Abstract
We have developed a transposon mutagenesis system for Vibrio fischeri ES114 that utilizes a hyperactive mutant Tn5 transposase (E54K and M56A) and optimized transposon ends. Using a conjugation-based procedure, we obtained independent single-insertion mini-Tn5 mutants at a rate of ∼10−6. This simple and inexpensive technique represents a significant improvement over previous methods for transposon mutagenesis of V. fischeri and should be applicable to many other bacteria.
Vibrio fischeri is a useful model for studies of bioluminescence (18, 35), pheromone-mediated signaling (39, 46), and beneficial animal-bacterium symbioses (32, 36, 47, 49). Other aspects of V. fischeri biology, including its motility (10, 16, 23, 26, 27, 29, 30), metabolism (1, 13, 42), and ability to form biofilms (9, 14, 19, 53, 55, 56), are also active areas of study. Interest in the light-organ symbiosis between V. fischeri and the Hawaiian bobtail squid Euprymna scolopes has led several researchers to adopt strain ES114 (4), a wild-type isolate from an E. scolopes light organ, as the type strain of choice for many of these studies. The recently published genome sequence of ES114 (33) and the ongoing development of genetic tools for ES114 (10-12, 38, 40, 48) promise to advance each of the research areas described above.
The goal of this study was to develop a more effective method for transposon mutagenesis of V. fischeri ES114. Transposon mutagenesis systems have been used with V. fischeri, but these have important limitations. Initially, a Mu-based transposon was used with V. fischeri (16), but in this system, the delivery plasmid was maintained at a low selective temperature, and multiple insertions were more common than single insertions at a higher temperature. Furthermore, insertions by the Mu d1 1681 transposon were not cloned or localized in the V. fischeri genome (16). A mini-Tn10-based system was then developed and used with V. fischeri (48); however, transposition frequency is relatively low, and unwanted integrants of the plasmid delivery vector are common and can be identified only by molecular methods (K. Visick, personal communication). Mini-Tn5 systems have been used with V. fischeri, as well (17, 37), but in one case, we found insertion to be impractically infrequent for large-scale screens (37), and that investigation, along with another study, relied on mutagenizing an already mutant V. fischeri background (17, 37).
The reason a mutant strain was used as the parent for further mutagenesis stems from each of the systems described above relying on conjugation from Escherichia coli to deliver the transposon into V. fischeri, so to counterselect against E. coli donors, a spontaneous rifampin-resistant derivative of ES114, designated ESR1, has been used (16). Unfortunately, rifampin resistance often results in pleiotropic phenotypes (2, 3, 20, 54), and recently, ESR1 was found to be attenuated in both competitive host colonization (24) and in production of bioluminescence (5). We have introduced plasmids directly into wild-type ES114 by conjugation, using low temperature and high salt to enrich against E. coli donors (38), but this is effective only if the desired transconjugants occur frequently relative to the number and growth of E. coli donor cells on the selective medium (e.g., more frequently than the mini-Tn10 or mini-Tn5 insertions described above occur). Thus, in our experience, existing transposon mutagenesis systems have limited utility with wild-type V. fischeri ES114.
To improve transposon mutagenesis of V. fischeri, we exploited advances in the understanding of Tn5 by combining optimized Tn5 end sequences (58) and a hyperactive transposase allele (59). The wild-type Tn5 transposase gene has an alternate translational start site, giving rise to an Inh protein that inhibits transposase activity, but an M56A change removes this alternate start and prevents Inh translation (51). An additional E54K change yielded a much more active transposase with improved binding to the transposon ends (59). Moreover, the Tn5 transposase normally acts at inside end and outside end sequences of IS50 elements; however, the transposase proved to be more active with a hybrid of the inside end and outside end sequences, termed a mosaic end (31, 58). These improvements in transposition efficiency formed the foundation of strategies for in vitro mutagenesis of naked DNA with mini-Tn5 derivatives and the generation of mutants in vivo by electroporation of transposase-DNA complexes (31). However, transformation frequencies by electroporation are extremely low in V. fischeri (45), and we therefore sought to generate a conjugation-based system that makes use of these improvements in Tn5 efficiency. One such system generated and modified by others has been effective in diverse bacteria (22, 41), although as discussed below, we sought to construct a system with somewhat different attributes, particularly with respect to the selectable and screenable markers compatible with V. fischeri.
