Abstract
Enzymatic steps from two different biosynthetic pathways were combined in Escherichia coli, directing the synthesis of a new class of biomolecules—ubiquinones with prenyl side chains containing conjugated double bonds. This was achieved by the activity of a C30 carotenoid desaturase, CrtN, from Staphylococcus aureus, which exhibited an inherent flexibility in substrate recognition compared to other carotenoid desaturases. By utilizing the known plasticity of E. coli's native ubiquinone biosynthesis pathway and the unusual activity of CrtN, modified ubiquinone structures with prenyl side chains containing conjugated double bonds were generated. The side chains of the new structures were confirmed to have different degrees of desaturation by mass spectrometry and nuclear magnetic resonance analysis. In vivo 14C labeling and in vitro activity studies showed that CrtN desaturates octaprenyl diphosphates but not the ubiquinone compounds directly. Antioxidant properties of conjugated side chain ubiquinones were analyzed in an in vitro β-carotene-linoleate model system and were found to be higher than the corresponding unmodified ubiquinones. These results demonstrate that by combining pathway steps from different branches of biosynthetic networks, classes of compounds not observed in nature can be synthesized and structural motifs that are functionally important can be combined or enhanced.
Ubiquinone (UQ), or coenzyme Q, is a respiratory chain electron carrier and important cellular antioxidant composed of a benzoquinone ring, responsible for UQs radical scavenging activity (27), and an isoprene tail of various chain lengths (7, 22, 42) (Fig. 1 and 2). UQ has applications as a pharmaceutical and dietary supplement and as a cosmetic ingredient. For example, oral administration of UQ-10, the UQ species synthesized by humans, is used in the treatment of several conditions, such as cardiomyopathy, Alzheimer's and Parkinson's diseases, and diabetes (reviewed in reference 4). An increasing demand for UQ as an antioxidant supplement has led to renewed efforts in improving current microbial production processes by engineering hosts such as Escherichia coli for commercial fermentation processes (4).
FIG. 1.
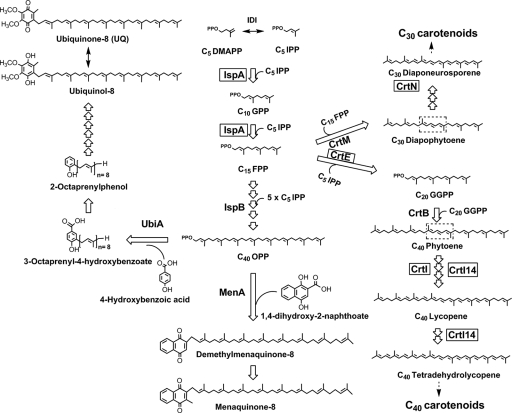
Simplified native E. coli UQ-8 and menaquinone-8 pathways and recombinant carotenoid pathways introduced into noncarotenogenic E. coli to direct C30 and C40 carotenoid biosynthesis are depicted. Recombinant enzymes overexpressed in this study in E. coli are boxed. Carotenoid desaturases were overexpressed in E. coli to investigate the production of UQs with conjugated prenyl side chains from E. coli's native UQ-8 pathway. C20 GGPP synthase (CrtE) and C15 FPP synthase (IspA) (boxed), in addition to C25 FGPP synthase (Fgs), and C50 DPP synthase (Dds) (not depicted), were coexpressed with C30 carotenoid desaturase CrtN in E. coli to investigate synthesis of UQs with shorter and longer desaturated prenyl chains. The central three CDBs characteristic for carotenoids are highlighted. Isoprenoid enzymes are as follows: IDI, IPP isomerase; IspA, FPP synthase; IspB, OPP synthase. Carotenoid enzymes are as follows: CrtM, diapophytoene synthase; CrtN, diapophytoene desaturase; CrtE, GGPP synthase; CrtB, phytoene synthase; CrtI, phytoene desaturase; CrtI14, in vitro-evolved phytoene desaturase. Quinone enzymes are as follows: UbiA, p-hydroxybenzoic acid octaprenyl transferase; MenA, 1,4-dihydroxy-2-naphthoate octaprenyltransferase. GPP, geranyl diphosphate.
FIG. 2.
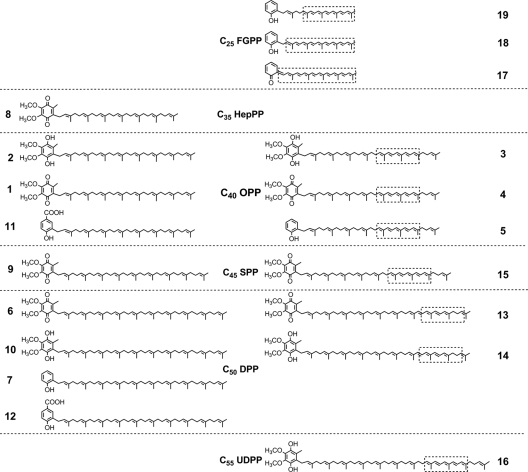
Structures of natural and unnatural hybrid UQs and UQ derivatives synthesized in E. coli from different linear isoprene chain precursors (center). Desaturated hybrid UQ derivatives (CDBs highlighted) identified in this study are shown on the right side. HepPP, heptaprenyl diphosphate; SPP, solanesyl diphosphate; UDPP, undecaprenyl diphosphate.
Biosynthesis of UQ has been extensively studied in the two model organisms, E. coli and Saccharomyces cerevisiae (22, 37, 40). The hydrophobic isoprene tail of UQ is derived via the successive head-to-tail condensation of C5 isoprene units (isopentenyl diphosphate [IPP]) catalyzed by prenyl diphosphate synthases. The length of the isoprenoid chain varies among different organisms; for example, S. cerevisiae synthesizes UQ-6 (C30 is six times C5 IPP), E. coli produces UQ-8 (C40 is eight times C5 IPP), and humans synthesize UQ-10 (C50 is 10 times C5 IPP). In addition to UQ-8, E. coli produces the octaprenyl naphthoquinones menaquinone and demethylmenaquinone as respiratory quinones. Under aerobic growth, the UQ concentration (∼200 to 300 nmol g−1) is about five times greater than the menaquinone and demethylmenaquinone concentration. Menaquinone and demethylmenaquinone, however, constitute the major quinones under anaerobic conditions (37, 46, 50).
In E. coli, the UQ isoprenoid tail is synthesized by octaprenyl diphosphate (OPP) synthase IspB, and an additional eight Ubi enzymes are required for UQ synthesis as shown in Fig. 1. UbiA, p-hydroxybenzoic acid octaprenyltransferase, catalyzes the transfer of a prenyl side chain to the quinone ring precursor p-hydroxybenzoic acid. UbiA represents a key node in the UQ pathway where precursors from two pathways, the aromatic amino acid and isoprenoid pathways (shikimate/chorismate and 1-deoxy-xylulose-5-phosphate pathways in E. coli), converge. The demonstrated ability of UbiA to accept longer and shorter prenyl diphosphate molecules as substrates (29) offers opportunities for the biosynthesis of modified prenyl chains in engineered hosts. This would enable the synthesis of UQs with new properties, such as improved antioxidant properties.
The length of the prenyl chain can be manipulated by expressing heterologous prenyltransferases with different chain length specificities in E. coli (4, 36, 52, 53). However, additional modifications of the isoprenoid chain, such as chain desaturations, require the recruitment of enzyme activities from other biosynthetic pathways. Recruitment of enzyme activities is a frequently exploited mechanism in nature to create chemical diversity, and in particular, isoprenoid pathways are often linked to other pathways (2, 9, 15, 45, 49, 51). We therefore sought to explore the possibility of using enzymes that modify the backbone of carotenoid molecules to generate UQs with modified prenyl chains.
