Abstract
Aspergillus fumigatus is the main cause of invasive aspergillosis in immunocompromised patients, and only a limited number of drugs for treatment are available. A screening method for new antifungal compounds is urgently required, preferably an approach suitable for in vitro and in vivo studies. Bioluminescence imaging is a powerful tool to study the temporal and spatial resolutions of the infection and the effectiveness of antifungal drugs. Here, we describe the construction of a bioluminescent A. fumigatus strain by fusing the promoter of the glyceraldehyde-3-phosphate dehydrogenase gene from A. fumigatus with the luciferase gene from Photinus pyralis to control the expression of the bioluminescent reporter. A. fumigatus transformed with this construct revealed high bioluminescence under all tested growth conditions. Furthermore, light emission correlated with the number of conidia used for inoculation and with the biomass formed after different incubation times. The bioluminescent strains were suitable to study the effectiveness of antifungals in vitro by several independent methods, including the determination of light emission with a microplate reader and the direct visualization of light emission with an IVIS 100 system. Moreover, when glucocorticoid-treated immunosuppressed mice were infected with a bioluminescent strain, light emission was detected from infected lungs, allowing the visualization of the progression of invasive aspergillosis. Therefore, this new bioluminescence tool is suitable to study the in vitro effectiveness of drugs and the disease development, localization, and burden of fungi within tissues and may also provide a powerful tool to study the effectiveness of antifungals in vivo.
Aspergillus fumigatus can cause life-threatening invasive aspergillosis in immunocompromised patients (22). Patients at risk are mainly those who have undergone organ transplantation or who suffer from illnesses like leukemia, neutropenia, human immunodeficiency virus infection, or cystic fibrosis (9, 33). Although infections caused by A. fumigatus have been known to exist for many years, the factors deriving from both sides, host and pathogen, that are involved in establishment and manifestation of the fungal infection are barely understood. It is well accepted that reduced immune defense mechanisms on the host side are generally required to allow fungal outgrowth. This is also reflected in infection models, in which high doses of conidia applied to the respiratory tracts of immunocompetent individuals are well tolerated (1, 12, 25). Alveolar macrophages, dendritic cells, and neutrophils form an insuperable barrier, which efficiently inactivates and removes conidia. In turn, treatment of mice with immunosuppressive agents or irradiation leads to a strongly increased susceptibility for invasive aspergillosis (24, 37).
Few antifungal compounds are available to treat invasive fungal infections, and they belong mainly to the groups of polyene macrolides (such as amphotericin B), azoles, and echinocandins (10). Although these compounds are effective against fungi under in vitro conditions, the severe physical condition of most patients accompanied by the side effects caused by several of these compounds substantially limits their use (29). Therefore, new antifungals are urgently needed and large-scale screenings under in vitro and in vivo conditions are required to find such drugs.
In recent years, bioluminescence imaging (BLI) has evolved as a powerful technique to study the establishment and manifestation of infection by pathogens. It has provided new insights into the onset and dissemination of infections, because real-time monitoring of spatial and temporal resolutions of infection can be achieved with a single animal and without sacrificing the animal; therefore, not only a snapshot view of the infection process but also the progression can be monitored (18). BLI may provide new insights into the infection processes and may significantly reduce the number of animals studied in each cohort.
BLI has been performed for several gram-positive and gram-negative bacteria (15, 20, 36), for different viruses (18), and for eukaryotic parasites, such as Toxoplasma, Leishmania, and Plasmodium species (7, 14, 18, 31). Furthermore, BLI has been used to study the promoter activities of genes involved in the circadian rhythm of the ascomycete Neurospora crassa by using different versions of the codon-optimized firefly luciferase gene (16, 26). Recently, BLI has also been applied to the pathogenic yeast Candida albicans to investigate virulence in vaginal mucosal and systemic infections (11). However, studies of invasive and disseminated infections were hampered by strongly reduced light emission when C. albicans switched into the filamentous growth form. This was attributed to a reduced d-luciferin uptake by hyphae compared to that by yeast cells (11). Nevertheless, the investigation implied that BLI may also be used to study the infection processes of other fungi and the efficiency of antifungal drugs in the clearance of infections.
BLI can easily be performed on gram-negative and -positive bacteria by introducing (i) the luxCDABE operon from Photorhabdus luminescens or Xenorhabdus luminescens (18) or (ii) the luxABCDE operon with optimized binding sites for gram-positive bacterial ribosomes (13). Since the bacterial system has not been adapted to eukaryotes and viruses, the luciferase coding region from the firefly Photinus pyralis or from the sea pansy Renilla reniformis is generally cloned under the control of a promoter which is active during disease development. Light emission is enforced by intraperitoneal or intravenous injection of d-luciferin or coelenterazine, depending on the source of luciferase. d-Luciferin emits light at a wavelength higher than 560 nm, enabling detection from deeper tissues than possible with coelenterazine (light emission at wavelengths of 480 and 490 nm). In addition, d-luciferin is nontoxic for mammalian cells and highly water soluble, which makes the substrate available at all body sites. The only additional prerequisite for light production is the availability of oxygen, which is involved in the light-producing luciferase reaction (18).
In this study, we established, for the first time, BLI for the pathogenic filamentous fungus A. fumigatus by use of firefly luciferase controlled by the glyceraldehyde-3-phosphate dehydrogenase gene (gpdA) promoter. The resulting strains were used to correlate light emission with fungal growth, a prerequisite for in vitro assays of antifungal drug efficiency testing. In addition, we monitored the progression of pathogenesis by using one selected bioluminescent strain in a murine infection model with the aim of visualizing the onset of infection and disease development in a time-lapse study of individual animals.
MATERIALS AND METHODS
Growth of A. fumigatus and harvesting of conidia and mycelia.
A. fumigatus was generally maintained either on malt extract (Fluka, Taufkirchen, Germany) slant cultures or on Aspergillus minimal medium (AMM) (http://www.fgsc.net/methods/anidmed.html), with the addition of pyrithiamine (0.1 μg/ml final concentration) when required for the selection of transformants. If not indicated otherwise, cultures were incubated at 37°C. For determination of GpdA, citrate synthase, and luciferase activities after growth on different carbon sources, liquid minimal media were supplemented with 100 mM glucose, 100 mM ethanol, or a combination of 100 mM glucose with 1% peptone. In addition, medium containing RPMI 1640 cell culture medium (Invitrogen, Karlsruhe, Germany) supplemented with 10% fetal calf serum (RPMI complete) was used. Liquid media used for shake flask cultures (100 ml in 250-ml flasks or 200 ml in 500-ml flasks) were generally inoculated with a final concentration of 2 × 106 conidia/ml, except for the activity determination from a 10-h culture, which was inoculated with 5 × 106 conidia/ml to yield sufficient biomass for subsequent analyses. All liquid cultures were incubated aerobically on a rotary shaker at 220 rpm.
Conidia were harvested by scraping from agar slants in the presence of phosphate-buffered saline (PBS) with 0.1% Tween 20, and suspensions were filtered through 40-μm cell strainers (BD Biosciences, Erembodegem, Belgium). Dilutions were made in PBS without the addition of Tween 20. Mycelia were harvested over Miracloth filter gauze (Merck/Calbiochem, Darmstadt, Germany), followed by a short washing with water and removal of excess liquid by pressing the mycelium between dry tissues. Subsequently, the mycelia were directly shock-frozen in liquid nitrogen and stored at −80°C.
Cloning procedures.
For amplification of all genes and promoter sequences from A. fumigatus, genomic DNA of strain CBS144.89 was used. For amplification of the luciferase gene (luc), the plasmid pSP-luc+ (Promega, Mannheim, Germany) served as the template, and the proofreading polymerase Accuzyme (Bioline, Luckenwalde, Germany) or the Phusion polymerase (Biozym GmbH, Hessisch Oldendorf, Germany) was used. PCR products were cloned directly into the pJET1/blunt cloning vector (GeneJET PCR cloning kit; Fermentas GmbH, St. Leon-Rot, Germany). If not indicated otherwise, plasmids were transferred to chemically competent Escherichia coli DH5α cells and plasmid DNA was reisolated by use of an EZNA plasmid miniprep kit II (PeqLab, Erlangen, Germany). For DNA digestions, “fast digest” restriction endonucleases were used (Fermentas GmbH). Sequences of all oligonucleotides, together with their respective restriction sites and functions, are listed in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Name | Target gene | Sequence (5′ → 3′)a | Restriction site | Function |
|---|---|---|---|---|
| PgpdAAfNot_up | gpdA | GCG GCC GCA GAT TCT AGA AGT CCT G | NotI | Amplification of the gpdA promoter |
| PgpdAAfBglATG | gpdA | AGA TCTCAT TGT GTA GAT TCG TCT GGT AC | BglII | Amplification of the gpdA promoter |
| Anchored oligo(dT)20 | mRNA | NVT TTT TTT TTT TTT TTT TTT T | Generation of cDNA | |
| BglAfGPDcDNA_up | gpdA | AGA TCT GCC ACT CCC AAG GTT GG | BglII | Amplification of gpdA cDNA |
| HindAfGPDcDNA_do | gpdA | AAG CTT ACT GGG AAT CGA CCT TGG | HindIII | Amplification of gpdA cDNA |
| LucBgl_up | luc | AGA TCT GAC GCC AAA AAC ATA AAG AAA GG | BglII | Amplification of luc |
| LucHind_down | luc | AAG CTT ACA CGG CGA TCT TTC CGC | HindIII | Amplification of luc |
| LucMi_up | luc | GAT GTC CAC CTC GAT ATG TG | Control primer | |
| Not_ptrA_up_for | ptrA | GCG GCC GCT TGA TTA CGG GAT CCC ATT GG | NotI | Amplification of ptrA |
| Not_ptrA_dow_rev | ptrA | GCG GCC GCG TAT TAT ACT GTC TTT CTT GTT ACA | NotI | Amplification of ptrA |
| cDNAgpdAAf_up | gpdA | CGT CGA TGC TCC CTC CAA C | RT-PCR with gpdA | |
| cDNAgpdAAf_do | gpdA | GGA ATC GAC CTT GGC GAT G | RT-PCR with gpdA | |
| lucProbe_up | luc | GAT GGA ACC GCT GGA GAG C | luc probe/RT-PCR with luc | |
| lucProbe_mi_rev | luc | CCA AAA CCG TGA TGG AAT GGA AC | luc probe/RT-PCR with luc | |
| AfCitAcode_up | citA | GCA AGG TCA TCG GCG AGG | RT-PCR with citA | |
| AfCitAcode_do | citA | GTT GTC ACC GTA GCC GAG C | RT-PCR with citA | |
| act_Afum_for_II | act1 | GCT TTG CTA CGT CGC TCT CG | RT-PCR with act1 | |
| act_Afum_rev_II | act1 | GGA GGA GCA ATG ATC TTG ACC | RT-PCR with act1 |
Boldface type indicates restriction sites, and italic type indicates the ATG codon in reverse complement orientation.
