Abstract
Bacillus subtilis is capable of producing 2,3-butanediol from acetoin by fermentation, but to date, the gene encoding the enzyme responsible, acetoin reductase/2,3-butanediol dehydrogenase (AR/BDH), has remained unknown. A search of the B. subtilis genome database with the amino acid sequences of functional AR/BDHs from Saccharomyces cerevisiae and Bacillus cereus resulted in the identification of a highly similar protein encoded by the B. subtilis ydjL gene. A knockout strain carrying a ydjL::cat insertion mutation was constructed, which (i) abolished 2,3-butanediol production in early stationary phase, (ii) produced no detectable AR or BDH activity in vitro, and (iii) accumulated the precursor acetoin in early stationary phase. The ydjL::cat mutation also affected the kinetics of lactate but not acetate production during stationary-phase cultivation with glucose under oxygen limitation. A very small amount of 2,3-butanediol was detected in very-late-stationary-phase (96-hour) cultures of the ydjL::cat mutant, suggesting the existence of a second gene encoding a minor AR activity. From the data, it is proposed that the major AR/BDH-encoding gene ydjL be renamed bdhA.
Evidence that global petroleum production is nearing or has already passed its peak, an event predicted nearly a half-century ago, is mounting (11). As petroleum-based products become increasingly scarce and expensive, there is an urgent need to develop alternative fuels and chemical feedstocks. This looming need has long been recognized. More than a quarter-century ago, it was suggested that a major fraction of the U.S. chemical industry could be supplied by four feedstocks derived from biological fermentation: ethanol, isopropanol, n-butanol, and 2,3-butanediol (24). Since then, however, only bioethanol production has been actively developed into a mature industry (23). Thus, exploration of economic routes to the large-scale synthesis of other chemical feedstocks is a high priority.
Gram-positive clostridia and bacilli possess a number of metabolic pathways, making them useful for exploitation in the chemical industry; the classic historical example is the use of Clostridium acetobutylicum to produce acetone and n-butanol (13). Increasingly, Bacillus spp. are being explored and developed for their abilities to convert cellulosic biomass into fermentable sugars from which useful fermentation products can be produced (25, 26, 33). However, most environmental isolates of Bacillus spp. suffer from a lack of an amenable genetic and molecular biology toolkit. For this reason, the gram-positive model bacterium Bacillus subtilis is often utilized as a platform organism to engineer gram-positive pathways for industrial production of various enzymes, antibiotics, probiotics, and secondary metabolites (reviewed in references 8, 29, 31, and 32).
B. subtilis is a mixed-acid fermenter, converting pyruvate into lactate, acetate, ethanol, acetoin, and 2,3-butanediol as fermentative end products (19; reviewed in references 20 and 21). The fermentation route to 2,3-butanediol is of particular interest because this compound can be converted into industrial solvents and precursors in the manufacture of synthetic rubber and plastics (36). The initial portion of the pathway leading from pyruvate to 2,3-butanediol in B. subtilis has been well established and results from conversion of pyruvate to α-acetolactate by the enzyme α-acetolactate synthase, encoded by the alsS gene, followed by conversion of α-acetolactate to acetoin by alsD-encoded α-acetolactate decarboxylase (28). Adjacent to and divergent from the alsSD operon is located the alsR gene, encoding a positive transcriptional regulator of alsSD (28).
