Abstract
DNA interstrand cross-links (ICLs), widely used in chemotherapy, are cytotoxic lesions because they block replication and transcription. Repair of ICLs involves proteins from different repair pathways however the precise mechanism is still not completely understood. Here, we report that the 3-methyladenine DNA glycosylase (Aag), an enzyme that initiates base excision repair at a variety of alkylated bases, is also involved plays a role in the repair of ICLs. Aag−/− mouse embryonic stem cells were shown to be more sensitive to the cross-linking agent 4, 5’, 8-trimethylpsoralen than wild-type cells, but no more sensitive than wild-type to the psoralen derivative Angelicin that forms only monoadducts. We show that γ-H2AX foci formation, a marker for double strand breaks that are formed during ICL repair, is impaired in psoralen treated Aag−/− cells in both quantity and kinetics. However, in our in vitro system, purified human AAG can neither bind to the ICL nor cleave it. Taken together, our results suggest that Aag is important for the resistance of mouse ES cells to psoralen-induced ICLs. has a role in the repair of ICLs in mouse ES cells, but that its involvement may be indirect, perhaps mediated through interaction with other proteins, or by its action on an intermediate of ICL repair.
1. Introduction
Cells are constantly exposed to DNA damaging agents. To overcome this constant assault, many different pathways have evolved to repair the damage thus restoring normal replication and transcription. Approximately 150 genes participate in different pathways of damage repair or tolerance in humans [1]. For every type of DNA damage, there is at least one repair mechanism or pathway, and some kinds of damage can be acted upon by several different pathways.
The enzyme 3-methyladenine DNA glycosylase (AAG in human, Aag in mouse) is specialized in removing various types of modified bases from the DNA, such as 3-methyladenine, 7-methylguanine, hypoxanthine (Hx) and 1,N6-ethenoadenine, among others [2,3]. AAG recognizes the damaged base and initiates the base excision repair (BER) process by cleaving the N-glycosylic bond between the damaged base and the deoxyribose, creating an abasic site [2,4]. In its simplest form, BER is completed by the action of AP endonuclease (APE1 in human) which cleaves at the abasic site, DNA polymerase β which trims the 5’ end and fills in the missing nucleotide, and DNA ligase which seals the nick [5–7].
Mouse embryonic stem (ES) cells that lack Aag are more sensitive than wild-type to alkylating agents such as methyl methanesulfonate (MMS) [8,9]. Interestingly, it was shown that Aag−/− mouse ES cells are also sensitive to 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU) and mitomycin C (MMC), both of which are chemotherapeutic agents known to induce DNA interstrand cross-links (ICLs) [8,10]; both BCNU and MMC initially induce monoadducts, only some of which can further react to form ICLs. Although Aag had no apparent in vitro glycosylase activity on double stranded DNA containing a MMC ICL or N2-guanine monoadduct [10], the sensitivity of Aag−/− cells to MMC could be explained by a possible role in the repair of yet another in vivo monoadduct formed by MMC. As for BCNU, it produces lesions at both the N7 and the O6 positions of guanine. O6-chloroethylguanine is normally repaired via direct reversal by the O6-methylguanine DNA methyltransferase (MGMT). However, when O6-chloroethylguanine escapes repair by MGMT it can go on to rearrange into an 1,O6-ethanoguanine lesion, which in turn goes on to react with the cytosine opposite, forming an ICL. 1,O6-ethanoguanine is structurally similar to 1,N6-ethenoadenine that is a known substrate for Aag. Therefore, Aag might protect ES cells against BCNU and MMC by repairing monoadducts that have the potential to form ICLs, rather than by repairing ICLs per se [10]. In addition, mutations in the yeast MAG1 gene, the functional homologue of Aag, render cells sensitive to nitrogen mustard treatment [11].
ICLs are very detrimental lesions to the cell, since they block fundamental processes required for cell survival – namely replication and transcription. The mechanisms for repair of ICLs in bacteria and yeast are somewhat understood, and appear to involve the nucleotide excision repair (NER) and homologous recombination (HR) pathways, as well as translesion synthesis (TLS) [12]. Likewise, ICL repair in mammalian cells is believed to involve some proteins from NER, HR and TLS pathways, along with other proteins [12–14]. The major repair pathway is believed to be both replication and recombination dependent, although two other minor repair pathways have been proposed [15–17]. According to most models, the major repair pathway for ICLs in mammals is initiated when the replication fork is stalled at the lesion, followed by strand cleavage on the fork side of the ICL, generating a collapsed replication fork with a one-sided double strand break (DSB) [13,14,18–21]. This cleavage is thought to be mediated by a structure-specific endonuclease, either Mus81-Eme1 [22] or XPF-ERCC1 [23]. Thereafter, XPF-ERCC1 cleaves the DNA on the other side of the cross-link, unhooking it from the dsDNA [14,23]. The requirement for only XPF-ERCC1 from the NER machinery for that step explains the hypersensitivity of XPF and ERCC1 mutants to ICLs agents, while other NER mutants exhibit only mild sensitivity [13,24]. After the lesion is unhooked and thus tethered to only one strand, the gap opposite can be filled via lesion bypass by a translesion polymerase. Once the gap opposite the ICL is filled, a simple NER process can excise the unhooked lesion and the gap will be filled by a polymerase, restoring the continuity of the DNA. The one sided DSB that was formed at the replication fork at the beginning of the process then needs to participate in replication fork restoration, probably by the action of the homologous recombination machinery. Strong evidence supports the involvement of homologous recombination in ICL repair, since mutations in the XRCC2, XRCC3, RAD51C, and RAD51D genes result in severe sensitivity to ICL inducing agents [13,25–27].
Additional proteins from other repair pathways have been shown to be involved in ICL repair. hMutSβ appears to be required for the recognition and uncoupling of psoralen ICLs in vitro [28]. Moreover, MMR deficient cells (MSH2 mutants) are hypersensitive to psoralen ICLs, but do not have lower frequencies of cross-link-induced mutations, suggesting that MMR may be involved in a relatively error-free mechanism to process ICLs [29]. The Fanconi Anemia proteins are thought to have a role both in the regulation of ICL repair, and in the actual repair reaction through FANCM [30,31] and FANCJ [11,32,33] (reviewed in [18] and [34]). BRCA2, which plays a role in homologous recombination [35] is the Fanconi Anemia gene FANCD1 [36]. Using an in vitro assay it was shown that BRAC2 participates in the repair of DSBs generated when replication forks encounter ICLs [37]. Monoubiquitination of FANCD2 promotes BRCA2 loading into chromatin complexes, that are required for normal homology-directed DNA repair [38,39]. In addition, it was suggested that the pre-mRNA spliceosome complex Pso4 together with the Werner helicase are required for ICL processing, in coordination with BRAC1 [40,41].