(i) Cloning of the gpdA promoter from A. fumigatus.
The promoter sequence of the glyceraldehyde-3-phosphate dehydrogenase gene (PgpdA) from A. fumigatus was selected for luciferase gene expression. The gpdA gene sequence was identified by a BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/) using the Aspergillus nidulans GpdA protein sequence (NCBI accession no. P20445) as a template. The putative A. fumigatus GpdA enzyme (locus tag AFUA_5G01970) displayed 83% identity to the A. nidulans enzyme, and a 970-bp fragment upstream of the ATG start codon was selected from the CADRE database (http://www.aspergillus.org.uk/indexhome.htm?secure/sequence_info/index.php∼main) and used as the promoter. The promoter was amplified with primers PgpdAAfNot_up and PgpdAAfBglATG, purified from the gel, and subcloned into the pJET1/blunt cloning vector, resulting in plasmid pJETPgpdAAf. Plasmid DNA was subjected to a NotI and BglII double restriction to release the promoter region from the cloning vector.
(ii) Cloning of the gpdA gene from A. fumigatus.
For recombinant synthesis and functional analysis of the glyceraldehyde-3-phosphate dehydrogenase gene from A. fumigatus, RNA was isolated from a peptone-grown mycelium, and total cDNA was prepared by use of anchored oligo(dT)20 primers as described previously (19). The gene was amplified using oligonucleotides BglAfGPDcDNA_up and HindAfGPDcDNA_do, cloned into the pJET1/blunt vector, and sequenced from both strands. For recombinant enzyme production, the gpdA gene was excised by BglII/HindIII restriction and subcloned into the BglII/HindIII-restricted pET43.1bHis6 vector (Novagen/Merck KGaA, Darmstadt, Germany). The resulting plasmid, pETgpdAAFHis6, was transferred to E. coli BL21(DE3) Rosetta2 cells for overproduction of GpdA.
(iii) Subcloning of the Photinus pyralis luciferase gene.
The firefly luciferase gene was amplified from plasmid pSP-luc+ by use of the primers LucBgl_up and LucHind_down. The resulting 1,656-bp PCR fragment was gel purified and cloned into pJET1/blunt. E. coli DH5α cells harboring positive clones were preselected by colony PCR with primers LucBgl_up and LucMi_up. Independent HindIII and BglII restrictions were used to check the orientation of the insert. HindIII-digested plasmids, which were devoid of a 270-bp fragment containing a vector-specific BglII restriction site, were gel purified and self-ligated. The resulting plasmid, pJETluc-Bgl, was reisolated and used either for the fusion with the gpdA promoter or for recombinant protein production in E. coli. For recombinant production, the luc gene was excised by BglII and HindIII digestion and cloned into the restricted pET43.1bHis6 vector as described for the gpdA gene. The resulting plasmid, pETlucHis6, was transferred to E. coli BL21(DE3) Rosetta2 cells.
(iv) Construction of the PgpdA::luc fusion and cloning of the pyrithiamine (ptrA) resistance cassette.
Plasmid pJETluc-Bgl was restricted with BglII and NotI and fused in frame with the ATG start codon downstream from the gpdA promoter region. This region was excised from pJETPgpdAAf as described above, and ligation resulted in plasmid PgpdA::luc/pJET. The plasmid DNA was purified and restricted with NotI for subsequent ligation of the pyrithiamine resistance gene (ptrA). The ptrA gene was amplified from pSK275 (kindly provided by S. Krappmann, Georg-August-Universität Goettingen, Germany) with the primers Not_ptrA_up_for and Not_ptrA_dow_rev. The amplicon was then ligated into the pJET1/blunt cloning vector, resulting in plasmid NotptrA/pJET. After release of the ptrA gene by NotI restriction, the resistance cassette was subcloned into the previously NotI-restricted vector PgpdA::luc/pJET. Positive clones were identified by colony PCR amplification of the ptrA gene, and plasmid DNA (PgpdA::luc/pJET/ptrA) was purified for transformation of A. fumigatus.
Transformation of A. fumigatus, initial screening, and Southern analysis.
Two independently constructed plasmids of PgpdA::luc/pJET/ptrA (for construction, see above) were used for transformation of the A. fumigatus wild-type strain CBS144.89. This A. fumigatus strain was chosen because it served as the origin of the recently sequenced strain CEA10 (8). The transformation was performed by standard protoplast transformation, as described previously (39). In brief, conidia were inoculated in 200 ml of AMM with 50 mM glucose as the carbon source and incubated at 220 rpm on a rotary shaker at 37°C for 18 h. After being washed, the mycelium was incubated in Glucanex (Gloesmann-Rohstoffe, Steinakirchen, Austria)-containing buffer (400 mg in 20 ml) for 2 h at 30°C. Protoplasts were filtered, washed, counted, and resuspended in transformation buffer. For each transformation, 6 μg of unrestricted plasmid DNA was used. The transformation mix was transferred to top agar containing AMM with glucose and 2% agar, 0.6 M KCl and 0.1 μg/ml pyrithiamine were added, and the mix was poured onto plates of equal medium compositions. Plates were incubated at 37°C for 5 days, and conidia from the colonies were transferred several times to pyrithiamine-containing media to yield pure clones. Sixteen clones were randomly selected, and some conidia were scraped from the colonies and transferred to 200 μl of AMM with glucose in white 96-well plates (Nunc GmbH, Wiesbaden, Germany). After incubation for 14 h at 37°C, 90 μl of luciferase assay buffer (50 mM Tricine [pH 8.0], 0.5 mM coenzyme A, 2 mM dithiothreitol, 5 mM MgCl2, 4 mM ATP) and 10 μl of d-luciferin (20 mM; Synchem OHG, Felsberg/Altenburg, Germany) were added. Wells were observed with the naked eye in the dark for green-light emission. The six strongest light-emitting clones were selected for further analysis. Conidia were inoculated in 250-ml culture flasks containing 50 ml of minimal medium with either glucose or acetate as the carbon source. All cultures were incubated at 37°C and at 220 rpm on a rotary shaker. Glucose cultures were harvested after 20 h of growth, and acetate cultures were harvested after 30 h of growth. The mycelium was ground under liquid nitrogen and resuspended in luciferase assay buffer. After centrifugation, the protein content in the cell extracts was determined by the Bradford test (3a) (Bio-Rad protein assay concentrate; Bio-Rad GmbH, Munich, Germany). Wells of a 96-well plate were loaded with 100 and 300 μg of protein from each extract, and the volume was adjusted to 190 μl. Light emission was started by the addition of 10 μl of d-luciferin (15 mM), and bioluminescence was recorded with a microplate reader. Purified recombinant luciferase and a crude extract of A. fumigatus CBS144.89 served as controls. In addition to the luminescence detection by the microplate reader, a photograph was taken with a digital camera (Minolta Dimage 7i) in a dark room (4-s exposure time). Three of the strains were selected for Southern blot analysis using digoxigenin-labeled probes directed against the luciferase coding region. Genomic DNA was isolated by use of a MasterPure yeast DNA purification kit (Biozym, Hessisch Oldendorf, Germany) and independently restricted with different endonucleases. Blotting and analysis were performed as described by the manufacturer of the Southern reagents (Roche, Mannheim, Germany).
Recombinant production of GpdA and luciferase in E. coli.
For overproduction of recombinant GpdA (EC 1.2.1.59) and luciferase (EC 1.13.12.7), BL21(DE3) Rosetta2 cells harboring the respective plasmids were grown for 22 h at 28°C in Overnight Express Instant TB medium (Novagen/Merck KGaA, Darmstadt, Germany). Cells were resuspended in 50 mM Tricine buffer (pH 8.0) (for GpdA) or 50 mM HEPES buffer (for luciferase), each containing 150 mM NaCl and 20 mM imidazole. Sonication was used to disrupt cells. The soluble fraction of the crude extract was loaded onto a Ni-chelate column (bed volume, 14 ml) previously equilibrated with Tricine or HEPES buffer, depending on the recombinant enzyme. After a stringency wash in the presence of 30 mM imidazole, proteins were eluted at an imidazole concentration of 200 mM and checked for purity by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on a NuPage 4 to 12% Bis-Tris gradient gel (Invitrogen, Karlsruhe, Germany). The resulting protein bands were excised and subjected to tryptic digestion as described by the manufacturer (sequencing-grade modified trypsin; Promega, Mannheim, Germany). Peptides were mixed with α-cyano-4-hydroxycinnamic acid and analyzed by matrix-assisted laser desorption ionization-time of flight analysis; some peptides were selected for sequencing by matrix-assisted laser desorption ionization-time of flight tandem mass spectrometry analysis. For storage, the eluted enzymes were desalted and concentrated via centrifugal filter devices (30-kDa cutoff; Millipore, Schwalbach, Germany). After the addition of glycerol to a final concentration of 50%, samples were stored at −20°C.
