Abstract
The aryl hydrocarbon receptor (AhR) is a ligand-activated transcription factor that controls the expression of a diverse set of genes. The toxicity of the potent AhR ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin is almost exclusively mediated through this receptor. However, the key alterations in gene expression that mediate toxicity are poorly understood. It has been established through characterization of AhR-null mice that the AhR has a required physiological function, yet how endogenous mediators regulate this orphan receptor remains to be established. A picture as to how the AhR/ARNT heterodimer actually mediates gene transcription is starting to emerge. The AhR/ARNT complex can alter transcription both by binding to its cognate response element and through tethering to other transcription factors. In addition, many of the coregulatory proteins necessary for AhR-mediated transcription have been identified. Cross talk between the estrogen receptor and the AhR at the promoter of target genes appears to be an important mode of regulation. Inflammatory signaling pathways and the AhR also appear to be another important site of cross talk at the level of transcription. A major focus of this review is to highlight experimental efforts to characterize nonclassical mechanisms of AhR-mediated modulation of gene transcription.
Keywords: TCDD, dioxin, Ah receptor, estrogen, AP-1, NFκB, inflammation
I. INTRODUCTION
Most transcription factors, by and large, have been defined for their ability to modulate or control a particular biology or physiological response. For example, the estrogen receptor has been associated with reproduction, thyroid, and glucocorticoid receptors with growth and inflammation, and NF-κB with apoptosis and inflammation. The aryl hydrocarbon receptor (AhR) along with its heterodimeric partner the aryl hydrocarbon receptor nuclear translocator (ARNT), together referred to as the aryl hydrocarbon receptor complex (AhRC), have long been associated with an organism’s response to environmental contaminants, most of which are man-made and irrelevant to normal eukaryotic biology. Indeed, for much of the last 40 years, the bulk of experimental effort invested into understanding the function of the AhR has focused on its activation by a wide variety of exogenous compounds, offering little or no insight into the role the AhR might play in normal physiological homeostasis. However, the advent of genetically engineered mice and the identification and emerging importance of nonclassical mechanisms of receptor function offer new avenues of research and may not only help us better define how AhR mediates its toxic effects, but may also shed light on the true physiological role of the receptor.
This review aims to provide a brief overview of nearly six decades of experimental investigation into the nature of aryl hydrocarbon receptor activity and the toxicities associated with the exogenous chemicals that activate it. In doing so, we hope to provide some historical perspective and outline the motivating factors that have driven research in this area in the past. Additionally, we hope to illuminate emerging areas of interest, the potential for discovery, and how new technologies offer an opportunity to explore the AhR function. However, any attempt to summarize data derived from decades of research presents many challenges. As those of us who have been in the field for many years understand, we often struggle with conflicting data derived from experimental systems that were the best available at the time. Yet, we increasingly find these systems rather inadequate to describe the complex interactions that exist between multiple signaling pathways as they converge on transcription. To our colleagues, we would like to apologize in advance if some work is not cited or if our conclusions seem critical. To those new to the field, we hope this serves as something of a road map. With the mapping of the human genome and the advent of DNA array technology, we are able to record changes in gene expression profiles in response to environmental cues. Applications such as chromatin immuno-precipitation (ChIP), fluorescence resonance energy transfer (FRET), small inhibitory RNAs (siRNAs) and real-time PCR allow us to study transcription factor function within the cell and measure associated changes in target gene expression. These advances afford us the opportunity to study the mechanistic determinants of AhR function and, potentially, to uncover the nature of AhR’s true physiological role. Future work may also aid in the discovery of a therapeutic use for AhR activation.
II. BACKGROUND AND HISTORICAL PERSPECTIVE
Organisms are challenged incessantly by potentially hazardous substances of anthropogenic origin. Xenobiotics, such as pesticides, solvents, and many other industrial products, are a major source of environmental pollution and public health concern. Throughout history several events of occupational and accidental exposure to halogenated and nonhalogenated aromatic hydrocarbons (HAHs and PAHs, respectively) revealed many of the health risks associated with exposure to these chemicals.1
In 1949, the accidental release of HAHs, including TCDD, from Monsanto’s chemical plant in Nitro, West Virginia, resulted in several medical cases of chloracne, liver disease, blood diseases, tumors, and alleged exposure-associated deaths.2 In 1957, Sandermann described the discovery and synthesis of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), one of the most potent non-genotoxic tumor-promoting substances known. Unfortunately, Sandermann’s laboratory assistant “mysteriously” suffered from chloracne, a painful and disfiguring skin condition, only days after an accidental exposure to the TCDD being synthesized in the laboratory. Despite the earlier association of chloracne with arochlor intoxication,3 it was not until later that it could also be associated with toxic exposure to TCDD. TCDD toxicity is primarily mediated through the aryl hydrocarbon receptor (AhR).4 In the post-TCDD discovery era, from 1962 through 1967, the United States introduced the term “chemical warfare” when the compound Agent Orange, containing residual TCDD, was utilized to defoliate large forested areas in Vietnam during Operation Trail Dust and Operation Ranch Hand. Surprisingly, despite the large number of experimental animal data suggesting TCDD as a potent carcinogen, a direct link to human cancer remains a topic of active research.5
TCDD is now known to be a trace by-product in the synthesis of the phenoxy-herbicide 2,4,5-trichlorophenoxyacetic acid and chlorophenol.6 In 1964 Dow Industrials released the first report on the possible dangers of TCDD exposure after several employees who worked in the production of Agent Orange developed chloracne. In 1976 a severe industrial accident took place at the Industrie Chimiche Meda Societa Azionaria in Seveso, Italy.7 Roughly up to kilogram quantities of TCDD and additional TCDD-like chemicals were dispersed into the air with adverse health consequences for the local population.8 Thousands of animals died and others were sacrificed to prevent further contamination of the food chain. Thanks to the foresight of Mocaralli (see Ref. 7) blood samples were collected and frozen from each patient for future analysis. A high proportion of human females were apparently born to couples exposed to highly contaminated areas in Seveso. However, this assertion continues to be debated.9,11 Nevertheless, a convincing role for AhR ligands and the receptor itself in estrogen receptor (ER) function, degradation, and abnormal sex hormone metabolism has been suggested by recent research.12–16 The increased awareness about the dangers of TCDD and related compounds prompted the industry to improve their production guidelines to protect their employees from chemical exposure and minimize the release of harmful substances into the environment.
In recent history, the Ukraine president Viktor Yushchenko became a widely recognized victim of TCDD poisoning.17 Unfortunately, an effective treatment for TCDD poisoning and from similar substances has yet to be established, although the use of potato chips fried in the indigestible synthetic fat Olestra™ (Procter & Gamble Company, Cincinnati, OH) has shown promise in this regard.18 Olestra cannot be absorbed in the gut and it is therefore believed to serve as a carrier of TCDD excreted from the body through feces.19 The half-life of TCDD in humans is estimated to be around seven years, although genetic and environmental factors are expected to minimize or prolong its clearance from human tissues.
III. IDENTIFICATION AND CLONING OF THE AHR
During the late 1950s it became clear that the efficacy or adverse effects of therapeutic drugs often could be associated with simple inheritable genetic differences (polymorphisms) between individuals. Despite hints dating as far back as Garrod’s 1906 publication entitled Inborn Errors of Metabolism,20 what apparently delayed this realization was the confounding influence of the environment (e.g., lifestyle) on the phenotypic manifestation of genetic traits, as well as the fact that most responses to xenobiotics involved several nonlinked genetic loci. Nevertheless, through advances in statistical and genetic analyses, it became widely recognized that at least some of these metabolic differences between individuals had simple traceable genetic origins that segregated in Mendelian frequencies.
Similarly, differences in susceptibility to HAHs, PAHs, and other xenobiotics, even between organisms of the same species, also could be traceable to their genetic makeup. These polymorphic genetic loci were recognized to play a role in the controlled expression and/or stability of xenobiotic metabolizing enzymes, receptors, transporters, and perhaps unidentified accessory/regulatory factors. Preceding the identification and cloning of the aryl hydrocarbon receptor the upregulation of aryl hydrocarbon hydroxylase (AHH) activities, measured in vitro by the formation of 3-hydroxybenzo[a]pyrene from the parent compound benzo[a]pyrene (B[a]P), could only be readily detected in liver extracts and some extra-hepatic tissues from so-called “responsive” mice.21 The crossing and backcrossing of multiple inbred strains of mice further led to the identification of the Ah (aryl hydrocarbon) genetic locus, suspected to control the inducible expression of the spectrally distinct cytochrome P1-450 (today known as CYP1A1)22,23 as well as other AHH activities. This genetic locus segregated primarily as an autosomal dominant trait in specific crosses (e.g., C57BL × DBA mice) where the dominant responsive allele was denoted as Ahb (in C57BL/6 and B6 strains) and the recessive nonresponsive as Ahd (in DBA/2 strains). As expected, the complex inheritance pattern of AHH responsiveness observed between a wider screen of wild-type and inbred mice strains strongly suggested the additional contribution of nonlinked loci to the inducibility of AHH/P1-450 activity.24 Regardless, the rescue of “unresponsive” DBA mice with the potent AHH inducer TCDD, for which AHH activity approached that of 3-MC-treated C57BL/6 mice, sparked a new theory, namely, perhaps the Ah locus encoded a gene product, such as a receptor protein, central to the downstream induction of AHH/P1-450.21 Thus, a mutated AhR in the DBA mice strain was suspected as the culprit for a failure to recognize the 3-MC signal.25–27 This hypothesis was validated only after the AhR was cloned. A comparison between the AhR from C57BL and DBA strains of mice indicated that an alanine to valine substitution at position 375 (381 in human AhR) in C57BL AhR and a mutation at the stop codon resulted in an elongated carboxyl terminus in the AhRd allelic variant with reduced affinity for [3H]TCDD.28
Several research observations reinforced the AhR theory. For example, the peritoneal injection of C57BL/6 mice with [3H]TCDD resulted in high levels of radioligand retention in the liver, especially when compared to the nearly absent levels in the nonresponsive DBA/2 strain. Crosses between C57BL/6J × DBA/2J also resulted in offspring (B6D2F1/J) displaying intermediate binding/responses, an observation that correlated with the segregation of a simple autosomal dominant trait controlling AHH inducibility.27 Extensive in vitro and comparative animal toxicology studies emphasizing structure-activity relationships further suggested that a soluble cytosolic receptor protein with varying affinities for TCDD-like compounds was likely responsible for the induction of AHH/P1-450 activity.27,29–32 A model began to emerge resembling that of the already established steroid receptor pathway.33 In brief, the cytosolic AhR, upon binding of TCDD, would translocate into the nucleus and activate genes controlled by the Ah genetic locus. However, until better methods (e.g., immunofluorescence34–36) were available, the location of the AhR complex proteins in nonstimulated cells remained a controversial issue for several years.37,38
The AHH/P1-450 “inducer/receptor-complex” was later confirmed to undergo translocation into the nucleus of responsive cells following the binding of radioligand, an event that clearly preceded the induction of AHH/P1-450 activity.39,40 A comparative study of the nonresponsive VERO and HTC mammalian cell culture lines and the responsive H-4-II-E and Hepa-1 cell lines also indicated that the translocation of the AhR into the nucleus, as in HTC cells, does not guarantee that AHH/P1-450 induction would proceed.41 These results further highlighted that both structural and regulatory genes other than the proposed AhR were likely controlled by the Ah locus. Work conducted by Okey and colleagues,40 and later revisited by Hanna and coworkers,42 identified the presence of ~ 6 S (at high ionic strength) and ~ 9 S (at low ionic strength) receptor complexes, respectively. Both complexes had a measurable [3H]TCDD binding capacity that was virtually absent in extracts from DBA/2N mice. These complexes also displayed sensitivity to proteases but not to DNases and RNases. [3H]TCDD binding could be competitively displaced with known P1-450 inducers but not with established steroid hormones or other classic P-450 activity inducers (e.g., phenobarbital). In addition, only known inducers of P1-450 (CYP1A1) and unlabeled TCDD could effectively compete for binding to the receptor complex.40
The lessons learned about the glucocorticoid receptor family were being progressively applied to the rapidly growing AhR field. For example, the use of 20–30 mM molybdate in sucrose gradient fractionation of the glucocorticoid receptor (GR) was found to stabilize the ligand-binding conformation of the receptor.43 However, the murine and rat AhRs appeared to benefit only partially from this treatment by showing increased thermostability, as well as retaining their ligand-binding activity under high ionic strength.44 However, the human AhR did appear to greatly benefit from enhanced stability in vitro by molybdate treatment.45
A key defining moment toward the future cloning and characterization of the AhR was the synthesis of 2-azido-3-iodo-7,8-dibromodibenzo-p-dioxin,46 a photoaffinity ligand capable of covalently binding the AhR when excited in the ultraviolet frequency. At first, two polypeptides with a mass of 70 kDa and a 95 kDa were suspected to represent the Ah receptor, given that the photoaffinity ligand could be displaced with known Ah agonists from both polypeptides (e.g., TCDD). However, the 70 kDa polypeptide was found to be no more than a proteolytic fragment of the 95 kDa polypeptide in Hepa-1 cells.47 The suspected protease had similar characteristics to the Ca+2-dependent calpain II and was ruled as the culprit of this isolation “artifact.” Hence, the addition of EDTA was recommended to stabilize the AhR during purifications. With the advent of a new molecular photoaffinity probe, the AhR field exploded with numerous research findings. For instance, the characterization of several inbred mice strains with the new photoaffinity radioligand led to the identification of two AhR allelic forms such as the 95 kDa (Ahb−1 allele) from the C57, C58, and MA/MyJ mice strains, and the 104 kDa (Ahb−2 allele) from the C3H/HeJ, BALB/cByJ, and A/J strains.48 Ligand competition-binding protocols with the photoaffinity ligand were also being established, which helped estimate the binding affinity and characteristics of other suspected AhR ligands.29,49 In 1988, Perdew and Poland described the partial purification of the AhR from C57BL/6J mice.50 This event led to the eventual purification, N-terminal sequencing, and production of antibodies against the AhR.51,52 The generation of antibodies capable of immunoprecipitating the glucocorticoid receptor-associated hsp90 53–55 and the AhR helped identify the hsp90 as part of the unliganded AhR complex with two different approaches.56,57
Before the cloning of the AhR, a subunit of the liganded complex was identified and thought to be required for the translocation of the AhR into the nucleus.58,59 Briefly, a cell line expressing a “functional” AhR but uninducible for P1-450 (CYP1A1) was systematically transfected with a cDNA library. The expression of one of these cDNA constructs restored the translocation of the AhR and the expression of CYP1A1. The product of this gene was therefore named ARNT (Ah receptor nuclear translocator), although the name itself is a misnomer, for ARNT is not directly involved in the AhR “translocation” event per se. ARNT was found to share homology with other known Drosophila proteins such as Per and Sim and contained a suspected DNA-binding/dimerizing domain termed basic helix-loop-helix.58 Together the Per, ARNT, and Sim proteins are the representative members of the PAS domain superfamily. PAS domain proteins have been associated with important functions in transcription by relaying environmental signals (e.g., light, oxygen, and xenobiotics) to the cell60 (for an excellent recent review see Ref. 61). The AhR appeared as a unique member in the PAS family because it could be activated by exogenous ligands. However, a putative high-affinity endogeous ligand for the AhR has yet to be identified and thus remains classified as an orphan receptor.
A distinction between the cytosolic 9 S and nuclear 6 S forms of the AhR was later established through chemical cross-linking studies.62,63 The AhR complex was found as a tetrameric complex containing the AhR, two molecules of hsp90, and an unidentified ~ 43 kDa protein that sedimented at 9 S in sucrose gradients. This tetrameric complex could be transformed into a dimeric 6 S complex in the presence of ligand.62 The ligand-activated 6 S AhR complex was also paradoxically composed of a heterodimer containing the signature 95 kDa AhR and an “unknown” 85 kDa polypeptide (ARNT). Often, the 9 S form of the AhR is referred to as the “latent” form and the 6 S form as the “transformed” form of the receptor.
Numerous inbred strains of mice were further characterized with the photoaffinity ligand. This work led to the identification of four polymorphic alleles, namely, the 95-kDa (Ahb−1), 104-kDa (Ahb−2), 105-kDa (Ahb−3), and the 104-kDa (Ahd) allelic variants. Given the availability of antibodies and a preestablished AhR purification protocol, the N-terminal sequence of the AhRb−1 was determined. This aided the creation of degenerate primers used in the cloning of the AhRb−1 gene.64 The AhRb−1 was found to share many features with the previously cloned ARNT. For instance, it contained a bHLH region followed by two 51-amino acid Per-ARNT-Sim (PAS) A-B repeats60 and an N-terminal glutamine-rich region.58,65 The similarities between the AhR and ARNT, the presence of a bHLH, and previous observations on their interaction with enhancer DNA regions at the CYP1A1 promoter led to the hypothesis that the AhR and ARNT could be heterodimeric partners. In addition, the covalent binding of the photoaffinity ligand to the A–B region suggested that this region could be structurally part of a ligand-binding pocket that serves as a switch for the transformation of the AhR into a DNA-binding conformation.64 Finally, the glutamine-rich region was suspected to serve as a transcription activation domain given its presence in other transcription factors.66,67 The human AhR was subsequently cloned by the screening of a cDNA library generated from the hepatoma cell line Hep-G2.68 In comparison with the murine AhRb−1, the human receptor was 6 kDa larger. Most of the differences between the murine and human receptors were found in the carboxyl terminus. Sequence comparison revealed < 60% conservation between species, whereas the basic region, the helix-loop-helix, and PAS domains displayed about 100%, 98%, and 87% levels of conservation, respectively.
The creation of a photoaffinity ligand and antibodies for the AhR facilitated the use of numerous biochemical approaches that accelerated our understanding of AhR biology. At the same time, differences between responsive and nonresponsive animals to xenobiotics and the low degree of conservation between the mouse and human AhR were quickly being recognized as major obstacles, especially for the extrapolation of animal toxicological data to humans.
IV. CHARACTERIZATION OF AHR AND ARNT FUNCTIONAL DOMAINS
Following the cloning of the AhR and ARNT, an extensive effort to map their functional and interaction domains was initiated.68–73 Detailed structure-function relationships were carried out in the laboratory of Oliver Hankinson.74,75 Briefly, through immunoprecipitations of the murine AhR, deletion mutants of [35S]ARNT were monitored for their ability to interact with the AhR in the presence or absence of TCDD.75 The bHLH together with the PAS-A and PAS-B domains (amino acids 70–474 of the murine ARNT) were all required for optimal heterodimerization to occur in the presence of TCDD. Deletion of the basic domain of ARNT had virtually no effect on the dimerization process. The ability of the AhR:ARNT heterodimer to bind double-stranded DNA oligos containing the consensus XRE sequence also required the basic region of ARNT. Interestingly, an ARNT construct containing only the region from the bHLH through the PAS-A domain was unable to bind its XRE while still being able to heterodimerize with the AhR. Conversely, a construct bearing the bHLH, PAS-A, and PAS-B domains (bHLHAB) was able to restore XRE binding. It was therefore hypothesized that the PAS-B domain may help the ARNT protein fold in a way to maximize the interaction between the basic region of ARNT and DNA. The presence of an oligo containing a xenobiotic responsive element alone did not catalyze the heterodimerization of the AhR with ARNT. Thus, a ligand seemed to be required for AhR:ARNT heterodimerization to occur. Finally, a construct containing only the bHLH, PAS-A, and PAS-B domains could not restore in vivo function of ARNT. Therefore, other regions of ARNT were suspected to contribute to its transcriptional activity, namely, the region containing amino acids 474–627 of the murine ARNT.75
To characterize the transactivation domains of the AhR and ARNT, chimeras were initially generated with the DNA binding domain of the glucocorticoid receptor.76 Since the bHLH region of the AhR and ARNT are required for their heterodimerization, this fusion-protein approach seemed like a proper approach to study the individual transactivation potential of the AhR and ARNT. In this study the glucocorticoid DNA binding domain was fused to ARNT and the AhR, generating GRDBD-ARNT and GRDBD-AhR chimeras. In general, GRDBD-ARNT chimeras were more transcriptionally active than the GRDBD-AhR variety. Furthermore, in the case of ARNT, its transactivation domain potency varied between cell lines, suggesting that cell- and promoter- specific activities are possible for this transcription factor. However, an AhR chimeric construct containing residues 83–593 reduced the ligand-induced activity while a chimeric construct based off residues 594–805 led to a constitutively active AhR that was as potent as the GRDBD-ARNT on an MMTV promoter. Therefore, a region close to the PAS homology/ligand-binding region was suspected to impart regulatory control to the AhR. For instance, the construction of a GRDBD-AhR containing residues 340–805 restored the repressive effect and the interaction between the AhR and the hsp90. Based on this and previous observations indicating that an AhR stripped from hsp90 could not bind ligand,77 a possible role for hsp90 in repressing constitutive AhR activity was suggested. In this model, the hsp90 protein would maintain the AhR in a latent state in the absence of a stimulus. Upon binding of a ligand, a conformational change would cause the concerted dissociation of AhR from the hsp90 complex, its heterodimerization with ARNT, and subsequent activation of target genes.
The use of chimeric AhR and ARNT containing the basic DNA binding domain of GR to study the transactivation domains was later criticized. 74 As a result, a complementary analysis of the AhR domains was performed by Fukunaga and colleagues,74 similar to previous domain-deletion studies with ARNT.75 The ligand-binding region of the AhR was mapped to the PAS B domain. Earlier, through the use of a photoaffinity ligand, the actual ligand-binding pocket was found to encompass the PAS B domain.64 Both the bHLH and PAS B domains of the AhR were required for hsp90 binding. Although ARNT could enhance the dissociation of the AhR from hsp90, ligand alone was sufficient to facilitate the dissociation of the AhR from hsp90 in vitro.74 Whether the release of AhR from hsp90 in the presence of ligand happens in vivo has yet to be fully established, although evidence suggests that dissociation of the AhR from hsp90 may only be required for its heterodimerization with ARNT.78 Indeed, the presence of monomeric AhR may not stably exist in the cell. The regions of the AhR responsible for its interaction with the hsp90 were further mapped to amino acids 1–166 and 289–347,79 whereas the use of previously published deletion constructs of the hsp90 80 suggested that amino acids 272–617 (“the middle portion”) of the hsp90 were involved in AhR binding.81
V. BASIC ASPECTS OF AHR-MEDIATED GENE REGULATION
The nuclear uptake of the AhR was a phenomenon observed after the treatment of C57BL/6J mice and responsive cell cultures with [3H]TCDD.39,40,82 It seemed evident that ligand binding was required for the AhR nuclear translocation event,83 whereas limited proteolysis of the receptor complex suggested the presence of a DNA binding component of the AhR complex.84 Indeed, subsequent studies involving mutant mice harboring an expressing nuclear localization-deficient form of the receptor were found to be resistant to the toxic effects of TCDD.85 However, these nonspecific DNA binding studies only provided limited information about the DNA motif(s) or response element(s) recognized by the activated AhR. Hence, another group took a genomic approach to determine the regions of the P1-450 (CYP1A1) promoter that contained TCDD-responsive elements. HAV (highly expressing variant) cells expressing high levels of the P1-450 gene were selected for a genomic screen. Several clones containing P1-450 DNA were screened and one containing a 2.58 kb fragment was subcloned upstream of a bacterial chloramphenicol acetyl-transferase gene (CAT) reporter.86 In this manner the 5′-end of the P1-450 gene was found to contain at least two elements that were responsive to several AhR ligands in responsive cell lines and contained a suspected cycloheximide-sensitive repressor binding site. Through a deletion analysis of the P1-450 promoter, an AhR/TCDD-dependent enhancer region was identified.87,88 This enhancer region was later documented as the dioxin responsive element (DRE),89 although the term XRE (xenobiotic responsive element) or AhRE (aryl hydrocarbon responsive element) has also been used to describe the DNA elements recognized by the activated AhR. A third DRE was later identified and the consensus DNA sequence for the binding of the AhR to its DRE was suggested. 90,91 This DRE sequence was suggested to be 5′-TA/TGCGTG-3′, a sequence that was present in all 3 DREs described to date. The asymmetrical nature of the DRE also differed from the typical palindromic responsive elements recognized by steroid receptors. The DRE flanking sequences in the enhancer/promoter appeared to be equally important because the subcloning of oligos containing the DRE sequence alone was not sufficient to restore inducible expression of a reporter gene.90,91 The core DRE sequence recognized by the AhR complex was further refined through methylation protection and interference studies to 5′-T/GCGTG-3′, and four copies of this element were mapped to the P1-450 promoter.92,93
The AhR was initially suspected to bind as a monomer to XRE sequences on double-stranded DNA. This was based on the notion that the response elements for steroid receptors were symmetrical (palindromic) and the steroid receptors bound DNA as homodimers. However, several lines of evidence suggested a heterodimeric structure for the DNA-binding AhR complex. Among these were that the transformation of the AhR into a DNA binding complex required a non–ligand-binding component.94 Through cross-linking studies and limited proteolysis studies, a ligand binding component and an accessory component of the activated AhR were identified.95 Finally, following the cloning of the protein ARNT and the AhR itself, the Hankinson group identified ARNT as a component of the activated AhR in nuclear extracts.96 In vivo DNA footprinting demonstrated that the AhR bound a DNA element with the sequence 5′ CACGCNA/T 3′ and another component interacted with a G-rich region.97 ARNT was found to bind a 5′-GTG-3′ half-site that resembled that of the E-box element utilized by other members of the helix-loop-helix family of transcription factors. Since nucleotides flanking the core DRE sequence had an impact on AhR complex affinity for its DNA element, the putative XRE was suggested to be 5′-TTGCGTGAGAA-3′, where the AhR binds the second 5′ thymine and ARNT the third 5′ thymine.98 With the identification of these elements and the cloning of the AhR and ARNT, efforts turned toward the identification of novel AhR-regulated genes. The list of genes directly regulated by the AhR continues to grow. Some examples of AhR-regulated genes are listed in Table 1.
