurred during sample storage and/or preparation.
Figure 2.
SDS-PAGE of MdSGHV stained with Coomassie blue. Molecular masses (kDa) of the Bio-Rad markers are denoted on the left side of the gel. The letters represent the excised gel sections that were subjected to GeLC-MS/MS analysis. Adjacent numbers represent the different ORFs that were detected in the denoted gel sections. The numbers in the parentheses represent the number of unique peptides that identified the designated ORFS.
Genome sequence analysis
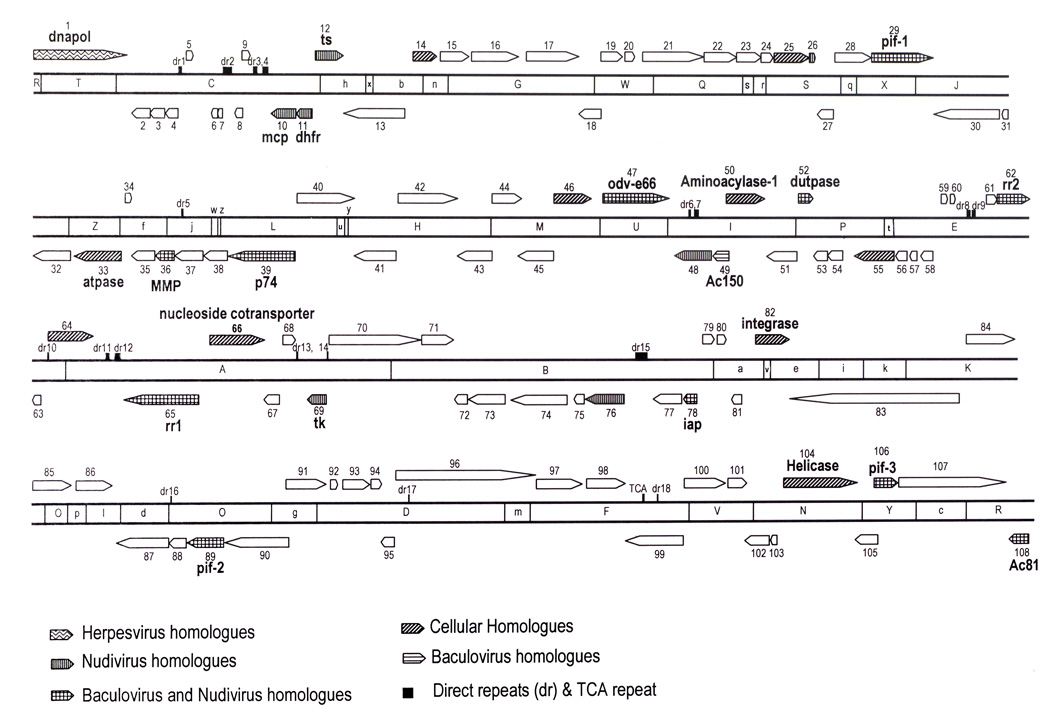
A total of 60.5 µg of DNA was extracted from a sample containing an estimated 2.5 mg of purified virus. End-labeling of undigested MdSGHV DNA was unsuccessful, suggesting a circular genome structure. This result was further confirmed when the genome was completely sequenced, showing that the double-stranded circular DNA of MdSGHV was 124,279 bp in size, somewhat smaller than the originally described 137 kbp, estimated by restriction endonuclease analysis (Coler et al., 1993). The end-labeled fragments of the digested DNA showed this viral preparation, to be more homogeneous (derived from a single gland) than the virus extracted from the pooled glands preparation previously analyzed (Coler et al., 1993). The sequence analysis, however, confirmed that our viral preparation was still polymorphic. At least 66 bases were found to be different when sequences from several DNA fragment clones were aligned. The differences at a given nucleotide position were always found to be nucleotide transitions. The most abundant base (present in the majority of the clones sequenced) was chosen as the consensus. The location of each polymorphic base was mapped in the genome (data not shown) as potential targets for assessing polymorphism of field-collected viral samples. The MdSGHV genome is smaller than the 185 kbp estimated for GpSGHV (Abd-Alla et al., 2007). It should be noted that the estimated genome sizes reported among the nudiviruses range from 96 to 228 kb (Wang et al., 2007a). The G+C content of MdSGHV was 43.5%, which is very similar to several baculoviruses, HzNV-1 and OrNV, but it is higher than GbNV, which has only a 28% G+C content (Cheng et al., 2002; Wang et al., 2007b, 2007c). The computer generated EcoRI digestion of the MdSGHV genome resulted in 52 fragments ranging from 10,219 bp (EcoRI-A) to 30 bp (EcoRI-z). From those, 47 fragments have been cloned, resulting in the construction of 90% of the MdSGHV EcoRI genome library. The physical map is shown in Fig. 3.
Figure 3.
Linear representation of the MdSGHV genome and EcoRI physical map. The genome location, relative size and transcriptional direction of each putative ORF is indicated by the arrows. Arrow patterns and color indicate homology to organisms found in the current protein database. Dark boxes indicate location of direct repeats.
Nucleotide 1 of the MdSGHV genome was considered to be the A of the ATG initiation codon for the DNA polymerase gene (dnapol). This gene was chosen as the start site in an effort to find a common gene present in all DNA viruses, so that other unclassified DNA viruses could be easily compared. A total of 108 putative ORFs were identified in the MdSGHV genome (Table 1, Fig. 3). There were 53 ORFs (49%) transcribed in the clockwise direction and 55 (51%) in the opposite direction. Interestingly, there were several clusters of ORFs transcribing in one direction. A balance in transcriptional direction is often found in baculoviruses and nudiviruses. A total of 22 ORFs overlapped adjacent ORFs with a maximum of 158 nucleotides between ORFs 105 and 106. These two ORFs had opposite transcriptional orientations and were over 240 aa in size. No biological activity could be assigned to ORF 105, since there was no protein homology, nor known motifs, while ORF 106 had homology to the baculovirus pif-3 gene. Elimination of ORF 105 just to avoid overlapping was considered unsupported at this stage in a poorly known virus, so it was accepted as a putative ORF.
Table 1.
MdSGHV putative ORFs
| Intergene distance | Size |
Best BlastP match |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF* | position | (bp)‡ | nt. | aa | kDa | ORF or protein encoded | Species | aa identity# | E-value | Motifs | |
| 1 | 1 > | 2934 | 153 | 2934 | 977 | 114 | DNA Polymerase | retroperitoneal fibromatosis-associated herpesvirus | 27% (766) 1013 | 5.00E-60 | DNA_pol_B, DNA_pol_B_exo |
| 2 | 3088 < | 3669 | −10 | 582 | 193 | 23 | |||||
| S 3 | 3660 < | 4130 | 26 | 471 | 156 | 19 | TM | ||||
| 4 | 4157 < | 4552 | 231 | 396 | 131 | 15 | |||||
| 5 | 4784 > | 4993 | 547 | 210 | 69 | 8 | |||||
| 6 | 5541 < | 5759 | −1 | 195 | 72 | 8 | |||||
| 7 | 5759 < | 5920 | 390 | 162 | 53 | 6 | |||||
| 8 | 6311 < | 6562 | −30 | 252 | 83 | 10 | |||||
| 9 | 6533 > | 6787 | 703 | 255 | 84 | 10 | SP | ||||
| 10 | 7491 < | 8261 | −16 | 771 | 256 | 29 | Mitochondrial carrier protein | Oryctes rhinoceros nudivirus | 38% (261) 262 | 7.00E-46 | 5TM, SP, Mito_carr |
| 11 | 8246 < | 8758 | 64 | 513 | 170 | 20 | Dihydrofolate reductase | Heliothis zea nudivirus 1 | 26% 163) 264 | 2.00E-09 | SP, Dihydrofolate Reductase |
| 12 | 8823 > | 9707 | 106 | 885 | 294 | 34 | Thymidylate synthase | Oryctes rhinoceros nudivirus | 53% (293) 321 | 9.00E-88 | Thymidylat_synt |
| S 13 | 9814 < | 11748 | 185 | 1935 | 644 | 75 | |||||
| S 14 | 11934 > | 12683 | 73 | 750 | 249 | 29 | Molybdopterin oxidoreductase | Mycobacterium sp | 28% (194) 743 | 7.00E-02 | |
| 15 | 12757 > | 13656 | 95 | 900 | 299 | 35 | |||||
| S 16 | 13752 > | 15251 | 271 | 1500 | 499 | 58 | |||||
| 17 | 15523 > | 17184 | −4 | 1662 | 553 | 64 | TM, SP | ||||
| 18 | 17181 < | 17882 | −22 | 702 | 233 | 26 | |||||
| 19 | 17861 > | 18538 | 65 | 678 | 225 | 26 | SP | ||||
| S 20 | 18604 > | 18918 | 264 | 315 | 104 | 11 | 2TM | ||||
| 21 | 19183 > | 21117 | 10 | 1935 | 644 | 72 | |||||
| S 22 | 21128 > | 22159 | −40 | 1032 | 343 | 40 | |||||
| S 23 | 22120 > | 22890 | −11 | 771 | 256 | 29 | |||||
| 24 | 22880 > | 23266 | 27 | 387 | 128 | 16 | |||||
| S 25 | 23294 > | 24424 | −8 | 1131 | 376 | 41 | Hypothetical repeat containing protein | Tetrahymena thermophila | 26% (120) 2640 | 6.00E-04 | 2TM |
| 26 | 24417 > | 24599 | 112 | 183 | 60 | 7 | |||||
| 27 | 24712 < | 25221 | 39 | 510 | 169 | 20 | |||||
| S 28 | 25261 > | 26406 | 5 | 1146 | 381 | 42 | |||||
| 29 | 26412 > | 28346 | 28 | 1935 | 644 | 74 | per os infectivity factor (pif-1) | Neodiprion abietis NPV | 24% (357) 537 | 1.00E-08 | TM, SP |
| 30 | 28375 < | 30453 | 43 | 2079 | 692 | 79 | |||||
| 31 | 30497 < | 30697 | 12 | 201 | 66 | 7 | TM/SP | ||||
| S 32 | 30710 < | 31891 | 101 | 1182 | 393 | 45 | |||||
| S 33 | 31993 < | 33486 | 95 | 1494 | 497 | 56 | Putative vacuolar sorting ATPase | Candida albicans | 26% (268) 439 | 1.00E-12 | AAA ATPASE |
| 34 | 33582 > | 33776 | 44 | 195 | 64 | 7 | TM, SP | ||||
| 35 | 33821 < | 34549 | 15 | 729 | 242 | 27 | TM, SP | ||||
| 36 | 34565 < | 35155 | 9 | 591 | 196 | 23 | Matrix metalloproteinase (MMP) | Invertebrate iridescent virus 6 | 34% (101) 264 | 1.00E-07 | TM, SP, metalloproteases (ZnMc_MMP) |
| 37 | 35165 < | 36049 | 9 | 885 | 294 | 33 | TM, SP | ||||