Determination of enzymatic activities. (i) GpdA activity.
GpdA activity was determined as described previously, with minor modifications (4). A typical assay buffer with a final volume of 1 ml contained 50 mM Tricine (pH 8.0), 10 mM NAD, 4 mM fructose-1,6-bisposphate, 2 mM dithiothreitol, 20 mM potassium arsenate (pH 8.0), 5 mM potassium phosphate (pH 8.0), 0.8 U aldolase (from rabbit muscle; Roche Diagnostics GmbH, Mannheim Germany), and GpdA-containing enzyme preparations. Generation of NADH was monitored spectrophotometrically (Lambda 25 UV/Vis double-beam spectrophotometer; PerkinElmer, Rodgau-Jügesheim, Germany) at 340 nm, and the specific activity was calculated as follows: μmol NADH oxidation × min−1 × mg−1 protein. Arsenate was also replaced by 45 mM potassium phosphate, which yielded 80% of the maximum activity. The latter arsenate replacement assay was used when GpdA activities were determined from fungal extracts.
(ii) Luciferase activity.
Luciferase activity was tested in vitro (crude cell extracts or purified luciferase) by a modification of the luciferase assay described by BD Biosciences (Erembodegem, Belgium). The double-concentrated assay buffer contained 30 mM Tricine, 15 mM MgCl2, 10 mM dithiothreitol, 5 mM ATP, and 1 mM free coenzyme A (pH 7.8). Various amounts of extracts were transferred to white 96-well plates and mixed with assay buffer to result in a volume of 190 μl, and the reaction was started by the addition of 10 μl d-luciferin (15 mM stock; Synchem OHG, Felsberg/Altenburg, Germany). The assay results were read with a microplate reader (FLUOstar Optima; BMG Labtech, Offenburg, Germany) with a signal gain of 1,000. The specific conditions for testing luciferase activities of intact fungal cultures or activities during infection are described below.
(iii) Citrate synthase activity.
Citrate synthase activity was determined by measuring the release of coenzyme A during the condensation reaction of acetyl-coenzyme A with oxaloacetate, as described previously (5).
Semiquantitative reverse transcription-PCR (RT-PCR).
For the analysis of transcript levels of act1 (actin gene), gpdA, citA (citrate synthase gene), and luc, total RNA was isolated from mycelia or from an infected mouse lung by use of a RiboPure-Yeast kit as described by the manufacturer (Ambion, Darmstadt, Germany), except that the material was first ground to a fine powder under liquid nitrogen prior to RNA extraction. Purified RNA was checked for genomic DNA contamination by standard PCR with oligonucleotides act_Afum_for_II and act_Afum_rev_II. Only RNA preparations that revealed no amplification products were used for subsequent cDNA synthesis. cDNA from mRNA was prepared by using anchored oligo(dT)20 primers and SuperScript III reverse transcriptase, as suggested by the manufacturer (Invitrogen, Karlsruhe, Germany). cDNA was purified by alkaline RNA hydrolysis, and nucleotides were removed by ethanol precipitation in the presence of small amounts of glycogen. The precipitate was washed and dissolved in 35 μl of a 10 mM Tris-HCl buffer, pH 8.0, and subsequently quantified using a NanoPhotometer (Implen GmbH, Munich, Germany). All subsequent PCR amplifications were performed with 10-μl reaction mixtures using GoTaq polymerase (Promega, Mannheim, Germany), gene-specific oligonucleotides (Table 1), and a speed cycler (Analytik Jena, Jena, Germany). The PCR program used for all amplifications consisted of (i) an initial denaturation at 95°C for 90 s, (ii) 35 cycles of 95°C for 2 s, 62°C for 2 s, and 72°C for 15 s, and (iii) a final elongation at 72°C for 120 s. For standardization of the cDNA template amounts and to yield equal quantities of act1 transcripts, different amounts of cDNA were added to PCRs with the act1 primers (Table 1). A standard curve revealed that approximately 10 ng of template cDNA was suitable to avoid saturation of act1 amplification. By use of these same quantities of cDNA for the amplification of gpdA, citA, and luc transcripts, amplification reaction mixtures were likely to be saturated, but a dilution factor of 2 was found to give reliable readouts. For determination of transcript levels from the infected mouse lung, 125 ng and 250 ng cDNA were tested, but 125 ng resulted in extremely faint bands, which made quantification difficult. Therefore, only the 250-ng values were used for analysis. All PCR products were separated on 1.2% agarose gels stained with ethidium bromide. Bands were quantified using the band detection and analysis tools of Quantity One software (Bio-Rad Laboratories GmbH, Munich, Germany), and all images were recorded at an exposure yielding no pixel saturation. For comparison of the band intensities from each gel and exposure time, the Trace Quantity tool was used. This method quantifies a band as measured by the area under its intensity profile curve (units = intensity × mm).
Antifungal drug efficiency testing using a microplate reader.
In an initial experiment, the luciferase-producing strain C3 was chosen for testing the growth-inhibitory effect of the model antibiotic cycloheximide (also toxic for humans) in order to correlate fungal growth with light emission. White, sterile, 96-well plates were used for inoculation and subsequent measurement of luciferase activity. Each well contained 50,000 conidia and cycloheximide in concentrations ranging from 0 μg/ml to 50 μg/ml. AMM with 50 mM glucose and 0.5% (wt/vol) peptone served as the growth medium, and the final volume within each well was adjusted to 200 μl. Plates were incubated for 15 h at 37°C, and luciferase activity was determined with a microplate reader. Ten microliters of d-luciferin stock solution (20 mM) was added to each well, and measurement was started after 5 min of preincubation. The signal gain was set to 3,800, and a 5-s shaking period was introduced before each cycle was started. Luminescence was recorded in three cycles with a photon detection time of 5 s each, and the sum of the three cycles was calculated from the raw data of the output analysis report. Results for negative controls containing medium with and without the addition of cycloheximide and not inoculated with conidia were subtracted from the raw data. All experiments were performed in triplicate, and standard deviations were calculated. We observed a decrease in maximum light emission over time, which made the determination of light emission from the control reaction mixture without cycloheximide extremely difficult. Accordingly, the method was changed in further testing by injecting the d-luciferin in a computer-controlled manner into the well before each measurement was started. This method was used to calculate the growth-inhibitory effect of the antifungal compound nystatin, which was tested in a range from 0 to 3 μg/ml. The same growth parameters were applied, but 20 μl of 10 mM d-luciferin solution was injected into the wells prior to measurement. For each well, 18 intervals of 2 s were recorded and intervals 3 to 6, which showed the most stable light emission, were used for data evaluation.
Antifungal drug efficiency testing using the IVIS 100 system.
An independent method for antifungal drug efficiency testing was based on the detection of light emission by the IVIS 100 system (Xenogen Corporation/Caliper Life Sciences, Alameda, CA). In this approach, 12-well plates containing 1 ml of complete RPMI 1640 cell culture medium with 10% fetal calf serum were inoculated with 2 × 105 conidia in the presence of various concentrations (2 to 200 μg) of a Triflucan solution (2 mg/ml fluconazole in saline for perfusion; Pfizer, Paris, France). After incubation at 37°C for 9 and 11 h, d-luciferin was added to a final concentration of 10 mM. The reaction mixtures were preincubated for 10 min at room temperature. Measurements were performed with the IVIS 100 system (1-min exposure time) using a binning of four, which increases the sensitivity by improving the signal-to-noise ratio without severely compromising the spatial resolution. Quantification was performed by use of Living Image software, version 3.0 (Xenogen).
Mouse infection.
The bioluminescent A. fumigatus strain C3 was subcultured on 2% malt extract agar slants for 8 days at room temperature. Conidia were harvested by scraping the surface of the slant cultures with 2 ml of PBS supplemented with 0.1% Tween 20. The suspension was filtered through a 40-μm cell strainer (BD Biosciences, Erembodegem, Belgium) to separate conidia from the contaminating mycelium. Male BALB/cJ mice (20 to 24 g, 6 weeks old) were supplied by the Centre d'Elevage R. Janvier (Le Genest Saint-Isle, France) and infected at about 7 to 8 weeks of age. Mice were fed normal mouse chow and water ad libitum and were reared and housed under standard conditions with air filtration. Mice were cared for in accordance with the Institut Pasteur guidelines and in compliance with European animal welfare regulation. Intranasal infection and immunosuppression were performed as previously described (32). In brief, mice were immunosuppressed with two single doses of 25 mg cortisone acetate (Sigma) injected intraperitoneally 3 days before and immediately prior to infection with conidia (day 0). Before infection, mice were anesthetized with an intramuscular injection of 0.1 ml of a solution containing 10 μg/ml ketamine (Merial, Lyon, France) and 2 μg/ml xylazine (Bayer AG, Leverkusen, Germany) per mouse. Conidia at a concentration of 2 × 106 in 25 μl of PBS-0.1% Tween 20 were applied to the nares of the mice by use of an automatic pipetting device. Weight loss of mice was determined daily. For BLI, 100 μl of PBS containing 3.33 mg d-luciferin was injected intraperitoneally before each measurement.
Imaging of bioluminescence from animals, organs, and fungal cultures.
