Abstract
Geobacter sulfurreducens reduced Ag(I) (as insoluble AgCl or Ag+ ions), via a mechanism involving c-type cytochromes, precipitating extracellular nanoscale Ag(0). These results extend the range of metals known to be reduced by Geobacter species and offer a method for recovering silver from contaminated water as potentially useful silver nanoparticles.
Silver is an element that has been used widely in industrial processes as diverse as photographic processing, catalysis, mirror production, electroplating, alkaline battery production, and jewelry making. Against the backdrop of increasing antibiotic resistance in pathogenic “superbugs,” it has also attracted much recent interest as a biocide (3, 6, 18). It has no known physiological functions and can exist in several oxidation states, although it is most commonly encountered in its elemental [Ag(0)] and monovalent (Ag+) forms. Although nanoscale elemental Ag(0) is finding increasing use as a biocide, for example in wound dressings and as an antimicrobial coating on consumer products, little is known about its mode of toxicity. Ionic Ag(I), in contrast, is known to disrupt the respiratory chain in Escherichia coli (1) and inhibit the exchange of phosphate and its uptake (16). It has also been linked to copper metabolism in Escherichia coli, potentially competing with copper binding sites on the cell surface and subsequent copper transport into the cell (4). The toxicity of silver is not limited to prokaryotes, however, as long-term exposure in humans can cause argyria, impaired night vision, and abdominal pain (13, 14). Thus, the treatment of waters contaminated with silver is important to both recover this valuable metal and also prevent environmental damage and potential uptake into the food chain.
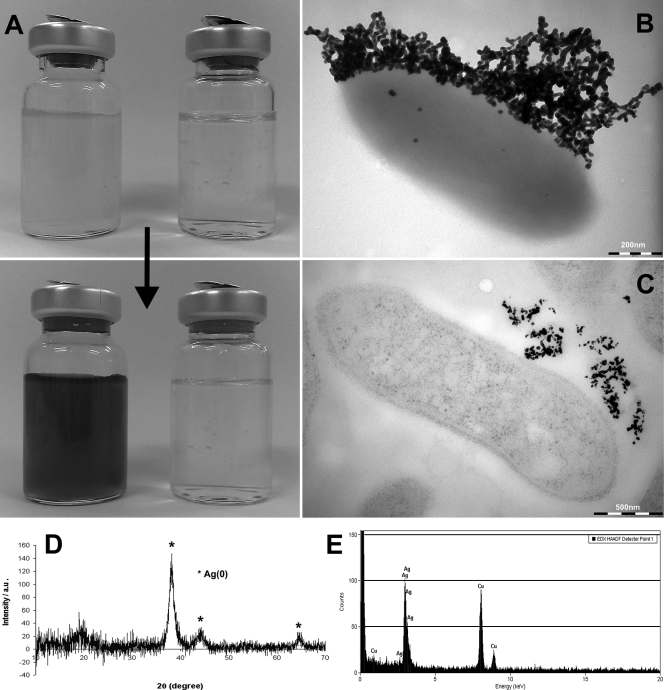
Despite the current interest in microbial interactions with silver (6, 9, 19, 20), there has been little work on the microbial redox cycling of this metal. This is surprising given the obvious potential of metal-reducing microorganisms, such as Fe(III)-reducing bacteria of the genus Geobacter, to reduce Ag(I) to Ag(0), capturing the metal from solution. Thus, to gain insight into the biological reduction of Ag(I), late-exponential-phase cultures of Geobacter sulfurreducens, grown anaerobically at 30°C in defined medium with 25 mM acetate and 40 mM fumarate as the electron donor and acceptor, respectively (11), were supplemented with 200 μM Ag(I) as AgNO3. Within seconds of adding the silver, there was a marked change in color of the cultures, which turned from pale red to dark brown/black (Fig. 1A), irrespective of ambient light intensity, suggesting that photoreduction of Ag(I) was not involved. Collection of the cells and black precipitate by centrifugation (14,400 × g for 4 min) and analysis using a Bruker D8 Advance X-ray diffractometer (Bruker AXS Ltd., Coventry, United Kingdom) with Cu Kα radiation, a step size of 0.02° and a scan time of 5 to 10 s, identified the black precipitate as elemental silver (Fig. 1D). Black precipitates of elemental silver were not formed when Ag(I) was added to autoclaved control cultures or culture supernatants (Fig. 1A), confirming that the reduction of Ag(I) to Ag(0) was enzymatically catalyzed by the living cells and was not mediated by an abiotic mechanism. Interestingly, in fresh or spent medium, a white