SUMMARY
MicroRNAs (miRNAs) regulate the expression of target mRNAs in plants and animals [1]. Plant miRNA targets have been predicted based on their extensive and often conserved complementarity to the miRNAs [2-4], as well as from miRNA over-expression experiments [5]; many of these target predictions have been confirmed by isolating the products of miRNA-directed cleavage. Here, we present a transcriptome-wide experimental method, called “degradome sequencing,” to directly detect cleaved miRNA targets without relying on predictions or over-expression. The 5' ends of poly-adenylated, uncapped mRNAs from Arabidopsis were directly sampled resulting in an empirical snapshot of the degradome. miRNA-mediated cleavage products were easily discerned from an extensive background of degraded mRNAs, which collectively covered the majority of the annotated transcriptome. Many previously known Arabidopsis miRNA targets were confirmed and several novel targets were also discovered. Quantification of cleavage fragments revealed that those derived from TAS transcripts, which are unusual in their production of abundant secondary siRNAs, accumulated to very high levels. A subset of secondary siRNAs are also known to direct cleavage of targets in trans [6]; degradome sequencing revealed many cleaved targets of these trans-acting siRNAs (ta-siRNAs). This empirical method is broadly applicable to discover and quantify cleaved targets of small RNAs without a priori predictions.
RESULTS AND DISCUSSION
The Arabidopsis degradome generally reflects transcript abundance
Although plant miRNAs can interact with target transcripts without directing their hydrolysis [7, 8], the most common outcome of pairing to known targets is miRNA-directed cleavage at the tenth nucleotide of complementarity relative to the guiding miRNA [9, 10]. The resulting 3' target fragments have a 5' monophosphate and a 3' polyA tail and can be recovered by RNA ligase-mediated 5' rapid amplification of cDNA ends (RLM 5'-RACE [9, 10]). These experiments require an a priori miRNA target prediction and are limited to testing one candidate target at a time. In an alternative approach, a primer designed to match presumed miRNA complementary sites is used to identify targets without predictions, but this method is also limited by low throughput [11].
We modified the RLM 5'-RACE method to globally sample RNAs with a 5' monophosphate and a 3' polyA tail. An RNA adapter was engineered to contain a 5' MmeI restriction site and ligated directly to polyA+ RNA; this selected against capped, full-length mRNAs, which lacked the free 5' monophosphate required for ligation. After reverse transcription and second-strand synthesis, MmeI digestion left a 20−21nt tag attached to the 5' adapter to which a 3' dsDNA adapter was attached. Amplified libraries of these “degradome” 5' tags were then sequenced. In most cases, the 20−21nt signatures were sufficient to unambiguously identify the transcripts of origin, with the junction between the signature and the 5' adapter indicating the 5' terminus of the original RNA.
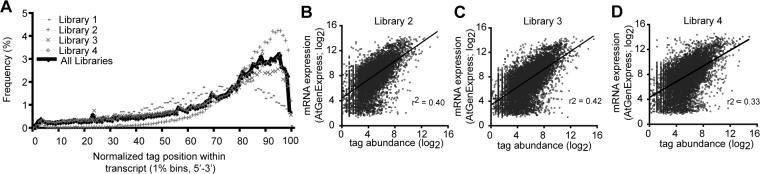
Four distinct libraries were sequenced from either inflorescence or seedling specimens of wild-type Arabidopsis (Supplemental Table 1). In total, 835,902 unique 20−21nt signatures (represented by 5,092,568 reads) were found to exactly match the sense strand of one or more annotated transcripts (Supplemental Table 1). The majority (∼73%) of annotated Arabidopsis transcripts had at least one unambiguously matched tag. The libraries of degradome tags were biased towards the 3' ends of annotated transcripts (Figure 1A). This bias persisted in libraries where random hexamers replaced oligo(dT) as the reverse-transcription primer. Thus, to a large extent, the global 3' bias likely reflected the distribution of in vivo polyadenylated degradation intermediates. The repeat-normalized total abundance of all degradome tags for each transcript was summed. Comparison of these degradome abundances to the AtGenExpress-derived expression values [12] for the relevant tissue samples revealed a positive correlation between degradome tag density and steady-state levels of mRNAs (Figures 1B-D). Thus, degradome tag abundance generally reflected the abundance of the respective mRNAs.
Figure 1. Degradome tags were 3' biased and correlated with transcript abundance.
