Abstract
A new immunologically based flow cytometry (IFCM) technique was developed to enumerate Aureococcus anophagefferens, a small pelagophyte alga that is the cause of “brown tides” in bays and estuaries of the mid-Atlantic states along the U.S. coast. The method utilizes a monoclonal antibody conjugated to fluorescein isothiocyanate (FITC-MAb) to label the surface of A. anophagefferens cells which are then detected and enumerated by using a flow cytometer. Optimal conditions for FITC-MAb staining, including solution composition, incubation times, and FITC-MAb concentrations, were determined. The FITC-MAb method was tested for cross-reactivity with nontarget, similarly sized, photoautotrophic protists, and the method was compared to an enzyme-linked immunosorbent assay (ELISA) using the same MAb. Comparisons of the IFCM technique to traditional microscopy enumeration of cultures and spiked environmental samples showed consistent agreement over several orders of magnitude (r2 > 0.99). Comparisons of the IFCM and ELISA techniques for enumerating cells from a predation experiment showed a substantial overestimation (up to 10 times higher) of the ELISA in the presence of consumers of A. anophagefferens, presumably due to egested cell fragments that retained antigenicity, using the ELISA method, but were not characterized as whole algal cells by the IFCM method. Application of the IFCM method to environmental “brown-tide” samples taken from the coastal bays of Maryland demonstrated its efficacy in resolving A. anophagefferens abundance levels throughout the course of a bloom and over a large range of abundance values. IFCM counts of the brown-tide alga from natural samples were consistently lower than those obtained using the ELISA method and were equivalent to those of the polyclonal immunofluorescence microscopy technique, since both methods discriminate intact cells. Overall, the IFCM approach was an accurate and relatively simple technique for the rapid enumeration of A. anophagefferens in natural samples over a wide range of abundance values (103 to 106 cells ml−1).
Harmful algal blooms have become an increasingly serious problem for coastal communities and ecosystems worldwide, during the last several decades. These events have been shown to negatively affect local economies and ecosystem functions, with annual economic losses totaling $47 million (27). Ecological effects have included poisoning of numerous species in aquatic food webs from a variety of phytotoxins, mortality of benthic communities due to direct and indirect effects, fish kills due to hypoxia, and overall declines in water quality (1, 4, 5, 10, 14, 19, 22, 28, 29, 35). Increasing occurrence and awareness of harmful algal bloom events in recent years have led to research efforts to better understand the initiation and termination of blooms, the development of more effective monitoring approaches, and ultimately proposals for the remediation or prevention of these events (41a, 43, 55).
The pelagophyte Aureococcus anophagefferens is the cause of recurring “brown tides” in estuaries along the mid-Atlantic coast of the United States, from Rhode Island to Maryland. Since the first recorded brown tides in 1985 in Narragansett Bay, RI, and inland bays and estuaries of Long Island, NY, numerous and widespread ecological and economic impacts have been recorded in the ecosystems afflicted with these blooms (6, 22, 33, 44). A. anophagefferens is often present in these environments at background levels (<103 cells ml−1), but the alga can grow rapidly to abundance levels of >106 cells ml−1 under favorable environmental conditions. These brown tides have been shown to have deleterious effects on eelgrass communities, local shellfish populations, and protozooplankton (5, 10, 14, 34). Economic losses resulting from brown tides in New York alone have been estimated at >$3 million annually, mainly as a result of dramatically reduced shellfish harvests (16, 22, 27, 37, 47, 52).
Establishment of A. anophagefferens as the causative agent of brown tides of the mid-Atlantic United States was initially difficult, and monitoring efforts for this organism have remained challenging mainly because the cells are small (2 to 3 μm), round, and nonflagellated and, thus, morphologically nondescript. They are virtually impossible to distinguish from many other minute algae, using traditional light microscopy. As a result, several techniques have been developed to study these events and specifically enumerate A. anophagefferens cells in natural water samples, including an immunofluorescence microscopy technique utilizing polyclonal antibodies (2, 3), an enzyme-linked immunosorbent assay (ELISA) using a monoclonal antibody (MAb) (7), and a genetic approach using quantitative real-time PCR (qPCR) (41).
These various methods each have advantages and limitations. The polyclonal immunofluorescence technique has been employed most commonly for natural samples and is accurate over a range of several orders of magnitude to a lower limit of detection of approximately 100 cells ml−1 (3). However, this microscopy-based approach is time consuming and prone to variability among operators and the reactivity and specificity of the polyclonal antisera will vary with each batch. The ELISA for A. anophagefferens (7) improved specificity and reduced variability by using a standard 96-well plate format and a MAb developed against cell surface antigens. This design also allows for rapid and simultaneous analysis of numerous samples. However, the practical limit of detection for the ELISA is higher than the polyclonal method (≈5,000 cells ml−1), and some technical expertise and specialized equipment are required. More recently, a qPCR approach has been developed to enumerate A. anophagefferens cells at low levels in natural samples (41). This method is based on a TaqMan molecular probe targeting an 18S ribosomal DNA sequence unique to A. anophagefferens and is the most sensitive of the current techniques (lower limit of detection, ≈1 cell ml−1). However, the technique is most applicable at abundance levels of the brown-tide alga of ≤104 cells ml−1, whereas the bloom abundance level of A. anophagefferens during a brown tide can exceed 106 cells ml−1. Additionally, the qPCR technique operates under the assumption of an invariant gene number among culture- and field-based specimens, an assumption that may not be accurate for natural populations.
The combined use of flow cytometry and immunofluorescence was investigated as a means of obviating some of the issues with extant methods for counting A. anophagefferens cells in natural samples. Flow cytometry (FCM) has become increasingly useful for the characterization of aquatic microbial assemblages. It has been used to detect and quantify bacteria, cyanobacteria, eukaryotes (fungi, protozoa, algae), and viruses (11, 15, 38, 39, 42, 48). The approach has been especially useful in the discrimination and characterization of photoautotrophic microbes in natural samples, using the autofluorescence of photosynthetic pigments as specific markers. These approaches have occasionally been augmented with the application of cell stains, genetic probes, and fluorescently labeled antibodies to increase the accuracy of FCM (15, 42, 46, 49-51, 53). Information about cell size based on light scattering provides additional information for resolving differences among microbial cells.
An immunofluorescence FCM (IFCM) approach for enumerating A. anophagefferens cells was developed using a highly specific MAb previously developed against A. anophagefferens (7). The antibody was directly conjugated to the fluorophore fluorescein isothiocyanate (FITC), chosen because its emission peak at 530 nm is a wavelength not commonly observed as natural fluorescence in most seawater samples. The IFCM method was used with laboratory cultures of A. anophagefferens to establish the cytometric parameters for accurately distinguishing and counting the algal cells. Cross-reactivity tests were conducted with nontarget but similarly sized algal cells. A comparison of the new method with an existing ELISA approach using cultures of A. anophagefferens in the presence and absence of protistan consumers of the brown-tide alga indicated significant overestimates of cell concentration by the ELISA method, presumably due to the egestion by the consumer of cell fragments that retained antigenicity. These results imply that the IFCM approach may provide more accurate cell counts in environmental samples, particularly for samples collected at the termination of brown tides, when cell fragments from viral lysis or protistan egestion might constitute a significant fraction of the total antigenic signal. Application of the new IFCM technique to environmental samples demonstrated the efficacy of this method for counting A. anophagefferens cells across a wide range of abundance levels in nature.
MATERIALS AND METHODS
Flow cytometry.