TABLE 1.
AhR-Regulated Genes
| Gene | References |
|---|---|
| CYP1A1 | 97 |
| CYP1A2 | 99–101 |
| CYP1B1 | 102, 103 |
| aldehyde dehydrogenase 3 | 104, 105 |
| UGT1A1 | 106 |
| Ya subunit of GST | |
| CYP2S1 | 107 |
| NAD(P)H:Quinone-oxireductase | 108 |
| murine epiregulin | 109 |
| ecto-ATP | 110 |
| δ-aminolevulinic acid synthase | 111 |
| Prostaglandin endoperoxide H synthase 2 | 112 |
| MDR1 and BRCP | 113, 114 |
| AhRR | 115 |
| p27kip1 | 116 |
VI. AHR ACTIVATION
The AhR is considered an orphan receptor, given that no putative high-affinity endogenous ligand has been identified to date. An endogenous role for the AhR has only been suspected from its ability to control the expression of drug-metabolizing enzymes when stimulated by synthetic substances belonging to the HAH and PAH families as well as numerous dietary substances. As such, a large number of AhR ligands can induce their own metabolism and clearance from the body by inducing drug-metabolizing enzymes of the phase I (oxidation) and phase II (conjugation), and transporters of the phase III (excretion) metabolic pathways.117 Unfortunately, the metabolism of some compounds (e.g., B[a]P) can lead to the formation of nucleophilic derivatives that bind covalently to DNA and proteins. Occasionally these adducts may form in a tissue-specific manner.118 The formation of DNA and protein chemical adducts has been associated with an increased risk for carcinogenesis.
Some widely recognized synthetic inducers of the AhR pathway include compounds such as B[a]P, 3-methyl cholanthrene, β-naphthoflavone, TCDD, 2,3,7,8-tetrachlorodibenzofuran, and 3,4,3′,4,′5-pentachlorobiphenyl.119 However, numerous studies have suggested that dietary substances can also readily activate AhR-regulated genes. Some examples include indole-3-carbinol,120 curcumin,121 and quercetin.122 Some ligands can also exert both agonist and antagonist activities at different concentrations, such as resveratrol123 and galangin.124 It appears logical that the high ligand promiscuity of the AhR may be indicative of its proposed role in the adaptation of organisms to environmental chemical challenges. On the other hand, putative endogenous ligands for the AhR have been suspected to exist for numerous reasons. For example, hydrodynamic shearing of cells results in the formation of arachidonic acid metabolites suspected to induce CYP1A1 in an AhR-dependent manner.125 A rat model of supplemental oxygen treatment for lung insufficiency revealed that hyperoxia treatment of rats led to AhR-dependent CYP1A1 induction.126 The incubation of epidermal cells in a methylcellulose suspension activated CYP1A1 expression and this effect could be prevented by treatment with the AhR antagonist α-naphtoflavone.127 UV irradiation could also possibly lead to an AhR-dependent CYP1A1 induction by the formation of active tryptophan oxidation products.128 There are also examples of substances (e.g., omeprazole) that can readily activate the AhR, yet are not direct AhR ligands.129,130 This leaves open the possibility that the AhR can be activated in a ligand-independent manner. In fact, AhR mutants that fail to bind ligand were used to demonstrate that they can still heterodimerize with ARNT and mediate transcription in the absence of ligand binding.131 This offers formal proof that the AhR does not require ligand binding to be transcriptionally active. Developmental studies of AhR “knockout” mice have revealed important roles for the AhR in the development of the liver, closure of the ductus venosus,132 the immune system,133 and the control of many “ligand-independent” cellular activities. 134 These observations strongly indicate a function for the AhR beyond the adaptive and toxic responses to xenobiotics. For instance, the affinity for exogenous ligands is not conserved between certain mouse strains and between species. Most notably there is a tenfold difference in affinity between C57BL/6 mice and human AhR.135 This lack of conservation may suggest that either a ligand-independent activation of the AhR is important for endogenous AhR function, or a key endogenous ligand’s ability to activate the AhR is conserved.
VII. THE LATENT AHR MULTI-SUBUNIT COMPLEX
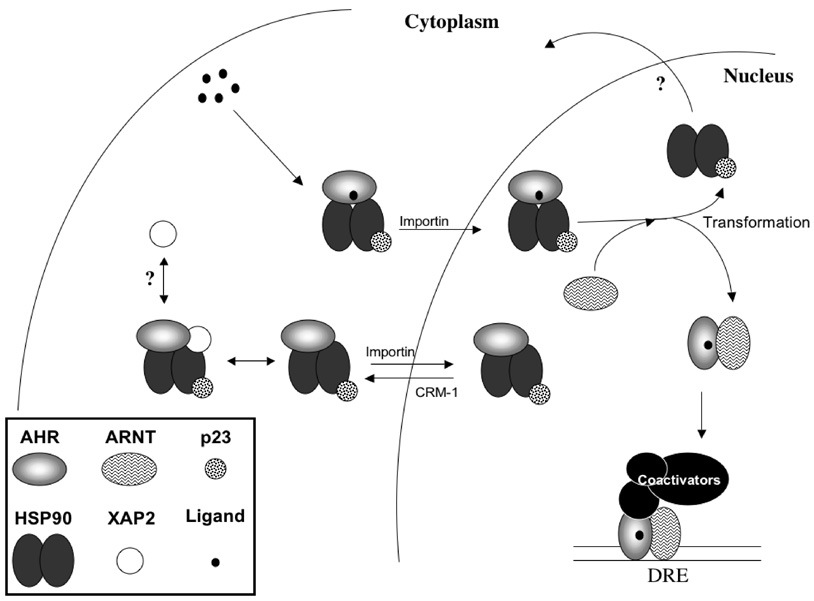
Prior to the cloning of the AhR and its designation as a bHLH-PAS protein,64 it was considered possible that the AhR, due to its analogous biochemical properties, was a member of the steroid receptor family.56,77,136 Although the AhR shares no sequence homology, it is functionally similar to some steroid receptors with respect to its regulation by chaperone proteins, principally HSP90. Glucocorticoid (GR) and progesterone receptor (PR) maturation has been well studied using purified proteins, and many of the conclusions obtained from these experimental systems have been extrapolated to pertain to the AhR. In the early 1990s, the Toft and Pratt laboratories demonstrated that rabbit reticulocyte lysate contained all the molecular components necessary to transform incompetent forms the PR and GR into competent hormone-binding receptors.137,138 Subsequently, the presence of the chaperone heat shock proteins (HSPs) 40, 70, and 90, as well as the cochaper-ones HOP/p60 and p23 were determined to be sufficient for achieving optimal hormone-binding status, although only HSP70 and HSP90 are absolutely required to form a less efficient binding-competent receptor.139,140 Of the five above-mentioned proteins required for GR and PR maturation, HSP90 and p23 are known components of the mature latent AhR complex. However, it is possible, if not likely, that the other three proteins are also involved in regulating the early steps of AhR maturation. The following sections will highlight several aspects of AhR regulation with respect to its association with HSP90, p23, and the immunophilin-like XAP2 protein. It is essential to recognize that activation of the AhR transcriptional complex does not stem simply from a bimolecular interaction of receptor and ligand, but rather is regulated through the interplay between the AhR and numerous ancillary factors. The chaperone and cochaperone components serve to maintain proper folding, ligand-binding competency, and overall transcriptional effectiveness of the AhR. An overview of our current understanding of the transformation of the latent AhR to an active transcriptional complex is outlined in Figure 1.
FIGURE 1.
Transformation of the latent AhR tetramer to an activated transcriptional complex.
VIII. 90 KDA HEAT SHOCK PROTEIN
As alluded to above, early biochemical characterization of the AhR complex was performed using the GR as a model due to the “virtually undistinguishable” physiochemical properties between the two receptors.141 As a result of these comparative studies, the inactive AhR, like the GR, was demonstrated to exist in a heterocomplex with HSP90.56,57 The association of HSP90 with the AhR is representative of the transcriptionally inactive receptor since the AhR, when bound to HSP90, is unable to heterodimerize with its DNA-binding partner ARNT. HSP90 and ARNT both interact with the AhR via the receptors HLH and PAS domains, thereby making their association with the AhR mutually exclusive.74,75,79,142 Similar to a number of other HSP90 substrates, the AhR interacts with HSP90 through the middle portion of the chaperone.81
Upon ligand activation, the human and mouse AhR translocate into the nucleus still associated with HSP90, demonstrating that the cytoplasmic AhR complex does not have to shed its chaperone machinery before nuclear uptake.78,143 It is within the nucleus that ARNT heterodimerizes with the AhR. Whether ARNT actively displaces HSP90 from the AhR or other factors stimulate HSP90 dissociation is not yet understood. In vitro, however, HSP90 is released from the ligand-bound AhR when coincubated with cell extracts from ARNT-containing but not ARNT-deficient mouse hepatoma cells.144 This study suggests that ARNT, or a component functionally tied to ARNT, is involved in the ligand-induced displacement of HSP90 from the AhR.
The GR and PR require the association with HSP90 in order to maintain hormone-binding competence 145 and references within). Analogously, AhR-HSP90 complex formation is important to produce a receptor that has maximal ligand-binding capacity.146,147 In a manner analogous to the GR, the monomeric AhR (isolated via high salt stripping of receptor-associated proteins) derived from mouse hepatoma cells has no ligand-binding capacity. However, the supplementation of the monomeric AhR with rabbit reticulocyte lysates, containing HSP90, partially restores ligand binding while HSP90-negative wheat germ lysate has no effect.77 The AhRs dependence on HSP90 association for ligand binding appears to vary between species, however, because the rat AhR is more sensitive to HSP90 disruption than the guinea pig and rabbit forms of the receptor.148 Additionally, disruption of the stable association between the AhR and HSP90 in living cells results in the rapid degradation of the receptor,149 highlighting the importance of the AhR-HSP90 interaction in receptor signaling.
HSP90 is a known phosphoprotein, but it is unclear to what extent phosphorylation of HSP90 serves to regulate AhR function. A recent study suggests that phosphorylation of the constitutively expressed HSP90β isoform on two serine residues modulates the affinity of HSP90 for the AhR. The authors suggest that the unphosphorylated HSP90β isoform has a high affinity for the AhR and subsequently higher transcriptional activity. 150 In yeast, it appears that the AhR may preferentially utilize the constitutive HSP90β isoform over the more inducible HSP90α chaperone.151 Further studies need to be performed to corroboratethe importance of these findings.
VIX. THE HSP90-ASSOCIATED CO-CHAPERONE
The phosphoprotein p23 is a small, acidic protein that is ubiquitously expressed in virtually all tissues. 152 It interacts with and stabilizes the ATP-bound conformation of HSP90. It is through this interaction that p23 incorporates into numerous steroid receptor complexes153 and references within). Although the amino acid residues required for HSP90-p23 interaction have not been determined, it is known that it binds at the amino-terminal portion of HSP90 154, and this interaction can be disrupted by agents that compete at the ATP binding site, such as geldanamycin.155 The entry of p23 into steroid receptor complexes occurs near the end of their maturation process, and it is likely that this is also true for the AhR. Experimental evidence shows that p23 is capable of modulating steroid receptor function by stabilizing the mature complex and altering its hormone binding capacity 153 and references within). Additionally, p23 has been demonstrated to regulate receptor function by enhancing the hormone-stimulated substrate release from HSP90 in an ATP-dependent manner.156 In vivo, p23 is essential for the perinatal survival of the developing mouse, and mortality associated with the loss of p23 expression appears to be due, at least in part, to defective GR signaling as evidenced by the disruption of glucocorticoid-stimulated lung development. 157
To date, the number of studies performed to assess p23’s role in regulating AhR function are limited, but they universally find that p23 acts to stimulate AhR activity. The ability of p23 to enter into the AhR complex was first demonstrated in vitro by receptor complex reconstitution experiments using individually purified components.158 Later studies using in vitro translated AhR demonstrated that p23 presence in the AhR complex is necessary to prevent the spontaneous formation of an AhR-ARNT heterodimer in the absence of ligand.159 This spontaneous heterodimer formation could be blocked by the addition of molyb-date, which is known to stabilize, or mimic, the ATP-bound (i.e., p23-bound) conformation of HSP90 complexes.160 Additionally, the increased formation of AhR-ARNT DNA-bound complexes in the presence of p23 has been reported, as determined by mobility gel shift assay.161 AhR activity is elevated in yeast reporter systems containing expression vectors for human p23 or the yeast p23 homolog SBA1 in strains null for the homolog.162 Finally, p23 has been hypothesized to enhance nuclear uptake of the AhR following ligand treatment by increasing the ability of the receptor to recognize the import protein importin β.163 Taken together, the limited number of experiments performed suggests that p23 acts as a stimulatory factor in regulating AhR activity.
X. THE HEPATITIS B VIRUS X-ASSOCIATED PROTEIN 2
An additional class of proteins known to be highly involved in regulating steroid receptor function is the immunophilins. Immunophilins derive their name from their ability to bind immunosuppressive drugs. The large immunophilins, FK506 binding proteins (FKBP) 51 and 52, as well as cyclophilin-40 have all been established to exist in multiple steroid receptor complexes (Ref. 164 and references within). Cochaperones, like p23, enter into steroid receptor complexes once the receptor achieves its mature folded state and therefore do not appear to be a component of the receptor folding machinery. The GR has been a model complex for a number of mechanistic studies performed to understand immunophilin-mediated regulation of steroid receptor function by FKBP51 and 52. Interestingly, although they are closely related proteins, FKBP51 acts as a repressor of the GR, whereas the presence of FKBP52 in the GR complex enhances receptor transcriptional activity.165,166 Also, it has been suggested that hormone binding to the GR stimulates the switching of the binding status of receptor from an FKBP51-bound form to an FKBP52-bound form, thereby enhancing nuclear uptake of the GR.167 The recent creation of the Fkbp52−/− mouse for the first time has demonstrated that the immunophilin class of cochaper-one proteins play a significant role in regulating the functional properties of certain nuclear receptors, like the PR168 and androgen receptor.169 The loss of the FKBP52 protein in the null mouse line results in both male and female infertility due to severe deficiencies in androgen or progesterone responsiveness, respectively.
The latent AhR complex contains an immunophilin-like component termed the hepatitis B virus X-associated protein 2 (XAP2). XAP2 is a 37 kDa phosphorylated protein170 that was first identified through its ability to interact with the hepatitis B virus X protein.171 Cross-linking studies performed by Perdew in 1992 showed that, in addition to two molecules of HSP90, the AhR tetrameric complex contains a ~ 46 kDa protein. 172 Five years later, three separate laboratories identified that the murine, human, and simian-forms of XAP2 (also known as AIP or ARA9) are components of the latent AhR complex.173–175 XAP2 displays 28.9% amino acid identity to the immunophilin FKBP52 in its amino terminus (amino acids 9–90).176 These residues of XAP2 overlap with the region in FKBP52 that harbors the peptidyl-prolyl isomerase activity associated with immunophilins. So far, however, XAP2 has not been demonstrated to possess this enzymatic activity. Also, unlike the FKBPs, XAP2 does not possess the ability to bind the immunosuppressant drugs FK506 or rapamycin,177 and XAP2 is therefore by definition not an immunophilin but is referred to as immunophilin-like. Like the immunophilins, XAP2 interacts with the receptor complex through the utilization of tetratricopeptide repeat (TPR) domains. XAP2 has three TPR motifs in its carboxy-terminal half. Its carboxy-terminal–most TPR displays homology to the TPRs in FKBP52, and is important in mediating the direct interaction between XAP2-AhR and XAP2-HSP90.81,177 In addition to the TPR motif in XAP2, amino acid residues in the extreme carboxy-terminus of XAP2 are also needed to mediate its interaction with the AhR. Alanine substitution in any of the last four amino acids in XAP2 results in the complete loss of interaction with the AhR, while HSP90 binding is still observed even when the last five amino acids are completely deleted.178
Overexpression of the mouse AhR in a cell culture causes a shift in receptor localization from the cytoplasm to the nucleus. However, the receptor can be redistributed into the cytoplasm on co-expression of XAP2, implying that AhR cellular localization may rely on the presence of XAP2 in the complex.179,180 When these cells are treated with the nuclear export inhibitor leptomycin B, the AhR remains in the cytoplasm, suggesting that XAP2 blocks the rapid nucleo-cytoplasmic shuttling of the AhR complex that normally occurs.181 A recent study by the Pollenz laboratory, however, has shown that silencing XAP2 expression in mouse hepatoma cells does not result in the nuclear accumulation of AhR, nor does it appear to alter AhR nucleo-cytoplasmic shuttling, confounding previous observations.182 To further complicate matters, overexpression of the human AhR in the absence of XAP2 coexpression does not result in nuclear accumulation of the receptor without XAP2 coexpression highlighting one of the numerous functional differences between the two species.183
It has been observed that increasing amounts of XAP2 expression in a cell culture significantly raises the total cellular concentration of the unliganded AhR.81,184 One report has attributed this effect to XAP2-mediated protection of the AhR against ubiquination, and subsequent reduced levels of proteosome-mediated degradation.185 Ectopic expression of XAP2 has been demonstrated to enhance AhR transcriptional activity. The exact level of enhancement, however, can be difficult to assess. In the presence of high XAP2 levels, elevated constitutive and induced expression of reporter genes has been observed using chimeric AhR fusion constructs with alternate DNA binding motifs, such as GAL4 or LEXA.177,179 In a yeast model system utilizing a β-galactosidase reporter gene to monitor AhR activity, the ED50 of the dose-response curve is decreased by approximately fivefold when XAP2 is expressed along with a LEXA-AhR fusion protein.177 When utilizing endogenous AhR levels in mouse hepatoma cells, however, XAP2 appears to cause a more modest (~ twofold) increase in ligand-induced AhR-driven luciferase activity, while the basal activity remains essentially unaltered.175 XAP2-mediated enhancement of AhR activity is generally attributed to the elevated cytosolic AhR levels achieved in the presence of high amounts of XAP2, thereby increasing the available ligand-binding sites in a cell. Contrasting reports show that XAP2 can act as a transcriptional repressor or have only a slight effect on AhR transcriptional activity. The Perdew lab has demonstrated that coexpression of XAP2 with the AhR results in an overall repression of human and mouse AhR activity. 181,183 It should be noted that under conditions in which coexpression of XAP2 results in repression of AhR signaling, little increase in overall AhR levels is observed.183,186 Additionally, the mutation of a single amino acid residue (tyrosine 408) in the mouse AhR disrupts XAP2 binding while maintaining HSP90, p23, and the ligand-binding potential. This mutant AhR displays increased basal and ligand-induced transcriptional activity relative to wild-type AhR.186 Furthermore, silencing of XAP2 expression in cell lines expressing different AhR alleles demonstrates that AhR stability is independent (Ahb−2) of or only modestly dependent (Ahb−1) on XAP2 expression to maintain AhR cellular levels.187 These later studies suggest that enhancement of AhR signaling by XAP2 in cell culture may be an experimental artifact due to the use of overexpression systems. This idea is supported by the recent creation of transgenic mouse lines that express high levels of hepatocyte-specific XAP2. In these mice, no increase in cellular AhR levels is observed, nor is there any change in ligand-induced AhR activity compared to wild-type animals.188 Clearly, future work must be performed to reconcile the published discrepancies with respect to AhR regulation by XAP2.
XI. TRANSCRIPTIONAL COACTIVATION OF THE AHRC
Shortly after the cloning of AhR and ARNT, several investigators set about the task of defining the functional domains of the receptor complex. In particular, attention was focused on the DNA-binding, dimerization, and transactivation capabilities of AhR and ARNT, with the latter involving studies utilizing heterologous reporter systems. These systems included chloramphenicol acetyl-transferase (CAT) and luciferase reporter plasmids serving as transcriptional readouts and, as such, represent unchromatinized templates. Gal4 fusions of AhR and ARNT and deletion mutation analysis allowed Jain and colleagues to map the basic transactivation function of both proteins to their carboxy-terminal regions.69 For AhR, this region contained three modular domains associated with transcription, namely, an acidic region, a glutamine-rich region, and a region rich in proline/serine/threonine residues, all hallmarks of previously described transactivation domains (TADs). As would be expected, these domains seem to serve different functions. For instance, the glutamine-rich region of the human receptor is necessary for the recruitment of SRC-1.189 If we take the examples of other modular transactivation domains such as those found in Sp1, Oct1, and myogenin,190,191 it can be inferred that the complexity of the AhR TAD would allow for extreme flexibility in transactivation potential. This could include allowing for the formation of a functional transcription complex at elements varying in distance from the proximal promoter of a given gene and the ability to confer a degree of specificity with regard to activating transcription from the regulatory regions of different target genes. Similarly, the carboxy-terminal portion of ARNT harbored both glutamine- and proline/ serine/threonine-rich residues.64,69 These observations were extended shortly thereafter by Sogawa and colleagues, who demonstrated the transactivation function of the three putative transactivation subdomains of AhR.192 Additionally, they suggested that the glutamine-rich region of ARNT had no transactivation function, but the 34 carboxy-terminal amino acids were essential for activity. Most importantly, the requirement of the carboxy-terminal transactivation function of ARNT, in the context of the AhR/ARNT heterodimer, was established with truncated mutants of ARNT using ARNT-negative mouse c4 Hepa1 cells.75 The glutamine and acidic domains of other transcription factors, including Sp1 and MyoD,193 have inherent transactivation functions that are capable of interacting directly with basal transcription factors.194,195 Not surprisingly, it was later demonstrated that AhR could interact directly with TFIIB, TFIID/TBP, and TFIIF,10,196 and that ARNT was capable of interacting with TFIIB and TFIIF. However, it has never been demonstrated that AhR and ARNT alone were sufficient to activate transcription in vitro in reconstituted systems with general transcription factors.