The complexity, the different possible repair pathways, and the variety of proteins involved in ICL repair, raise several questions: how is ICL repair initiated? How are the different types of ICLs recognized in the cell, and which proteins are involved in this recognition? Here, we show that Aag−/− mouse ES cells are more sensitive than wild-type cells to the interstrand cross-linking agent 4, 5’, 8-trimethylpsoralen (TMP), while they have similar sensitivity to Angelicin, a psoralen derivative which forms mainly monoadducts and no cross-links. The formation of γ-H2AX foci, markers for DSBs, was delayed and less robust in Aag−/− cells compared to wild-type cells following treatment with TMP, while there was no difference following treatment with Angelicin. Caspase 3 activation, a marker for apoptosis, was higher in Aag−/− than in wild-type cells following the cross-linking treatment. Taken together, our results suggest an important role for Aag in cellular resistance to repair of psoralen ICLs.
2. Material and methods
2.1. Reagents and proteins
Cells were treated with either MMS, 4, 5’, 8-trimethylpsoralen (TMP) or Angelicin (Sigma-Aldrich). TMP and Angelicin treatments were followed by UVA irradiation using a UVA lamp at 365 nm (UVP, CA). The truncated form of human 3-methyladenine DNA glycosylase (Δ80 AAG) and the full length human AAG were purified. Human AP endonuclease (APE1) was from Trevigen. Monoclonal anti-phospho-Histone H2AX antibody was from Upstate Technology (NY). Alexa fluor 594 goat anti mouse, Alexa fluor 647 goat anti rabbit, and ProLong Gold with DAPI were from Invitrogen. Rabbit anti-active Caspase-3 was from BD Pharmingen. Giemsa staining solution (0.02% w/v), 10x TBS, Triton-X were from Sigma. 16% solution EM grade paraformaldehyde was from Electron Microscopy Sciences. 10% Twin 20 and blotting grade blocker non-fat dry milk, and 40% polyacrylamide solution, were from Bio-Rad.
Cell culture reagents: DMEM, L-Glutamine, Penicillin-Streptomycin, 2-mercaptoethanol, Trypsin-EDTA were from Invitrogen, Fetal Bovine Serum (FBS) was from Hyclone.
2.2. Cells
AB1 ES cells and their Aag−/− derivative [8] were cultured on SNL76/7 feeder cells that were mitotically inactivated by gamma irradiation, in DMEM, supplemented with 15% FBS, 50 U/ml penicillin, 50 µg/ml streptomycin, 2 mM L-glutamine, and 0.1 mM beta-mercaptoethanol. ES cells that were grown without feeder cells were maintained in an undifferentiated state in the presence of LIF (leukemia inhibitory factor) purified according to Mereau et al. [42].
2.3. Drug sensitivity testing
Various dilutions of ES cells were plated onto 24 well feeder-coated plates. After 16 hours, cells were incubated for 1 hour with MMS, TMP, or Angelicin in serum free media at 37°C in the dark. For TMP and Angelicin treatments, the cells were then irradiated with UVA (20 KJ/m2). After treatment the cells were washed and supplemented with media containing serum. 7–9 days later dried colonies were air-dried, stained with Giemsa, and counted. All survival curves were performed at least 4 times. For TMP and Angelicin treatments, survival was calculated by comparing psoralen plus UVA treated cells, to cells treated with UVA alone.
2.4. DNA substrates
The oligonucleotides used in this study were as follows: Hx oligonucleotide: 5’-GCAATCTAGCCAXGTCGATGTATGC-3’ where X=Hx, and its complementary strand: 5’-GCATACATCGACTTGGCTAGATTGC-3’. The target oligonucleotides for TMP: 5’-CTCGTCTGTACACCGAAGAGC-3’, and its complementary: 5’-ACCGGCTCTTCGGTGTACAGACGAG-3’ (the Ts that are target for the TMP cross-link are boldface underlined letters). All the oligonucleotides were synthesized by Invitrogen, and purified from a denaturing urea (8M) polyacrylamide (15%) gel. For Hx DNA substrate, the oligonucleotide containing the Hx was labeled with [32P]-γATP (6000 Ci/mmol, Perkin Elmer) using polynucleotide kinase (NEB), annealed with its complimentary oligonucleotide at 80°C for 10 minutes, allowed to cool to room temperature, and then purified from the unincorporated radionucleotides using G-25 column (GE Healthcare). In order to make the TMP cross-linked substrate, the two oligonucleotides were annealed as described above. TMP (5 mg/ml in water) was added to the substrate at a 1:1 volume ratio, incubated for 5 minutes in the dark, and then irradiated at a UVA dose of 12 mW/cm2 for 20 minutes. The efficiency of the cross-linking reaction was about 30%, and the cross-linked product was purified from a 15% denaturing PAGE using UV shadowing (supplementary Fig. 1). Then, the TMP cross-linked DNA was 32P radiolabeled as described for the Hx DNA.
2.5. Glycosylase activity assay
80 fmol of substrate DNA, either containing Hx or TMP cross-link, was incubated for 1 hour at 37°C with 500 nM human full length AAG, in a final volume of 10 µl. The reaction conditions contained 20 mM Tris.Cl pH=7.5, 100 mM KCl, 5 mM EDTA, and 5 mM beta-mercaptoethanol. In order to visualize AAG activity, the abasic-site that is formed by the glycosylase activity should be cleaved. Since alkaline conditions might disrupt the TMP cross-link, we used human APE1 (70 units) together with 10 mM MgCl2. The reactions were stopped by the addition of formamide supplemented with 10 mM EDTA, Xylene Cyanol, and Bromophenol Blue, and loaded onto a 15% denaturing Urea-PAGE. The gels were exposed to storage phosphor screens (Packard) and bands visualized using a Cyclone instrument (Packard) and the program OptiQuant.