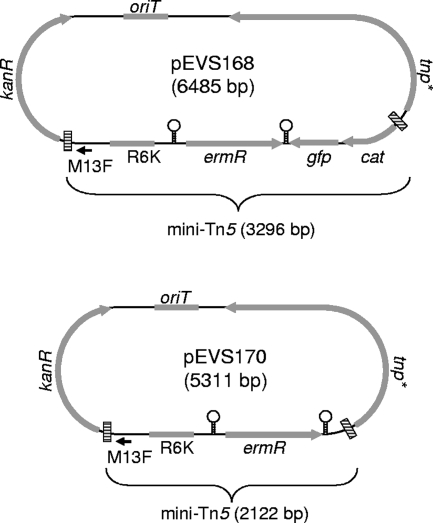
We combined the optimized transposase allele and end elements with an erythromycin resistance gene (ermR) to generate mini-Tn5 delivery vectors pEVS168 and pEVS170 (Fig. 1). Each of these vectors contains the RP4 origin of transfer (oriT), thereby allowing their mobilization from E. coli into V. fischeri (38). The transposon in pEVS168 also has promoterless chloramphenicol resistance (cat) and green fluorescent protein (gfp) genes, such that insertion in the correct orientation in actively transcribed regions can generate transcriptional fusions to these selectable (cat) and readily screened (gfp) markers.
FIG. 1.
Schematic representation of the mini-Tn5-ermR delivery vectors pEVS168 and pEVS170. The tnp* gene encodes a Tn5 transposase with mutations leading to E54K and M56A substitutions (59). Cross-hatched boxes indicate a mosaic transposon end (5′-CTG TCT CTT ATA CAC ATC T-3′) that serves as a more frequent substrate for transposition (31, 58). R6K denotes the origin of replication, oriT indicates the RP4 origin of conjugative transfer, and the kanR and ermR genes encode resistance to kanamycin and erythromycin, respectively. Stem loops indicate the locations of bidirectional transcriptional terminators, and M13F shows the location of an M13 forward priming site for sequencing. Restriction sites (presented 5′-3′) absent from both transposons include BglII (A/GATCT), EcoRI (G/AATTC), HhaI (GCG/C), XbaI (T/CTAGA), and XhoI (C/TCGAG).
We constructed pEVS168 and pEVS170, using standard cloning techniques and employing reagents and procedures described previously (1). Details of pEVS168 and pEVS170 construction were as follows. The R6K origin, ermR, cat, and gfp genes of pEVS127 (see supplemental material in reference 12) were PCR amplified using primers EVS90 and EVS91 (Table 1); BglII sites introduced on the primers were digested with this enzyme, and the product was self-ligated to form pEVS155. Optimized 19-bp Tn5 mosaic ends were incorporated into primers EVS90 and EVS91, along with restriction sites, such that most of pEVS155 was within a transposable unit with only the unique BglII and SalI sites outside the mini-transposon. Plasmid pEVS129 was generated by deleting the XbaI-SpeI fragment of pEVS118 (11), and pEVS129 was linearized with BglII and fused to BamHI-digested pRZ5412 (59), which encodes a hyperactive mutant transposase. The transposase gene on the resulting plasmid fusion was PCR amplified using primers EVS62 and EVS92 (Table 1), this amplicon was digested with BamHI, and the fragment was cloned into the BglII site of pEVS155, generating pEVS157. The inside-end-containing SalI fragment of pEVS157 was deleted and subsequently replaced with the oriT- and kanR-containing SalI fragment of pEVS76 (38), resulting in pEVS167. pEVS168 was then generated by annealing oligonucleotides M13LKF and M13LKR (Table 1) together and cloning this linker into the XbaI site of pEVS167, thereby introducing a unique ApaI site and a complement for the M13 forward sequencing primer into one end of the transposon. pEVS168, which is similar to pEVS170 (Fig. 1) but contains a promoterless cat-gfp transcriptional reporter on the transposon distal to the M13 forward site, was digested with BspEI and BsrGI, the overhangs were filled with Klenow fragment, and the vector was self-ligated to delete gfp along with most of cat, generating pEVS170. Translational stop codons were incorporated into primer EVS91 (Table 1), so that insertions of the transposon from pEVS168 could generate transcriptional, but not translational, fusions of chromosomal genes to cat and gfp.
TABLE 1.