All analyses were performed with a FACSCalibur flow cytometer (FCM; Becton Dickinson, San Jose, CA) equipped with a 15-mW 488-nm air-cooled blue argon-ion laser and a 635-nm red diode laser. Cytometric measurements included forward-angle light scatter, side scatter (SSC), and four channels for the detection of emitted light: FL1 (green light, 500 to 560 nm), FL2 (orange light, 543 to 627 nm), FL3 (dark red light, >670 nm), and FL4 (red light, 645 to 677 nm). Instrument settings were optimized based on A. anophagefferens cultures, and detectors were set to log mode, providing a range of several orders of magnitude. Seawater filtered through 0.22-μm-pore-sized filters was used as sheath fluid when undiluted seawater samples were analyzed. The manufacturer-supplied FACSFlow sheath fluid (product no. 342003; Becton Dickinson) was used for samples that were diluted in a phosphate-buffered saline Tween 20 (PBS-T) solution (product no. P3563; Sigma), a commonly used buffered saline solution with salinity of ∼11 practical salinity units (PSU). The use of different sheath fluids was meant to minimize complications due to sample-sheath fluid refractive index incompatibilities (12). The FACSCalibur has three flow rate settings of approximately 12, 35, and 60 μl min−1. Samples were analyzed at dilutions and flow rates that maintained the event rate below the maximum of 1,000 events s−1 recommended for the cytometer. All data files were acquired and analyzed using manufacturer-supplied Cell Quest software (Becton Dickinson).
A gravimetric technique was employed to determine the volume of sample examined during each FCM analysis (42). Sample tubes were weighed using an analytical balance prior to and immediately following FACSCalibur analysis. The change in mass was used as an estimate of the volume run. Estimated volumes and appropriate dilution factors were used to calculate the abundance of algal cells for each cytometric analysis performed. An alternative bead method for volume estimation can be substituted for this step with comparable volume estimations (42).
Cultures.
A. anophagefferens (strain CCMP1794) was obtained from the Culture Collection of Marine Phytoplankton (Boothbay Harbor, ME). The strain was isolated from Barnegat Bay, New Jersey, and cultured in modified f/2 medium (25) at 20°C and a light intensity of 200 μE m−2 s−1 on a 12:12-h light/dark cycle. Cells were harvested in late exponential phase for all cytometric analyses described herein, preserved with a final concentration of 1% glutaraldehyde (from a 10% solution prepared with natural seawater and filtered through 0.22-μm-pore-size filters), and stored at 4°C in dark glass.
The immunocytometry method for enumerating A. anophagefferens was developed using algae of a size similar to the brown-tide alga, to serve as negative controls for immunocytometry cross-reactivity tests. Cultures of Isochrysis galbana, Pelagococcus subviridis (CCMP 1429), Chlorella stigmatophora, Micromonas pusilla (CCMP 487), and Minutocellus polymorphus were employed. In addition, two unidentified golden coccoid algae (not A. anophagefferens) obtained during a brown tide in Great South Bay, NY, in November 1986 (strains BT3 and BT8) were tested. All of these algal strains were cultured, preserved, and stored in the same manner employed for A. anophagefferens, with the exception that silica was added to the culture medium for the diatom M. polymorphus. The culture of Pseudopedinella sp. used in the experiment examining predation on A. anophagefferens was isolated from Long Island, NY, and maintained in the laboratory on a medium consisting of early to mid-exponential-phase A. anophagefferens culture.
Development of conjugated FITC-MAb.
A monoclonal mouse immunoglobulin G MAb specific to A. anophagefferens was developed by Maine Biotechnology Inc. (Portland, ME) for use in an ELISA for enumerating brown-tide cells (7). For the FCM method, the antibody was conjugated directly to FITC for a final concentration of 0.85 mg ml−1 by Maine Biotechnology Inc. Aliquots of 50 μl FITC-MAb were dispensed and stored frozen and protected from light. Aliquots were thawed for several minutes at room temperature prior to dilution and use in the IFCM procedure.
Solution, incubation time, and FITC-MAb concentration.
Reactivity of A. anophagefferens with FITC-MAb was determined using preserved algal cells in seawater (salinity of 35 PSU), and algal cells resuspended in 10 mM PBS-T. The latter suspensions were prepared by collecting algal cells on a 0.2-μm polycarbonate filter and resuspending the cells in PBS-T. FITC-MAb was added to both types of cell suspensions at a final concentration of 0.1% (0.85 μg ml−1) and incubated for 10 to 15 min. Incubations were performed at room temperature (20°C), and samples were protected from light. Unstained samples of A. anophagefferens cells in seawater and in PBS-T were analyzed by FCM to produce baseline cytometric profiles of the light scatter and fluorescent properties of the cells. A gated region was established that encompassed the unstained population of A. anophagefferens on plots of SSC and FL3 (indicative of chlorophyll) fluorescence. Stained samples of A. anophagefferens cells were then analyzed, and the algal cells were identified on plots of SSC and FL3, using the gated region established for the unstained cells. The FL1 signals (green fluorescence) of the stained and unstained cells were then compared to determine the degree of MAb labeling.
An alternative to the filtration and resuspension of A. anophagefferens cells in PBS-T was examined to facilitate application of the technique to natural, preserved samples of seawater. Preserved, cultured cells in seawater were diluted 1:5 or 1:10 in 10 mM PBS-T (resulting salinity of 13 PSU) and stained with FITC-MAb diluted 1:100 for 15 min. Relative increases in the FL1 signal, as described above, were compared to determine if dilution was a suitable alternative to the labor-intensive and error-prone process of filtration and resuspension.
Optimal incubation time and FITC-MAb concentration were determined empirically using preserved, cultured A. anophagefferens cells (approximately 105 cells ml−1) diluted 1:10 in 10 mM PBS-T. Optimal incubation time was determined by staining cells with FITC-MAb diluted 1:100. Samples were incubated for 10, 20, 30, 40, 50, or 60 min, and differences in the FL1 signal were used to determine staining intensity. Optimal FITC-MAb concentration was determined over a range of dilutions from 1:100 to 1:20,000. FITC-MAb was added to samples and incubated for 30 min. Differences in mean FL1 intensity were statistically examined using a bootstrap-t analysis of means of replicate runs for each sample (54). This method was used in place of a Student's t test due to its increased power, even with low sample size, and because it does not rely on assumptions of normality.
Antibody cross-reactivity with other photoautotrophs.
Samples from the following phototrophic cultures were preserved in 1% glutaraldehyde and individually analyzed by FCM to determine baseline characteristics, including SSC, FL1, and FL3 signals for unstained cells: BT3, BT8, M. polymorphus, I. galbana, P. subviridis, C. stigmatophora, and M. pusilla. These test samples served as controls to examine nonspecific binding of the MAb. Algal abundance values in the cultures were estimated by light microscopy to be <106 cells ml−1. Three mixtures of the cultures were prepared as follows and preserved with a 1% final concentration of glutaraldehyde: 250 μl of A. anophagefferens plus 1.75 ml of filtered seawater; 250 μl of each phototrophic culture, except A. anophagefferens, plus 250 μl filtered seawater; and 250 μl of each phototrophic culture plus 250 μl of A. anophagefferens cells. Aliquots of each mixture were filtered onto a 0.2-μm Millipore filter and resuspended in 10 mM PBS-T. Samples were analyzed unstained and then after staining for 30 min with FITC-MAb diluted 1:20,000 (the latter parameters were based on previous optimization steps). Fluorescence signals in the FL1 channel from the unstained cells were used as a baseline to determine if algae other than A. anophagefferens were stained by the antibody.