It is now well established that all transcription factors, irrespective of structure or function, appear to recruit protein complexes consisting of common pools of coactivator/corepressor proteins197,198 often described as coregulators or master regulators of gene transcription. The intrinsic functions of transcription factors are therefore modulated by the recruitment of these ancillary factors. Furthermore, they appear to regulate a wide variety of functions including chromatin remodeling, stabilization of GTFs, RNA elongation and processing, translation, DNA break formation, and transcription termination.199–202
Upon DNA binding, it is clear that the AhRC, like NRs, is capable of recruiting CBP/p300,203–205 the p160/bHLH-PAS coactivators SRC-1,189,206 NCoA2/GRIP1/TIF2, and p/CIP,206 as well as other transcriptional coactivators, including RIP140,189,207 components of ATP-dependent chromatin remodeling complexes, such as Brahma-related gene 1 (BRG-1), components of the mediator complex,208,209 and P-TEFb and RNA elongation factor.210 These proteins are incorporated into multimeric complexes, which interact with and modulate the activity of the core transcriptional machinery, as well as modifying local chromatin structure.211 However, the identity of many factors, and the mechanisms in which they are recruited by the AhRC to its cognate response element, are largely unknown. A recent review by Hankinson provides a comprehensive overview of coactivator function and AhRC-dependent gene activation and we will not attempt to reiterate it here.212 However, there have been a few additions to the pantheon of putative coregulators for AhRC transcriptional activity. Table 2 provides a list of most, if not all, of the putative transcriptional coregulators known to interact with AhR and/or ARNT. It is important to note that most of these studies have been performed in the context of either synthetic DRE concatomers or the CYP1A1 promoter.
TABLE 2.
AhR- and ARNT-Interacting Proteins
| Name | Function | Reference |
|---|---|---|
| CBP/p300 | HAT | 205 |
| SRC-1/NCoA1 | 189, 206 | |
| NCoA-2/GRIP1/SRC-2 | 206 | |
| P/CIP,SRC-3/AIB1/ACTR | 206 | |
| ERAP-140 | 213 | |
| Brg-1 | Chromatin remodeling | 209 |
| Med220 | 208 | |
| CDK8 | 208 | |
| TRIP230 | Coactivation/Unknown | 214 |
| CoCoA | 215 | |
| GAC63 | 216 | |
| NcoA-4 | 217 | |
| BRCA1 | 218 | |
| Rb | 219 | |
| Mybbp1a | 220 | |
| PML | 221 | |
| Nedd8 | 222 | |
| SMRT | Corepressor/coactivator | 213, 223 |
| RIP140 | 207 | |
| P-TEFb | Elongation factor | 210 |
| TFIIB | General transcription factor | 196 |
| TFIID/TBP | 224 | |
| TAF4 | 224 | |
| TAF6 | 224 | |
| Sp1 | Transcription factor | 225 |
| ERα/β | 226–229 | |
| ERRα1 | 228 | |
| COUP-TF1 | 228 | |
| NF-κB | 230 | |
| UBC9 | Sumo E3 ligase | 221 |
| CUL4B | 13 |
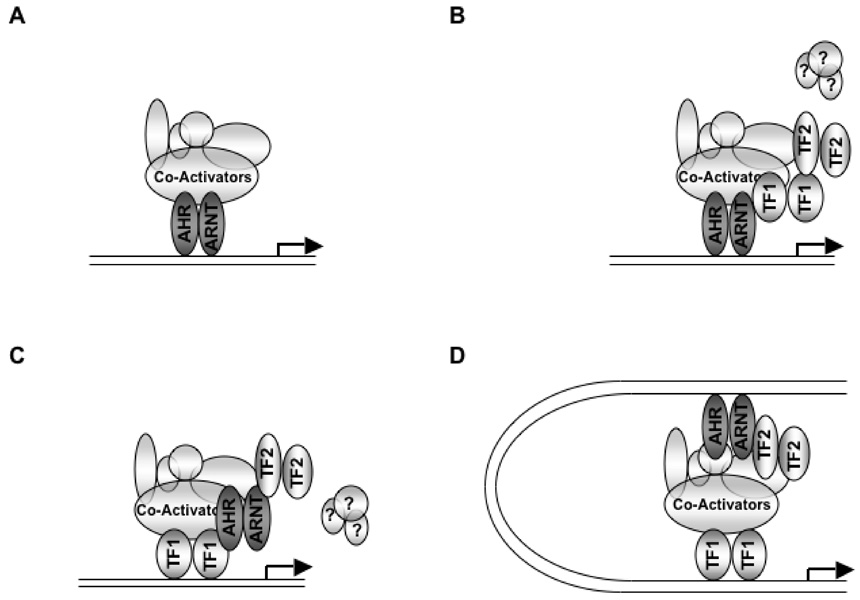
Because of its heterodimeric nature, the AhRC is capable of building multimeric complexes on multiple transcriptional activation platforms, namely, AhR and ARNT. Indeed, many coactivators are selectively recruited into this complex by ARNT, including CBP, BRCA1, and TRIP230.205,214,218 The gene for thyroid hormone receptor (TR)/retinoblastoma protein (Rb) interacting protein 230 (TRIP230) was originally identified independently by two different labs based on its ability to interact with TR231 and Rb,232 respectively. In addition, it was demonstrated that Rb negatively regulated the TRIP230 coactivation function of TR-regulated transcription.232 The manner in which Rb facilitates repression is less clear, although it may involve sequestration of TRIP230.232 However, this does illustrate a salient point, which is that transcription is tightly regulated at many levels. It may be that Rb attenuates the coactivator function of TRIP230 in order to ensure that transcription of TRIP230-regulated genes proceeds at an appropriate rate and in response to appropriate temporal or contextual signals. As was the case with TR and Rb, TRIP230 was identified as an ARNT- interacting protein by the yeast two-hybrid assay.214 Microinjection of Hepa-1 cells with affinity purified antibodies and transient transfection with siRNAs against TRIP230 demonstrated that this coregulator was essential for both TCDD and hypoxia responsive transcription. The dependency of TRIP230 on Rbs ability to modulate AhRC function has not yet been thoroughly investigated. However, a direct interaction between AhR and Rb has been demonstrated, 219 and the ability of the AhRC to drive transcription in rat 5L hepatoma cells is somewhat dependent on Rb.233 For an overview of AhR/Rb function, particularly with regard to AhR cross talk with factors governing cell cycle, readers are referred to reviews by Puga and colleagues234 and Cornelius Elferink.235
Similarly, BRCA1 is recruited to the AhRC complex by virtue of its interaction with ARNT.218 Thus, prompted by the observation that BRCA1 regulates numerous genes involved in xenobiotic stress responses,236 investigators in the laboratory of Insoo Bae carried out a series of insightful experiments to determine its effect on TCDD-responsive CYP1A1 and CYP1B1 transcription. Using conventional structure-activity approaches and siRNA, they demonstrated that BRCA1 is indispensable for AhRC transcriptional activity and that this coactivation function is mediated by ARNT.218 Furthermore, this interaction is mediated through a domain of BRCA1 not associated with its own intrinsic transactivation function. Finally, the non-p160-related coactivator NcoA4/ARA70 has been demonstrated to enhance AhRC-driven transcription in a heterologous reporter system.217 Interestingly, two splice variants of NcoA4 are expressed, with the shorter isoform being the dominant species in most cancer cell lines.237,238 Furthermore, Kollara and colleagues demonstrate that the short isoform’s (NcoA4β) ability to coactivate AhRC-dependent transcription is significantly diminished.217 The fact that BRCA-1, NcoA4, and Rb are deregulated, or functionally ablated in some fashion, in many cancers/cancer cell lines is intriguing.
As mentioned above, these early studies relied heavily on unchromatinized reporter plasmids as a readout of transcription. Therefore, it is unlikely that these systems would recognize interactions with proteins whose contribution to transactivation is based on the ability to modify chromatin structure or whose activity is dependent on a chromatin environment. The lone study regarding AhR on a chromatinized template focused solely on the acidic domain of AhR239 and did not address the requirement of cofactors in a chromatinized setting. Although all putative coactivators of AhR function enhance DRE-driven reporter constructs in mammalian systems, it is clear that not all coactivators mediate their effects purely in an AhR and ARNT-TAD dependent fashion. Studies to determine the interaction domain of the coiled-coiled coactivator, CoCoA, GAC63 and the p160 steroid receptor coactivator, SRC-1, revealed that both the b-HLH-PAS and Helix1 domain of AhR were essential for these interactions, respectively.206,215,216 Furthermore, microinjection assays with antibodies to SRC-1 revealed that SRC-1 was absolutely essential for the transcriptional activity of the AhRC.206 Therefore, although CoCoA, GAC63, and SRC-1 are capable of forming complexes with other coactivators that are dependent on AhR’s TAD for protein-protein interactions (i.e., CBP, GRIP1, and p/CIP), it is apparent that transiently transfected heterologous reporter plasmids cannot adequately describe the contribution of all factors to a receptor’s transactivation potential.
The transcriptional activation domains of AhR and ARNT are structurally different from nuclear receptors and several other classes of transcription factors, with at least three distinct domains representing novel platforms on which transcriptional machinery can be assembled. In the case of nuclear receptors, separate amino and carboxy terminal TADs have been identified, termed AF-1 and AF-2.240 Furthermore, these domains are characterized by the presence of alpha-helical motifs. Presumably, these structural differences allow for the assembly of different complexes conferring different specificities, although one might note that these structurally divergent receptors share a common pool of coactivators. In fact, a coactivator that is recruited to the AhR and not to an NR has not been described. Therefore, it seems likely that coactivators can be recruited in different fashions to exploit different properties. The nature in which the coiled-coiled coactivator, CoCoA, is recruited to different classes of transcription factor may be a good illustration of this point. Nuclear receptor recruitment of CoCoA is mediated by the p160 coactivator, GRIP1, and this is by virtue of a direct interaction with the bHLH-PAS domain of GRIP1.241 In addition, it appears that CoCoA is necessary for the synergy that exists between GRIP1, CARM1, and p300241 in the context of nuclear receptor-mediated transcription. In direct contrast to this, the AhRC recruits CoCoA in a p160-independent process and the requirement for CoCoA and CARM1 function in this context is unclear.215 In the case of β-catenin, CoCoA may serve as the scaffold on which GRIP1 is recruited to β-catenin–regulated genes.242 Therefore, it is apparent that many of these master regulatory proteins are multifunctional and the diversity of transcription factor structure allows for the formation of transcriptional machinery utilizing these factors in multiple configurations. Thus, examination of the coactivator-AhR literature reveals that the AhRC has much in common with nuclear receptors.
XII. TRANSCRIPTION FACTORS AND OFF-TARGET EFFECTS
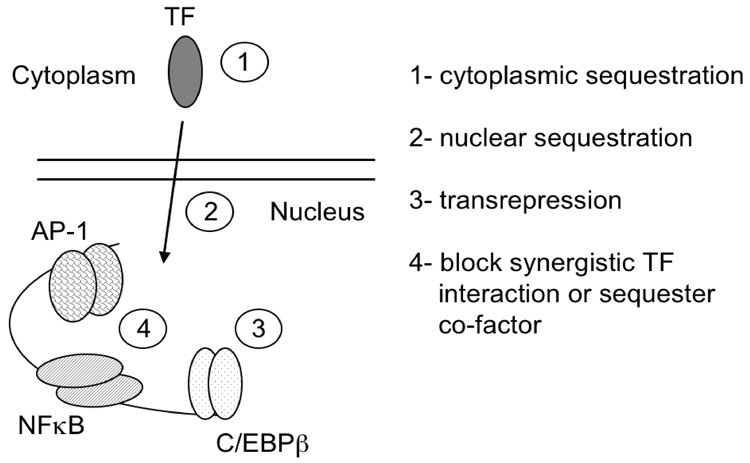
An intrinsic quality of many transcription factors’ function, whose importance is increasingly being recognized, is the ability to modulate transcription in a DNA-binding independent fashion. As with most transcription factors, nuclear receptors are thought to manifest their main biological functions by transducing the transcriptional information of their cognate response elements. This has been questioned over the past decade by several independent findings. Investigators in Gunther Schutz’s laboratory made the startling observation that DNA-binding/dimerization–deficient glucocorticoid receptor (GR) mutant mice were viable,243 whereas null mutations were lethal.244 These observations demonstrated unequivocally, for the first time, that the DNA-binding capability of a transcription factor is not essential for survival but that the non-DNA binding properties are.243 Furthermore, AP-1 regulation of interstitial collagenase was repressed by GR through a direct protein-protein interaction between GR and AP-1, a phenomenon termed “transrepression245–248 GR-mediated transrepression of NF-κB signaling is also well documented.249–251 Subsequently, other nuclear hormone receptors, including the thyroid hormone (TR) and estrogen receptor (ER), were shown to be able to repress AP-1 and NF-κB activity via direct protein-protein interactions.252–254 The androgen receptor (AR), on the other hand, appears to transrepress Smad and Ets-mediated transactivation,255,256 and to be transrepressed itself by AP-1.257 Therefore, it seems plausible that the intrinsic transrepression and coactivator functions of some transcription factors are as important as their respective DNA-binding functions. Furthermore, and certainly in the case of ER, the effects of endocrine disruptors and compounds that mimic the activities of endogenous hormones affect not only estrogen response element (ERE)-regulated genes, but also genes transrepressed/coactivated by ER. However, the relative importance of each phenomenon has yet to be established.
The nature of repression by tethering of ligand activated NRs is not well understood. The classic model of NR activation by ligand suggests that ligand facilitates an exchange of NCoR/SMRT/Sin3/HDAC corepressor complexes for coactivator complexes with ATP-dependent chromatin remodeling and histone acetyl-transferase activities.197,198 It has been suggested that this happens only in the context of a receptor’s own cognate positive response element. Studies regarding the repression of thyrotropin b gene by TR suggest that this switch does not occur in the context of a negative TRE.258 Other studies have suggested that GR-mediated repression of the IL-8 gene occurs through a direct protein-protein interaction with the NF-κB heterodimeric transcription factor, and is independent of GR binding to its cognate DNA response element.259 They present evidence that GR recruits GRIP1 but a steric change in the tethered complex unmasks GRIP1’s intrinsic repressor function.260 This, in turn, may play a role in the GR-mediated inhibition of phosphorylation of the carboxy-terminal catalytic domain of RNA polymerase II.261 Whether AhR exhibits similar properties is just beginning to be explored.
XIII. AHR AS A MEDIATOR OF ENDOCRINE DISRUPTION
Several indirect lines of evidence exist that would argue that AhR may mediate off-target or non-DNA binding dependent transcription. Arguably, the strongest support for this theory is provided by the ability of diverse classes of AhR ligands to perturb endocrine function. Even a cursory survey of the literature will make it readily apparent that different investigations and observations across species and tissues are often conflicting or paradoxical. This should serve to reinforce the reality that effects are contextual and highlight the need for investigations into the molecular nature of cross talk between AhR and other signaling pathways. Nevertheless, some of the effects mediated by AhR activators on nuclear receptor–regulated systems are well established.
TCDD and certain poly-chlorinated biphenyls are potent repressors of thyroid function. Rats administered single doses of TCDD were observed to have decreased thyroxine levels,262 and this is apparently accompanied by a concomitant increase in levels of thyroid stimulating hormone (TSH).263 A study involving U.S. Air Force veterans who had been exposed to TCCD in Vietnam also revealed that they had abnormally high levels of TSH.264 TCDD and glucocorticoids at high doses are known teratogens, although they elicit their effects through different mechanisms and, in the case of cleft palate in mice, the defects are morphologically different (for review, see 265). Studies investigating the synergistic effects of glucocorticoids and TCDD in the formation of cleft palate in mice revealed that glucocorticoids upregulate AhR message and protein levels, while TCDD upregulates GR.266 This effect of TCDD has also been observed in the rat ovary.267
Peroxisome proliferator–activated receptors (PPARs) also are affected by dioxin and dioxin-like chemicals in an AhR-dependent fashion. Dioxin downregulates the expression of PPARγ in 3T3 cells during adipogenesis.268 Subsequently, researchers in the laboratory of Jefcoate, using wild-type and immortalized AhR−/− mouse embryonic fibroblasts, determined that this is likely due to AhR-dependent repression of adipogenesis.269 Concentrations of the PPARα activator WY-14643 that do not increase AhR mRNA levels potentiate CYP1A1 expression by 3-MC,270 while, paradoxically, clofibrate represses AhR-dependent CYP1A1 and CYP1A2 expression in rat liver.271 Certainly, multiple diverse mechanisms play a significant role in the effects described above, including alterations in ligand metabolizing or synthesizing enzymes, direct activation or repression of target genes, and alterations in mRNA stability and protein turnover. The remainder of this review will focus on attempts to describe the molecular events that occur in response to activation of AhR and how direct protein-protein interactions affect the activity of the AhRC and other transcription factors.
XIV. AHR AND ER
By far the most extensive studies involving cross talk between AhR and another transcription factor are those involving the estrogen receptor alpha (ERα). The antiestrogenic properties of TCDD have been documented repeatedly over the last 20 years, beginning with the observations that TCDD repressed estradiol function in rat uterus and liver.272–276 Subsequently, HAHs and PAHs were implicated in the repression of ER-driven transcriptional activity. However, part of this phenomenon may be due to structural similarities that exist between PAHs and estrogenic compounds, and some overlap in the spectra of their respective activities may exist. Diethylstilbestrol (DES), one of the earliest clinically useful synthetic estrogens was originally synthesized from a polycyclic aromatic hydrocarbon277 and is linked to an increased incidence of vaginal and cervical adenocarcinoma.278,279
The means by which AhR activation inhibits ER-regulated gene expression pathways have been the subject of much study in the past and are relatively well understood. TCDD inhibits the expression of several E2-inducible genes, including pS2,16 cathepsin D,280–282 c-fos,283 and cyclin D1,284 among others. The first explanation proposed for these phenomena was that it was due to increased metabolism of E2 mediated by the TCDD-induced expression of CYP1A1 and 1B1.285–287 Subsequently, evidence to support other theories has emerged. For example, TCDD-mediated inhibition of E2-induced pS2 expression in BG-1 cells was blocked by the presence of the protein translation inhibitor cycloheximide.288 This suggests that TCDD induces the expression of another inhibitory factor of E2-mediated gene induction, such as activation of proteasomes leading to increased ER degradation.15 A direct transcriptional mechanism for TCDD-induced repression of E2 signaling was proposed with the identification of an inhibitory DRE in the upstream region of the cathepsin D (catD) gene.289 More recently, a direct interaction between AhR and ER, or transrepression, was proposed.229 However, this was confounded by the observation that 3-methylcholanthrene at relatively high concentrations (10 µM) could induce both ERE-driven luciferase activity and BrdU incorporation in the glandular epithelium of mouse uterus.229 This observation was supported by other studies that identified several AhR agonists, including 3MC and B[a]P, as ligand activators of ERα.290 Finally, the most simplistic explanation for TCDD-mediated inhibition of E2 activity is the competition for a common pool of coactivators, although this hypothesis has not been pursued rigorously.
Pre-natal, peri-natal, and long-term exposure to estrogens, phyto-estrogens, and endocrine disruptors such as TCDD, put individuals at an increased risk of multiple forms of cancer and developmental defects of the urogenital tract.291–294 Many testicular cancer etiologies implicate the activation of the AhR294–296 or conversely, the disruption of the estrogen signaling pathways by these compounds and other endocrine disruptors such as diethylstilbestrol. Subsequent studies continue to reiterate these findings.297,298 Furthermore, clinical applications of ER agonist and antagonist therapies are associated with many ER-mediated side effects, including breast and endometrial cancer, thrombosis, sexual dysfunction, CNS effects, and estrogen-independent tumors.299 As a result, cross talk between AhR and ER pathways have been implicated, but studies investigating the molecular mechanisms underlying AhR-ER cross talk are scarce. GST-pull-down and in vivo coimmunoprecipitation studies have demonstrated a direct interaction between AhR and ER,226,228,229 as well as ARNT and ER.227 Furthermore, dominant negative studies have shown that the AhR TAD and the N-terminal region of ER can reciprocally repress transcription from ERE- and DRE-driven luciferase constructs, respectively.300 TCDD exposure causes a robust increase in CYP1A1 mRNA and DRE-driven reporter gene activity, which are almost completely abrogated by cotreatment with E2 in Hepa-1 and MCF-7 cells.301 Results similar to these were obtained in the human endometrial carcinoma cell line, ECC-1.302 A more recent study showed no effect of E2 on 3-methylcholanthrene-inducible AhR activity, but failed to investigate the effects of E2 on HAH-mediated AhRC activity.229 We have demonstrated that estradiol-activated ERα directly represses AhRC activity at the transcriptional level via tethering to the AhRC in MCF-7 cells.226 However, others have reported the E2-inducible enhancement of TCDD-dependent CYP1A1 expression in T47D human breast cancer and Huh7 liver cell lines, while having little or no effect in MCF-7 cells.303 This disparity could reflect differences in cell culture conditions or of relative abundance of essential cofactors in different cell lines. Most studies concerning endocrine disruption by AhR ligands have focused on the impact of AhR activation on ER target gene induction using transiently transfected cell lines, transcription reporter systems, and ex vivo approaches.227,228,283,288,300–306 However, a recent study using AhR “knockout” mice implicates AhR in the regulation of Cyp19 and ultimately in estrogenesis, suggesting that the disruptive effects of dioxin and other exogenous AhR ligands on endocrine physiology may be due to the untoward activation of AHR target genes, and may not be due to a direct effect on ER function.307 In addition, AhR knockout mice do not reproduce easily.308 Therefore, it seems imperative to elucidate the molecular mechanisms involved in AhR transcriptional processes and, in particular, ER-AhR cross talk. It is also worthy to note that most experimental efforts to characterize the biochemical pathways involved in ER-AhR cross talk have focused on the TCDD-inducible repression of ER target genes.
Other studies have placed ARNT227–229 and AhR229 at the promoter elements of E2-responsive genes, again with disparate results. In one study ARNT was directed to the pS2 and c-fos promoters only in response to 3MC in MCF-7 cells,229 whereas in another study ARNT was implicated as an ER coactivator associating with the pS2 promoter in an E2-dependent fashion in T47D cells.227 These discrepancies may reflect differences in the cell lines used and the experimental techniques used to gauge each parameter. One possible explanation is that these studies relied heavily on synthetic response elements to drive heterologous reporters for transcriptional readout. Nevertheless, the in vitro and ex vivo observations of Ohtake and colleagues were, to a great extent, validated by their in vivo observations in wild-type and mutant mouse models.229 Their observation that 3-MC alone could activate ER target genes was confirmed by several other independent studies that established that 3-MC and other PAHs are weak agonists of ERα.290,309,310 The Kato group later expanded this work with the report that AhR is part of a ligand activated E3 ubiquitin ligase complex.13 In turn, this cullin 4B-dependent process is responsible for the ultimate degradation of ERα.
Cotransfection of ERα with a CYP1A1 promoter–driven luciferase vector in Hepa1 and MCF-7 cells treated with TCDD, with or without E2, leads to an E2-mediated repression of TCDD-dependent luciferase activity.226 However, heterologous reporter systems are inadequate for studying the molecular mechanisms of receptor “cross talk” and transrepression. These systems cannot distinguish between direct transcriptional events at the target and secondary transcriptional events such as the transcriptional activation/repression of other regulatory genes. Therefore, in order to assess the direct transcriptional effect of ER activation on AhRC-dependent gene transcription, studies employed reverse-transcription/real-time PCR of the AhRC target genes CYP1A1 and CYP1B1 in the presence and absence of the protein translation inhibitor, cycloheximide. TCDD caused a significant increase in CYP1A1 gene transcription in the human breast cancer cell line, MCF-7, whereas 100 nM E2 did not.226 However, E2 (like TCDD alone) caused a significant increase in CYP1B1 gene transcription, consistent with reports that the 5′’ regulatory region of the CYP1B1 gene harbors a functional ERE.311 The addition of 100 nM estradiol significantly abrogated TCDD-mediated CYP1A1 gene induction in the presence or absence of cycloheximide.226 The observation that maximal CYP1A1 gene induction was repressed by E2 by approximately 50%, with or without cycloheximide, suggests that direct transcriptional repression of CYP1A1 is responsible for the decreased mRNA and protein levels observed, and is not due to another downstream posttranscriptional mechanism(s). Furthermore, E2 is capable of repressing TCDD-inducible CYP1A1 protein levels in a dose-dependent fashion (Beischlag and Perdew, unpublished data).226 Reciprocally, TCDD repressed E2-inducible pS2 and PR mRNA levels in MCF-7 cells (Beischlag et al., unpublished data), a phenomenon observed with other ER-regulated genes and reporter constructs.302,306,310 These models are represented schematically in Figure 2.
FIGURE 2.