Carotenoids are isoprenoid-derived pigments with functions in photosynthesis and as antioxidants. Linear isoprenoid chains are synthesized by the sequential addition of IPP to allylic diphosphates of various lengths in head-to-tail reactions catalyzed by prenyl diphosphate synthases (Fig. 1). Carotenoid biosynthesis, however, proceeds via the head-to-head condensation of two linear isoprenoid diphosphate molecules, providing a symmetrical polyene backbone. Although most natural carotenoids have a C40 backbone resulting from the condensation of two C20 geranylgeranyl diphosphate (GGPP) molecules, some gram-positive bacteria are known to produce C30 carotenoids by the condensation of two C15 farnesyl diphosphate (FPP) molecules. Carotenoid desaturase enzymes subsequently generate a chromophore system of conjugated double bonds (CDBs) necessary for antioxidant function (Fig. 1). Additional structural diversity is generated by a diverse array of modifying enzymes, many of which can accept alternative substrates, a characteristic which has been exploited to direct the synthesis of novel, unnatural carotenoid structures in heterologous hosts (17, 43). Recent studies have also shown some promiscuity of carotenoid desaturase activity (17, 32, 34, 35, 44, 47).
Here we report on the successful generation of UQs with prenyl side chains containing CDBs (hereafter the term “desaturated” will be used to describe UQs with additional double bonds in their prenyl side chains not present in the native structures), not observed in nature, by the C30 carotenoid desaturase CrtN from Staphylococcus aureus (31, 32). Further combinatorial biosynthesis provided longer polyprenyl diphosphate side chains, which were also readily desaturated and incorporated in the UQ biosynthetic pathway, thus generating additional structural diversity (Fig. 2). We also show that these desaturated UQ structures have improved antioxidant activities by examining the antioxidant properties of the modified structures in an in vitro β-carotene-linoleate assay.
MATERIALS AND METHODS
Chemicals and materials.
IPP, FPP, GGPP, [14C]IPP (55 Ci/mol), and [3H]FPP (20.5 Ci/mol) were purchased from Sigma. UQ-6, -9, and -10 were purchased from Sigma-Aldrich. Chloroform d (99.9%) for nuclear magnetic resonance (NMR) studies was purchased from Aldrich.
Cloning and culture growth.
Cloning of genes encoding carotenoid desaturases CrtI, CrtI14, and CrtN into the constitutive expression vector pUCmod has been previously described (17, 34). Cloning of genes encoding different prenyl diphosphate synthases IspA, Fgs, and CrtE into pUCmod has been previously described (16, 18, 34). The gene encoding the decaprenyl diphosphate synthase (Dds) from Rhodobacter sphaeroides (DSM 158) was amplified from genomic DNA with sequence-specific primers introducing an XbaI site followed by an optimized Shine-Dalgarno sequence (34) at its 5′ end and a EcoRI site at its 3′ end. The digested PCR product was cloned into the corresponding sites of pUCmod to facilitate constitutive expression from a modified lac promoter (34). The gene encoding OPP synthase IspB from E. coli was amplified from genomic DNA with sequence-specific primers and cloned in the same way as Dds into pUCmod. For constitutive coexpression of carotenoid desaturase CrtN with different prenyl diphosphate synthases, crtN along with its lac promoter was subcloned into the compatible plasmid pACmod essentially as described previously (34).
For UQ production, recombinant E. coli JM109 was cultivated for 24 to 36 h in the dark at 30οC in Terrific broth (TB) medium supplemented with the appropriate selective antibiotics chloramphenicol (50 μg/ml) (pACmod) and/or carbenicillin (100 μg/ml) (pUCmod).
Isolation of quinone compounds.
Wet cells from a 250-ml (∼50 mg) or 4-liter (∼10 g) culture were washed with distilled water and repeatedly extracted at 4°C with a total volume of 30 ml or 400 ml acetone. After centrifugation (4°C and 6,000 rpm), the organic phase was pooled, centrifuged again, filtered (nylon membrane, 0.2 μm; Whatman) to remove fine particles, evaporated under N2 gas or by rotary evaporator until dry, and finally resuspended with 5 to 50 ml acetone. The acetone extract was kept at −80°C for 1 day to form a white precipitate and then was filtered through a 0.2-μm nylon membrane to remove the precipitate. The resulting extracts were reextracted with an equal volume of ethyl acetate or hexane after the addition of a 1/2 volume of saltwater (15% NaCl). The organic phase, which contained quinone compounds, was collected and washed again with distilled water. The collected organic phase was completely evaporated in a vacuum until dry at room temperature, resuspended with 0.5 to 5 ml hexane, applied to silica gel chromatography (2.5 by 100 cm), and eluted stepwise with increasing amounts of acetone in hexane (0% acetone to 10% acetone). The fractions were then dried under nitrogen gas or in a vacuum and dissolved in 1 to 2 ml hexane. For further purification of UQ compounds, preparative thin-layer chromatography (TLC) and preparative high-performance liquid chromatography (HPLC) were performed as described previously (17).
HPLC, LC-MS, and NMR spectroscopy.
For the analysis of quinone compounds, 10 to 20 μl of the crude extract and the collected fractions was applied to a Zorbax SB-C18 column (4.6 by 250 mm, 5 μm; Agilent Technologies, Palo Alto, CA), and typically eluted under isocratic conditions with solvent system A containing ethanol/H2O (95:5) or solvent system B containing acetonitrile/methanol/N-propanol (80:15:5) at a flow rate of 1 ml/min using an Agilent 1100 HPLC system equipped with a photodiode array detector. Gradient conditions with solvent A (an acetonitrile-to-H2O ratio of 85:15) and solvent B (100% methanol) were also used for the elution of polar UQs (0 to 5 min for solvent A; 5 to 30 min of 100% solvent A to 100% solvent B; and 30 to 50 min for solvent B). For structural elucidation, quinones were identified by a combination of HPLC retention times, UV/visible light (UV/Vis) absorption spectra and LC-mass spectroscopy (LC-MS). Authentic UQ-6, -9, and -10 were used for comparison. Mass fragmentation spectra were monitored at an m/z of 100 or 200 to 1,000 on an LCQ MS equipped with a positive or negative atmospheric pressure chemical ionization or electron spray ionization interface (Thermo Finnigan). Parent molecular ions were further fragmented by LC-tandem MS (MS-MS) using an atmospheric pressure chemical ionization (positive) interface at different collision-induced dissociation energy levels (15 to 30%). For NMR studies, 720 μg of compound 3 was purified by a series including silica gel chromatography, preparative TLC, and preparative HPLC as described above and in reference 17. Commercially available UQ-10 was used as a control. NMR spectra were acquired on a 600 MHz Varian NMR spectrometer, and chemical shifts were reported relative to nondeuterated solvent signals. All samples were dissolved in deuterated chloroform for analysis.
Quantification of UQ compounds.
For quantification, UQs and desaturated side chain UQs were extracted and purified as described above. Compound peaks of HPLC chromatograms recorded at 275 nm (for UQs), 290 nm (for ubiquinols), and 330 nm (for desaturated side chain UQs) were integrated and compared to known concentrations of commercial (UQ-10) or isolated standard compounds. Reduced ubiquinol-10 standard was prepared by reduction of authentic UQ-10 with 0.25% (wt/vol) NaBH4 (26). Since authentic UQ-8 and ubiquinol-8 are not commercially available, both compounds were purified from E. coli and dissolved in ethanol for spectroscopic quantification (E275 equals 15,000 M−1 cm−1 for UQ-8 in ethanol, E290 equals 4,000 M−1 cm−1 for ubiquinol-8 in ethanol) (21) and used as standard compounds. Similarly, modified UQ compounds with five CDBs were purified from E. coli cells and dissolved in hexane for spectroscopic quantification using the extinction coefficient of phytofluene, which has five CDBs and a comparable UV/Vis spectrum (E348 equals 85,500 M−1 cm−1 in hexane) (1).
Construction and affinity purification of His6-tagged CrtN and IspB.