Images were acquired using an IVIS 100 system (Xenogen Corporation, Alameda, CA) according to the manufacturer's instructions. Analysis and acquisition were performed using Living Image software, version 2.6 (Xenogen). During image acquisition, mice were anesthetized using a constant flow of 2.5% isoflurane mixed with oxygen by means of an XGI-8 gas anesthesia system (Xenogen), which allowed control over the duration of anesthesia. Images were acquired for 5 min with a binning of four luminescent signals from the exteriors of mice, whereas luminescence of internal organs during dissection or from cultures in plates was integrated for 1 min. All other photographic parameters were held constant. Quantification of photons per second emitted by each organ was performed by defining regions of interest corresponding to the respective organs of interest, using the Xenogen software Living Image, version 3.0. The presence or absence of A. fumigatus within the lungs of dead animals was verified by bioluminescence of organs directly injected with d-luciferin and by histopathologic analyses.
Histopathology.
Lungs were removed from mice and immediately fixed in 4% neutral-buffered formaldehyde, embedded in paraffin, and cut into 5-μm-thick sections. Serial sections were stained with hematoxylin and eosin for histopathologic examination and with Grocott's methenamine silver for fungal detection and examined microscopically (34).
Accession numbers.
The corrected coding sequence of the gpdA gene has been assigned accession number AM999768 and the protein sequence has been assigned accession no. CAQ53727 in the NCBI database.
RESULTS
Determination of the gpdA open reading frame and recombinant synthesis of glyceraldehyde-3-phosphate dehydrogenase.
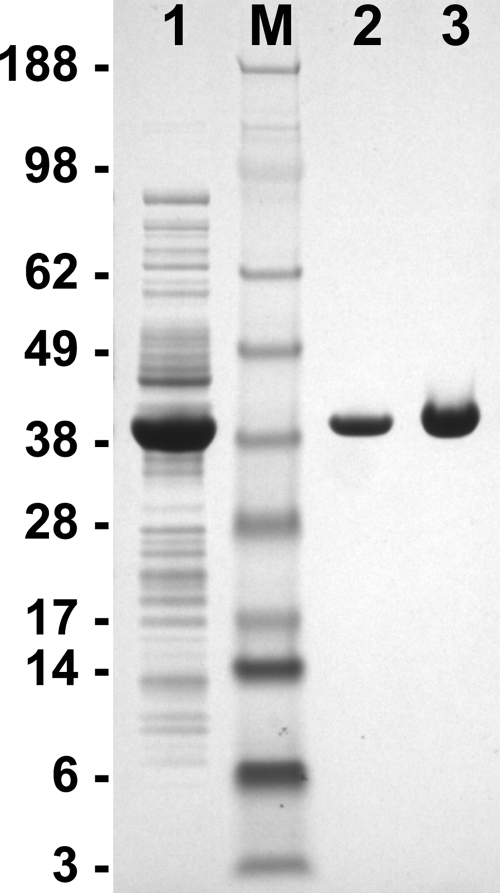
The gpdA coding region was amplified from A. fumigatus cDNA, which resulted in a single band of 1,021 bp. Analysis of sequencing reactions revealed that the open reading frame of the gpdA gene deposited in the CADRE database was not assigned correctly. Intron no. 2 of the gpdA gene actually contained a 14-bp exon flanked by two 61-bp introns. In addition, the second of these two introns ended 14 nucleotides further downstream than predicted for intron 2 of AFUA_5G01970, leading to exactly the same transcript size but an altered amino acid sequence at the N-terminal region. Furthermore, we identified a base exchange at position 816 of the coding sequence, a conservative exchange in a wobble position (AAA instead of AAG, both coding for lysine), which may be strain dependent. The gpdA cDNA was subcloned into a pET expression vector. BL21(DE3) Rosetta2 cells were used for enzyme production, and a strain with an empty vector served as a control. After growth of the cells in Overnight Express Instant TB medium, crude extracts were prepared and tested for GpdA activity. The control cells displayed a significant activity of 0.92 U/mg, which was attributed to glyceraldehyde-3-phosphate dehydrogenase from E. coli. This activity increased to 22.3 U/mg in the clones containing the gpdA expression construct. After purification by Ni-chelate chromatography, the purified protein displayed a specific activity of 27.55 U/mg. When arsenate was omitted from the assay, only 80% of the maximum activity was achieved; this result is in agreement with observations for other glyceraldehyde-3-phosphate dehydrogenases (4). Electrophoresis of the samples in a 4 to 12% SDS-polyacrylamide gradient gel gave a single band with an apparent molecular mass of approximately 35 kDa (Fig. 1). Tryptic digestion and peptide analysis identified the A. fumigatus GpdA from the NCBI database with a sequence coverage of 41.1%, and the protein was unambiguously identified by sequencing of a C-terminal peptide. Therefore, we conclude that the predicted gpdA gene indeed codes for glyceraldehyde-3-phosphate dehydrogenase, although the annotation of the AFUA_5G01970 open reading frame from the CADRE database needs correction.
FIG. 1.
SDS-polyacrylamide gel electrophoresis showing the purification of recombinant glyceraldehyde-3-phosphate dehydrogenase (GpdA) from A. fumigatus. Lane 1, 25 μg of E. coli crude extract overproducing A. fumigatus GpdA; lane 2, 2.4 μg purified GpdA; lane 3, 3.5 μg purified GpdA; lane M, molecular mass standard. Masses (in kilodaltons) are shown on the left.
Generation of firefly luciferase-producing A. fumigatus strains.
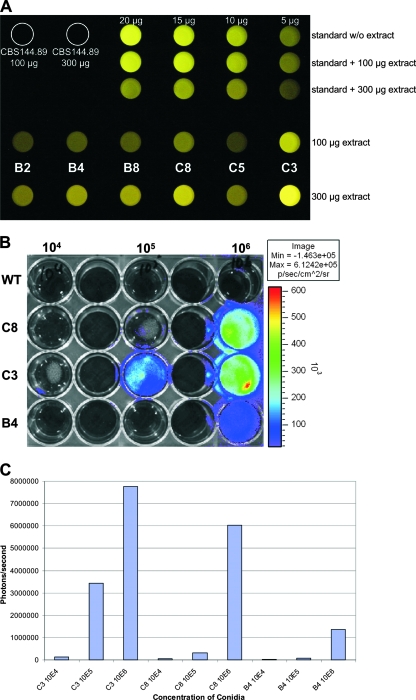
For the generation of luciferase-producing A. fumigatus strains, we cloned the luciferase gene from the firefly Photinus pyralis under the control of the gpdA promoter from A. fumigatus. The ptrA resistance cassette was used as a marker for transformation, and more than 100 colonies were obtained. In an initial screening, 32 transformants were streaked repeatedly on pyrithiamine-containing media to ensure the selection of strains without contamination of wild-type nuclei. From these 32 strains, 16 clones were randomly selected and tested for luciferase production by observation with the naked eye after adaptation to the dark. Six clones that revealed a very faint light emission were selected for further analysis. Mycelia were grown in glucose minimal media; cell extracts were used for a luciferase activity screen, and purified luciferase and an extract from the parental A. fumigatus wild-type strain CBS144.89 served as controls. Luminescence detection by a microplate reader and a photograph taken with a digital camera (Minolta Dimage 7i; 4-s exposure time) correlated well (Fig. 2A). Intensity of light emission was in the following order: C3 > C8 > B4 ≥ B8 > B2 > C5. To prove the in vivo activity of light emission, the parental wild-type strain and the strains C8, C3, and B4 were inoculated in 24-well plates and incubated for 8 h at 37°C in RPMI medium to allow the formation of germ tubes. d-Luciferin was added to each well, and luminescence was counted by an IVIS light detection system (Fig. 2B) and quantified with Living Image software (Fig. 2C). In agreement with the determination of luminescence from crude extracts, the light emission was stronger with strains C3 and C8 than with strain B4. Furthermore, light emission correlated with the number of conidia used to inoculate the media. Therefore, readouts by the IVIS system give a quantitative measure for the fungal biomass, which is important for the evaluation of the fungal burden in infected tissues and for the determination of growth in the presence of antifungals. To correlate the intensity of light emission with the number of integrations of the luciferase construct, a Southern blot analysis of the luciferase coding region was performed. Independent restrictions with EcoRV, PstI, and SmaI revealed two ectopic integrations for B4, three for C8, and four for C3 (data not shown). Therefore, the increase in light emission among transformants directly correlated with the copy number of luciferase genes introduced. None of the strains displayed an obvious abnormal growth phenotype, which made the strains suitable for subsequent investigations.
FIG. 2.
In vitro and in vivo bioluminescence of selected A. fumigatus strains producing firefly luciferase. (A) Digital picture showing light emission from crude extracts of the transformants B2, B4, B8, C8, C5, and C3 after growth on glucose. The parental wild-type strain CBS144.89 and recombinant luciferase (in the range of 5 to 20 μg) with and without (w/o) the addition of wild-type crude extract served as controls. The addition of crude extract slightly quenched the light emission from the luciferase control. (B) Determination of light emission by use of the IVIS 100 system. Different conidium concentrations (1 × 104, 1 × 105, and 1 × 106) of the wild-type strain (WT) and the three transformants C8, C3, and B4 were inoculated in RPMI medium and incubated for 8 h at 37°C. Light emission was induced by the addition of d-luciferin to the medium, and photons were collected for 1 min. (C) Quantification of light emission of wells shown in panel B by use of Living Image software, version 3.0.
Activity determination and expression analyses of gpdA, citA, and luc.
