Abstract
The horizontal transfer of the bacterium Wolbachia pipientis between invertebrate hosts hinges on the ability of Wolbachia to adapt to new intracellular environments. The experimental transfer of Wolbachia between distantly related host species often results in the loss of infection, presumably due to an inability of Wolbachia to adapt quickly to the new host. To examine the process of adaptation to a novel host, we transferred a life-shortening Wolbachia strain, wMelPop, from the fruit fly Drosophila melanogaster into a cell line derived from the mosquito Aedes albopictus. After long-term serial passage in this cell line, we transferred the mosquito-adapted wMelPop into cell lines derived from two other mosquito species, Aedes aegypti and Anopheles gambiae. After a prolonged period of serial passage in mosquito cell lines, wMelPop was reintroduced into its native host, D. melanogaster, by embryonic microinjection. The cell line-adapted wMelPop strains were characterized by a loss of infectivity when reintroduced into the original host, grew to decreased densities, and had reduced abilities to cause life-shortening infection and cytoplasmic incompatibility compared to the original strain. We interpret these shifts in phenotype as evidence for genetic adaptation to the mosquito intracellular environment. The use of cell lines to preadapt Wolbachia to novel hosts is suggested as a possible strategy to improve the success of transinfection in novel target insect species.
Wolbachia pipientis is a maternally transmitted obligate intracellular bacterium that chronically infects thousands of insect species, as well as a range of other arthropods and filarial nematodes (13). Wolbachia bacteria can induce various reproductive abnormalities in hosts, such as cytoplasmic incompatibility (CI), that promote the bacteria's vertical transmission and spread (14). The discordance of host and Wolbachia phylogenies indicates that these bacteria have moved between host lineages on multiple occasions during their evolutionary history (3, 42, 48), although the mechanisms that facilitate the transfer of Wolbachia are not well understood. The success of such host shifts is inherently reliant on the ability of the bacteria to adapt to new intracellular environments.
The experimental transfer of Wolbachia between host species (transinfection) has proved technically challenging, and the success of such experiments is difficult to predict. Despite an increasing number of reports that document Wolbachia transinfection, many attempts to experimentally infect host species are unsuccessful due to poor maternal transmission rates in the novel host (40). In some cases, transferred strains are extremely stable and maternally inherited at very high rates. This situation occurs primarily when Wolbachia is transferred within or between closely related species in a family or genus (6, 45, 47). In other cases, the infecting strain appears to be poorly adapted to its new host, showing fluctuating infection densities and various degrees of transovarial transmission. The result is often the loss of infection within a few host generations. Not surprisingly, Wolbachia infections tend to be more susceptible to loss when they have been transferred between phylogenetically distant hosts (17, 35). Similarly, those species that do not naturally harbor Wolbachia can be especially challenging to successfully transinfect (10, 36).
Understanding the process of Wolbachia adaptation to new hosts is central to gaining insight into the current distribution of Wolbachia bacteria among species and the evolutionary success of the genus. It may also facilitate the use of Wolbachia in an applied setting to introduce desirable traits into insect populations. For example, it has been proposed previously that the life-shortening Wolbachia strain wMelPop from the fruit fly Drosophila melanogaster might be introduced into populations of mosquito disease vectors in order to shift the population age structure and reduce pathogen transmission to humans (9, 38). The success of these strategies is predicated on the successful transfer of Wolbachia strains between host species in the laboratory.
To examine the process of Wolbachia adaptation to a new host, we transinfected a cell line from the mosquito Aedes albopictus with the wMelPop strain native to D. melanogaster (23) and maintained the line for ∼240 serial passages. We then used the cell line-adapted wMelPop to infect both Aedes aegypti and Anopheles gambiae cell lines. Both lines were maintained for an additional 60 passages before D. melanogaster was transinfected with the A. aegypti-adapted Wolbachia by embryonic microinjection. We report on the phenotypic outcomes of the long-term serial passage of wMelPop in mosquito cell lines as demonstrated by direct comparisons of the growth kinetics, life-shortening abilities, and levels of CI expression of the cell line-adapted wMelPop and original wMelPop strains in D. melanogaster.
MATERIALS AND METHODS
Cell lines and maintenance.
Three cell lines were used in this study: (i) Aa23.T, a tetracycline-treated cell line derived from A. albopictus embryos and referred to herein as Aa23 when infected with Wolbachia (27); (ii) RML-12, derived from A. aegypti larvae (19; C. E. Yunker, personal communication); and (iii) MOS-55, derived from A. gambiae larvae (20). All these cell lines were confirmed by PCR to be negative for Wolbachia infection prior to this study, as outlined below. Aa23.T and RML-12 cell lines were maintained in growth medium consisting of equal volumes of Mitsuhashi-Maramorosch (24) [1 mM CaCl2, 0.2 mM MgCl2, 2.7 mM KCl, 120 mM NaCl, 1.4 mM NaHCO3, 1.3 mM NaH2PO4, 22 mM d-(+)-glucose, 6.5 g of lactalbumin hydrolysate/liter, and 5.0 g of yeast extract/liter] and Schneider's insect medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10% heat-inactivated fetal bovine serum. MOS-55 was maintained in Schneider's insect medium supplemented with 20% heat-inactivated fetal bovine serum. Both media also contained penicillin (50 U/ml) and streptomycin (50 μg/ml). For routine maintenance, cells were grown in 25-cm2 plastic tissue culture flasks containing 5 ml of medium at 26°C without CO2 incubation. Cells were passaged every 3 to 4 days by vigorous shaking of the flask and the seeding of a new flask with 20% of the resuspended cells in 5 ml of fresh medium.