Genes encoding CrtN and IspB were amplified with a forward primer containing at its 5′ end a XbaI site and a reverse primer containing at its 5′ end an EcoRI site, followed by (GTG)6, flanking the gene. The PCR products were then purified, digested with the restriction enzymes (XbaI and EcoRI), and subcloned into the corresponding sites of pUCmod to construct pUC-CrtN_hisc and pUC-IspB_hisc, which constitutively express C-terminal His6-tagged CrtN and IspB, respectively. The C-terminal His6-tagged CrtN and IspB were expressed in E. coli strain JM109 and purified with a packed BD Talon affinity resin (Clontech, CA) as described previously (16).
In vitro assays.
In vitro desaturation of UQs was examined using purified CrtN under conditions essentially described in references 31 and 32. Briefly, the reaction (500 μl) contained 0.1 M sodium phosphate buffer (pH 7.0), 0.1% (wt/vol) bile salt, 1 mM flavin adenine dinucleotide, 2.5 μg substrate (UQ-6, -8, -9, -10), and 10 μg purified CrtN. As a control reaction, diapophytoene, a natural substrate for CrtN, was substituted for UQ. Diapophytoene was purified and quantified (E286 equals 49,800 M−1 cm−1 in hexane) (1) as described above. The reactions were incubated for 16 h at 30°C with gentle agitation and extracted three times with 0.5 ml hexane. Products were separated and monitored by HPLC as described above.
Preparation of lyophilized cells and in vivo labeling with [14C]IPP.
Lyophilized recombinant E. coli cells overexpressing IspB or CrtN were prepared as described previously with slight modifications (11). Briefly, the cells were grown in TB medium at 30°C (an optical density at 600 nm of ∼2.0), harvested, and washed once with 0.1 M potassium phosphate buffer (pH 7.5). The wet cells (∼500 mg) were suspended in 1 ml of 3% sodium glutamate solution, frozen in liquid nitrogen, and lyophilized in a VirTis freeze dryer shelf unit (Virtis Company Inc., NY).
The lyophilized cells (∼50 mg dry cell weight) were rehydrated by adding 0.5 ml of 50 mM potassium phosphate buffer (pH 7.5) containing nonlabeled FPP (5 nmol) and [14C]IPP (2.2 × 106 dpm). The cells were incubated for 20 min at 30οC, harvested, washed, resuspended in TB medium, and grown at 30°C for a further 2 h.
Radio-HPLC.
In vitro- and in vivo-labeled compounds were applied to a C18 column (4.6 by 150 mm, 5 μm; Research Products International Corp.) and eluted under gradient conditions with 100% acetonitrile and 25 mM NH4HCO3 at a flow rate of 1 ml min−1 using a Waters 600 multisolvent delivery system equipped with a radiochemical detector (β-RAM model 3; IN/US Systems). A linear gradient was used for the separation of phosphorylated isoprenoids (0 to 10 min, 60% 25 mM NH4HCO3; 10 to 50 min, 40 to 100% acetonitrile; and 50 to 60 min, 100% acetonitrile), and the eluate was automatically mixed with a liquid scintillation solution (1:2 [vol/vol]) and monitored in the radiodetector. Labeled OPP was used as an authentic standard. Because OPP is not commercially available, purified IspB was utilized for generating [3H]OPP from [3H]FPP and IPP using the conditions previously described for IspB (14).
Analysis of antioxidant activity.
Antioxidant activity of desaturated UQs was investigated using the β-carotene-linoleate model system as described before (3). A 1-ml aliquot of β-carotene (0.3 mg ml−1) was added to a flask containing linoleic acid (4 mg) and Tween 40 (40 mg). After the addition of 50 ml distilled water (saturated with air for 45 min), the resulting mixture was split into five portions supplemented with 18.7 μM of isolated UQ compounds (except for the control) to be analyzed. Bleaching of β-carotene in the presence of UQ compounds was then followed at 470 nm over 2 h in 12-s intervals at 42°C with a microplate reader (SpectraMax 384 Plus; Molecular Devices). All experiments were performed in triplicate.
RESULTS
In vivo activity of carotenoid desaturase enzymes on UQs.
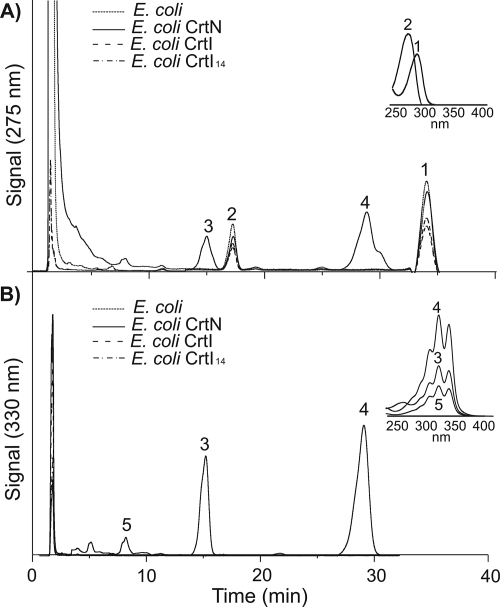
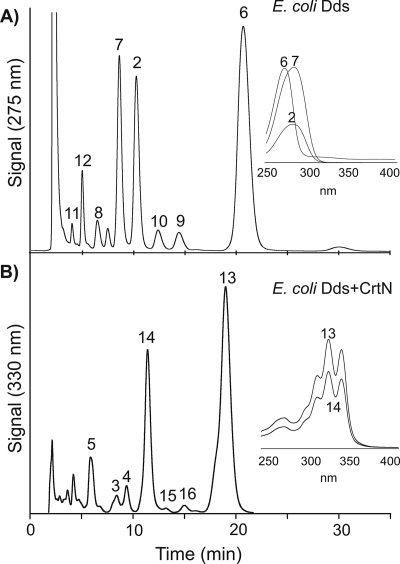
We examined the activity of three different carotenoid desaturases (CrtN, CrtI, and CrtI14) on the native UQ-8 biosynthetic pathway in E. coli. The enzymes CrtN (C30 carotenoid desaturase) (17), CrtI (C40 carotenoid desaturase) (34), and CrtI14 (in vitro-evolved C40 carotenoid desaturase) (34) were selected to provide catalytic diversity (Fig. 1). E. coli strain JM109 harboring the plasmids pUCMod (control), pUC-CrtN, pUC-CrtI, or pUC-CrtI14 was cultured aerobically, and organic extracts of cell pellets were examined for the presence of altered UQ products. HPLC analysis (Fig. 3A) detected native E. coli UQ-8 in oxidized (UQ-8, structure 1) (Fig. 2) and reduced (ubiquinol-8, structure 2) (Fig. 2) forms in all four E. coli transformants. However, acetone extracts of E. coli pUC-CrtN demonstrated three additional peaks designated compounds 3, 4, and 5, shown in Fig. 2 and 3A and B (structures 3 to 5; see subsequent analysis below). Compounds 3, 4, and 5 have different retention times but identical UV/Vis spectra with a main absorption band absorption maximum (λmax) at 341 nm and two additional distinct peaks at 327 and 359 nm (Table 1). In contrast, native UQ-8 compounds exhibit only one absorption band at shorter wavelengths (λmax at 274 or 290 nm). These results suggest that compounds 3, 4, and 5 are new metabolites with the λmax shifted to longer wavelengths (Fig. 3B), resulting from the interaction between CrtN and the UQ pathway in E. coli. The total amount of compounds 3, 4, and 5 was 12.6 μg/g dry cell weight compared to the 43 μg/g dry cell weight of UQ-8/ubiquinol-8.
FIG. 3.