In contrast, the metabolic step from acetoin to 2,3-butanediol in B. subtilis has been relatively uncharacterized. The reversible enzyme catalyzing this reaction is known alternatively as acetoin reductase (AR; forward reaction) or 2,3-butanediol dehydrogenase (BDH; reverse reaction). In representative 2,3-butanediol-producing gram-negative bacterial species, such as Klebsiella and Enterobacter spp., the gene encoding AR/BDH is found as part of the budRABC operon, including the genes encoding α-acetolactate decarboxylase (budA), α-acetolactate synthase (budB), and AR/BDH (budC) (2), under the control of a transcriptional activator encoded by the divergent budR gene (17). In B. subtilis, downstream from the analogous alsRSD operon is located ywrO, a gene that encodes a putative NAD(P)-dependent oxidoreductase; however, this gene does not appear to encode AR/BDH (see below). One report describing the isolation of B. subtilis mutants lacking AR/BDH activity exists (15), but these mutants were not mapped and are not available. Searches of the B. subtilis genome sequence database SubtiList (http://genolist.pasteur.fr/SubtiList/) (14) failed to provide clear identification of any putative genes encoding AR/BDH, suggesting that the AR/BDH-encoding gene may have been misannotated in this database. However, search of another B. subtilis genome database, BSORF (http://bacillus.genome.jp/), indicated that the putative ydjL gene exhibited amino acid sequence similarity to AR/BDH. Reexamination of the SubtiList database revealed that ydjL is annotated as similar to l-iditol 2-dehydrogenase. In support of the notion that the ydjL gene may encode AR/BDH, experiments measuring the response of the B. subtilis transcriptome (37) or proteome (4) revealed that ydjL transcripts and YdjL protein were upregulated in response to growth under fermentative conditions.
Therefore, a body of circumstantial evidence indicates that the putative B. subtilis ydjL gene may encode AR/BDH. In order to positively identify the gene encoding AR/BDH in B. subtilis, the literature was searched for “wet laboratory” studies in which AR/BDH-encoding genes were experimentally linked to established AR/BDH enzymatic activity. In this communication, the deduced amino acid sequences of two authentic AR/BDH-encoding genes, from Saccharomyces cerevisiae and Bacillus cereus, were used to query the SubtiList database to identify putative AR/BDH-encoding B. subtilis genes for further analysis. The B. subtilis ydjL gene was identified as a likely candidate for the gene encoding AR/BDH activity, and its identity was confirmed definitively by analysis of an engineered ydjL::cat knockout strain.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. subtilis and Escherichia coli strains used in this study are listed in Table 1. Cells were routinely grown in Luria-Bertani (LB) medium (18), either lacking or containing glucose (1% [wt/vol] final concentration) as appropriate. Competent E. coli and B. subtilis cells were prepared for transformation as described previously (3, 30). The antibiotics used were ampicillin (Ap) and chloramphenicol (Cm), at final concentrations of 100 and 5 μg/ml, respectively. Optical density (OD) was monitored using a Klett-Summerson colorimeter fitted with the no. 66 (red) filter or using a spectrophotometer (Pharmacia Ultrospec 3000) set at 660 nm. One hundred Klett Units was determined to equal 1 OD660 unit.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Source (reference) |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| 168 | trpC2 | Laboratory stock |
| WN1038 | Spontaneous Trp+ revertant of 168 | Laboratory stocka |
| WN1063 | yycR::cat; Cmr | pWN1057 → WN1038 tfb |
| WN1075 | ydjL::cat; Cmr | pWN1071 → WN1038 tf |
| E. coli | ||
| DH5α | deoR endA1 gyrA96 hsdR17 (rk− mk+) recA1 relA1 supE44 thi-1 Δ(lacZYA-argFV169) φ80lacZΔM15 F− λ− | H. Jost (9) |
| WN1057 | DH5α (pWN1057); Apr | This study |
| WN1071 | DH5α (pWN1071); Apr | This study |
| Plasmids | ||
| pBGSC-6 | Replicates in E. coli (Apr); integration vector in B. subtilis (Cmr) | BGSCb (5) |
| pWN1057 | pBGSC-6 with internal yycR fragment | This study |
| pWN1071 | pBGSC-6 with internal ydjL fragment | This study |
Strain WN1038 was donated to the BGSC as strain 1A874.
Abbreviations: BGSC, Bacillus Genetic Stock Center; tf, transformation.
Mutant construction and genetic techniques.