Comparison of IFCM and hemacytometer counts.
FITC-MAb flow cytometry counts and direct microscopy counts of cultures of A. anophagefferens were compared to determine the accuracy of the IFCM method for enumerating brown-tide cells. A. anophagefferens cells from a culture in exponential growth phase were counted using light microscopy following preservation with glutaraldehyde (1% final concentration). The preserved culture was serially diluted in filtered seawater (0.2-μm-pore filter) to produce a range of abundance values from 1.4 × 103 to 1.4 × 106 cells ml−1, and each dilution was counted using standard practices with a hemacytometer at a magnification of ×400. Each sample was then diluted 1:10 in 10 mM PBS-T for enumeration by IFCM and analyzed in duplicate before and after staining with FITC-MAb at a dilution of 1:1,000 for 30 min.
A comparison was also conducted between IFCM and microscopy-based enumeration of A. anophagefferens cells in natural seawater samples amended with known aliquots of cultured brown-tide algae. Natural seawater was collected from Long Beach, CA (where A. anophagefferens is not known to occur). The samples contained a wide variety of small photoautotrophs. The sample was filtered through 10-μm Nitex mesh to remove large phytoplankton and zooplankton and preserved with glutaraldehyde (1% final concentration). Cultured A. anophagefferens cells of known concentration were added to the sample, and the sample was then serially diluted to achieve final abundance values of 2 × 102 to 6.7 × 105 cells ml−1. The spiked environmental samples were then diluted 1:10 in 10 mM PBS-T and run on the flow cytometer in duplicate after staining with FITC-MAb, added at 1:1,000 and incubated for 30 min.
IFCM versus ELISA enumeration of A. anophagefferens subjected to protistan predation.
An experiment was conducted to compare the accuracy of the IFCM and the ELISA approaches for estimating the abundance of A. anophagefferens in situations where grazing on the cells is a potentially active source of mortality. A culture of A. anophagefferens was grown to late exponential growth phase in modified f/2 medium at 20°C at 50 to 100 μE m−2 s−1. Aliquots of the culture were removed and subjected to the following conditions in duplicate: A. anophagefferens in a light/dark cycle; A. anophagefferens in continuous darkness; A. anophagefferens and Pseudopedinella sp. in a light/dark cycle; A. anophagefferens and Pseudopedinella sp. in continuous darkness. The light/dark cycle was 12:12 h, and treatments kept in the dark were covered with aluminum foil. This experimental design also allowed us to test hypotheses that the mixotrophic Pseudopedinella sp. is an obligate phototroph, requiring light even in the presence of ample prey. Samples of 2 ml were taken twice daily, preserved with glutaraldehyde (1% final concentration), and stored at 4°C in the dark until ELISA and IFCM analyses. Samples for IFCM analysis were diluted 1:10 in 10 mM PBS-T, incubated with 1:1,000 FITC-MAb for 30 min, and analyzed for FL1 fluorescence to determine the abundance of A. anophagefferens cells present based on previously established gating parameters. The ELISA analysis was conducted following the protocol described in Caron et al. (7).
IFCM and ELISA enumeration of A. anophagefferens in environmental samples.
An additional comparison between the ELISA and IFCM methods was made using environmental brown-tide samples collected from several locations in Maryland's coastal bays as part of the State of Maryland's' harmful algae monitoring program. Staff from Assateague National Seashore collected samples during a bloom in 2004 at the following sites: Public Landing, Taylor's Landing, Newport Bay, Tingle's Island, Sinnickson (VA), Green Point, Trappe Creek, and Ferry Landing. Samples were preserved with glutaraldehyde (1% final concentration) and stored and shipped in the dark at 4°C. For each sample the target cells were counted by both the IFCM and ELISA methods.
For the IFCM enumerations, samples were diluted 1:10 in 10 mM PBS-T and consecutive analyses were performed using consistent instrument settings optimized for A. anophagefferens detection. To determine baseline FL1 fluorescence, a gated region was defined using the unstained SSC-FL1 cytogram of an individual sample such that >99% of events were included. Samples were then stained with FITC-MAb at a dilution of 1:1,000, incubated for 30 min, and analyzed for cells exhibiting increased FL1 fluorescence, indicative of stained A. anophagefferens. Cells were identified and counted as A. anophagefferens if they fell within the SSC-FL1 region expected for stained cells (based on cultures) and outside of the unstained region defined for each sample.
RESULTS
Optimizing MAb reactivity and specificity.
Seawater is not a common medium for conducting antibody reactivity tests, and therefore, development of the IFCM method was initiated by comparing the reactivity of FITC-MAb to A. anophagefferens in seawater relative to reactivity in solutions that are commonly employed (e.g., PBS-T). The FL1 fluorescent signal of cultured A. anophagefferens stained with FITC-MAb in seawater was not markedly different from the signal observed for the alga in the absence of antibody in either seawater or PBS-T (Fig. 1A to C). In contrast, a nearly 30-fold increase in FL1 fluorescence was observed for cells resuspended in PBS-T and exposed to the FITC-MAb (compare Fig. 1D to A through C).
FIG. 1.
Efficacy of staining A. anophagefferens cells with FITC-MAb in seawater and PBS-T. Bivariate dot plots of side scatter and green fluorescence are shown. (A) A. anophagefferens cells in seawater without FITC-MAb. (B) A. anophagefferens cells in seawater and stained for 15 min with 1:1,000 FITC-MAb. (C) A. anophagefferens cells removed from seawater by filtration and resuspended in 10 mM PBS-T without FITC-MAb. (D) A. anophagefferens cells removed from seawater by filtration, resuspended in 10 mM PBS-T, and stained for 15 min with 1:1,000 FITC-MAb. Average FL1 signal strength (FITC-MAb) for cells within the gated regions (outlined) is indicated in each panel.
Filtration and resuspension of A. anophagefferens in PBS-T yielded a high level of reactivity to the antibody, but this procedure was not readily applicable to natural water samples because it was cumbersome and time-consuming and resulted in the loss of some cells during recovery and resuspension (centrifugation also resulted in loss of cells). Therefore, we examined the effect of directly diluting seawater samples in PBS-T as an alternative to filtration/centrifugation. A 1:10 dilution of cultured cells in PBS-T yielded FITC staining that was equivalent to that of filtered, resuspended cells (data not shown) and was employed in all further testing as an acceptable alternative.
Incubation of A. anophagefferens cultures with FITC-MAb for 10, 20, 30, 45, or 60 min showed little variation of staining intensity between 10 and 30 min. The mean FL1 signal increased slightly with prolonged incubation times (40 to 60 min), but the increase was not significant (bootstrap-t analysis, P > 0.05), and stained cells were easily distinguishable on flow cytograms from unstained cells, even with shorter incubation times (Table 1). A standard incubation time of 30 min was therefore chosen for subsequent assays.
TABLE 1.
Effect of incubation time on staining of A. anophagefferensa
| Incubation time (min) | Mean green fluorescence (arbitrary units) |
|---|---|
| 10 | 290 |
| 20 | 329 |
| 30 | 341 |
| 40 | 337 |
| 50 | 329 |
| 60 | 325 |
A. anophagefferens cells were diluted 1:10 in 10 mM PBS-T and tested over a range of incubation times. All incubations were performed with a 1:100 FITC-MAb dilution in the dark at room temperature. Values represent the means of green fluorescence signal for A. anophagefferens.