Analysis of UQ synthesis in E. coli cells overexpressing carotenoid desaturases CrtN, CrtI, and CrtI14. HPLC profiles of extracts from E. coli cells transformed with the control plasmid pUCMod and the carotenoid desaturases CrtN, CrtI, or CrtI14 on plasmid pUC-CrtN, pUC-CrtI, or pUC-CrtI14. Signals were recorded at 275 nm (A) and 330 nm (B) simultaneously for each of the four extracts to monitor all UQ compounds at 275 nm and show peaks corresponding to desaturated UQ compounds at 330 nm. Peaks are labeled according to the compound numbers shown in Fig. 2. Insets show recorded UV/Vis spectra for individual compound peaks.
TABLE 1.
Properties of structures identified in this study
| Structure | Compound description | Mass (Da) | λmax (nm) | CDB(s) |
|---|---|---|---|---|
| 1 | UQ-8 | 726.4 | 274 | 0 |
| 2 | UQ-8 | 728.4 | 290 | 0 |
| 3 | UQ-8 derivative | 724.4 | 327, 341, 359 | 5 |
| 4 | UQ-8 derivative | 722.4 | 327, 341, 359 | 5 |
| 5 | 2-Octaprenylphenol derivative | 634.5 | 327, 341, 359 | 5 |
| 6 | UQ-10 | 862.5 | 274 | 0 |
| 7 | Decaprenylphenol | 774.5 | 290 | 0 |
| 8 | UQ-7 | 659.4 | 274 | 0 |
| 9 | UQ-9 | 794.6 | 274 | 0 |
| 10 | UQ-10 | 864.5 | 290 | 0 |
| 11 | Octaprenyl-4-hydroxybenzoic acid | 682.5 | 284 | 0 |
| 12 | Decaprenyl-4-hydroxybenzoic acid | 818.5 | 284 | 0 |
| 13 | UQ-10 derivative | 858.5 | 327, 341, 359 | 5 |
| 14 | UQ-10 derivative | 860.5 | 327, 341, 359 | 5 |
| 15 | UQ-9 derivative | 790.5 | 327, 341, 359 | 5 |
| 16 | UQ-11 derivative | 926.5 | 327, 341, 359 | 5 |
| 17 | 2-Pentaprenylphenol derivative | 424.2 | 435, 457, 485 | 10 |
| 18 | 2-Pentaprenylphenol derivative | 426.3 | 408, 429, 456 | 9 |
| 19 | 2-Pentaprenylphenol derivative | 428.3 | 389, 413, 437 | 8 |
Structural characterization of UQ-8-derived products with desaturated prenyl side chains.
The distinctive fine structure observed in the UV/Vis spectra of compounds 3, 4, and 5 suggests the presence of a system of CDBs similar to the chromophores of acyclic carotenoids (1). Comparison of the compounds' λmax (327, 341, and 367 nm) with known UV/Vis spectra from acyclic carotenoids indicates the presence of five CDBs as found in the linear carotenoid phytofluene (λmax at 331, 348, and 367 nm) (1).
The LC-MS and MS-MS of compounds 3 and 4 showed that the parent ions [M + H+] were at an m/z of 725.4 (M = 724.4) for compound 3 and an m/z of 723.4 (M = 722.4) for compound 4, respectively (Table 1; see Fig. S1 in the supplemental material). These molecular ions indicate that they are ubiquinol-8 (reduced form; M = 728.4) and UQ-8 (oxidized form; M = 726.4) derivatives devoid of four hydrogen atoms (i.e., an addition of two double bonds) (20). This is consistent with the five predicted CDBs for these compounds based on their UV/Vis spectra.
Compound 3 was further characterized by NMR spectroscopy to confirm the presence of the new CDB system. A combination of preparative TLC, silica gel column chromatography, and preparative HPLC afforded 720 μg of purified compound 3 from E. coli cell extracts for NMR studies. The 1H NMR spectrum showed that compound 3 was present in a 2:1 ratio with compound 4 as determined by integration of several distinct, isolated proton peaks (CH2-7, OCH3-2′, and OCH3-3′) (see Fig. S2 in the supplemental material). Compound 3 seemed therefore to be spontaneously oxidized into compound 4 during acquisition. Proton peaks clearly indicating the new CDB regions were present in the downfield region of the spectrum (δ 5.8 to 6.8) compared to the spectrum of the nonconjugated control compound UQ-8 (structure 1) in which most of the isolated vinyl proton signals are in a single overlapping multiplet (δ 5.0 to 5.2) (see Fig. S2 in the supplemental material). This new CDB region was highly complex due to both the mixture of the two forms of the compound as well as an apparent degradation of the double bond region over time. 2D COSY, gTOCSY, HMQC, and gHMBC NMR experiments were used to assign the proton and carbon peaks corresponding to each compound. Although there was a significant overlap, particularly in the prenyl side chain signals, which prevented complete assignment of all protons and carbons, all of the quinone and quinol ring signals as well as the CDB region of structure 3 were determined, providing support for the structural assignments.
By comparing the fragmentation patterns of compounds 3 and 4 from LC-MS, MS-MS (1, 8), and NMR spectroscopic analyses, we propose that the five CDBs extend from the penultimate isoprenoid unit of the octaprenyl tail of compounds 1 and 2 (structures 1 and 2) as shown in Fig. 2. Although compound 5 was present in much smaller yields than compounds 3 and 4, a parent mass [M + H+] of 635.5 was obtained by LC-MS. This is 4 Da less than the UQ-8 intermediate 2-octaprenylphenol (M = 638.5) (6), suggesting that compound 5 (structure 5; M = 634.5) is a 2-octaprenylphenol derivative with two additional double bonds (Fig. 2).
Consequently, based on the UV/Vis spectra, molecular masses, mass fragmentation patterns, and NMR spectra, we propose that compounds 3, 4, and 5 are UQ-8 derivatives with desaturated prenyl side chains (Table 1; Fig. 2).
Analysis of prenyl side chain desaturation.
Synthesis of these desaturated UQ-8 compounds may either be the result of UbiA accepting a desaturated prenyl group of OPP synthesized by CrtN or a direct desaturation of UQ-8/ubiquinol-8 by CrtN. Considering that hydrophobic UQ compounds are sequestered into the center of lipid bilayer membranes (12) upon synthesis by the integral membrane protein UbiA (23) (Fig. 1), it appears unlikely that the membrane-associated but soluble desaturase CrtN (31) can access UQ-8. UbiA, on the other hand, is known to have relaxed substrate specificity for its isoprenoid diphosphate substrate (22), suggesting that UbiA may accept a desaturated octaprenyl group synthesized by CrtN.
To examine whether CrtN can desaturate UQ compounds in vitro, the purified His6-tagged CrtN was incubated with commercially available UQ-6, -9, -10, commercially unavailable UQ-8 isolated from E. coli, and diapophytoene (CrtN′s native substrate) isolated from recombinant E. coli overexpressing diapophytoene synthase CrtM from S. aureus (Fig. 1). HPLC analysis of the in vitro reaction extracts did not detect any new modified forms of UQs. However, diapophytoene was sequentially desaturated into diaponeurosporene (Fig. 1; see Fig. S3 in the supplemental material), suggesting that the purified CrtN was active but did not desaturate the prenyl side chain of UQs in vitro.
Similarly, in vitro assays for desaturation of the OPP by CrtN were carried out. Because OPP is not commercially available, purified His6-tagged IspB was utilized to obtain [3H]OPP from [3H]FPP and IPP (see Fig. S4 in the supplemental material). When [3H]OPP was added to the assay, no desaturation of OPP was detected by radio-HPLC, indicating that there was no CrtN activity on this substrate in vitro. In addition, coincubation of purified His6-tagged IspB and His6-tagged CrtN did not desaturate the prenyl side chain of [3H]OPP in the reaction mixtures (data not shown). These results suggest that a more sophisticated reaction environment, such as that found inside the cell where the enzyme is thought to oligomerize at the membrane, is probably required for efficient desaturation of prenyl diphosphates by CrtN. Even with its native substrate diapophytoene, in vitro product formation by CrtN was low and CrtN′s activity under in vitro conditions with prenyl substrates was likely too low to be detected. Carotenoid desaturases, including CrtN, have been described to have poor activity under in vitro conditions, requiring extended assay times (12 to 16 h) for measurable product formation (32).