| S 38 | 36059 < | 36811 | 8 | 753 | 250 | 29 | |||||
| S 39 | 36820 < | 38943 | 53 | 2124 | 707 | 82 | p74 protein | Heliothis zea nudivirus 1 | 22% (316) 682 | 1.00E-07 | TM, Baculo_p74_N |
| S 40 | 38997 > | 40823 | 31 | 1827 | 608 | 69 | |||||
| 41 | 40855 < | 42180 | −17 | 1326 | 441 | 52 | |||||
| 42 | 42164 > | 44047 | 12 | 1884 | 627 | 72 | |||||
| 43 | 44060 < | 45169 | −52 | 1110 | 369 | 44 | |||||
| S 44 | 45118 > | 46053 | −24 | 936 | 311 | 36 | SP | ||||
| 45 | 46030 < | 47172 | −61 | 1143 | 380 | 45 | N/A | ||||
| 46 | 47112 > | 48299 | 128 | 1188 | 395 | 46 | hypothetical protein | Bradyrhizobium sp. | 28% (140) 457 | 2.00E-05 | |
| S 47 | 48428 > | 50530 | 465 | 2103 | 700 | 81 | Occlusion-derived virus envelope protein (ODV-E66) | Glossina pallidipes SGHV | 35% (346) 346 | 7.00E-42 | TM, SP, Baculo_E66 |
| 48 | 50996 < | 52168 | 16 | 1173 | 390 | 43 | Hypothetical protein LNTAR 25495 | Lentisphaera araneosa | 26% (246) 794 | 1.00E-03 | SP |
| 49 | 52185 < | 52688 | −147 | 504 | 167 | 19 | Ac ORF 150-like | Autographa californica NPV | 30% (98) 99 | 3.00E-06 | Chitin binding perotrophin A domain (CBM-14) |
| S 50 | 52542 > | 53756 | 31 | 1215 | 404 | 46 | aminoacylase 1 | Sus scrofa | 28% (413) 407 | 2.00E-37 | AMINOACYLASE-1, Zn-dependent exopeptidases |
| 51 | 53788 < | 54741 | 86 | 954 | 317 | 36 | |||||
| S 52 | 54828 > | 55280 | 111 | 453 | 150 | 16 | dUTPase | grouper iridovirus | 51% (141) 159 | 7.00E-33 | dUTPase |
| 53 | 55392 < | 55832 | −46 | 441 | 146 | 18 | |||||
| 54 | 55787 < | 56272 | 118 | 486 | 161 | 19 | |||||
| 55 | 56391 < | 57641 | 36 | 1251 | 416 | 49 | hypothetical protein LNTAR_10481 | Lentisphaera araneosa | 36% (84) 234 | 6.00E-04 | |
| 56 | 57678 < | 58025 | 178 | 348 | 115 | 13 | |||||
| 57 | 58204 < | 58425 | 71 | 222 | 73 | 8 | N/A | TM/SP | |||
| 58 | 58497 < | 58892 | 309 | 396 | 131 | 16 | N/A | ||||
| 59 | 59202 > | 59399 | 173 | 198 | 65 | 7 | N/A | SP | |||
| 60 | 59573 > | 59728 | 905 | 156 | 51 | 6 | N/A | ||||
| 61 | 60634 > | 60957 | 21 | 324 | 107 | 12 | |||||
| 62 | 60979 > | 62010 | 102 | 1032 | 343 | 40 | Ribonucleoside diphosphate reductase small subunit (rr2) | Lymantria dispar NPV | 72% (337) 348 | 4.00E-142 | TM, Ribonuc_red_sm (RNRR2) |
| 63 | 62113 < | 62385 | 205 | 273 | 90 | 10 | |||||
| 64 | 62591 > | 64018 | 974 | 1428 | 475 | 52 | zinc finger protein | Danio rerio | 44% (49) 450 | 4.00E-03 | zinc finger (C3HC4), RING |
| 65 | 64993 < | 67350 | 303 | 2358 | 785 | 89 | ribonucleoside diphosphate reductase large subunit (rr1) | Spodoptera litura NPV | 61% (786) 770 | 0 | Ribonuc_red_lgC (RNR_1) |
| 66 | 67654 > | 69393 | −12 | 1740 | 579 | 64 | sodium/nucleoside cotransporter | Aedes aegypti | 24% (451) 602 | 4.00E-32 | 13TM, Sodium/Nucleoside cotransporter (Nucleos_tra 2_C) |
| 67 | 69382 < | 69870 | 79 | 489 | 162 | 19 | |||||
| 68 | 69950 > | 70348 | 388 | 399 | 132 | 15 | 2TM | ||||
| 69 | 70737 < | 71348 | 71 | 612 | 203 | 24 | Thymidine kinase (tk) | Spodoptera frugiperda ascovirus | 29% (195) 210 | 1.00E-12 | Deoxynucleoside kinase (dNK) |
| 70 | 71420 > | 74323 | 3 | 2904 | 967 | 111 | |||||
| S 71 | 74327 > | 75328 | 57 | 1002 | 333 | 37 | N/A | ||||
| S 72 | 75386 < | 75796 | 42 | 411 | 136 | 16 | TM | ||||
| 73 | 75839 < | 77011 | 23 | 1173 | 390 | 46 | |||||
| 74 | 77035 < | 79131 | 46 | 2097 | 698 | 79 | Galactose-binding domain-like | ||||
| 75 | 79178 < | 79504 | 20 | 327 | 108 | 12 | |||||
| 76 | 79525 < | 80673 | 881 | 1149 | 382 | 43 | OrNV ORF C3 | Oryctes rhinoceros nudivirus | 38% (386) 448 | 6.00E-63 | PIN domain-like |
| 77 | 81555 < | 82451 | 24 | 897 | 298 | 33 | |||||
| 78 | 82476 < | 82904 | 52 | 429 | 142 | 17 | inhibitor of apoptosis (iap) | ORF MSV248 Melanoplus sanguinipes ento mopoxvirus | 33% (140) 150 | 2.00E-19 | IAP, BIR |
| 79 | 82957 > | 83325 | 265 | 369 | 122 | 14 | TM, SP | ||||
| 80 | 83591 > | 83875 | 259 | 285 | 94 | 11 | N/A | ||||
| 81 | 84135 < | 84419 | 192 | 285 | 94 | 10 | |||||
| 82 | 84612 > | 85691 | 63 | 1080 | 359 | 43 | integrase/recombinase protein | Sulfolobus acidocaldarius | 24% (153) 287 | 4.00E-02 | Phage_integrase, DNA breaking & rejoining (INT_REC_C) |
| 83 | 85755 < | 91097 | 156 | 5343 | 1780 | 206 | |||||
| S 84 | 91254 > | 92783 | 60 | 1530 | 509 | 60 | |||||
| S 85 | 92844 > | 94055 | 154 | 1212 | 403 | 46 | N/A | ||||
| S 86 | 94210 > | 95355 | 70 | 1146 | 381 | 43 | |||||
| 87 | 95426 < | 97060 | 42 | 1635 | 544 | 64 | |||||
| 88 | 97103 < | 97642 | 25 | 540 | 179 | 21 | N/A | 2TM, SP | |||
| 89 | 97668 < | 98807 | 56 | 1140 | 379 | 42 | per os infectivity factor 2 (pif-2) | Ecotropis obliqua NPV | 26% (186) 392 | 3.00E-10 | TM, SP, Baculo_44 |
| S 90 | 98864 < | 100882 | −59 | 2019 | 672 | 78 | |||||
| S 91 | 100824 > | 102095 | 125 | 1272 | 423 | 48 | |||||
| 92 | 102221 > | 102442 | 126 | 222 | 73 | 8 | N/A | TM, SP | |||
| 93 | 102569 > | 103420 | 64 | 852 | 283 | 32 | TM, SP | ||||
| S 94 | 103485 > | 103820 | −4 | 336 | 111 | 13 | N/A | TM | |||
| 95 | 103817 < | 104239 | 20 | 423 | 140 | 16 | |||||
| S 96 | 104260 > | 108681 | 37 | 4422 | 1473 | 165 | N/A | ||||
| S 97 | 108719 > | 110167 | 162 | 1449 | 482 | 57 | |||||
| 98 | 110330 > | 111544 | −18 | 1215 | 404 | 47 | 9TM | ||||
| 99 | 111527 < | 113365 | 18 | 1839 | 612 | 68 | N/A | ||||
| 100 | 113384 > | 114751 | 6 | 1368 | 455 | 50 | |||||
| 101 | 114758 > | 115354 | −42 | 597 | 198 | 23 | N/A | ||||
| S 102 | 115313 < | 116086 | 30 | 774 | 257 | 30 | |||||
| 103 | 116117 < | 116320 | 161 | 204 | 67 | 8 | N/A | ||||
| 104 | 116482 > | 118794 | −14 | 2313 | 770 | 89 | helicase-like | Parabacteroides | 34% (72) 708 | 8.00E-02 | p-loop containing triphosphate hydrolases |
| 105 | 118781 < | 119506 | −158 | 726 | 241 | 28 | TM, SP | ||||
| 106 | 119349 > | 120077 | 1 | 729 | 242 | 28 | per os infectivity factor 3 (pif-3) | Spodoptera litura GV | 25% (153) 215 | 5.00E-03 | TM, SP, DUF666 |
| 107 | 120079 > | 123456 | 188 | 3378 | 1125 | 131 | |||||
| 108 | 123645 < | 124262 | 16 | 618 | 205 | 24 | Ac ORF 81-like | Neodiprion lecontei NPV | 32% (108) 175 | 1.00E-03 | 2TM |
S corresponds to ORFs identified as structural proteins. See Table 2 for details
- indicates number of nucleotides overlapping between adjacent ORFs
% of amino acid identity (number of aa used in the identity comparison) total number of aa from the compared protein
MdSGHV gene content analysis
From the putative 108 ORFs found in MdSGHV, only 30 had significant homology to proteins from the current database (Table 1, Fig. 3). From those, 11 had homology to proteins coded by both baculovirus and nudivirus genes, seven to nudivirus, one to baculovirus, one to herpesvirus, and ten to cellular genes. From the 12 ORFs homologous with proteins encoded by baculovirus genes, only five were from the conserved core of 29 genes (Garcia-Maruniak et al., 2004). The highest number of homologous ORFs was found with the nudiviruses (18 in total), although just seven of those are present in all of the nudiviruses completely sequenced to date: HzNV-1 virus (Cheng et al., 2002), GbNV (Wang et al., 2007b), and HzNV-2 virus (information obtained from Wang et al., 2007a).