Establishment of wMelPop-infected cell lines.
wMelPop was purified from D. melanogaster w1118 embryos (23) and established in an uninfected A. albopictus cell line (Aa23.T) by using the shell vial technique (11). Embryos were collected every 45 min onto molasses agar plates covered with live yeast paste and dechorionated using freshly prepared 50%-diluted bleach (2.1% sodium hypochlorite final concentration; White King, Victoria, Australia) for 2 min. Embryos were then rinsed several times in sterile distilled water, immersed in 70% ethanol for 15 s, and rinsed three times in sterile phosphate-buffered saline (PBS), pH 7.4. Approximately 20 mg of surface-sterilized embryos (∼50 to 100 μl of packed embryos) was transferred into a mini Dounce tissue homogenizer (Wheaton) and suspended in 400 μl of PBS. Embryos were then homogenized for 2 to 3 min with a tight pestle. Two wells of 80% confluent Aa23.T cells in a 12-well cell culture plate prepared 24 h earlier were overlaid with 200 μl of homogenate each. The plate was centrifuged at 2,000 × g for 1 h at 15°C. Cells were then incubated at 26°C for 24 h, and the contents of each well were transferred into individual 25-cm2 cell culture flasks with 5 ml of fresh medium. After a confluent monolayer had formed, cells were split 1:5 and passaged as usual.
To establish the infection in A. aegypti RML-12 and A. gambiae MOS-55 cell lines, wMelPop was purified from Aa23 cells as described below and introduced into these cell lines by using the shell vial technique.
Characterization of wMelPop in cell lines.
Wolbachia infections in cell lines were characterized using (i) PCR screening and sequencing and (ii) electron microscopy. For each assay, naturally uninfected or tetracycline-cured derivatives of each cell line were used as negative controls.
(i) PCR screening and sequencing.
To monitor the infection status of cells, DNA was extracted from cultures as described previously (11) and amplified using the general Wolbachia surface protein (wsp) gene primers 81F and 691R or the diagnostic wsp primer set for wMelPop, 308F and 691R (48). To confirm the presence of wMelPop in these three cell lines, fragments of the Wolbachia 16S rRNA and wsp genes were PCR amplified, cloned, and sequenced. DNA was extracted from cells using a DNeasy tissue kit (Qiagen) and amplified as described previously using the diagnostic primers 99F and 994R for the Wolbachia 16S rRNA gene (26) and the primers 81F and 691R for the wsp gene (48). Total DNA from cell lines was also PCR amplified using the general eubacterial 16S rRNA primers 10F/1507R (21) and 968F/R1401R (25). The resulting PCR products were cloned into the pGEM-T Easy vector (Promega), and for each product, four clones from each infected cell line were randomly picked and sequenced. The presence of wMelPop and no other contaminating bacteria in cell lines was verified by denaturing gradient gel electrophoresis using a general primer set targeting eubacterial 16S rRNA genes (F-968-GC and R-1401) (25) by methods described previously (29).
(ii) Electron microscopy.
Insect cells were washed in PBS and rapidly fixed with microwave processing in 2.5% glutaraldehyde solution containing 0.1% CaCl2 and 1% sucrose in 0.1 M Na cacodylate, enrobed in 2% agarose, and postfixed in 1% osmium tetroxide in 0.1 M Na cacodylate buffer. Samples were then dehydrated in a sequence of increasing ethanol concentrations and, in a final step, in acetone (100%) and then embedded in epoxy resin (Epon 812) by using microwave processing (12, 27). Ultrathin sections (50 to 80 nm) prepared on an Ultracut T ultramicrotome (Leica Inc.) were placed onto copper grids and stained with 2% uranyl acetate followed by Reynolds lead citrate. The sections were examined in a JEOL-1010 electron microscope operated at 80 kV.
Purification of Wolbachia from cell culture for embryonic microinjection.
Insect cells from the confluent monolayers of two 175-cm2 flasks were harvested and centrifuged in 50-ml conical flasks at 1,000 × g for 5 min at 4°C, and the cell culture medium was discarded. The cellular pellet was then washed in SPG buffer (218 mM sucrose, 3.8 mM KH2PO4, 7.2 mM K2HPO4, and 4.9 mM l-glutamate, pH 7.2), and the centrifugation and wash steps were repeated. After being washed, the pellet was resuspended in 5 ml of SPG buffer on ice and sonicated twice for 10 s at 12.5 W with a Fisher Scientific model 60 sonic dismembranator (3-mm microtip diameter) to lyse the cells. This suspension was then centrifuged at 1,000 × g for 5 min at 4°C to pellet cellular debris. The supernatant was passed through a 5-μm Acrodisc syringe filter (Pall Life Sciences), and the filtrate was collected in 1.5-ml microcentrifuge tubes. These tubes were centrifuged at 12,000 × g for 15 min at 4°C to pellet Wolbachia cells. The supernatant was discarded, and the pellets were combined and resuspended in 400 μl of SPG buffer and centrifuged at 300 × g for 5 min to remove any remaining debris (44). The supernatant was then transferred into a clean tube and stored on ice until being used for injection (<3 h).
Embryonic microinjection.
Purified Wolbachia from RML-12 was microinjected into embryos of the D. melanogaster line w1118.T, which had previously been cured of infection by tetracycline treatment (23). Prior to microinjection, this line was confirmed to be free of Wolbachia by PCR using primers specific for the wMelPop IS5 repeat: IS5-FWD1 (5′-GTATCCAACAGATCTAAGC) and IS5-REV1 (5′-ATAACCCTACTCATAGCTAG). IS5 is a multicopy insertion element and, as such, is a much more sensitive target for determining infection status than single-copy genes such as wsp. For microinjection, early (preblastoderm)-stage embryos were collected every 30 min by using molasses agar plates with live yeast paste. Purified Wolbachia was microinjected into the posterior poles of embryos within 30 min of collection by standard techniques (1, 6, 44). After hatching, larvae were transferred into a standard cornmeal-based Drosophila rearing medium (2) and incubated at 24°C.
Drosophila rearing and PCR screening for infection status.