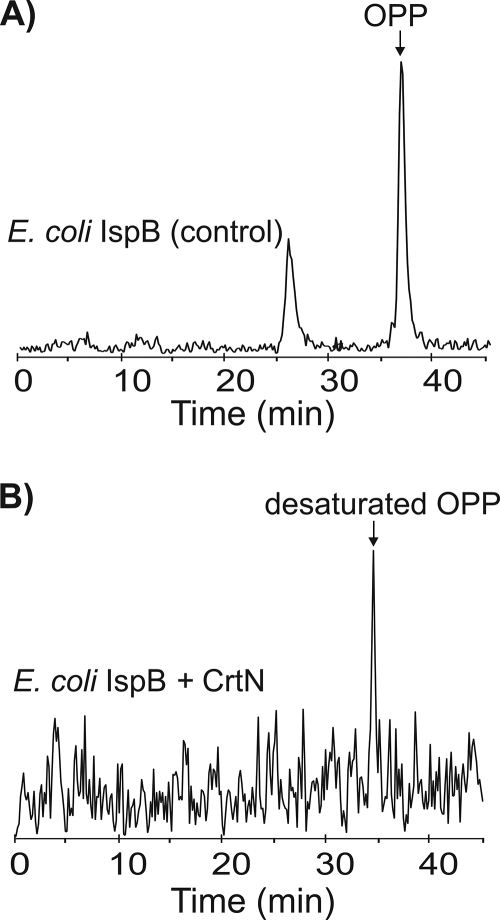
We next attempted to isolate isoprenoid diphosphate compounds from E. coli cells overexpressing IspB alone (control) or together with CrtN to detect desaturated OPP compounds in cells overexpressing CrtN. However, the free isoprenoid diphosphate pool in E. coli was too small to allow isolation of the compound at quantities that are detectable by standard HPLC. We therefore carried out in vivo metabolic tracing experiments with [14C]IPP and nonlabeled FPP to produce labeled [14C]OPP in E. coli that can be detected by radio-HPLC. Although exogenous IPP and FPP cannot normally enter E. coli cells due to the negative charge of the diphosphate group, IPP, geranyl diphosphate, and FPP can be transported into lyophilized wild-type E. coli cells at the rehydration stage (11). This approach was applied to our recombinant E. coli cells overexpressing IspB alone or together with CrtN. Rehydrated E. coli cells overexpressing IspB synthesized [14C]OPP from the exogenously fed [14C]IPP and FPP after a 30-min incubation (Fig. 4A). The [14C]OPP compound peak from this reaction showed the same retention time as a [14C]OPP standard prepared with purified IspB (see Fig. S3 in the supplemental material). Other labeled peaks were also detected, which may be shorter prenyl diphosphates. However, the radio-HPLC of E. coli cells overexpressing CrtN showed a single peak different from that of [14C]OPP (Fig. 4B). The new compound eluted earlier than [14C]OPP, which would be expected for an OPP derivative with additional double bonds. No additional peaks, such as [14C]OPP or shorter prenyl diphosphates, were detected. For unknown reasons, the quantities of radiolabeled desaturated [14C]OPP isolated from several E. coli cultures coexpressing IspB and CrtN were always much lower than the quantities of [14C]OPP obtained from control E. coli cultures expressing only IspB. Desaturated OPP may be converted faster into downstream metabolites or is degraded more rapidly than natural OPP. Nevertheless, the appearance of a new [14C]OPP derivative in cells coexpressing CrtN and IspB suggests that CrtN desaturates OPP and that desaturated UQ-8 compounds are the result of UbiA accepting a desaturated form of OPP.
FIG. 4.
Metabolic tracing of CrtN activity with [14C]IPP. Lyophilized E. coli cells overexpressing IspB (OPP synthase) alone (A) and together with carotenoid desaturase CrtN (B) were radiolabeled with [14C]IPP and nonlabeled FPP. Labeled cells were harvested after 30 min of incubation, extracted, and analyzed by radio-HPLC. A new peak with a shorter retention time than OPP appears in labeled cells expressing CrtN (B), suggesting synthesis of desaturated OPP by CrtN.
We obtained additional results that support the proposed reaction sequence by identifying desaturated derivatives of the naphthoquinone menaquinone-8 in a ubiCA deletion strain (E. coli strain RKP4152 [38, 39]) expressing CrtN (unpublished data). Since OPP is a shared precursor of both the UQ-8 and menaquinone pathways in E. coli (Fig. 1), desaturated derivatives of menaquinone-8 would be synthesized from desaturated OPP in E. coli the same way as in UQ.
Biosynthesis of UQ-10-derived products with desaturated prenyl side chains.
Previous reports have demonstrated that the E. coli UQ-8 pathway enzyme UbiA is able to utilize C50 decaprenyl diphosphate (DPP) as a substrate and synthesize UQ-10 (22). Therefore, we attempted to create a biosynthetic pathway in which CrtN desaturates this decaprenyl side chain, producing desaturated UQ-10 derivatives. The gene encoding C50 Dds was cloned from Rhodobacter sphaeroides (pUC-Dds) and overexpressed in E. coli to supply the UQ-10 pathway precursor C50 DPP. HPLC analysis of extracts of E. coli pUC-Dds demonstrated that C50 DPP supplied by Dds was efficiently converted by the native UQ-8 pathway enzymes into UQ-10 (structure 6; M = 862.5) along with ubiquinol-8 (structure 2) and the UQ-10 intermediate decaprenylphenol (structure 7; M = 774.5) as major products (Fig. 5A). Small amounts of UQ-7 (structure 8; M = 659.4), UQ-9 (structure 9; M = 794.6), and ubiquinol-10 (structure 10; M = 864.5) (41) were also detected along with pathway intermediates octaprenyl-4-hydroxybenzoic acid (structure 11; M = 682.5) and decaprenyl-4-hydroxybenzoic acid (structure 12; M = 818.5) (Fig. 2; Table 1).
FIG. 5.
HPLC analysis of extracts from E. coli cells overexpressing the UQ-10 enzyme C50 Dds alone (A) and together with the carotenoid desaturase CrtN (B). Signals shown were recorded at 275 nm (A) to detect unmodified UQ compounds and at 330 nm (B) to show peaks corresponding to desaturated UQ compounds that absorb at longer wavelengths. Peaks are labeled according to the compound numbers shown in Fig. 2. Insets show recorded UV/Vis spectra for major compound peaks (see Table 1 for λmax of other compounds).
This strain was then transformed with pAC-CrtN in order to direct the synthesis of UQ-10 derivatives with desaturated prenyl side chains. HPLC analysis of the organic extract of E. coli pAC-CrtN plus pUC-Dds showed that two new compounds (structures 13 and 14) were present with other minor peaks (Fig. 5B). The main two peaks corresponding to compounds 13 and 14 have UV/Vis spectra (λmax = 327, 341, and 359) identical to compounds 4, 5, and 6, indicating that compounds 13 and 14 have the same chromophore system of five CDBs in their structures. LC-MS showed that the parent ion [M + H+] of compound 13 had an m/z of 859.5 (structure 13; M = 858.5) (see Fig. S1 in the supplemental material). The mass difference between compound 13 (M = 858.5) and UQ-10 (M = 862.5) is 4 Da (two double bonds), confirming that compound 13 is a UQ-10 derivative with five CDBs. Similarly, the observed mass of compound 14 was at an m/z of 861.5 (structure 14; M = 860.5), a loss of 4 Da compared to ubiquinol-10 (M = 864.5), indicating compound 14 is a ubiquinol-10 derivative modified by the addition of two double bonds (Fig. 2; Table 1).