Analysis of structural proteins
A total of 29 MdSGHV ORFs encoding structural proteins were identified from the GeLC-MS/MS analysis of the SDS-PAGE gel of the purified virus preparation (Fig. 2; Table 2). The number of proteins detected was less than that detected in enveloped baculovirus (Braunagel et al., 2003; Perera et al., 2007). For example, Perera and colleagues, using the GeLC-MS/MS approach, identified 44 structural proteins in the nucleopolyhedrovirus of Culex nigripalpus. In many cases in our analysis, the calculated molecular mass of the gel band contained peptide data that corresponded to an ORF(s) that encoded for a similar-sized peptide. For example, the major band b (~145 kDa) contained peptides that accounted for 67% coverage of ORF 96 (165 kDa), whereas band h (~38 kDa) contained peptides that accounted for 61% and 70% coverage of ORF 86 (43 kDA) and ORF 22 (40 kDa) respectively. Additionally, the LC-MS/MS analysis identified peptide fragments that originated from an array of ORFs that had molecular masses different from the calculated values of the gel fragments. For example, similar motifs identified from ORFs 96 and 86 were found in all gel fragments, suggesting a breakdown of certain protein complexes. Alternatively, motifs from ORF 94 (13 kDa) were detected in bands h (43 kDa) and i (35 kDa), suggesting an incomplete solubilization of the structural peptides. Of the 29 structural ORFs, only five ORFs (13, 16, 39, 50, and 91) had less than 10% coverage. Fourteen ORFs had 10–50% coverage, and ten ORFs had coverage greater than 50% (Table 2). It should be noted that all 29 ORFs identified as bona fide structural peptides contained unique peptide fragments, ranging from 6 to 27 amino acids that had a 100% match to the translated MdSGHV putative ORFs. The structural ORFs appeared to be clustered loosely on the genome; a total of 17 structural ORFs were located between MdSGHV013 and MdSGHV052, and eight structural ORFs were located between MdSGHV084 and MdSGHV097.
Table 2.
Structural proteins identified from MdSGHV by mass spectrum analysis. The percent coverage and number of unique fragments represents the pooled amino acid sequence attained from all gel slices analyzed using the GeLC-MS/MS analysis.
| MdSGHV ORF | MdSGHV predicted peptide length (aa) | No. of unique peptide fragments with 100% identity to MdSGHV predicted proteins | Coverage of total MdSGHV predicted protein (%) |
|---|---|---|---|
| 3 | 156 | 5 | 33 |
| 13* | 644 | 13 | 5 |
| 14 | 249 | 18 | 76 |
| 16* | 499 | 3 | 7 |
| 20 | 104 | 3 | 20 |
| 22 | 343 | 25 | 70 |
| 23 | 256 | 16 | 75 |
| 25 | 376 | 4 | 14 |
| 28 | 381 | 7 | 20 |
| 32 | 393 | 4 | 11 |
| 33 | 497 | 25 | 64 |
| 38 | 250 | 11 | 60 |
| 39* | 707 | 5 | 7 |
| 40 | 608 | 29 | 65 |
| 44 | 311 | 10 | 34 |
| 47 | 700 | 30 | 55 |
| 50* | 404 | 2 | 4 |
| 52 | 150 | 3 | 27 |
| 71 | 333 | 6 | 19 |
| 72 | 136 | 5 | 43 |
| 84 | 509 | 5 | 10 |
| 85 | 403 | 5 | 16 |
| 86 | 381 | 29 | 70 |
| 90 | 672 | 10 | 19 |
| 91* | 423 | 3 | 9 |
| 94 | 111 | 8 | 47 |
| 96 | 1473 | 82 | 71 |
| 97 | 482 | 19 | 59 |
| 102 | 257 | 8 | 30 |
Indicates MdSGHV ORFs with protein coverage of less than 10%
MdSGHV proteins homologous to proteins involved in DNA replication
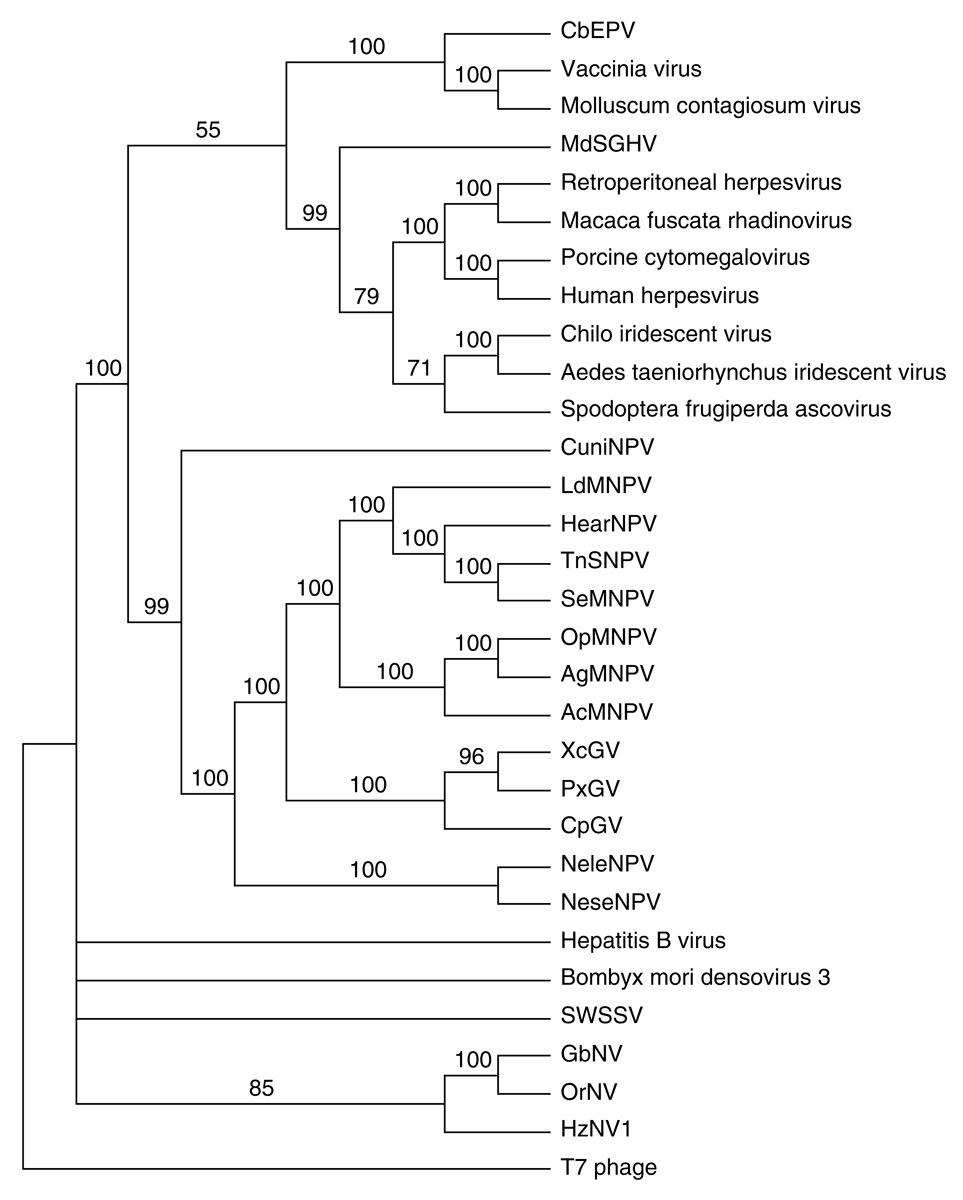
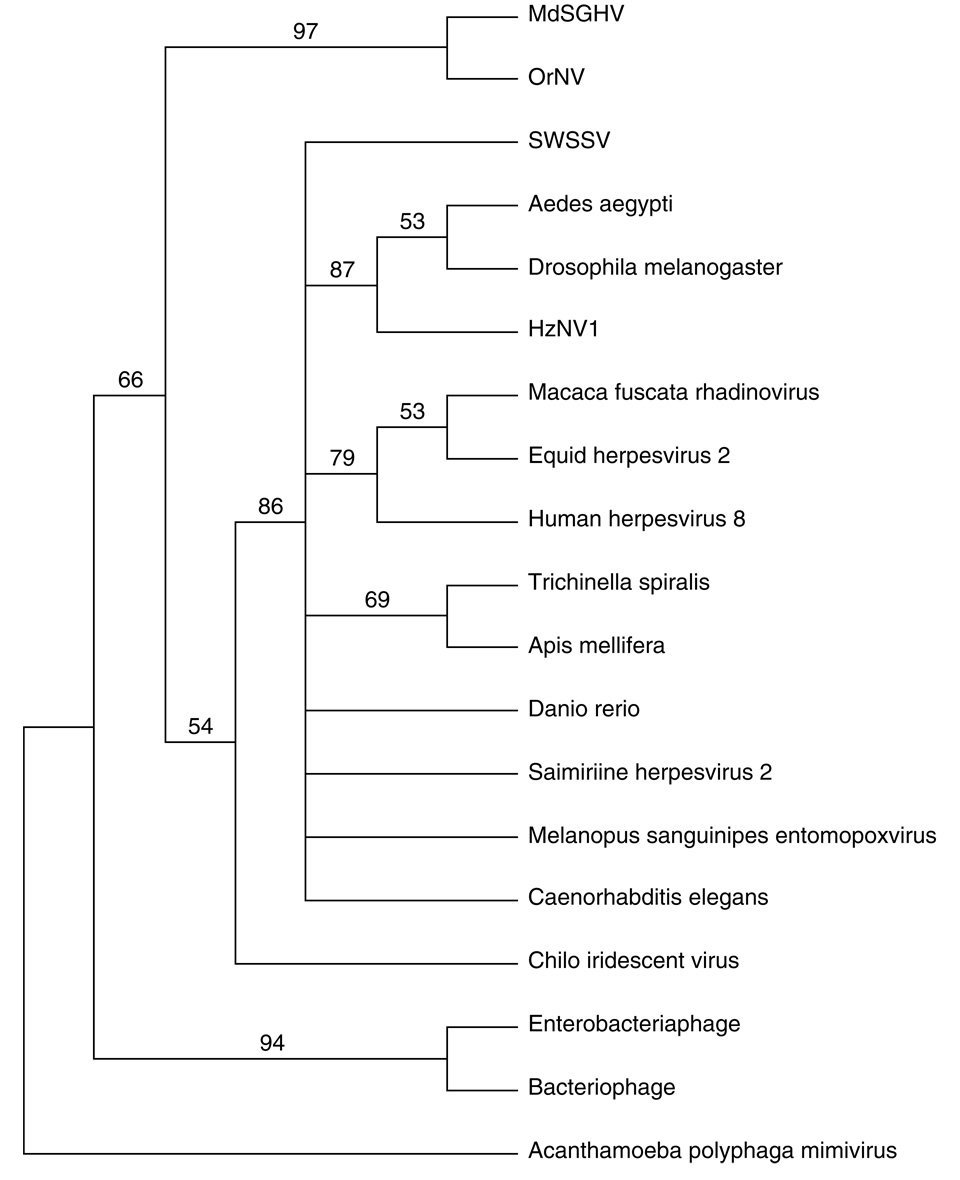
Two ORFs, MdSGHV001 and MdSGHV104, had homology to the DNA polymerase and helicase enzymes, respectively (Table 1), and both are known to be involved in DNA replication. The best match of MdSGHV001 to viral genes was to a herpesvirus DNA polymerase with 22.8% amino acid identity. No significant BLASTP matches were obtained with either the baculovirus or nudivirus DNA polymerases, even when the comparison was done with the local database containing only nudiviruses ORFs. The DNA polymerase of MdSGHV had homology to a delta 1 catalytic subunit similar to the DNA-directed DNA polymerase type B family that has a DNA binding polymerase and a 3'–5' exonuclease activity (Shamoo and Steitz, 1999). The conserved nucleotide binding site K(3x)NSxYG(2x)G motif at the C terminus was found almost intact in the MdSGHV DNA polymerase putative peptide (KLCANAIYGLLG starting at aa 631). In order to asses the relationship of MdSGHV to other DNA virus families, a phylogenetic analysis was done on the DNA polymerase gene. The results show that MdSGHV DNA polymerase was closer related to the baculovirus DNA polymerase than to the nudivirus DNA polymerase (Fig. 4A). MdSHGV001 clustered close to Herpesviridae, iridoviruses and ascoviruses. The baculovirus, poxvirus and herpesvirus families appeared to have monophyletic DNA polymerase lineages. Without additional family members, viruses such as iridescent virus, ascoviruses, densoviruses, shrimp white spot syndrome virus (SWSSV), nudiviruses and MdSGHV do not show direct descent and origins of their DNA polymerases.