precipitate which was subsequently identified as AgCl by X-ray diffraction (data not shown) formed upon the addition of Ag+. Given the relatively high concentrations of chloride ions (15.5 mM) in the medium used in this study (11) and the very-low-solubility product of AgCl (Ksp = 1.8 × 10−10), it is probable that the cells reduced insoluble AgCl in our initial experiments. Indeed this may explain why we were able to grow G. sulfurreducens in medium supplemented with up to 2 mM Ag(I) without a negative impact on growth of the organism; previous studies have also noted enhanced silver resistance in other organisms in the presence of chloride ions (5, 19, 20), presumably reflecting decreased bioavailability of the metal due to the formation of insoluble AgCl.
FIG. 1.
Reduction of 200 μM Ag(I) by late-exponential-phase cultures of G. sulfurreducens. (A) Cultures (top left) and spent supernatant (top right) before addition of Ag(I) and 2-min incubation (bottom) in the dark after addition of Ag(I). TEM of whole-mount (B) and ultrathin (C) section. (D) XRD profile of the cells and black precipitate, confirming the formation of Ag(0). (E) EDS spectrum of extracellular precipitates from unstained sectioned samples.
To confirm that G. sulfurreducens is indeed able to reduce insoluble AgCl, the white precipitate from the interaction of AgNO3 and Cl− was collected. It was washed and resuspended at a concentration of 200 μM Ag in 30 mM bicarbonate buffer (pH 7.0) (11) supplemented with 10 mM acetate. The insoluble AgCl was converted to Ag(0) and identified by X-ray diffraction (XRD) (results not shown) in a matter of minutes when cell suspensions of G. sulfurreducens, harvested at late exponential phase and washed in anaerobic 30 mM bicarbonate buffer (pH 7.0), were added to a final concentration of 0.17 mg ml−1 cellular protein.
To study the morphology and localization of the elemental silver particles formed in the late-exponential-phase cultures supplemented with 200 μM Ag(I) (presumably as AgCl), samples were viewed using transmission electron microscopy (TEM). Cells and precipitate were first collected by centrifugation (14,400 × g for 4 min), fixed using 2.5% glutaraldehyde, and then placed onto a Formvar- and carbon-coated copper TEM grid and air dried prior to viewing using an FEI Tecnai 12 Biotwin TEM (FEI, Eindhoven, The Netherlands) at 100 kV. These “whole mount” preparations showed that the silver was precipitated outside the cell as nanoscale deposits, approximately 30 nm in diameter, and often associated with extracellular material in discrete locations (Fig. 1B). Thin sections were also prepared by dehydration in an ethanol series prior to embedding in LR white resin (London Resin Company, Theale, Berkshire, United Kingdom) and sectioning to 60- to 80-nm thickness. To identify cellular structures, sections were stained by floating the section on 2% (wt/vol) uranyl acetate and 0.2% (wt/vol) lead citrate prior to TEM examination as described by Schooling and Beveridge (15), and unstained sections were used for energy-dispersive X-ray spectroscopy (EDS) using an FEM Tecnai F30 TEM. Viewing these thin sections by TEM confirmed that the silver was precipitated exclusively outside the cells with no intracellular, electron-dense silver precipitates apparent (Fig. 1C), and EDS confirmed that these extracellular precipitates were Ag rich (Fig. 1E).