Optimal staining of A. anophagefferens cells in 30-min incubations was determined by examining a range of FITC-MAb concentrations (Table 2). Staining efficacy decreased more than 20-fold over the range of dilutions tested, as indicated by a reduction in mean FL1 fluorescence of the A. anophagefferens cells. Mean FL1 fluorescence values of labeled cells for three dilutions (1:100, 1:1,000, 1:10,000) were significantly greater than fluorescence values at the next higher dilutions (1:500, 1:5,000, and 1:20,000, respectively; P < 0.05). No significant differences were observed between the dilutions 1:500 versus 1:1,000 and 1:5,000 versus 1:1,000 (Table 2). Furthermore, the background fluorescence of FITC-MAb in PBS-T within the gated region of the SSC-FL1 cytogram that was characteristic of A. anophagefferens cells constituted 28% of the total fluorescence events at a dilution of 1:100 of the FITC-MAb, whereas the 1:1,000 dilution resulted in no events within the gated region. Therefore, a final FITC-MAb concentration of 1:1,000 was chosen for all subsequent analyses as a compromise between completeness of staining, low background fluorescence, and conservation of the expensive monoclonal antibody.
TABLE 2.
Effect of MAb concentration on staining of A. anophagefferensa
| FITC-MAb concn (MAb:sample) | Mean green fluorescence (arbitrary units)b |
|---|---|
| 1:100 | 698* |
| 1:500 | 382 |
| 1:1,000 | 302* |
| 1:5,000 | 66 |
| 1:10,000 | 43* |
| 1:20,000 | 29 |
A. anophagefferens cells were diluted 1:10 in 10 mM PBS-T and tested over a range of FITC-MAb concentrations. All incubations were performed for 30 min in the dark at room temperature. Values represent the means of the green fluorescence (FL1) signal for the A. anophagefferens gated region.
Asterisks indicate concentrations at which units of mean green fluorescence of stained cells differed from that of more dilute concentrations based on pair-wise bootstrap-t analysis of means (P < 0.05).
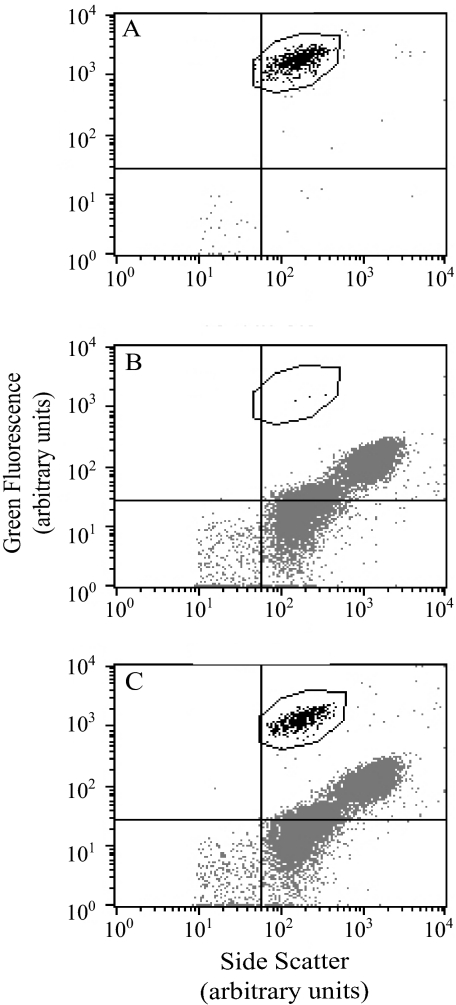
Extensive cross-reactivity tests of the antibody were previously conducted in an ELISA format (7), and the antibody was found to be highly specific in that format. Cross-reactivity of the MAb for nontarget algae in the flow cytometry method was also found to be exceptionally low and easily detected from reactivity with A. anophagefferens culture at a dilution of 1:20,000 (Fig. 2). A cytogram of SSC-FL1 exhibited a strong FL1 signal from stained A. anophagefferens cells (Fig. 2A). The same plot of a stained sample containing seven similarly sized phytoplankton, exclusive of A. anophagefferens (Fig. 2B), showed very few cells within the A. anophagefferens region (three gated events) but a large population of cells displaying a wide range of side scatter with very low FL1 signal. Stained samples containing the seven phytoplankton species together with A. anophagefferens exhibited two distinct populations, one within the A. anophagefferens gated region and another culture corresponding to the nontarget mixture of phytoplankton species (Fig. 2C). Identical volumes of A. anophagefferens culture were dispensed into the samples represented in Fig. 2B and C. The abundance of A. anophagefferens cells in the mixed sample corresponded to approximately 88% of that in the sample containing only A. anophagefferens, when corrected for the volume of sample analyzed.
FIG. 2.
Cross-reactivity of FITC-MAb with A. anophagefferens cells and a mixture of seven nontarget algae. (A) Dot plot of side scatter and green fluorescence for A. anophagefferens stained for 30 min with 1:20,000 FITC-MAb in PBS-T. The gated region (outlined) was used in subsequent plots to determine nonspecific staining. (B) A mixture of seven nontarget algae stained for 30 min with 1:20,000 FITC-MAb in PBS-T. (C) A mixture of seven nontarget algae and A. anophagefferens stained for 30 min with 1:20,000 FITC-MAb in PBS-T.
Comparison of flow cytometry and direct microscopy.
The accuracy of the flow cytometry approach for enumerating A. anophagefferens cells was examined by comparison with direct microscopy in a dilution series of pure cultures of the alga and in natural seawater samples spiked with cultured A. anophagefferens. Standard curves generated for both types of samples showed excellent agreement with microscopy counts over more than three orders of magnitude (r2 > 0.99; Fig. 3). Accurate assessment of the sample volume analyzed by the flow cytometer was essential for obtaining comparable numerical abundance values between the two methods. The use of known concentrations of fluorescent beads to determine the volume analyzed was examined in this study, but the gravimetric approach provided a faster, yet equally accurate, estimate of the volume analyzed, particularly with natural samples.
FIG. 3.
Standard curves of flow cytometry analysis and direct microscopy counts of A. anophagefferens. Samples from a serially diluted culture (A) and natural seawater spiked with cultured A. anophagefferens cells (B) were counted using a hemacytometer and standard methods. Subsamples of the same dilution series were stained with FITC-MAb at a concentration of 1:1,000 for 30 min and analyzed using flow cytometry. Slope and r2 values were determined from linear regression.
IFCM versus ELISA during protistan predation.
The purpose of the grazing experiment was twofold: first, to compare counts of A. anophagefferens using ELISA and IFCM and the same monoclonal antibody when populations are exposed to grazing pressure; second, to test growth of the mixotrophic Pseudopedinella sp. under light and dark culture conditions, thus determining if the organism is an obligate or facultative phototroph. The results indicated marked differences between the two counting methods under some culture conditions. Estimates by the two methods of the abundance values of A. anophagefferens in grazed cultures differed by approximately an order of magnitude in the light (Fig. 4A), as growth of the phagotrophic consumer progressed (Fig. 4C). The ELISA and IFCM counts from the grazed cultures exposed to light diverged at approximately 130 h after the addition of the phagotrophs, and these differences became more dramatic until 225 h when the Pseudopedinella sp. approached stationary growth phase. In contrast, both methods yielded similar estimates for cultures of A. anophagefferens when cultured without the Pseudopedinella sp. in the light. Slight increases in the abundance values of the brown-tide alga were observed over the course of the experiment (Fig. 4A).
FIG. 4.