2.6. Gel mobility shift assay
20 fmol DNA was incubated in a final volume of 10 µl with various concentrations of Δ80 truncated, or full length human AAG at 16°C for 15 minutes, in a reaction mixture containing 4 mM Tris.Cl PH=7.8, 20 mM KCl, 5 mM MgCl2, 0.4 mM EDTA, 1 mM beta-mercaptoethanol, 50 ng sonicated salmon sperm DNA (Stratagene), and 10% glycerol. The samples were loaded immediately on a native 5% PAGE and run for 3 hours at 130V at 4°C. The bands were visualized as described above.
2.7. Immunofluorescence microscopy
For microscopy experiments the cells were grown on coverslips at a density of 1×105 per coverslip without feeder cells, supplemented with LIF. After treatment with either 20 KJ/m2 UVA alone, 0.3 ng/ml TMP+UVA, or with 1 mg/ml Angelicin+ UVA, cells were fixed at the indicated time points in 4% paraformaldehyde for 20 minutes, washed in Tris buffer saline with 0.1% Tween (TBST), and permeabilized in 0.5% Triton X in Tris buffer saline (TBS) for 30 minutes at room temperature. Blocking was done by gentle shaking in the presence of 5% milk in TBST for 1 hour at room temperature followed by incubation with rabbit anti-serum. Primary anti phospho-H2AX was diluted in milk to 1.6 mg/ml (1:625) and incubated for 1 hour at 37°C followed by washes with TBST and incubation with 1:200 dilution of Alexa fluor 594 in milk. The coverslips were mounted on ProLong gold with DAPI for 2–3 hours, and analyzed using Nikon Eclipse E800 Microscope, with Volocity4 / Improvision software for Image Acquisition. The γ-H2AX foci were counted throughout the volume of each nucleus at ×600 magnification. For every cell line and time point at least 100 cells were analyzed, and each time point was examined in triplicate.
2.8. Caspase-3 activation assay
For measuring apoptosis by Caspase-3 cleavage, cells were grown without feeder cells, supplemented with LIF, and 2×105–2.5×106 cells were plated on 60 mm2 dishes. Cells were treated with 0.5 ng/ml TMP + UVA (20 kJ/m2), fixed at the indicated time points with paraformaldehyde, and permeabilized in methanol for flow cytometry analysis. Cells were labeled with rabbit anti-active Caspase-3 (1:200) followed by Alexa Fluor 647 (1:500) and analyzed using BD FACSCalibur system (BD Biosciences). A target of 20,000 cells per data point was set. However, due to cell killing some data points represent a minimum of 2,000 cells. The experiment was done four times, with different number of starting cells per plate. The P value (p=0.0339) was calculated by two-way-ANOVA for the different genotypes.
3. Results
3.1. Aag−/− ES cells are more sensitive than wild-type to psoralen induced cross-links but not to psoralen induced monoadducts
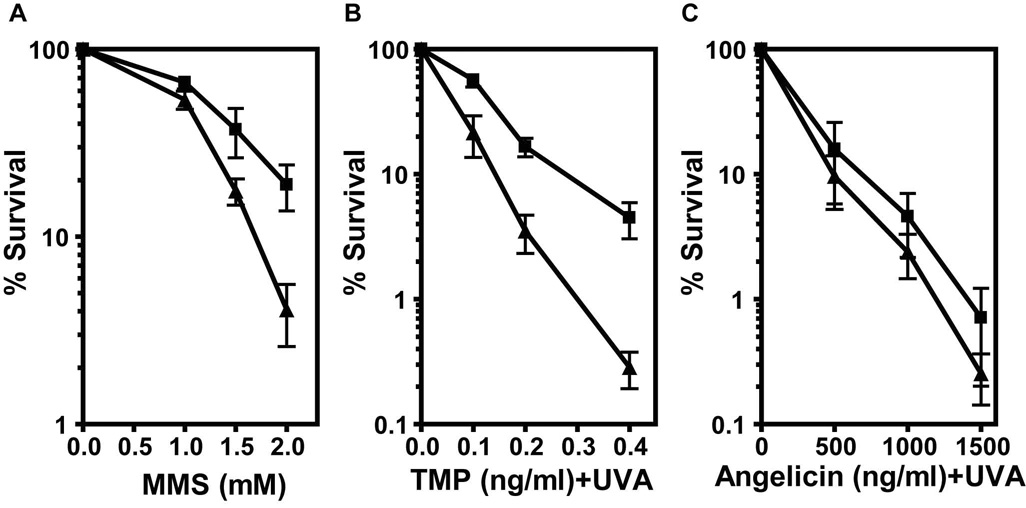
Aag is known to initiate the BER pathway at 3-methyladenine DNA replication blocking lesions, and Aag−/− ES cells are sensitive to killing by the alkylating agent MMS that efficiently induces such lesions (Fig. 1A and [8]). In order to test the involvement of Aag in the repair of cross-links, we treated Aag−/− and wild-type mouse ES cells with 4, 5’, 8-trimethylpsoralen (TMP) plus UVA irradiation to induce DNA ICLs, and compared their sensitivities. TMP acts by intercalating between DNA base pairs, and upon UVA irradiation the psoralen moiety becomes covalently linked to bases on opposite strands forming the ICL and linking the two DNA strands together. TMP creates a higher fraction of ICLs than other cross-linking agents such as BCNU, cisplatin or MMC [12], and the cross-link formation is controlled by the co-treatment with UVA. A major advantage of a psoralen cross-linking agent is that it has a chemical analogue, Angelicin that allows comparison between the biological effects of monoadducts versus ICLs. Cells were incubated with different doses of TMP for one hour in the dark, and then irradiated with 20 KJ/m2 UVA. Using a colony forming survival assay we found that Aag−/− cells are more sensitive than wild-type to TMP+UVA treatment (Fig. 1B). Surprisingly, the difference in survival between the wild-type and Aag−/− cells was even more pronounced than that following MMS treatment (Fig. 1A, and [8]). TMP treatment of wild-type and Aag−/− cells without UVA did not result in any cell killing, and UVA alone induced minimal cell death (data not shown). Importantly, no difference in survival was detected between wild-type and Aag−/− cells after UVA treatment (data not shown).
Figure 1. Aag−/− cells are sensitive to the cross-linking treatment with TMP+UVA.
Cell survival of wild-type (▯) and Aag−/− (▯) mouse ES cells was measured following treatments with MMS (A), TMP+UVA (B) or Angelicin+UVA (C). Each data point represents the mean ± SEM of at least 4 independent experiments.