In order to confirm that the gpdA promoter is constitutively active during growth on various carbon sources, we cultivated the A. fumigatus wild-type strain CBS144.89 and the luciferase-producing strain C3 on liquid media with different nutrient compositions. Cells were grown for 20 h on RPMI complete cell culture medium or on minimal media containing glucose, ethanol, or a mixture of glucose and peptone. For the last composition, we were interested not only in the expression of gpdA at a single time point but also in the expression profile over time. Therefore, mycelia grown on glucose-peptone were harvested after 10, 20, and 30 h. We first determined the specific activities of GpdA, CitA, and luciferase, as shown in Table 2. Comparison of the activities of GpdA from the 20-h cultures revealed that the activity was weakest on RPMI medium but rather constant on ethanol and glucose and slightly elevated on glucose-peptone medium. The biomass formed on RPMI medium made up only approximately one-third of that obtained from glucose or glucose-peptone after 20 h, and determination of the residual glucose concentration revealed a total consumption of this substrate. This indicates that, due to the large number of conidia used for inoculation (2 × 106/ml), the mycelium was faced with glucose starvation. Nevertheless, GpdA, luciferase, and citrate synthase activities were still relatively high. Interestingly, GpdA activity increased over time, as determined when the 10-, 20-, and 30-h glucose-peptone cultures from both strains were compared (glucose was still present after 30 h). This was assumed to be due to either an increased gene expression in later growth states or a high stability of GpdA, which might accumulate during growth. In contrast, luciferase activity varied only slightly over time and correlated quite well with the GpdA activity determined from the fixed 20-h values. Citrate synthase is also known to be a constitutively produced enzyme of A. fumigatus but is additionally induced on ethanol, because it also serves as part of the glyoxylate cycle within the glyoxysomes. However, in contrast to GpdA activity, citrate synthase activity decreased slightly in later growth phases, indicating that the production level of this enzyme is not constant.
TABLE 2.
Determination of GdpA, CitA, and luciferase activities from the A. fumigatus wild-type strain and the luciferase-producing strain C3a
| Carbon source, growth timeb | Activity (mU/mg)
|
Light emission (RRU/mg)e
|
||||
|---|---|---|---|---|---|---|
| GpdA from WTc | GpdA from C3 | CSd from WT | CS from C3 | Luciferase from WT | Luciferase from C3 | |
| RPMI, 20 h | 587 ± 30 | 608 ± 32 | 307 ± 8 | 311 ± 5 | NDf | 14,906 ± 750 |
| EtOH, 20 h | 986 ± 150 | 906 ± 55 | 958 ± 34 | 785 ± 28 | ND | 31,520 ± 1,938 |
| Gluc, 20 h | 990 ± 58 | 1,019 ± 3 | 343 ± 17 | 364 ± 12 | ND | 32,207 ± 1,032 |
| Gluc+Pep, 10 h | 849 ± 41 | 689 ± 9 | 350 ± 20 | 305 ± 7 | ND | 74,610 ± 2,816 |
| Gluc+Pep, 20 h | 1,339 ± 103 | 1,169 ± 24 | 242 ± 10 | 221 ± 5 | ND | 51,010 ± 2,643 |
| Gluc+Pep, 30 h | 1,671 ± 96 | 1,316 ± 20 | 203 ± 8 | 143 ± 4 | ND | 52,437 ± 127 |
All data are mean values derived from triplicate measurements, and standard deviations are given.
EtOH, ethanol; Gluc, glucose; Pep, peptone; Gluc+Pep, glucose-peptone.
WT, wild type.
CS, citrate synthase.
RRU/mg denotes relative response units of light emission per mg of total protein. All light emission values were within the range of a standard curve recorded with purified luciferase.
No light emission was observed for the wild type (ND, not detectable).
To correlate the observed enzymatic activities with gene expression, we harvested mycelia from glucose-peptone-grown cultures and isolated RNA, which was reverse transcribed into cDNA. Additionally, cDNA was prepared from a mouse lung that was infected with the A. fumigatus wild type and was sacrificed 5 days after infection because of severe physical distress and strong signs of dyspnea.
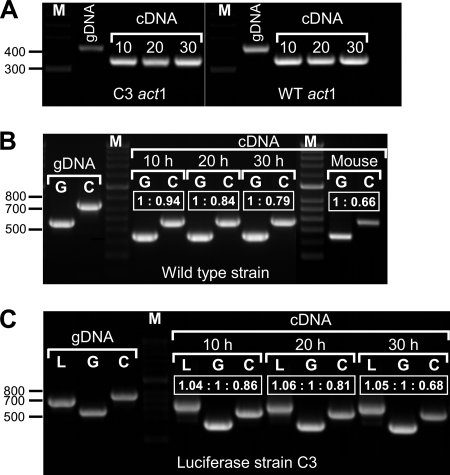
We assumed that the actin gene would serve as a suitable marker to compare levels of gene expression, since all cultures were still increasing in biomass, making the continued synthesis of actin necessary. Therefore, the cDNA levels were normalized to the transcript level of the act1 gene (Fig. 3A), but this standardization was not possible for the cDNA isolated from the mouse lung, because even 250 ng template cDNA yielded only a very faint band. Presumably, this was due to the small proportion of cDNA of fungal origin in this lung sample.
FIG. 3.
Determination of the relative abundance of transcript levels of gpdA, luc, and citA. cDNA from a glucose-peptone-grown mycelium, incubated for 10, 20, and 30 h, and cDNA from an infected mouse lung were studied. (A) Standardization of the cDNAs against the actin gene act1. Numbers above the cDNA amplification products denote the incubation times (in hours) of the respective strains. Genomic DNA (gDNA) served as the control to visualize the size shift in the cDNA amplification. The fragment sizes were 432 bp for gDNA and 354 bp for cDNA. WT, wild type. (B) Determination of gpdA (G) and citA (C) transcript levels on standardized amounts of cDNA from the wild type and on cDNA from an infected mouse lung. gDNA served as a control. The transcript sizes were 564 bp for gpdA gDNA, 451 bp for gpdA cDNA, 754 bp for citA gDNA, and 575 bp for citA cDNA. Boxed values denote the ratios of band intensities as calculated by trace quantity determinations. The gpdA transcript levels stayed constant, whereas those of citA declined slightly in later growth phases. The gpdA transcript from the infected mouse lung is much more pronounced than that of citA, resembling a late growth phase of the fungus in severe infection. (C) Same as for panel B, but the luciferase-producing strain C3 was studied and the luc gene (L) was included. Fragment sizes for both gDNA and cDNA are identical (688 bp), since the luc gene does not contain an intron. Boxed values denote the ratios of band intensities as calculated by trace quantity determinations. Transcript levels for luc and gpdA stayed constant, whereas that of citA steadily declined. Lane M, molecular size standard. Sizes (in base pairs) are shown on the left.
Standardized cDNA was used to amplify the gpdA and citA transcripts from the wild type (approximately 5 ng template) and from the infected mouse lung (250 ng). The luc, gpdA, and citA transcripts from the luciferase-producing strain C3 were investigated (Fig. 3B and C). In order to calculate the ratio of transcript levels, we calculated the trace quantities of each band (area under the intensity profile curve) (data not shown) and correlated the resulting data (ratios are given in Fig. 3). This analysis revealed that indeed the citA transcript level decreased in the older mycelium whereas the gpdA expression remained constant and, furthermore, that the ratio between the luc and the gpdA transcripts stayed constant. Quantification of the transcript levels from the infected mouse lung showed a high proportion of gpdA transcripts, whereas those of citA revealed a much weaker signal, which correlates with a mycelium in a later growth phase.
These data clearly showed that (i) gpdA is a constitutively expressed gene, although some variations may occur depending on the carbon source; (ii) luc expression correlates well with gpdA expression; (iii) expression of gpdA and luc is constant over time on a given carbon source; and (iv) gpdA is still expressed when the mycelium is growing invasively within infected mouse tissues. We conclude that the gpdA promoter is indeed a suitable tool for constitutive luciferase production on a given nutrient.
Suitability of the luciferase-producing A. fumigatus strains for drug efficiency testing.
To test the suitability of the luciferase-producing A. fumigatus strain C3 for monitoring the in vitro effectiveness of antifungal compounds, we initially evaluated cycloheximide as a model antibiotic. Because cycloheximide inhibits protein synthesis of all eukaryotic cells, we expected a growth-inhibitory response dependent on the drug concentration. A strong light emission was observed when no cycloheximide was present, and nearly all conidia were germinated and formed branched hyphae (Table 3). The addition of small amounts of cycloheximide (2.5 μg/ml) still allowed germination of conidia, but hyphal length was decreased and branching was observed less frequently. Further increase of the cycloheximide concentration revealed a dose-dependent inhibition of the germination rate.
TABLE 3.
