Abstract
An oligodeoxynucleotide (ODN) containing a 2′-aminoalkylmercaptouridine was ligated chemically with another ODN containing a 2′-carboxyalkylmercaptouridine to produce a 2′,2′-amide linked conjugate. Templation by a DNA hairpin resulted in ligation yields of > 95%. At room temperature, similar yields were obtained for amide ligations between pendent nucleotides located adjacent to each other or separated by three nucleotides. However, at 4 °C, ligation was vastly favored for the case where the pendent amine and carboxylate were attached to adjacent residues (i.e., at the 3′- and 5′-termini of the two ODNs). No reaction was observed to occur either at 4 °C or at room temperature between pendent nucleotides separated by 9 nucleotides. Denaturing gel electrophoresis, MALDI-TOF MS and full nuclease digestion coupled with LC-MS were used to characterize the reaction products.
Keywords: Nucleic acids, Templated chemical ligation, Reaction regioselectivity, Thermal control, Positional Synthesis
1. Introduction
Non-enzymatic oligodeoxynucleotide (ODN) ligation with complementary DNA as a template offers potential applications in nucleic acid-based biomedicine and nanomaterials construction.1, 2, 3, 4 Since natural DNA is tethered through the 3′- and 5′-positions of ribose by phosphodiester bonds, most reported template-directed chemical approaches ligate two ODNs between the 3′- and 5′-positions of nucleosides, either by a native phosphodiester bond 5 or an unnatural linkage.6 Templated ligation through nucleobases has also been achieved by photochemical reaction.7 Sawai and coworkers reported ligation reactions between the 2′- and 5′-positions of oligonucleotides on either 2′,5′- or 3′,5′-linked complementary templates. 8 A rare case is the templated ligation of TNA (threofuranosyl-oligonucleotides);9 owing to the unique structure of TNA, ligation involves the formation of 2′- to 3′-linkages.
We have reported previously the attachment of alkylcarboxyl and alkylamino groups to 2′-mercapto-ribonucleotides.10 The side chains were condensed to form short segments of a nylon-like polymer tethered to the nucleic acid backbone only through the 2′-position of individual nucleotide units (nylon nucleic acid). Success with these intramolecular reactions has encouraged us to explore the possibility of intermolecular ligation of ODNs through 2′-pendent amines and carboxylates. Linkage via available 2′-positions would leave the 3′- and 5′-positions free for further ligation or modification. The resulting ODN conjugates could then be incorporated into more complex (e.g., multiply-connected or topologically intricate) structures. Another benefit of such a 2′,2′-linkage technology would be the addition of a new ligation strategy for the assembly of DNA nanostructures, in addition to enzymatic or chemical phosphodiester ligation.11 Thus, the goal of this study was to investigate the binding fidelity of two ODNs terminating in 2′-modified nucleotides to their complementary DNA template and the consequent impact on ligation efficiency and selectivity through the 2′-ribose positions of the two ODNs.
Here, we report the results of a study of DNA template-directed ODN 2′,2′-ligation via amide formation. Optimized ligation conditions afforded > 95% yield. The ligation of ODNs with modified nucleotides incorporated in different positions of the sequence was examined so as to study the regioselectivity of the templated coupling.
2. Results and Discussion
2.1. Template-directed ligation
Our notation is to indicate a pendent alkylamine by a lowercase ‘n’, and a pendent carboxylic acid by a lowercase ‘c’ (Fig. 1B). The syntheses of modified uridine phosphoramidite monomers, Un and Uc, were as described earlier.10a All ODN sequences are listed in Table 1. ODN 4 was synthesized on the Universal Support II Resin (Glen Research, Sterling, VA) by an automated oligonucleotide synthesizer to obtain the strand with modified Un anchored at the 3′-end. All other ODNs were synthesized following standard solid phase oligonucleotide synthesis procedures with modifications that we have described elsehere.10a After routine purification by HPLC or by gel electrophoresis, both the ligation precursor strands and the complementary hairpin were desalted by reverse-phase HPLC and the modified ODNs were characterized by MALDI-TOF MS (Table 1).
Figure 1.