Insertional inactivation of the ydjL and yycR genes was accomplished as follows. From the SubtiList database (http://genolist.pasteur.fr/SubtiList/), the following oligonucleotide primers were designed and built to amplify internal coding regions of the ydjL and yycR genes: ydjL-F (5′-GCGAAGCTTGAATTCTCCGGTGAAGTTGTCG-3′), ydjL-R (5′-CCCGGATCCTTTTCCCAAATGCTGACGAT-3′), yycR-F (5′-GTTGAATTCGGCTTACGGCTACGTCGATA-3′), and yycR-R (CATGGATCCGAAGCGGATTTTTAATGA-3′) (the recognition sequences for HindIII, EcoRI, and BamHI, respectively, are underlined). These primers were used to amplify by PCR 620- and 618-bp amplicons corresponding to internal coding fragments of the ydjL and yycR genes, respectively, from chromosomal DNA of B. subtilis strain 168. The ydjL and yycR amplicons were double digested with HindIII/BamHI or EcoRI/BamHI, respectively, and cloned into the appropriately cleaved, dephosphorylated integration plasmid pBGSC-6 as described previously (5). The resulting plasmids, pWN1071 (ydjL) and pWN1057 (yycR), were introduced into competent cells of B. subtilis strain WN1038, a spontaneous Trp+ revertant of strain 168, with selection for Cmr transformants. Strains WN1075 (ydjL::cat) and WN1063 (yycR::cat) were chosen for further characterization.
BDH assays.
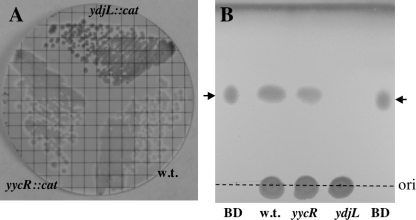
For screening purposes, a filter assay for BDH (15) was used. Briefly, cells were picked or streaked onto gridded sterile membrane filters (47-mm diameter, 0.45-μm pore size; Whatman) placed on the surfaces of LB plates and incubated overnight. The filters were carefully floated, colony side up, on 1 ml of a solution of the redox dye 2,6-dichlorophenolindophenol (DCPIP; 4 mg/ml [wt/vol] in 0.1 M potassium phosphate buffer, pH 7.0) and incubated at room temperature until colonies were stained blue (5 to 15 min). Excess DCPIP was blotted from the filter and the filter floated on 1 ml of 2,3-butanediol (0.1 M in 0.1 M potassium phosphate buffer, pH 7.0). BDH-positive colonies turned white within 1 to 2 min, whereas BDH-negative colonies remained blue (Fig. 1A).
FIG. 1.
(A) BDH activity staining of parental B. subtilis strains WN1038 (wt), WN1063 (yycR::cat), and WN1075 (ydjL::cat). Colonies were prestained with DCPIP and photographed at 3 min after incubation with 2,3-butanediol. (B) Silica gel TLC of 2,3-butanediol markers (BD, black arrows) and 30 μl of culture supernatants from the following B. subtilis strains: WN1038 (wt), WN1063 (yycR::cat), and WN1075 (ydjL::cat). The dark band at the top of the figure is the solvent front. The dashed line denotes the origin (ori).
A quantitative assay for BDH activity from cell extracts was developed using the reverse reaction 2,3-butanediol + NAD+ → acetoin + NADH to monitor the 2,3-butanediol-dependent reduction of NAD+. Cell extracts were prepared as follows. Cells were harvested from 1-ml culture samples by centrifugation for 1 min in a microcentrifuge and lysed, and cell extracts prepared in Z buffer (18) as described previously (22). Cell extracts were assayed for BDH activity by a method described for assaying glucose dehydrogenase activity (6, 22), with the exception that 0.1 M 2,3-butanediol was used as the substrate. Reduction of NAD+ to NADH was monitored spectrophotometrically by the rate of 2,3-butanediol-dependent A340 increase. Under these conditions, 1 U of BDH activity was defined as 1 μmol NADH produced per minute per ml per OD660 of culture. To verify that the assay detected authentic BDH, in vitro BDH activity reactions were also assayed for the reaction product acetoin by a quantitative Voges-Proskauer assay, as described below.
AR assay.
A quantitative assay for AR activity from cell extracts was developed using the reaction acetoin + NADH → 2,3-butanediol + NAD+ to monitor the acetoin-dependent consumption of NADH. Cell extracts were prepared and assayed for AR activity as described above for the BDH assay, with the exception that 50 mM acetoin and 0.2 mM NADH were used in the assay reactions. Oxidation of NADH to NAD+ was monitored spectrophotometrically by rate of acetoin-dependent A340 decrease. Under these conditions, 1 U of AR activity was defined as 1 μmol NADH consumed per minute per ml per OD660 of culture.