Effects of antibiotics on growth and light emission from the luciferase-producing A. fumigatus strain C3
| Growth condition and concn (μg/ml) | Avg value of fungal outgrowtha | RRUb | Relative activityc (%) |
|---|---|---|---|
| Cycloheximide | |||
| 0 | 4.94 | 1.05 × 106 ± 5.22 × 104 | 100 ± 5.0 |
| 2.5 | 4.00 | 5.84 × 105 ± 5.80 × 104 | 56 ± 5.1 |
| 5.0 | 3.02 | 4.02 × 105 ± 2.01 × 104 | 38 ± 2.2 |
| 10 | 2.57 | 1.88 × 105 ± 1.02 × 104 | 18 ± 0.9 |
| 25 | 1.48 | 8.31 × 103 ± 4.54 × 102 | 0.8 ± 0.03 |
| 50 | 1.04 | 8.21 × 102 ± 1.97 × 102 | 0.08 ± 0.02 |
| Nystatin | |||
| 0 | 4.84 | 3.02 × 105 ± 7.94 × 103 | 100 ± 2.6 |
| 0.5 | 4.78 | 2.54 × 105 ± 1.34 × 104 | 84 ± 4.8 |
| 1.0 | 4.34 | 1.91 × 105 ± 2.81 × 103 | 63 ± 1.2 |
| 1.5 | 2.32 | 9.54 × 104 ± 4.32 × 103 | 32 ± 1.0 |
| 2.0 | 1.90 | 6.35 × 104 ± 5.89 × 102 | 21 ± 0.2 |
| 3.0 | 1.08 | 4.10 × 104 ± 3.17 × 103 | 14 ± 0.6 |
Microscopic growth analysis after incubation in the presence of antibiotics. Classification was performed by removing an aliquot and evaluating at least 100 conidia grown under each condition at a ×400 magnification. Values are classified as follows: 5, branching hyphae; 4, elongated hyphae without branching; 3, germ tubes; 2, swollen conidia; 1, resting conidia. All experiments were performed in triplicate.
For experiments with cycloheximide, relative response units (RRU) are given as the sums of three cycles of 5 s each, calculated after 5 min of preincubation. For experiments with nystatin, relative response units are given as the averages from 2-s intervals, determined between 4 and 10 s directly after d-luciferin addition. All experiments were performed in triplicate, and standard deviations are given.
All experiments were performed in triplicate, and standard deviations are given.
At 50 μg/ml, hardly any conidium was swollen or germinated. Paralleling this effect, light emission strongly decreased, and at 50 μg/ml the luciferase activity was reduced by a factor of 1,000. In this experiment, luciferin was added to all samples prior to measurement. Although three independent experiments with three replicate wells were conducted, we found it extremely difficult to determine the maximum light emission when no drug was added. We attributed this to the decline in light emission due to the rapid consumption of luciferin, which was more pronounced at a high level of luciferase activity. Therefore, we changed the strategy of luciferin addition in subsequent determinations by automatic injection of the substrate directly prior to measurement.
In this second approach we tested the antifungal drug nystatin, which is frequently used to combat superficial fungal infections because of its nonabsorption by the skin and the intestine. After the computer-controlled injection of d-luciferin into wells, data were recorded at 2-s intervals. Comparison of the data from 16 intervals revealed that the highest and most stable luminescence signals were obtained between 4 and 10 s after injection of d-luciferin, and the average of these four measurements was taken for comparative analyses. Table 3 shows the correlation between fungal growth and light emission. Only a minor effect on growth and light emission was observed in the presence of 0.5 or 1.0 μg/ml of nystatin. However, an increased nystatin concentration strongly delayed fungal germination, and at 3 μg/ml nearly all conidia stayed in the resting state, as indicated by a strong decrease in the luminescence signal.
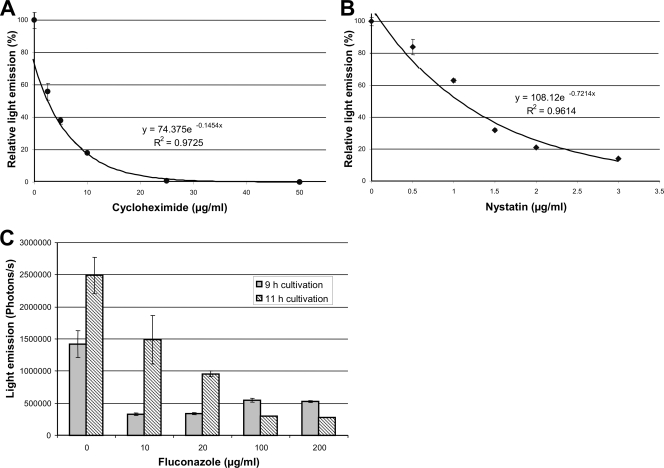
From these experiments, we conclude that luciferase production correlates with the growth-inhibitory effects mediated by both cycloheximide and nystatin. Furthermore, this method allows MIC determinations, which can be performed by graphical analysis of the luminescence signals (Fig. 4A and B). By defining 1% of residual light emission in comparison to that of the control as a suitable MIC, it would require 29.6 μg/ml of cycloheximide and 6.5 μg/ml of nystatin to achieve this goal. In an independent investigation, 18 out of 36 A. fumigatus isolates showed an MIC for nystatin of ≤8.0 μg/ml. Thus, the value determined here seems to lie in a realistic range (27).
FIG. 4.
Graphical analysis of drug efficiency by luminescence detection. (A) Light emission from wells containing 50,000 conidia of A. fumigatus strain C3 with the addition of different amounts of cycloheximide. Measurements were performed with a microplate reader after 15 h of incubation at 37°C. Standard deviations were calculated from three independent wells for each concentration. A fitted trend line with the corresponding formula and the correlation coefficient is given. (B) Same as for panel A, but nystatin was used as the antifungal drug. (C) Determination of light emission from strain C3 by the IVIS 100 system after growth in the presence of different concentrations of fluconazole. Light emission was detected after 9 and 11 h of incubation in RPMI complete medium at 37°C. Average values from two independent wells are given, and standard deviations are shown by error bars.
Drug efficiency screening by use of an IVIS 100 system.
In addition to using the microplate reader approach, we investigated whether readouts could be performed using the IVIS 100 system. This system has the advantage that light intensity maps of the observed object are shown directly. In our approach, we used the antifungal drug fluconazole to test its efficiency to inhibit growth and light emission of the A. fumigatus strain C3 cultured in complete RPMI medium. Concentrations ranged from 0 to 200 μg/ml, and light emission was determined after 9 and 11 h. Figure 4C shows the effect of fluconazole concentration and time dependency. Although the germination of conidia can be retarded even by the addition of 10 μg/ml of fluconazole, at least 100 μg/ml is required to suppress growth after continued incubation. These results are in agreement with the low effectiveness of fluconazole against molds, including A. fumigatus (30). Therefore, we conclude that screening for antifungal drug efficiency is also possible by using the IVIS 100 system, which gives a rapid readout of the data.
Intranasal infection of corticosteroid-treated BALB/cJ mice with the bioluminescent A. fumigatus strain C3.
For mouse infection, the bioluminescent transformant C3 was used. Mice were immunosuppressed by corticosteroids, which made them susceptible to invasive aspergillosis. For determination of light emission, we injected d-luciferin intraperitoneally, which led to a rapid distribution of the luciferase substrate to all organs.
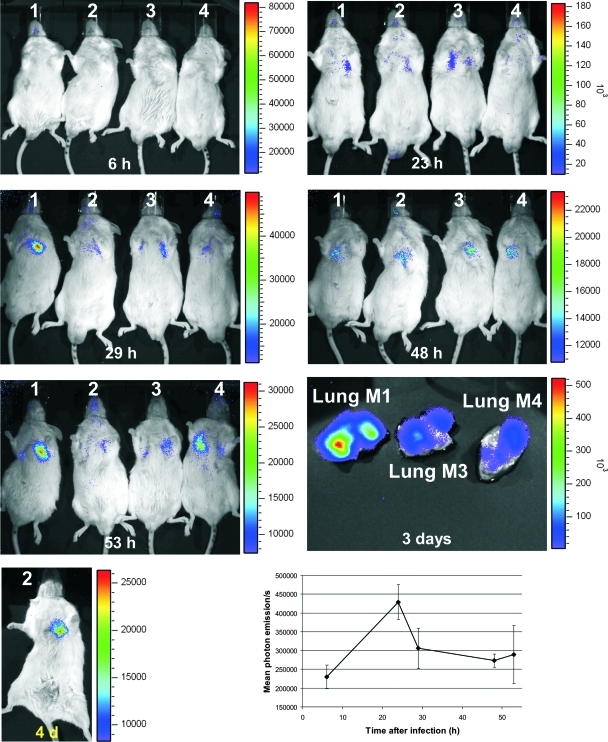
Uninfected control mice gave a weak background signal, which was visible only in high-sensitivity measurements. No specific organ was highlighted in this experiment. The histopathologic analysis did not reveal any significant lesions (data not shown). In contrast, infected mice developed invasive pulmonary aspergillosis and died within 3 to 4 days after infection. This killing rate by strain C3 was comparable to that of the parental A. fumigatus strain CBS144.89 and confirmed that the luciferase-producing strain was not attenuated in virulence (data not shown). Bioluminescence of mice infected with strain C3 revealed bioluminescence from the chest (lung), which peaked at around 23 h after infection (Fig. 5). No organs other than the lungs showed any light emission, indicating that invasive aspergillosis was restricted to the lower respiratory tract. Interestingly, light emissions from the lungs decreased slightly after reaching the maximum 1 day postinfection. This was an interesting observation since weight loss and physical distress continued, indicating a progression of infection. Furthermore, gpdA expression analysis of a mouse infected with the wild-type strain showed that, even in the late stage of the infection process, gpdA transcripts were present (Fig. 3B). d-Luciferin injected into the lungs after the lungs were removed from dead animals revealed an extremely high luminescence, confirming the presence of viable fungal cells (Fig. 5).
FIG. 5.
Time-lapse study of luminescence emission from BALB/cJ mice intranasally infected with 2 × 106 conidia of strain C3. All mice displayed luminescence from their lungs 23 h after infection, when d-luciferin was injected intraperitoneally. Luminescence stayed visible until the death of the animals but declined steadily. The lungs from mouse no. 1 (M1), M3, and M4, which were removed postmortem, displayed strong luminescence after the direct injection of d-luciferin. Light emission from live animals was recorded for 5 min, whereas the exposure time for organs ex vivo was set to 1 min. 4 d, 4 days. The lower-right graph shows the mean levels of light emission from the chests of mice as determined by data analysis performed with Living Image software, version 3.0. Standard deviations are shown by error bars.