Comparison of IFCM and ELISA techniques for the enumeration of A. anophagefferens cells in a grazing experiment. Duplicate cultures of A. anophagefferens were exposed to grazing by the phagotrophic alga Pseudopedinella sp. under conditions of light (A) or continuous dark (B). Samples were taken from all treatments at regular intervals and preserved with 1% glutaraldehyde. The abundance of A. anophagefferens cells was determined by using IFCM (circles) and ELISA (triangles) in the presence (filled symbols) and absence (open symbols) of the phagotrophic protist. The abundance of Pseudopedinella sp. in each of the grazed treatments was determined by IFCM (C). Vertical bars indicate the range for duplicate cultures.
Differences between the two methods were less dramatic for algal cultures maintained in continuous darkness (Fig. 4B). The two methods yielded similar estimates of A. anophagefferens abundance when cultured in continuous darkness in the absence of the predator (Fig. 4B) and showed minor decreases in the abundance of the brown-tide alga over the experimental period. In cultures with consumers present, enumerations from the two methods agreed closely until approximately 200 h, when IFCM counts in the grazed cultures (Fig. 4B) yielded consistently lower cell concentrations of A. anophagefferens than the ELISA technique. Pseudopedinella sp. grew at a substantially faster rate and attained much higher densities when grown in the light, indicating obligate phototrophy for population growth of this phagotrophic alga (Fig. 4C).
A. anophagefferens in environmental samples using IFCM and ELISA.
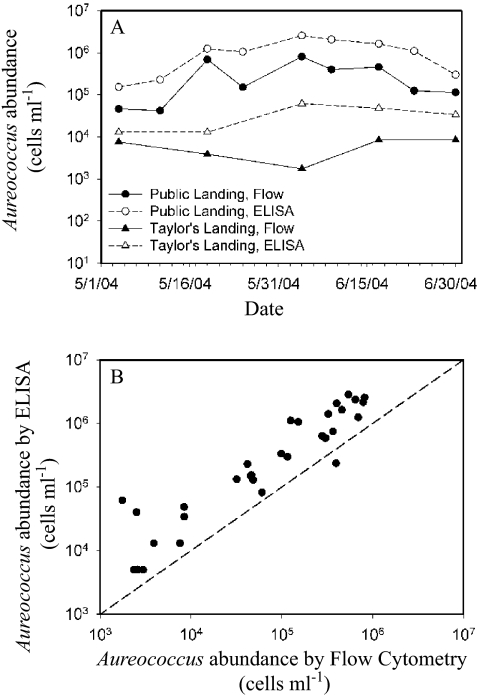
Natural seawater samples collected from several coastal locations in Maryland during the summer of 2004 were analyzed using IFCM and ELISA. Representative counts for two locations (Public Landing, Taylor Landing) indicated that estimates of A. anophagefferens abundance for both sites using ELISA were consistently higher than those obtained using the IFCM method, although the magnitude of differences varied with site and date (Fig. 5A). At the Public Landing site, both enumeration methods yielded a similar overall pattern, and all paired absolute cell densities were within an order of magnitude of each other. A similar result was observed for the Taylor Landing site, with one exception on 4 June 2004, when the estimates from the ELISA and those of the IFCM method showed a substantial difference (the IFCM estimate was more than an order of magnitude lower). A plot of paired data from all sites and sampling dates (38 data points in total) indicated a consistent deviation from the expected 1:1 relationship between the two methods (Fig. 5B).
FIG. 5.
Comparison of IFCM and ELISA techniques for the enumeration of natural brown-tide samples collected from a variety of coastal sites by the Maryland Department of Natural Resources during the summer of 2004. (A) Samples from two sites with distinctly different abundance levels of A. anophagefferens (Public Landing and Taylor's Landing, triangles) were preserved in 1% glutaraldehyde and analyzed for A. anophagefferens abundance by IFCM (filled symbols) and ELISA (open symbols). (B) The counts from both techniques were compared relative to an expected 1:1 relationship (dashed line).
DISCUSSION
Eukaryotic algae less than 10 μm in size from freshwater and marine ecosystems have historically been problematic to identify and enumerate. These species typically possess few useful criteria for morphological identification. Therefore, traditional light microscopy methods commonly applied in ecological studies have proven ineffective for distinguishing among these species or for obtaining accurate estimates of their abundance. Toward this goal, several techniques have been developed during the past few decades to enumerate and identify minute algae in natural seawater samples. These methods have relied increasingly on genetic and immunological approaches, often combined with instruments for improved visualization, rapid enumeration, and high sample throughput.
A. anophagefferens, the cause of harmful “brown tides” in estuaries of the mid-Atlantic United States, is no exception to this trend. The detection and enumeration of A. anophagefferens have received wide attention and research effort during the past two decades due to the considerable ecological and economic impacts of blooms of this tiny pelagophyte (6, 16, 19, 20, 22, 24, 32, 33, 37, 52). The accurate and efficient detection of this small, morphologically nondescript unicellular alga has remained a challenge for both research and monitoring efforts.
Several molecular techniques have been developed for the identification and enumeration of A. anophagefferens. These approaches have included immunology-based (3, 7, 30) and genetics-based detection protocols (41), and each method has inherent advantages, disadvantages, and appropriate ecological applications. The immunofluorescence microscopy method of Anderson et al. (3) provided the first method to accurately identify and count A. anophagefferens organisms in natural algal assemblages through the use of immunofluorescence microscopy. This method employs a polyclonal antiserum, and it has been widely employed. It has a reasonably low limit of detection (approximately a few hundred cells ml−1) but is extremely time consuming and inconsistent (it depends on the reactivity level of the serum, the amount of serum used, and differences in individual microscope work). These drawbacks were addressed in a subsequent immunological approach that employed a highly specific monoclonal antibody in an ELISA format. The ELISA technique has a high rate of sample throughput relative to the polyclonal microscopy method but also has a somewhat higher limit of detection (approximately 5,000 cells ml−1). A recent application of the same monoclonal antibody enabled single-cell detection by atomic force microscopy (30). This approach demonstrates the potential to build sensors with single-cell resolution, but its application to natural water samples is not yet technically feasible. Finally, the use of real-time qPCR provides extremely high sensitivity, a wide dynamic range, and relatively high sample throughput (41). However, the qPCR method relies upon an assumption of invariant gene copy number to enable quantification of cell abundance in natural samples, an assumption which is not always accurate across time and space. For example, temporal changes in 18S copy number have been observed for cultures of Lingulodinium polyedrum (36). It is an issue similar to that of the ELISA methods, in which the antigenicity is assumed to be the same between cultures and field populations (and that this correlates to “whole” cells) without the ability to physically resolve individual target cells.
We sought to address the aforementioned methods' disadvantages by developing a method that would be easy to use (i.e., would require a low level of technical training), have a relatively low limit of detection (≈103 cells ml−1), allow enumeration over a wide range of algal abundance levels, and alleviate the use of highly variable proxies (antigenicity, gene copy number). IFCM was chosen because of the availability of the highly specific monoclonal antibody previously produced for the ELISA method (7) and because flow cytometry has become a common approach for assessing assemblages of minute phytoplankton in marine and freshwater ecosystems (31, 40). Additionally, flow cytometry facilitates identification of cells based on multiple parameters simultaneously, allowing for the combination of species-specific identifiers, whether molecular or immunological, and more basic morphological characteristics to accurately determine cell abundance levels.