(A) Histogram displaying the 5' positions of degradome tags from the indicated libraries relative to normalized transcript position. Tags were counted in one percent bins. For clarity, one tag that was extremely abundant in libraries three and four was omitted.
(B-D) Relationships between microarray-derived estimates of transcript abundance (y-axis) and degradome tag abundance (x-axis) for the indicated libraries. RMA-normalized array values were from [12] and were matched according to the tissue source (Inflorescence, ATGE_29, B and C; Seedlings, ATGE_96, D). Transcripts with degradome tag abundances equal to or less than one were omitted. Lines represent best-fit linear regressions; r2 values represent correlation coefficients.
Discovery of miRNA targets
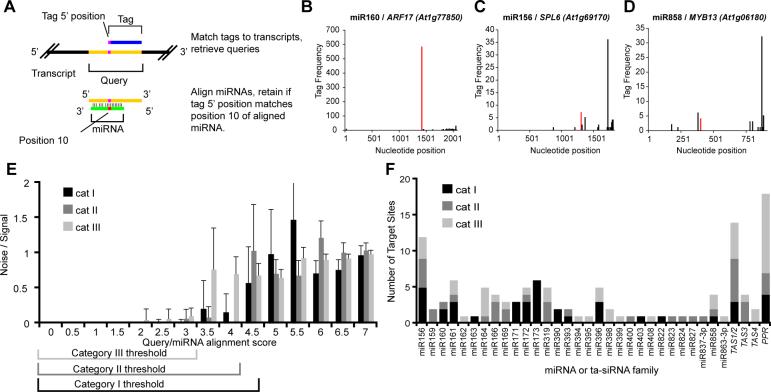
The 5' ends of tags derived from miRNA cleavage fragments would precisely correspond to the tenth nucleotide of miRNA complementary sites. To find such tags, we first extracted the potential complementary sites by retrieving query sequences extended 15nts upstream from the tag 5' end (Figure 2A). These sequences were aligned with confidently annotated Arabidopsis miRNAs and were scored according to previously described methods for Arabidopsis miRNA target predictions [2, 13]. All alignments with mismatch scores of seven or lower were initially retained. Alignments where the degradome tag 5' end was precisely opposite the tenth nucleotide of the miRNA constituted potential evidence of miRNA-mediated cleavage (Figure 2A). When such alignments were found, neighboring tags whose 5' ends aligned with the ninth or eleventh positions were also scored as evidence for miRNA-mediated cleavage; this accounts for the occasional positional heterogeneity seen for mature miRNAs [13] and their cleavage products [3, 14].
Figure 2. Experimental identification of cleaved miRNA targets without predictions.
(A) Schematic of methodology to find evidence of miRNA-mediated cleavage from degradome tags.
(B) Density of 5' position of degradome tags corresponding to ARF17, a category I target. Tags aligned with the ninth through eleventh nucleotides of a miR160 complementary site were combined and shown in red.
(C) As in B for SPL6, a category II target of miR156.
(D) As in B for MYB13, a category III target of miR858.
(E) Histogram displaying mean ratio of targets found using 30 cohorts of randomly permuted miRNAs to the number found using the annotated mature miRNA query data set at different alignment scores. Error bars represent one standard deviation. Alignment score thresholds are indicated for category I, II, and III targets.
(F) Summary of 121 cleaved miRNA and/or ta-siRNA target sites found using degradome analyses. Details in Supplementary Table 2.
Examination of potential targets revealed a spectrum of evidence for target cleavage. For many targets, degradome tags indicative of miRNA-mediated cleavage were the most abundant tags matching the transcript (Figure 2B); these were classified as category I. For others, tags diagnostic of cleavage were not the most abundant tags matching the transcript, but still formed a clear peak at the complementary site – these were classified as category II (Figure 2C). miRNA-aligned tags which formed minor peaks were deemed category III targets (Figure 2D). As a control, alignments were also performed against 30 cohorts of randomized miRNA sequences; the resulting signal-to-noise ratios were used to define appropriate cutoffs for confident target identification (Figure 2E).