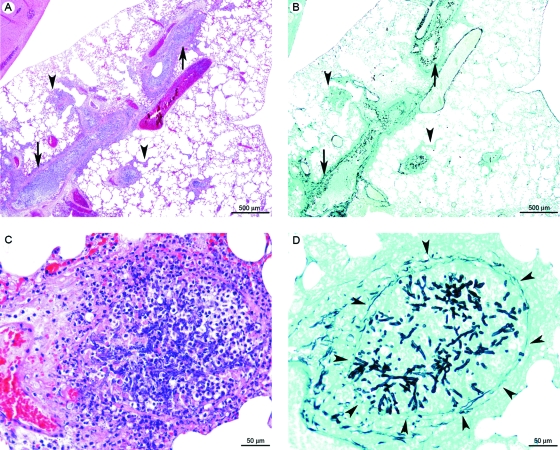
Infected-mouse necropsy revealed suppurative and hemorrhagic bronchopneumonia in all animals. By histology, we observed a multifocal inflammatory lesion, centered on bronchi and bronchioles. This was characterized by destruction of the overlying epithelium and filling of the lumen by an acidophilic amorphous material containing cell debris (necrosis), karyorrhectic neutrophils (suppurative lesion), extravasated erythrocytes (hemorrhage), and fungi, which were differentiated by using Grocott's methenamine silver method (Fig. 6).
FIG. 6.
Lung histopathology of BALB/cJ mice intranasally infected with the bioluminescent A. fumigatus strain C3. (A and B) Multifocal inflammatory lesion, generally centered on bronchi and bronchioles (arrows) and rarely extending to alveoli and blood vessels (veins and arteries) (arrowheads), characterized by infiltration of karyorrhectic neutrophils (suppuration) and erythrocytes (hemorrhage) (C), destruction of the bronchial/bronchiolar overlying epithelium (necrosis) (C), and the presence of intralesional fungal hyphae (D). These hyphae generally did not cross the bronchiolar wall (arrowheads in panel D). Panels A and C show hematoxylin and eosin staining, and panels B and D show Grocott's methenamine silver staining.
Altogether, these results explain the clinical signs of respiratory distress observed from day 2 until the death of the animals. They also confirm previous experiments in which corticosteroid treatments led to hyperinflammation caused by neutrophils at the site of infection (19).
DISCUSSION
In this study, we constructed a bioluminescent pathogenic A. fumigatus strain to monitor the manifestation of invasive aspergillosis in live animals and to use this strain in different assay systems to test antifungal drug efficiency. Since bacterial luciferase operons have not been adapted for use in eukaryotic systems, we utilized the luc gene from the firefly Photinus pyralis to construct a strain constitutively producing luciferase. Since only limited data on the nutrient consumption of pathogenic fungi during tissue invasion are available, the promoter region of the glyceraldehyde-3-phosphate dehydrogenase gene (gpdA) was selected. GpdA functions in glycolysis and gluconeogenesis and catalyzes the interconversion of glyceraldehyde-3-phosphate and 1,3-bisphosphoglycerate. Therefore, GpdA is assumed to be indispensable for the pathogen. This is also supported by investigations of the pathogenic yeast C. albicans. Although enzymes involved in both glycolysis and gluconeogenesis are differentially regulated during pathogenesis of C. albicans, they have been shown to be essential for full virulence, confirming the importance of both pathways during pathogenesis (2).
To confirm the functionality of GpdA from A. fumigatus, we produced the enzyme heterologously in E. coli. The purified protein showed significant activity in the conversion of glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate, which ensured that the selected promoter region indeed belonged to a functional gpdA gene. The fact that the promoter was active during invasive growth, as shown by the detection of the gpdA transcript from an infected mouse lung, also implies the importance of glycolysis/gluconeogenesis during the infection process of A. fumigatus.
Light emission from different A. fumigatus transformants correlated with the number of ectopic DNA integrations. Strain C3 was judged most suitable for subsequent investigations, since no phenotypic growth effects became visible and attenuation in virulence was not observed. In addition, luciferase was produced under all growth conditions tested, although some variations, depending on the available nutrients, were observed. All strains displayed light emission not only from cell extracts but also from whole cells, which confirms that the cells take up d-luciferin efficiently. This observation reveals an advantage for the A. fumigatus strain over the luciferase-producing C. albicans strain, which was assumed to take up d-luciferin efficiently only in the yeast growth state but not in the hyphal growth state (11). In our model, the A. fumigatus strain allowed the in vivo monitoring of luciferase production in relation to the fungal growth state and the quantification of the biomass produced. These parameters are essential prerequisites for monitoring the antifungal efficiencies of different drugs in our experiments.
Antifungal drug testing requires an easy and efficient readout system. Different standardized methods, such as the M38-A test, agar diffusion assays, the Etest, and others, are currently used to determine the susceptibility of filamentous fungi against antifungals (21). All of these tests are quite laborious or expensive and are therefore difficult for the application of screening attempts on drug libraries. Although our luciferase-based assay system is limited to a single A. fumigatus strain, the construction of additional luciferase-producing fungal strains is under way. These luciferase-producing strains may be suitable to prescreen drug libraries, at least in vitro, for the presence of new antifungal compounds.
One of the most interesting features of the luciferase-based system is the high correlation between light emission and fungal growth and the possibility of gleaning information related to the mode of action of an antifungal drug. In both treatments, cycloheximide at 50 μg/ml and nystatin at 3 μg/ml, we observed similar fungal growth indexes of 1.04 and 1.08, respectively. However, light emission and thus luciferase activity were strongly reduced with cycloheximide compared to levels with nystatin. This difference may reflect the different modes of action of these compounds.
In contrast to cycloheximide, which inhibits the de novo synthesis of proteins, nystatin forms ion channels in the fungal membrane by an interaction with ergosterol similar to the action of the antifungal amphotericin B (17). This mode of action may prevent the growth of the fungus but may leave the synthesis of proteins unaffected. Therefore, nystatin allows some de novo synthesis of luciferase, which explains the significantly increased luminescence signal compared to that with cycloheximide.
Another positive task of the luciferase-based system was the ability to screen the cultures several times, as shown in the fluconazole experiment performed with the IVIS 100 system. Samples were measured 9 and 11 h after inoculation, and results revealed that fluconazole was able to delay spore germination at low concentrations but prevented germination only at high concentrations. Therefore, the luciferase-based system seems suitable to study the actions of different antifungals in time-lapse studies.
A further application of our system was to monitor the in vivo development of invasive aspergillosis by using a small number of living animals. In the corticosteroid-treated mouse infection model, the ability of alveolar macrophages to kill conidia is reduced (6), leading to the early swelling and germination of conidia and the rapid onset of invasive growth. This was confirmed in vivo by using the bioluminescent strain. Light emission was strongly visible 1 day after infection, indicating that conidia started to swell and germinate within the lung. From 24 h after inoculation of conidia until the time of death, a marked decrease in lung luminescence was observed, although a high number of branched fungi were present in histology at the end of the experiments. This phenomenon could be explained by two factors: (i) the poor general condition of the mice and (ii) the pulmonary lesions.
Oxygen and d-luciferin are essential for the light-producing reaction. From 24 h postinfection, mice started to display severe clinical distresses (tachypnea, dyspnea, tachycardia, weight loss, and apathy) that inevitably led to death due to respiratory failure. These distresses were most probably responsible for the significant decrease in the efficacy of the systemic distribution of intraperitoneally injected d-luciferin but also for the decrease in the oxygen uptake. Moreover, pulmonary lesions (in particular, the filling of bronchial/bronchiolar spaces with a suppurative and necrotic exudate) were presumably severe enough to restrict oxygen dispersion in the bronchoalveolar tree. The low oxygen and d-luciferin supplies within necrotic areas are also supported by the fact that anaerobic bacteria causing pleuropulmonary infections are generally found within necrotic tissues (3, 23) and by the observation of a strong increase in light emission when d-luciferin was directly injected ex vivo into the mouse lungs (Fig. 5). Altogether, these clinical and pathologic observations were severe enough to explain the luminescence decrease while a high density of fungi could be observed in histology.
In further studies, we will use this new tool to compare the impacts of different immunosuppression conditions on disease development. Furthermore, investigations of the systemic dissemination of fungi will be studied using different routes of inoculation. In our corticosteroid pretreatments, we never observed dissemination of fungal elements, indicating that mice died from their pulmonary lesions before fungal dissemination occurred. Since antifungal drug efficiency is frequently tested by use of disseminated infection models (28, 35, 38), the suitability of our luciferase-based system as an in vivo screening method in disseminated aspergillosis has to be tested. Last but not least, we will also generate bioluminescent fungi from other filamentous fungal species responsible for invasive fungal diseases in immunocompromised patients to broaden the spectrum of fungi for which antifungals can be tested.
Acknowledgments
We express our thanks to M. Huerre and T. Hohl for their advice and helpful suggestions. In addition, we express our gratitude to T. Angelique for his assistance with the animal facilities.
This work was supported by grants from the Hans Knoell Institute (M.B.), a Bourse Roux Fellowship from the Institut Pasteur (G.J.), and funding from the Institut Pasteur through a Programme Tansversal de Recherche (O.I.-G.).
Footnotes
Published ahead of print on 26 September 2008.