Angelicin is a psoralen derivative that together with UVA produces mainly monoadducts and very few, if any, cross-links. The molar concentration of Angelicin required to induce 95% cell death in wild-type cells was more than 3,000 fold higher than that for TMP (Fig. 1B, 1C), underscoring the toxicity of ICLs compared to monoadducts. When ES cells were treated with Angelicin+UVA, the wild-type and Aag−/− cells showed similar sensitivity (Fig. 1C). We infer from these results that Aag protects specifically against the ICLs produced by the TMP+UVA treatment, and not against the monoadducts that are formed by both TMP and Angelicin.
We next tested the ability of purified human AAG protein to either cleave or bind a short double stranded DNA oligonucleotide with a site specific TMP cross-link. Although AAG was able to cleave and bind a control oligonucleotide DNA containing Hx very efficiently, no cleavage or binding was seen to the cross-linked DNA (supplemental figure 1); suggesting that Aag’s role in conferring protection against psoralen-induced ICLs is either indirect or involves a DNA substrate that is an intermediate of the repair process.
3.2. Human AAG is unable to cleave or bind a TMP cross-link in a short dsDNA
One explanation for the role that Aag plays in protecting cells against TMP induced ICLs is that the enzyme cleaves the glycosylic bond of one or more of the two crossed-linked bases, as it does in the normal process of base excision repair. To test the possibility that Aag plays such a direct role in cross-link repair we prepared a short dsDNA with a site specific TMP cross-link (supplementary Fig. 1). We purified full length human AAG, which shares significant sequence homology and substrate specificity with the mouse Aag enzyme, and tested its glycosylase activity on the TMP containing DNA (Fig. 2A). As a positive control we used a short dsDNA with a site specific Hx, a known substrate for AAG. The structure of AAG bound either to DNA containing an abasic site analogue or to DNA containing εA was previously solved, and it was shown that AAG flips out the damaged nucleotide into its active site, whereupon the glycosylic bond is cleaved. One would expect that the presence of an ICL in the DNA will prevent the flipping out of nucleotides into AAG active site. Nevertheless, we incubated the DNA containing the TMP ICL with AAG, and added APE1 to complete the cleavage of the putative abasic site reaction product. While AAG was able to cleave the Hx adduct with great efficiency, no cleavage at the TMP cross-link was detected.
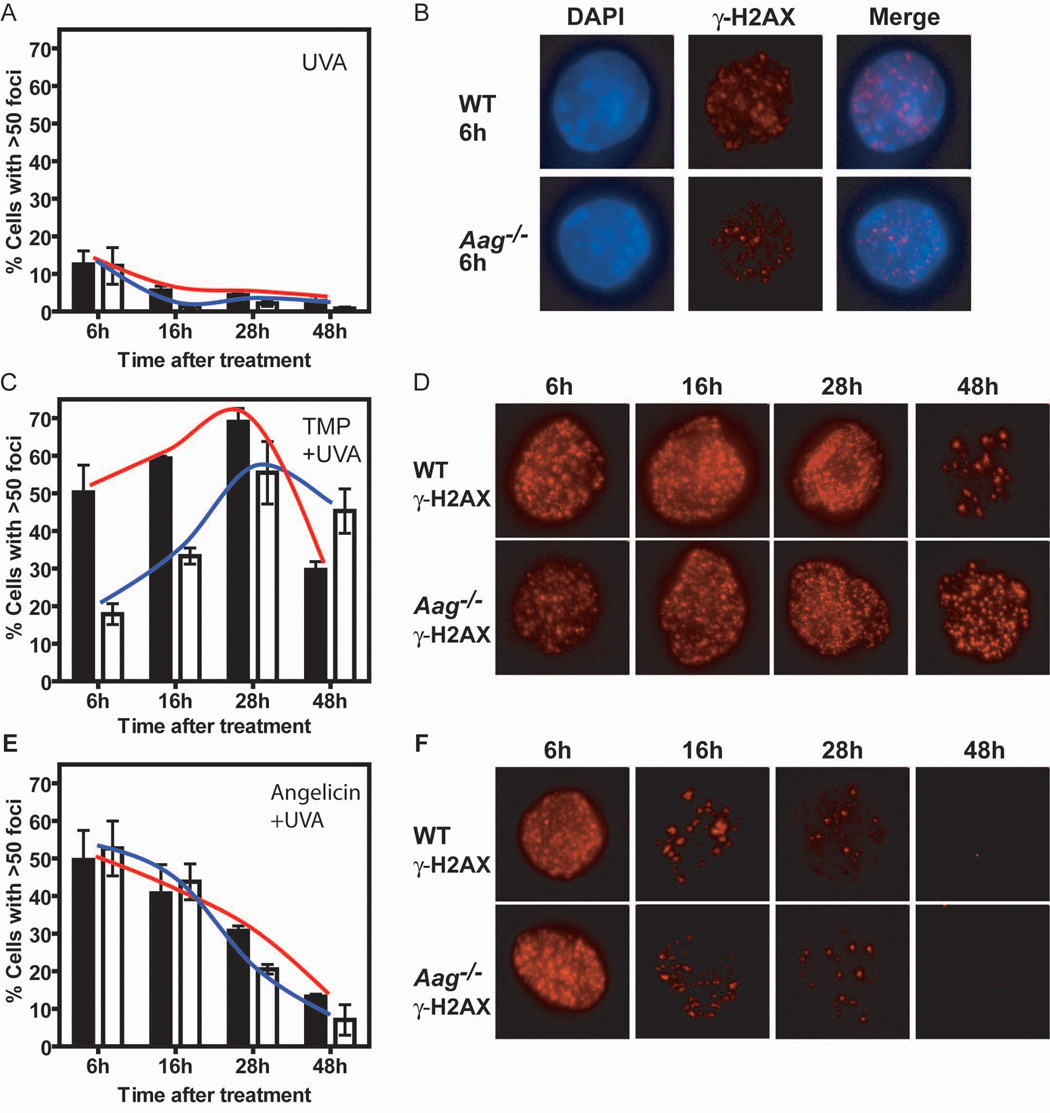
Figure 2. Aag−/− cells are impaired for the induction of γ-H2AX foci following TMP+UVA treatment. Panels A, C and E.