Figure 4.
Phylogenetic analysis of selected MdSGHV putative proteins: A. DNA Polymerase, B. Combined analysis of p74, pif-1, pif-2 and pif-3. C. Thymidylate synthase. The trees were inferred using neighbor-joining in PAUP*, and a bootstrap analysis with 1000 replicates was performed to measure support for the branches. Organisms included in this analysis with abbreviated names: Anticarsia gemmatalis nucleopolyhedrovirus (AgMNPV), Autographa californica MNPV (AcMNPV), Choristoneura biennis entomopoxvirus (CbEPV), Culex nigripalpus NPV (CuniNPV), Cydia pomonella granulovirus (CpGV), Gryllus bimaculatus nudivirus (GbNV), Helicoverpa armigera NPV (HearNPV), Heliothis zea nudivirus 1 (HzNV1), Lymantria dispar MNPV (LdMNPV), Musca domestica salivary gland hypertrophy virus (MdSGHV), Neodiprion lecontei NPV (NeleNPV), Neodiprion sertifer NPV (NeseNPV), Orgyia pseudotsugata MNPV (OpMNPV), Oryctes rhinoceros nudivirus (OrNV), Plutella xylostella GV (PxGV), Shrimp white spot syndrome virus (SWSSV), Spodoptera exigua MNPV (SeMNPV), Trichoplusia ni SNPV (TnSNPV), Xestia c-nigrum GV (XcGV).
The best match of MdSGHV104 was to a Parabacteroides helicase-like protein, although the homology was low. Helicase is one of the 29 conserved baculovirus core genes, and ORFs with homology to helicase-2 have been described in HzNV-1, HzNV-2 and OrNV (Wang et al., 2007a). MdSGHV104, however, presented no significant homology to either the baculovirus or the nudivirus peptides. The viral replication genes from MdSGHV seem to be distinct from other insect DNA viruses. Interestingly, no baculovirus transcriptional regulator gene homologues were found in the MdSGHV genome. Also, there was no homology to any of the early transcriptional genes, to the late transcriptional activators, nor to the genes encoding for known viral RNA polymerases. Analysis of the DNA replication and transcriptional genes of other SGHVs could help decipher the strategy used by these viruses.
MdSGHV proteins homologous to baculovirus structural proteins
Six MdSGHV ORFs had homology to known baculovirus structural proteins (Table 1): MdSGHV029 (pif-1), MdSGHV039 (p74), MdSGHV047 (odv-e66), MdSGHV049 (Ac150), MdSGHV089 (pif-2) and MdSGHV106 (pif-3). While pif-1, pif-2, pif-3 and the p74 genes belong to the core of conserved baculovirus genes and are present in all completely sequenced nudiviruses, odv-e66 is present only in the nucleopolyhedrovirus (alphabaculovirus) and granulovirus (betabaculovirus) that infect Lepidoptera hosts (Jehle et al., 2006) and in OrNV (Wang et al., 2007c). On the other hand, Ac150-like is present in all GVs but not all NPVs and is not present in nudiviruses. The per os infectivity factor (pif) proteins seem to be associated with the baculovirus occlusion-derived viruses (ODVs), are released from occlusion bodies by the alkaline environment of the insect midgut, and have been reported to be responsible for both oral infectivity and host-to-host dispersal of these viruses. It has been shown that p74 (Kuzio et al., 1989), pif-1 (Kikhno et al., 2002), pif-2 (Pijlman et al., 2003), and pif-3 (Ohkawa et al., 2005) are essential proteins for oral infection. The phylogenetic analysis of the combined p74, pif-1, pif-2 and pif-3 proteins showed a distant relationship between MdSGHV and the baculoviruses (Fig. 4B), and that there is no common ancestor with the nudiviruses with the current number of viruses with pif proteins analyzed. There appears to be a monophyletic relationship amongst the baculovirus combined pifs including lepidopteran, hymenopteran and dipteran viruses. The HzNV-1 and GbNV pifs appear to branch together, but the MdSGHV pifs are basal, as are the nudiviruses to the baculovirus pif proteins.
MdSGHV049 had homology to the Autographa californica NPV (AcMNPV) ORF 150 (Ac150). Transmission electron microscopy coupled with immunogold labeling of Ac150 show this protein is associated mainly with enveloped virions before and after they had been occluded within the nuclei of infected Sf-21 cells, and it is also found in budded virus (Lapointe et al., 2004). Deletion of Ac150 decreases oral infectivity, but has no effect when the budded viruses (BV) are injected into the insect hemocoel indicating that Ac150, can be considered a per os infection factor that mediates, but is not essential for oral infection (Zhang et al., 2005). The C terminus of MdSGHV049 (from aa 98 to 152) had a chitin-binding peritrophin-A (CBM-14) domain similar to Ac150. This domain is found in many chitin-binding proteins including peritrophic matrix proteins of insects and animal chitinases (Tellan et al., 1999).
MdSGHV does not produce occlusion bodies, so there is no ODV form of the virus. Although six putative genes were found in the MdSGHV genome that had significant amino acid identity to known structural genes (Table 1), the mass spectrometry analysis of the structural proteins separated by an acrylamide gel found only two genes coding for the p74 (MdSGHV039) and odv-e66 (MdSGHV047) proteins associated with the enveloped MdSGHV (Table 2). Possibly, the MdSGHV pif proteins either could be produced in nondetectable quantities as reported for Spodoptera littoralis NPV (SpliNPV) pif-1 (Gutierrez at al., 2004), or may not be structural proteins. If pif-1, pif-2, pif-3 and Ac150-like proteins are expressed and are important in the oral infection of MdSGHV they may be delivered in the salivary secretions to the next host. The predicted transmembrane domains of the MdSGHV pif-1, pif-2, pif-3 and odv-e66 were localized in the N-terminal region, while the p74 transmembrane motif was found in the C-terminus of the putative peptides, similar to those proteins in the baculoviruses.
Although pif-1, pif-2, pif-3 and p74 have been described in the genomes of other nudiviruses (Wang et al., 2007a), there was no significant amino acid homology between MdSGHV pif-3 and the nudiviruses pif-3. MdSGHV odv-e66 had the best amino acid homology (35% identity) with GpSGHV. This gene has also been found in the genome of OrNV (Wang et al., 2007c), but not in the other two nudiviruses (HzNV-1 and GbNV).
MdSGHV proteins homologous to auxiliary and undefined baculovirus proteins
MdSGHV036 is homologous to the zinc-dependant matrix metalloproteinase (MMP) of several viruses including the granuloviruses, HzNV-1, entomopoxvirus and the insect iridescent virus. MMPs are responsible for pericellular proteolysis of extracellular matrix and cell surface molecules and, in many instances, they are anchored to cell membranes via trans-membrane domains (Baumann et al., 1993). MdSGHV036 had a transmembrane domain in its N-terminus (Table 1), although it had not been detected by the mass spectrum analysis of MdSGHV as a structural protein. In granuloviruses this protein is believed to disrupt the peritrophic matrix or the basement membranes that help hold tissues together thus assisting in virus dissemination (Ko et al., 2000).
MdSGHV052 had a high homology to a dUTP pyrophosphatase (dUTPase) of the iridovirus (51% aa identity). This gene, also present in several baculoviruses and in HzNV-1, is involved with nucleotide transport and metabolism. UTPase hydrolyzes the triphosphate DNA polymerase substrate dUTP to non-substrate monophosphate (dUMP) to prevent incorporation of uracil into the DNA while providing the substrate for thymidine synthesis (Harris et al., 1999). The protein coded by MdSGHV052 was detected by mass spectrometry to be a structural protein (Table 2).
The highest amino acid homology was found for MdSGHV062 and MdSGHV065 to Lymantria dispar NPV (LdMNPV) and Spodoptera litura NPV (SpltNPV) ribonucleotide reductase small (rr2) and large (rr1) subunits, with 72% and 61% aa identities, respectively (Table 1). These two MdSGHV ORFs were separated by almost 3 kbp and had opposite transcriptional orientations. The genes coding for rr1 and rr2 are present in viruses including nudiviruses and baculoviruses, as well as eukaryotes and many prokaryotes.
MdSGHV078 encoded an inhibitor of apoptosis (iap) homologue. The iaps were first discovered in baculoviruses and are present in various animal species (Clem, 1997; Uren, et al., 1998). They have one to three consensus amino-terminal regions named BIR (baculoviral iap repeat) domains, essential to inhibit apoptosis by acting as a direct inhibitor of the caspase family of protease enzymes (Miller, 1999). The iaps also have a carboxy-terminal zinc-binding motif named ring finger, which appears to be involved in ubiquitination of the iap itself and potentially, any caspase bound to it (Wilson et al., 2002). The MdSGHV078 encodes a 142 aa peptide that is smaller than the majority of the baculovirus iaps, but very similar in size to the Melanoplus sanguinipes entomopoxvirus iap which had the highest homology to MdSGHV078, although baculoviruses and nudiviruses iap-3 also had significant homology. MdSGHV078 had only one BIR motif in its N-terminus with the three conserved cysteines (aa position 46, 49 and 73) and one histidine (aa 66) in the expected conserved locations, but did not have the zinc finger domain at its C-terminus. Other baculoviruses have iap proteins with only one BIR motif, and since not all iap are capable of preventing cell death (Maguire et al., 2000), the function of this ORF will need to be confirmed experimentally.
A homologue of AcMNPV ORF 81(Ac81) present in all the baculoviruses and nudiviruses was found in the MdSGHV genome. The best homology was found with the Neodiprion lecontei NPV Ac-81 like protein. MdSGHV108 has two transmembrane domains at its C-terminus. Although this gene belongs to the core of baculovirus conserved genes, its functions has not been defined (Slack and Arif, 2007).