Many miRNA hairpins produce positional variants of the annotated mature miRNA, abundant miRNA* sequences, and in some cases multiple miRNA/miRNA* duplexes from the same hairpin [13, 15-17]. Some of these variants might also direct target cleavage. To find cleavage targets that may have been missed when considering only annotated mature miRNA sequences, we expanded the search to include all experimentally identified small RNAs between 20 and 22nts in length [13, 18] matching one or more Arabidopsis miRNA hairpin. Randomizations using this expanded set of queries indicated a need for more stringent cutoffs than those used for annotated mature miRNAs only (Supplemental Figure 1A). Seven additional targets of three miRNA families were found; because one target had two cleavage sites these represented eight additional distinct cleavage sites (Supplemental Table 2). This analysis was critical to identify the PPR targets of miR161 and the previously predicted DC1 target of miR822. Altogether, these two analyses confirmed 57 out of 103 previously validated miRNA targets, 14 which were previously predicted but had not been validated, and six for which there was no previous experimental evidence or predictions (Figure 2F, Supplemental Table 2).
The identified miRNA cleavage targets were biased towards conserved miRNA families: 18 of the 29 miRNA families for which one or more target was found have been annotated in at least one other plant species besides Arabidopsis [19]. This is consistent with the general correlation between miRNA conservation, expression level, and number of verifiable targets [13, 14]. Newly confirmed or discovered targets were mostly related to already known targets of the same miRNA. For instance, SPL5, −6, −9, −13, and −15 were confirmed as miR156 targets and MYB13, −20, and −111 were confirmed as miR858 targets (Supplemental Table 2). Seven MYB targets of miR159 have been described [20], but regulation of only two, MYB33 and MYB65, accounts for the phenotypes observed in mir159ab loss of function mutants [21]. That only MYB33 and MYB65 were confirmed in our experiments suggests that the degradome technique might have a propensity to identify targets of higher in vivo relevance.
Discovery of ta-siRNA targets
Known Arabidopsis secondary siRNA-producing loci include four families of TAS genes, a set of PPR genes targeted by multiple miRNAs and ta-siRNAs, as well as the cleaved targets of miR393 and miR168 [7, 13, 22, 23]. A subset of these secondary siRNAs direct the cleavage of targets distinct from their precursors, and thus are trans-acting siRNAs (ta-siRNAs; [6]). Cleaved ta-siRNA targets were identified by using all secondary siRNAs between 20 and 22nts in length whose expression has been documented by sequencing [13, 18] as queries to find degradome tags indicative of cleavage. Alignments indicative of cis interactions (where an siRNA directs cleavage of its precursor transcript) were excluded. After applying empirically determined thresholds (Supplemental Figure 1B), 38 ta-siRNA directed cleavage events were confidently identified within 24 distinct transcripts (Figure 2F; Supplemental Table 2). These 24 transcripts included most of those previously confirmed by gene-specific 5'-RACE (11 out of 14). The dense network of PPR-derived ta-siRNAs directing cleavage of other closely related PPR genes was particularly striking in our analysis and supports the notion of a self-reinforcing cascade of PPR repression initiated by miR173, miR161, and TAS1/2 family ta-siRNAs ([22, 24]; Supplemental Table 2).
TAS cleavage products accumulate to high levels
Degradome tags corresponding to cleaved TAS transcripts were among the most abundant (Figure 3). TAS genes are unusual miRNA targets in that miRNA-mediated cleavage serves to stimulate the production of abundant secondary siRNAs from one of the two resulting fragments [2, 25]. High accumulation levels of 3' TAS cleavage products were observed regardless of whether the 3' product directly contributes to downstream ta-siRNA formation (TAS1a-c and TAS2), or not (TAS3a and TAS3c; [2, 25]). High accumulation of TAS cleavage products could reflect initially high TAS transcript levels and/or a specific stabilization after miRNA-mediated processing. The observation that SGS3, a factor required for post-transcriptional gene silencing and ta-siRNA accumulation [26-28], is required to stabilize TAS1a and TAS2 cleavage products [25] argues in favor of selective stabilization. One recent model for secondary siRNA biogenesis from the TAS3a locus invokes specific stabilization of the 5' cleavage product, which serves as a template for siRNA production [29]. How this might connect to the high abundance of the 3' TAS3a cleavage product (Figure 3), which does not serve as a template for siRNA production, is unclear. Perhaps high accumulation of a miRNA cleavage product is a hallmark of the secondary siRNA pathway. In cases like TAS1a-c and TAS2, for which other factors known to promote secondary siRNA formation such as multiple miRNA complementary sites [7, 22] or AGO7 dependency [29-32] are not evident, high cleavage product accumulation may promote entrance to the secondary siRNA pathway.
Figure 3.