In addition, desaturated derivatives of UQ-8 (structure 3), UQ-9 (structure 15; M = 790.5), and ubiquinol-11 (structure 16; M = 926.5) were also identified based on HPLC data, UV/Vis spectra, and LC-MS. It is well known that prenyl diphosphate synthases often generate small quantities of shorter and longer chain products, as the mechanism for determining product size is not perfect (29). These results suggest that the inherent flexibility of E. coli's native UbiA allows the incorporation of shorter and longer prenyl diphosphate side chains and that CrtN readily desaturates these side chains precursors.
Biosynthesis of UQ-5-derived products with desaturated prenyl side chains.
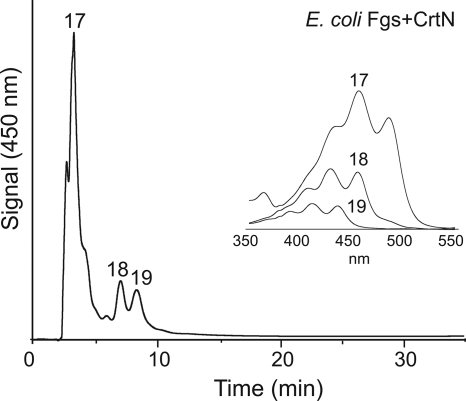
In order to further investigate the ability of UbiA to accept unnatural substrates and generate UQ derivatives with shorter, desaturated prenyl side chains, a number of recombinant short-chain prenyl diphosphate synthases were coexpressed with pUC-CrtN. It was thought that overexpression of these shorter chain prenyl diphosphate synthases might provide more suitable substrates for the C30 carotenoid desaturase CrtN and perhaps direct the synthesis of extended CDB systems. The prenyl diphosphate synthases selected were E. coli C15 FPP synthase (IspA, to increase the FPP pool in E. coli) (18), Erwinia uredovora C20 GGPP diphosphate synthase (CrtE) (34), and Aeropyrum pernix C25 farnesylgeranyl diphosphate (FGPP) synthase (Fgs) (16). Extracts of E. coli expressing CrtE plus CrtN and Fgs plus CrtN were yellow, suggesting the formation of products with an extended CDB system. Three new products (structures 17 to 19) could be detected by HPLC analysis in extracts from these two strains, and each compound peak had a carotenoid-like UV/Vis spectrum (HPLC analyses of compounds from E. coli Fgs plus CrtN are shown in Fig. 6). Both strains produced similar ratios of the products. Analysis of the colorless extract of E. coli expressing IspA and CrtN identified no new UQ products in addition to those produced by E. coli cells expressing CrtN alone.
FIG. 6.
HPLC analysis of partially purified extracts from E. coli cells coexpressing C25 FGPP synthase Fgs and carotenoid desaturase CrtN. Peaks are labeled according to compound numbers shown in Fig. 2. Inset shows recorded UV/Vis spectra for individual compound peaks.
The UV/Vis spectrum of compound 17 (λmax = 435, 457, and 485) suggested a system of 10 CDBs. The 10-CDB system of compound 17 appears to extend to the quinone ring structure based on the relatively low fine-structure index (percent III/II) of 31, characteristic for carotenoids where the conjugated acyclic isoprenoid backbone extends to a cyclic end group (1). LC-MS showed that the parent molecular ion [M + H+] of compound 17 is at an m/z of 425.3 (M = 424.3) (see Fig. S1 in the supplemental material). In comparison with fragmentation patterns of carotenoid compounds (1, 8), we propose that compound 17 is a 10-CDB-containing 2-pentaprenylphenol derivative (structure 17 is shown in Fig. 2). Compound 17 is therefore a new metabolite with a modified, desaturated prenyl side chain but an incompletely functionalized quinone ring. Similarly, the proposed structure of compound 18 (M = 426.3) is a 2-pentaprenylphenol derivative with a system of nine CDBs (λmax = 408, 429, and 456) that does not extend to the ring structure (structure 18) (Fig. 2). Finally, the proposed structure of compound 19 is a 2-pentaprenylphenol derivative with a system of eight CDBs based on the spectral data (λmax = 389, 413, and 437) and mass data (M = 428.3) (structure 19 is shown in Fig. 2).
Synthesis of compounds 17, 18, and 19 suggests that CrtN is able to catalyze the extended desaturation of shorter chain polyprenyl substrates. Unexpectedly, modified UQ-5 derivatives with complete quinone ring structures were not observed. However, the additional double bonds in the side chains of these compounds may render further pathway derivatives inaccessible to other UQ pathway enzymes.
Antioxidant activity of UQs with desaturated side chains.
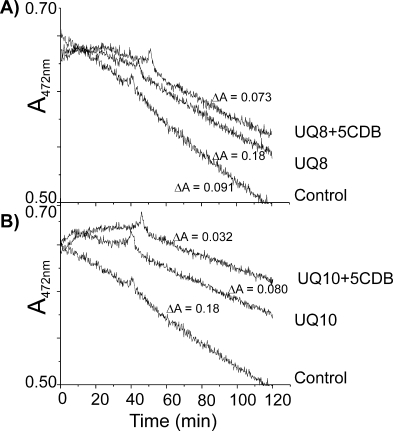
It is well established that the benzoquinone rings of UQs play an important role in radical-scavenging activity and that the CDBs of carotenoids confer antioxidant activity. We therefore reasoned that UQ derivatives with CDB-containing prenyl side chains might prove to be more effective antioxidants than unmodified UQs. To explore this possibility, antioxidant activities of isolated UQ-8 and UQ-10 as well as their corresponding five CDB-containing derivatives against peroxyl radicals were investigated with the β-carotene-linoleate assay (3) over a fixed time period (2 h at 42°C).
In the β-carotene-linoleate assay, β-carotene is subjected to peroxyl radical oxidation, causing pigment bleaching. Compounds with antioxidant activities reduce β-carotene oxidation and thus slow down chromophore destruction. Isolated UQ-8 and UQ-10 with unmodified and desaturated (five CDBs) prenyl side chains were individually tested in this model system at a concentration of 18.7 μM (Fig. 7A and B). Decreasing absorbance at 470 nm was monitored over a time period of 2 h. All tested UQ compounds reduced β,β-carotene oxidation, but the presence of a five-CDB system increased the protective activities of both UQ-8 and UQ-10. Carotenoid bleaching was lowest in the presence of the two desaturated UQ-10 (ΔA470 = 0.032) or UQ-8 (ΔA470 = 0.072) derivatives, followed by unmodified UQ-10 (ΔA470 = 0.080), UQ-8 (ΔA470 = 0.091), and the control (ΔA470 = 0.18). These results show that the introduction of CDBs into the isoprenoid side chain of UQs increases their antioxidant activities in vitro.
FIG. 7.
Antioxidant activities of four isolated UQs were measured by monitoring β-carotene bleaching at 470 nm over 2 h at 42°C compared to a control without UQ. (A) UQ-8 and UQ-8 with five CDBs. (B) UQ-10 and UQ-10 with five CDBs.
DISCUSSION
There are numerous evolutionary mechanisms by which chemical diversity is generated in nature in order to provide a selective advantage (10). Many of these mechanisms, like the evolution of new protein functions and combination of genes from related pathways into new biosynthetic reaction sequences, have been adapted as laboratory approaches to direct the biosynthesis of novel metabolites (17, 24, 25, 30, 33, 34, 48). There are, however, few examples of separate biosynthetic pathways being combined in engineered cells to generate new classes of natural products. The potential of this approach is dictated by the availability and compartmentalization of substrates as well as the capacity for metabolic pathway enzymes to accept unnatural substrates (28). By combining pathway steps from different branches of biosynthetic networks, classes of compounds not observed in nature can be synthesized and structural motifs that are functionally important can be combined or enhanced.