Quantitative acetoin assay.
The Voges-Proskauer test for acetoin (35) was modified for use as a quantitative colorimetric assay for acetoin. Pure acetoin (Sigma) dissolved in water exhibited an Amax at 560 nm and a molar extinction coefficient of 2.941 × 103 liters/mol·cm (data not shown). The assay was performed as follows. To 200 μl of acetoin standard solution or appropriately diluted culture supernatant was added sequentially 140 μl of creatine (0.5% [wt/vol] in water), 200 μl of α-naphthol (5% [wt/vol] in 95% ethanol), and 200 μl KOH (40% [wt/vol] in water). Samples were vortexed after each addition. The samples were incubated for 15 min at room temperature and vortexed again before measurement of A560 in the spectrophotometer. The assay was linear over the entire absorbance range of the spectrophotometer (0 to 3 A560 units), and 1 A560 unit was equal to 340 μM acetoin.
TLC.
For screening of AR-negative mutants, 2,3-butanediol was assayed from culture supernatants by thin-layer chromatography (TLC) on precoated silica gel G plates (20 cm by 20 cm) (Analtech, Wilmington, DE), run in n-heptane-ethyl acetate (1:5). Butanediol was visualized after the plates were sprayed with modified Seebach solution [0.1 g phosphomolybdic acid, 0.05 g ammonium cerium(IV) sulfate dihydrate, 0.6 ml concentrated H2SO4 dissolved in 9.2 ml water] and incubated in an oven at 150°C. Under these conditions, 2,3-butanediol migrated at an Rf of 0.5.
HPLC.
High-performance liquid chromatography (HPLC) analyses of fermentation products were performed as described previously (34). Briefly, cells were removed from culture samples by centrifugation in a microcentrifuge, culture supernatants were passed through a 0.22-μm filter, and H2SO4 was added to give a 4 mM final concentration. Fermentation products were analyzed from treated culture supernatants by using a Hewlett Packard 1090 HPLC fitted with an Aminex HPX-87H (Bio-Rad Laboratories) column, a filter photometric detector (210 nm), and a refractive index detector in series. The column temperature was 45°C, and the flow rate of eluant (4 mM H2SO4) was 0.4 ml/min. The chromatograms were processed using Chemstation software, and the concentration of each analyte was determined after the peak area was integrated, which was done after the compounds were identified on the basis of retention times of pure standards.
Statistical analyses.
All assays were performed with either duplicate or triplicate cultures. Basic statistical parameters and analyses of variance (ANOVA) were performed using commercial statistical software (Kaleidagraph, version 3.6.2; Synergy Software, Reading, PA). Differences with P values of <0.05 were considered statistically significant.
RESULTS AND DISCUSSION
Identification of putative B. subtilis AR/BDH genes.
A search of the literature resulted in identification of two AR/BDH-encoding genes that were experimentally linked to their corresponding AR/BDH enzymatic activities: the YAL060W gene product from S. cerevisiae, an NAD+-dependent BDH (7), and an acetylacetoin reductase II (AACRII)/BDH-encoding gene from B. cereus strain YUF-4 (10). Both of these proteins belong to the alcohol dehydrogenase superfamily (12, 27). Alignment of the two protein sequences by use of ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) showed that they were clearly related, with 38.6% identical amino acids and 64.1% overall highly conserved amino acids (data not shown).
The S. cerevisiae YAL060W and B. cereus AACRII/BDH protein sequences (GenBank accession numbers AAT92941 and AB063194, respectively) were used to search the SubtiList database with the BLASTP function (Table 2). A number of B. subtilis proteins exhibiting high similarity scores were identified, including the alcohol dehydrogenases AdhA and AdhB, threonine dehydrogenase (Tdh), and sorbitol dehydrogenase (GutB) (Table 2). High-scoring hits to these proteins would be expected, as they all belong to the Adh superfamily (12, 27). Interestingly, three proteins of unknown function (YdjL, YjmD, and YycR) were also identified by BLASTP (Table 2). These proteins were annotated in the SubtiList database as similar to l-iditol 2-dehydrogenase, alcohol dehydrogenase, and formaldehyde dehydrogenase, respectively. YdjL is encoded by the monocistronic ydjL gene located at kb 678.5 on the B. subtilis 168 sequence. The yjmD gene is the fourth cistron within a 10-cistron operon involved in utilization of hexuronate (16), and yycR is a monocistronic gene located at kb 4136 on the B. subtilis sequence. Of all B. subtilis proteins, the YdjL protein scored as most closely related to the S. cerevisiae and B. cereus AR/BDHs (E values of 7 × 10−46 and 1 × 10−114, respectively) (Table 2).