Virgin females resulting from injected embryos (generation 0 [G0]) were placed in vials with three w1118.T males to establish isofemale lines. After egg laying, G0 females were sacrificed and DNA was extracted using the Holmes-Bonner DNA extraction protocol (16). Wolbachia was detected in samples using PCR primers specific for the IS5 repeat element in wMelPop. The quality of the insect DNA was assessed using the primer set 12SA1 and 12SB1, which amplifies the D. melanogaster 12S rRNA gene (26). The amplification of DNA was carried out in a 20-μl reaction volume which included 2.0 μl of 10× buffer (New England Biolabs, Beverly, MA), 25 μM deoxynucleoside triphosphates, 0.5 μM forward and reverse primers, 0.75 U of Taq polymerase (New England Biolabs, Beverly, MA), and 1.0 μl of the DNA template. PCR conditions were as follows: denaturation at 94°C for 3 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min; and a final 10-min extension step at 72°C.
To select for a stable infection, only offspring from females that tested positive for Wolbachia by PCR screening were used as parental stock. Each generation, 25 to 50 females from each line were isolated as virgins, placed into individual vials, and outcrossed with three w1118.T males. Females that tested negative for Wolbachia were discarded along with their progeny. This selection regime was maintained for three generations, after which the lines were closed. The two resulting lines, those carrying the wMelPopCLA-1 and wMelPopCLA-2 (wMelPop cell line adapted) strains, were then monitored periodically by PCR to confirm the infection status. The selection regime was again repeated at G14 due to fluctuations in infection frequencies in both lines.
Life span assays.
The life spans of wMelPopCLA-1-, wMelPopCLA-2-, and wMelPop-infected lines were compared to those of tetracycline-cured derivatives of each line created by the addition of tetracycline (0.3 mg/ml) to the adult diet according to standard methods (15). Treated flies were reared on tetracycline for two generations and then transferred to a normal diet for a minimum of five generations before being used in experiments. To reduce the effects of genetic drift that may have occurred in these lines during tetracycline treatment, 100 females from each fly line (including infected lines) were backcrossed with 100 males from the same w1118.T stock line and the progeny were combined to form the next generation. This procedure was repeated for five generations (G23 to G28). Longevity assays were then conducted at G31, G33, and G35. To control for any crowding effects or size variability, the larval density in each stock bottle used to obtain flies was standardized (200 larvae/bottle) prior to longevity assays. Stock bottles were kept at 24°C until adult eclosion 9 to 10 days later, when flies were sexed as virgins and separated. In each assay, six sets of 20 flies for each sex were maintained at 29°C in standard cornmeal food vials without additional live yeast. Each day, the number of new deaths was recorded. Flies were moved into fresh food vials every 5 days. Survival curves for the various treatment groups were compared using a mixed-effects Cox proportional hazard model of survival analysis with the kinship package of the R suite of statistical software (www.r-project.org).
CI tests.
CI tests were conducted at G36 and G38 posttransinfection by using the previously backcrossed lines. To standardize rearing conditions for CI tests, larvae in fly stock bottles were grown under low-density conditions (150 to 200 larvae/bottle) at 24°C with a 12-h light-dark cycle. To obtain offspring for CI crosses, stock bottles were seeded with a set density of 200 eggs per 40 ml of food. After eclosion, flies were sexed, separated as virgins, and aged until the CI tests. Male flies were collected on day 2 of emergence and were used within 24 h of eclosion (33, 46). The female flies used were 5 to 7 days old. For each cross, single mating pairs (n = 40) were introduced into plastic bottles with molasses plate lids. Pairs were given 24 h to mate, and then the males were removed and the females were allowed to lay eggs. Eggs were collected onto molasses agar plates dotted with a live yeast suspension every 24 h for 3 days. Females that laid <50 eggs total across the three plates were discarded from the experiment. The plates were then placed at 24°C for a further 36 to 48 h, and then the numbers of total and unhatched eggs were determined. The statistical significance of hatch rates for various crosses was determined using a Mann-Whitney U test. A Bonferroni correction was used to compensate for multiple comparisons.
qPCR and density determination.
To examine if the density of Wolbachia bacteria in D. melanogaster had changed after long-term serial passage in mosquito cell lines, infection densities in head tissues of w1118 flies carrying the wMelPopCLA-1, wMelPopCLA-2, or wMelPop strain were monitored over the life spans of the flies by quantitative PCR (qPCR). Heads were selected for qPCR as wMelPop infection densities had previously been shown to increase rapidly in nervous tissue with adult age (22, 23). The density of bacteria of the closely related nonvirulent Wolbachia strain wMel was also examined after introgression for three generations from yw67c23 into the w1118 genetic background. qPCR assays were conducted at G46 posttransinfection. Flies reared as described for life span assays were collected at 4-day intervals (from days 4 to 32 postemergence) until all the flies in a line were dead, and tissue samples were stored at −80°C before analysis. Total DNA was extracted from dissected head tissues by using the DNeasy tissue kit protocol (Qiagen). To estimate the relative abundance of Wolbachia bacteria in each sample, we compared the abundance of the single-copy Wolbachia ankyrin repeat gene WD_0550 to that of the single-copy D. melanogaster gene Act88F. The forward primer 5′-CAGGAGTTGCTGTGGGTATATTAGC and the reverse primer 5′-TGCAGGTAATGCAGTAGCGTAAA were used to amplify a 74-bp amplicon from WD_0550, and the forward primer 5′-ATCGAGCACGGCATCATCAC and the reverse primer 5′-CACGCGCAGCTCGTTGTA were used to amplify a 78-bp amplicon from Act88F. For each treatment, 12 biological replicates per time point were examined. For each sample, qPCR amplification of DNA was performed in triplicate using a Rotor-Gene 6000 system (Corbett Research, Australia). Amplification was carried out in a 10-μl reaction volume which included 5 μl of Platinum Sybr green I supermix (Invitrogen, CA), 1 μM forward and reverse primers, and 1 ng of the DNA template. The PCR conditions were 50°C for 2 min; 95°C for 2 min; 40 cycles of 95°C for 5 s, 60°C for 5 s, and 72°C for 10 s; and a melting curve from 67 to 95°C. A standard calibrator was used to normalize between qPCR runs, and the specificity of PCR products was determined by melting-curve analysis. Crossing threshold (CT) and amplification efficiency values for each sample were calculated using Corbett Rotor-Gene (version 1.7.75) software. The relative abundance of Wolbachia bacteria in each sample was then determined using the method discussed by Pfaffl (28). Regression analysis was used to detect trends in the density of Wolbachia bacteria over the lifetimes of flies of individual lines. An analysis of covariance was then employed to examine the relationship between density and the covariates of fly age and bacterial strain. All abundance data were log transformed prior to analysis. A Bonferroni correction was used to compensate for multiple comparisons.