Oligonucleotides designed for this study
| Primer name | Oligonucleotide sequence (5′-3′)a |
|---|---|
| EVS62 | CTT CAG ATC CTC TAC GCC GGA CGC |
| EVS90 | GGG AGA TCT GTC GACCTG TCT CTT ATA CAC ATC TGC GGC CGC TCT AGA ACT AGT GGA TCC |
| EVS91 | GGG AGA TCT GTC TCT TAT ACA CAT CTA AGG TAA TCA GGA GCT AAG GAA GCT AAA ATG G |
| EVS92 | CCC GGA TCC GTA GCG TCC TGA ACG GAA CCT TTC CCG |
| M13LKF | CTA GGG GCC CTG TAA AAC GAC GGC CAG TC |
| M13LKR | CTA GGA CTG GCC GTC GTT TTA CAG GGC CC |
Optimized 19-bp Tn5 mosaic ends (5′-CTG TCT CTT ATA CAC ATC T-3′) are indicated by single underlines in EVS90 and EVS91. The M13 Forward priming site (5′-TGT AAA ACG ACG GCC AGT-3′) and its complement formed by annealing M13LKF and M13LKR together are underlined in those sequences. Translational stops (5′-TAA-3′), present in all three potential reading frames in EVS91, are shown in boldface. Key restriction enzyme recognition sequences are in italics, including ApaI (5′-GGGCC/C-3′) in M13LKF and M13LKR, BamHI (5′-G/GATCC-3′) in EVS92, BglII (5′-A/GATCT-3′) in EVS90 and EVS91, and SalI (5′-G/TCGAC-3′) in EVS90.
This transposon system proved highly effective with V. fischeri. We used a simple mating procedure (38) to introduce pEVS170 into V. fischeri ES114 and found that typically, ∼104 mini-Tn5-ermR mutants were selected as erythromycin resistant from ∼1010 total recipients in each small-scale mating, yielding a transposon mutagenesis frequency of 10−6. It is worth noting that erythromycin selection, which was carried out on LBS medium (5) supplemented with 5 μg ml−1 erythromycin, resulted in a clearer distinction between Tn mutants and other cells in the mating mixture than did the chloramphenicol selection associated with some previous transposons, and this may have contributed to our success. Erythromycin-resistant transconjugants were recovered about 104-fold more frequently upon introduction of the stable ermR plasmid pS44S (11), and assuming that conjugative transfer frequency is similar for pEVS170 and pS44S, which each have the same oriT, the frequency of transposition once pEVS170 has entered V. fischeri can be estimated as ∼10−4. We identified mutants resulting from plasmid integration, rather than mini-Tn5-ermR transposition, by screening for kanamycin resistance, which we observed with ∼10% of the erythromycin-resistant mutants. Similar frequencies were obtained when the mini-transposon contained promoterless cat and gfp genes, and epifluorescence microscopy revealed that about a quarter of these insertion mutants visibly expressed GFP, with different mutants varying in their degree of green fluorescence (data not shown). This result is consistent with nonspecific insertions generating transcriptional fusions to the cat-gfp reporter, assuming that (i) only a fraction of the genome will be transcribed significantly during growth on LBS plates, (ii) approximately half of the insertions in transcribed regions will be oriented in the direction of transcription, and (iii) levels of transcription will vary between loci.
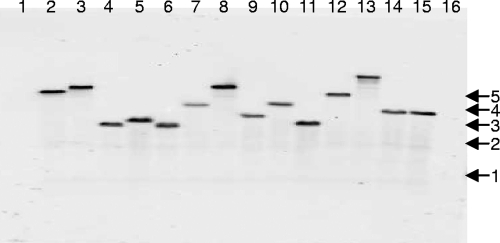
Although the recovery of transposon insertion mutants was more efficient than we and others (K. Visick, personal communication) have observed with other Tn5- and Tn10-based systems in V. fischeri, we were concerned that the hyperactive transposase on pEVS170 might yield multiple mini-Tn5-ermR insertions in mutants. However, Southern blotting of over 50 mutants revealed that this is not the case. Chromosomal DNA was purified from the transposon insertion mutants using Easy-DNA (Invitrogen, Carlsbad, CA), the DNA was restriction digested, and 5 ng of each digested DNA sample was separated by gel electrophoresis. Digested DNA was transferred from the gel onto nitrocellulose paper and then prepared for imaging using a digoxigenin High Prime DNA labeling and detection starter kit II (Roche Applied Science, Mannheim, Germany). Southern blot analysis revealed a single Tn insertion in each strain examined (e.g., Fig. 2).