Proposed mechanisms of AhR/ARNT-mediated transcriptional regulation. (A) Ligand-activated AhR-ARNT heterodimers bind their cognate response element in the regulatory regions of their target genes, assemble coactivator machinery, and facilitate transcriptional activation. (B) In combination with other stimuli, transcription factor tethering (TF1) to AhR, including the combinatorial recruitment of other classes of transcription factors (TF2), or (C) AhR/ARNT tethering to transcription factors at their cognate response elements. (D) DNA bending allowing AhR/ARNT DRE-bound complexes to interact with other DNA-bound transcription factors.
Studies performed in vitro using glutathione-s-transferase pull-down assays were able to demonstrate an interaction of ERα with either AhR or ARNT.226,228 Furthermore, one study demonstrated that the ERα interaction domain within AhR resides within the P/S/T region of AhR’s TAD.226 However, in this study, 100 nM E2 abrogated both wild-type AhR and AhRΔP/S/T TCDD-inducible reporter gene activity. Therefore, the ER-ARNT interaction may be more physiologically relevant to the transrepression phenomena than the ER-AhR interaction. In addition, transcriptional activity driven by a GAL4-ARNT chimera was repressed dramatically by overexpression of ERα in an E2-dependent fashion, further suggesting that ERα may transduce its repressor function via its interaction with ARNT. This functionality is further supported, if only tangentially, by the observation that ER may recruit ARNT for a coactivator function during E2-responsive transcription.227
Recently, two separate models involving ER and AhR function demonstrated the complexity of this interaction. Hockings and colleagues have identified a second level of complexity of AhR-ER interaction, which further expands our understanding of transcription factor function.312 Their model proposes that the unliganded AhRC would tether to ligand-activated ER, which in turn would be capable of being recruited to Fos/Jun heterodimers at AP1-responsive elements in a CBP/p300-dependent fashion. They also provide evidence that this scaffolding is XRE dependent; that is to say, DNA bending allows for the AhRC to bind its cognate response element and interact directly with ER tethered to AP1 (Fig. 2D). Studies using FRET and ChIP have demonstrated that ligand activated AhR disrupts the synergy between ERα and Sp1 at the CAD gene promoter.313 Whether this is a promoter-specific phenomenon is unclear. We have observed Sp1 at the pS2 promoter in MCF-7 cells in response to E2, but this seems unaffected by cotreatment with TCDD (Beischlag et al., unpublished data). Whether either of these phenomena is mediated by the CUL4B or another E3 uquibitin ligase complex remains unknown, nor have other potential repressor mechanisms been explored. The interaction between the nuclear corepressor SMRT and AhR may offer some insights into potential mechanisms of AhR mediated cross talk with NRs. Widerak and colleagues have demonstrated that RXR-heterodimer–responsive genes (e.g., TR and RAR) are activated by TCDD by sequestration of SMRT.314 However, nuclear receptors that form homodimers (i.e., ER and PR) are repressed by TCDD. The notion that AhR may recruit a repressor function to NR homodimers is as plausible as it is provocative. Clearly, additional studies are needed to determine the importance of AhR-ER tethering to the overall phenomenon of gene regulation.
XV. CROSS TALK BETWEEN AHR AND INFLAMMATORY SIGNALING PATHWAYS
Transcriptional control of inflammatory signaling is mediated by a variety of transcription factors, including NFκB, AP-1, STAT3, and C/EBP. Many genes that are activated during inflammation contain promoter regions with multiple regulatory elements that often are synergistically activated by a combination of regulatory proteins bound to their cognate response elements. Transcriptional regulation of these genes is further complicated by the combinatorial complexity of dimer formation by various members of the NFκB and AP-1 families of transcription factors, as well as by their posttranscriptional modification status. The NFκB family consists of p65 (RelA), c-Rel, Rel-B, p50, and p52.315 Both p65 and p50 are maintained in the cytoplasm through the ability of IκB to sterically hinder importin binding to its nuclear localization sequence.316 An increase in NFκB translocation into the nucleus occurs through an induction of toll-like receptor (TLR) signaling by bacterial products such as lipopolysacharide (LPS) or cytokine production. TLR activation leads to an increase in IκB kinase (IKK) activity that in turn causes phosphorylation of IκB on serines 32 and 36. These modifications mediate proteolytic turnover of IκB, release of p65/p50, and its subsequent translocation into the nucleus. The transcriptionally competent p65/p50 dimer binds to its cognate element, often displacing a p50 homodimer, which is in effect a switch from a repressive to a actively remodeled chromatin state on the proximal promoter of NFκB-regulated genes.317
The AP-1 family of transcription factors are key mediators of oxidant stress and inflammatory signaling.318 Members of this family include c-Fos, FosB, Fra1, c-Jun, JunB, JunD, and ATF2. These factors can form both homo- and heterodimers and bind to several related core DNA response elements. The C/EBP family of transcription factors also frequently participates in the regulation of genes targeted by inflammatory signaling. These basic-leucine zipper proteins bind to their cognate DNA response elements as homodimers. In particular, C/EBPβ and C/EBPδ are dramatically upregulated by IL-6 or TNF-α and appear to play an important role in sustaining the transcriptional response to inflammation.319,320 An excellent example of combinatorial transcriptional regulation of inflammatory signaling can be observed on the enhancer/promoter of the IL-8 gene.321 Within the proximal promoter, there are functional response elements for AP-1, C/EBPβ, and NFκB. These factors act together to synergistically activate IL-8 mRNA production. Another example of synergistic regulation of inflammation-mediated gene expression can be seen on the promoter of the COX2 gene. Both C/EBPβ and NFκB are required for high-level induction of COX2 mRNA expression.322
For several decades it has been shown that inflammation leads to repression of drug metabolism and, in particular, expression of a number of cytochrome P-450s is suppressed.323 The ability of IL-1β, IL-6, TNF-α, and LPS to suppress constitutive and AhR ligand-inducible CYP1A1 activity and protein levels in Hepa 1 cells has been firmly established.324–326 Cytokines also reduce constitutive and AhR ligand-mediated CYP1A1 mRNA levels and enzymatic activity in human hepatocytes.327,328 Reduction in CYP1A1 expression has also been observed in the brain after cytokine exposure.329 Whether these effects are due to direct transcriptional repression or changes in mRNA stability has been addressed in rat hepatocytes.330 Recombinant IL-1 suppressed the rate of transcription of both CYP1A1 and CYP1A2, as measured by nuclear run on assays. One possible mechanism for this repression was explored through studies examining the role of H2O2 and NF1 in CYP1A1 gene regulation.331 Oxidation of NF1 decreases CYP1A1 activity, as determined using a CYP1A1 enhancer/promoter in reporter assays. Whether this mechanism occurs on short-term treatment with cytokines needs to be tested. Another possible mechanism would include the ability of cytokine-activated transcription factors to bind to the AhR/ARNT heterodimer at the CYP1A1 promoter. Support for this mechanism includes the ability of RelA to physically associate with the AhR in Hepa 1 cell lysates.230 ChIP assays have revealed that LPS and TNF-α treatment of Hepa 1 cells in the presence of TCDD leads to a decrease in histone H4 acetylation, yet does not alter the amount of AhR/ARNT bound to the promoter.332 Expression of a mutant form of IκB, which acts as a potent repressor of NFκB, leads to rescue of TNF-α–induced repression of liganded AhR-mediated transcriptional activity. Taken together, these results would suggest that RelA/p50 tethers at the CYP1A1, leading to transcriptional repression. However, the presence of RelA at the CYP1A1 promoter has not yet been established. Thus, additional studies are needed to determine the exact transcriptional mechanism of cytokine-mediated repression of CYP1A1. Another question that also needs to be addressed is whether NFκB can repress other genes regulated by the AhR. Investigators are currently utilizing the combination of quantitative RT-PCR, siRNA, and ChIP assays to obtain additional mechanistic information to aid in our understanding of the transcriptional mechanisms involved.
Numerous reports have demonstrated that phorbol esters lead to repression of AhR-TCDD–mediated CYP1A1 expression. The phorbol ester 12-O-tetradecanoylphorbol acetate (PMA) has been utilized as a potent activator of protein kinase C (PKC), which ultimately leads to down-regulation of PKC. Studies both in cell culture and in mice have suggested that the repression of AhR ligand-induced CYP1A1 expression after PMA treatment is due to the loss of PKC activity.333,334 Indeed, studies suggest that AhR transcriptional activity is repressed on inhibition of PKC activity as measured in cell-based DRE-driven luciferase reporter studies.335 However, there are also other possible mechanisms that may explain the ability of PMA to repress CYP1A1, and that is through its ability to induce an immediate early gene response. PMA is able to dramatically induce AP-1 activity both through the transcriptional upregulation of the Fos and Jun genes and direct phosphorylation of these transcription factors.336 In addition, oxidant stress levels are increased by PMA treatment, leading to NFκB activation.337 Thus, it is quite plausible that PMA-directed repression of CYP1A1 may be mediated through a mechanism similar to what has been observed with the cytokines discussed previously.
XVI. EVIDENCE FOR AHR-MEDIATED REPRESSION OF NFκB AND AP-1 TRANSCRIPTIONAL ACTIVITY
Sustained activation of the AhR by coplanar PCBs, TCDD, and PAH exposure has been linked to an enhancement of inflammatory signaling in numerous cell culture and in vivo models.338–340 Much of this data is based on relatively high dose exposure to AhR ligands that would lead to highly elevated and prolonged cytochrome P4501A1 activity and other DRE-mediated gene expressions. Recently, a number of reports have challenged this view, and may actually indicate that conditions such as transient or constitutively low levels of AhR activity may actually inhibit cytokine production in certain cell types in the presence of an inflammatory signal. For example, the ability of AhR agonist to repress LPS-mediated IL-6 mRNA transcription has been demonstrated to occur in bone marrow stromal cells.341 In the stromal cell system, a reduction in p65/p50 DNA-binding activity was observed on cotreatment with an AhR ligand and LPS, compared to LPS alone. Treatment of C6 glioma cells with dbcAMP/theophyline leads to enhanced IL-6 gene expression that appears to be mediated, at least in part, by STAT3 activation. Cotreatment with β-naphthoflavone, an AhR agonist, represses dbcAMP/theophyline-mediated activation of the IL-6 promoter.342 However, this report did not firmly establish that the effect of β-naphthoflavone on IL-6 transcription was mediated by the AhR. Support for the ability of the AhR to inhibit NFκB activity can be found in cell-based reporter studies that demonstrate that activation of the AhR leads to repression of NFκB-mediated transcription.230 Furthermore, biochemistry studies have shown that the AhR can associate with Rel A in Hepa 1 and dendritic cells.230,343 In the dendritic cell line DC2.4, the AhR inhibited Rel A/p50 nuclear translocation. However, it is important to note that neither study demonstrated that activation of the AhR repressed transcription of an NFκB target gene. Further support for a role of the AhR in repressing inflammatory/immune signaling can be seen in studies performed in AhR null mice. The lung of AhR-null mice exposed to cigarette smoke or endotoxin exhibit greater neutrophilia in vivo and enhanced TNF-α and IL-6 levels in bron-cholveolar lavage cells in vitro.344 Also, the absence of AhR expression can cause premature Rel B degradation, which may explain the heightened response to inflammatory stimuli observed in the lung. In another study, examination of in vitro differentiation of Th cells revealed heightened expression of IL-4 and IFN-γ.345 Thus, the expression of the AhR appears to be important in the regulation of immune signaling.
The possible interplay between the AhR and other transcription factors, such as AP-1 and C/EBP, has been explored to a limited extent. Dioxin has been shown to induce c-Jun, jun-B, jun-D, and c-Fos in both an AhR-dependent and AhR-independent manner in Hepa 1 cells maintained under basal cell culture conditions.346 However, studies in CH12LX B cells revealed that activation of the AhR resulted in a marked repression of c-jun mRNA expression and AP-1 DNA-binding activity.347 These results appear to indicate that the relationship between the AhR and AP-1 may be cell-type specific. There are no reports that reveal that the AhR can directly interact with AP-1 or its components. However, it is quite possible that in the context of a specific promoter they may then interact. For example, both AP-1 and the AhR have been shown to directly regulate CYP1A2 gene expression, although whether these factors interact at the promoter of the CYP1A2 gene has not been reported.100,348 It has been established that the AhR can interact directly with the estrogen receptor.226,228 This fact, coupled with the ability of ER to bind directly to AP-1, may indicate that the AhR could be present at occupied AP-1 response elements through its interaction with ER.349 In fact, this model has been proposed to occur at the BRCA-1 promoter in the presence of estrogen.312 Neither the AhR nor ARNT has been shown to directly interact with C/EBP. However, the presence and functional interplay between C/EBP and AhRC has been established at the promoter of glutathione S-transferase Ya (GST Ya) and CYP1A1 genes.350,351 In the case of the GST Ya promoter, there are functionally over-lapping C/EBPα and AhR response elements, suggesting that C/EBPα and the AhR may physically interact. Interestingly, induction of prostaglandin endoperoxide H synthase-2 by TCDD in rat hepatocytes appears to be mediated by both the AhR and by the ability of TCDD to increase overall C/EBP levels.352
The ability of nuclear receptors and NFκB signaling to mutually repress transcription mediated by these factors has been firmly established 353–355 To gain insight into how the AhR and NFκB may interact at gene promoters, it is useful to examine the nature of the cross talk between nuclear receptors and NFκB. Numerous nuclear receptors have been shown to interact with RelA/p50, including, GR, ER, PPARγ, AR, and PXR. Perhaps the most studied nuclear receptor in terms of repression of NFκB transcriptional activity is GR. Over the years a number of mechanisms have been proposed to explain GR-mediated repression of NFκB, and indeed probably multiple mechanisms mediate the overall repression. However, it has recently been recognized that transrepression or tethering of GR to NFκB at the promoter of its target genes may be the primary means of gene repression. The key question to address is whether and how the AhR interferes with transcription mediated by transcription factors activated by inflammatory signaling. Many genes regulated by NFκB are also regulated by CEBP/β, AP-1, and/or STAT3 on exposure to cytokines and other inflammatory signals. Several possible mechanisms that could lead to repression of inflammatory signaling are outlined in Figure 3. The ability of the liganded AhR to activate or repress genes involved in inflammatory signaling probably will be cell-type and promoter-context specific. Thus, the exact transcriptional mechanism(s) that mediate these effects warrants further investigation and may lead to new therapeutic approaches.
FIGURE 3.
Possible sites of AhR repression of inflammation-mediated transcription.
XVII. CONCLUSION
As with any field of research, certain discrepancies in the literature cannot be resolved immediately. Undoubtedly, this reflects the complexities of biological systems and the inadequacies of the experimental techniques that we continue to employ. Cloning of the human genome, the availability of new technologies that allow us to study protein function and gene expression in vivo, and examples of mechanistic phenomena in other biological systems have afforded us insights into AhR function that were previously unrecognized or underappreciated. Nonclassical mechanisms of gene expression modulation represents a relatively new avenue for transcription factor research in general, and may be the next appreciable step forward in our understanding of AhR function. Certainly, the field has suffered from a lack of attention given to the ability of the AhR to attenuate the activity of other transcription factors. Future challenges will include elucidating the mechanisms and associated enzymatic activities of cofactor recruitment and identification of target genes whose expression is modified in response to complex mixtures/combinations of xenobiotics and endogenous signals. Almost certainly, proteomics and DNA array technology will play a large role in these endeavors. The absolute number and the identity of genes whose expression is regulated by the AhR is only partially known and conventional DNA array technologies are likely not sensitive enough to detect these genes. However, the combination of sophisticated bioinformatics algorithms, along with chromatin I.P./DNA array strategies such as chromatin immunoprecipitation on tiled arrays (ChIPotle)356 and ChIP-DNA selection and ligation357 (DSL), provide researchers with powerful tools for identifying target genes whose expression is modified in response to transcription factor tethering.
The challenge that we continue to face is identifying the targets of AhR action that are vulnerable to regulatory perturbations and put an individual at risk to the development of cancer. Undoubtedly, attention must be given not just to genes whose expression is enhanced or repressed, but to other potential biomarkers as well, such as micro-RNAs or regions of chromatin susceptible to epigenetic modification in response to combinatorial stimuli. This would also include related factors such as imprinting that may further explain the maternal and generational effects observed in the progeny of exposed individuals. Certainly the identification of biomarkers could provide the impetus for the rational development of pharmaceuticals based on studies with selective aryl hydrocarbon modulators (SARMs), selective estrogen receptor modulators (SERMs), and other nonclassical ligands. Already, the benefits of some SERMs are being recognized for their potential to elicit nonclassical estrogenic effects without the often negative, direct target effects associated with ER target gene activation.358–360 The potential benefits of AhR ligands that could be exploited for antiestrogenic properties, yet not produce the toxic side effects of PAHs and HAHs, would be enormous. The methyl-substituted diindolyl-methanes recently characterized in Stephen Safe’s laboratory are two such compounds.361 In addition, the identification of an endogenous ligand(s) for AhR, if any, would prove a huge boon to the field and our understanding of AhR function.
It is likely that the AHR and ARNT Are integral components of endocrine function, cytokine signaling, and, undoubtedly, many other signal transduction pathways as well. therefore, in light of the evidence reviewed herein, it may be that AHR’S endogenous physiological function, at least in part, is mediated through a DNA-binding independent mechanism(s), such as tethering or some other function heretofore unrecognized. it is quite plausible that realization of the true impact of tethering may not be appreciated until the activation of the AhR is studied in combination with other stimuli. AhR target gene activation may be only one mode of function. in fact, activation by exogenous compounds may only serve to disrupt normal AhR function. as such, nonclassical receptor activities may be a key aspect of AhR’S true physiological role, as has been observed with GR.
ACKNOWLEDGMENTS
This work was supported by NIEHS Grants No. ES04869 and No. ES011834 awarded to Gary H. Perdew. We also thank Marcia H. Perdew for editorial assistance.