RESULTS
Several initial attempts to establish wMelPop in the A. albopictus embryonic cell line Aa23 were unsuccessful. Typically, infection was lost after several passages or lines were discontinued due to a complete loss of confluence or growth of mosquito cells. This situation mirrors that observed when wMelPop purified from Drosophila is injected into mosquitoes, with large fluctuations in infection density eventually leading to the loss of infection (E. A. McGraw and S. L. O'Neill, unpublished data). In total, only 2 (3%) of 68 independent attempts to establish the wMelPop infection in Aa23 cells were successful.
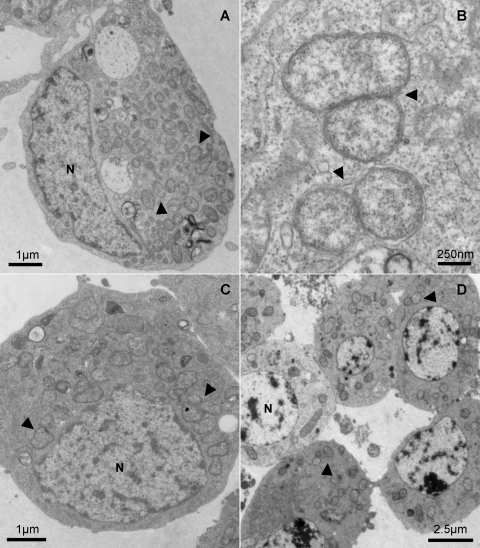
Once established in Aa23, wMelPop was serially passaged for 237 passages (∼2.5 years) before being transferred into the A. aegypti cell line RML-12 and the A. gambiae cell line MOS-55. The stable establishment of wMelPop in these two cell lines occurred much more easily than the initial infection of Aa23, with two of two independent attempts for each cell line yielding stable wMelPop infections. Partial sequences of the Wolbachia 16S rRNA and wsp genes from the three cell lines used were all identical to the sequence from wMelPop, confirming that infections were not the result of contamination with other strains. Infection in mosquito cells was also confirmed using transmission electron microscopy (TEM). TEM micrographs of the three infected mosquito cell lines show that representative cells from each line were heavily infected with wMelPop (Fig. 1).
FIG. 1.
Electron microscopy analysis of wMelPop in mosquito cell lines. (A) Low-magnification transmission electron micrograph showing a large number of Wolbachia bacteria (examples are marked with arrowheads) dispersed throughout the cytoplasm of an A. aegypti RML-12 cell. N, nucleus. (B) High-magnification micrograph of four Wolbachia cells presumably undergoing the process of cell division in RML-12 cells (arrowheads). (C) Low-magnification micrograph showing the presence of several Wolbachia bacteria in the cytoplasm of an A. albopictus Aa23 cell. (D) Cluster of A. gambiae MOS-55 cells each infected with multiple Wolbachia bacteria.
wMelPop was purified from the A. aegypti RML-12 cell line and reintroduced into its native host, D. melanogaster w1118, which had been previously cured of its natural wMelPop infection by tetracycline treatment. At the time of reintroduction, wMelPop had been maintained for over 3 years outside its native host, through 237 passages in Aa23 cells and 60 passages in RML-12 cells. In total, 446 embryos were microinjected, giving rise to 108 G0 larvae (24% hatch rate). All 10 surviving G0 females were PCR positive for Wolbachia. Of these, eight produced offspring and two produced PCR-positive G1 isofemale lines. The Wolbachia strains in these two independent isofemale lines were named wMelPopCLA-1 and wMelPopCLA-2.
The infection frequencies in wMelPopCLA-infected lines were then monitored periodically over time (Fig. 2). Both wMelPopCLA strains were initially observed to display variable maternal transmission rates in the original Drosophila host, reflected in fluctuating infection frequencies in the absence of experimental selection. During an initial period of experimental selection for increased infection (G1 to G3 posttransinfection), infection frequencies as detected by PCR were observed to increase in both the wMelPopCLA-1-infected line (58 to 87%) and the wMelPopCLA-2-infected line (55 to 100%). In the absence of experimental selection from G4 onwards, infection frequencies in both lines initially were stable or fluctuated but then rapidly decreased such that by G14 posttransinfection only 32% of individuals carrying wMelPopCLA-1 and 24% of those carrying wMelPopCLA-2 remained infected. Selection was repeated again at G14, and after one additional generation, infection frequencies in both lines increased to 100% and remained fixed until G46, when last assayed.
FIG. 2.
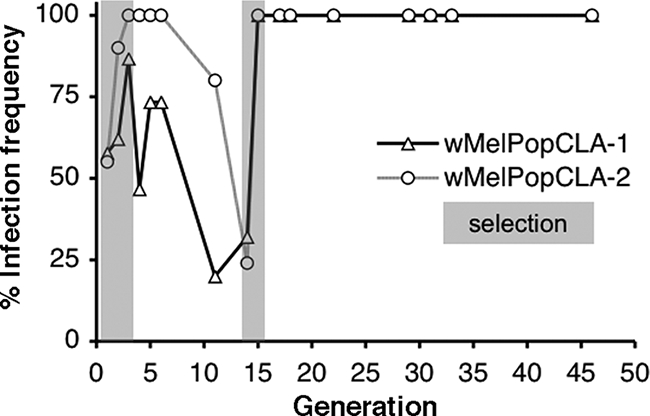
Frequencies of Wolbachia infection in D. melanogaster wMelPopCLA-1- and wMelPopCLA-2-infected lines posttransinfection (G0). Gray shaded regions represent periods of experimental selection for infection.