Immunofluorescent staining of protistan cells has been conducted without the transfer of cells to a defined buffer (13), but initial tests with A. anophagefferens cells indicated a complete loss of reactivity when the cells were probed in seawater (Fig. 1). Preliminary investigations suggested that this effect was not merely a result of salinity per se but a result of the specific ionic composition of the seawater (data not shown). Additional tests indicated that resuspension of A. anophagefferens in PBS-T demonstrated strong reactivity with the MAb. Furthermore, dilution of seawater samples in PBS-T was adopted as an easier and faster approach to restoring MAb reactivity than filtration/resuspension of A. anophagefferens cells. Dilution resulted in a somewhat reduced sensitivity of the method (a practical lower limit of detection of 103 cells ml−1) but this value is still 3 orders of magnitude less than bloom abundance values of the alga in nature and five times more sensitive than the ELISA method.
The specificity and accuracy of the IFCM method were established through cross-reactivity tests and comparisons of IFCM with traditional microscopy counts and the ELISA method using cultures and natural samples amended with known concentrations of cultured A. anophagefferens. Conjugation of an antibody to another molecule may alter its reactivity; yet the MAb remained highly reactive and specific to A. anophagefferens after conjugation to FITC (Fig. 2). The highly linear relationships between counts of the brown-tide alga by IFCM and other methods demonstrated the efficacy of the method under these conditions (Fig. 3).
Nevertheless, a very significant disparity was observed between the abundance levels of A. anophagefferens obtained using the IFCM method and estimates based on the ELISA technique during the grazing experiment with Pseudopedinella sp. (Fig. 4). The ELISA method yielded abundance estimates that were approximately an order of magnitude higher than the IFCM estimates during the latter half of the grazing experiment, when many of the algal cells had been ingested. This disparity may be explained by the retention of antigenic character by fragments of A. anophagefferens cells remaining after consumption, digestion, and elimination by the phagotrophic consumer. It is unlikely that egested material would be recorded as A. anophagefferens cells by the IFCM method because size and granularity contribute to the identification as well as the immunofluorescent signal (Fig. 2A), and the size of such small particulates would likely fall substantially outside of the gated regions established using whole cells. However, egested particulate material would be collected along with intact cells on filters during the ELISA technique, and any antigenic properties remaining in the egested material would result in an overestimation of the number of A. anophagefferens cells. This interpretation is consistent with the observation that the two methods yielded comparable estimates of abundance in the absence of consumers (Fig. 4A and B) and for most of the experimental period in continuous darkness when consumers were relatively inactive (Fig. 4B).
The results of the grazing experiment have potentially important implications for the interpretation of estimates of A. anophagefferens in field samples. Natural populations of A. anophagefferens appear to experience reduced grazing pressure by microbial grazers during the early phases of the development of brown tides (8, 21, 23). In these situations, one could expect close agreement between abundances estimated by the ELISA and IFCM methods. As a bloom progresses, however, consumption by microbial predators and/or lysis by viruses may have significant impacts on the number of viable A. anophagefferens organisms in the water (8, 9, 17, 21, 23, 45). These trophic interactions could significantly reduce the number of intact cells that would be observed using IFCM, but cell fragments could retain sufficient antigenic character that the ELISA method might overestimate the abundance of intact cells. Therefore, IFCM counts during bloom demise should provide more accurate estimates of abundance than the ELISA method.
Interestingly, the ELISA method also provided higher estimates of the abundance levels of A. anophagefferens than the immunofluorescence microscopy method described by Anderson et al. (3), which uses a polyclonal antiserum, when applied to natural samples from Long Island estuaries (7). Reduced or variable reactivity of different polyclonal antisera may explain some of the differences between the method of Anderson et al. and the ELISA method when analyzing natural water samples. However, like the IFCM method, the immunofluorescence microscopy method also relies on visual recognition of whole, labeled cells. The presence of cell fragments that retained antigenic quality (resulting from protistan grazing or viral lysis) may explain differences between estimates of the abundances of A. anophagefferens by ELISA and IFCM. Slightly higher abundances of the brown-tide alga were also observed for the ELISA method compared to counts performed using a hemacytometer in that study, in accordance with the above explanation. Hence the IFCM method is more compatible with results from the polyclonal method and thus more comparable to the thresholds of biological impacts that have been documented in the literature and used by managers to assess potential brown-tide impacts (18).
A comparison of the IFCM and ELISA techniques for enumerating A. anophagefferens organisms in environmental samples from Maryland's coastal bays further indicated a consistent overestimation of the abundance levels of A. anophagefferens by the ELISA method (Fig. 5). The largest differences observed between the two methods among the field samples from Maryland estuaries were consistent with the differences observed when analyzing the grazing experiment with Pseudopedinella (approximately 1 order of magnitude). This showed that the IFCM method was an accurate and relatively simple technique for the rapid enumeration of A. anophagefferens cells in natural samples over a wide range of abundances.
The IFCM technique for counting A. anophagefferens described here has several advantages for application to natural samples. Beyond the cost of the instrument, the cost to analyze individual samples is modest. However, numerous laboratories and research groups studying phytoplankton have access to flow cytometers, and many cytometers are now remarkably user friendly, providing straightforward instrument controls, powerful imaging software, and statistical analyses with a minimal amount of training. The IFCM method provides rapid analyses (30-min incubation period and 60-s analysis sample−1), and many samples can be incubated at the same time and analyzed in a single session on the cytometer. In contrast, the polyclonal method of Anderson et al. (3) requires a full day of analysis to processes approximately 10 samples. The ELISA method of Caron et al. (7) provides higher throughput (tens of samples h−1), but it is cost effective only in large batches. The practical lower limit of detection of the IFCM method is approximately 103 cells ml−1 (sufficient to characterize subbloom abundance levels of the alga), and measurements of lower abundance levels are possible with extended-analysis cytometric run times or simple concentration procedures. Finally, the IFCM approach is amenable to modifications for use in monitoring programs and research efforts on other ecologically important phototrophic and heterotrophic protists for which specific antibodies exist. Overall, the IFCM method for characterizing A. anophagefferens populations in natural water samples has applicability to a number of research and monitoring needs and contributes to the growing cadre of tools available for studying this and other important phytoplankton species.
Acknowledgments
We gratefully acknowledge Tim McLean for initial antibody testing and helpful discussions on method development.
Funding for this work was provided by National Science Foundation grant CCR-0120778 (to D.A.C.).
Footnotes
Published ahead of print on 26 September 2008.