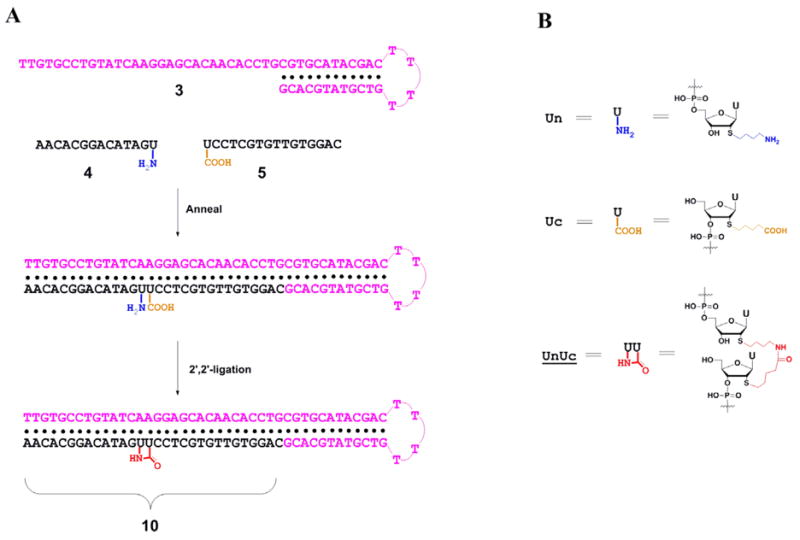
Schematic depiction of the template-mediated 2′,2′-ligation of two ODNs. (A) Two end-modified ODNs are aligned together by forming duplex with a DNA hairpin template and then proximal amine and carboxylate are connected using chemical ligation. (B) Symbols and chemical structures representing ligation units.
Table 1.
Oligonucleotides used in this study and their analysis by MALDI-TOF MS. 1 is the unmodified DNA control strand for strand 4; 2 is the control strand for 5 ~ 8; 3 is the hairpin template; 9 is the control for nuclease digestion, UnUc represents two uridine residues linked by both 3′,5′-phosphate diester linkage and 2′,2′-amide linkage; 10 is ligated product from 4 and 5, UnUc represents two uridine residuals linked by only 2′,2′-amide linkage. (For structures UnUc and UnUc of see Fig. 4)
| Mass
|
|||
|---|---|---|---|
| ODNs | Sequence | Calculated | Found |
| 1 | 5′-AACACGGACATAGT-3′ a | -- | -- |
| 2 | 5′-TCCTCGTGTTGTGGAC-3′ a | -- | -- |
| 3 | 5′-GCACGTATGCTGTTTTCAGCATACGTGCG--TCCACAACACGAGGAACTATGTCCGTGTT-3′ a | -- | -- |
| 4 | 5′-AACACGGACATAGUn-3′ | 4371.0 | 4370.9 |
| 5 | 5′-UcCCTCGTGTTGTGGAC-3′ | 4998.4 | 4996.2 |
| 6 | 5′-TCCUcCGTGTTGTGGAC-3′ | 4998.4 | 4995.4 |
| 7 | 5′-TCCTCGUcGTTGTGGAC-3′ | 4998.4 | 4997.4 |
| 8 | 5′-TCCTCGTGTUcGTGGAC-3′ | 4998.4 | 4997.9 |
| 9 | 5′-GCATAGTTUnUcTTGTCTAC-3′ b | 5660.9 | 5661.3 |
| 10 | 5′-AACACGGACATAGUnUcCCTCGTGTTGTGGAC-3′ | 9348.4 | 9347.7 |
Strands 1, 2 and 3 were obtained from Integrated DNA Technologies (Coralville, IA) and purified by denaturing gel electrophoresis.
For synthesis and thermal denaturing study of strand 9 see reference.13
For ligation experiments, ODNs 4, 5 and hairpin template 3 were first annealed in 1 M NaCl, 0.1 M MOPS buffer at a 1 : 1 : 1 ratio before applying the coupling reagent, 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMTMM, Fig. 1A). After incubation at room temperature, the reaction was quenched by adding 1 M ammonium acetate solution. After desalting and concentration, the final solutions were subjected to denaturing gel electrophoresis analysis and the ligation yield was quantified by measuring the band intensities using Kodak Image Station Gel Logic 200.
The ligations via 2′,2′-amide formation were tested at 0.06, 0.2, 1 and 4 _M duplex concentrations. 12, 6c, 7a Analytical gel electrophoresis showed almost identical ligation yields across this concentration range (85%~90%) after 24 h of reaction time (Supporting Information S1). The observed lack of dependence on concentration indicates that the reaction occurred at high effective molarity within the pre-annealed duplex. The template hairpin brings these reaction partners into close proximity, which also increases the reaction rate. These observations are consistent with other ODN-templated ligation reports.6e, 6f Eventually it was found that extending the reaction time to 48 h increased the yield above 95% (Fig. 2).