REFERENCES
- 1.Vanden Heuvel JP, Lucier G. Environmental toxicology of polychlorinated dibenzop-dioxins and polychlorinated dibenzofurans. Environ. Health Perspect. 1993;100:189–200. doi: 10.1289/ehp.93100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zack JA, Suskind RR. The mortality experience of workers exposed to tetrachlorodibenzodioxin in a trichlorophenol process accident. J Occup Med. 1980;22(1):11–14. [PubMed] [Google Scholar]
- 3.Meigs JW, Albom JJ, Kartin BL. Chloracne from an unusual exposure to arochlor. J Am Med Assoc. 1954;154(17):1417–1418. doi: 10.1001/jama.1954.02940510017007. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Salguero PM, Hilbert DM, Rudikoff S, Ward JM, Gonzalez FJ. Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol Appl Pharmacol. 1996;140(1):173–179. doi: 10.1006/taap.1996.0210. [DOI] [PubMed] [Google Scholar]
- 5.Mukerjee D. Health impact of polychlorinated dibenzo-p-dioxins: a critical review. J Air Waste Manag Assoc. 1998;48(2):157–165. doi: 10.1080/10473289.1998.10463655. [DOI] [PubMed] [Google Scholar]
- 6.Muranyi-Kovacs I, Rudali G, Imbert J. Bioassay of 2,4,5-trichlorophenoxyacetic acid for carcinogenicity in mice. Br J Cancer. 1976;33(6):626–633. doi: 10.1038/bjc.1976.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mocarelli P, Marocchi A, Brambilla P, Gerthoux P, Young DS, Mantel N. Clinical laboratory manifestations of exposure to dioxin in children. A six-year study of the effects of an environmental disaster near Seveso, Italy. Jama. 1986;256(19):2687–2695. [PubMed] [Google Scholar]
- 8.Cerlesi S, Di Domenico A, Ratti S. 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) persistence in the Seveso (Milan, Italy) soil. Ecotoxicol Environ Saf. 1989;18(2):149–164. doi: 10.1016/0147-6513(89)90076-6. [DOI] [PubMed] [Google Scholar]
- 9.Kogevinas M. Human health effects of dioxins: cancer, reproductive and endocrine system effects. Hum Reprod Update. 2001;7(3):331–339. doi: 10.1093/humupd/7.3.331. [DOI] [PubMed] [Google Scholar]
- 10.Rowlands JC, McEwan IJ, Gustafsson JA. Transactivation by the human aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator proteins: direct interactions with basal transcription factors. Mol Pharmacol. 1996;50(3):538–548. [PubMed] [Google Scholar]
- 11.Yoshimura T, Kaneko S, Hayabuchi H. Sex ratio in offspring of those affected by dioxin and dioxin-like compounds: the Yusho, Seveso, and Yucheng incidents. Occup Environ Med. 2001;58(8):540–541. doi: 10.1136/oem.58.8.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchanan DL, Sato T, Peterson RE, Cooke PS. Antiestrogenic effects of 2,3,7,8–tetrachlorodibenzo-p-dioxin in mouse uterus: critical role of the aryl hydrocarbon receptor in stromal tissue. Toxicol Sci. 2000;57(2):302–311. doi: 10.1093/toxsci/57.2.302. [DOI] [PubMed] [Google Scholar]
- 13.Ohtake F, Baba A, Takada I, Okada M, Iwasaki K, Miki H, et al. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature. 2007;446(7135):562–566. doi: 10.1038/nature05683. [DOI] [PubMed] [Google Scholar]
- 14.Safe S, Wormke M, Samudio I. Mechanisms of inhibitory aryl hydrocarbon receptor-estrogen receptor crosstalk in human breast cancer cells. J Mammary Gland Biol Neoplasia. 2000;5(3):295–306. doi: 10.1023/a:1009550912337. [DOI] [PubMed] [Google Scholar]
- 15.Wormke M, Stoner M, Saville B, Walker K, Abdelrahim M, Burghardt R, et al. The aryl hydrocarbon receptor mediates degradation of estrogen receptor alpha through activation of proteasomes. Mol Cell Biol. 2003;23(6):1843–1855. doi: 10.1128/MCB.23.6.1843-1855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zacharewski TR, Bondy KL, McDonell P, Wu ZF. Antiestrogenic effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on 17 beta-estradiol-induced pS2 expression. Cancer Res'. 1994;54(10):2707–2713. [PubMed] [Google Scholar]
- 17.Sterling JB, Hanke CW. Dioxin toxicity and chloracne in the Ukraine. J Drugs Dermatol. 2005;4(2):148–150. [PubMed] [Google Scholar]
- 18.Geusau A, Tschachler E, Meixner M, Sandermann S, Papke O, Wolf C, et al. Olestra increases faecal excretion of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Lancet. 1999;354(9186):1266–1267. doi: 10.1016/S0140-6736(99)04271-3. [DOI] [PubMed] [Google Scholar]
- 19.Moser GA, McLachlan MS. A non-absorbable dietary fat substitute enhances elimination of persistent lipophilic contaminants in humans. Chemosphere. 1999;39(9):1513–1521. doi: 10.1016/s0045-6535(99)00219-2. [DOI] [PubMed] [Google Scholar]
- 20.Bearn AG. Inborn errors of metabolism: Garrod's legacy. Mol Med. 1996;2(3):271–273. [PMC free article] [PubMed] [Google Scholar]
- 21.Poland AP, Glover E, Robinson JR, Nebert DW. Genetic expression of aryl hydrocarbon hydroxylase activity. Induction of monooxygenase activities and cytochrome P1-450 formation by 2,3,7,8-tetrachlorodibenzo-p-dioxin in mice genetically "nonresponsive" to other aromatic hydrocarbons. J Biol Chem. 1974;249(17):5599–5606. [PubMed] [Google Scholar]
- 22.Gielen JE, Goujon FM, Nebert DW. Genetic regulation of aryl hydrocarbon hydroxylase induction. II. Simple Mendelian expression in mouse tissues in vivo. J Biol Chem. 1972;247(4):1125–1137. [PubMed] [Google Scholar]
- 23.Gielen JE, Nebert DW. Aryl hydrocarbon hydroxylase induction in mammalian liver cell culture. I. Stimulation of enzyme activity in non-hepatic cells and in hepatic cells by phenobarbital, polycyclic hydrocarbons, and 2,2-bis(p-chlorophenyl)-1,1,1-trichloroethane. J Biol Chem. 1971;246(17):5189–5198. [PubMed] [Google Scholar]
- 24.Robinson JR, Considine N, Nebert DW. Genetic expression of aryl hydrocarbon hydroxylase induction. Evidence for the involvement of other genetic loci. J Biol Chem. 1974;249(18):5851–5859. [PubMed] [Google Scholar]
- 25.Nebert DW, Robinson JR, Niwa A, Kumaki K, Poland AP. Genetic expression of aryl hydrocarbon hydroxylase activity in the mouse. J Cell Physiol. 1975;85(2 Pt 2 Suppl 1):393–414. doi: 10.1002/jcp.1040850407. [DOI] [PubMed] [Google Scholar]
- 26.Poland A, Glover E, Kende AS. Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J Biol Chem. 1976;251(16):4936–4946. [PubMed] [Google Scholar]
- 27.Poland A, Kende A. 2,3,7,8-Tetrachlorodibenzo-p-dioxin: environmental contaminant and molecular probe. Fed Proc. 1976;35(12):2404–2411. [PubMed] [Google Scholar]
- 28.Ema M, Ohe N, Suzuki M, Mimura J, Sogawa K, Ikawa S, et al. Dioxin binding activities of polymorphic forms of mouse and human arylhydro-carbon receptors. J Biol Chem. 1994;269(44):27337–27343. [PubMed] [Google Scholar]
- 29.Bradfield CA, Poland A. A competitive binding assay for 2,3,7,8-tetrachlorodibenzo-p-dioxin and related ligands of the Ah receptor. Mol Pharmacol. 1988;34(5):682–688. [PubMed] [Google Scholar]
- 30.McKinney JD, Singh P. Structure-activity relationships in halogenated biphenyls: unifying hypothesis for structural specificity. Chem Biol Interact. 1981;33(2–3):271–283. doi: 10.1016/0009-2797(81)90046-6. [DOI] [PubMed] [Google Scholar]
- 31.Poland A, Glover E. Chlorinated biphenyl induction of aryl hydrocarbon hydroxylase activity: a study of the structure-activity relationship. Mol Pharmacol. 1977;13(5):924–938. [PubMed] [Google Scholar]
- 32.Safe SH. Comparative toxicology and mechanism of action of polychlorinated dibenzo-p-dioxins and dibenzofurans. Annu Rev Pharmacol Toxicol. 1986;26:371–399. doi: 10.1146/annurev.pa.26.040186.002103. [DOI] [PubMed] [Google Scholar]
- 33.Gorski J, Gannon F. Current models of steroid hormone action: a critique. Annu Rev Physiol. 1976;38:425–450. doi: 10.1146/annurev.ph.38.030176.002233. [DOI] [PubMed] [Google Scholar]
- 34.Hord NG, Perdew GH. Physicochemical and immunocytochemical analysis of the aryl hydrocarbon receptor nuclear translocator: characterization of two monoclonal antibodies to the aryl hydrocarbon receptor nuclear translocator. Mol Pharmacol. 1994;46(4):618–626. [PubMed] [Google Scholar]
- 35.Perdew GH, Hord N, Hollenback CE, Welsh MJ. Localization and characterization of the 86- and 84-kDa heat shock proteins in Hepa 1c1c7 cells. Exp Cell Res. 1993;209(2):350–356. doi: 10.1006/excr.1993.1320. [DOI] [PubMed] [Google Scholar]
- 36.Pollenz RS, Sattler CA, Poland A. The aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator protein show distinct subcellular localizations in Hepa 1c1c7 cells by immunofluorescence microscopy. Mol Pharmacol. 1994;45(3):428–438. [PubMed] [Google Scholar]
- 37.Gudas JM, Karenlampi SO, Hankinson O. Intracellular location of the Ah receptor. J Cell Physiol. 1986;128(3):441–448. doi: 10.1002/jcp.1041280313. [DOI] [PubMed] [Google Scholar]
- 38.Whitlock JP, Jr, Galeazzi DR. 2,3,7,8-Tetrachlorodibenzo-p-dioxin receptors in wild type and variant mouse hepatoma cells. Nuclear location and strength of nuclear binding. J Biol Chem. 1984;259(2):980–985. [PubMed] [Google Scholar]
- 39.Greenlee WF, Poland A. Nuclear uptake of 2,3,7,8-tetrachlorodibenzo-p-dioxin in C57BL/6J and DBA/2J mice. Role of the hepatic cytosol receptor protein. J Biol Chem. 1979;254(19):9814–9821. [PubMed] [Google Scholar]
- 40.Okey AB, Bondy GP, Mason ME, Kahl GF, Eisen HJ, Guenthner TM, et al. Regulatory gene product of the Ah locus. Characterization of the cytosolic inducer-receptor complex and evidence for its nuclear translocation. J Biol Chem. 1979;254(22):11636–11648. [PubMed] [Google Scholar]
- 41.Okey AB, Bondy GP, Mason ME, Nebert DW, Forster-Gibson CJ, Muncan J, et al. Temperature-dependent cytosol-to-nucleus translocation of the Ah receptor for 2,3,7,8-tetrachlorodibenzo-p-dioxin in continuous cell culture lines. J Biol Chem. 1980;255(23):11415–11422. [PubMed] [Google Scholar]
- 42.Hannah RR, Nebert DW, Eisen HJ. Regulatory gene product of the Ah complex. Comparison of 2,3,7,8-tetrachlorodibenzo-p-dioxin and 3-methylcholanthrene binding to several moieties in mouse liver cytosol. J Biol Chem. 1981;256(9):4584–4590. [PubMed] [Google Scholar]
- 43.Okret S, Wikstrom AC, Gustafsson JA. Molybdate-stabilized glucocorticoid receptor: evidence for a receptor heteromer. Biochemistry. 1985;24(23):6581–6586. doi: 10.1021/bi00344a041. [DOI] [PubMed] [Google Scholar]
- 44.Denison MS, Vella LM, Okey AB. Hepatic Ah receptor for 2,3,7,8-tetrachlorodibenzo-p-dioxin. Partial stabilization by molybdate. J Biol Chem. 1986;261(22):10189–10195. [PubMed] [Google Scholar]
- 45.Manchester DK, Gordon SK, Golas CL, Roberts EA, Okey AB. Ah receptor in human placenta: stabilization by molybdate andcharacterization of binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin, 3-methylcholanthrene, and benzo(a)pyrene. Cancer Res. 1987;47(18):4861–4868. [PubMed] [Google Scholar]
- 46.Poland A, Glover E, Ebetino FH, Kende AS. Photoaffinity labeling of the Ah receptor. J Biol Chem. 1986;261(14):6352–6365. [PubMed] [Google Scholar]
- 47.Poland A, Glover E. Ca2+-dependent proteolysis of the Ah receptor. Arch Biochem Biophys. 1988;261(1):103–111. doi: 10.1016/0003-9861(88)90109-9. [DOI] [PubMed] [Google Scholar]
- 48.Poland A, Glover E, Taylor BA. The murine Ah locus: a new allele and mapping to chromosome 12. Mol Pharmacol. 1987;32(4):471–478. [PubMed] [Google Scholar]
- 49.Bradfield CA, Kende AS, Poland A. Kinetic and equilibrium studies of Ah receptor-ligand binding: use of [125I 2-iodo-7,8-dibromodibenzo-p-dioxin. Mol Pharmacol. 1988;34(2):229–237. [PubMed] [Google Scholar]
- 50.Perdew GH, Poland A. Purification of the Ah receptor from C57BL/6J mouse liver. J Biol Chem. 1988;263(20):9848–9852. [PubMed] [Google Scholar]
- 51.Bradfield CA, Glover E, Poland A. Purification and N-terminal amino acid sequence of the Ah receptor from the C57BL/6J mouse. Mol Pharmacol. 1991;39(1):13–19. [PubMed] [Google Scholar]
- 52.Poland A, Glover E, Bradfield CA. Characterization of polyclonal antibodies to the Ah receptor prepared by immunization with a synthetic peptide hapten. Mol Pharmacol. 1991;39(1):20–26. [PubMed] [Google Scholar]
- 53.Catelli MG, Binart N, Feramisco JR, Helfman DM. Cloning of the chick hsp 90 cDNA in expression vector. Nucleic Acids Res. 1985;13(17):6035–6047. doi: 10.1093/nar/13.17.6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Catelli MG, Binart N, Jung-Testas I, Renoir JM, Baulieu EE, Feramisco JR, et al. The common 90-kd protein component of non-transformed '8S' steroid receptors is a heat-shock protein. Embo J. 1985;4(12):3131–3135. doi: 10.1002/j.1460-2075.1985.tb04055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanchez ER, Toft DO, Schlesinger MJ, Pratt WB. Evidence that the 90-kDa phosphoprotein associated with the untransformed L-cell glucocorticoid receptor is a murine heat shock protein. J Biol Chem. 1985;260(23):12398–12401. [PubMed] [Google Scholar]
- 56.Denis M, Cuthill S, Wikstrom AC, Poellinger L, Gustafsson JA. Association of the dioxin receptor with the Mr 90,000 heat shock protein: a structural kinship with the glucocorticoid receptor. Biochem Biophys Res Commun. 1988;155(2):801–807. doi: 10.1016/s0006-291x(88)80566-7. [DOI] [PubMed] [Google Scholar]
- 57.Perdew GH. Association of the Ah receptor with the 90-kDa heat shock protein. J Biol Chem. 1988;263(27):13802–13805. [PubMed] [Google Scholar]
- 58.Hoffman EC, Reyes H, Chu FF, Sander F, Conley LH, Brooks BA, et al. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 1991;252(5008):954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 59.Johnson EF. A partnership between the dioxin receptor and a basic helix-loop-helix protein. Science. 1991;252(5008):924–925. doi: 10.1126/science.1852074. [DOI] [PubMed] [Google Scholar]
- 60.Huang ZJ, Edery I, Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature. 1993;364(6434):259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 61.Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36(2):189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 62.Chen HS, Perdew GH. Subunit composition of the heteromeric cytosolic aryl hydrocarbon receptor complex. J Biol Chem. 1994;269(44):27554–27558. [PubMed] [Google Scholar]
- 63.Perdew GH. Chemical cross-linking of the cytosolic and nuclear forms of the Ah receptor in hepatoma cell line 1c1c7. Biochem Biophys Res Commun. 1992;182(1):55–62. doi: 10.1016/s0006-291x(05)80111-1. [DOI] [PubMed] [Google Scholar]
- 64.Burbach KM, Poland A, Bradfield CA. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci U S A. 1992;89(17):8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nambu JR, Lewis JO, Wharton KA, Jr, Crews ST. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67(6):1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- 66.Courey AJ, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 67.Laurent BC, Treitel MA, Carlson M. The SNF5 protein of Saccharomyces cerevisiae is a glutamine-and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol Cell Biol. 1990;10(11):5616–5625. doi: 10.1128/mcb.10.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dolwick KM, Schmidt JV, Carver LA, Swanson HI, Bradfield CA. Cloning and expression of a human Ah receptor cDNA. Mol Pharmacol. 1993;44(5):911–917. [PubMed] [Google Scholar]
- 69.Jain S, Dolwick KM, Schmidt JV, Bradfield CA. Potent transactivation domains of the Ah receptor and the Ah receptor nuclear translocator map to their carboxyl termini. J Biol Chem. 1994;269(50):31518–31524. [PubMed] [Google Scholar]
- 70.Ma Q, Dong L, Whitlock JP., Jr Transcriptional activation by the mouse Ah receptor. Interplay between multiple stimulatory and inhibitory functions. J Biol Chem. 1995;270(21):12697–12703. doi: 10.1074/jbc.270.21.12697. [DOI] [PubMed] [Google Scholar]
- 71.Poland A, Palen D, Glover E. Analysis of the four alleles of the murine aryl hydrocarbon receptor. Mol Pharmacol. 1994;46(5):915–921. [PubMed] [Google Scholar]
- 72.Whitelaw M, Pongratz I, Wilhelmsson A, Gustafsson JA, Poellinger L. Ligand-dependent recruitment of the Arnt coregulator determines DNA recognition by the dioxin receptor. Mol Cell Biol. 1993;13(4):2504–2514. doi: 10.1128/mcb.13.4.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whitelaw ML, Gottlicher M, Gustafsson JA, Poellinger L. Definition of a novel ligand binding domain of a nuclear bHLH receptor: co-localization of ligand and hsp90 binding activities within the regulable inactivation domain of the dioxin receptor. Embo J. 1993;12(11):4169–4179. doi: 10.1002/j.1460-2075.1993.tb06101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fukunaga BN, Probst MR, Reisz-Porszasz S, Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor. J Biol Chem. 1995;270(49):29270–29278. doi: 10.1074/jbc.270.49.29270. [DOI] [PubMed] [Google Scholar]
- 75.Reisz-Porszasz S, Probst MR, Fukunaga BN, Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor nuclear translocator protein (ARNT) Mol Cell Biol. 1994;14(9):6075–6086. doi: 10.1128/mcb.14.9.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Whitelaw ML, Gustafsson JA, Poellinger L. Identification of transactivation and repression functions of the dioxin receptor and its basic helix-loop-helix/PAS partner factor Arnt: inducible versus constitutive modes of regulation. Mol Cell Biol. 1994;14(12):8343–8355. doi: 10.1128/mcb.14.12.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pongratz I, Mason GG, Poellinger L. Dual roles of the 90-kDa heat shock protein hsp90 in modulating functional activities of the dioxin receptor. Evidence that the dioxin receptor functionally belongs to a subclass of nuclear receptors which require hsp90 both for ligand binding activity and repression of intrinsic DNA binding activity. J Biol Chem. 1992;267(19):13728–13734. [PubMed] [Google Scholar]
- 78.Heid SE, Pollenz RS, Swanson HI. Role of heat shock protein 90 dissociation in mediating agonist-induced activation of the aryl hydrocarbon receptor. Mol Pharmacol. 2000;57(1):82–92. [PubMed] [Google Scholar]
- 79.Perdew GH, Bradfield CA. Mapping the 90 kDa heat shock protein binding region of the Ah receptor. Biochem Mol Biol Int. 1996;39(3):589–593. doi: 10.1080/15216549600201651. [DOI] [PubMed] [Google Scholar]
- 80.Young JC, Obermann WM, Hartl FU. Specific binding of tetratricopeptide repeat proteins to the C-terminal 12-kDa domain of hsp90. J Biol Chem. 1998;273(29):18007–18010. doi: 10.1074/jbc.273.29.18007. [DOI] [PubMed] [Google Scholar]
- 81.Meyer BK, Perdew GH. Characterization of the AhR-hsp90-XAP2 core complex and the role of the immunophilin-related protein XAP2 in AhR stabilization. Biochemistry. 1999;38(28):8907–8917. doi: 10.1021/bi982223w. [DOI] [PubMed] [Google Scholar]
- 82.Poellinger L, Kurl RN, Lund J, Gillner M, Carlstedt-Duke J, Hogberg B, et al. High-affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin in cell nuclei from rat liver. Biochim Biophys Acta. 1982;714(3):516–523. doi: 10.1016/0304-4165(82)90162-3. [DOI] [PubMed] [Google Scholar]
- 83.Carlstedt-Duke JM, Harnemo UB, Hogberg B, Gustafsson JA. Interaction of the hepatic receptor protein for 2,3,7,8-tetrachlorodibenzo-rhodioxin with DNA. Biochim Biophys Acta. 1981;672(2):131–141. doi: 10.1016/0304-4165(81)90386-x. [DOI] [PubMed] [Google Scholar]
- 84.Hannah RR, Lund J, Poellinger L, Gillner M, Gustafsson JA. Characterization of the DNA-binding properties of the receptor for 2,3,7,8-tetrachlorodibenzo-p-dioxin. Eur J Biochem. 1986;156(2):237–242. doi: 10.1111/j.1432-1033.1986.tb09573.x. [DOI] [PubMed] [Google Scholar]
- 85.Bunger MK, Moran SM, Glover E, Thomae TL, Lahvis GP, Lin BC, et al. Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. J Biol Chem. 2003;278(20):17767–17774. doi: 10.1074/jbc.M209594200. [DOI] [PubMed] [Google Scholar]
- 86.Jones PB, Galeazzi DR, Fisher JM, Whitlock JP., Jr Control of cytochrome P1-450 gene expression by dioxin. Science. 1985;227(4693):1499–1502. doi: 10.1126/science.3856321. [DOI] [PubMed] [Google Scholar]
- 87.Jones PB, Durrin LK, Fisher JM, Whitlock JP., Jr Control of gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Multiple dioxin-responsive domains 5′-ward of the cytochrome P1-450 gene. J Biol Chem. 1986;261(15):6647–6650. [PubMed] [Google Scholar]
- 88.Jones PB, Durrin LK, Galeazzi DR, Whitlock JP., Jr Control of cytochrome P1-450 gene expression: analysis of a dioxin-responsive enhancer system. Proc Natl Acad Sci USA. 1986;83(9):2802–2806. doi: 10.1073/pnas.83.9.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Durrin LK, Jones PB, Fisher JM, Galeazzi DR, Whitlock JP., Jr 2,3,7,8-Tetrachlorodibenzo-p-dioxin receptors regulate transcription of the cytochrome P1-450 gene. J Cell Biochem. 1987;35(2):153–160. doi: 10.1002/jcb.240350208. [DOI] [PubMed] [Google Scholar]
- 90.Denison MS, Fisher JM, Whitlock JP., Jr The DNA recognition site for the dioxin-Ah receptor complex. Nucleotide sequence and functional analysis. J Biol Chem. 1988;263(33):17221–17224. [PubMed] [Google Scholar]
- 91.Denison MS, Fisher JM, Whitlock JP., Jr Inducible, receptor-dependent protein-DNA interactions at a dioxin-responsive transcriptional enhancer. Proc Natl Acad Sci USA. 1988;85(8):2528–2532. doi: 10.1073/pnas.85.8.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Denison MS, Fisher JM, Whitlock JP., Jr Protein-DNA interactions at recognition sites for the dioxin-Ah receptor complex. J Biol Chem. 1989;264(28):16478–16482. [PubMed] [Google Scholar]
- 93.Shen ES, Whitlock JP., Jr The potential role of DNA methylation in the response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem. 1989;264(30):17754–17758. [PubMed] [Google Scholar]
- 94.Henry EC, Rucci G, Gasiewicz TA. Characterization of multiple forms of the Ah receptor: comparison of species and tissues. Biochemistry. 1989;28(15):6430–6440. doi: 10.1021/bi00441a041. [DOI] [PubMed] [Google Scholar]
- 95.Elferink CJ, Gasiewicz TA, Whitlock JP., Jr Protein-DNA interactions at a dioxin-responsive enhancer. Evidence that the transformed Ah receptor is heteromeric. J Biol Chem. 1990;265(33):20708–20712. [PubMed] [Google Scholar]
- 96.Reyes H, Reisz-Porszasz S, Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992;256(5060):1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- 97.Watson AJ, Hankinson O. Dioxin- and Ah receptor-dependent protein binding to xenobiotic responsive elements and G-rich DNA studied by in vivo footprinting. J Biol Chem. 1992;267(10):6874–6878. [PubMed] [Google Scholar]