Graphs depict the percentage of cells that showed a significant induction of γ-H2AX foci (more than 50 foci per cell) following UVA at 20 KJ/m2 (A), TMP (0.3 ng/ml) + UVA (C), or Angelicin (1 mg/ml) + UVA (E) treatment. Black bars and red line – wild-type; White bars and blue line – Aag−/−. Panel B: Representative images of UVA-treated cells, 6 hours after treatment. Nuclei are evidenced by DAPI staining (blue) and γ-H2AX foci by labeling with an antibody for γ-H2AX (red). The merged image is also shown. Panels D and F: Representative cells at 6, 16, 28, and 48 hours after treatment with TMP+UVA (D), or Angelicin+UVA (F) are shown stained with antibody for γ-H2AX only. For every cell line and time point at least 100 cells were analyzed, and every time point was examined 3 times. Pictures were taken using a magnification of 600x.
Although no cleavage activity was detected using purified human AAG on a dsDNA with TMP cross-link, we reasoned that AAG might bind the TMP cross-link and serve to either stimulate or inhibit the binding of other proteins at the site of the lesion. In order to test whether AAG binds the TMP cross-link, we performed a gel shift mobility assay, using the same dsDNA with a site specific TMP (Fig. 2B). In our assay conditions we were unable to detect binding of full length AAG to Hx DNA. Therefore, we used an N-terminal truncated form of AAG (Δ80 AAG in this study) known to bind Hx DNA very well (Fig. 2B). However, no specific binding of either the full length AAG or the truncated protein to the TMP cross-linked DNA was observed.
3.2. Induction of γ-H2AX foci following UVA irradiation
We next monitored whether Aag influenced the formation and disappearance of γ-H2AX foci. γ-H2AX, a phosphorylated form of the histone variant H2AX, is known to localize at DSBs (reviewed in [43]), that are known intermediates of the major pathway for ICL repair [14,19,20,22]. Formation of γ-H2AX foci occurs at the beginning of the ICL repair process, and their disappearance likely indicates the completion of ICL repair. ES cells were plated on coverslips, treated, and fixed at various time points after treatment. More than 95% of untreated cells had either no foci at all, or <10 foci per cell (Supplemental Fig. 2). Since the cross-linking treatment requires both TMP and UVA, we looked first at UVA induced γ-H2AX foci. Cells treated with UVA alone developed a significant number of γ-H2AX foci (Fig. 2, panels A and B; Supplemental Figure 3A). UVA induction of γ-H2AX foci formation was previously shown [44] and is probably the consequence of oxidized DNA lesions [45,46] creating sites of single strand DNA and/or stalled replication forks that could also be sites for γ-H2AX foci formation [47–50]. UVA treated wild-type and Aag−/− cell cultures had many cells with 11–30 foci per cell by 6 hours after treatment (data not shown) but very few cells with >50 foci per cell (12% for both wild-type and Aag−/−; Fig. 2A). 16 and 28 hours after treatment the number of foci diminished, with most cells showing <10 foci per cell, and fewer than 6% of the cells showing >50 foci per cell. 48 hours after treatment only 3% of the wild-type cells and 1% of the Aag−/− cells had >50 foci per cell. Thus, the lesions that induced γ-H2AX foci following UVA irradiation were mostly repaired by 16 hours after treatment, and by 48 hours repair was complete. Moreover, there was no difference between wild-type and Aag−/− cells 6 hours after treatment, and very small differences at later time points, indicating that the lesions formed by UVA do not require Aag for their repair.
3.4. γ-H2AX foci formation is delayed in Aag−/− cells following TMP+UVA treatment
Treatment with TMP+UVA (Fig. 2, panels C and D; Supplemental Figure 3B) resulted in a much higher induction of γ-H2AX foci than treatment with UVA alone (Fig. 2, panels A and B). In order to minimize the background effect of UVA alone, we set the cutoff for a significant induction of foci after TMP+UVA treatment at >50 foci per cell. 6 hours after treatment with TMP+UVA most wild-type cells had between 30–70 foci per cell, while most Aag−/− cells had between 11–50 foci per cell (Supplemental Fig. 4, left panel). The fraction of wild-type cells with >50 foci was high at 6 hours after TMP+UVA treatment (50%) and continued to increase at 16 hours and 28 hours (~60% and ~70% respectively); by 48 hours there was a sharp drop in the fraction of cells with >50 foci, that presumably reflects resolution of DSBs and the completion of ICL repair in many cells (Fig. 2C). In contrast, the fraction of Aag−/− cells with >50 foci was much lower than that of wild-type at 6 and 16 hours after treatment (18% and 33% respectively) and we thus observed that the extent of initiating ICL repair is both diminished and delayed in the absence of Aag. At 28 hours after treatment both wild-type and Aag−/− cells reach the maximum γ-H2AX foci induction, although the induction in the Aag−/− cells was somewhat smaller than the maximal induction of wild-type cells (56% versus 69%). A modest drop in the fraction of Aag−/− cells with >50 foci at 48 hours indicates that at least some ICLs can be resolved in Aag−/− cells, albeit delayed compared to wild-type cells. The lengthy kinetics of γ-H2AX foci induction in both wild-type and Aag−/− cells fits an ICL repair mechanism that requires the replication fork to encounter the lesion. To summarize, after TMP+UVA treatment we observed a diminished and delayed induction and disappearance of γ-H2AX foci in Aag−/− versus wild-type cells, suggesting that Aag contributes to the efficiency of ICL repair.
3.5. γ-H2AX foci formation and disappearance is similar in wild-type and Aag−/− cells following Angelicin+UVA treatment
To test whether the difference in γ-H2AX foci induction between wild-type and Aag−/− cells is indeed due to ICL repair, we monitored γ-H2AX foci following treatment with Angelicin+UVA. This treatment forms DNA monoadducts that are most probably repaired by NER. As shown in Figure 2, panels E and F (and Supplemental Fig. 3C and 4) Angelicin+UVA treatment led to significant foci induction. As for some other DNA damaging agents, the formation of monoadducts can block replication forks and γ-H2AX foci will be formed at these sites [47–49]. Another possibility is that closely opposed single strand breaks that arise from processing of closely opposed monoadducts may go on to form a DSB. Although we used a much higher molar concentration of Angelicin than TMP, γ-H2AX foci induction following Angelicin+UVA treatment was less extensive than that following TMP+UVA treatment. The percentage of cells showing significant induction was maximal 6 hours after Angelicin+UVA treatment for both wild-type and Aag−/− cells and the fraction of cells with >50 foci steadily decreased over the next 42 hours. These kinetics fit repair of monoadducts by NER that does not require the lesion to first be encountered by the replication fork. At later time points a small difference appeared between wild-type and Aag−/− cells, but overall, there was no major difference between them, suggesting that Aag is not required for the repair of the monoadducts induced by Angelicin.