Here we show that the C30 carotenoid desaturase CrtN from Staphylococcus aureus (32) acts on prenyl substrates to produce UQs with conjugated prenyl chains when heterologously expressed in E. coli. Overexpression of CrtN in E. coli cells led to synthesis of desaturated UQ-8 compounds with octaprenyl side chains containing CDBs. This activity was not observed with C40 carotenoid desaturases CrtI (C40 carotenoid desaturase) (34) and CrtI14 (in vitro-evolved C40 carotenoid desaturase) (34). All carotenoid desaturases, including CrtN, are thought to recognize the essential three CDBs at the center of C40 phytoene or C30 diapophytoene (Fig. 1) precursors and then initiate and continue desaturation reactions in either direction (19). However, we observed CrtN activity on prenyl substrates without this structural motif, indicating that the mechanism of CrtN′s substrate recognition is more complex.
Coexpression of CrtN in recombinant E. coli with heterologous prenyl diphosphate showed that CrtN can also act in a similar manner on prenyl side chains that are longer (C50) or shorter (C25) than the octaprenyl chains produced by wild-type E. coli cells. Desaturated prenyl side chains of various lengths were readily accepted by UbiA for the synthesis of novel UQs (and also menaquinones [data not shown]), confirming previous results that the E. coli UQ pathway, in particular the prenyltransferase UbiA, has considerable metabolic flexibility (22). Other enzymes of the UbiA prenyltransferase family catalyze prenyl group transfer in the biosynthesis of hemes, chlorophylls, vitamin E, shikonin, and archael isoprenoid lipids (13). These prenyltransferases may also accept prenyl chains in various lengths and degrees of desaturation, thus making it possible to apply an approach similar to the one described in this study for the biosynthesis of diverse prenylated products with new biological properties in engineered cells. UQ derivatives with conjugated prenyl side chains generated in this study have increased antioxidant activities in vitro. Introduction of crtN into commercial UQ-10 microbial producer strains or metabolically engineered strains (reviewed in reference 4) could yield UQ-10 compounds with superior antioxidant properties for the treatment of various medical conditions, provided the additional double bonds do not negatively impact other biological functions and/or activities of the UQs.
In conclusion, by combining biosynthetic steps from two pathways synthesizing separate classes of metabolites, we generated recombinant strains of E. coli that produce structurally new antioxidants. This “crossing over” of downstream biosynthetic steps from related but divergent metabolic pathways is thought to be an important evolutionary mechanism by which natural chemical diversity is created (5). Applying this in an engineered system exploits the plasticity of biosynthetic pathway steps. Although this is not a characteristic inherent in all enzymes, the enzymatic diversity accessible as a result of genomics and the ability to alter enzymes by rational engineering or directed evolution will assist in overcoming this obstacle.
Supplementary Material
Acknowledgments
We gratefully acknowledge support from the Defense Advanced Research Projects Agency (DARPA-BIOS N66001-02-1-8928) and the David and Lucile Packard Foundation (grant no. 2001-18996).
We thank Jerry D. Cohen and Lana Barkawi for assistance with the radiochemical experiments and Beverly Ostrowski for assistance with NMR.
Footnotes
Published ahead of print on 26 September 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Britton, G., S. Liaaen-Jensen, and H. Pfander. 1995. Carotenoids: spectroscopy, vol. 1B. Birkhauser, Basel, Switzerland.
- 2.Brown, K. R., B. M. Allan, P. Do, and E. L. Hegg. 2002. Identification of novel hemes generated by heme A synthase: evidence for two successive monooxygenase reactions. Biochemistry 41:10906-10913. [DOI] [PubMed] [Google Scholar]
- 3.Burda, S., and W. Oleszek. 2001. Antioxidant and antiradical activities of flavonoids. J. Agric. Food Chem. 49:2774-2779. [DOI] [PubMed] [Google Scholar]
- 4.Cluis, C. P., A. M. Burja, and V. J. Martin. 2007. Current prospects for the production of coenzyme Q10 in microbes. Trends Biotechnol. 25:514-521. [DOI] [PubMed] [Google Scholar]
- 5.Copley, S. D. 2000. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: the patchwork approach. Trends Biochem. Sci. 25:261-265. [DOI] [PubMed] [Google Scholar]
- 6.Cox, G. B., I. G. Young, L. M. McCann, and F. Gibson. 1969. Biosynthesis of ubiquinone in Escherichia coli K-12: location of genes affecting the metabolism of 3-octaprenyl-4-hydroxybenzoic acid and 2-octaprenylphenol. J. Bacteriol. 99:450-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crane, F. L. 2001. Biochemical functions of coenzyme Q10. J. Am. Coll. Nutr. 20:591-598. [DOI] [PubMed] [Google Scholar]
- 8.Enzell, C. R., G. W. Francis, and S. Liaaen-Jensen. 1969. Mass spectrometric studies of carotenoids. 2. A survey of fragmentation reactions. Acta Chem. Scand. 23:727-750. [DOI] [PubMed] [Google Scholar]
- 9.Facchini, P. J. 2001. Alkaloid biosynthesis in plants: biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52:29-66. [DOI] [PubMed] [Google Scholar]
- 10.Firn, R. D., and C. G. Jones. 2003. Natural products—a simple model to explain chemical diversity. Nat. Prod. Rep. 20:382-391. [DOI] [PubMed] [Google Scholar]
- 11.Fujisaki, S., T. Nishino, and H. Katsuki. 1986. Biosynthesis of isoprenoids in intact cells of Escherichia coli. J. Biochem. (Tokyo) 99:1137-1146. [DOI] [PubMed] [Google Scholar]
- 12.Hauss, T., S. Dante, T. H. Haines, and N. A. Dencher. 2005. Localization of coenzyme Q10 in the center of a deuterated lipid membrane by neutron diffraction. Biochim. Biophys. Acta 1710:57-62. [DOI] [PubMed] [Google Scholar]
- 13.Hemmi, H., Y. Takahashi, K. Shibuya, T. Nakayama, and T. Nishino. 2005. Menaquinone-specific prenyl reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus. J. Bacteriol. 187:1937-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kainou, T., K. Okada, K. Suzuki, T. Nakagawa, H. Matsuda, and M. Kawamukai. 2001. Dimer formation of octaprenyl-diphosphate synthase (IspB) is essential for chain length determination of ubiquinone. J. Biol. Chem. 276:7876-7883. [DOI] [PubMed] [Google Scholar]
- 15.Kuzuyama, T., J. P. Noel, and S. B. Richard. 2005. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature 435:983-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, P. C., B. N. Mijts, R. Petri, K. T. Watts, and C. Schmidt-Dannert. 2004. Alteration of product specificity of Aeropyrum pernix farnesylgeranyl diphosphate synthase (Fgs) by directed evolution. Prot. Eng. Des. Sel. 17:771-777. [DOI] [PubMed] [Google Scholar]
- 17.Lee, P. C., A. Z. Momen, B. N. Mijts, and C. Schmidt-Dannert. 2003. Biosynthesis of structurally novel carotenoids in Escherichia coli. Chem. Biol. 10:453-462. [DOI] [PubMed] [Google Scholar]
- 18.Lee, P. C., R. Petri, B. N. Mijts, K. T. Watts, and C. Schmidt-Dannert. 2005. Directed evolution of Escherichia coli farnesyl diphosphate synthase (IspA) reveals novel structural determinants of chain length specificity. Metab. Eng. 7:18-26. [DOI] [PubMed] [Google Scholar]
- 19.Lee, P. C., and C. Schmidt-Dannert. 2002. Metabolic engineering towards biotechnological production of carotenoids in microorganisms. Appl. Microbiol. Biotechnol. 60:1-11. [DOI] [PubMed] [Google Scholar]
- 20.Lee, P. T., A. Y. Hsu, H. T. Ha, and C. F. Clarke. 1997. A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J. Bacteriol. 179:1748-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenaz, G. 1985. Coenzyme Q: biochemistry, bioenergetics and clinical applications of ubiquinone. John Wiley & Sons, New York, NY.