To assess the effect of continuous cell line culture on the ability of the Wolbachia strain wMelPop to colonize Drosophila, we compared infection densities in flies that contained wMelPopCLA strains with those in flies carrying the original wMelPop strain by qPCR. Since it is known that wMelPop densities increase rapidly in adult flies held at 29°C, we assessed Wolbachia densities across the adult fly life span. As populations of flies aged, Wolbachia densities in head tissues of wMelPop-infected flies rapidly increased (Fig. 3). The densities of Wolbachia bacteria in wMelPopCLA-1- and wMelPopCLA-2-infected flies also increased as the flies aged, although these increases were noticeably less than those of the wMelPop densities. Wolbachia densities in wMelPop-infected flies were roughly fourfold higher than those in wMelPopCLA-1- or wMelPopCLA-2-infected flies at day 12 postemergence. Flies infected with the non-life-shortening wMel strain had the lowest levels of infection, which increased only slightly over the life spans of the flies. Overall, there were significant effects of fly age (F1,275 = 41.92; P < 0.001) and bacterial strain (F3,275 = 678.37; P < 0.001) on the Wolbachia density for all fly lines. This outcome was reflected by significant differences in the effects of strain and age after pairwise comparisons between lines (P < 0.001 for all comparisons), except for the wMelPopCLA-1- and wMelPopCLA-2-infected lines, for which strain effects were not significantly different from each other (F1,144 = 0.09; P > 0.05).
FIG. 3.
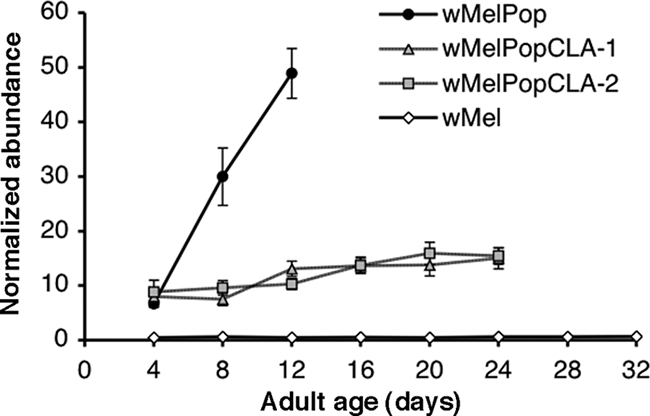
Mean relative Wolbachia densities in fly heads (±SE; n = 12 per datum point) as determined by real-time qPCR for four lines of infected flies collected at various ages over their life spans and maintained at 29°C. Fly samples were collected at 4-day intervals until the flies were dead.
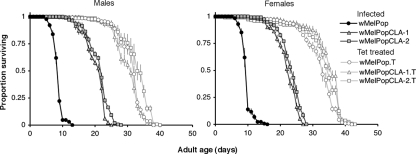
To test whether the ability of wMelPop to induce life-shortening infection had changed during long-term serial passage, we conducted a series of longevity assays at G31, G33, and G35 posttransinfection. For these experiments, the survival rates of infected flies from the wMelPopCLA-1-, wMelPopCLA-2-, and wMelPop-carrying lines were compared with those of the corresponding uninfected tetracycline-treated derivates maintained at 29°C. Survival curves for males and females of each treatment group were measured independently. In all assays, male and female flies from the wMelPop-infected line demonstrated more pronounced life span reductions than flies from the wMelPopCLA lines relative to the life spans of the tetracycline-treated controls (Fig. 4). The life spans of wMelPopCLA-1- and wMelPopCLA-2-infected lines appeared to be intermediate relative to those of the wMelPop-infected line but were shortened relative to those of the tetracycline-treated controls. For example, at G31 posttransinfection, the mean time to death (± standard error [SE]) for wMelPop-infected females (9.8 ± 0.1 days) was noticeably shorter than that for wMelPopCLA-1-infected females (22.2 ± 0.3 days) or wMelPopCLA-2-infected females (23.4 ± 0.3 days). The mean time to death was increased for tetracycline-treated control lines, with the life spans of tetracycline-treated females derived from the wMelPop-infected line (wMelPop.T females; 32.1 ± 0.5 days), wMelPopCLA-1.T females (34.6 ± 0.5 days), and wMelPopCLA-2.T females (33.4 ± 0.6 days) all being longer than the life spans of infected counterparts. For females, the proportional hazard of death associated with carrying infection was significantly greater for individuals with wMelPop (relative risk ratio, 135.7; 95% confidence interval, 40.3 to 456.5) than for those carrying either wMelPopCLA-1 (relative risk ratio, 30.0; 95% confidence interval, 15.4 to 58.5) or wMelPopCLA-2 (relative risk ratio, 17.7; 95% confidence interval, 10.5 to 30.7) (P < 0.001 for all comparisons to wMelPop-infected flies). The same trends were also observed for males. These results were consistent with those obtained from measurements at G33 and G35 posttransinfection (data not shown).
FIG. 4.
Survival curves of populations of male and female flies from wMelPop- and wMelPopCLA-infected lines at G31 posttransinfection. Shaded symbols represent infected flies, and unshaded symbols represent uninfected tetracycline (Tet)-treated counterparts. Error bars on curves represent SEs. Adult flies were maintained at 29°C.
In order to examine the effects of long-term cell culture on CI expression, we established test crosses between uninfected and infected flies and examined hatch rates of the resulting eggs. Results from incompatible test crosses indicated that wMelPop.T females mated with wMelPop males produced embryos with a mean hatch rate of 24%, which was significantly lower than that for embryos from the corresponding cross with wMelPopCLA-1-infected males or wMelPopCLA-2-infected males (P < 0.001; Mann-Whitney) (Fig. 5). A statistically significant difference in the mean hatch rate for crosses with wMelPopCLA-1-infected males and those with wMelPopCLA-2-infected males (P < 0.001) was also observed. In rescue tests, mean hatch rates of embryos produced from crosses between wMelPop-infected males and wMelPop-infected females, wMelPopCLA-1-infected females, or wMelPopCLA-2-infected females were not significantly different from one another. Thus, wMelPopCLA strains had reduced abilities to induce CI compared to that of wMelPop. In contrast, the abilities of the cell-adapted strains to rescue an incompatible cross appeared to be unchanged.