Figure 2.
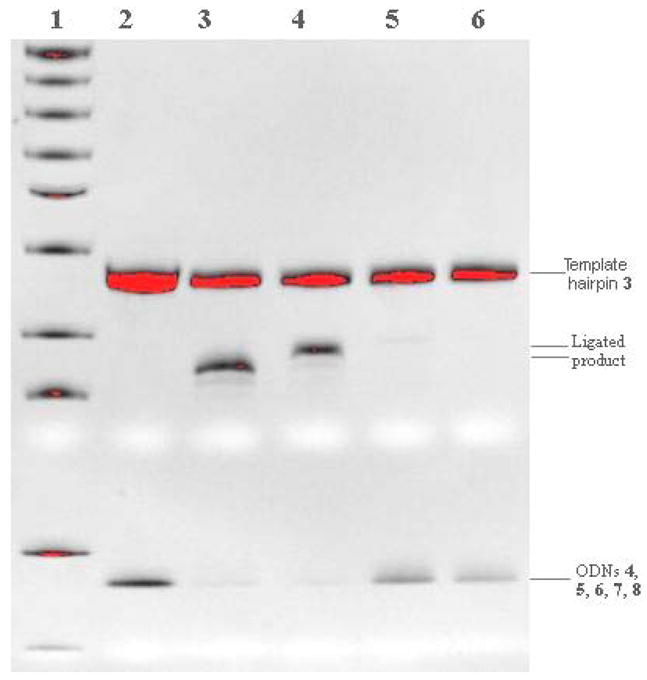
Denaturing gel electrophoresis assay of regioselectivity of templated ligation at room temperature (48 h, 200 nM duplex concentration). Lane 1: 10 base pairs marker; Lane 2: ODNs 5, 4 and template 3 in absence of coupling reagent DMTMM as references; Lane 3: ODNs 5, 4 and template 3; Lane 4: ODNs 6, 4 and template 3; Lane 5: ODNs 7, 4 and template 3; Lane 6: ODNs 8, 4 and template 3.
2.2. Characterization of the reaction products
The 2′,2′-ligated product 10 was purified by denaturing gel electrophoresis and characterized by MALDI-TOF MS [M - H calcd, 9348.4; Found, 9347.7] (Fig 3). There is a possibility that side reactions may result in undesired ligated products with the same mass as the desired product, such as cross-linking the pendent 2′-carboxyl group of ODN 5 with nucleobases in 4 or pendent 2′-amino group of 4 with the phosphate backbone of 5. To verify the desired ligation through 2′-pendent amino and carboxyl groups, two control experiments were performed. Ligation of unmodified DNA 1 (control strand corresponding to 4) with ODN 5 and ligation of DNA strand 2 (control strand corresponding to 5) with ODN 4 under the same conditions. In both experiments, 1 : 5 : 3 and 2 : 4 : 3, no ligated product was detected (Supporting Information S2), excluding the possibility of undesired cross coupling.
Figure 3.
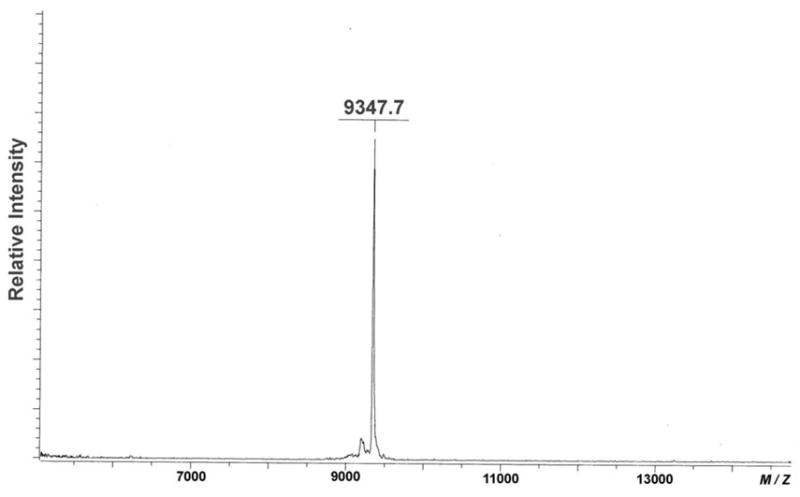
MALDI-TOF MS of the ligated product 10 from templated ligation of ODN 4 and 5.