MdSGHV proteins homologous to nudivirus proteins
Seven ORFs (MdSGHV ORFs 10, 11, 12, 26, 48, 69 and 76) had homology to nudivirus putative proteins not found in any baculovirus genome (Fig. 3, Table 1). Five of those are cellular genes, the majority of which have been well studied in eucaryotes (see below). MdSGHV010 has homology with a mitochondrial carrier protein present in OrNV (ORF C7). The mitochondrial carrier protein family comprises a variety of small proteins (28 to 34 kDa) that catalyze the exchange of substrates across the inner mitochondrial membrane (IMM), and have three tandemly repeated mitochiondria carrier domains of approximately 100 aa each (Walker and Runswick, 1993). MdSGHV010 had a predicted molecular weight of 29 KDa and two mitochondria carrier domains at its C terminus (between aa 83–179 and 176– 256, respectively).
MdSGHV011 was homologous to HzNV-1 dihydrofolate reductase (DHFR). DHFR is a ubiquitous enzyme that plays a key role in DNA replication and cell division, since the enzyme is essential in providing purines and pyrimidine precursors for the biosynthesis of DNA, RNA and amino acids (Miller and Benkovick, 1998).
MdSGHV012 was homologous to a putative thymidylate synthase (TS) found in the genomes of OrNV, HzNV-1, HzNV-2, SWSSV, herpesviruses and other viruses. The TS is involved in the biosynthesis of DNA precursors and, hence, it is one of the most conserved enzymes across species and phyla (Perryman et al., 1986). The phylogenetic analysis of several viral and cellular TS grouped MdSGHV012 closer to OrNV than to HzNV-1 TS (Fig. 4C). The lack of monophyletic structure in the tree suggested that this gene was acquired from the host at multiple independent occasions rather than originating from a common DNA virus ancestor. This is further corroborated by the tree positions found between HzNV-1 and insects, as well as an entomopoxvirus and herpesviruses basal with insects and a nematode. In several protozoa, a single polypeptide chain codes for both, DHFR) and TS, forming a bifunctional enzyme (DHFR-TS), possibly through gene fusion at a single evolutionary point. Interestingly, MdSGHV ORFs 11 and 12 were adjacent to each other although with opposite transcriptional directions, and their functions (if confirmed to be active proteins) are interconnected. The size of both putative proteins was similar to those of active proteins. As mentioned before, the DNA polymerase and helicase genes from MdSGHV are distantly related to those of the baculoviruses and other insect viruses. Potentially, different genes, not even present in the baculoviruses are key players in the DNA replication of MdSGHV.
MdSGHV069 was a 203 aa putative protein homologous to the deoxynucleoside kinase (dNK) family protein. The best match was found to Spodoptera frugiperda ascovirus 1a thymidine kinase (tk). This protein is produced by several cellular organisms and viruses such as HzNV-1 (ORF 70). The are key enzymes in the salvage of deoxyribonucleosides originating from extra-or intracellular breakdown of DNA (Johansson et al., 2001). MdSGHV069 contained the conserved substrate-binding sites of this protein family (cd01673).
MdSGHV026 was a 60 aa long, serine-rich putative protein. This ORF had homology only to GbNV ORF 73 (Wang et al., 2007b). MdSGHV048 presented a low homology to a cellular hypothetical gene and also to HzNV-1 ORF 153. Lastly, MdSGHV076 was homologous to unknown proteins detected in nudivirus genes including OrNV-C3, GbNV ORF 65, HzNV-1 ORF 68 and HzNV-2 ORF 70 (Cheng et al., 2002; Wang et al., 2007a, 2007b, 2007c).
MdSGHV ORFs with homology to cellular genes
Ten ORFs had homology to cellular genes (Fig. 3, Table 1). MdSGHV014 had a low homology to a molybdopterin oxidoreductase from Mycobacterium. MdSGHV033, was similar to the putative vacuolar sorting ATPase from Candida albicans. It had an AAA-ATPase motif that belongs to the AAA-superfamily of ATPases, associated with a wide variety of cellular activities, including membrane fusion, proteolysis, and DNA replication (Patel and Latterich, 1998). MdSGHV050 was similar to a cellular aminoacylase-1. Mass spectral analysis showed that these three ORFs code for structural proteins of MdSGHV (Table 2). MdSGHV064 had a RING-, and a zinc-finger (C3HC4) motifs. The RING-finger domain is a specialized type of a Zinc-finger of 40 to 60 residues that binds two atoms of zinc; it is defined by the 'cross-brace' motif C-X2-C-X(9-39)-C-X(1-3)- H-X(2-3)-(N/C/H)-X2-C-X(4-48)C-X2-C. This C3HC4 motif was present between aa 214 – 251 of MdSGHV064. Proteins carrying this motif have been identified with a wide range of functions such as viral replication, signal transduction, and probably are involved in mediating proteinprotein interactions (Saurin et al., 1996). MdSGHV066 is a 579 aa putative sodium dependent nucleoside transporter protein. The conserved motif of the nucleoside transporter was in the C-terminal end of MdSGHV066, and a total of 13 transmembrane domains were found in this ORF. A similar sized protein having 14 transmembrane motifs, capable of selectively transporting pyrimidine nucleoside and adenine has been studied in mammal jejunal epithelium (Huang et al., 1994). MdSGHV082 had homology to an integrase/recombinase protein that is involved in DNA rearrangements by site-specific DNA recombination (Grainge and Jayaram, 1999). The conserved integrase/recombinase motif was found at the C-terminal end of MdSGHV082. Although GbNV also has an integrase protein (ORF 57) (Wang et al., 2007b), there was not significant homology to MdSGHV082. MdSGHV035, MdSGHV046 and MdSGHV055 had homology to functionally unknown cellular proteins. Finally, MdSGHV104 with low homology to helicase gene has been discussed previously (under homologues to proteins involved in DNA replication).
Direct Repeats
Eighteen tandem direct repeats (drs) were found distributed throughout the genome. The size of the repeated sequences ranged from 149 bp (dr15) to only 9 bp long (dr18), and the number of copies of the drs ranged from 1.9 to 7.4, making the total sizes of the drs range from 30 bp to 380 bp long (Table 3). Unlike the homologous regions (hrs) found in large, circular invertebrate dsDNA viruses that have imperfect palindromic sequences within the repeated region (Cochran, and Faulkner, 1983; Garcia-Maruniak et al., 1996), no palindromes were found in the MdSGHV direct repeats. The MdSGHV drs appeared similar to those found in HzNV-1, GbNV, and OrNV and Neodiprion sertifer NPV (NeseNPV) (Cheng et al. 2002; Garcia-Maruniak et al., 2004; Wang et al., 2007b, 2007c), however the AT percentage was not as high as GbNV drs. Seven drs (dr5, 6, 7, 10, 16, 17 and 18) were located inside putative ORFs. Baculovirus hrs are located between genes and act as transcriptional enhancers (Guarino and Summers, 1986) and/or origin of DNA replication (Pearson et al., 1992). Another feature of the hrs is that their sequences have homology within the same genome. This feature was not present in the MdSGHV drs, as no significant homology was observed between the 18 drs. A 45 bp TCA repeat was located inside ORF 101, similar to the 3 base repeats reported in GbNV (Wang et al., 2007b).
Table 3.
MdSGHV direct repeat (dr) sequences
| Name | Repeat size (bp) × N. of copies | dr sequence and genomic position | AT% | Total size (bp) |
|---|---|---|---|---|
| dr1 | 21 × 3.3 | 4559 CGAGTTTGTCTTTTAATATGAAC 4581 | 65 | 72 |
| 4582 CGAGTTTG-CTTT-GATGTGTAC 4602 | ||||
| 4603 CGAGTTTG-TTTT-AATATGAAC 4623 | ||||
| 4624 CGAGTTT 4630 | ||||
| dr2 | 64 × 4.3 | 5933 TGTTTGTGTGTCACTCCGATTTTA--T--TTAATAGACTGTTCCGCCTAGT-CCCATAAGAAATATACA 5996 | 63 | 279 |
| 5997 TGTTTGTGTGTCACTCCGATTTTATTTGGTTGGTATGGACTATTAGTGGGT-CCCATAAGAAATATACA 6064 | ||||
| 6065 TGTTTGTGTGTCACTCCGATTTCA--T--TTAATAGACTGTTTCGTTTGGT-CCCATATAAAATATACA 6128 | ||||
| 6129 TGTTTGTCTGTCACTCCGATTTTAA-T---TAGTGGACCGTTTCGTTTGGTCCCCATAAGAAATGTACA 6193 | ||||
| 6194 TGTTTGTGTGTCACTCCG 6211 | ||||
| dr3 | 53 × 2.3 | 6912 TGTAATTTGGGAGCAGAACTTGACCGACAGAACTCAGCCTGTATAGTTGTTAG 6964 | 59 | 126 |
| 6965 TGTAATTTGGGAGCAGAATTTAACCGACAGAACTCAGCCTGCATTGTTGTTAG 7017 | ||||
| 7018 TGTAATTTGTGGAAGCAGAA 7037 | ||||
| dr4 | 26 × 7.4 | 7173 TTGTTTTTGTAATCCAG-CCTGCATAG 7198 | 64 | 192 |
| 7199 TTGTTTTTGTACTCTAG-CCTGTATAG 7224 | ||||
| 7225 TTG-TTTTGTACTCGAG-CCTGTATAG 7249 | ||||
| 7250 TTGTTTTTGTACTCGAG-CCTGCATAG 7275 | ||||
| 7276 TTGTTTTTGTACCCATG-TCTGTATAG 7301 | ||||
| 7302 TTGTTAGTGTAATTC-G-CTACATGAAA 7327 | ||||
| 7328 TTGTTTTTGTAATC-AGGCCTGCATAG 7353 | ||||
| 7354 TTGTTTTTGTA 7364 | ||||
| dr5 | 24 × 2.7 | 35734 GTGGTGCCT-GGGACGGAGCTTCCT 35757 | 34 | 64 |
| 35758 GTGGTGGTGGGGATGGAGG-TTCCT 35781 | ||||
| 35782 GTGGTGCTGGGGATGG 35797 | ||||
| dr6 | 30 × 2.6 | 51725 TTGATCTCCTGGGGCTTCGGTTGGGGCAGA 51754 | 44 | 77 |