3.6. TMP+UVA induction of apoptosis is greater in Aag−/− versus wild-type ES cells
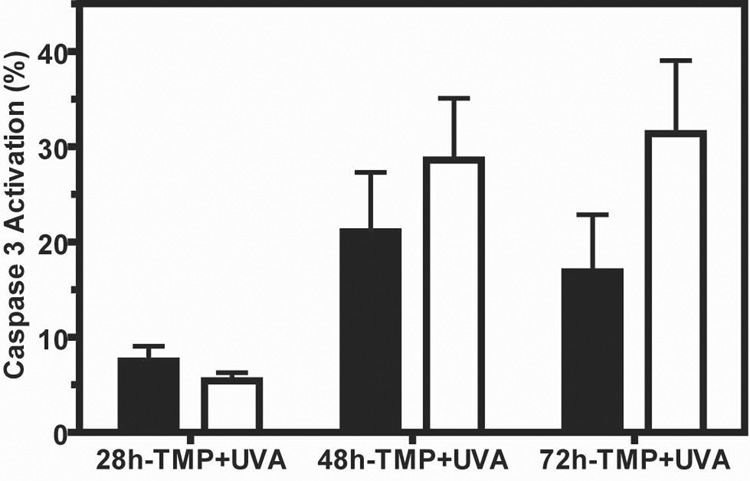
We showed that Aag−/− ES cells are sensitive to the toxic effects of TMP+UVA, and that their γ-H2AX foci induction is both delayed and diminished compared to wild-type cells. We hypothesized that the delay in ICL repair indicated by delayed γ-H2AX foci formation is responsible for the increased cytotoxicity in Aag−/− versus wild-type cells. Caspase-3 activation is a known marker for apoptotic programmed cell death [51,52]. To investigate whether the delay in γ-H2AX foci induction is accompanied by increased apoptosis, we measured Caspase-3 activation following treatment with TMP+UVA (Fig. 3). Caspase-3 activation was very low and similar in untreated and UVA treated cells (data not shown). However, TMP+UVA induced ~2-fold more Caspase-3 activation in Aag−/− versus wild-type cells at 72 hours after treatment (31.4%, compared to 16.8%). Thus in Aag−/− cells, that showed delayed and reduced repair of ICLs (Fig. 2, panels C and D), we observed increased apoptosis (Fig. 3), which presumably contributes to the decreased survival of these cells (Fig. 1).
Figure 3. Enhanced apoptosis in Aag−/− cells following TMP+UVA treatment.
Cells were treated with TMP (0.5 ng/ml) + UVA and tested for Caspase-3 activation using flow cytometry. Black bars - wild-type; White bars - Aag−/−. For each data point a minimum of 2,000 cells was analyzed, and a mean of 4 experiments ± SEM is presented. The P value (p=0.0339) was calculated by two-way-ANOVA for the different genotypes.
4. Discussion
ICL repair is a complex process that involves proteins from a variety of DNA repair pathways. Here we show that the Aag 3-methyladenine DNA glycosylase, an enzyme that initiates the base excision repair pathway, is involved in the repair of psoralen ICLs. This is based on the evidence that mouse ES Aag−/− cells are more sensitive than wild-type cells to the cross-linking treatment with TMP+UVA, show a delayed induction and resolution of γ-H2AX foci formation, and undergo enhanced apoptosis.
In principle, Aag could protect against ICL-induced cell death either by preventing the conversion of TMP-induced monoadducts into ICLs, or by contributing to the increased efficiency of ICL repair. Two lines of evidence rule out the possibility that Aag acts upon, or binds to the psoralen monoadducts to prevent ICL formation. First, Angelicin produces mainly monoadducts that are efficiently repaired by NER, and the presence or absence of Aag does not influence sensitivity to Angelicin-induced cell death; we infer from this that Aag does not significantly bind to or repair psoralen monoadducts. Second, we see that in the presence of Aag there is a more robust induction of DSBs than in the absence of Aag, as evidenced by the formation of γ-H2AX foci; since DSBs are induced after the formation of ICLs it seems quite clear that Aag does not prevent ICL formation. It therefore seems likely that Aag has a role in the process of ICL repair, rather than preventing ICL formation.
Recently, Couve-Privat et. al [53] reported that psoralen induced DNA monoadducts are substrates for NEIL1, a human DNA glycosylase that removes oxidized bases. They thus showed that BER can provide an alternative to NER for repairing these bulky DNA adducts. Couve-Privat et. al reported that in vitro, NEIL1 cleaved the monoadduct produced by 8-methoxypsoralen (8-MOP) but could not cleave the ICLs. Moreover, they showed that human AAG could not cleave monoadducts produced by 8-MOP. Although we did not detect any cleavage of TMP ICL by human AAG (Sup. Fig. 1), we specifically show that Aag nevertheless provides in vivo protection against ICLs, and not against monoadducts, by showing that wild-type and Aag−/− cells are equally susceptible to Angelicin induced cell killing and γ-H2AX foci formation.
Does Aag play a direct role in the repair process? When assayed on a short dsDNA with a site specific TMP ICL lesion, human AAG was unable to cleave any bases at the vicinity of the lesion. This is not surprising, since AAG acts by flipping out the target base into its active site in order to cleave the glycosylic bond. Since the ICL connects the two DNA strands, there is no way for the enzyme to flip the damaged base into its active site. Still, AAG might be able to act on an intermediate repair product, after the cross-link is unhooked from the DNA. However, there is room for only one nucleotide in the AAG active site, and so exonuclease action would be required around the cross-link in order for it to be flipped into the AAG active site.
We reasoned that if Aag does not have a direct role in the cleavage at the cross-link site, it might have a role in the recognition of the lesion and in assisting other repair proteins to process it. Using a short dsDNA with a site specific TMP cross-link, we were unable to detect any specific binding of either the full length or the truncated AAG protein to the cross-linked DNA. Again, it may be that AAG is able to bind an intermediate of ICL repair rather than the ICL per se. Alternatively, AAG might bind the lesion via another repair protein, or accelerate the action of another repair protein by indirect interaction with the lesion.