FIG. 5.
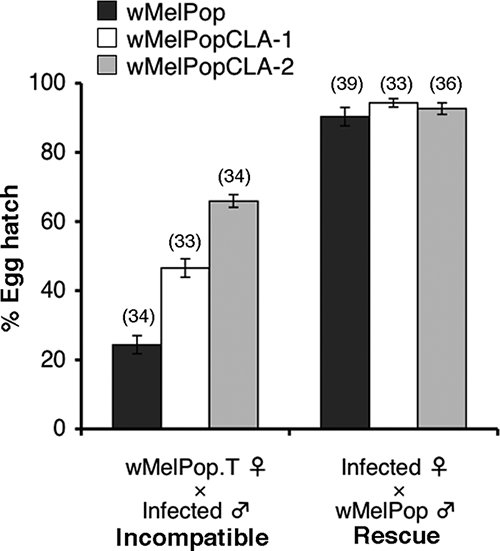
Abilities of wMelPop and wMelPopCLA strains to induce and abolish CI. Shown are mean percentages of hatching eggs (±SE) for wMelPop.T females mated with infected males (incompatible cross) and infected females mated with wMelPop-infected males (rescue cross). Values in parentheses above error bars represent the numbers of replicate crosses.
DISCUSSION
The use of an in vitro cell culture system provided an ideal means to examine the adaptation of Wolbachia to a novel host cell environment. This approach contrasts with the direct transfer of Wolbachia between insects, a method in which selective forces are presumably different and more complex and in which longer insect generation times, vertical transmission, and the labor-intensive nature of rearing live insects make selection for transinfected lines challenging.
The initial difficulty in establishing a wMelPop infection in the A. albopictus cell line Aa23 demonstrated that wMelPop was not naturally preadapted for growth in mosquito cells. Following the stable infection of Aa23 and serial passage for several years, wMelPop was successfully established in the RML-12 and MOS-55 cell lines from A. aegypti and A. gambiae, respectively, two species that are not naturally infected with Wolbachia (10, 18, 32, 34, 39). The transfer of wMelPop between Aa23 and these two mosquito cell lines occurred much more readily than the initial transfer from D. melanogaster to Aa23, potentially due to (i) the use of a higher infective dose of wMelPop purified from Aa23 for transfer than of that from Drosophila and/or (ii) a smaller divergence in intracellular environments among these mosquito cell lines than between those of Drosophila and Aa23. Our ability to establish stable wMelPop infections in MOS-55 was also consistent with previous reports that Wolbachia can be established in A. gambiae cell lines (31).
As observed by TEM, infection densities in the three cell lines, particularly in RML-12, closely resembled those previously described for somatic and nervous tissue in D. melanogaster, with individual mosquito cells heavily infected with bacteria (23). Whether wMelPop in Aa23, RML-12, and MOS-55 was exhibiting tropism for cell types similar to or different from D. melanogaster is unknown. The morphology of cells within Aa23 (derived from embryos) (27) and RML-12 and MOS-55 (derived from larvae) (C. E. Yunker, personal communication) (20) appeared to be heterogeneous, and the tissue-specific origin of cell lineages within these lines has not been identified.
When wMelPop was reintroduced into Drosophila after long-term serial passage in mosquito cell lines, initial establishment in wMelPopCLA-1- and wMelPopCLA-2-infected lines was problematic due to unstable fluctuations in infection frequency in the absence of selection. Unstable maternal transmission and variable infection frequencies are often commonly observed when Wolbachia is moved between distantly related hosts (17, 35, 40). After two rounds of selection for infection, populations progressed to fixation for infection and have remained stable (∼2 years).
Longevity assays revealed that wMelPopCLA strains had become partially attenuated in virulence compared to the original wMelPop strain in D. melanogaster. This phenotypic shift in life-shortening ability may be related to the decreased replication rates of both wMelPopCLA strains in the head tissues of infected lines relative to that of wMelPop. The life-shortening phenotype of wMelPop is thought to result from pathology induced by the uncontrolled replication of bacteria in muscle and nervous tissue (22, 23). Therefore, it is likely that the decreased replication rates of the Wolbachia wMelPopCLA strains were directly correlated with decreased abilities to induce life shortening in the original host.
In addition to reductions in the abilities of cell line-adapted Wolbachia strains to induce life shortening, similar reductions in the abilities of these strains to induce CI were noted. Presumably, this phenotypic shift was also linked to the reduced replication rate of the cell line-adapted Wolbachia in its original host. In several insects, decreased Wolbachia densities in developing sperm cells have been correlated with decreased levels of CI expression (4, 5, 7, 8, 30, 41). It is also possible that the tropism of Wolbachia for different host cell types may have been altered during long-term passage in cell lines since CI induction was clearly distorted but CI rescue was not. This idea suggests that the densities of infecting bacteria in some tissues may be changed more dramatically than those in others.
In summary, the wMelPop strain was initially difficult to transfer into cell lines, but a small number of infected lines could eventually be established. The strain was subsequently much more easily transferred into cell lines derived from other mosquito species. The cell line-adapted Wolbachia displayed reduced infectivity and maternal transmission rates when injected back into its original host. It grew to lower densities and showed phenotypic shifts for both life shortening and CI expression. Taken together, our results provide evidence for the active genetic adaptation of wMelPop to mosquito cell lines during long-term serial passage.