FIG. 2.
Southern blotting of mutants, using a probe generated from pEVS170. Arrows and corresponding numbers at right indicate DNA fragment sizes in kb. Lanes 1 and 16 were loaded with a 1-kb DNA ladder. Lanes 2 to 14 were loaded with HhaI-digested chromosomal DNA from mutant strains NL1, NL2, NL3, NL4, NL5, NL6, NL8, EMH5, EMH6, EMH7, EMH9, EMH11, EMH12, and EMH13, respectively. Fourteen representative mutants of more than 50 examined are shown.
The composition of the mini-Tn5-ermR mutant from pEVS170 allowed us to clone it readily from insertion mutants and to identify insertion locations in the V. fischeri genome. The R6K plasmid origin of replication within the mini-transposon does not function in V. fischeri, but it does replicate in E. coli strains harboring the pir gene (21), such as DH5α-λpir (11). Chromosomal DNA from mutants was digested with HhaI, which does not cut within the transposon, the fragments were self-ligated, and transposons along with flanking chromosomal DNA were transformed as circularized plasmids into DH5α-λpir. The M13 forward sequencing primer was then used to sequence across transposon-chromosome junctions, and we localized the point of insertion by comparisons to the genome sequence (33). HhaI digests DNA at a 4-bp recognition sequence (5′-GCG/C-3′), and virtually all HhaI fragments of the ES114 genome are small enough to be recovered readily by this cloning procedure yet large enough to allow unambiguous identification of the genomic location.
Multiple lines of evidence suggest that our mini-Tn5-ermR transposons exhibit a lack of target specificity, similar to that of other Tn5 derivatives (15, 34), which generally insert with some deviation from complete randomness but not enough to present a practical barrier to mutagenesis strategies. For example, Fig. 2 illustrates a lack of target specificity with the mini-Tn5-ermR inserting into different-sized chromosomal fragments. In addition, we sequenced the location of more than 60 insertions, and we found them distributed on both V. fischeri chromosomes. Moreover, we have never identified an insertion in the same site from more than one independent mating. Finally, we illustrated this transposon mutagenesis system's practical utility recently when we used it to identify the cellobiose utilization cel gene cluster in V. fischeri ES114 (1). Some deviation from random insertion may be intimated in that study by an apparently disproportionate number of insertions in celK, which coincidentally has a lower GC content (31.8%) than other genes of the cluster (35.3 to 39.6%); however, insertions in all six cel genes were recovered without difficulty (1).
Despite the use of ES114 for nearly 2 decades, to our knowledge, the use of pEVS170 (Fig. 1) to identify cellobiose utilization mutants (1) was the first report of transposon mutagenesis in this wild-type strain and not a pleiotropic and marked derivative such as ESR1. Other laboratories are currently using pEVS168, pEVS170, or their derivatives to make new discoveries in V. fischeri. In a particularly exciting development, Cheryl Whistler (personal communication) has proposed utilizing this system to generate a comprehensive transposon mutant library with insertions in every nonessential gene of the V. fischeri genome.
The tools we describe here are similar to pRL27 (22), a conjugation-based mini-Tn5 delivery vector that exploits the same Tn5 mosaic end sequences and hyperactive transposase mutations that are similar to those we used in pEVS168 and pEVS170; however, our constructs differ somewhat from that system. For example, the hyperactive transposase in pRL27 contains an additional L372P allele, which apparently minimizes nonproductive multimerization of transposase and improves transposition frequency when the transposase gene is in trans to the transposon ends (50). However, the L372P allele had no effect on transposition frequency in cis (50), and it therefore would not be expected to influence systems like those described here or in pRL27, where the transposase gene is delivered on the same vector as the mini-Tn5. For our purposes, a more important difference is the use of an erythromycin resistance gene cassette within the transposon, rather than the kanamycin resistance gene in pRL27. We have never observed spontaneous resistance to erythromycin in V. fischeri, but we do observe spontaneous resistance to kanamycin at a frequency rivaling that of transposition, making the erythromycin resistance marker superior for primary selection of transposon hops. The kanamycin resistance gene functions well as a screenable marker in V. fischeri, and by placing it outside the transposon, we distinguished transposon hops from vector integrants. There is no such marker outside the transposon in pRL27. For unknown reasons, we observe a large number of integrants (∼10% of Tn-harboring clones) in V. fischeri, and the ability to screen for and discard vector integrants is important, as they may contain the transposase gene and therefore could accumulate multiple mini-Tn5 insertions. Finally, our constructs contain other advantageous features, including promoterless selectable and screenable markers (cat-gfp) in pEVS168, a universal primer site near the transposon end, and a unique ApaI site within each mini-Tn5-ermR construct that can be used to further engineer this system.