It is well established that a DSB is formed in the process of unhooking the ICL from the DNA [14,18–22]. γ-H2AX foci are known markers for DSBs. However, γ-H2AX foci have also been shown to be induced by agents that do not cause DSBs directly, such as UVA [44], MMS [47–49,54,55], and Angelicin [19]. In these cases it is believed that either the formation of a single strand break generated during the processing of oxidized or alkylated DNA bases subsequently lead to DSBs [55], or that γ-H2AX foci can be formed at sites of ssDNA, and/or at stalled replication forks [47–50]. In our experiments we see a maximal induction of γ-H2AX foci 28 hours after TMP+UVA treatment that would be expected for DSB formation as a part of the ICL repair mechanism [14].
In the absence of Aag, the formation of γ-H2AX foci following TMP+UVA treatment was decreased and delayed compared to wild-type cells, suggesting that Aag plays a yet unidentified role in the initial steps of the cross-link processing. Slower induction of γ-H2AX foci might indicate that fewer TMP cross-links were recognized in order to initiate the repair process. Moreover, the significant foci induction in the wild-type cells diminishes to less than half of the maximum induction by 48 hours after treatment, while the foci induced in the Aag−/− cells do not drop by half, but instead by only 18%. This could indicate that resolving the DSBs and completion of the repair reaction is more efficient in wild-type cells than in Aag−/− cells. Such inefficient ICL processing presumably accounts for the greater apoptosis seen by Caspase-3 activation in Aag−/− cells 72 hours after treatment with TMP+UVA, and eventually to their sensitivity. The kinetics of the γ-H2AX foci and apoptosis induction might suggest a role for Aag in both an early and a later step of the ICL repair reaction. After the cross-link is unhooked from the DNA, it is believed that a translesion polymerase bypasses the lesion, and that homologous recombination restores the replication fork. If Aag is bound directly or via another protein to the unhooked lesion, it could either inhibit or stimulate the action of TLS polymerase and/or recombination proteins.
Psoralen combined with UVA irradiation (PUVA) is a common treatment for the skin disorders Psoriasis and Vitiligo [56,57] despite the fact that one of the side effects is increased skin cancer [56–58]. Both diseases are believed to result from abnormalities of skin cells (epidermal keratinocytes for Psoriasis, melanocytes for Vitiligo) in a process that could be controlled by the immune system. Our finding that Aag is involved has a role in the reaction of cells to TMP+UVA might suggest that AAG’s level in human cells can affect the efficiency of PUVA treatment for these disorders. It is possible that by knowing or controlling the level of AAG in cells, a lower dose of PUVA might be required in order to get the same treatment result, thus reducing the probability of skin cancer. Moreover, understanding the exact mechanism of ICL repair will assist in planning chemotherapy treatments using PUVA and other DNA cross-linking agents that are used to treat cancer.
Supplementary Material
Acknowledgments
We thank Catherine Moroski for excellent technical assistance. This work was supported by NIH grants ES02109, CA075576, CA055042 (to L.D.S.) and a Damon Runyon Cancer Research Foundation fellowship (to A.M.S.). L.D.S. is an American Cancer Society Research Professor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
REFERENCES
- 1.Wood RD, Mitchell M, Lindahl T. Human DNA repair genes 2005. Mutat Res. 2005;577:275–283. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Wyatt MD, Allan JM, Lau AY, Ellenberger TE, Samson LD. 3-methyladenine DNA glycosylases: structure, function, and biological importance. Bioessays. 1999;21:668–676. doi: 10.1002/(SICI)1521-1878(199908)21:8<668::AID-BIES6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien PJ, Ellenberger T. Human alkyladenine DNA glycosylase uses acid-base catalysis for selective excision of damaged purines. Biochemistry. 2003;42:12418–12429. doi: 10.1021/bi035177v. [DOI] [PubMed] [Google Scholar]
- 4.Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell. 1998;95:249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 5.Fortini P, Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 2007;6:398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Dianov GL, Parsons JL. Co-ordination of DNA single strand break repair. DNA Repair (Amst) 2007;6:454–460. doi: 10.1016/j.dnarep.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Wilson DM, 3rd, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Engelward BP, Dreslin A, Christensen J, Huszar D, Kurahara C, Samson L. Repair-deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing. EMBO Journal. 1996;15:945–952. [PMC free article] [PubMed] [Google Scholar]
- 9.Engelward BP, Weeda G, Wyatt MD, Broekhof JLM, De Wit J, Donker I, Allan JM, Gold B, Hoeijmakers JHJ, Samson LD. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc. Natl. Acad. Sci. USA. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allan JM, Engelward BP, Dreslin AJ, Wyatt MD, Tomasz M, Samson LD. Mammalian 3-methyladenine DNA glycosylase protects against the toxicity and clastogenicity of certain chemotherapeutic DNA cross-linking agents. Cancer Research. 1998;58:3965–3973. [PubMed] [Google Scholar]
- 11.McHugh PJ, Gill RD, Waters R, Hartley JA. Excision repair of nitrogen mustard-DNA adducts in Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:3259–3266. doi: 10.1093/nar/27.16.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 13.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Peterson CA, Zheng H, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol Cell Biol. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng H, Wang X, Warren AJ, Legerski RJ, Nairn RS, Hamilton JW, Li L. Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol Cell Biol. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng H, Wang X, Legerski RJ, Glazer PM, Li L. Repair of DNA interstrand cross-links: interactions between homology-dependent and homology-independent pathways. DNA Repair (Amst) 2006;5:566–574. doi: 10.1016/j.dnarep.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Mogi S, Oh DH. gamma-H2AX formation in response to interstrand crosslinks requires XPF in human cells. DNA Repair (Amst) 2006;5:731–740. doi: 10.1016/j.dnarep.2006.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothfuss A, Grompe M. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2004;24:123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang N, Liu X, Li L, Legerski R. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair (Amst) 2007;6:1670–1678. doi: 10.1016/j.dnarep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. Embo J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fisher LA, Bessho M, Bessho T. Processing of a psoralen DNA interstrand cross-link by XPF-ERCC1 complex in vitro. J Biol Chem. 2007 doi: 10.1074/jbc.M708072200. [DOI] [PubMed] [Google Scholar]