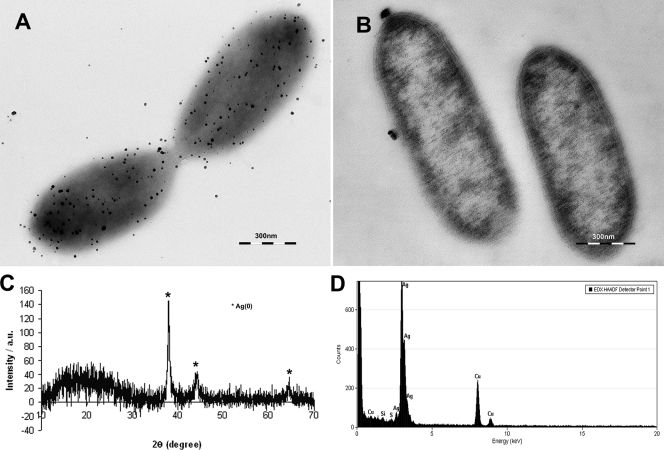
To determine the fate of Ag+ ions (rather than insoluble AgCl) added to cultures, experiments were also carried out in chloride-free 30 mM bicarbonate buffer (pH 7.0) supplemented with 10 mM acetate and 200 μM Ag(I). Similar black Ag(0) precipitates were also formed in around 10 min when suspensions of G. sulfurreducens, harvested at late exponential phase and washed in anaerobic 30 mM bicarbonate buffer (pH 7.0), were added at a concentration corresponding to 0.1 mg ml−1 total cellular protein. TEM, XRD, and EDS analyses showed precipitation of nanoscale Ag particles on the surface of the cell (Fig. 2), but these were more widely dispersed than in the previous experiments, possibly because the extracellular template tethering nanoparticles of Ag(0) had been removed during the washing steps. To measure the rate of Ag(I) reduction in these experiments, samples were taken at regular intervals and the concentrations of soluble silver were quantified by inductively coupled plasma atomic emission spectroscopy (Optima 5300DV; Perkin-Elmer, Salem, MA).
FIG. 2.
TEM of a whole-mount (A) and ultrathin (B) section of washed cell suspensions in chloride-free buffer supplemented with acetate (electron donor) and 200 μM Ag(I). (C) XRD profile of the cells and black precipitate, confirming the formation of Ag(0). (D) EDS spectrum of extracellular precipitates from unstained sectioned samples.
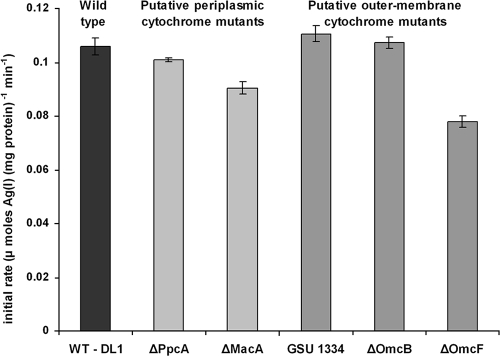
The use of washed cell suspensions in chloride-free buffer gave insight into the mechanism and kinetics of electron transfer events leading up to the precipitation of extracellular Ag(0). By varying the concentrations of Ag(I) between 100 and 500 μM and measuring the initial rate of Ag removal as Ag(0), the maximum rate of reduction (Vmax) for this transformation was estimated as 7.26 μmol of Ag (I)/(mg protein [dry weight]·h) with a Km of approximately 90 μM. In addition, the initial rate of reduction and removal of Ag(I) was also measured over the first 10 min of incubation (sampling at 0, 5, and 10 min) in a range of G. sulfurreducens knockout mutants kindly donated by D. R. Lovley and colleagues at the University of Massachusetts, Amherst, MA. Strains lacking the structural genes for two periplasmic c-type cytochromes (PpcA and MacA) (2, 11) and three outer membrane c-type cytochromes (OmcB and OmcF, in addition to the cytochrome annotated in the genome as GSU 1334) (7, 10, 17) were selected (Fig. 3). Similar rates of reduction were noted in all cultures except those lacking the structural genes for MacA and OmcF. MacA is an inner membrane-bound c-type cytochrome (2) which is located in the periplasm and is proposed to transfer electrons from the inner membrane to the periplasm. It has been shown to be important in the reduction of Fe(III) citrate and U(VI) (19), both of which could potentially enter the periplasm, and MacA could be involved in electron transfer to outer membrane cytochromes, including those on the surface of the cell, where the Ag(0) accumulated in these experiments. OmcF is an outer membrane c-type cytochrome and is thought to play a role in the reduction of extracellular electron acceptors (7), although the omcF mutant that we have used also has impaired expression of two additional outer membrane cytochromes (OmcB and OmcC) and overexpression of a third, OmcS (7). No mutants were completely impaired with respect to their abilities to reduce Ag(I), presumably due to the possibility of multiple electron transfer paths to Ag(I) or the pleiotropic nature of some mutations. However, these data would seem to suggest a role for an electron transfer chain traversing the periplasm and the outer membrane, consistent with the extracellular localization of reduced Ag(0) nanoparticles.