Given that there is growing interest in the potential to use Wolbachia strains in an applied context (37, 45, 47), the preadaptation of strains to particular host intracellular environments may facilitate the subsequent transfer of these symbionts into hosts that are difficult to transinfect. Such an application may involve the use of life-shortening wMelPop adapted to mosquito cell lines as a source of material for experiments with the transinfection of A. aegypti and Anopheles mosquitoes, as part of an applied strategy to alter the mosquito population age structure to reduce the transmission of pathogens such as dengue virus and Plasmodium parasites to humans (9, 38). Furthermore, given the availability of genome sequence information for this bacterium (43), this system may allow for the underlying genetic mechanisms responsible for adaptation to novel hosts to be identified.
Acknowledgments
We gratefully acknowledge the gift of RML-12 and MOS-55 cell lines from Robert B. Tesh, University of Texas Medical Branch. We also thank Jasmin Howie, Geoff Pittman, Jenny Gough, and Manni Sidhu for technical assistance; Simon Blomberg and Catriona Condon for assistance with statistics; and members of the O'Neill lab for providing helpful technical advice and comments on the manuscript.
This work was supported by a grant from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health Initiative of the Bill and Melinda Gates Foundation.
Footnotes
Published ahead of print on 3 October 2008.
REFERENCES
- 1.Ashburner, M. 1989. Drosophila: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 2.Ashburner, M., and J. Roote. 2000. Laboratory culture of Drosophila, p. 585-599. In W. Sullivan, M. Ashburner, and R. S. Hawley (ed.), Drosophila protocols. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Baldo, L., N. A. Ayoub, C. Y. Hayashi, J. A. Russell, J. K. Stahlhut, and J. H. Werren. 2008. Insight into the routes of Wolbachia invasion: high levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol. Ecol. 17:557-569. [DOI] [PubMed] [Google Scholar]
- 4.Bordenstein, S. R., M. L. Marshall, A. J. Fry, U. Kim, and J. J. Wernegreen. 2006. The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog. 2:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourtzis, K., A. Nirgianaki, G. Markakis, and C. Savakis. 1996. Wolbachia infection and cytoplasmic incompatibility in Drosophila species. Genetics 144:1063-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle, L., S. L. O'Neill, H. M. Robertson, and T. L. Karr. 1993. Interspecific and intraspecific horizontal transfer of Wolbachia in Drosophila. Science 260:1796-1799. [DOI] [PubMed] [Google Scholar]
- 7.Breeuwer, J. A., and J. H. Werren. 1993. Cytoplasmic incompatibility and bacterial density in Nasonia vitripennis. Genetics 135:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bressac, C., and F. Rousset. 1993. The reproductive incompatibility system in Drosophila simulans: DAPI-staining analysis of the Wolbachia symbionts in sperm cysts. J. Invertebr. Pathol. 61:226-230. [DOI] [PubMed] [Google Scholar]
- 9.Cook, P. E., C. J. McMeniman, and S. L. O'Neill. 2008. Modifying insect population age structure to control vector-borne disease, p. 126-140. In S. Aksoy (ed.), Transgenesis and the management of vector-borne disease. Landes Biosciences, Austin, TX. [DOI] [PubMed]
- 10.Curtis, C. F., and S. P. Sinkins. 1998. Wolbachia as a possible means of driving genes into populations. Parasitology 116(Suppl.):S111-S115. [DOI] [PubMed] [Google Scholar]
- 11.Dobson, S. L., E. J. Marsland, Z. Veneti, K. Bourtzis, and S. L. O'Neill. 2002. Characterization of Wolbachia host cell range via the in vitro establishment of infections. Appl. Environ. Microbiol. 68:656-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feinberg, M. D., K. M. Szumowski, and K. M. Harris. 2001. Microwave fixation of rat hippocampal slices. In R. T. Giberson and R. S. Demaree, Jr. (ed.), Microwave protocols for microscopy. Humana Press, Totowa, NJ.
- 13.Hilgenboecker, K., P. Hammerstein, P. Schlattmann, A. Telschow, and J. H. Werren. 2008. How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol. Lett. 281:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann, A. A., and M. Turelli. 1997. Cytoplasmic incompatibility in insects, p. 42-80. In S. L. O'Neill, A. A. Hoffmann, and J. H. Werren (ed.), Influential passengers: inherited microorganisms and arthropod reproduction. Oxford University Press, Oxford, United Kingdom.
- 15.Hoffmann, A. A., M. Turelli, and G. M. Simmons. 1986. Unidirectional incompatibility between populations of Drosophila simulans. Evolution 40:692-701. [DOI] [PubMed] [Google Scholar]
- 16.Holmes, D. S., and J. Bonner. 1973. Preparation, molecular weight, base composition, and secondary structure of giant nuclear ribonucleic acid. Biochemistry 12:2330-2338. [DOI] [PubMed] [Google Scholar]
- 17.Kang, L., X. Ma, L. Cai, S. Liao, L. Sun, H. Zhu, X. Chen, D. Shen, S. Zhao, and C. Li. 2003. Superinfection of Laodelphax striatellus with Wolbachia from Drosophila simulans. Heredity 90:71-76. [DOI] [PubMed] [Google Scholar]
- 18.Kittayapong, P., K. J. Baisley, V. Baimai, and S. L. O'Neill. 2000. Distribution and diversity of Wolbachia infections in Southeast Asian mosquitoes (Diptera: Culicidae). J. Med. Entomol. 37:340-345. [DOI] [PubMed] [Google Scholar]
- 19.Kuno, G. 1983. Cultivation of mosquito cell lines in serum-free media and their effects on dengue virus replication. In Vitro 19:707-713. [DOI] [PubMed] [Google Scholar]
- 20.Marhoul, Z., and M. Pudney. 1972. A mosquito cell line (MOS. 55) from Anopheles gambiae larva. Trans. R. Soc. Trop. Med. Hyg. 66:183-184. [DOI] [PubMed] [Google Scholar]