| 51755 TTGATCTCCTGGGGCTTCGGTTGGGGCAGA 51784 | ||||
| 51785 TTGATCTTCTGAGGCTT 51801 | ||||
| dr7 | 54 × 2.7 | 51918 TGATCTTCTGGGACTTCGTTAACGGACTGATCTCCTTGGGCCTCGTAAACGGAC 51971 | 49 | 148 |
| 51972 TGATCTCCTGGGTTTTCGTTAACGGATTGATCTCCTTGGGCCTCGTCAACGGAC 52025 | ||||
| 52026 TGATCTCCTGGGTTTTCGTTAACGGATTGATCTCCTTGGG 52065 | ||||
| dr8 | 43 × 2.9 | 60302 GACGGTCGACAGAGAAAACACTAACAACAAGTACAGGG-TATAT 60344 | 54 | 122 |
| 60345 GACGGTCGGCAGAGAAAACACTAACAA-AA-TACATGGGGCTTAT 60387 | ||||
| 60388 GGTGG-GGGCAGCGAAAACACTAACAACAAGTACAGG 60423 | ||||
| dr9 | 44 × 3.4 | 60422 GGCTTGCGAATATCGACAGCGGGTTACAAAAACAAAATACATGG 60465 | 62 | 151 |
| 60466 GGCTTGTGGATATCGACAGAGGGTTACAAAAACAAAATATACAG 60509 | ||||
| 60510 GCCTTATGAATATCGACAGAGGGTTACAAAAACAAAATATACAG 60553 | ||||
| 60554 ACCTTATGAATATCGACAG 60572 | ||||
| dr10 | 18 × 2 | 62619 AGGAACCAAGTATCCAGG 62636 | 50 | 36 |
| 62637 AGGAACCAAGTATCCAGG 62654 | ||||
| dr11 | 59 × 1.9 | 64420 TATTTTTCATTTTCATGTTAGTGTATATTTGGACTTAGTATTTTTCAAGTTCGCGAGAA 64478 | 71 | 115 |
| 64479 TATTTTTCATTTTCATCTTCGCATATATTGGGACTTAGTATTTTTCAAGTTCGCGA 64534 | ||||
| dr12 | 60 × 3.0 | 64704 TTTTCATTTCAGACGAGTAGGATATTTTTT-ATTATCAAGTCCGCATAT-GAC--TTGTGC 64760 | 67 | 177 |
| 64761 TTTTCATTTTCAGATGAGTAGGGTATTTTTCATTTTCAAGACCACATATTGAGACTTTGTA 64821 | ||||
| 64822 TTTTCGTTTCAG-TCAAGTAGGGTATTTTTCATTTTCAAGTCCGCATATTGAGACTTGTG 64880 | ||||
| dr13 | 23 × 2.0 | 70445 ACCCAAGTCCCCATACGAAGAGC 70467 | 46 | 46 |
| 70468 ACCCAAGTCCCCATACAAAGAGC 70490 | ||||
| dr14 | 17 × 2.0 | 71349 TTTTGGTTTTTGGTTGG 71365 | 68 | 34 |
| 71366 TTTTGATTTTTGGTTGG 71382 | ||||
| dr15* | 149 × 2.6 | 81117 TTCA-AACCAAGTTGAAACCTGGCCAGCGAAAAATGAAAAATTGTCTAGCAAAAATTTCTGAAAA 81180 | 56 | 380 |
| 81265 TTCACAACCAAGTTGAAACCCGGCCAGCGAAAAATGAAAAATTG-CAGAGTAAAAATTTCTGAAA 81328 | ||||
| 81409 TTCACAACCAAGTTGAAACCCACCCAGTGAAAAATGAAAAATTGTCGAGCAAAAAATTTCTGAAA 81473 | ||||
| 81181 AGCAAGCATGATCGGGCATGCACACTCTCTCGTTAGGTATCAGAGGGAGAGGGGTGATGGGGGTG 81245 | ||||
| 81329 AACC-GGACATGATCGAGCATGCACACT-T-ACGATA-G-ACTCA-AGGGAG-GGGATGATGGGA 81386 | ||||
| 81474 AATCAAGCATGATCGAGCATGCA 81496 | ||||
| 81246 AGGAGGGA-–T-CGATCTACAA 81264 | ||||
| 81387 GAGCGGGGGTTCCGATCCACAA 81408 | ||||
| dr16 | 12 × 2.5 | 97194 TCATCGTCATCA 97205 | 57 | 30 |
| 97206 TCATCGTCATCA 97217 | ||||
| 97218 TCATCG 97223 | ||||
| dr17 | 15 × 2.0 | 104632 GGTGGAAGTGGTGGA 104646 | 43 | 30 |
| 104647 GGTGGAAGTGATGGA 104661 | ||||
| TCA repeat | 3 × 15 | 112083 TCA TCA TCA TCA TCG TCA TCA TCC TCA TCC TCA TCA CCA TCA TCA 112128 | 58 | 45 |
| dr18 | 9 × 3.8 | 112519 CATCACCAT 112527 | 50 | 34 |
| 112528 CATCAACAC 112536 | ||||
| 112537 CATCACCAC 112545 | ||||
| 112546 CATCACC 112552 |
sequence of each 149 bp repeat is presented in 3 different rows
In this study, we have determined the complete genome sequence and provided the first comprehensive analysis of the MdSGHV genome. Completion of the genome sequence has provided a foundation to examine the biology of a unique insect virus. This virus, with the ability to sterilize female flies, has the potential of being developed into an adult biopesticide. The sequence data will play an important role in elucidating the mechanisms underlying its ability to shut down reproductive behaviors and will provide a platform to engineer improved biopesticides against pest fly populations. The sequence data will also be used to characterize genetic variants isolated from feral housefly populations and determine which polymorphisms are associated with the epizootic or the enzootic profiles exhibited by this virus in fly populations. Importantly, related SGHVs have been isolated from tsetse fly vectors of African trypanosomiasis. These SGHVs have a larger genome than MdSGHV, show wider tissue tropism, are vertically transmitted, and are known to persist asymptomatically in tsestse flies. Comparisons among the different SGHV sequences will provide opportunities to assign genotypic profiles to similar and dissimilar viral phenotypes.
MATERIAL AND METHODS
Virus production and purification
As an alternative to cloning the virus (no cell lines are known to support MdSGHV replication), a single hypertrophic gland was dissected from an MdSGHV infected feral adult house fly. This gland was homogenized in 1 ml of phosphate buffered saline (PBS), filter-sterilized (Ultrafree-MC, Millipore Corp., Billerica, CA), and injected (2 µl/fly) into 200 newly emerged adult house flies. Injected house flies were placed in cages, provided with food and water, and incubated at 26 °C. After 5 days, the injected flies were immobilized on ice, and infected salivary glands were dissected, pooled, and frozen at −70 °C. Glands from 100 infected flies were homogenized in 5 mM Tris-HCl (pH 7.5) containing 0.2 M sucrose, 0.13 M NaCl, 3 mM KCl, and 3 mM EDTA, subjected to low-speed centrifugation (1,500 g, 20 min) to remove tissue debris, applied to a 10–50% isotonic linear gradient of Nycodenz (Accurate Chemicals & Scientific Corp., Westbury, NY), and centrifuged at 30,000 rpm (~100,000 g) for 90 min. The virus band was collected and subjected to another high-speed centrifugation for 30 min to remove residual gradient material. Purified virus was suspended in TE buffer (10mM TRIS, 1 mM EDTA), aliquoted and stored at −20°C. The quality of the virus fraction was determined by electron microscopy. A dilute viral preparation was applied to formvar + carbon-coated grids, fixed with 2% glutaraldehyde in 0.1M cacodylate, water washed, dried, and stained with aqueous uranyl acetate and lead citrate. Grids were examined with an Hitachi H-7000 TEM operating at 100 KV and images captured with a Soft-Imaging System MegaView III with AnalySIS.
Analysis and identification of viral structural proteins
An aliquot of the purified virus preparation was solubilized in SDS-β mercaptoethanol and electrophoresed onto a 5 to 15% gradient polyacrylamide gel electrophoresis (PAGE) gel (Laemmli, 1970). The Coomassie R-250 blue-stained gel was recorded with the Bio-Rad gel imaging XRS system. Precision Plus Protein unstained standards (Bio-Rad Laboratories, Inc., Hercules, CA) were used to calculate a standard curve (R-square = 0.987) and subsequent molecular weights of viral peptide bands were calculated using the Quantity One software (Bio-Rad Laboratories, Inc.).
Coomassie R-250 blue-stained protein bands and interband gel regions were excised and digested in-gel with trypsin (Link et al., 1999). Capillary reversed-phase HPLC separation of protein digests was performed on a 15-cm-by-75-µm (inner diameter) TARGA C18 column (Higgins Analytical, Mountain View, CA) in combination with an Ultimate Capillary HPLC system (LC Packings, Sunnyvale, CA) operated at a flow rate of 200 nl/min. Mass spectrometry analysis of the trypsin-digested peptides derived from the one-dimensional gel (GeLC-MS/MS) was accomplished by a hybrid quadrupole time-of-flight MS instrument (QSTAR; Applied Biosystems, Foster City, CA) equipped with a nanoelectrospray source at the Proteomics Core Facility from the Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville, Florida. Amino acid sequence data generated by the QSTAR from the MS/MS were searched against the National Center for Biotechnology Information non-redundant sequence database using the Mascot (Matrix Science, Boston, MA) database search algorithm. Mascot was set up to search the MdSGHV ORF database. Scaffold (version Scaffold-01_06_19, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Proteins identified by Mascot were considered significant if individual ion scores exceeded the threshold value calculated for identity or extensive homology (P ≤ 0.05). The percent coverage of each protein by peptide fragments identified was calculated using only the unique peptide fragments with 100% amino acid identity (i.e., amino acid sequences from recurring peptide fragments or overlapping amino acid sequences were counted only once).
MdSGHV genome purification and EcoRI library construction
Virus was treated with 1.5% sarcosyl and 250 µg/ml proteinase K and the DNA was purified by phenol/ether extractions as described by Garcia-Maruniak et al. (2004). To determine if the genome was circular or linear, 400 ng of undigested DNA was end-labeled with 2 µCi 32P-dATP and DNA polymerase I large fragment (Klenow, New England BioLabs, Beverly, MA). MdSGHV genomic DNA (200 ng) digested separately with EcoRI, HindIII, BamHI and XhoI was also end-labeled and run in a 0.8% agarose gel. To construct an EcoRI library, 1 µg of DNA was digested, ligated into 350 ng pGEM 7Z plasmid (Promega Corp., Madison, WI), and used to transform MAX efficiency DH5-α competent cells (Invitrogen, Carlsbad, CA). Multiple colonies were screened to confirm fragment size. Fragments not cloned during by this approach were cut out of the agarose gel and individually cloned.