REFERENCES
- 1.Anderson, D. M., P. M. Glibert, and J. M. Burkholder. 2002. Harmful algal blooms and eutrophication: nutrient sources, composition, and consequences. Estuaries 25:704-726. [Google Scholar]
- 2.Anderson, D. M., B. A. Keafer, D. M. Kulis, R. M. Waters, and R. Nuzzi. 1993. An immunofluorescent survey of the brown tide chrysophyte Aureococcus anophagefferens along the northeast coast of the United States. J. Plankton Res. 15:563-580. [Google Scholar]
- 3.Anderson, D. M., D. M. Kulis, and E. M. Cosper. 1989. Immunofluorescent detection of the brown tide organism Aureococcus anophagefferens, p. 265-294. In E. M. Cosper, V. M. Bricelj, and E. J. Carpenter (ed.), Novel phytoplankton blooms: causes and impacts of recurrent brown tides and other unusual blooms, vol. 35. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 4.Azanza, R. V., Y. Fukuyo, L. G. Yap, and H. Takayama. 2005. Prorocentrum minimum bloom and its possible link to a massive fish kill in Bolinao, Pangasinan, Northern Philippines. Harmful Algae 4:519-524. [Google Scholar]
- 5.Bologna, P. A. X., M. L. Fetzer, S. McDonnell, and E. M. Moody. 2005. Assessing the potential benthic-pelagic coupling in episodic blue mussel (Mytilus edulis) settlement events within eelgrass (Zostera marina) communities. J. Exp. Mar. Biol. Ecol. 316:117-131. [Google Scholar]
- 6.Bricelj, V. M. 1997. Aureococcus anophagefferens: causes and ecological consequences of brown tides in US mid-Atlantic coastal waters. Limnol. Oceanogr. 42:1023-1038. [Google Scholar]
- 7.Caron, D. A., M. R. Dennett, D. M. Moran, R. A. Schaffner, D. J. Lonsdale, C. J. Gobler, R. Nuzzi, and T. I. McLean. 2003. Development and application of a monoclonal-antibody technique for counting Aureococcus anophagefferens, an alga causing recurrent brown tides in the mid-Atlantic United States. Appl. Environ. Microbiol. 69:5492-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caron, D. A., C. J. Gobler, D. J. Lonsdale, R. M. Cerrato, R. A. Schaffner, J. M. Rose, N. J. Buck, G. Taylor, K. R. Boissonneault, and R. Mehran. 2004. Microbial herbivory on the brown tide alga, Aureococcus anophagefferens: results from natural ecosystems, mesocosms and laboratory experiments. Harmful Algae 3:439-457. [Google Scholar]
- 9.Caron, D. A., E. L. Lim, H. Kunze, E. M. Cosper, and D. M. Anderson. 1989. Trophic interactions between nano- and microzooplankton and the “Brown Tide,” p. 265-294. In E. M. Cosper, V. M. Bricelj, and E. J. Carpenter (ed.), Novel phytoplankton blooms: causes and impacts of recurrent brown tides and other unusual blooms, vol. 35. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 10.Cosper, E. M., W. C. Dennison, E. J. Carpenter, V. M. Bricelj, J. G. Mitchell, S. H. Kuenstner, D. Colflesh, and M. Dewey. 1987. Recurrent and persistent brown tide blooms perturb coastal marine ecosystem. Estuaries 10:284-290. [Google Scholar]
- 11.Cucci, T. L., S. E. Shumway, W. S. Brown, and C. R. Newel. 1989. Using phytoplankton and flow cytometry to analyze grazing by marine organisms. Cytometry 10:659-669. [DOI] [PubMed] [Google Scholar]
- 12.Cucci, T. L., and M. E. Sieracki. 2001. Effects of mismatched refractive indices in aquatic flow cytometry. Cytometry 44:173-178. [DOI] [PubMed] [Google Scholar]
- 13.Delauney, A., G. Gargala, X. Li, L. Favennec, and J. J. Ballet. 2000. Quantitative flow cytometric evaluation of maximal Cryptosporidium parvum oocyst infectivity in a neonate mouse model. Appl. Environ. Microbiol. 66:4315-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dennison, W., G. Marshall, and C. Wigand. 1989. Effect of “brown tide” shading on eelgrass (Zostera marina L.) distributions, p. 675-692. In E. M. Cosper, V. M. Bricelj, and E. J. Carpenter (ed.), Novel phytoplankton blooms: causes and impacts of recurrent brown tides and other unusual blooms. Springer-Verlag, Berlin, Germany.
- 15.Fouchet, P., C. Jayat, Y. Hechard, M.-H. Ratinaud, and G. Frelat. 1993. Recent advances of flow cytometry in fundamental and applied microbiology. Biol. Cell 78:95-109. [DOI] [PubMed] [Google Scholar]
- 16.Gastrich, M. D., R. Lathrop, S. Haag, M. P. Weinstein, M. Danko, D. A. Caron, and R. Schaffner. 2004. Assessment of brown tide blooms, caused by Aureococcus anophagefferens, and contributing factors in New Jersey coastal bays: 2000-2002. Harmful Algae 3:305-320. [Google Scholar]
- 17.Gastrich, M. D., J. A. Leigh-Bell, C. J. Gobler, O. R. Anderson, S. W. Wilhelm, and M. Bryan. 2004. Viruses as potential regulators of regional brown tide blooms caused by the alga, Aureococcus anophagefferens. Estuaries 27:112-119. [Google Scholar]
- 18.Gastrich, M. D., and C. E. Wazniak. 2002. A Brown Tide Bloom Index based on the potential harmful effects of the brown tide alga, Aureococcus anophagefferens. AEHM 5:435-441. [Google Scholar]
- 19.Glibert, P. M., R. Magnien, M. W. Lomas, J. Alexander, C. L. Fan, E. Haramoto, M. Trice, and T. M. Kana. 2001. Harmful algal blooms in the Chesapeake and coastal bays of Maryland, USA: comparison of 1997, 1998, and 1999 events. Estuaries 24:875-883. [Google Scholar]
- 20.Gobler, C. J., G. E. Boneillo, C. J. Debenham, and D. A. Caron. 2004. Nutrient limitation, organic matter cycling, and plankton dynamics during an Aureococcus anophagefferens bloom. Aquatic Microb. Ecol. 35:31-43. [Google Scholar]
- 21.Gobler, C. J., S. Deonarine, J. Leigh-Bell, M. D. Gastrich, O. R. Anderson, and S. W. Wilhelm. 2004. Ecology of phytoplankton communities dominated by Aureococcus anophagefferens: the role of viruses, nutrients, and microzooplankton grazing. Harmful Algae 3:471-483. [Google Scholar]
- 22.Gobler, C. J., D. J. Lonsdale, and G. L. Boyer. 2005. A review of the causes, effects, and potential management of harmful brown tide blooms caused by Aureococcus anophagefferens (Hargraves et Sieburth). Estuaries 28:726-749. [Google Scholar]
- 23.Gobler, C. J., M. J. Renaghan, and N. J. Buck. 2002. Impacts of nutrients and grazing mortality on the abundance of Aureococcus anophagefferens during a New York brown tide bloom. Limnol. Oceanogr. 47:129-141. [Google Scholar]
- 24.Gobler, C. J., and S. A. Sanudo-Wilhelmy. 2001. Temporal variability of groundwater seepage and brown tide blooms in a Long Island embayment. Marine Ecol. Prog. Ser. 217:299-309. [Google Scholar]
- 25.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 29-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Publishing, New York, NY.
- 26.Reference deleted.