Combining optimizing elements in an in vivo conjugation-based mini-Tn5 mutagenesis system has opened up new research opportunities in V. fischeri, and we expect our constructs will be useful in other bacteria for which existing transposon mutagenesis strategies are inefficient. The similar pRL27 system devised by Larsen et al. (22) has proven useful in many bacteria, including Xanthobacter autotrophicus (22), Pseudomonas stutzeri (22) and other Pseudomonas strains (28, 57), Alcaligenes faecalis (22, 52), E. coli (6), Bdellovibrio bacteriovorus (44), Prochlorococcus sp. strain MIT9313 (43), and Synechococcus sp. strain WH8102 (25), while modified versions of pRL27 were also successfully employed with V. parahaemolyticus (41), Citrobacter rodentium (8), and Francisella tularensis (7). It therefore seems likely that our constructs will have similarly broad utility and the different features of our system will extend the range of organisms in which conjugation-based mini-Tn5 mutagenesis is a practical tool.
Acknowledgments
We thank Erica M. Hall and Alecia N. Septer for technical assistance and William Reznikoff for providing pRZ5412.
This material is based upon work supported by the National Science Foundation (NSF) under grant CAREER MCB-0347317 to E.V.S. and by National Institutes of Health (NIH) grant AI50661 to M. McFall-Ngai. S.L.V. was supported by an NSF Research Experience for Undergraduates site award (DBI-0453353).
Genomic information was provided by the Vibrio fischeri Genome Project, supported by the W. M. Keck Foundation.
Footnotes
Published ahead of print on 19 September 2008.
REFERENCES
- 1.Adin, D. M., K. L. Visick, and E. V. Stabb. 2008. Identification of a cellobiose utilization gene cluster with cryptic beta-galactosidase activity in Vibrio fischeri. Appl. Environ. Microbiol. 74:4059-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Björkman, J., D. Hughes, and D. I. Andersson. 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 31:3949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc-Potard, A., E. Gari, F. Spirito, N. Figueroa-Bossi, and L. Bossi. 1995. RNA polymerase (rpoB) mutants selected for increased resistance to gyrase inhibitors in Salmonella typhimurium. Mol. Gen. Genet. 247:680-692. [DOI] [PubMed] [Google Scholar]
- 4.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bose, J. L., C. S. Rosenberg, and E. V. Stabb. 2008. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch. Microbiol. 190:169-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradshaw, J. S., and A. Kuzminov. 2003. RdgB acts to avoid chromosome fragmentation in Escherichia coli. Mol. Microbiol. 48:1711-1725. [DOI] [PubMed] [Google Scholar]
- 7.Buchan, B. W., M. K. McLendon, and B. D. Jones. 2008. Identification of differentially regulated Francisella tularensis genes by use of a newly developed Tn5-based transposon delivery system. Appl. Environ. Microbiol. 74:2637-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coulthurst, S. J., S. Clare, T. J. Evans, I. J. Foulds, K. J. Roberts, M. Welch, G. Dougan, and G. P. Salmond. 2007. Quorum sensing has an unexpected role in virulence in the model pathogen Citrobacter rodentium. EMBO Rep. 8:698-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darnell, C. L., E. A. Hussa, and K. L. Visick. 2008. The putative hybrid sensor kinase SypF coordinates biofilm formation in Vibrio fischeri by acting upstream of two response regulators, SypG and VpsR. J. Bacteriol. 190:4941-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLoney-Marino, C. R., A. J. Wolfe, and K. L. Visick. 2003. Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl. Environ. Microbiol. 69:7527-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn, A. K., M. O. Martin, and E. V. Stabb. 2005. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid 54:114-134. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, A. K., D. S. Millikan, D. M. Adin, J. L. Bose, and E. V. Stabb. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn, A. K., and E. V. Stabb. 2008. Genetic analysis of trimethylamine N-oxide reductases in the light organ symbiont Vibrio fischeri ES114. J. Bacteriol. 190:5814-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geszvain, K., and K. L. Visick. 2008. The hybrid sensor kinase RscS integrates positive and negative signals to modulate biofilm formation in Vibrio fischeri. J. Bacteriol. 190:4437-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goryshin, I. Y., J. A. Miller, Y. V. Kil, V. A. Lanzov, and W. S. Reznikoff. 1998. Tn5/IS50 target recognition. Proc. Natl. Acad. Sci. USA 95:10716-10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graf, J., P. V. Dunlap, and E. G. Ruby. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 176:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graf, J., and E. G. Ruby. 2000. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol. Microbiol. 37:168-179. [DOI] [PubMed] [Google Scholar]