TABLE 2.
High-scoring B. subtilis proteins with similarity to the S. cerevisiae YAL060W BDH or the B. cereus AACRII/BDH
| Gene product | Annotated function (from SubtiList) | E value
|
|
|---|---|---|---|
| YAL060W | AACRII/BDH | ||
| YdjL | Unknown; similar to l-iditol 2-dehydrogenase | 7e−46 | e−114 |
| GutB | Sorbitol dehydrogenase | 1e−28 | 4e−64 |
| YjmD | Unknown; similar to alcohol dehydrogenase | 1e−25 | 6e−46 |
| Tdh | Threonine 3-dehydrogenase | 1e−19 | 6e−43 |
| AdhB | Alcohol dehydrogenase | 1e−12 | 4e−21 |
| AdhA | NADP-dependent alcohol dehydrogenase | 4e−11 | 3e−17 |
| YycR | Unknown; similar to formaldehyde dehydrogenase | 3e−11 | 6e−14 |
| YogA | Unknown; similar to alcohol dehydrogenase | NDa | 3e−9 |
ND, similarity not detected.
Alignment of the deduced B. subtilis YdjL amino acid sequence with those of YAL060W and AACRII/BDH revealed overall 218/346 (63%) identical and 278/346 (80%) highly conserved amino acids, and the three proteins were of similar lengths, 346 to 349 amino acids (data not shown). From these observations, it appeared that the ydjL gene was the best candidate for a test for the capacity to encode AR/BDH. In addition, because the B. cereus YUF-4 AACRII/BDH gene had been found to be monocistronic (10) rather than part of a bud operon, as in the gram-negative Klebsiella or Enterobacter spp. (2), the monocistronic yycR gene was also considered a possible candidate for the gene encoding AR/BDH.
Insertional inactivation of ydjL and yycR.
Fragments internal to the coding sequences of ydjL and yycR were PCR amplified, cloned into integration plasmid pBGSC-6, and introduced into the chromosome of wild-type (wt) B. subtilis strain WN1038 by transformation. By use of DCPIP staining, it was ascertained that all Cmr ydjL::cat integrants tested negative for BDH activity (i.e., colonies retained the blue DCPIP stain), while the wt strain WN1038 and all Cmr yycR::cat integrants tested BDH positive (data not shown). Two Cmr integrants, strains WN1063 (yycR::cat) and WN1075 (ydjL::cat), were streak purified and chosen for further characterization. Qualitative BDH activity staining of streak-purified strains by use of the DCPIP filter assay showed that WN1075 (ydjL::cat) retained its BDH-negative phenotype, while the wt parent and yycR::cat strains were clearly BDH positive (Fig. 1A).
Production of 2,3-butanediol by the wt and mutant strains.
In order to visualize accumulation of 2,3-butanediol, strains WN1038 (wt), WN1063 (yycR::cat), and WN1075 (ydjL::cat) were grown for 24 h at 37°C with limited aeration in LB medium containing 1% glucose, and 30 μl of each culture supernatant was run on silica gel TLC and stained for 2,3-butanediol as described in Materials and Methods. The results clearly showed that the WN1038 (wt) and WN1063 (yycR::cat) strains produced 2,3-butanediol, while ydjL::cat mutant strain WN1075 did not (Fig. 1B). Thus, the ydjL::cat mutation abolished both in vivo BDH activity (Fig. 1A) and AR activity (Fig. 1B) in strain WN1075.
In vitro assay of AR and BDH activities in wt and mutant strains.