Genome sequencing and analysis
Both Sanger (Sanger, et al., 1977) and pyrophosphate-based (454) sequencing techniques were used to sequence the MdSGHV genome. Initially, 192 EcoRI clones were used for cycle sequencing using T7 and SP6 primers and BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) and sent to the University of Florida Interdisciplinary Center for Biotechnology Research (ICBR). Secondly, an aliquot of 20 µg of total DNA was submitted to 454 sequencing. This was performed as described in the supplementary material and methods to Margulies et al. (2005) with slight modifications from the specifications (454 Life Sciences, Branford, CT). Briefly, high molecular weight DNA from the RCA reactions was sheared by nebulization to a size range of 300 to 800 bp. DNA fragment ends were repaired and phosphorylated using T4 DNA polymerase and T4 polynucleotide kinase. Adaptor oligonucleotides “A” and “B” supplied with the 454 Life Sciences sequencing reagent kit were ligated to the DNA fragments using T4 DNA ligase. Purified DNA fragments were hybridized to DNA capture beads and clonally amplified by emulsion PCR (emPCR). DNA capture beads containing amplified DNA were deposited in individual lanes of a 40 × 75 mm PicoTiter plate and DNA sequences determined using the GS-FLX instrument. DNA sequence information from the initial and supplementary runs was combined in a single assembly using Newbler sequence assembly software. Some PCR amplifications followed by sequencing of the DNA products were needed to close every gap of the genome. The contigs assembled by Newbler and the electropherograms obtained from the EcoRI clones or PCR products were further aligned using Sequencher 4.8 (Gene Codes Corp., Ann Arbor, MI). Regions of nucleotide polymorphism were mapped. Putative open reading frames containing at least 50 amino acids, ATG start sites, and presenting minimal overlapping in the genome were identified with ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and Gene Finding in Viruses program from SoftBerry (http://www.softberry.com/berry.phtml). The translated amino acid sequence of each ORF was compared to the GenBank database by protein BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) against the default and virus-only databases. A local database with only the nudivirus protein sequences was created and used to compare all MdSGHV ORFs. Significant homology was arbitrarily set at a BlastP E-value of 0.01 or less. Conserved domains and putative functional protein sites were searched for each ORF using Interproscan (http://www.ebi.ac.uk/interpro/) (Quevillon et al., 2005). Tandem-repeat sequences were found with the help of Tandem Repeat software (http://tandem.bu.edu/trf/trf.html) (Benson, 1999).
Phylogenetic analysis
Amino acid sequences were aligned with ClustalX 2.0 (Thompson et al., 1997). Alignments were edited, when needed, with MacClade 4.0 PPC (Madison and Madison, 2000) and the phylogenetic trees were constructed using PAUP* 4.0b (Swofford, 2003). Distance analysis with neighbor joining was done. The robustness of the tree was tested using bootstrap and 1000 replicates. For the construction of the combined tree of p74, pif-1, pif-2, and pif-3, a file containing the concatamer of the four genes was created and analyzed as described before.
Acknowledgements
We acknowledge the support of Chris Geden (USDA/ARS), Hilary Lauzon (Canadian Forest Service, Ontario, Canada) and Khuong Nguyen (University of Florida, Department of Entomology) and extend our gratitude to Tamer Salem, Verena Lietze (University of Florida, Department of Entomology), Adly Abd-Alla and Andrew Parker (Entomology Unit, FAO/IAEA laboratory, Vienna, Austria) for critical reviews of the manuscript. The excellent technical support provided by Scott McClung and Sophie Alvarez (ICBR Proteomics Core), Savita Shanker and Regina Shaw (ICBR Sequencing Core), and Karen Kelly (ICBR Electron Microscopy Core). We gratefully acknowledge the financial support provided by the National Institute of Health (NIAID R21 AI073501-01) and USDA/NRI (2007-35302-18127).
Footnotes
Sequence data for this article have been deposited in GenBank accession number EU522111
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Alejandra Garcia-Maruniak, Email: alemar@ufl.edu.
James E. Maruniak, Email: jem@ifas.ufl.edu.
William Farmerie, Email: wgf@biotech.ufl.edu.
Drion G. Boucias, Email: pathos@ufl.edu.
REFERENCES
- Abd-Alla A, Bossin H, Cousserans F, Parker A, Begoin M, Robinson A. Development of a non-destructive PCR method for detection of the salivary gland hypertrophy virus (SGHV) in tsetse flies. J. Virol. Methods. 2007;139:143–149. doi: 10.1016/j.jviromet.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Abd-Alla AMM, Couserans F, Parker AG, Jehle JA, Parker NJ, Vlack JM, Robinson AS, Bergoin M. Genome analysis of a Glossina pallidipes salivary gland hypertrophy virus (GpSGHV) reveals a novel large double stranded circular DNA virus. J. Virol. 2008 doi: 10.1128/JVI.02588-07. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amargier A, Lyon JP, Vago C, Meynadier G, Veyrunes JC. Mise en evidence et purification d'un virus dans la proliferation monstreuse glandulaire d'insectes. Étude sur Merodon equestris (Diptera, Syrphidae) Note. Comp. Rend. Seanc. Acad. Sci. Ser. D Sci. Natur. 1979;289:481–484. [PubMed] [Google Scholar]
- Baumann U, Wu S, Flaherty KM, McKay DB. Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two-domain protein with a calcium binding parallel beta roll motif. EMBO J. 1993;12:3357–3364. doi: 10.1002/j.1460-2075.1993.tb06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucias DG, Maruniak JE, Pendland JC. Characterization of a non-occluded baculovirus (subgroup C) from the field cricket, Gryllus rubens. Arch. Virol. 1989;106:93–102. doi: 10.1007/BF01311041. [DOI] [PubMed] [Google Scholar]
- Braunegel SC, Russell WK, Rosas-Acosta G, Russell DH, Summers MD. Determination of the protein composition of the occlusion-derived virus of Autographa californica. Proc. Natl. Acad. Sci. USA. 2003;100:9797–9802. doi: 10.1073/pnas.1733972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtt E. Hypertrophied salivary glands in Glossina: evidence that G. pallidipes with this abnormality is peculiarly suited to trypanosome infection. Ann. Trop. Med. Parasit. 1945;39:11–13. [Google Scholar]
- Cheng CH, Liu MS, Chow TY, Hsiao YY, Wang DP, Huang JJ, Chen HH. Analysis of the complete genome sequence of the Hz-1 virus suggests that it is related to members of the Baculoviridae. J. Virol. 2002;76:9024–9034. doi: 10.1128/JVI.76.18.9024-9034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem RJ. Regulation of programmed cell death by baculoviruses expression. In: Miller LK, editor. The Baculoviruses. New York: Plenum Press; 1997. pp. 237–266. [Google Scholar]
- Cochran MA, Faulkner P. Location of homologous DNA sequences interspersed at five regions in the baculovirus AcMNPV Genome. J. Virol. 1983;45:961–970. doi: 10.1128/jvi.45.3.961-970.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coler RR, Boucias DG, Frank JH, Maruniak JE, Garcia-Canedo A, Pendland JC. Characterization and description of a virus causing salivary gland hyperplasia in the housefly, Musca domestica. Med. Vet. Entomol. 1993;7:275–282. doi: 10.1111/j.1365-2915.1993.tb00688.x. [DOI] [PubMed] [Google Scholar]
- Ellis DS, Maudlin I. Salivary gland hyperplasia in wild caught tsetse from Zimbabwe. Entomol. Exp. Appl. 1987;45:167–173. [Google Scholar]
- Feldmann U, Barnor H, Acs R. Abweichungen in der Reproduktion von Glossina morsitans submorsitans Newstead (Diptera: Glossinidae): Untersuchungen zur Bestimmung eines gestörten Geschlechterverhältnisses und zum Übertragungensweg von fertilitäts-reduzierenden Viren an die Nachkommen. Mitteilungen der Deutschen Gesellschaft fuer Allgemeine und Angewandte Entomologie. 1992;8:248–251. [Google Scholar]
- Garcia-Maruniak A, Maruniak JE, Zanotto PM, Doumbouya AE, Liu JC, Merritt TM, Lanoie JS. Sequence analysis of the genome of the Neodiprion sertifer nucleopolyhedrovirus. J. Virol. 2004;78:7036–7051. doi: 10.1128/JVI.78.13.7036-7051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Maruniak A, Pavan OHO, Maruniak JE. A variable region of Anticarsia gemmatalis nuclear polyhedrosis virus contains tandemly repeated DNA sequences. Virus Res. 1996;41:123–132. doi: 10.1016/0168-1702(95)01264-8. [DOI] [PubMed] [Google Scholar]
- Geden CJ, Lietze V, Boucias DG. Seasonal prevalence and transmission of salivary gland hyperplasia virus of house flies, Musca domestica L. (Diptera:Muscidae) J. Med. Entomol. 2008;45:42–51. doi: 10.1603/0022-2585(2008)45[42:spatos]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Gouteux JP. Prevalence of enlarged salivary glands in Glossina palpalis, G. pallicera, G. nigrofusca (Diptera: Glossinidae) from the Vavoua area, Ivory Coast. J. Med. Entom. 1987;24:268. doi: 10.1093/jmedent/24.2.268. [DOI] [PubMed] [Google Scholar]
- Grainge I, Jayaram M. The integrase family of recombinase: organization and function of the active site. Mol. Microbiol. 1999;33:449–456. doi: 10.1046/j.1365-2958.1999.01493.x. [DOI] [PubMed] [Google Scholar]
- Gutierrez S, Kikhno I, Ferber ML. Transcription and promoter analysis of pif, an essential but low-expressed baculovirus gene. J. Gen. Virol. 2004;85:331–341. doi: 10.1099/vir.0.19623-0. [DOI] [PubMed] [Google Scholar]
- Guarino LA, Summers MD. Interspersed homologous DNA of Autographa californica nuclear polyhedrosis virus enhances delayed-early gene expression. J. Virol. 1986;60:215–223. doi: 10.1128/jvi.60.1.215-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JM, McIntosh EM, Muscat GE. Structure/function analysis of a dUTPase: catalytic mechanism of a potential chemotherapeutic target. J. Mol. Biol. 1999;288:275–287. doi: 10.1006/jmbi.1999.2680. [DOI] [PubMed] [Google Scholar]
- Huang QQ, Yao SY, Ritzel MW, Paterson AR, Cass CE, Young JD. Cloning and functional expression of a complementary DNA encoding a mammalian nucleoside transport protein. J. Biol. Chem. 1994;269:17757–17760. [PubMed] [Google Scholar]
- Jaenson TGT. Virus-like rods associated with salivary gland hyperplasia in tsetse, Glossina pallidipes. Trans. Roy. Soc. Trop. Med. Hyg. 1978;72:234–238. doi: 10.1016/0035-9203(78)90200-6. [DOI] [PubMed] [Google Scholar]