Analysis of nucleoside fragments generated from nuclease full digestion of ODNs can provide precise information about base identity, composition and structure of nucleic acids. Therefore, ligation product 10 was digested using snake venom phosphodiesterase (SVP) and bacterial alkaline phosphatase (BAP), and the digests were analyzed by LCMS. In Figure 4, U2 designates the 2′,2′-amide linked uridine dimer from the digestion of strand 10. Compound 9,13 containing a modified uridine dimer joined by both 3′,5′-phosphodiester and 2′,2′-alkylamide linkages, should give the same digested components as 10, i.e. dC, dA, dG, T and U2. Thus, 9 served as a control for enzyme digestion analysis of 10. Both digests show five peaks in UV chromatograms (Fig. 4A, 4C). The peaks are identified as dC, dA, dG, T and U2 in eluent order.14 Analysis of 9 and 10 gave identical retention time, mass, and isotopic pattern for each nucleoside. The mass data agreed with theory. Integration ratios from the UV chromatograms were measured, and the relative ratios of five components were consistent with the base composition of both strands (Supporting Information S3).
Figure 4.
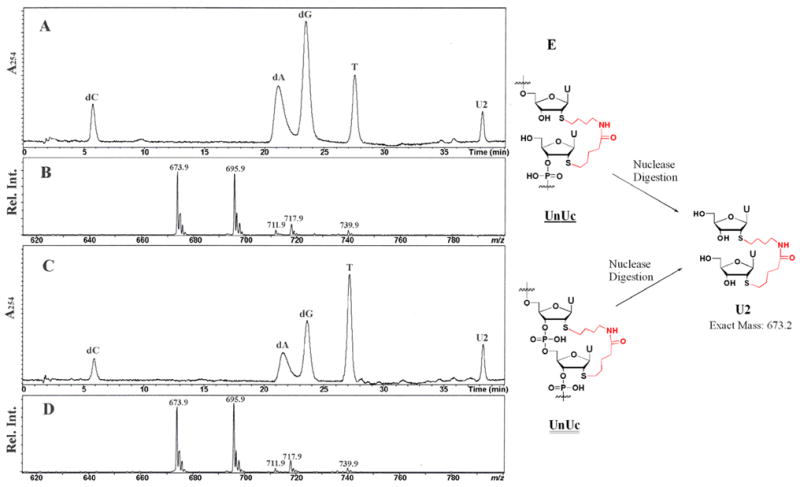
LCMS analysis of nuclease full digestion products of ligation product 10 and control compound 9. (A) UV chromatogram of 10 digestion products, (B) mass spectrum of fragment U2 from 10 digestion, (C) UV chromatogram of 9 digestion products, (D) mass spectrum of fragment U2 from 9 digestion, (E) structures of UnUc in 10, UnUc in 9 and U2. LCMS eluent: A, water (0.1% formic acid); B, methanol (0.1% formic acid). Gradient: 0 min, 95% A; 2 min, 95% A; 5 min, 65% A; 40 min, 15% A; 45 min, 0% A.
To examine duplex formation between 2′,2′-ligated product ODN 10 and complementary natural DNA, thermal denaturation experiments were performed by absorbance spectrophotometry (Fig. 5). The melting curve showed a two-state (helix-coil) transition, which resulted from the cooperative dissociation/association of ODN 10 from complementary DNA. The Tm value (67.7 °C) of ODN 10 was 10 °C higher than pre-ligated ODN 4 and 5 (Tm = 57.1 °C) but somewhat lower than the unmodified DNA control (Tm = 73.2 °C). These results indicated that ODN 10 can form a stable duplex with complementary natural DNA. The 5°C decrease of Tm relative to the unmodified DNA control may result from the greater flexibility associated with the amide 2′,2′-linkage and/or the known destabilizing affect of the 2′-alkylmercapto substituents.15 Interestingly, a larger Tm suppression was observed in our previous experiment involving the 2′,2′- and 3′,5′-double-linked strand 9.13 Overall, the nicked, 2′,2′-conjugated duplex was thus found to be somewhat less stable than the analogous duplex with all phosphodiester bonds intact. For pre-ligated strands 4 : 5, 4 : 6 and 4 : 7, the melting curves showed duplex formation with complementary DNA at room temperature, which verified that ligation occurred under the direction of DNA template (Supporting Information S4).