- 98.Bacsi SG, Reisz-Porszasz S, Hankinson O. Orientation of the heterodimeric aryl hydrocarbon (dioxin) receptor complex on its asymmetric DNA recognition sequence. Mol Pharmacol. 1995;47(3):432–438. [PubMed] [Google Scholar]
- 99.Black VH, Quattrochi LC. Molecular cloning of the guinea pig CYP1A2 gene 5′-flanking region: identification of functional aromatic hydrocarbon response element and characterization of CYP1A2 expression in GPC16 cells. Drug Metab Dispos. 2004;32(6):595–602. doi: 10.1124/dmd.32.6.595. [DOI] [PubMed] [Google Scholar]
- 100.Quattrochi LC, Tukey RH. The human cytochrome Cyp1A2 gene contains regulatory elements responsive to 3-methylcholanthrene. Mol Pharmacol. 1989;36(1):66–71. [PubMed] [Google Scholar]
- 101.Quattrochi LC, Vu T, Tukey RH. The human CYP1A2 gene and induction by 3-methylcholanthrene. A region of DNA that supports AH-receptor binding and promoter-specific induction. J Biol Chem. 1994;269(9):6949–6954. [PubMed] [Google Scholar]
- 102.Eltom SE, Zhang L, Jefcoate CR. Regulation of cytochrome P-450 (CYP) 1B1 in mouse Hepa-1 variant cell lines: A possible role for aryl hydrocarbon receptor nuclear translocator (ARNT) as a suppressor of CYP1B1 gene expression. Mol Pharmacol. 1999;55(3):594–604. [PubMed] [Google Scholar]
- 103.Zhang L, Savas U, Alexander DL, Jefcoate CR. Characterization of the mouse Cyp1B1 gene. Identification of an enhancer region that directs aryl hydrocarbon receptor-mediated constitutive and induced expression. J Biol Chem. 1998;273(9):5174–5183. doi: 10.1074/jbc.273.9.5174. [DOI] [PubMed] [Google Scholar]
- 104.Asman DC, Takimoto K, Pitot HC, Dunn TJ, Lindahl R. Organization and characterization of the rat class 3 aldehyde dehydrogenase gene. J Biol Chem. 1993;268(17):12530–12536. [PubMed] [Google Scholar]
- 105.Asman DC, Takimoto K, Pitot HC, Lindahl R. Preliminary characterization of the rat class 3 aldehyde dehydrogenase gene. Adv Exp Med Biol. 1993;328:81–86. doi: 10.1007/978-1-4615-2904-0_10. [DOI] [PubMed] [Google Scholar]
- 106.Yueh MF, Huang YH, Hiller A, Chen S, Nguyen N, Tukey RH. Involvement of the xenobiotic response element (XRE) in Ah receptor-mediated induction of human UDP-glucuronosyltransferase 1A1. J Biol Chem. 2003;278(17):15001–15006. doi: 10.1074/jbc.M300645200. [DOI] [PubMed] [Google Scholar]
- 107.Rivera SP, Saarikoski ST, Hankinson O. Identification of a novel dioxin-inducible cytochrome P450. Mol Pharmacol. 2002;61(2):255–259. doi: 10.1124/mol.61.2.255. [DOI] [PubMed] [Google Scholar]
- 108.Favreau LV, Pickett CB. Transcriptional regulation of the rat NAD(P)H:quinone reductase gene. Identification of regulatory elements controlling basal level expression and inducible expression by planar aromatic compounds and phenolic antioxidants. J Biol Chem. 1991;266(7):4556–4561. [PubMed] [Google Scholar]
- 109.Patel RD, Kim DJ, Peters JM, Perdew GH. The aryl hydrocarbon receptor directly regulates expression of the potent mitogen epiregulin. Toxicol Sci. 2006;89(1):75–82. doi: 10.1093/toxsci/kfi344. [DOI] [PubMed] [Google Scholar]
- 110.Gao L, Dong L, Whitlock JP., Jr A novel response to dioxin. Induction of ecto-ATPase gene expression. J Biol Chem. 1998;273(25):15358–15365. doi: 10.1074/jbc.273.25.15358. [DOI] [PubMed] [Google Scholar]
- 111.Poland A, Glover E. 2,3,7,8-Tetrachlorodibenzo-p-dioxin: a potent inducer of -aminolevulinic acid synthetase. Science. 1973;179(72):476–477. doi: 10.1126/science.179.4072.476. [DOI] [PubMed] [Google Scholar]
- 112.Kraemer SA, Arthur KA, Denison MS, Smith WL, DeWitt DL. Regulation of prostaglandin endoperoxide H synthase-2 expression by 2,3,7,8,-tetrachlorodibenzo-p-dioxin. Arch Biochem Biophys. 1996;330(2):319–328. doi: 10.1006/abbi.1996.0259. [DOI] [PubMed] [Google Scholar]
- 113.Jigorel E, Le Vee M, Boursier-Neyret C, Parmentier Y, Fardel O. Differential regulation of sinusoidal and canalicular hepatic drug transporter expression by xenobiotics activating drug-sensing receptors in primary human hepatocytes. Drug Metab Dispos. 2006;34(10):1756–1763. doi: 10.1124/dmd.106.010033. [DOI] [PubMed] [Google Scholar]
- 114.Mathieu MC, Lapierre I, Brault K, Raymond M. Aromatic hydrocarbon receptor (AhR).AhR nuclear translocator- and p53-mediated induction of the murine multidrug resistance mdr1 gene by 3-methylcholanthrene and benzo(a)pyrene in hepatoma cells. J Biol Chem. 2001;276(7):4819–4827. doi: 10.1074/jbc.M008495200. [DOI] [PubMed] [Google Scholar]
- 115.Baba T, Mimura J, Gradin K, Kuroiwa A, Watanabe T, Matsuda Y, et al. Structure and expression of the Ah receptor repressor gene. J Biol Chem. 2001;276(35):33101–33110. doi: 10.1074/jbc.M011497200. [DOI] [PubMed] [Google Scholar]
- 116.Kolluri SK, Weiss C, Koff A, Gottlicher M. p27(Kip1) induction and inhibition of proliferation by the intracellular Ah receptor in developing thymus and hepatoma cells. Genes Dev. 1999;13(13):1742–1753. doi: 10.1101/gad.13.13.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu C, Li CY, Kong AN. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. 2005;28(3):249–268. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- 118.Nerurkar PV, Schut HA, Anderson LM, Riggs CW, Fornwald LW, Davis CD, et al. Ahr locus phenotype in congenic mice influences hepatic and pulmonary DNA adduct levels of 2-amino-3-methylimidazo[4,5-f]quinoline in the absence of cytochrome P450 induction. Mol Pharmacol. 1996;49(5):874–881. [PubMed] [Google Scholar]
- 119.Safe S. Polychlorinated biphenyls (PCBs), dibenzo-p-dioxins (PCDDs), dibenzofurans (PCDFs), and related compounds: environmental and mechanistic considerations which support the development of toxic equivalency factors (TEFs) Crit Rev Toxicol. 1990;21(1):51–88. doi: 10.3109/10408449009089873. [DOI] [PubMed] [Google Scholar]
- 120.Heath-Pagliuso S, Rogers WJ, Tullis K, Seidel SD, Cenijn PH, Brouwer A, et al. Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry. 1998;37(33):11508–11515. doi: 10.1021/bi980087p. [DOI] [PubMed] [Google Scholar]
- 121.Ciolino HP, Daschner PJ, Wang TT, Yeh GC. Effect of curcumin on the aryl hydrocarbon receptor and cytochrome P450 1A1 in MCF-7 human breast carcinoma cells. Biochem Pharmacol. 1998;56(2):197–206. doi: 10.1016/s0006-2952(98)00143-9. [DOI] [PubMed] [Google Scholar]
- 122.Ciolino HP, Daschner PJ, Yeh GC. Dietary flavonols quercetin and kaempferol are ligands of the aryl hydrocarbon receptor that affect CYP1A1 transcription differentially. Biochem J. 1999;340(Pt 3):715–722. [PMC free article] [PubMed] [Google Scholar]
- 123.Singh SU, Casper RF, Fritz PC, Sukhu B, Ganss B, Girard B, Jr, et al. Inhibition of dioxin effects on bone formation in vitro by a newly described aryl hydrocarbon receptor antagonist, resveratrol. J Endocrinol. 2000;167(1):183–195. doi: 10.1677/joe.0.1670183. [DOI] [PubMed] [Google Scholar]
- 124.Ciolino HP, Yeh GC. The flavonoid galangin is an inhibitor of CYP1A1 activity and an agonist/antagonist of the aryl hydrocarbon receptor. Br J Cancer. 1999;79(9–10):1340–1346. doi: 10.1038/sj.bjc.6690216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mufti NA, Shuler ML. Possible role of arachidonic acid in stress-induced cytochrome P450IA1 activity. Biotechnol Prog. 1996;12(6):847–854. doi: 10.1021/bp960067j. [DOI] [PubMed] [Google Scholar]
- 126.Couroucli XI, Welty SE, Geske RS, Moorthy B. Regulation of pulmonary and hepatic cytochrome P4501A expression in the rat by hyperoxia: implications for hyperoxic lung injury. Mol Pharmacol. 2002;61(3):507–515. doi: 10.1124/mol.61.3.507. [DOI] [PubMed] [Google Scholar]
- 127.Monk SA, Denison MS, Rice RH. Transient expression of CYP1A1 in rat epithelial cells cultured in suspension. Arch Biochem Biophys. 2001;393(1):154–162. doi: 10.1006/abbi.2001.2475. [DOI] [PubMed] [Google Scholar]
- 128.Helferich WG, Denison MS. Ultraviolet photoproducts of tryptophan can act as dioxin agonists. Mol Pharmacol. 1991;40(5):674–678. [PubMed] [Google Scholar]
- 129.Lesca P, Peryt B, Larrieu G, Alvinerie M, Galtier P, Daujat M, et al. Evidence for the ligand-independent activation of the AH receptor. Biochem Biophys Res Commun. 1995;209(2):474–482. doi: 10.1006/bbrc.1995.1526. [DOI] [PubMed] [Google Scholar]
- 130.Quattrochi LC, Tukey RH. Nuclear uptake of the Ah (dioxin) receptor in response to omeprazole: transcriptional activation of the human CYP1A1 gene. Mol Pharmacol. 1993;43(4):504–508. [PubMed] [Google Scholar]
- 131.Murray IA, Reen RK, Leathery N, Ramadoss P, Bonati L, Gonzalez FJ, et al. Evidence that ligand binding is a key determinant of Ah receptor-mediated transcriptional activity. Arch Biochem Biophys. 2005;442(1):59–71. doi: 10.1016/j.abb.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 132.Walisser JA, Bunger MK, Glover E, Bradfield CA. Gestational exposure of Ahr and Arnt hypomorphs to dioxin rescues vascular development. Proc Natl Acad Sci USA. 2004;101(47):16677–16682. doi: 10.1073/pnas.0404379101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fernandez-Salguero P, Pineau T, Hilbert DM, McPhail T, Lee SS, Kimura S, et al. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science. 1995;268(5211):722–726. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- 134.Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol Pharmacol. 2006;69(1):140–153. doi: 10.1124/mol.105.018705. [DOI] [PubMed] [Google Scholar]
- 135.Ramadoss P, Perdew GH. Use of 2-azido-3[125I]iodo-7,8-dibromodibenzo-p-dioxin as a probe to determine the relative ligand affinity of human versus mouse aryl hydrocarbon receptor in cultured cells. Mol Pharmacol. 2004;66(1):129–136. doi: 10.1124/mol.66.1.129. [DOI] [PubMed] [Google Scholar]
- 136.Cuthill S, Poellinger L, Gustafsson JA. The receptor for 2,3,7,8-tetrachlorodibenzo-p-dioxin in the mouse hepatoma cell line Hepa 1c1c7. A comparison with the glucocorticoid receptor and the mouse and rat hepatic dioxin receptors. J Biol Chem. 1987;262(8):3477–3481. [PubMed] [Google Scholar]
- 137.Scherrer LC, Hutchison KA, Sanchez ER, Randall SK, Pratt WB. A heat shock protein complex isolated from rabbit reticulocyte lysate can reconstitute a functional glucocorticoid receptor-Hsp90 complex. Biochemistry. 1992;31(32):7325–7329. doi: 10.1021/bi00147a017. [DOI] [PubMed] [Google Scholar]
- 138.Smith DF, Schowalter DB, Kost SL, Toft DO. Reconstitution of progesterone receptor with heat shock proteins. Mol Endocrinol. 1990;4(11):1704–1711. doi: 10.1210/mend-4-11-1704. [DOI] [PubMed] [Google Scholar]
- 139.Dittmar KD, Banach M, Galigniana MD, Pratt WB. The role of DnaJ-like proteins in glucocorticoid receptor.hsp90 heterocomplex assembly by the reconstituted hsp90.p60. hsp70 foldosome complex. J Biol Chem. 1998;273(13):7358–7366. doi: 10.1074/jbc.273.13.7358. [DOI] [PubMed] [Google Scholar]
- 140.Dittmar KD, Hutchison KA, Owens-Grillo JK, Pratt WB. Reconstitution of the steroid receptor. hsp90 heterocomplex assembly system of rabbit reticulocyte lysate. J Biol Chem. 1996;271(22):12833–12839. doi: 10.1074/jbc.271.22.12833. [DOI] [PubMed] [Google Scholar]
- 141.Cuthill S, Wilhelmsson A, Mason GG, Gillner M, Poellinger L, Gustafsson JA. The dioxin receptor: a comparison with the glucocorticoid receptor. J Steroid Biochem. 1988;30(1–6):277–280. doi: 10.1016/0022-4731(88)90106-9. [DOI] [PubMed] [Google Scholar]
- 142.Antonsson C, Whitelaw ML, McGuire J, Gustafsson JA, Poellinger L. Distinct roles of the molecular chaperone hsp90 in modulating dioxin receptor function via the basic helix-loop-helix and PAS domains. Mol Cell Biol. 1995;15(2):756–765. doi: 10.1128/mcb.15.2.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Perdew GH. Comparison of the nuclear and cytosolic forms of the Ah receptor from Hepa 1c1c7 cells: charge heterogeneity and ATP binding properties. Arch Biochem Biophys. 1991;291(2):284–290. doi: 10.1016/0003-9861(91)90136-7. [DOI] [PubMed] [Google Scholar]
- 144.McGuire J, Whitelaw ML, Pongratz I, Gustafsson JA, Poellinger L. A cellular factor stimulates ligand-dependent release of hsp90 from the basic helix-loop-helix dioxin receptor. Mol Cell Biol. 1994;14(4):2438–2446. doi: 10.1128/mcb.14.4.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Picard D. Chaperoning steroid hormone action. Trends Endocrinol Metab. 2006;17(6):229–235. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 146.Carver LA, Jackiw V, Bradfield CA. The 90-kDa heat shock protein is essential for Ah receptor signaling in a yeast expression system. J Biol Chem. 1994;269(48):30109–30112. [PubMed] [Google Scholar]
- 147.Whitelaw ML, McGuire J, Picard D, Gustafsson JA, Poellinger L. Heat shock protein hsp90 regulates dioxin receptor function in vivo. Proc Natl Acad Sci U S A. 1995;92(10):4437–4441. doi: 10.1073/pnas.92.10.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Phelan DM, Brackney WR, Denison MS. The Ah receptor can bind ligand in the absence of receptor-associated heat-shock protein 90. Arch Biochem Biophys. 1998;353(1):47–54. doi: 10.1006/abbi.1997.0614. [DOI] [PubMed] [Google Scholar]
- 149.Chen HS, Singh SS, Perdew GH. The Ah receptor is a sensitive target of geldanamycin-induced protein turnover. Arch Biochem Biophys. 1997;348(1):190–198. doi: 10.1006/abbi.1997.0398. [DOI] [PubMed] [Google Scholar]
- 150.Ogiso H, Kagi N, Matsumoto E, Nishimoto M, Arai R, Shirouzu M, et al. Phosphorylation analysis of 90 kDa heat shock protein within the cytosolic arylhydrocarbon receptor complex. Biochemistry. 2004;43(49):15510–15519. doi: 10.1021/bi048736m. [DOI] [PubMed] [Google Scholar]
- 151.Cox MB, Miller CA., 3rd Pharmacological and genetic analysis of 90-kDa heat shock isoprotein-aryl hydrocarbon receptor complexes. Mol Pharmacol. 2003;64(6):1549–1556. doi: 10.1124/mol.64.6.1549. [DOI] [PubMed] [Google Scholar]
- 152.Freeman BC, Felts SJ, Toft DO, Yamamoto KR. The p23 molecular chaperones act at a late step in intracellular receptor action to differentially affect ligand efficacies. Genes Dev. 2000;14(4):422–434. [PMC free article] [PubMed] [Google Scholar]
- 153.Felts SJ, Toft DO. p23, a simple protein with complex activities. Cell Stress Chaperones. 2003;8(2):108–113. doi: 10.1379/1466-1268(2003)008<0108:paspwc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chadli A, Bouhouche I, Sullivan W, Stensgard B, McMahon N, Catelli MG, et al. Dimerization and N-terminal domain proximity underlie the function of the molecular chaperone heat shock protein 90. Proc Natl Acad Sci U S A. 2000;97(23):12524–12529. doi: 10.1073/pnas.220430297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Johnson JL, Toft DO. A novel chaperone complex for steroid receptors involving heat shock proteins, immunophilins, and p23. J Biol Chem. 1994;269(40):24989–24993. [PubMed] [Google Scholar]
- 156.Young JC, Hartl FU. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. Embo J. 2000;19(21):5930–5940. doi: 10.1093/emboj/19.21.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Grad I, McKee TA, Ludwig SM, Hoyle GW, Ruiz P, Wurst W, et al. The Hsp90 cochaperone p23 is essential for perinatal survival. Mol Cell Biol. 2006;26(23):8976–8983. doi: 10.1128/MCB.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nair SC, Toran EJ, Rimerman RA, Hjermstad S, Smithgall TE, Smith DF. A pathway of multi-chaperone interactions common to diverse regulatory proteins: estrogen receptor, Fes tyrosine kinase, heat shock transcription factor Hsf1, and the aryl hydrocarbon receptor. Cell Stress Chaperones. 1996;1(4):237–250. doi: 10.1379/1466-1268(1996)001<0237:apomci>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kazlauskas A, Poellinger L, Pongratz I. Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (Aryl hydrocarbon) receptor. J Biol Chem. 1999;274(19):13519–13524. doi: 10.1074/jbc.274.19.13519. [DOI] [PubMed] [Google Scholar]
- 160.Sullivan WP, Owen BA, Toft DO. The influence of ATP and p23 on the conformation of hsp90. J Biol Chem. 2002;277(48):45942–45948. doi: 10.1074/jbc.M207754200. [DOI] [PubMed] [Google Scholar]
- 161.Shetty PV, Bhagwat BY, Chan WK. P23 enhances the formation of the aryl hydrocarbon receptor-DNA complex. Biochem Pharmacol. 2003;65(6):941–948. doi: 10.1016/s0006-2952(02)01650-7. [DOI] [PubMed] [Google Scholar]
- 162.Cox MB, Miller CA., 3rd The p23 co-chaperone facilitates dioxin receptor signaling in a yeast model system. Toxicol Lett. 2002;129(1–2):13–21. doi: 10.1016/s0378-4274(01)00465-9. [DOI] [PubMed] [Google Scholar]
- 163.Kazlauskas A, Sundstrom S, Poellinger L, Pongratz I. The hsp90 chaperone complex regulates intracellular localization of the dioxin receptor. Mol Cell Biol. 2001;21(7):2594–2607. doi: 10.1128/MCB.21.7.2594-2607.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Davies TH, Sanchez ER. Fkbp52. Int J Biochem Cell Biol. 2005;37(1):42–47. doi: 10.1016/j.biocel.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 165.Denny WB, Valentine DL, Reynolds PD, Smith DF, Scammell JG. Squirrel monkey immunophilin FKBP51 is a potent inhibitor of glucocorticoid receptor binding. Endocrinology. 2000;141(11):4107–4113. doi: 10.1210/endo.141.11.7785. [DOI] [PubMed] [Google Scholar]
- 166.Riggs DL, Roberts PJ, Chirillo SC, Cheung-Flynn J, Prapapanich V, Ratajczak T, et al. The Hsp90-binding peptidylprolyl isomerase FKBP52 potentiates glucocorticoid signaling in vivo. Embo J. 2003;22(5):1158–1167. doi: 10.1093/emboj/cdg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Davies TH, Ning YM, Sanchez ER. A new first step in activation of steroid receptors: hormone-induced switching of FKBP51 and FKBP52 immunophilins. J Biol Chem. 2002;277(7):4597–4600. doi: 10.1074/jbc.C100531200. [DOI] [PubMed] [Google Scholar]
- 168.Tranguch S, Cheung-Flynn J, Daikoku T, Prapapanich V, Cox MB, Xie H, et al. Cochaperone immunophilin FKBP52 is critical to uterine receptivity for embryo implantation. Proc Natl Acad Sci U S A. 2005;102(40):14326–14331. doi: 10.1073/pnas.0505775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cheung-Flynn J, Prapapanich V, Cox MB, Riggs DL, Suarez-Quian C, Smith DF. Physiological role for the cochaperone FKBP52 in androgen receptor signaling. Mol Endocrinol. 2005;19(6):1654–1666. doi: 10.1210/me.2005-0071. [DOI] [PubMed] [Google Scholar]
- 170.Dull AB, Carlson DB, Petrulis JR, Perdew GH. Characterization of the phosphorylation status of the hepatitis B virus X-associated protein 2. Arch Biochem Biophys. 2002;406(2):209–221. doi: 10.1016/s0003-9861(02)00444-7. [DOI] [PubMed] [Google Scholar]
- 171.Kuzhandaivelu N, Cong YS, Inouye C, Yang WM, Seto E. XAP2, a novel hepatitis B virus X-associated protein that inhibits X transactivation. Nucleic Acids Res. 1996;24(23):4741–4750. doi: 10.1093/nar/24.23.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Perdew GH. Chemical cross-linking of the cytosolic and nuclear forms of the Ah receptor in hepatoma cell line 1c1c7. Biochem Biophys Res Commun. 1992;182(1):55–62. doi: 10.1016/s0006-291x(05)80111-1. [DOI] [PubMed] [Google Scholar]
- 173.Carver LA, Bradfield CA. Ligand-dependent interaction of the aryl hydrocarbon receptor with a novel immunophilin homolog in vivo. J Biol Chem. 1997;272(17):11452–11456. doi: 10.1074/jbc.272.17.11452. [DOI] [PubMed] [Google Scholar]
- 174.Ma Q, Whitlock JP., Jr A novel cytoplasmic protein that interacts with the Ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem. 1997;272(14):8878–8884. [PubMed] [Google Scholar]
- 175.Meyer BK, Pray-Grant MG, Vanden Heuvel JP, Perdew GH. Hepatitis B virus X-associated protein 2 is a subunit of the unliganded aryl hydrocarbon receptor core complex and exhibits transcriptional enhancer activity. Mol Cell Biol. 1998;18(2):978–988. doi: 10.1128/mcb.18.2.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kazlauskas A, Poellinger L, Pongratz I. Two distinct regions of the immunophilin-like protein XAP2 regulate dioxin receptor function and interaction with hsp90. J Biol Chem. 2002;277(14):11795–11801. doi: 10.1074/jbc.M200053200. [DOI] [PubMed] [Google Scholar]
- 177.Carver LA, LaPres JJ, Jain S, Dunham EE, Bradfield CA. Characterization of the Ah receptor-associated protein, ARA9. J Biol Chem. 1998;273(50):33580–33587. doi: 10.1074/jbc.273.50.33580. [DOI] [PubMed] [Google Scholar]
- 178.Bell DR, Poland A. Binding of aryl hydrocarbon receptor (AhR) to AhR-interacting protein. The role of hsp90. J Biol Chem. 2000;275(46):36407–36414. doi: 10.1074/jbc.M004236200. [DOI] [PubMed] [Google Scholar]
- 179.LaPres JJ, Glover E, Dunham EE, Bunger MK, Bradfield CA. ARA9 modifies agonist signaling through an increase in cytosolic aryl hydrocarbon receptor. J Biol Chem. 2000;275(9):6153–6159. doi: 10.1074/jbc.275.9.6153. [DOI] [PubMed] [Google Scholar]
- 180.Petrulis JR, Hord NG, Perdew GH. Subcellular localization of the aryl hydrocarbon receptor is modulated by the immunophilin homolog hepatitis B virus X-associated protein 2. J Biol Chem. 2000;275(48):37448–37453. doi: 10.1074/jbc.M006873200. [DOI] [PubMed] [Google Scholar]
- 181.Petrulis JR, Kusnadi A, Ramadoss P, Hollingshead B, Perdew GH. The hsp90 Cochaperone XAP2 alters importin beta recognition of the bipartite nuclear localization signal of the Ah receptor and represses transcriptional activity. J Biol Chem. 2003;278(4):2677–2685. doi: 10.1074/jbc.M209331200. [DOI] [PubMed] [Google Scholar]
- 182.Pollenz RS, Wilson SE, Dougherty EJ. Role of endogenous XAP2 protein on the localization and nucleocytoplasmic shuttling of the endogenous mouse Ahb-1 receptor in the presence and absence of ligand. Mol Pharmacol. 2006;70(4):1369–1379. doi: 10.1124/mol.106.027672. [DOI] [PubMed] [Google Scholar]
- 183.Ramadoss P, Petrulis JR, Hollingshead BD, Kusnadi A, Perdew GH. Divergent roles of hepatitis B virus X-associated protein 2 (XAP2) in human versus mouse Ah receptor complexes. Biochemistry. 2004;43(3):700–709. doi: 10.1021/bi035827v. [DOI] [PubMed] [Google Scholar]
- 184.Meyer BK, Petrulis JR, Perdew GH. Aryl hydrocarbon (Ah) receptor levels are selectively modulated by hsp90-associated immunophilin homolog XAP2. Cell Stress Chaperones. 2000;5(3):243–254. doi: 10.1379/1466-1268(2000)005<0243:aharla>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kazlauskas A, Poellinger L, Pongratz I. The immunophilin-like protein XAP2 regulates ubiquitination and subcellular localization of the dioxin receptor. J Biol Chem. 2000;275(52):41317–41324. doi: 10.1074/jbc.M007765200. [DOI] [PubMed] [Google Scholar]