REFERENCES
- 1.Balloy, V., M. Huerre, J. P. Latgé, and M. Chignard. 2005. Differences in patterns of infection and inflammation for corticosteroid treatment and chemotherapy in experimental invasive pulmonary aspergillosis. Infect. Immun. 73:494-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barelle, C. J., C. L. Priest, D. M. Maccallum, N. A. Gow, F. C. Odds, and A. J. Brown. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol. 8:961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett, J. G. 1993. Anaerobic bacterial infections of the lung and pleural space. Clin. Infect. Dis. 16(Suppl. 4):S248-S255. [DOI] [PubMed] [Google Scholar]
- 3a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brenneman, F. N., and W. A. Volk. 1959. Glyceraldehyde phosphate dehydrogenase activity with triphosphopyridine nucleotide and with diphosphopyridine nucleotide. J. Biol. Chem. 234:2443-2447. [PubMed] [Google Scholar]
- 5.Brock, M., and W. Buckel. 2004. On the mechanism of action of the antifungal agent propionate. Eur. J. Biochem. 271:3227-3241. [DOI] [PubMed] [Google Scholar]
- 6.Brummer, E., A. Maqbool, and D. A. Stevens. 2001. In vivo GM-CSF prevents dexamethasone suppression of killing of Aspergillus fumigatus conidia by bronchoalveolar macrophages. J. Leukoc. Biol. 70:868-872. [PubMed] [Google Scholar]
- 7.Dellacasa-Lindberg, I., N. Hitziger, and A. Barragan. 2007. Localized recrudescence of Toxoplasma infections in the central nervous system of immunocompromised mice assessed by in vivo bioluminescence imaging. Microbes Infect. 9:1291-1298. [DOI] [PubMed] [Google Scholar]
- 8.d'Enfert, C. 1996. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr. Genet. 30:76-82. [DOI] [PubMed] [Google Scholar]
- 9.Denning, D. W. 1994. Treatment of invasive aspergillosis. J. Infect. 28(Suppl. 1):25-33. [DOI] [PubMed] [Google Scholar]
- 10.Dockrell, D. H. 2008. Salvage therapy for invasive aspergillosis. J. Antimicrob. Chemother. 61(Suppl. 1):i41-i44. [DOI] [PubMed] [Google Scholar]
- 11.Doyle, T. C., K. A. Nawotka, C. B. Kawahara, K. P. Francis, and P. R. Contag. 2006. Visualizing fungal infections in living mice using bioluminescent pathogenic Candida albicans strains transformed with the firefly luciferase gene. Microb. Pathog. 40:82-90. [DOI] [PubMed] [Google Scholar]
- 12.Dubourdeau, M., R. Athman, V. Balloy, M. Huerre, M. Chignard, D. J. Philpott, J. P. Latgé, and O. Ibrahim-Granet. 2006. Aspergillus fumigatus induces innate immune responses in alveolar macrophages through the MAPK pathway independently of TLR2 and TLR4. J. Immunol. 177:3994-4001. [DOI] [PubMed] [Google Scholar]
- 13.Francis, K. P., D. Joh, C. Bellinger-Kawahara, M. J. Hawkinson, T. F. Purchio, and P. R. Contag. 2000. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect. Immun. 68:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franke-Fayard, B., A. P. Waters, and C. J. Janse. 2006. Real-time in vivo imaging of transgenic bioluminescent blood stages of rodent malaria parasites in mice. Nat. Protoc. 1:476-485. [DOI] [PubMed] [Google Scholar]
- 15.Glomski, I. J., A. Piris-Gimenez, M. Huerre, M. Mock, and P. L. Goossens. 2007. Primary involvement of pharynx and Peyer's patch in inhalational and intestinal anthrax. PLoS Pathog. 3:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gooch, V. D., A. Mehra, L. F. Larrondo, J. Fox, M. Touroutoutoudis, J. J. Loros, and J. C. Dunlap. 2008. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot. Cell 7:28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helrich, C. S., J. A. Schmucker, and D. J. Woodbury. 2006. Evidence that nystatin channels form at the boundaries, not the interiors of lipid domains. Biophys. J. 91:1116-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutchens, M., and G. D. Luker. 2007. Applications of bioluminescence imaging to the study of infectious diseases. Cell. Microbiol. 9:2315-2322. [DOI] [PubMed] [Google Scholar]
- 19.Ibrahim-Granet, O., M. Dubourdeau, J. P. Latgé, P. Ave, M. Huerre, A. A. Brakhage, and M. Brock. 2008. Methylcitrate synthase from Aspergillus fumigatus is essential for manifestation of invasive aspergillosis. Cell. Microbiol. 10:134-148. [DOI] [PubMed] [Google Scholar]
- 20.Karsi, A., and M. L. Lawrence. 2007. Broad host range fluorescence and bioluminescence expression vectors for Gram-negative bacteria. Plasmid 57:286-295. [DOI] [PubMed] [Google Scholar]
- 21.Lass-Flörl, C., and S. Perkhofer. 2008. In vitro susceptibility-testing in Aspergillus species. Mycoses 51:437-446. [DOI] [PubMed] [Google Scholar]
- 22.Latgé, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levison, M. E. 2001. Anaerobic pleuropulmonary infection. Curr. Opin. Infect. Dis. 14:187-191. [DOI] [PubMed] [Google Scholar]
- 24.Liebmann, B., T. W. Mühleisen, M. Müller, M. Hecht, G. Weidner, A. Braun, M. Brock, and A. A. Brakhage. 2004. Deletion of the Aspergillus fumigatus lysine biosynthesis gene lysF encoding homoaconitase leads to attenuated virulence in a low-dose mouse infection model of invasive aspergillosis. Arch. Microbiol. 181:378-383. [DOI] [PubMed] [Google Scholar]
- 25.Montagnoli, C., F. Fallarino, R. Gaziano, S. Bozza, S. Bellocchio, T. Zelante, W. P. Kurup, L. Pitzurra, P. Puccetti, and L. Romani. 2006. Immunity and tolerance to Aspergillus involve functionally distinct regulatory T cells and tryptophan catabolism. J. Immunol. 176:1712-1723. [DOI] [PubMed] [Google Scholar]
- 26.Morgan, L. W., A. V. Greene, and D. Bell-Pedersen. 2003. Circadian and light-induced expression of luciferase in Neurospora crassa. Fungal Genet. Biol. 38:327-332. [DOI] [PubMed] [Google Scholar]
- 27.Oakley, K. L., C. B. Moore, and D. W. Denning. 1999. Comparison of in vitro activity of liposomal nystatin against Aspergillus species with those of nystatin, amphotericin B (AB) deoxycholate, AB colloidal dispersion, liposomal AB, AB lipid complex, and itraconazole. Antimicrob. Agents Chemother. 43:1264-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odds, F. C., F. Van Gerven, A. Espinel-Ingroff, M. S. Bartlett, M. A. Ghannoum, M. V. Lancaster, M. A. Pfaller, J. H. Rex, M. G. Rinaldi, and T. J. Walsh. 1998. Evaluation of possible correlations between antifungal susceptibilities of filamentous fungi in vitro and antifungal treatment outcomes in animal infection models. Antimicrob. Agents Chemother. 42:282-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasqualotto, A. C., and D. W. Denning. 2008. New and emerging treatments for fungal infections. J. Antimicrob. Chemother. 61(Suppl. 1):i19-i30. [DOI] [PubMed] [Google Scholar]
- 30.Sabatelli, F., R. Patel, P. A. Mann, C. A. Mendrick, C. C. Norris, R. Hare, D. Loebenberg, T. A. Black, and P. M. McNicholas. 2006. In vitro activities of posaconazole, fluconazole, itraconazole, voriconazole, and amphotericin B against a large collection of clinically important molds and yeasts. Antimicrob. Agents Chemother. 50:2009-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeij, J. P., J. P. Boyle, M. E. Grigg, G. Arrizabalaga, and J. C. Boothroyd. 2005. Bioluminescence imaging of Toxoplasma gondii infection in living mice reveals dramatic differences between strains. Infect. Immun. 73:695-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schöbel, F., O. Ibrahim-Granet, P. Ave, J. P. Latgé, A. A. Brakhage, and M. Brock. 2007. Aspergillus fumigatus does not require fatty acid metabolism via isocitrate lyase for development of invasive aspergillosis. Infect. Immun. 75:1237-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh, N., and D. L. Paterson. 2005. Aspergillus infections in transplant recipients. Clin. Microbiol. Rev. 18:44-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha, B. K., D. P. Monga, and S. Prasad. 1988. A combination of Gomori-Grocott methenamine silver nitrate and hematoxylene and eosin staining technique for the demonstration of Candida albicans in tissue. Quad. Sclavo Diagn. 24:129-132. [PubMed] [Google Scholar]
- 35.Sionov, E., S. Mendlovic, and E. Segal. 2006. Efficacy of amphotericin B or amphotericin B-intralipid in combination with caspofungin against experimental aspergillosis. J. Infect. 53:131-139. [DOI] [PubMed] [Google Scholar]
- 36.Sjölinder, H., and A. B. Jonsson. 2007. Imaging of disease dynamics during meningococcal sepsis. PLoS ONE 2:e241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens-Romero, S. D., A. J. Mednick, and M. Feldmesser. 2005. The pathogenesis of fatal outcome in murine pulmonary aspergillosis depends on the neutrophil depletion strategy. Infect. Immun. 73:114-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takemoto, K., Y. Yamamoto, Y. Ueda, Y. Sumita, K. Yoshida, and Y. Niki. 2004. Comparative studies on the efficacy of AmBisome and Fungizone in a mouse model of disseminated aspergillosis. J. Antimicrob. Chemother. 53:311-317. [DOI] [PubMed] [Google Scholar]
- 39.Weidner, G., C. d'Enfert, A. Koch, P. C. Mol, and A. A. Brakhage. 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33:378-385. [DOI] [PubMed] [Google Scholar]