Figure 5.
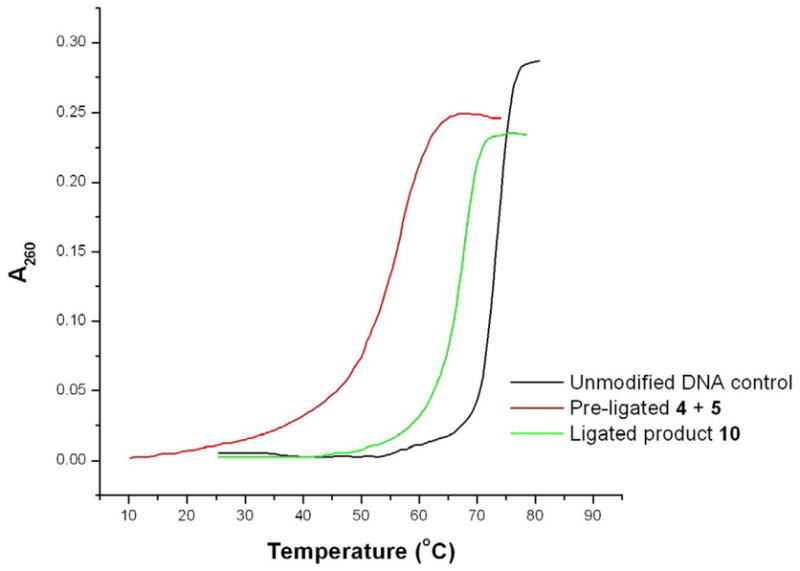
Melting curves of DNA control, pre-ligated 4 : 5 and ligated strand 10 with complementary natural DNA. A260 = [A260(To) - A260(T)]/A260(To). (A260(To) is the absorbance at 25 oC for DNA control and ligated strand 10, the absorbance at 10 °C for pre-ligated 4 : 5). Unmodified DNA control: 5′-AACACGGACATAGTTCCTCGTGTTGTGGAC-3′. Complementary DNA: 5′-GTCCACAACACGAGGAACTATGTCCGTGTT-3′.
2.3. Positional control of ligation
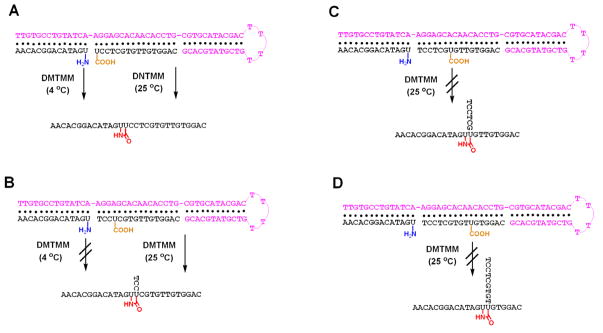
After successful templated ligation of ODNs 4 and 5, new sequences were designed and synthesized to examine the regioselectivity of this ligation reaction. Instead of anchoring at the 5′-end of ODN 5, the modified uridine-containing pendent carboxyl group was incorporated into the sequences as the fourth (ODN 6), the seventh (ODN 7) and the tenth (ODN 8) nucleotide, respectively. The ligations of 5 – 8 with the amine-containing strand 4 were tested under ambient temperature templated conditions (Fig. 2). Examination of the reaction products by denaturing gel electrophoresis showed that almost no ligation occurred for 4 : 7 and 4 : 8 after 48 h reaction time, in contrast to the high ligation yield obtained for the reaction between strands 4 and 5. Unexpectedly, significant ligation product from ODN 4 and 6 was formed. The ligation product had a different electrophoretic mobility from 10, most likely resulting from the presence of a branch in its backbone primary structure. Apparently, over the extended course of the reaction, the modified uridine in ODN 6, only four nucleotides from the 5′-end, enjoys sufficient flexibility at room temperature that it can make contact with the terminal residue of ODN 4, and then react when this occurs. For ODNs 7 and 8, the greater number of nucleotides between the modified uridine and 5′-end fixes the modified uridine into a less flexible duplex DNA structure, keeping it remote from the pendent amine of ODN 4 and preventing reaction. These observations are consistent with the frayed nature of the termini of nucleic acid duplexes compared with internal residues.16
The ligation reactions between 4 : 5 and 4 : 6 were examined more closely as a function of time at room temperature. However, similar ligation rates and yields were obtained (Supporting Information S5). Since low temperature can increase the stability of duplex DNA and minimize the fraying of strand termini, higher regioselectivity of ligation was expected. Reaction between 4 : 5 and 4 : 6 was carried out at 4 °C (Fig. 6). Denaturing gel electrophoresis showed a significant difference in ligation rates between 4 : 5 and 4 : 6. For 4 : 5, the reaction was 50% complete within 5 min; product increased considerably reaching 71% yield at 4 h. In contrast, no ligation was detected for 4 : 6 over the same time interval. After 24 h, the ligation yield of 4 : 5 was higher than 85%, while that of 4 : 6 remained below 35%. Given enough time (71 h), both ligations afforded more than 90% ligation as shown by quantitative analysis of the electrophorograms. Thus, positional control of 2′,2′-templated ligation can be achieved by decreasing both reaction temperature and reaction time (Fig. 7). Temperature dependence of regioselectivity is commonly observed in organic synthesis.17 Here, temperature can affect the stability and dynamics of the templated ligation precursor strands and can control further ligation regioselectivity.