- 22.Meganathan, R. 2001. Ubiquinone biosynthesis in microorganisms. FEMS Microbiol. Lett. 203:131-139. [DOI] [PubMed] [Google Scholar]
- 23.Melzer, M., and L. Heide. 1994. Characterization of polyprenyldiphosphate: 4-hydroxybenzoate polyprenyltransferase from Escherichia coli. Biochim. Biophys. Acta 1212:93-102. [DOI] [PubMed] [Google Scholar]
- 24.Mijts, B. N., P. C. Lee, and C. Schmidt-Dannert. 2005. Identification of a carotenoid oxygenase synthesizing acyclic xanthophylls: combinatorial biosynthesis and directed evolution. Chem. Biol. 12:453-460. [DOI] [PubMed] [Google Scholar]
- 25.Mijts, B. N., and C. Schmidt-Dannert. 2003. Engineering of secondary metabolite pathways. Curr. Opin. Biotechnol. 14:597-602. [DOI] [PubMed] [Google Scholar]
- 26.Motchnik, P. A., B. Frei, and B. N. Ames. 1994. Measurement of antioxidants in human blood plasma. Methods Enzymol. 234:269-279. [DOI] [PubMed] [Google Scholar]
- 27.Mukai, K., A. Tokunaga, S. Itoh, Y. Kanesaki, K. Ohara, S. Nagaoka, and K. Abe. 2007. Structure-activity relationship of the free-radical-scavenging reaction by vitamin E (alpha-, beta-, gamma-, delta-tocopherols) and ubiquinol-10: pH dependence of the reaction rates. J. Phys. Chem. B 111:652-662. [DOI] [PubMed] [Google Scholar]
- 28.Müller, M. 2004. Chemical diversity through biotransformations. Curr. Opin. Biotechnol. 15:591-598. [DOI] [PubMed] [Google Scholar]
- 29.Okada, K., Y. Kamiya, X. Zhu, K. Suzuki, K. Tanaka, T. Nakagawa, H. Matsuda, and M. Kawamukai. 1997. Cloning of the sdsA gene encoding solanesyl diphosphate synthase from Rhodobacter capsulatus and its functional expression in Escherichia coli and Saccharomyces cerevisiae. J. Bacteriol. 179:5992-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petri, R., and C. Schmidt-Dannert. 2004. Dealing with complexity: evolutionary engineering and genome shuffling. Curr. Opin. Biotechnol. 15:298-304. [DOI] [PubMed] [Google Scholar]
- 31.Raisig, A., and G. Sandmann. 1999. 4,4′-diapophytoene desaturase: catalytic properties of an enzyme from the C-30 carotenoid pathway of Staphylococcus aureus. J. Bacteriol. 181:6184-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raisig, A., and G. Sandmann. 2001. Functional properties of diapophytoene and related desaturases of C-30 and C-40 carotenoid biosynthetic pathways. Biochim. Biophys. Acta 1533:164-170. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt-Dannert, C. 2001. Directed evolution of single proteins, metabolic pathways and viruses. Biochemistry 40:13125-13136. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt-Dannert, C., D. Umeno, and F. H. Arnold. 2000. Molecular breeding of carotenoid biosynthetic pathways. Nat. Biotechnol. 18:750-753. [DOI] [PubMed] [Google Scholar]
- 35.Schneider, C., P. Boger, and G. Sandmann. 1997. Phytoene desaturase: heterologous expression in an active state, purification, and biochemical properties. Prot. Expr. Purif. 10:175-179. [DOI] [PubMed] [Google Scholar]
- 36.Seo, M. J., E. M. Im, J. H. Hur, J. Y. Nam, C. G. Hyun, Y. R. Pyun, and S. O. Kim. 2006. Production of coenzyme Q10 by recombinant E. coli harboring the decaprenyl diphosphate synthase gene from Sinorhizobium meliloti. J. Microbiol. Biotechnol. 16:933-938. [Google Scholar]
- 37.Soballe, B., and R. K. Poole. 1999. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology 145:1817-1830. [DOI] [PubMed] [Google Scholar]
- 38.Soballe, B., and R. K. Poole. 1998. Requirement for ubiquinone downstream of cytochrome(s) b in the oxygen-terminated respiratory chains of Escherichia coli K-12 revealed using a null mutant allele of ubiCA. Microbiology 144:361-373. [DOI] [PubMed] [Google Scholar]
- 39.Soballe, B., and R. K. Poole. 2000. Ubiquinone limits oxidative stress in Escherichia coli. Microbiology 146:787-796. [DOI] [PubMed] [Google Scholar]
- 40.Tran, U. C., and C. F. Clarke. 2007. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 7(Suppl.):S62-S71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trumpower, B. L., A. S. Aiyar, C. E. Opliger, and R. E. Olson. 1972. Studies on ubiquinone. The isolation and identification of 5-demethoxyubiquinone-9 as an intermediate in biosynthesis of ubiquinone-9 in the rat. J. Biol. Chem. 247:2499-2511. [PubMed] [Google Scholar]
- 42.Turunen, M., J. Olsson, and G. Dallner. 2004. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta 1660:171-199. [DOI] [PubMed] [Google Scholar]
- 43.Umeno, D., A. V. Tobias, and F. H. Arnold. 2005. Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol. Mol. Biol. Rev. 69:51-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umeno, D., A. V. Tobias, and F. H. Arnold. 2002. Evolution of the C30 carotenoid synthase CrtM for function in a C40 pathway. J. Bacteriol. 184:6690-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valentin, H. E., and Q. Qi. 2005. Biotechnological production and application of vitamin E: current state and prospects. Appl. Microbiol. Biotechnol. 68:436-444. [DOI] [PubMed] [Google Scholar]
- 46.Wallace, B. J., and I. G. Young. 1977. Role of quinones in electron transport to oxygen and nitrate in Escherichia coli. Studies with a ubiA-menA-double quinone mutant. Biochim. Biophys. Acta 461:84-100. [DOI] [PubMed] [Google Scholar]
- 47.Wang, C. W., and J. C. Liao. 2001. Alteration of product specificity of Rhodobacter sphaeroides phytoene desaturase by directed evolution. J. Biol. Chem. 276:41161-41164. [DOI] [PubMed] [Google Scholar]
- 48.Watts, K. T., B. N. Mijts, and C. Schmidt-Dannert. 2005. Current and emerging approaches for natural product biosynthesis in microbial cells. Adv. Synth. Catal. 347:927-940. [Google Scholar]
- 49.Willows, R. D. 2003. Biosynthesis of chlorophylls from protoporphyrin IX. Nat. Prod. Rep. 20:327-341. [DOI] [PubMed] [Google Scholar]
- 50.Wissenbach, U., D. Ternes, and G. Unden. 1992. An Escherichia coli mutant containing only demethylmenaquinone, but no menaquinone: effects on fumarate, dimethylsulfoxide, trimethylamine N-oxide and nitrate respiration. Arch. Microbiol. 158:68-73. [DOI] [PubMed] [Google Scholar]
- 51.Yazaki, K., M. Kunihisa, T. Fujisaki, and F. Sato. 2002. Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon. Cloning and characterization of a key enzyme in shikonin biosynthesis. J. Biol. Chem. 277:6240-6246. [DOI] [PubMed] [Google Scholar]
- 52.Zahiri, H. S., S. H. Yoon, J. D. Keasling, S. H. Lee, S. Won Kim, S. C. Yoon, and Y. C. Shin. 2006. Coenzyme Q10 production in recombinant Escherichia coli strains engineered with a heterologous decaprenyl diphosphate synthase gene and foreign mevalonate pathway. Metab. Eng. 8:406-416. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, D. W., B. Shrestha, Z. P. Li, and T. W. Tan. 2007. Ubiquinone-10 production using Agrobacterium tumefaciens dps gene in Escherichia coli by coexpression system. Mol. Biotechnol. 35:1-14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.