- Jehle JA, Blissard GW, Bonning BC, Cory JS, Herniou EA, Rohrmann GF, Theilmann DA, Thiem SM, Vlak JM. On the classification and nomenclature of baculoviruses: a proposal for revision. Arch. Virol. 2006;151:1257–1266. doi: 10.1007/s00705-006-0763-6. [DOI] [PubMed] [Google Scholar]
- Johansson K, Ramaswamy S, Ljungcrantz C, Knecht W, Piskur J, Munch-Petersen B, Eriksson S, Eklund H. Structural basis for substrate specificities of cellular deoxyribonucleoside kinases. Nat. Struct. Biol. 2001;8:616–620. doi: 10.1038/89661. [DOI] [PubMed] [Google Scholar]
- Jura WGZO, Odhiambo TR, Otieno LH, Tabu NO. Gonadal lesions in virus-infected male and female tsetse, Glossina pallidipes (Diptera: Glossinidae) J. Invertebr. Pathol. 1988;52:1–8. doi: 10.1016/0022-2011(88)90095-x. [DOI] [PubMed] [Google Scholar]
- Jura WGZO, Otieno LH, Chimtawi M. Ultrastructural evidence for trans-ovum transmission of the DNA virus of tsetse Glossina pallidipes (Diptera: Glossinidae) Current Microb. 1989;18:1–4. [Google Scholar]
- Jura WGZO, Zdarek J, Otieno LH. A simple method for artificial infection of tsetse, Glossina morsitans morsitans larvae with the DNA virus of G. pallidipes. Insect Science Appl. 1993;14:383–387. [Google Scholar]
- Ko E, Okano K, Maeda S. Structural and functional analysis of the Xestia c-nigrum granulovirus matrix metalloproteinase. J. Virol. 2000;74:11240–11246. doi: 10.1128/jvi.74.23.11240-11246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokwaro ED, Nyindo M, Chimtawi M. Ultrastructural changes in salivary glands of tsetse, Glossina morsitans morsitans, infected with virus and rickettsia-like organisms. J. Invertebr. Pathol. 1990;56:337–346. doi: 10.1016/0022-2011(90)90120-u. [DOI] [PubMed] [Google Scholar]
- Kokwaro ED, Otieno LH, Chimtawi M. Salivary glands of the tsetse Glossina pallidipes Austen infected with Trypanosoma brucei and virus particles: an ultrastructural study. Insect Science Appl. 1991;12:661–669. [Google Scholar]
- Kikhno I, Gutierrez S, Croizier L, Croizier G, Ferber ML. Characterization of pif, a gene required for the per os infectivity of Spodoptera littoralis nucleopolyhedrovirus. J. Gen. Virol. 2002;83:3013–3022. doi: 10.1099/0022-1317-83-12-3013. [DOI] [PubMed] [Google Scholar]
- Kuzio J, Jaques R, Faulkner P. Identification of p74, a gene essential for virulence of baculovirus occlusion bodies. Virology. 1989;173:759–763. doi: 10.1016/0042-6822(89)90593-x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:681–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lapointe R, Popham HJ, Straschil U, Goulding D, O’Reilly DR, Olszewski JA. Characterization of two Autographa californica nucleopolyhedrovirus proteins, Ac145 and Ac150, which affect oral infectivity in a host-dependent manner. J. Virol. 2004;78:6438–6448. doi: 10.1128/JVI.78.12.6439-6448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauzon HA, Lucarotti CJ, Krell PJ, Feng Q, Retnakaran A, Arif BM. Sequence and organization of the Neodiprion lecontei nucleopolyhedrovirus genome. J. Virol. 2004;78:7023–7035. doi: 10.1128/JVI.78.13.7023-7035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietze V, Geden CJ, Blackburn P, Boucias DG. Effects of MdSGHV infection on the reproductive behavior of the house fly, Musca domestica. Appl. Environ. Microbiol. 2007;73:6811–6818. doi: 10.1128/AEM.02694-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link A, Eng JJ, Schieltz DM, Carmack E, Mize EJ, Morris DR, Garvik BM, Yates JR. Direct analysis of protein complexes using mass spectrometry. Nature Biotechnology. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. MacClade 4:0 analysis of phylogeny and character evolution, version 4.06 . Sunderland, Massachusetts, USA: Sinauer; 2000. [Google Scholar]
- Maguire T, Harrison P, Hyink O, Kalmakoff J, Ward VK. The inhibitor of apoptosis of Epiphyas postvittana nucleopolyedrovirus. J. Gen. Virol. 2000;81:2803–2811. doi: 10.1099/0022-1317-81-11-2803. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XM, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GP, Benkovic SJ. Stretching exercises - flexibility in dihydrofolate reductase catalysis. Chem. Biol. 1998;5:R105–R113. doi: 10.1016/s1074-5521(98)90616-0. [DOI] [PubMed] [Google Scholar]
- Miller LK. An exegesis of IAPs: salvation and surprises from BIR motifs. Trends Cell. Biol. 1999;9:323–328. doi: 10.1016/s0962-8924(99)01609-8. [DOI] [PubMed] [Google Scholar]
- Minter-Goedbloed E, Minter DM. Salivary gland hyperplasia and trypanosome infection of Glossina in two areas of Kenya. Trans. Royal Soc. Trop. Med. Hyg. 1989;83:640–641. doi: 10.1016/0035-9203(89)90380-5. [DOI] [PubMed] [Google Scholar]
- Odindo MO. Incidence of salivary gland hypertrophy in field populations of the tsetse Glossina pallidipes on the south Kenyan coast. Insect Science Appl. 1982;3:59–64. [Google Scholar]
- Odindo MO, Sabwa DM, Amutalla PA, Otieno WA. Preliminary tests on the transmission of virus-like particles to the tsetse Glossina pallidipes. Insect Science Appl. 1981;2:219–221. [Google Scholar]
- Odindo MO, Payne CC, Crook NE, Jarrett P. Properties of a novel DNA virus from the tsetse fly, Glossina pallidipes. J. Gen. Virol. 1986;67:527–536. doi: 10.1099/0022-1317-67-3-527. [DOI] [PubMed] [Google Scholar]
- Ohkawa T, Washburn JO, Sitapara R, Sid E, Volkman LE. Specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to midgut cells of Heliothis virescens larvae is mediated by products of pif genes Ac119 and Ac022 but not by Ac115. J. Virol. 2005;79:15258–152764. doi: 10.1128/JVI.79.24.15258-15264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otieno LH, Kokwaro ED, Chimtawi M, Onyango P. Prevalence of enlarged salivary glands in wild populations of Glossina pallidipes in Kenya, with a note on the ultrastructure of the affected organ. J. Invertebr. Pathol. 1980;36:113–118. [Google Scholar]
- Patel S, Latterich M. The AAA team: related ATPases with diverse functions. Trends Cell. Biol. 1988;8:65–71. [PubMed] [Google Scholar]
- Payne CC. The isolation and characterization of a virus from Oryctes rhinoceros. J. Gen Virol. 1974;25:105–116. doi: 10.1099/0022-1317-25-1-105. [DOI] [PubMed] [Google Scholar]
- Pearson M, Bjornson R, pearson G, Rohrmann G. The Autographa californica baculovirus genome: evidence for multiple replication origins. Science. 1992;257:1382–1384. doi: 10.1126/science.1529337. [DOI] [PubMed] [Google Scholar]
- Perera O, Green TB, Stevens SM, White S, Becnel. J. Proteins associated with the Culex nigripalpus nucleopolyhedrovirus occluded virions. J. Virol. 2007;81:4585–4590. doi: 10.1128/JVI.02391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perryman SM, Rossana C, Deng TL, Vanin EF, Johnson LF. Sequence of a cDNA for mouse thymidylate synthase reveals striking similarity with the prokaryotic enzyme. Mol. Biol. Evol. 1986;3:313–321. doi: 10.1093/oxfordjournals.molbev.a040400. [DOI] [PubMed] [Google Scholar]
- Pijlman GP, Pruijsser AJ, Vlak JM. Identification of pif-2, a third conserved baculovirus gene required for per os infection of insects. J. Gen. Virol. 2003;84:2041–2049. doi: 10.1099/vir.0.19133-0. [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang RC, Jura WGZO, Otieno LH, Mwangi RW. The effects of a DNA virus infection on the reproductive potential of female tsetse flies, Glossina morsitans centralis and Glossina morsitans morsitans (Diptera: Glossinidae) Mem. Inst. Oswaldo Cruz. 1998;93:861–864. doi: 10.1590/s0074-02761998000600030. [DOI] [PubMed] [Google Scholar]
- Sang RC, Jura WGZO, Otieno LH, Mwangi RW, Ogaja P. The effects of a tsetse DNA virus infection on the functions of the male accessory reproductive gland in the host fly Glossina morsitans centralis (Diptera; Glossinidae) Curr. Microbiol. 1999;38:349–354. doi: 10.1007/pl00006815. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain termination inhibitors. Proc. Natl. Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Borden KL, Boddy MN, Freemont PS. Does this have a familiar RING? Trends Biochem. Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- Shaw MK, Moloo SK. Virus-like particles in Rickettsia within the midgut epithelial cells of Glossina morsitans centralis and Glossina brevipalpis. J. Invertebr. Path. 1993;61:162–166. [Google Scholar]
- Slack JM, Arif BM. The baculoviruses occlusion-derived virus: virion, structure and function. Adv. Virus Res. 2007;69:99–165. doi: 10.1016/S0065-3527(06)69003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland Massachusetts, USA: Sinauer Associates; 2003. [Google Scholar]
- Tellam RL, Wijffels G, Willadsen P. Peritrophic matrix proteins. Insect. Biochem. Mol. Biol. 1999;29:87–101. doi: 10.1016/s0965-1748(98)00123-4. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Coulson EJ, Vaux DL. Conservation of baculovirus inhibitor of apoptosis repeat proteins (BIRPs) in viruses, nematodes, vertebrates and yeasts. Trends Biochem. Sci. 1998;23:159–162. doi: 10.1016/s0968-0004(98)01198-0. [DOI] [PubMed] [Google Scholar]
- Walker JE, Runswick MJ. The mitochondrial transport protein superfamily. J. Bioenerg. Biomembr. 1993;25:435–446. doi: 10.1007/BF01108401. [DOI] [PubMed] [Google Scholar]
- Wang Y, Burand JP, Jehle JA. Nudivirus genomics: diversity and classification. Virologica Sinica. 2007a;22:128–136. [Google Scholar]
- Wang Y, Kleespies RG, Huger AM, Jehle JA. The genome of Gryllus bimaculatus nudivirus indicates an ancient diversification of baculovirus-related nonoccluded nudiviruses of insects. J. Virol. 2007b;81:5395–5406. doi: 10.1128/JVI.02781-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, van Oers MM, Crawford AM, Vlak JM, Jehle JA. Genomic analysis of Oryctes rhinoceros virus reveals genetic relatedness to Heliothis zea virus 1. Arch. Virol. 2007c;152:519–531. doi: 10.1007/s00705-006-0872-2. [DOI] [PubMed] [Google Scholar]
- Whitnall AMB. The trypanosome infections of Glossina pallidipes in Umfolosi game reserve, Zuhuland. Onderstepoot J. Anim. Ind. 1934;11:7–21. [Google Scholar]
- Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Seller H, Meier P. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nature Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Ohkawa T, Washburn JO, Volkman LE. Effects of Ac150 on virulence and pathogenesis of Autographa californica multiple nucleopolyhedrovirus in noctuid hosts. J. Gen. Virol. 2005;86:1619–1627. doi: 10.1099/vir.0.80930-0. [DOI] [PubMed] [Google Scholar]