High accumulation levels of TAS cleavage products. Histogram displays abundance of degradome tags corresponding to miRNA-mediated cleavage sites; red indicates TAS genes, as labeled.
Confirmation of novel targets
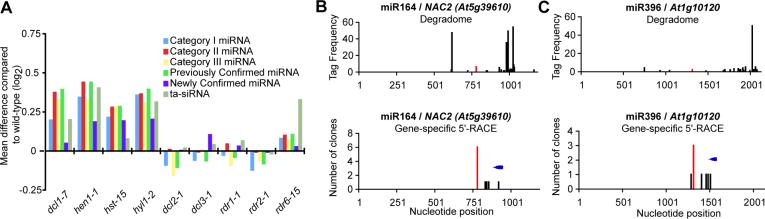
The accumulation of many Arabidopsis miRNA targets increases in mutants with defects in miRNA biogenesis, but remains unchanged in siRNA mutants [2]. Similarly, the accumulation of targets identified through degradome sequencing generally increased in mutants affecting miRNAs (dcl1−7, hen1−1, hst-15, and hyl1−2) but not in siRNA mutants (dcl2−1, dcl3−1, rdr1−1, and rdr6−15; Figure 4A). This trend was evident for all three categories of degradome tag-identified miRNA targets, as well as when comparing previously confirmed to newly confirmed miRNA targets. As expected, ta-siRNA target levels were generally increased in both miRNA mutants as well as the rdr6−15 mutant (Figure 4A).
Figure 4. Independent confirmation of cleavage targets identified in the degradome.
(A) Accumulation of confirmed miRNA and ta-siRNA targets, separated by category or by previous experimental knowledge, in various miRNA (dcl1−7, hen1−1, hst-15, hyl1−2) and siRNA (dcl2−1, dcl3−1, rdr1−1, rdr2−1, rdr6−15) mutants (microarray data from [2]).
(B) Top: Density of 5' position of degradome tags corresponding to the NAC2 transcript. Tags aligned with the ninth through eleventh nucleotides of a miR164 complementary site were combined and shown in red. Bottom: Density of clones obtained via gene-specific 5'-RACE. Blue arrow indicates position of gene-specific oligo sequences.
(C) As in B for At1g10120 and miR396.
We employed signal-to-noise analyses to determine empirical cutoffs for confident target identification using degradome data (Figure 2E, Supplemental Figure 1). Potential targets that exceeded these cutoffs were not all necessarily false positives – in these cases, however, additional data are required to confidently demonstrate targeting. Gene-specific 5'-RACE was used to individually test three cases involving potential targets that exceeded the degradome confidence thresholds: One which had previously been predicted but not confirmed (miR164/NAC2 [3]) and two novel, previously unpredicted miRNA/target interactions (miR390/AT3G24660, miR396/AT1G10120). In two of three cases, gene-specific 5'-RACE supported the hypothesis developed from degradome data (Figures 4B-C). Thus, potential targets that cannot be confidently confirmed using degradome data alone can still be validated by secondary, focused experiments. Further supporting this conclusion, we found another three transcripts with at least one tag matching a previously validated or predicted miRNA cleavage site in contexts that exceeded the confidence thresholds for degradome-based target identification (Supplemental Table 2). Taken together with the ta-siRNA targets, our sampling of the Arabidopsis degradome discerned 121 miRNA and ta-siRNA cleavage sites within 99 distinct transcripts (Figure 2F; Supplemental Table 2).
Conclusions
Direct sequencing of degradome tags derived from the 5' ends of uncapped mRNAs delivered an empirical overview of cleaved miRNA and ta-siRNA targets without computational predictions or overexpression. This methodology is likely to be broadly applicable to small RNA target discovery and quantification for other organisms in which target cleavage is a frequent mode of repression. Degradome sequencing also provides information on the relative abundance of cleaved targets. The observation that Arabidopsis TAS cleavage products accumulated to high levels implies that stabilization and/or high-level expression of ta-siRNA precursors might play a role in promoting ta-siRNA biogenesis.