- 186.Hollingshead BD, Petrulis JR, Perdew GH. The aryl hydrocarbon (Ah) receptor transcriptional regulator hepatitis B virus X-associated protein 2 antagonizes p23 binding to Ah receptor-Hsp90 complexes and is dispensable for receptor function. J Biol Chem. 2004;279(44):45652–45661. doi: 10.1074/jbc.M407840200. [DOI] [PubMed] [Google Scholar]
- 187.Pollenz RS, Dougherty EJ. Redefining the role of the endogenous XAP2 and C-terminal hsp70-interacting protein on the endogenous Ah receptors expressed in mouse and rat cell lines. J Biol Chem. 2005;280(39):33346–33356. doi: 10.1074/jbc.M506619200. [DOI] [PubMed] [Google Scholar]
- 188.Hollingshead BD, Patel RD, Perdew GH. Endogenous hepatic expression of the hepatitis B virus X-associated protein 2 is adequate for maximal association with aryl hydrocarbon receptor-90-kDa heat shock protein complexes. Mol Pharmacol. 2006;70(6):2096–2107. doi: 10.1124/mol.106.029215. [DOI] [PubMed] [Google Scholar]
- 189.Kumar MB, Perdew GH. Nuclear receptor coactivator SRC-1 interacts with the Q-rich subdomain of the AhR and modulates its transactivation potential. Gene Expr. 1999;8(5–6):273–286. [PMC free article] [PubMed] [Google Scholar]
- 190.Escher D, Bodmer-Glavas M, Barberis A, Schaffner W. Conservation of glutamine-rich transactivation function between yeast and humans. Mol Cell Biol. 2000;20(8):2774–2782. doi: 10.1128/mcb.20.8.2774-2782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Schwarz JJ, Chakraborty T, Martin J, Zhou JM, Olson EN. The basic region of myogenin cooperates with two transcription activation domains to induce muscle-specific transcription. Mol Cell Biol. 1992;12(1):266–275. doi: 10.1128/mcb.12.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sogawa K, Iwabuchi K, Abe H, Fujii-Kuriyama Y. Transcriptional activation domains of the Ah receptor and Ah receptor nuclear translocator. J Cancer Res Clin Oncol. 1995;121(9–10):612–620. doi: 10.1007/BF01197779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251(4995):761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 194.Gill G, Pascal E, Tseng ZH, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci U S A. 1994;91(1):192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Heller H, Bengal E. TFIID (TBP) stabilizes the binding of MyoD to its DNA site at the promoter and MyoD facilitates the association of TFIIB with the preinitiation complex. Nucleic Acids Res. 1998;26(9):2112–2119. doi: 10.1093/nar/26.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Swanson HI, Yang JH. The aryl hydrocarbon receptor interacts with transcription factor IIB. Mol Pharmacol. 1998;54(4):671–677. [PubMed] [Google Scholar]
- 197.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14(2):121–141. [PubMed] [Google Scholar]
- 198.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108(4):465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 199.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312(5781):1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 200.Ju BG, Rosenfeld MG. A breaking strategy for topoisomerase IIbeta/PARP-1-dependent regulated transcription. Cell Cycle. 2006;5(22):2557–2560. doi: 10.4161/cc.5.22.3497. [DOI] [PubMed] [Google Scholar]
- 201.O'Malley BW. Coregulators: from whence came these "master genes.". Mol Endocrinol. 2007;21(5):1009–1013. doi: 10.1210/me.2007-0012. [DOI] [PubMed] [Google Scholar]
- 202.Yu C, York B, Wang S, Feng Q, Xu J, O'Malley BW. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol Cell. 2007;25(5):765–778. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Arany Z, Huang LE, Eckner R, Bhattacharya S, Jiang C, Goldberg MA, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci USA. 1996;93(23):12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kallio PJ, Okamoto K, O'Brien S, Carrero P, Makino Y, Tanaka H, et al. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. Embo J. 1998;17(22):6573–6586. doi: 10.1093/emboj/17.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kobayashi A, Numayama-Tsuruta K, Sogawa K, Fujii-Kuriyama Y. CBP/p300 functions as a possible transcriptional coactivator of Ah receptor nuclear translocator (Arnt) J Biochem (Tokyo) 1997;122(4):703–710. doi: 10.1093/oxfordjournals.jbchem.a021812. [DOI] [PubMed] [Google Scholar]
- 206.Beischlag TV, Wang S, Rose DW, Torchia J, Reisz-Porszasz S, Muhammad K, et al. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol Cell Biol. 2002;22(12):4319–4333. doi: 10.1128/MCB.22.12.4319-4333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kumar MB, Tarpey RW, Perdew GH. Differential recruitment of coactivator RIP140 by Ah and estrogen receptors. Absence of a role for LXXLL motifs. J Biol Chem. 1999;274(32):22155–22164. doi: 10.1074/jbc.274.32.22155. [DOI] [PubMed] [Google Scholar]
- 208.Wang S, Ge K, Roeder RG, Hankinson O. Role of mediator in transcriptional activation by the aryl hydrocarbon receptor. J Biol Chem. 2004;279(14):13593–13600. doi: 10.1074/jbc.M312274200. [DOI] [PubMed] [Google Scholar]
- 209.Wang S, Hankinson O. Functional involvement of the Brahma/SWI2-related gene 1 protein in cytochrome P4501A1 transcription mediated by the aryl hydrocarbon receptor complex. J Biol Chem. 2002;277(14):11821–11827. doi: 10.1074/jbc.M110122200. [DOI] [PubMed] [Google Scholar]
- 210.Tian Y, Ke S, Chen M, Sheng T. Interactions between the aryl hydrocarbon receptor and P-TEFb. Sequential recruitment of transcription factors and differential phosphorylation of C-terminal domain of RNA polymerase II at cyp1a1 promoter. J Biol Chem. 2003;278(45):44041–44048. doi: 10.1074/jbc.M306443200. [DOI] [PubMed] [Google Scholar]
- 211.Carlson DB, Perdew GH. A dynamic role for the Ah receptor in cell signaling? Insights from a diverse group of Ah receptor interacting proteins. J Biochem Mol Toxicol. 2002;16(6):317–325. doi: 10.1002/jbt.10051. [DOI] [PubMed] [Google Scholar]
- 212.Hankinson O. Role of coactivators in transcriptional activation by the aryl hydrocarbon receptor. Arch Biochem Biophys. 2005;433(2):379–386. doi: 10.1016/j.abb.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 213.Nguyen TA, Hoivik D, Lee JE, Safe S. Interactions of nuclear receptor coactivator/ corepressor proteins with the aryl hydrocarbon receptor complex. Arch Biochem Biophys. 1999;367(2):250–257. doi: 10.1006/abbi.1999.1282. [DOI] [PubMed] [Google Scholar]
- 214.Beischlag TV, Taylor RT, Rose DW, Yoon D, Chen Y, Lee WH, et al. Recruitment of thyroid hormone receptor/retinoblastoma-interacting protein 230 by the aryl hydrocarbon receptor nuclear translocator is required for the transcriptional response to both dioxin and hypoxia. J Biol Chem. 2004;279(52):54620–54628. doi: 10.1074/jbc.M410456200. [DOI] [PubMed] [Google Scholar]
- 215.Kim JH, Stallcup MR. Role of the coiled-coil coactivator (CoCoA) in aryl hydrocarbon receptor-mediated transcription. J Biol Chem. 2004;279(48):49842–49848. doi: 10.1074/jbc.M408535200. [DOI] [PubMed] [Google Scholar]
- 216.Chen YH, Beischlag TV, Kim JH, Perdew GH, Stallcup MR. Role of GAC63 in transcriptional activation mediated by the aryl hydrocarbon receptor. J Biol Chem. 2006;281(18):12242–12247. doi: 10.1074/jbc.M512537200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kollara A, Brown TJ. Functional interaction of nuclear receptor coactivator 4 with aryl hydrocarbon receptor. Biochem Biophys Res Commun. 2006;346(2):526–534. doi: 10.1016/j.bbrc.2006.05.148. [DOI] [PubMed] [Google Scholar]
- 218.Kang HJ, Kim HJ, Kim SK, Barouki R, Cho CH, Khanna KK, et al. BRCA1 modulates xenobiotic stress-inducible gene expression by interacting with ARNT in human breast cancer cells. J Biol Chem. 2006;281(21):14654–14662. doi: 10.1074/jbc.M601613200. [DOI] [PubMed] [Google Scholar]
- 219.Ge NL, Elferink CJ. A direct interaction between the aryl hydrocarbon receptor and retinoblastoma protein. Linking dioxin signaling to the cell cycle. J Biol Chem. 1998;273(35):22708–22713. doi: 10.1074/jbc.273.35.22708. [DOI] [PubMed] [Google Scholar]
- 220.Jones LC, Okino ST, Gonda TJ, Whitlock JP., Jr Myb-binding protein 1a augments AhR-dependent gene expression. J Biol Chem. 2002;277(25):22515–22519. doi: 10.1074/jbc.M200740200. [DOI] [PubMed] [Google Scholar]
- 221.Tojo M, Matsuzaki K, Minami T, Honda Y, Yasuda H, Chiba T, et al. The aryl hydrocarbon receptor nuclear transporter is modulated by the SUMO-1 conjugation system. J Biol Chem. 2002;277(48):46576–46585. doi: 10.1074/jbc.M205987200. [DOI] [PubMed] [Google Scholar]
- 222.Antenos M, Casper RF, Brown TJ. Interaction with Nedd8, a ubiquitin-like protein, enhances the transcriptional activity of the aryl hydrocarbon receptor. J Biol Chem. 2002;277(46):44028–44034. doi: 10.1074/jbc.M202413200. [DOI] [PubMed] [Google Scholar]
- 223.Rushing SR, Denison MS. The silencing mediator of retinoic acid and thyroid hormone receptors can interact with the aryl hydrocarbon (Ah) receptor but fails to repress Ah receptor-dependent gene expression. Arch Biochem Biophys. 2002;403(2):189–201. doi: 10.1016/s0003-9861(02)00233-3. [DOI] [PubMed] [Google Scholar]
- 224.Watt K, Jess TJ, Kelly SM, Price NC, McEwan IJ. Induced alpha-helix structure in the aryl hydrocarbon receptor transactivation domain modulates protein-protein interactions. Biochem-istry. 2005;44(2):734–743. doi: 10.1021/bi0487701. [DOI] [PubMed] [Google Scholar]
- 225.Wang F, Wang W, Safe S. Regulation of constitutive gene expression through interactions of Sp1 protein with the nuclear aryl hydrocarbon receptor complex. Biochemistry. 1999;38(35):11490–11500. doi: 10.1021/bi982578f. [DOI] [PubMed] [Google Scholar]
- 226.Beischlag TV, Perdew GH. ER alpha-AHR-ARNT protein-protein interactions mediate estradiol-dependent transrepression of dioxin-inducible gene transcription. J Biol Chem. 2005;280(22):21607–21611. doi: 10.1074/jbc.C500090200. [DOI] [PubMed] [Google Scholar]
- 227.Brunnberg S, Pettersson K, Rydin E, Matthews J, Hanberg A, Pongratz I. The basic helix-loop-helix-PAS protein ARNT functions as a potent coactivator of estrogen receptor-dependent transcription. Proc Natl Acad Sci U S A. 2003;100(11):6517–6522. doi: 10.1073/pnas.1136688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Klinge CM, Kaur K, Swanson HI. The aryl hydrocarbon receptor interacts with estrogen receptor alpha and orphan receptors COUP-TFI and ERRalpha1. Arch Biochem Biophys. 2000;373(1):163–174. doi: 10.1006/abbi.1999.1552. [DOI] [PubMed] [Google Scholar]
- 229.Ohtake F, Takeyama K, Matsumoto T, Kitagawa H, Yamamoto Y, Nohara K, et al. Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature. 2003;423(6939):545–550. doi: 10.1038/nature01606. [DOI] [PubMed] [Google Scholar]
- 230.Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA. Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. J Biol Chem. 1999;274(1):510–515. doi: 10.1074/jbc.274.1.510. [DOI] [PubMed] [Google Scholar]
- 231.Lee JW, Choi HS, Gyuris J, Brent R, Moore DD. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9(2):243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 232.Chang KH, Chen Y, Chen TT, Chou WH, Chen PL, Ma YY, et al. A thyroid hormone receptor coactivator negatively regulated by the retinoblastoma protein. Proc Natl Acad Sci USA. 1997;94(17):9040–9045. doi: 10.1073/pnas.94.17.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Elferink CJ, Ge NL, Levine A. Maximal aryl hydrocarbon receptor activity depends on an interaction with the retinoblastoma protein. Mol Pharmacol. 2001;59(4):664–673. doi: 10.1124/mol.59.4.664. [DOI] [PubMed] [Google Scholar]
- 234.Puga A, Xia Y, Elferink C. Role of the aryl hydrocarbon receptor in cell cycle regulation. Chem Biol Interact. 2002;141(1–2):117–130. doi: 10.1016/s0009-2797(02)00069-8. [DOI] [PubMed] [Google Scholar]
- 235.Elferink CJ. Aryl hydrocarbon receptor-mediated cell cycle control. Prog Cell Cycle Res. 2003;5:261–267. [PubMed] [Google Scholar]
- 236.Bae I, Fan S, Meng Q, Rih JK, Kim HJ, Kang HJ, et al. BRCA1 induces antioxidant gene expression and resistance to oxidative stress. Cancer Res. 2004;64(21):7893–7909. doi: 10.1158/0008-5472.CAN-04-1119. [DOI] [PubMed] [Google Scholar]
- 237.Alen P, Claessens F, Schoenmakers E, Swinnen JV, Verhoeven G, Rombauts W, et al. Interaction of the putative androgen receptor-specific coactivator ARA70/ELE1alpha with multiple steroid receptors and identification of an internally deleted ELE1beta isoform. Mol Endocrinol. 1999;13(1):117–128. doi: 10.1210/mend.13.1.0214. [DOI] [PubMed] [Google Scholar]
- 238.Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93(11):5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Jones LC, Whitlock JP., Jr Dioxin-inducible transactivation in a chromosomal setting. Analysis of the acidic domain of the Ah receptor. J Biol Chem. 2001;276(27):25037–25042. doi: 10.1074/jbc.M102910200. [DOI] [PubMed] [Google Scholar]
- 240.Steinmetz AC, Renaud JP, Moras D. Binding of ligands and activation of transcription by nuclear receptors. Annu Rev Biophys Biomol Struct. 2001;30:329–359. doi: 10.1146/annurev.biophys.30.1.329. [DOI] [PubMed] [Google Scholar]
- 241.Kim JH, Li H, Stallcup MR. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol Cell. 2003;12(6):1537–1549. doi: 10.1016/s1097-2765(03)00450-7. [DOI] [PubMed] [Google Scholar]
- 242.Yang CK, Kim JH, Li H, Stallcup MR. Differential use of functional domains by coiled-coil coactivator in its synergistic coactivator function with beta-catenin or GRIP1. J Biol Chem. 2006;281(6):3389–3397. doi: 10.1074/jbc.M510403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, et al. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93(4):531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 244.Cole TJ, Blendy JA, Monaghan AP, Krieglstein K, Schmid W, Aguzzi A, et al. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 1995;9(13):1608–1621. doi: 10.1101/gad.9.13.1608. [DOI] [PubMed] [Google Scholar]
- 245.Jonat C, Rahmsdorf HJ, Park KK, Cato AC, Gebel S, Ponta H, et al. Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell. 1990;62(6):1189–1204. doi: 10.1016/0092-8674(90)90395-u. [DOI] [PubMed] [Google Scholar]
- 246.Konig H, Ponta H, Rahmsdorf HJ, Herrlich P. Interference between pathway-specific transcription factors: glucocorticoids antagonize phorbol ester-induced AP-1 activity without altering AP-1 site occupation in vivo. Embo J. 1992;11(6):2241–2246. doi: 10.1002/j.1460-2075.1992.tb05283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Schule R, Rangarajan P, Kliewer S, Ransone LJ, Bolado J, Yang N, et al. Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell. 1990;62(6):1217–1226. doi: 10.1016/0092-8674(90)90397-w. [DOI] [PubMed] [Google Scholar]
- 248.Yang-Yen HF, Chambard JC, Sun YL, Smeal T, Schmidt TJ, Drouin J, et al. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell. 1990;62(6):1205–1215. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]
- 249.Caldenhoven E, Liden J, Wissink S, Van de Stolpe A, Raaijmakers J, Koenderman L, et al. Negative cross-talk between RelA and the glucocorticoid receptor: a possible mechanism for the antiinflammatory action of glucocorticoids. Mol Endocrinol. 1995;9(4):401–412. doi: 10.1210/mend.9.4.7659084. [DOI] [PubMed] [Google Scholar]
- 250.Ray A, Prefontaine KE. Physical association and functional antagonism between the p65 subunit of transcription factor NF-kappa B and the glucocorticoid receptor. Proc Natl Acad Sci U S A. 1994;91(2):752–756. doi: 10.1073/pnas.91.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Scheinman RI, Gualberto A, Jewell CM, Cidlowski JA, Baldwin AS., Jr Characterization of mechanisms involved in transrepression of NF-kappa B by activated glucocorticoid receptors. Mol Cell Biol. 1995;15(2):943–953. doi: 10.1128/mcb.15.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL. Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J Biol Chem. 2001;276(17):13615–13621. doi: 10.1074/jbc.M008384200. [DOI] [PubMed] [Google Scholar]
- 253.Lee SK, Kim JH, Lee YC, Cheong J, Lee JW. Silencing mediator of retinoic acid and thyroid hormone receptors, as a novel transcriptional corepressor molecule of activating protein-1, nuclear factor-kappaB, and serum response factor. J Biol Chem. 2000;275(17):12470–12474. doi: 10.1074/jbc.275.17.12470. [DOI] [PubMed] [Google Scholar]
- 254.Tyree CM, Zou A, Allegretto EA. 17beta-Estradiol inhibits cytokine induction of the human E-selectin promoter. J Steroid Biochem Mol Biol. 2002;80(3):291–297. doi: 10.1016/s0960-0760(02)00022-5. [DOI] [PubMed] [Google Scholar]
- 255.Chipuk JE, Cornelius SC, Pultz NJ, Jorgensen JS, Bonham MJ, Kim SJ, et al. The androgen receptor represses transforming growth factor-beta signaling through interaction with Smad3. J Biol Chem. 2002;277(2):1240–1248. doi: 10.1074/jbc.M108855200. [DOI] [PubMed] [Google Scholar]
- 256.Schneikert J, Peterziel H, Defossez PA, Klocker H, Launoit Y, Cato AC. Androgen receptor-Ets protein interaction is a novel mechanism for steroid hormone-mediated down-modulation of matrix metalloproteinase expression. J Biol Chem. 1996;271(39):23907–23913. doi: 10.1074/jbc.271.39.23907. [DOI] [PubMed] [Google Scholar]
- 257.Sato N, Sadar MD, Bruchovsky N, Saatcioglu F, Rennie PS, Sato S, et al. Androgenic induction of prostate-specific antigen gene is repressed by protein-protein interaction between the androgen receptor and AP-1/c-Jun in the human prostate cancer cell line LNCaP. J Biol Chem. 1997;272(28):17485–17494. doi: 10.1074/jbc.272.28.17485. [DOI] [PubMed] [Google Scholar]
- 258.Sasaki S, Lesoon-Wood LA, Dey A, Kuwata T, Weintraub BD, Humphrey G, et al. Ligand-induced recruitment of a histone deacetylase in the negative-feedback regulation of the thyrotropin beta gene. Embo J. 1999;18(19):5389–5398. doi: 10.1093/emboj/18.19.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Rogatsky I, Zarember KA, Yamamoto KR. Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase-3 response element that mediates regulation by phorbol esters and hormones. Embo J. 2001;20(21):6071–6083. doi: 10.1093/emboj/20.21.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci USA. 2002;99(26):16701–16706. doi: 10.1073/pnas.262671599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14(18):2314–2329. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Viluksela M, Raasmaja A, Lebofsky M, Stahl BU, Rozman KK. Tissue-specific effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on the activity of 5′-deiodinases I and II in rats. Toxicol Lett. 2004;147(2):133–142. doi: 10.1016/j.toxlet.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 263.Nishimura N, Miyabara Y, Sato M, Yonemoto J, Tohyama C. Immunohistochemical localization of thyroid stimulating hormone induced by a low oral dose of 2,3,7,8-tetrachlorodibenzo-p-dioxin in female Sprague-Dawley rats. Toxicology. 2002;171(2–3):73–82. doi: 10.1016/s0300-483x(01)00559-5. [DOI] [PubMed] [Google Scholar]
- 264.Pavuk M, Schecter AJ, Akhtar FZ, Michalek JE. Serum 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) levels and thyroid function in Air Force veterans of the Vietnam War. Ann Epidemiol. 2003;13(5):335–343. doi: 10.1016/s1047-2797(02)00422-2. [DOI] [PubMed] [Google Scholar]
- 265.Abbott BD. Review of the interaction between TCDD and glucocorticoids in embryonic palate. Toxicology. 1995;105(2–3):365–373. doi: 10.1016/0300-483x(95)03234-7. [DOI] [PubMed] [Google Scholar]
- 266.Abbott BD, Perdew GH, Buckalew AR, Birnbaum LS. Interactive regulation of Ah and glucocorticoid receptors in the synergistic induction of cleft palate by 2,3,7,8-tetrachlorodibenzo-p-dioxin and hydrocortisone. Toxicol Appl Pharmacol. 1994;128(1):138–150. doi: 10.1006/taap.1994.1191. [DOI] [PubMed] [Google Scholar]
- 267.Mizuyachi K, Son DS, Rozman KK, Terranova PF. Alteration in ovarian gene expression in response to 2,3,7,8-tetrachlorodibenzo-p-dioxin: reduction of cyclooxygenase-2 in the blockage of ovulation. Reprod Toxicol. 2002;16(3):299–307. doi: 10.1016/s0890-6238(02)00024-2. [DOI] [PubMed] [Google Scholar]
- 268.Chen CL, Brodie AE, Hu CY. CCAAT/ enhancer-binding protein beta is not affected by tetra-chlorodibenzo-p-dioxin (TCDD) inhibition of 3T3-L1 preadipocyte differentiation. Obes Res. 1997;5(2):146–152. doi: 10.1002/j.1550-8528.1997.tb00655.x. [DOI] [PubMed] [Google Scholar]
- 269.Alexander DL, Ganem LG, Fernandez-Salguero P, Gonzalez F, Jefcoate CR. Aryl-hydrocarbon receptor is an inhibitory regulator of lipid synthesis and of commitment to adipogenesis. J Cell Sci. 1998;111(Pt 22):3311–3322. doi: 10.1242/jcs.111.22.3311. [DOI] [PubMed] [Google Scholar]
- 270.Fallone F, Villard PH, Decome L, Seree E, Meo M, Chacon C, et al. PPARα activation potentiates AhR-induced CYP1A1 expression. Toxicology. 2005;216(2–3):122–128. doi: 10.1016/j.tox.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 271.Shaban Z, El-Shazly S, Ishizuka M, Kimura K, Kazusaka A, Fujita S. PPARalpha-dependent modulation of hepatic CYP1A by clofibric acid in rats. Arch Toxicol. 2004;78(9):496–507. doi: 10.1007/s00204-004-0569-9. [DOI] [PubMed] [Google Scholar]
- 272.Astroff B, Rowlands C, Dickerson R, Safe S. 2,3,7,8-Tetrachlorodibenzo-p-dioxin inhibition of 17 beta-estradiol-induced increases in rat uterine epidermal growth factor receptor binding activity and gene expression. Mol Cell Endocrinol. 1990;72(3):247–252. doi: 10.1016/0303-7207(90)90149-3. [DOI] [PubMed] [Google Scholar]
- 273.Astroff B, Safe S. Comparative antiestrogenic activities of 2,3,7,8-tetrachlorodibenzo-p-dioxin and 6-methyl-1,3,8-trichlorodibenzofuran in the female rat. Toxicol Appl Pharmacol. 1988;95(3):435–443. doi: 10.1016/0041-008x(88)90361-4. [DOI] [PubMed] [Google Scholar]
- 274.Astroff B, Safe S. 2,3,7,8-Tetrachlorodibenzo-p-dioxin as an antiestrogen: effect on rat uterine peroxidase activity. Biochem Pharmacol. 1990;39(3):485–488. doi: 10.1016/0006-2952(90)90054-o. [DOI] [PubMed] [Google Scholar]
- 275.Gallo MA, Hesse EJ, Macdonald GJ, Umbreit TH. Interactive effects of estradiol and 2,3,7,8-tetrachlorodibenzo-p-dioxin on hepatic cytochrome P-450 and mouse uterus. Toxicol Lett. 1986;32(1–2):123–132. doi: 10.1016/0378-4274(86)90058-5. [DOI] [PubMed] [Google Scholar]
- 276.Romkes M, Piskorska-Pliszczynska J, Safe S. Effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on hepatic and uterine estrogen receptor levels in rats. Toxicol Appl Pharmacol. 1987;87(2):306–314. doi: 10.1016/0041-008x(87)90292-4. [DOI] [PubMed] [Google Scholar]