FIG. 3.
Rate of removal of Ag(I) from solution by G. sulfurreducens and c-type cytochrome deletion mutants. The values presented indicate the mean initial rates measured from triplicate cultures, and the error bars represent the standard deviation of the mean. WT - DL1 represents the wild-type parental strain.
In conclusion, building on earlier work (12) suggesting that the cytochrome content of Geobacter metallireducens could be oxidized by soluble silver ions, this study extends the range of metals known to be reduced efficiently by Geobacter species to include Ag(I), both as insoluble, extracellular AgCl and also potentially as soluble Ag+. The extracellular precipitation of the nanoscale Ag(0) product, in combination with physiological data from deletion mutants, would imply that Ag(I) is reduced on the surface of the cell, with c-type cytochromes traversing the periplasm and outer membrane implicated in electron transfer to the metal, in keeping with their involvement in the reduction of other extracellular electron acceptors (7, 10). In addition, nanoparticles of silver are currently fabricated using a variety of abiotic methods which are often ineffective and expensive (8, 9). The use of microbial cells to manufacture silver nanoparticles offers a useful alternative to these more conventional processes; in the case of Geobacter, silver is captured from waste industrial waste streams resulting in the synthesis of extracellular Ag(0) nanoparticles. Other bacteria, notably Pseudomonas stutzeri, are known to transform soluble silver to nanoscale silver particles (9), but these are precipitated within the cell in this organism and are not fully reduced to elemental silver, potentially complicating processing of the particles and making the bacteria less appealing for technological applications. Finally, as we were able to grow the organism in millimolar concentrations of Ag(I), concomitant with the formation of nanoscale silver precipitates, these studies suggest that silver nanoparticles are not necessarily toxic in moderately high concentrations to all gram-negative cells, despite their increasing use as a biocide.
Acknowledgments
We acknowledge financial support from the U.S. Department of Energy-ERSP (grant no. DE-FG02-02ER63422) and the United Kingdom EPSRC (grant no. EP/D058767/1) and thank the Wellcome Trust for its equipment grant support to the electron microscopy facility. The Geobacter deletion mutants used in this study were generated through support from the Office of Science (BER), U.S. Department of Energy grant no. DE-FC02-02ER63446.
We also thank the staff of the Life Sciences Electron Microscope Facility for their assistance. Derek Lovley and colleagues (UMASS, Amherst, MA) are also thanked for providing the range of Geobacter deletion mutants used in this study.
Footnotes
Published ahead of print on 22 August 2008.