Our initial analysis of the degradome data focused on miRNA and ta-siRNA targets. However, most of the tags represented degradation intermediates of messages that were not targets of these small RNAs. We anticipate that analyses of these tags with a focus on other degradation pathways might provide general insights into mRNA turnover in plants which can be extended to other species, including those that do not undergo small-RNA-dependent mRNA cleavage. For example, degradome analysis of mutants in RNA metabolism, such as the XRN family of 5'-to-3' exonucleases [33, 34] are likely to yield rich insights into RNA turnover in a wide variety of species. Of particular interest is the observation that the 3' bias of degradome tags persisted even when random hexamers were used to prime reverse transcription. This could have resulted from in vitro degradation prior to polyA enrichment. However, the most typical sources of such artifacts (spontaneous and RNAseA-catalyzed hydrolysis) leave fragments with 5' hydroxyls which are excluded from our 5'-monophosphate dependent protocol. Thus, while more experiments are clearly needed, our data hint at a role for endonucleases and/or frequent 3' stalling of 5'−3' exonucleases during mRNA turnover in Arabidopsis.
EXPERIMENTAL PROCEDURES
Degradome library construction and gene-specific 5'-RACE
Degradome libraries were constructed by ligation of polyA-enriched RNA samples to a custom RNA adapter containing a 3' MmeI site, followed by reverse transcription (RT), second-strand synthesis, MmeI digestion, ligation of a 3' dsDNA adapter, gel-purification, and PCR amplification. Different RT and second-strand synthesis were used during development of the protocol, as noted (Supplemental Experimental Procedures). Amplified degradome tag libraries were then sequenced using a 454/Roche GS20 genome analyzer or a Solexa/Illumina genome analyzer. Gene-specific RLM 5'-RACE was performed as described [15]; oligo sequences are listed in Supplemental Experimental Procedures.
Data analysis
Raw reads were processed to remove 5' and 3' adapter sequences; tags with sizes of 20 or 21nts (the sizes expected from MmeI cleavage) were retained. Tags that did not correspond to structural RNAs (rRNA, tRNA, snRNA, snoRNA) were then mapped to the sense polarity of annotated Arabidopsis transcripts (TAIR 7 release); the abundance of tags which matched more than one transcript was repeat-normalized. A 35−36nt extended signature was derived from each transcript-matched tag by adding 15nts of upstream sequence; these were then aligned to a set of annotated mature Arabidopsis miRNAs [35], an expanded set of expressed 20−22mers derived from miRNA hairpins, or a set of expressed 20−22mers derived from known producers of secondary siRNAs (Supplemental Experimental Procedures). Alignments where the 5' tag position aligned with the tenth nucleotide of a miRNA were retained and scored as in [2]. Control alignments were also performed with cohorts of permuted miRNA queries controlled for di- and tri-nucleotide composition [36]. Targets were categorized as I, II, or III as described in the Supplemental Experimental Procedures. Processed degradome tags have been deposited with NCBI GEO (GSE11007).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Chanseok Shin and Sumeet Gupta for assistance with Solexa/Illumina sequencing. This work was support by a grant from the NSF (0718051) to MJA and a grant from the NIH (GM067031) to DPB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 4.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 5.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 6.Vaucheret H. MicroRNA-dependent trans-acting siRNA production. Sci STKE. 2005;2005:pe43. doi: 10.1126/stke.3002005pe43. [DOI] [PubMed] [Google Scholar]
- 7.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 8.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 9.Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 10.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 11.Axtell MJ, Bartel DP. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17:1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 13.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talmor-Neiman M, Stav R, Frank W, Voss B, Arazi T. Novel micro-RNAs and intermediates of micro-RNA biogenesis from moss. Plant J. 2006;47:25–37. doi: 10.1111/j.1365-313X.2006.02768.x. [DOI] [PubMed] [Google Scholar]
- 18.Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Axtell MJ, Bowman J. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008 doi: 10.1016/j.tplants.2008.03.009. In Press. [DOI] [PubMed] [Google Scholar]
- 20.Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, Warthmann N, Allen E, Dezulian T, Huson D, et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell. 2007;13:115–125. doi: 10.1016/j.devcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Allen RS, Li J, Stahle MI, Dubroue A, Gubler F, Millar AA. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16371–16376. doi: 10.1073/pnas.0707653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- 24.Chen HM, Li YH, Wu SH. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3318–3323. doi: 10.1073/pnas.0611119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 27.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 29.Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 30.Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 31.Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 32.Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gy I, Gasciolli V, Lauressergues D, Morel JB, Gombert J, Proux F, Proux C, Vaucheret H, Mallory AC. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell. 2007;19:3451–3461. doi: 10.1105/tpc.107.055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell. 2004;15:173–183. doi: 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.