- 277.Okey AB. An Aryl Hydrocarbon Receptor Odyssey to the Shores of Toxicology: The Deichmann Lecture, International Congress of Toxicology-XI. Toxicol Sci. 2007;98(1):5–38. doi: 10.1093/toxsci/kfm096. [DOI] [PubMed] [Google Scholar]
- 278.Greenwald P, Barlow JJ, Nasca PC, Burnett WS. Vaginal cancer after maternal treatment with synthetic estrogens. N Engl J Med. 1971;285(7):390–392. doi: 10.1056/NEJM197108122850707. [DOI] [PubMed] [Google Scholar]
- 279.Herbst AL, Ulfelder H, Poskanzer DC. Adeno-carcinoma of the vagina. Association of maternal stilbestrol therapy with tumor appearance in young women. N Engl J Med. 1971;284(15):878–881. doi: 10.1056/NEJM197104222841604. [DOI] [PubMed] [Google Scholar]
- 280.Krishnan V, Safe S. Polychlorinated biphenyls (PCBs), dibenzo-p-dioxins (PCDDs), and dibenzofurans (PCDFs) as antiestrogens in MCF-7 human breast cancer cells: quantitative structure-activity relationships. Toxicol Appl Pharmacol. 1993;120(1):55–61. doi: 10.1006/taap.1993.1086. [DOI] [PubMed] [Google Scholar]
- 281.Krishnan V, Wang X, Safe S. Estrogen receptor-Sp1 complexes mediate estrogen-induced cathepsin D gene expression in MCF-7 human breast cancer cells. J Biol Chem. 1994;269(22):15912–15917. [PubMed] [Google Scholar]
- 282.Wang F, Samudio I, Safe S. Transcriptional activation of cathepsin D gene expression by 17beta-estradiol: mechanism of aryl hydrocarbon receptor-mediated inhibition. Mol Cell Endocrinol. 2001;172(1–2):91–103. doi: 10.1016/s0303-7207(00)00379-8. [DOI] [PubMed] [Google Scholar]
- 283.Duan R, Porter W, Samudio I, Vyhlidal C, Kladde M, Safe S. Transcriptional activation of c-fos protooncogene by 17beta-estradiol: mechanism of aryl hydrocarbon receptor-mediated inhibition. Mol Endocrinol. 1999;13(9):1511–1521. doi: 10.1210/mend.13.9.0338. [DOI] [PubMed] [Google Scholar]
- 284.Wang W, Smith R, 3rd, Safe S. Aryl hydrocarbon receptor-mediated antiestrogenicity in MCF-7 cells: modulation of hormone-induced cell cycle enzymes. Arch Biochem Biophys. 1998;356(2):239–248. doi: 10.1006/abbi.1998.0782. [DOI] [PubMed] [Google Scholar]
- 285.Spink DC, Eugster HP, Lincoln DW, 2nd, Schuetz JD, Schuetz EG, Johnson JA, et al. 17 beta-estradiol hydroxylation catalyzed by human cytochrome P450 1A1: a comparison of the activities induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin in MCF-7 cells with those from heterologous expression of the cDNA. Arch Biochem Biophys. 1992;293(2):342–348. doi: 10.1016/0003-9861(92)90404-k. [DOI] [PubMed] [Google Scholar]
- 286.Spink DC, Johnson JA, Connor SP, Aldous KM, Gierthy JF. Stimulation of 17 beta-estradiol metabolism in MCF-7 cells by bromochloro- and chloromethyl-substituted dibenzo-p-dioxins and dibenzofurans: correlations with antiestrogenic activity. J Toxicol Environ Health. 1994;41(4):451–466. doi: 10.1080/15287399409531856. [DOI] [PubMed] [Google Scholar]
- 287.Spink DC, Lincoln DW, 2nd, Dickerman HW, Gierthy JF. 2,3,7,8-Tetrachlorodibenzo-p-dioxin causes an extensive alteration of 17 beta-estradiol metabolism in MCF-7 breast tumor cells. Proc Natl Acad Sci USA. 1990;87(17):6917–6921. doi: 10.1073/pnas.87.17.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Rogers JM, Denison MS. Analysis of the antiestrogenic activity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in human ovarian carcinoma BG-1 cells. Mol Pharmacol. 2002;61(6):1393–1403. doi: 10.1124/mol.61.6.1393. [DOI] [PubMed] [Google Scholar]
- 289.Krishnan V, Porter W, Santostefano M, Wang X, Safe S. Molecular mechanism of inhibition of estrogen-induced cathepsin D gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in MCF-7 cells. Mol Cell Biol. 1995;15(12):6710–6719. doi: 10.1128/mcb.15.12.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Shipley JM, Waxman DJ. Aryl hydrocarbon receptor-independent activation of estrogen receptor-dependent transcription by 3-methylcholanthrene. Toxicol Appl Pharmacol. 2006;213(2):87–97. doi: 10.1016/j.taap.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 291.Hess-Wilson JK, Knudsen KE. Endocrine disrupting compounds and prostate cancer. Cancer Lett. 2006;241(1):1–12. doi: 10.1016/j.canlet.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 292.Mandal PK. Dioxin: a review of its environmental effects and its aryl hydrocarbon receptor biology. J Comp Physiol [B] 2005;175(4):221–230. doi: 10.1007/s00360-005-0483-3. [DOI] [PubMed] [Google Scholar]
- 293.McGregor DB, Partensky C, Wilbourn J, Rice JM. An IARC evaluation of polychlorinated dibenzo-p-dioxins and polychlorinated dibenzofurans as risk factors in human carcinogenesis. Environ Health Perspect. 1998;106 Suppl 2:755–760. doi: 10.1289/ehp.98106755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Moline JM, Golden AL, Bar-Chama N, Smith E, Rauch ME, Chapin RE, et al. Exposure to hazardous substances and male reproductive health: a research framework. Environ Health Perspect. 2000;108(9):803–813. doi: 10.1289/ehp.00108803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Foley S, Middleton S, Stitson D, Mahoney M. The incidence of testicular cancer in Royal Air Force personnel. Br J Urol. 1995;76(4):495–496. doi: 10.1111/j.1464-410x.1995.tb07755.x. [DOI] [PubMed] [Google Scholar]
- 296.Kazerouni N, Thomas TL, Petralia SA, Hayes RB. Mortality among workers exposed to cutting oil mist: update of previous reports. Am J Ind Med. 2000;38(4):410–416. doi: 10.1002/1097-0274(200010)38:4<410::aid-ajim6>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 297.Imaida K, Shirai T. Endocrine disrupting chemicals and carcinogenesis--breast, testis and prostate cancer. Nippon Rinsho. 2000;58(12):2527–2532. [PubMed] [Google Scholar]
- 298.Ohlson CG, Hardell L. Testicular cancer and occupational exposures with a focus on xenoestrogens in polyvinyl chloride plastics. Chemosphere. 2000;40(9–11):1277–1282. doi: 10.1016/s0045-6535(99)00380-x. [DOI] [PubMed] [Google Scholar]
- 299.Gustafsson JA. What pharmacologists can learn from recent advances in estrogen signalling. Trends Pharmacol Sci. 2003;24(9):479–485. doi: 10.1016/S0165-6147(03)00229-3. [DOI] [PubMed] [Google Scholar]
- 300.Reen RK, Cadwallader A, Perdew GH. The subdomains of the transactivation domain of the aryl hydrocarbon receptor (AhR) inhibit AhR and estrogen receptor transcriptional activity. Arch Biochem Biophys. 2002;408(1):93–102. doi: 10.1016/s0003-9861(02)00518-0. [DOI] [PubMed] [Google Scholar]
- 301.Kharat I, Saatcioglu F. Antiestrogenic effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin are mediated by direct transcriptional interference with the liganded estrogen receptor. Cross-talk between aryl hydrocarbon- and estrogen-mediated signaling. J Biol Chem. 1996;271(18):10533–10537. doi: 10.1074/jbc.271.18.10533. [DOI] [PubMed] [Google Scholar]
- 302.Wormke M, Castro-Rivera E, Chen I, Safe S. Estrogen and aryl hydrocarbon receptor expression and crosstalk in human Ishikawa endometrial cancer cells. J Steroid Biochem Mol Biol. 2000;72(5):197–207. doi: 10.1016/s0960-0760(00)00030-3. [DOI] [PubMed] [Google Scholar]
- 303.Matthews J, Wihlen B, Thomsen J, Gustafsson JA. Aryl Hydrocarbon Receptor-Mediated Transcription: Ligand-Dependent Recruitment of Estrogen Receptor {α} to 2,3,7,8-Tetrachlorodibenzo- p-Dioxin-Responsive Promoters. Mol Cell Biol. 2005;25(13):5317–5328. doi: 10.1128/MCB.25.13.5317-5328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Chen I, Hsieh T, Thomas T, Safe S. Identification of estrogen-induced genes downregulated by AhR agonists in MCF-7 breast cancer cells using suppression subtractive hybridization. Gene. 2001;262(1–2):207–214. doi: 10.1016/s0378-1119(00)00530-8. [DOI] [PubMed] [Google Scholar]
- 305.Porter W, Wang F, Duan R, Qin C, Castro-Rivera E, Kim K, et al. Transcriptional activation of heat shock protein 27 gene expression by 17beta-estradiol and modulation by antiestrogens and aryl hydrocarbon receptor agonists. J Mol Endocrinol. 2001;26(1):31–42. doi: 10.1677/jme.0.0260031. [DOI] [PubMed] [Google Scholar]
- 306.Wormke M, Stoner M, Saville B, Safe S. Crosstalk between estrogen receptor alpha and the aryl hydrocarbon receptor in breast cancer cells involves unidirectional activation of proteasomes. FEBS Lett. 2000;478(1–2):109–112. doi: 10.1016/s0014-5793(00)01830-5. [DOI] [PubMed] [Google Scholar]
- 307.Baba T, Mimura J, Nakamura N, Harada N, Yamamoto M, Morohashi K, et al. Intrinsic function of the aryl hydrocarbon (dioxin) receptor as a key factor in female reproduction. Mol Cell Biol. 2005;25(22):10040–10051. doi: 10.1128/MCB.25.22.10040-10051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Abbott BD, Schmid JE, Pitt JA, Buckalew AR, Wood CR, Held GA, et al. Adverse reproductive outcomes in the transgenic Ah receptor-deficient mouse. Toxicol Appl Pharmacol. 1999;155(1):62–70. doi: 10.1006/taap.1998.8601. [DOI] [PubMed] [Google Scholar]
- 309.Abdelrahim M, Ariazi E, Kim K, Khan S, Barhoumi R, Burghardt R, et al. 3-Methylcholanthrene and other aryl hydrocarbon receptor agonists directly activate estrogen receptor alpha. Cancer Res. 2006;66(4):2459–2467. doi: 10.1158/0008-5472.CAN-05-3132. [DOI] [PubMed] [Google Scholar]
- 310.Liu S, Abdelrahim M, Khan S, Ariazi E, Jordan VC, Safe S. Aryl hydrocarbon receptor agonists directly activate estrogen receptor alpha in MCF-7 breast cancer cells. Biol Chem. 2006;387(9):1209–1213. doi: 10.1515/BC.2006.149. [DOI] [PubMed] [Google Scholar]
- 311.Tsuchiya Y, Nakajima M, Kyo S, Kanaya T, Inoue M, Yokoi T. Human CYP1B1 is regulated by estradiol via estrogen receptor. Cancer Res. 2004;64(9):3119–3125. doi: 10.1158/0008-5472.can-04-0166. [DOI] [PubMed] [Google Scholar]
- 312.Hockings JK, Thorne PA, Kemp MQ, Morgan SS, Selmin O, Romagnolo DF, et al. The ligand status of the aromatic hydrocarbon receptor modulates transcriptional activation of BRCA-1 promoter by estrogen. Cancer Res. 2006;66(4):2224–2232. doi: 10.1158/0008-5472.CAN-05-1619. [DOI] [PubMed] [Google Scholar]
- 313.Khan S, Barhoumi R, Burghardt R, Liu S, Kim K, Safe S. Molecular mechanism of inhibitory aryl hydrocarbon receptor-estrogen receptor/Sp1 cross talk in breast cancer cells. Mol Endocrinol. 2006;20(9):2199–2214. doi: 10.1210/me.2006-0100. [DOI] [PubMed] [Google Scholar]
- 314.Widerak M, Ghoneim C, Dumontier MF, Quesne M, Corvol MT, Savouret JF. The aryl hydrocarbon receptor activates the retinoic acid receptoralpha through SMRT antagonism. Biochimie. 2006;88(3–4):387–397. doi: 10.1016/j.biochi.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 315.Karin M, Yamamoto Y, Wang QM. The IKK NF-kappa B system: a treasure trove for drug development. Nat Rev Drug Discov. 2004;3(1):17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 316.Beg AA, Ruben SM, Scheinman RI, Haskill S, Rosen CA, Baldwin AS., Jr I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: a mechanism for cytoplasmic retention. Genes Dev. 1992;6(10):1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 317.Voleti B, Agrawal A. Regulation of basal and induced expression of C-reactive protein through an overlapping element for OCT-1 and NF-kappaB on the proximal promoter. J Immunol. 2005;175(5):3386–3390. doi: 10.4049/jimmunol.175.5.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9(2):240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 319.Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem. 1998;273(45):29279–29282. doi: 10.1074/jbc.273.45.29279. [DOI] [PubMed] [Google Scholar]
- 320.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365(Pt 3):561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Roebuck KA. Regulation of interleukin-8 gene expression. J Interferon Cytokine Res. 1999;19(5):429–438. doi: 10.1089/107999099313866. [DOI] [PubMed] [Google Scholar]
- 322.Tsatsanis C, Androulidaki A, Venihaki M, Margioris AN. Signalling networks regulating cyclooxygenase-2. Int J Biochem Cell Biol. 2006;38(10):1654–1661. doi: 10.1016/j.biocel.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 323.Morgan ET. Regulation of cytochromes P450 during inflammation and infection. Drug Metab Rev. 1997;29(4):1129–1188. doi: 10.3109/03602539709002246. [DOI] [PubMed] [Google Scholar]
- 324.Gharavi N, El-Kadi AO. Down-regulation of aryl hydrocarbon receptor-regulated genes by tumor necrosis factor-alpha and lipopolysaccharide in murine hepatoma Hepa 1c1c7 cells. J Pharm Sci. 2005;94(3):493–506. doi: 10.1002/jps.20267. [DOI] [PubMed] [Google Scholar]
- 325.Jeong HG. Cytokine-mediated suppression of cytochrome P450 1A1 in Hepa-1c1c7 cells by pokeweed mitogen. Toxicol Lett. 2001;119(2):125–132. doi: 10.1016/s0378-4274(00)00310-6. [DOI] [PubMed] [Google Scholar]
- 326.Paton TE, Renton KW. Cytokine-mediated down-regulation of CYP1A1 in Hepa1 cells. Biochem Pharmacol. 1998;55(11):1791–1796. doi: 10.1016/s0006-2952(98)00028-8. [DOI] [PubMed] [Google Scholar]
- 327.Abdel-Razzak Z, Loyer P, Fautrel A, Gautier JC, Corcos L, Turlin B, et al. Cytokines down-regulate expression of major cytochrome P-450 enzymes in adult human hepatocytes in primary culture. Mol Pharmacol. 1993;44(4):707–715. [PubMed] [Google Scholar]
- 328.Muntane-Relat J, Ourlin JC, Domergue J, Maurel P. Differential effects of cytokines on the inducible expression of CYP1A1, CYP1A2, and CYP3A4 in human hepatocytes in primary culture. Hepatology. 1995;22(4 Pt 1):1143–1153. [PubMed] [Google Scholar]
- 329.Nicholson TE, Renton KW. Role of cytokines in the lipopolysaccharide-evoked depression of cytochrome P450 in the brain and liver. Biochem Pharmacol. 2001;62(12):1709–1717. doi: 10.1016/s0006-2952(01)00859-0. [DOI] [PubMed] [Google Scholar]
- 330.Barker CW, Fagan JB, Pasco DS. Interleukin-1 beta suppresses the induction of P4501A1 and P4501A2 mRNAs in isolated hepatocytes. J Biol Chem. 1992;267(12):8050–8055. [PubMed] [Google Scholar]
- 331.Morel Y, Mermod N, Barouki R. An autoregulatory loop controlling CYP1A1 gene expression: role of H(2)O(2) and NFI. Mol Cell Biol. 1999;19(10):6825–6832. doi: 10.1128/mcb.19.10.6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Ke S, Rabson AB, Germino JF, Gallo MA, Tian Y. Mechanism of suppression of cytochrome P-450 1A1 expression by tumor necrosis factor-alpha and lipopolysaccharide. J Biol Chem. 2001;276(43):39638–39644. doi: 10.1074/jbc.M106286200. [DOI] [PubMed] [Google Scholar]
- 333.Reiners JJ, Jr, Cantu AR, Scholler A. Phorbol ester-mediated suppression of cytochrome P450 Cyp1a-1 induction in murine skin: involvement of protein kinase C. Biochem Biophys Res Commun. 1992;186(2):970–976. doi: 10.1016/0006-291x(92)90841-8. [DOI] [PubMed] [Google Scholar]
- 334.Reiners JJ, Jr, Scholler A, Bischer P, Cantu AR, Pavone A. Suppression of cytochrome P450 Cyp1a-1 induction in murine hepatoma 1c1c7 cells by 12-O-tetradecanoylphorbol-13-acetate and inhibitors of protein kinase C. Arch Biochem Biophys. 1993;301(2):449–454. doi: 10.1006/abbi.1993.1170. [DOI] [PubMed] [Google Scholar]
- 335.Long WP, Pray-Grant M, Tsai JC, Perdew GH. Protein kinase C activity is required for aryl hydrocarbon receptor pathway-mediated signal transduction. Mol Pharmacol. 1998;53(4):691–700. doi: 10.1124/mol.53.4.691. [DOI] [PubMed] [Google Scholar]
- 336.Cochran BH. Regulation of immediate early gene expression. NIDA Res Monogr. 1993;125:3–24. [PubMed] [Google Scholar]
- 337.Remacle J, Raes M, Toussaint O, Renard P, Rao G. Low levels of reactive oxygen species as modulators of cell function. Mutat Res. 1995;316(3):103–122. doi: 10.1016/0921-8734(95)90004-7. [DOI] [PubMed] [Google Scholar]
- 338.Thurmond TS, Silverstone AE, Baggs RB, Quimby FW, Staples JE, Gasiewicz TA. A chimeric aryl hydrocarbon receptor knockout mouse model indicates that aryl hydrocarbon receptor activation in hematopoietic cells contributes to the hepatic lesions induced by 2,3,7, 8-tetrachlorodibenzo-p-dioxin. Toxicol Appl Pharmacol. 1999;158(1):33–40. doi: 10.1006/taap.1999.8681. [DOI] [PubMed] [Google Scholar]
- 339.Tian Y, Rabson AB, Gallo MA. Ah receptor and NF-kappaB interactions: mechanisms and physiological implications. Chem Biol Interact. 2002;141(1–2):97–115. doi: 10.1016/s0009-2797(02)00068-6. [DOI] [PubMed] [Google Scholar]
- 340.Vrzal R, Ulrichova J, Dvorak Z. Aromatic hydrocarbon receptor status in the metabolism of xenobiotics under normal and pathophysiological conditions. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2004;148(1):3–10. [PubMed] [Google Scholar]
- 341.Jensen BA, Leeman RJ, Schlezinger JJ, Sherr DH. Aryl hydrocarbon receptor (AhR) agonists suppress interleukin-6 expression by bone marrow stromal cells: an immunotoxicology study. Environ Health. 2003;2(1):16. doi: 10.1186/1476-069X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Takanaga H, Yoshitake T, Yatabe E, Hara S, Kunimoto M. Beta-naphthoflavone disturbs astrocytic differentiation of C6 glioma cells by inhibiting autocrine interleukin-6. J Neurochem. 2004;90(3):750–757. doi: 10.1111/j.1471-4159.2004.02681.x. [DOI] [PubMed] [Google Scholar]
- 343.Ruby CE, Leid M, Kerkvliet NI. 2,3,7,8-Tetrachlorodibenzo-p-dioxin suppresses tumor necrosis factor-alpha and anti-CD40-induced activation of NF-kappaB/Rel in dendritic cells: p50 homodimer activation is not affected. Mol Pharmacol. 2002;62(3):722–728. doi: 10.1124/mol.62.3.722. [DOI] [PubMed] [Google Scholar]
- 344.Thatcher TH, Maggirwar SB, Baglole CJ, Lakatos HF, Gasiewicz TA, Phipps RP, et al. Aryl hydrocarbon receptor-deficient mice develop heightened inflammatory responses to cigarette smoke and endotoxin associated with rapid loss of the nuclear factor-kappaB component RelB. Am J Pathol. 2007;170(3):855–864. doi: 10.2353/ajpath.2007.060391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Negishi T, Kato Y, Ooneda O, Mimura J, Takada T, Mochizuki H, et al. Effects of aryl hydrocarbon receptor signaling on the modulation of TH1/TH2 balance. J Immunol. 2005;175(11):7348–7356. doi: 10.4049/jimmunol.175.11.7348. [DOI] [PubMed] [Google Scholar]
- 346.Hoffer A, Chang CY, Puga A. Dioxin induces transcription of fos and jun genes by Ah receptor-dependent and -independent pathways. Toxicol Appl Pharmacol. 1996;141(1):238–247. doi: 10.1006/taap.1996.0280. [DOI] [PubMed] [Google Scholar]
- 347.Suh J, Jeon YJ, Kim HM, Kang JS, Kaminski NE, Yang KH. Aryl hydrocarbon receptor-dependent inhibition of AP-1 activity by 2,3,7,8-tetrachlorodibenzo-p-dioxin in activated B cells. Toxicol Appl Pharmacol. 2002;181(2):116–123. doi: 10.1006/taap.2002.9403. [DOI] [PubMed] [Google Scholar]
- 348.Quattrochi LC, Shih H, Pickwell GV. Induction of the human CYP1A2 enhancer by phorbol ester. Arch Biochem Biophys. 1998;350(1):41–48. doi: 10.1006/abbi.1997.0491. [DOI] [PubMed] [Google Scholar]
- 349.Jeffy BD, Hockings JK, Kemp MQ, Morgan SS, Hager JA, Beliakoff J, et al. An estrogen receptor-alpha/p300 complex activates the BRCA-1 promoter at an AP-1 site that binds Jun/Fos transcription factors: repressive effects of p53 on BRCA-1 transcription. Neoplasia. 2005;7(9):873–882. doi: 10.1593/neo.05256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Pimental RA, Liang B, Yee GK, Wilhelmsson A, Poellinger L, Paulson KE. Dioxin receptor and C/EBP regulate the function of the glutathione S-transferase Ya gene xenobiotic response element. Mol Cell Biol. 1993;13(7):4365–4373. doi: 10.1128/mcb.13.7.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Shin SM, Cho IJ, Kim SG. CCAAT/enhancer binding protein activation by PD98059 contributes to the inhibition of AhR-mediated 3-methylcholanthrene induction of CYP1A1. Xenobiotica. 2005;35(10–11):975–987. doi: 10.1080/00498250500354584. [DOI] [PubMed] [Google Scholar]
- 352.Vogel C, Boerboom AM, Baechle C, El-Bahay C, Kahl R, Degen GH, et al. Regulation of prostaglandin endoperoxide H synthase-2 induction by dioxin in rat hepatocytes: possible c-Src-mediated pathway. Carcinogenesis. 2000;21(12):2267–2274. doi: 10.1093/carcin/21.12.2267. [DOI] [PubMed] [Google Scholar]
- 353.De Bosscher K, Vanden Berghe W, Haegeman G. Cross-talk between nuclear receptors and nuclear factor kappaB. Oncogene. 2006;25(51):6868–6886. doi: 10.1038/sj.onc.1209935. [DOI] [PubMed] [Google Scholar]
- 354.Pascual G, Glass CK. Nuclear receptors versus inflammation: mechanisms of transrepression. Trends Endocrinol Metab. 2006;17(8):321–327. doi: 10.1016/j.tem.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 355.Zhou C, Tabb MM, Nelson EL, Grun F, Verma S, Sadatrafiei A, et al. Mutual repression between steroid and xenobiotic receptor and NF-kappaB signaling pathways links xenobiotic metabolism and inflammation. J Clin Invest. 2006;116(8):2280–2289. doi: 10.1172/JCI26283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Buck MJ, Nobel AB, Lieb JD. ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 2005;6(11):R97. doi: 10.1186/gb-2005-6-11-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, et al. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci U S A. 2007;104(12):4852–4857. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Booth EA, Marchesi M, Knittel AK, Kilbourne EJ, Lucchesi BR. The Pathway-Selective Estrogen Receptor Ligand WAY-169916 Reduces Infarct Size After Myocardial Ischemia and Reperfusion by an Estrogen Receptor Dependent Mechanism. J Cardiovasc Pharmacol. 2007;49(6):401–407. doi: 10.1097/FJC.0b013e3180544527. [DOI] [PubMed] [Google Scholar]
- 359.Chadwick CC, Chippari S, Matelan E, Borges-Marcucci L, Eckert AM, Keith JC, Jr, et al. Identification of pathway-selective estrogen receptor ligands that inhibit NF-kappaB transcriptional activity. Proc Natl Acad Sci USA. 2005;102(7):2543–2548. doi: 10.1073/pnas.0405841102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Steffan RJ, Matelan E, Ashwell MA, Moore WJ, Solvibile WR, Trybulski E, et al. Control of chronic inflammation with pathway selective estrogen receptor ligands. Curr Top Med Chem. 2006;6(2):103–111. doi: 10.2174/156802606775270279. [DOI] [PubMed] [Google Scholar]
- 361.McDougal A, Gupta MS, Morrow D, Ramamoorthy K, Lee JE, Safe SH. Methyl-substituted diindolylmethanes as inhibitors of estrogen-induced growth of T47D cells and mammary tumors in rats. Breast Cancer Res Treat. 2001;66(2):147–157. doi: 10.1023/a:1010608000074. [DOI] [PubMed] [Google Scholar]