Figure 6.
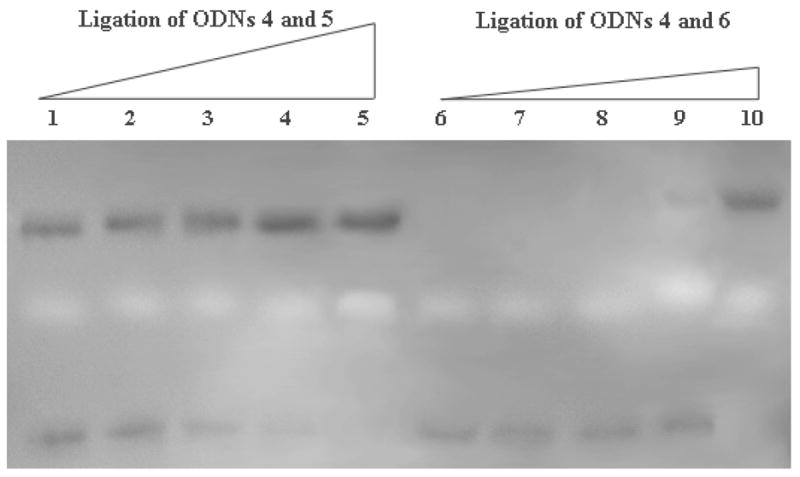
Denaturing gel electrophoresis assay of the time dependence of ligation at 4 °C (200 nM duplex concentration). (1) ODNs 5, 4 and template 3: Lane 1: 5 min; Lane 2: 1.5 h; Lane 3: 4 h; Lane 4: 24 h; Lane 5: 71 h; (2) ODNs 6, 4 and template 3: Lane 6: 5 min; Lane 7: 1.5 ; Lane 8: 4 h; Lane 9: 24 h; Lane 10: 71 h. (Template bands not shown.)
Figure 7.
Schematic illustration of positional control of 2′,2′-templated ligation by temperature. (A) 4 and 5; (B) 4 and 6; (C) 4 and 7; (D) 4 and 8.
Superficially, these results would seem to differ from prior studies indicating that spatial separation of reactive species has little effect on reaction yield in DNA-templated reactions. For example, reactions between two DNA strands, each containing a reactive functional group, are typically dominated by the rate of annealing and therefore are unlikely to offer regioselective reactivity as observed here. In annealing-controlled reactions, several architectures have been examined, all showing similar reactivity when the reagents were adjacent or up to 30 nucleotides apart.2, 18 In this study, natural, unmodified DNA was used to template the reaction: The three DNA molecules were annealed prior to introduction of the coupling reagent, so that the reaction rate depended on the structural parameters (including separation distance) of the reactive partners. In such a system, reactivity can be controlled by the positions of the pendent carboxylate and amine groups in a DNA assembly.
3. Conclusions
We have demonstrated that two ODNs can be 2′,2′-ligated by amide formation between proximal nucleosides contained in two separate DNA molecules, and templated by a complementary DNA hairpin. The ligation yield was > 95% under optimized reaction conditions. The reactivity was found to depend on the distance between the pendent amine and carboxylate groups. The positional control of the reaction was enhanced significantly at low temperatures and short times. In contrast to common ligation strategies that only create linear ODNs from short pieces, in this study ODNs were joined together at the 2′-positions of the ribose, leaving the both the 3′- and 5′-positions available for further ligation reactions or other modifications. This strategy may facilitate the design and synthesis of branched ODNs or other ODN modules, which can be used in nucleic acid-based nanotechnology or in biomedicine.