- 24.Hoy CA, Thompson LH, Mooney CL, Salazar EP. Defective DNA cross-link removal in Chinese hamster cell mutants hypersensitive to bifunctional alkylating agents. Can. Res. 1985;45:1737–1743. [PubMed] [Google Scholar]
- 25.Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, Narayana LS, Zhou ZQ, Adamson AW, Sorensen KJ, Chen DJ, Jones NJ, Thompson LH. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 26.Godthelp BC, Wiegant WW, van Duijn-Goedhart A, Scharer OD, van Buul PP, Kanaar R, Zdzienicka MZ. Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic Acids Res. 2002;30:2172–2182. doi: 10.1093/nar/30.10.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinz JM, Tebbs RS, Wilson PF, Nham PB, Salazar EP, Nagasawa H, Urbin SS, Bedford JS, Thompson LH. Repression of mutagenesis by Rad51D- mediated homologous recombination. Nucleic Acids Res. 2006;34:1358–1368. doi: 10.1093/nar/gkl020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang N, Lu X, Zhang X, Peterson CA, Legerski RJ. hMutSbeta is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol Cell Biol. 2002;22:2388–2397. doi: 10.1128/MCB.22.7.2388-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Q, Christensen LA, Legerski RJ, Vasquez KM. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6:551–557. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, Hoatlin M, Joenje H, de Winter JP, Wang W. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi Anemia Protein FANCM Can Promote Branch Migration of Holliday Junctions and Replication Forks. Mol Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 32.Gupta R, Sharma S, Sommers JA, Kenny MK, Cantor SB, Brosh RM., Jr FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007;110:2390–2398. doi: 10.1182/blood-2006-11-057273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Kennedy RD, D'Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 35.Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 36.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, Ikeda H, Fox EA, D'Andrea AD. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 37.Cipak L, Watanabe N, Bessho T. The role of BRCA2 in replication-coupled DNA interstrand cross-link repair in vitro. Nat Struct Mol Biol. 2006;13:729–733. doi: 10.1038/nsmb1120. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Andreassen PR, D'Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24:5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, D'Andrea AD. The interplay of Fanconi anemia proteins in the DNA damage response. DNA Repair (Amst) 2004;3:1063–1069. doi: 10.1016/j.dnarep.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Zhang N, Kaur R, Lu X, Shen X, Li L, Legerski RJ. The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J Biol Chem. 2005;280:40559–40567. doi: 10.1074/jbc.M508453200. [DOI] [PubMed] [Google Scholar]
- 41.Cheng WH, Kusumoto R, Opresko PL, Sui X, Huang S, Nicolette ML, Paull TT, Campisi J, Seidman M, Bohr VA. Collaboration of Werner syndrome protein and BRCA1 in cellular responses to DNA interstrand cross-links. Nucleic Acids Res. 2006;34:2751–2760. doi: 10.1093/nar/gkl362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mereau A, Grey L, Piquet-Pellorce C, Heath JK. Characterization of a binding protein for leukemia inhibitory factor localized in extracellular matrix. J Cell Biol. 1993;122:713–719. doi: 10.1083/jcb.122.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fillingham J, Keogh MC, Krogan NJ. GammaH2AX and its role in DNA double-strand break repair. Biochem Cell Biol. 2006;84:568–577. doi: 10.1139/o06-072. [DOI] [PubMed] [Google Scholar]
- 44.Lu C, Zhu F, Cho YY, Tang F, Zykova T, Ma WY, Bode AM, Dong Z. Cell apoptosis: requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol Cell. 2006;23:121–132. doi: 10.1016/j.molcel.2006.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 46.Besaratinia A, Kim SI, Bates SE, Pfeifer GP. Riboflavin activated by ultraviolet A1 irradiation induces oxidative DNA damage-mediated mutations inhibited by vitamin C. Proc Natl Acad Sci U S A. 2007;104:5953–5958. doi: 10.1073/pnas.0610534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nazarov IB, Smirnova AN, Krutilina RI, Svetlova MP, Solovjeva LV, Nikiforov AA, Oei SL, Zalenskaya IA, Yau PM, Bradbury EM, Tomilin NV. Dephosphorylation of histone gamma-H2AX during repair of DNA double-strand breaks in mammalian cells and its inhibition by calyculin A. Radiat Res. 2003;160:309–317. doi: 10.1667/rr3043. [DOI] [PubMed] [Google Scholar]
- 48.Nikiforov A, Svetlova M, Solovjeva L, Sasina L, Siino J, Nazarov I, Bradbury M, Tomilin N. DNA damage-induced accumulation of Rad18 protein at stalled replication forks in mammalian cells involves upstream protein phosphorylation. Biochem Biophys Res Commun. 2004;323:831–837. doi: 10.1016/j.bbrc.2004.08.165. [DOI] [PubMed] [Google Scholar]
- 49.Liu JS, Kuo SR, Melendy T. Comparison of checkpoint responses triggered by DNA polymerase inhibition versus DNA damaging agents. Mutat Res. 2003;532:215–226. doi: 10.1016/j.mrfmmm.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 50.O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 51.Li P, Nijhawan D, Wang X. Mitochondrial activation of apoptosis. Cell. 2004;116:S57–S59. doi: 10.1016/s0092-8674(04)00031-5. 52 p following S59. [DOI] [PubMed] [Google Scholar]
- 52.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 53.Couve-Privat S, Mace G, Rosselli F, Saparbaev MK. Psoralen-induced DNA adducts are substrates for the base excision repair pathway in human cells. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou C, Li Z, Diao H, Yu Y, Zhu W, Dai Y, Chen FF, Yang J. DNA damage evaluated by gammaH2AX foci formation by a selective group of chemical/physical stressors. Mutat Res. 2006;604:8–18. doi: 10.1016/j.mrgentox.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pascucci B, Russo MT, Crescenzi M, Bignami M, Dogliotti E. The accumulation of MMS-induced single strand breaks in G1 phase is recombinogenic in DNA polymerase beta defective mammalian cells. Nucleic Acids Res. 2005;33:280–288. doi: 10.1093/nar/gki168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morison WL. Psoralen ultraviolet A therapy in 2004. Photodermatol Photoimmunol Photomed. 2004;20:315–320. doi: 10.1111/j.1600-0781.2004.00125.x. [DOI] [PubMed] [Google Scholar]
- 57.Stern RS. Psoralen and ultraviolet a light therapy for psoriasis. N Engl J Med. 2007;357:682–690. doi: 10.1056/NEJMct072317. [DOI] [PubMed] [Google Scholar]
- 58.Rezaei N, Gavalas NG, Weetman AP, Kemp EH. Autoimmunity as an aetiological factor in vitiligo. J Eur Acad Dermatol Venereol. 2007;21:865–876. doi: 10.1111/j.1468-3083.2007.02228.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.