- 18.Hastings, J. W., and K. H. Nealson. 1977. Bacterial bioluminescence. Annu. Rev. Microbiol. 31:549-595. [DOI] [PubMed] [Google Scholar]
- 19.Hussa, E. A., C. L. Darnell, and K. L. Visick. 2008. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J. Bacteriol. 190:4576-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin, D. J., and C. A. Gross. 1989. Characterization of the pleiotropic phenotypes of rifampin-resistant rpoB mutants of Escherichia coli. J. Bacteriol. 171:5229-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 22.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-201. [DOI] [PubMed] [Google Scholar]
- 23.Lupp, C., and E. G. Ruby. 2005. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 187:3620-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCann, J., E. V. Stabb, D. S. Millikan, and E. G. Ruby. 2003. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl. Environ. Microbiol. 69:5928-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarren, J., and B. Brahamsha. 2005. Transposon mutagenesis in a marine Synechococcus strain: isolation of swimming motility mutants. J. Bacteriol. 187:4457-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millikan, D. S., and E. G. Ruby. 2002. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl. Environ. Microbiol. 68:2519-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millikan, D. S., and E. G. Ruby. 2004. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J. Bacteriol. 186:4315-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman, K. L., S. Chatterjee, K. A. Ho, and S. E. Lindow. 2008. Virulence of plant pathogenic bacteria attenuated by degradation of fatty acid cell-to-cell signaling factors. Mol. Plant Microbe Interact. 21:326-334. [DOI] [PubMed] [Google Scholar]
- 29.O'Shea, T. M., C. R. DeLoney-Marino, S. Shibata, S.-I. Aizawa, A. J. Wolfe, and K. L. Visick. 2005. Magnesium promotes flagellation of Vibrio fischeri. J. Bacteriol. 187:2058-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Shea, T. M., A. H. Klein, K. Geszvain, A. J. Wolfe, and K. L. Visick. 2006. Diguanylate cyclases control magnesium-dependent motility of Vibrio fischeri. J. Bacteriol. 188:8196-8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reznikoff, W. S., I. Y. Goryshin, and J. J. Jendrisak. 2004. Tn5 as a molecular genetics tool. In W. J. Miller and P. Capy (ed.), Mobile genetic elements: protocols and genomic applications, vol. 260. Humana Press Inc., Totowa, NJ.
- 32.Ruby, E. G. 1996. Lessons from a cooperative bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50:591-624. [DOI] [PubMed] [Google Scholar]
- 33.Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lohstroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shevchenko, Y., G. G. Bouffard, Y. S. Butterfield, R. W. Blakesley, J. L. Hartley, A. C. Young, M. A. Marra, S. J. Jones, J. W. Touchman, and E. D. Green. 2002. Systematic sequencing of cDNA clones using the transposon Tn5. Nucleic Acids Res. 30:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stabb, E. V. 2005. Shedding light on the bioluminescence “paradox.” ASM News 71:223-229. [Google Scholar]
- 36.Stabb, E. V. 2006. The Vibrio fischeri-Euprymna scolopes light organ symbiosis, p. 204-218. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington, DC.
- 37.Stabb, E. V., and E. G. Ruby. 1998. Construction of a mobilizable Tn5 derivative and its utility for mutagenizing, cloning, and mapping loci in the luminescent symbiont Vibrio fischeri, abstr. N-60, p. 375-376. Abstr. 98th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 38.Stabb, E. V., and E. G. Ruby. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 39.Stabb, E. V., A. Schaefer, J. L. Bose, and E. G. Ruby. 2008. Quorum signaling and symbiosis in the marine luminous bacterium Vibrio fischeri, p. 233-250. In S. C. Winans and B. L. Bassler (ed.), Chemical communication among microbes. ASM Press, Washington, DC.