4. Experimental Section
4.1. Materials
All buffer solutions were prepared from analytically pure chemicals and doubly distilled water and adjusted to desired pH values. Bacterial Alkaline Phosphatase (BAP) and 10×BAP buffer were purchased from Invitrogen (Carlsbad, CA). Snake Venom Phosphodiesterase (SVP) was purchased from USB Corp (Cleveland, OH). Universal Support II resin for the synthesis of ODN 4 to obtain the terminal 3′-Un sequence was purchased form Glen Research (Sterling, VA). The preparation of the modified phosphoramidite uridine monomers and synthesis of modified oligonucleotides on an ABI 394 automatic DNA synthesizer followed the procedures reported previously.10a The template hairpin DNA 3 and the unmodified DNA 1 and 2 were obtained from Integrated DNA Technologies (Coralville, IA). All commercial DNA strands and synthesized ODN strands were purified by denaturing gel electrophoresis (20 % acrylamide; running buffer contains 89 mM Tris.HCl, pH 8.0, 89 mM boric acid, and 2 mM EDTA) or by HPLC (20 mM phosphate buffer, pH 7.0/methanol). All strands were further desalted by HPLC (water/acetonitrile). Concentrations of oligonucleotides were determined by UV spectroscopy (OD260).
4.2. Template-mediated 2′,2′-ligation of ODNs
General procedure for templated ligation, using ODN 4 and 5 ligation as an example: ODNs 4, 5 and complementary hairpin DNA 3 (40 pmol each, 1 : 1 : 1 ratio) were dissolved in 100 μL MOPS buffer (0.1 M MOPS, 1.0 M NaCl, pH 7.0) in an Eppendorf vial. The resulting solution was annealed overnight from 90 °C to room temperature. The condensing agent DMTMM (13.8 mg, 0.05 mmol) was dissolved in 500 μL MOPS buffer. The quantity 100 μL of DMTMM solution (100 mM) was added into the solution of ODNs. Final concentrations were 200 nM ODN duplex, and 50 mM DMTMM. The reaction solution was mixed with a pipette and centrifuged before being incubated at room temperature for 48 h. After completion of the ligation, 11 μL ammonium acetate (1 M) was added to quench the reaction. The final solution was subjected to ethanol precipitation or filtration through a G25 cartridge in order to remove most of the salt before analysis by denaturing gel electrophoresis.
Time course of the templated ligation procedure, (using ODNs 4 and 5 ligation as an example): ODNs 4, 5 and complementary hairpin DNA 3 (240 pmol each) were dissolved in 600 μL MOPS buffer (0.1 M MOPS, 1.0 M NaCl, pH 7.0) in an Eppendorf vial. The resulting solution was annealed overnight from 90 °C to room temperature. The condensing agent DMTMM (41.4 mg, 0.15 mmol) was dissolved in 1.5 mL MOPS buffer. The quantity 600 μL of DMTMM solution (100 mM) was added into the ODN solution. Final concentrations were kept at 200 nM ODNs duplex and 50 mM DMTMM. The reaction solution was mixed with a pipette and centrifuged. At 5 min, 1.5, 4, 24 and 71 h, 200 μL of solution was removed and quenched with 11 μL of 1 M ammonium acetate. After desalting, all aliquot solutions were analyzed by denaturing gel electrophoresis.
4.3. Denaturing Polyacrylamide Gel Electrophoresis (PAGE) Analysis
The ligation mixtures were analyzed on 20% acrylamide; running buffer contained 89 mM Tris.HCl, pH 8.0, 89 mM boric acid, and 2 mM EDTA. The tracking dye contained buffer, 50% glycerol and a trace amount of Bromphenol Blue and Xylene Cyanol FF. The gel was stained with ethidium bromide and gel pictures were taken on Kodak Image Station Gel Logic 200 and yields were calculated by measuring the band intensity using Kodak Molecular Imaging Software.