- 27.Hoagland, P., D. M. Anderson, Y. Kaoru, and A. W. White. 2002. The economic effects of harmful algal blooms in the United States: estimates, assessment issues, and information needs. Estuaries 25:819-837. [Google Scholar]
- 28.Kemp, W. M., W. R. Boynton, J. E. Adolf, D. F. Boesch, W. C. Boicourt, G. Brush, J. C. Cornwell, T. R. Fisher, P. M. Glibert, J. D. Hagy, L. W. Harding, E. D. Houde, D. G. Kimmel, W. D. Miller, R. I. E. Newell, M. R. Roman, E. M. Smith, and J. C. Stevenson. 2005. Eutrophication of Chesapeake Bay: historical trends and ecological interactions. Marine Ecol. Prog. Ser. 303:1-29. [Google Scholar]
- 29.Kroger, K., J. P. A. Gardner, A. A. Rowden, and R. G. Wear. 2006. Long-term effects of a toxic algal bloom on subtidal soft-sediment macroinvertebrate communities in Wellington Harbour, New Zealand. Estuar. Coast. Shelf Sci. 67:589-604. [Google Scholar]
- 30.Lee, A. S., M. Mahapatro, D. A. Caron, A. A. G. Requicha, B. A. Stauffer, M. E. Thompson, and C. W. Zhou. 2006. Whole-cell sensing for a harmful bloom-forming microscopic alga by measuring antibody-antigen forces. IEEE Trans. Nanobioscience 5:149-156. [DOI] [PubMed] [Google Scholar]
- 31.Legendre, L., C. Courties, and M. Troussellier. 2001. Flow cytometry in oceanography 1989-1999: environmental challenges and research trends. Cytometry 44:164-172. [DOI] [PubMed] [Google Scholar]
- 32.Lomas, M. W., P. M. Glibert, D. A. Clougherty, D. R. Huber, J. Jones, J. Alexander, and E. Haramoto. 2001. Elevated organic nutrient ratios associated with brown tide algal blooms of Aureococcus anophagefferens (Pelagophyceae). J. Plankton Res. 23:1339-1344. [Google Scholar]
- 33.Lomas, M. W., and C. J. Gobler. 2004. Aureococcus anophagefferens research: 20 years and counting. Harmful Algae 3:273-277. [Google Scholar]
- 34.Lonsdale, D. J., E. M. Cosper, W.-S. Kim, M. Doall, A. Divadeenam, and S. H. Jonasdottir. 1996. Food web interactions in the plankton of Long Island bays, with preliminary observations on brown tide effects. Marine Ecol. Prog. Ser. 134:247-263. [Google Scholar]
- 35.MacIntyre, H. L., M. W. Lomas, J. Cornwell, D. J. Suggett, C. J. Gobler, E. W. Koch, and T. M. Kana. 2004. Mediation of benthic-pelagic coupling by microphytobenthos: an energy- and material-based model for initiation of blooms of Aureococcus anophagefferens. Harmful Algae 3:403-437. [Google Scholar]
- 36.Moorthi, S. D., P. D. Countway, B. A. Stauffer, and D. A. Caron. 2006. Use of quantitative real-time PCR to investigate the dynamics of the red tide dinoflagellate Lingulodinium polyedrum. Microbiol. Ecol. 52:136-150. [DOI] [PubMed] [Google Scholar]
- 37.Nuzzi, R., and R. A. Waters. 2004. Long-term perspective on the dynamics of brown tide blooms in Long Island coastal bays. Harmful Algae 3:279-293. [Google Scholar]
- 38.Olson, R. J., S. W. Chisholm, E. R. Zettler, M. A. Altabet, and J. A. Dusenberry. 1990. Spatial and temporal distributions of prochlorophyte picoplankton in the North Atlantic Ocean. Deep Sea Res. 37:1033-1051. [Google Scholar]
- 39.Olson, R. J., S. W. Chisholm, E. R. Zettler, and E. V. Armbrust. 1990. Pigments, size and distribution of Synechococcus in the North Atlantic and Pacific Oceans. Limnol. Oceanogr. 35:45-58. [Google Scholar]
- 40.Peperzak, L., E. G. Vrieling, B. Sandee, and T. Rutten. 2000. Immuno flow cytometry in marine phytoplankton research. Sci. Mar. 64:165-181. [Google Scholar]
- 41.Popels, L. C., S. C. Cary, D. A. Hutchins, R. Forbes, F. Pustizzi, C. J. Gobler, and K. J. Coyne. 2003. The use of quantitative polymerase chain reaction for the detection and enumeration of the harmful alga Aureococcus anophagefferens in environmental samples along the United States East Coast. Limnol. Oceanogr. Methods 1:92-102. [Google Scholar]
- 41a.Ramsdell, J. S., D. M. Anderson, and P. M. Glibert. 2005. HARRNESS, 2005. Harmful algal research and response: a national environmental science strategy 2005-2015. Ecological Society of America, Washington, DC.
- 42.Rose, J. M., A. David, M. E. Sieracki, and N. Poulton. 2004. Counting heterotrophic nanoplanktonic protists in cultures and aquatic communities by flow cytometry. Aquatic Microb. Ecol. 34:263-277. [Google Scholar]
- 43.Sengco, M. R., and D. M. Anderson. 2004. Controlling harmful algal blooms through clay flocculation. J. Eukaryot. Microbiol. 51:169-172. [DOI] [PubMed] [Google Scholar]
- 44.Sieburth, J. M., P. W. Johnson, and P. E. Hargraves. 1988. Ultrastructure and ecology of Aureococcus-anopha gefferens gen-et-sp-nov (Chrysophyceae): the dominant picoplankter during a bloom in Narragansett Bay, Rhode Island, summer 1985. J. Phycol. 24:416-425. [Google Scholar]
- 45.Sieracki, M. E., C. J. Gobler, T. L. Cucci, E. C. Thier, I. C. Gilg, and M. D. Keller. 2004. Pico- and nanoplankton dynamics during bloom initiation of Aureococcus in a Long Island, N.Y. bay. Harmful Algae 3:459-470. [Google Scholar]
- 46.Simon, N., L. Campbell, E. Ornolfsdottir, R. Groben, L. Guillou, M. Lange, and L. K. Medlin. 2000. Oligonucleotide probes for the identification of three algal groups by dot blot and fluorescent whole-cell hybridization. J. Eukaryot. Microbiol. 47:76-84. [DOI] [PubMed] [Google Scholar]
- 47.Tettelbach, S. T., and P. Wenczel. 1993. Reseeding efforts and the status of bay scallop Argopecten irradians (Lamarck, 1819) populations in New York following the occurrence of brown tide algal blooms. J. Shellfish Res. 12:423-431. [Google Scholar]
- 48.Troussellier, M., C. Courties, and A. Vaquer. 1993. Recent applications of flow cytometry in aquatic microbial ecology. Biol. Cell 78:111-121. [DOI] [PubMed] [Google Scholar]
- 49.Veal, D. A., D. Deere, B. Ferrari, J. Piper, and P. V. Attfield. 2000. Fluorescence staining and flow cytometry for monitoring microbial cells. J. Immunol. Methods 243:191-210. [DOI] [PubMed] [Google Scholar]
- 50.Vrieling, E. G., and D. M. Anderson. 1996. Immunofluorescence in phytoplankton research: applications and potential. J. Phycol. 32:1-16. [Google Scholar]
- 51.Vrieling, E. G., G. Vriezekolk, W. W. C. Gieskes, M. Veenhuis, and W. Harder. 1996. Immuno-flow cytometric identification and enumeration of the ichthyotoxic dinoflagellate Gyrodinium aureolum Hulburt in artificially mixed algal populations. J. Plankton Res. 18:1503-1512. [Google Scholar]
- 52.Wazniak, C. E., and P. M. Glibert. 2004. Potential impacts of brown tide, Aureococcus anophagefferens, on juvenile hard clams, Mercenaria mercenaria, in the coastal bays of Maryland, USA. Harmful Algae 3:321-329. [Google Scholar]
- 53.West, N. J., R. Bacchieri, G. Hansen, C. R. Tomas, P. Lebaron, and H. Moreau. 2006. Rapid quantification of the toxic alga Prymnesium parvum in natural samples by use of a specific monoclonal antibody and solid-phase cytometry. Appl. Environ. Microbiol. 72:860-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilcox, R. R. 2003. Applying contemporary statistical techniques. Academic Press, New York, NY.
- 55.Yu, Z. M., M. R. Sengco, and D. M. Anderson. 2004. Flocculation and removal of the brown tide organism, Aureococcus anophagefferens (Chrysophyceae), using clays. J. Appl. Phycol. 16:101-110. [Google Scholar]