- 40.Stabb, E. V., K. L. Visick, D. S. Millikan, A. A. Corcoran, L. Gilson, S. V. Nyholm, M. McFall-Ngai, and E. G. Ruby. 2001. The Vibrio fischeri-Euprymna scolopes symbiosis: a model marine animal-bacteria interaction, p. 269-277. In N. Saxena (ed.), Recent advances in marine science and technology 2000. PACON International, Honolulu, HI.
- 41.Stewart, B. J., and L. L. McCarter. 2003. Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 185:4508-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Studer, S. V., M. J. Mandel, and E. G. Ruby. 2008. AinS quorum sensing regulates the Vibrio fischeri acetate switch. J. Bacteriol. 190:5915-5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tolonen, A. C., G. B. Liszt, and W. R. Hess. 2006. Genetic manipulation of Prochlorococcus strain MIT9313: green fluorescent protein expression from an RSF1010 plasmid and Tn5 transposition. Appl. Environ. Microbiol. 72:7607-7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tudor, J. J., J. J. Davis, M. Panichella, and A. Zwolak. 2008. Isolation of predation-deficient mutants of Bdellovibrio bacteriovorus using transposon mutagenesis. Appl. Environ. Microbiol. 74:5436-5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visick, K. G., and E. G. Ruby. 1996. Construction and symbiotic competence of a luxA-deletion mutant of Vibrio fischeri. Gene 175:89-94. [DOI] [PubMed] [Google Scholar]
- 46.Visick, K. L. 2005. Layers of signaling in a bacterium-host association. J. Bacteriol. 187:3603-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visick, K. L., and M. J. McFall-Ngai. 2000. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J. Bacteriol. 182:1779-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Visick, K. L., and E. G. Ruby. 1997. New genetic tools for use in the marine bioluminescent bacterium Vibrio fischeri, p. 119-122. In J. W. Hastings, L. J. Kricka, and P. E. Stanley (ed.), Bioluminescence and chemiluminescence. John Wiley and Sons, New York, NY.
- 49.Visick, K. L., and E. G. Ruby. 2006. Vibrio fischeri and its host: it takes two to tango. Curr. Opin. Microbiol. 9:632-638. [DOI] [PubMed] [Google Scholar]
- 50.Weinreich, M. D., A. Gasch, and W. S. Reznikoff. 1994. Evidence that the cis preference of the Tn5 transposase is caused by nonproductive multimerization. Genes Dev. 8:2363-2374. [DOI] [PubMed] [Google Scholar]
- 51.Wiegand, T. W., and W. S. Reznikoff. 1992. Characterization of two hypertransposing Tn5 mutants. J. Bacteriol. 174:1229-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, M. M., and W. W. Metcalf. 2005. Genetic diversity and horizontal transfer of genes involved in oxidation of reduced phosphorus compounds by Alcaligenes faecalis WM2072. Appl. Environ. Microbiol. 71:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfe, A. J., D. S. Millikan, J. M. Campbell, and K. L. Visick. 2004. Vibrio fischeri σ54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 70:2520-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yanofsky, C., and V. Horn. 1981. Rifampicin resistance mutations that alter the efficiency of transcription termination at the tryptophan operon attenuator. J. Bacteriol. 145:1334-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yip, E. S., K. Geszvain, C. R. DeLoney-Marino, and K. L. Visick. 2006. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol. 62:1586-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yip, E. S., B. T. Grublesky, E. A. Hussa, and K. L. Visick. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and sigma-dependent biofilm formation by Vibrio fischeri. Mol. Microbiol. 57:1485-1498. [DOI] [PubMed] [Google Scholar]
- 57.Yun, J. I., K. M. Cho, J. K. Kim, S. O. Lee, K. Cho, and K. Lee. 2007. Mutation of rpoS enhances Pseudomonas sp. KL28 growth at higher concentrations of m-cresol and changes its surface-related phenotypes. FEMS Microbiol. Lett. 269:97-103. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, M., A. Bhasin, and W. S. Reznikoff. 1998. Molecular genetic analysis of transposase-end DNA sequence recognition: cooperativity of three adjacent base-pairs in specific interaction with a mutant Tn5 transposase. J. Mol. Biol. 276:913-925. [DOI] [PubMed] [Google Scholar]
- 59.Zhou, M., and W. S. Reznikoff. 1997. Tn5 transposase mutants that alter DNA binding specificity. J. Mol. Biol. 271:362-373. [DOI] [PubMed] [Google Scholar]