4.4. MALDI-TOF MS analysis
MALDI-TOF mass spectra were recorded on a Bruker OmniFLEX MALDI-TOF spectrometer. A 3-HPA matrix solution was prepared by dissolving 3-HPA (18 mg) in CH3CN (150 μL) and H2O (150 μL). An ammonium citrate co-matrix solution was prepared by dissolving ammonium citrate (35 mg) in H2O (1 mL). The working matrix solution was obtained by mixing 40 μL of 3-HPA matrix solution and 10 μL of ammonium citrate co-matrix solution. An oligonucleotide sample (20~100 μM, 2 μL) was mixed with the working matrix solution (2.5 μL) using a vortex mixer and then centrifuged before being loaded on a target. ODNs with known masses were used as calibrants in each measurement.
4.5. Full enzymatic digestion and LCMS analysis
ODN 10 (200 pmol) was dissolved in 15 μL of 1X BAP buffer (diluted from 10X BAP buffer) containing 16 mM MgCl2, 1 unit bacterial alkaline phosphatase (BAP) and 0.5 units of phosphodiesterase I (SVP). The digestion solution was incubated at 37 °C for 24 h. The final mixture solution was subjected directly to LCMS analysis without further purification. The analysis of digest by LCMS was conducted on an Agilent 1100 serial Capillary LCMSD Trap XCT system equipped with atmospheric pressure electrospray ionization source. A 2.1 mm x150 mm Zorbax SB-C18 column (particle size, 5 μm, Agilent Technologies) was used for the separation of the nuclease digestion products. A 6 μL portion of digestion solution was injected onto the analytical column thermostated at 30 °C (eluent A, water containing 0.1% formic acid; B, methanol containing 0.1% formic acid; gradient: 0 min, 95% A; 2 min, 95% A; 5 min, 65% A; 40 min, 15% A; 45 min, 0% A). The flow rate was 200 L/min. The eluent was monitored by UV absorption detection at 254 nm. Electrospray source conditions were 8 L/min drying gas flow rate, 40.0 psi nebulizer pressure and 350 °C drying temperature. Mass spectra were recorded in positive mode and all data were analyzed using the LC/MSD Trap Control 4.0 data analysis software.
4.6. Thermal denaturing studies
All complementary strands (0.83 μM each) were dissolved in a buffer (40 mM sodium cacodylate and 100 mM sodium chloride, pH 7.3) to a final volume of 700 μL, and annealed overnight from 90 °C to room temperature. The samples were transferred to quartz cuvettes with 1 cm path length, and the same cacodylate buffer was used as a blank. Thermal denaturation was monitored at 260 nm on a Spectronic Genesys 5 spectrophotometer, using a Neslab RTE-111 programmable circulating bath. At least two consecutive heating-cooling cycles were applied with a linear temperature gradient of 0.1 °C/min. Heating and cooling ramps were superimposable in all cases, indicating equilibrium conditions. Tm values were derived from the maxima of the first-derivative curves.
Supplementary Material
Concentration control of ligation analyzed by denaturing gel electrophoresis; ligation with unmodified DNA control analyzed by denaturing gel electrophoresis; relative integration data from UV chromatogram of LCMS analysis of nuclease digestion; melting curves of pre-ligated 4 : 5, 4 : 6 and 4 : 7 with complementary natural DNA; Denaturing gel electrophoresis assay of the time dependence of ligation at room temperature
Acknowledgments
We gratefully acknowledge support by NSF (CTS-0548774 and CTS-0608889) to N.C.S. and J.W.C., grant GM-076202 from NIGMS, CHE-0316589 from NSF, grants from Cancer Innovations, Inc. and the NYU Cancer Center to J.W.C., and by grants GM-29554 from NIGMS, grants DMI-0210844, EIA-0086015, CCF-0432009, CTS-0548774, CTS-0608889, CCF-0726378 and CCF-0523290 from the NSF, 48681-EL and MURI W911NF-07-1-0439 from ARO and DE-FG02-06ER64281 from DOE (Subcontract from the Research Foundation of SUNY), a grant from the W.M. Keck Foundation to N.C.S. The instrument facility was supported by Research Facilities Improvement Grant Number C06 RR-16572-01 from the National Center for Research Resources, National Institutes of Health. Instrumentation in the facility was purchased with support from NSF grants CHE-0234863 and MRI-0116222.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Concentration control of ligation analyzed by denaturing gel electrophoresis; ligation with unmodified DNA control analyzed by denaturing gel electrophoresis; relative integration data from UV chromatogram of LCMS analysis of nuclease digestion; melting curves of pre-ligated 4 : 5, 4 : 6 and 4 : 7 with complementary natural DNA; Denaturing gel electrophoresis assay of the time dependence of ligation at room temperature