To test quantitatively for AR and BDH activities, triplicate cultures of strains WN1038, WN1063, and WN1075 were cultivated for 24 h at 37°C with limited aeration in LB medium containing 1% glucose. Cells were harvested and lysed and their cell extracts assayed for AR and BDH activities as described in Materials and Methods. Cell extracts of strains WN1038 (wt) and WN1063 (yycR::cat) produced both similar amounts of AR activity, as measured by acetoin-dependent oxidation of NADH to NAD+, and similar amounts of BDH activity, as measured by 2,3-butanediol-dependent reduction of NAD+ to NADH (Table 3). In sharp contrast, neither AR nor BDH activity was detectable in cell extracts of strain WN1075 (ydjL::cat) (Table 3).
TABLE 3.
BDH and AR activity in B. subtilis strains
| Strain (mutation) | Replicate | BDH activity (Ua) (avg ± SD) | AR activity (Ub) (avg ± SD) |
|---|---|---|---|
| WN1038 (none) | A | 190.9 (172.8 ± 15.7) | 61.8 (49.7 ± 12.6) |
| B | 164.7 (172.8 ± 15.7) | 50.5 (49.7 ± 12.6) | |
| C | 162.8 (172.8 ± 15.7) | 36.7 (49.7 ± 12.6) | |
| WN1063 (yycR::cat) | A | 124.7 (127.9 ± 11.2) | 71.2 (88.5 ± 24.1) |
| B | 118.6 (127.9 ± 11.2) | 78.3 (88.5 ± 24.1) | |
| C | 140.4 (127.9 ± 11.2) | 116.0 (88.5 ± 24.1) | |
| WN1075 (ydjL::cat) | A | NDc | ND |
| B | ND | ND | |
| C | ND | ND |
One unit of BDH activity is defined as 1 μmol NADH produced per minute per ml per OD660 unit of culture.
One unit of AR activity is defined as 1 μmol NADH consumed per minute per ml per OD660 unit of culture.
ND, not detected. The lower limit of the assay was ∼1 U BDH or AR.
To confirm that BDH activity was indeed being measured in the above-mentioned in vitro reactions, the end product of the reaction, acetoin, was also measured. Cell extracts were prepared from triplicate cultures of strains WN1038 (wt) and WN1075 (ydjL::cat) grown for 24 h in LB medium plus 1% glucose under limited aeration, and in vitro BDH assays were performed at 37°C. At 5-min intervals, the reaction mixtures were sampled and assayed for acetoin by using the quantitative Voges-Proskauer assay as described in Materials and Methods. A time-dependent increase in acetoin concentration was observed from the cell extracts of strain WN1038 (wt) assayed in vitro but not from those of WN1075 (ydjL::cat) (Fig. 2). Taken together, the results shown in Table 3 and Fig. 2 strongly indicated that the ydjL gene encodes AR/BDH in B. subtilis.
FIG. 2.
Production of acetoin from 2,3-butanediol in vitro by cell extracts of B. subtilis strains WN1038 (wt, open circles) and WN1075 (ydjL::cat, filled circles). Data are averages ± standard deviations (n = 3).
In vivo acetoin accumulation by the ydjL::cat strain.
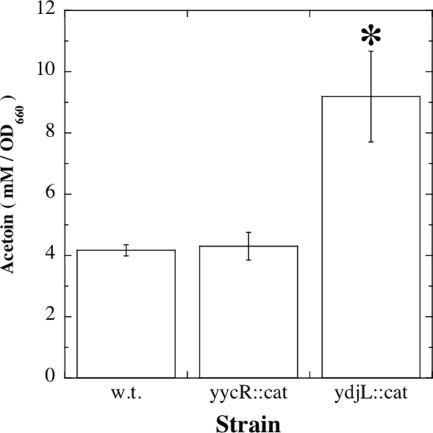
In vivo, the loss of AR activity would be predicted to lead to an accumulation of elevated amounts of the metabolic precursor acetoin in culture supernatants of the ydjL::cat mutant. This notion was tested by assaying acetoin concentrations in culture supernatants of strains WN1038 (wt), WN1063 (yycR::cat), and WN1075 (ydjL::cat), each grown for 24 h at 37°C with limited aeration in LB medium containing 1% glucose. Acetoin was assayed using the quantitative Voges-Proskauer test as described in Materials and Methods. Acetoin concentrations were indeed observed to be ∼2-fold greater in ydjL::cat mutant culture supernatants than in either wt or yycR::cat mutant culture supernatants (Fig. 3). The increased concentration of acetoin accumulated by the ydjL::cat mutant was highly significant by ANOVA (P = 0.004), whereas the wt and the yycR::cat mutant produced essentially the same concentration of acetoin (P = 0.636; no significant difference by ANOVA).