- 21.Mateos, M., S. J. Castrezana, B. J. Nankivell, A. M. Estes, T. A. Markow, and N. A. Moran. 2006. Heritable endosymbionts of Drosophila. Genetics 174:363-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGraw, E. A., D. J. Merritt, J. N. Droller, and S. L. O'Neill. 2002. Wolbachia density and virulence attenuation after transfer into a novel host. Proc. Natl. Acad. Sci. USA 99:2918-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Min, K. T., and S. Benzer. 1997. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc. Natl. Acad. Sci. USA 94:10792-10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitsuhashi, J., and K. Maramorosch. 1964. Leafhopper tissue culture: embryonic, nymphal and imaginal tissues from aseptic insects. Contrib. Boyce Thompson Inst. 22:435-460. [Google Scholar]
- 25.Nubel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Neill, S. L., R. Giordano, A. M. Colbert, T. L. Karr, and H. M. Robertson. 1992. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. USA 89:2699-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Neill, S. L., M. M. Pettigrew, S. P. Sinkins, H. R. Braig, T. G. Andreadis, and R. B. Tesh. 1997. In vitro cultivation of Wolbachia pipientis in an Aedes albopictus cell line. Insect Mol. Biol. 6:33-39. [DOI] [PubMed] [Google Scholar]
- 28.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pittman, G. W., S. M. Brumbley, P. G. Allsopp, and S. L. O'Neill. 2008. “Endomicrobia” and other bacteria associated with the hindgut of Dermolepida albohirtum larvae. Appl. Environ. Microbiol. 74:762-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poinsot, D., K. Bourtzis, G. Markakis, C. Savakis, and H. Mercot. 1998. Wolbachia transfer from Drosophila melanogaster into D. simulans: host effect and cytoplasmic incompatibility relationships. Genetics 150:227-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasgon, J. L., X. Ren, and M. Petridis. 2006. Can Anopheles gambiae be infected with Wolbachia pipientis? Insights from an in vitro system. Appl. Environ. Microbiol. 72:7718-7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasgon, J. L., and T. W. Scott. 2004. An initial survey for Wolbachia (Rickettsiales: Rickettsiaceae) infections in selected California mosquitoes (Diptera: Culicidae). J. Med. Entomol. 41:255-257. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds, K. T., L. J. Thomson, and A. A. Hoffmann. 2003. The effects of host age, host nuclear background and temperature on phenotypic effects of the virulent Wolbachia strain popcorn in Drosophila melanogaster. Genetics 164:1027-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricci, I., G. Cancrini, S. Gabrielli, S. D'Amelio, and G. Favia. 2002. Searching for Wolbachia (Rickettsiales: Rickettsiaceae) in mosquitoes (Diptera: Culicidae): large polymerase chain reaction survey and new identifications. J. Med. Entomol. 39:562-567. [DOI] [PubMed] [Google Scholar]
- 35.Riegler, M., S. Charlat, C. Stauffer, and H. Mercot. 2004. Wolbachia transfer from Rhagoletis cerasi to Drosophila simulans: investigating the outcomes of host-symbiont coevolution. Appl. Environ. Microbiol. 70:273-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rigaud, T., P. S. Pennings, and P. Juchault. 2001. Wolbachia bacteria effects after experimental interspecific transfers in terrestrial isopods. J. Invertebr. Pathol. 77:251-257. [DOI] [PubMed] [Google Scholar]
- 37.Sinkins, S. P., and F. Gould. 2006. Gene drive systems for insect disease vectors. Nat. Rev. Genet. 7:427-435. [DOI] [PubMed] [Google Scholar]
- 38.Sinkins, S. P., and S. L. O'Neill. 2000. Wolbachia as a vehicle to modify insect populations, p. 271-287. In A. M. Handler and A. A. James (ed.), Insect transgenesis: methods and applications. CRC Press, London, United Kingdom.
- 39.Tsai, K. H., J. C. Lien, C. G. Huang, W. J. Wu, and W. J. Chen. 2004. Molecular (sub) grouping of endosymbiont Wolbachia infection among mosquitoes of Taiwan. J. Med. Entomol. 41:677-683. [DOI] [PubMed] [Google Scholar]
- 40.Van Meer, M. M., and R. Stouthamer. 1999. Cross-order transfer of Wolbachia from Muscidifurax uniraptor (Hymenoptera: Pteromalidae) to Drosophila simulans (Diptera: Drosophilidae). Heredity 82:163-169. [DOI] [PubMed] [Google Scholar]
- 41.Veneti, Z., M. E. Clark, S. Zabalou, T. L. Karr, C. Savakis, and K. Bourtzis. 2003. Cytoplasmic incompatibility and sperm cyst infection in different Drosophila-Wolbachia associations. Genetics 164:545-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werren, J. H., W. Zhang, and L. R. Guo. 1995. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. R. Soc. Lond. B 261:55-63. [DOI] [PubMed] [Google Scholar]
- 43.Wu, M., L. V. Sun, J. Vamathevan, M. Riegler, R. Deboy, J. C. Brownlie, E. A. McGraw, W. Martin, C. Esser, N. Ahmadinejad, C. Wiegand, R. Madupu, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, A. S. Durkin, J. F. Kolonay, W. C. Nelson, Y. Mohamoud, P. Lee, K. Berry, M. B. Young, T. Utterback, J. Weidman, W. C. Nierman, I. T. Paulsen, K. E. Nelson, H. Tettelin, S. L. O'Neill, and J. A. Eisen. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xi, Z., and S. L. Dobson. 2005. Characterization of Wolbachia transfection efficiency by using microinjection of embryonic cytoplasm and embryo homogenate. Appl. Environ. Microbiol. 71:3199-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xi, Z., C. C. Khoo, and S. L. Dobson. 2005. Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science 310:326-328. [DOI] [PubMed] [Google Scholar]
- 46.Yamada, R., K. D. Floate, M. Riegler, and S. L. O'Neill. 2007. Male development time influences the strength of Wolbachia-induced cytoplasmic incompatibility expression in Drosophila melanogaster. Genetics 177:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zabalou, S., M. Riegler, M. Theodorakopoulou, C. Stauffer, C. Savakis, and K. Bourtzis. 2004. Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc. Natl. Acad. Sci. USA 101:15042-15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, W., F. Rousset, and S. L. O'Neill. 1998. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. R. Soc. Lond. B 265:509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]