REFERENCES
- 1.Bragg, P. D., and D. J. Rainnie. 1974. Effect of silver ions on respiratory-chain of Escherichia coli. Can. J. Microbiol. 20:883-889. [DOI] [PubMed] [Google Scholar]
- 2.Butler, J. E., F. Kaufmann, M. V. Coppi, C. Nunez, and D. R. Lovley. 2004. MacA a diheme c-type cytochrome involved in Fe(III) reduction by Geobacter sulfurreducens. J. Bacteriol. 186:4042-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao, Y., and R. Cranston. 2008. Recent advances in antimicrobial treatments of textiles. Textile Res. J. 78:60-72. [Google Scholar]
- 4.Ghandour, W., J. A. Hubbard, J. Deistung, M. N. Hughes, and R. K. Poole. 1988. The uptake of silver ions by Escherichia coli K12: toxic effects and interaction with copper ions. Appl. Microbiol. Biotechnol. 28:559-565. [Google Scholar]
- 5.Haefeli, C., C. Franklin, and K. Hardy. 1984. Plasmid-determined silver resistance in Pseudomonas stutzeri isolated from a silver mine. J. Bacteriol. 158:389-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermans, M. H. 2006. Silver-containing dressings and the need for evidence. Am. J. Nurs. 106(12):60-68. [DOI] [PubMed] [Google Scholar]
- 7.Kim, B. C., C. Leang, Y. H. R. Ding, R. H. Glaven, M. V. Coppi, and D. R. Lovley. 2005. OmcF, a putative c-type monoheme outer membrane cytochrome required for the expression of other outer membrane cytochromes in Geobacter sulfurreducens. J. Bacteriol. 187:4505-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klaus, T., R. Joerger, E. Olsson, and C. G. Granqvist. 1999. Silver-based crystalline nanoparticles, microbially fabricated. Proc. Nat. Acad. Sci. USA 96:13611-13614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klaus-Joerger, T., R. Joerger, E. Olsson, and C. G. Granqvist. 2001. Bacteria as workers in the living factory: metal-accumulating bacteria and their potential for materials science. Trends Biotechnol. 19:15-20. [DOI] [PubMed] [Google Scholar]
- 10.Leang, C., M. V. Coppi, and D. R. Lovley. 2003. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 185:2096-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lloyd, J. R., C. Leang, A. L. H. Myerson, M. V. Coppi, S. Cuifo, B. Methe, S. J. Sandler, and D. R. Lovley. 2003. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369:153-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovley, D. R., S. J. Giovannoni, D. C. White, J. E. Champine, E. J. P. Phillips, Y. A. Gorby, and S. Goodwin. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336-344. [DOI] [PubMed] [Google Scholar]
- 13.Rosenman, K. D., A. Moss, and S. Kon. 1979. Argyria: clinical implications of exposure to silver nitrate and silver oxide. J. Occup. Environ. Med. 21:430-435. [PubMed] [Google Scholar]
- 14.Rosenman, K. D., N. Seixas, and I. Jocobs. 1987. Potential nephrotoxic effects of exposure to silver. Br. J. Indust. Med. 44:267-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schooling, S. R., and T. J. Beveridge. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188:5945-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schreurs, W. J. A., and H. Rosenberg. 1982. Effect of silver ions on transport and retention of phosphate by Escherichia coli. J. Bacteriol. 152:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shelobolina, E. S., M. V. Coppi, A. A. Korenevsky, L. N. DiDonato, S. A. Sullivan, H. Konishi, H. F. Xu, C. Leang, J. E. Butler, B. C. Kim, and D. R. Lovley. 2007. Importance of c-type cytochromes for U(VI) reduction by Geobacter sulfurreducens. BMC Microbiol. 7:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silver, S. 2003. Bacterial silver resistance: molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 27:341-353. [DOI] [PubMed] [Google Scholar]
- 19.Slawson, R. M., E. M. Lohmeiervogel, H. Lee, and J. T. Trevors. 1994. Silver resistance in Pseudomonas stutzeri. Biometals 7:30-40. [DOI] [PubMed] [Google Scholar]
- 20.Slawson, R. M., J. T. Trevors, and H. Lee. 1992. Silver accumulation and resistance in Pseudomonas stutzeri. Arch. Microbiol. 158:398-404. [Google Scholar]