FIG. 3.
Acetoin concentrations (mM), corrected for OD660, in the culture supernatants of strains WN1038 (wt), WN1063 (yycR::cat), and WN1075 (ydjL::cat). The asterisk denotes a significant difference from the wt and the yycR::cat mutant as determined by ANOVA (n = 3). See text for details.
Time course of fermentation in the wt and ydjL::cat strains.
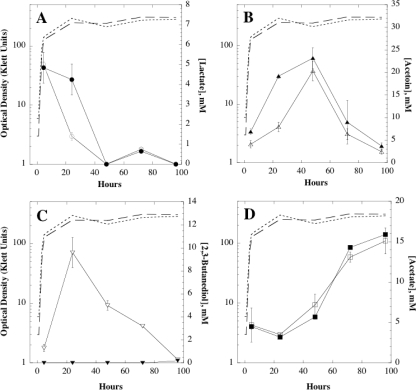
In order to assess the effect of the ydjL::cat mutation on production of the major fermentation products in B. subtilis, strains WN1038 (wt) and WN1075 (ydjL::cat) were cultivated for 96 h in LB medium containing 1% glucose with limited aeration. Samples were removed at 24-hour intervals and analyzed by HPLC as described in Materials and Methods and a previous study (34).
Lactate.
Examination of the HPLC results revealed that in both wt and ydjL::cat mutant cultures, lactate was produced at its highest concentration during exponential growth and then disappeared from the medium to very low levels by 48 h, where it remained at low concentrations until 96 h. Lactate disappearance during the first 24 h of fermentation was much slower in the ydjL::cat mutant than in the wt, but both strains showed similar lactate levels by 48 h (Fig. 4A).
FIG. 4.
Time course of production of fermentation products. Concentrations of lactate (A, circles), acetoin (B, upward triangles), 2,3-butanediol (C, downward triangles), and acetate (D, squares) produced by strain WN1038 (wt, open symbols) and WN1075 (ydjL::cat, filled symbols) are shown. In each case, the concentration of the product is corrected for OD. The growth levels of WN1038 and WN1075 are denoted by short- and long-dashed lines, respectively. Data are averages ± standard deviations for duplicate cultures.
Acetoin.
In the wt strain, acetoin production was low during exponential growth but increased during stationary phase, peaked at 48 h, and then declined again to low levels by 96 h (Fig. 4B). The ydjL::cat mutant accumulated a greater concentration of acetoin than the wt at 24 h, as had been observed (Fig. 3), but acetoin production in the ydjL::cat mutant peaked at essentially the wt level by 48 h, and its decline paralleled that of the wt from 48 to 96 h (Fig. 4B).
2,3-Butanediol.
When 2,3-butanediol production was examined, it was found to peak in the wt strain by 24 h and then steadily declined to nearly undetectable levels by 96 h (Fig. 4C). As expected, 2,3-butanediol was not detected in culture supernatants of the ydjL::cat mutant at any time point out to 72 h. Surprisingly, however, a very small amount of 2,3-butanediol (0.2 ± 0.14 mM) was detected in the ydjL::cat mutant culture supernatant at 96 h (Fig. 4C), suggesting the existence of a second gene encoding BDH activity present at low levels very late in stationary phase.
Acetate.
The levels of acetate production in the wt and the ydjL::cat mutant strains were very similar, remaining low until 24 h and then rising steadily until reaching a maximum at 96 h (Fig. 4D).
Other fermentation products.
Ethanol was not detected in culture supernatants of either strain, but very low concentrations (1 to 2 mM) of 1,2-propanediol were detected at 72 and 96 h in both wt and ydjL::cat mutant culture supernatants (data not shown). In addition to the known fermentation products, HPLC analysis showed that both the wt and the ydjL::cat mutant strains produced four unidentified fermentation products, the concentrations of which became elevated in late time points (data not shown); identification of these compounds is ongoing at this time.
The ydjL gene encodes AR/BDH in B. subtilis.
The results presented in this communication demonstrate that the ydjL gene encodes the major AR/BDH in B. subtilis and support earlier experiments measuring the global responses of the B. subtilis transcriptome (37) or proteome (4), which revealed that both ydjL mRNA and YdjL protein levels were indeed upregulated in response to fermentative growth (reviewed in reference 21). The observations reported here underscore the importance of performing wet laboratory experiments to confirm the identities of putative genes rather than relying solely upon modeling and automated annotation programs (1).
From the above data, I propose that the ydjL gene be renamed bdhA. Now that the identity of B. subtilis bdhA is known, a search of the National Center for Biotechnology Information (NCBI) database with the amino acid sequence of B. subtilis BdhA protein revealed at least 100 protein sequences with highly significant E values (<10−66) to which various putative functions had previously been described based on automated annotation; some are correctly annotated as AR/BDHs; most are annotated as some type of alcohol dehydrogenase, generic dehydrogenase, or oxidoreductase; and a few are annotated as related to sorbitol-, threonine-, or l-iditol dehydrogenases (data not shown). In many cases, these highly related proteins could likely also be safely reannotated as AR/BDHs.
In conclusion, identification of the bdhA gene function as encoding AR/BDH in the model organism B. subtilis is a significant advance in the engineering of gram-positive biochemical pathways leading directly from cellulosic substrates to the industrial-scale production of 2,3-butanediol. Current studies are concentrated on extending our understanding of the regulation of the fermentation pathway to 2,3-butanediol, with the aim of optimizing carbon flow to this important chemical feedstock compound.
Acknowledgments
I thank Andy Schuerger for helpful discussions, Lanfang Levine for assistance with silica gel TLC, Helen Jost for generous donation of strains, K. T. Shanmugam for performing HPLC on time course samples, M. Patrick Sanou for performing in vitro AR assays, and the anonymous reviewers for helpful advice.
This work was supported by a grant from the U.S. Department of Agriculture through the Florida Agricultural Experiment Station (FLA-MCS-04602).
Footnotes
Published ahead of print on 26 September 2008.
REFERENCES
- 1.Bender, R. A., D. Downs, P. Kiley, R. A. LaRossa, A. L. Sonenshein, and G. Storz. 2008. Bridges and chasms: summary of the IMAGE2 meeting in Montreal, Canada, 30 April to 3 May 2007. J. Bacteriol. 190:792-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blomqvist, K., M. Nikkola, P. Lehtovaara, P., M. Suihko, U. Airaksinon, K. Straby, J. K. Knowles, and M. E. Penttila. 1993. Characterization of the genes of the 2,3-butanediol operons from Klebsiella terrigena and Enterobacter aerogenes. J. Bacteriol. 175:1392-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boylan, R. J., N. Mendelson, D. Brooks, and F. E. Young. 1972. Regulation of the bacterial cell wall: analysis of a mutant of Bacillus subtilis defective in biosynthesis of teichoic acid. J. Bacteriol. 110:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clements, L. D., U. N. Streips, and B. S. Miller. 2002. Differential proteomic analysis of Bacillus subtilis nitrate respiration and fermentation in defined medium. Proteomics 2:1724-1734. [DOI] [PubMed] [Google Scholar]
- 5.Fajardo-Cavazos, P., C. Salazar, and W. L. Nicholson. 1993. Molecular cloning and characterization of the Bacillus subtilis spore photoproduct lyase (spl) gene, which is involved in repair of ultraviolet radiation-induced DNA damage during spore germination. J. Bacteriol. 175:1735-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita, Y., R. Ramaley, and E. Freese. 1977. Location and properties of glucose dehydrogenase in sporulating cells and spores of Bacillus subtilis. J. Bacteriol. 132:282-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.González, E., M. R. Fernández, C. Larroy, L. Sola, M. A. Pericas, X. Parés, and J. A. Biosca. 2000. Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene product. Disruption and induction of the gene. J. Biol. Chem. 275:35876-35885. [DOI] [PubMed] [Google Scholar]
- 8.Graumann, P. (ed.). 2007. Bacillus: cellular and molecular biology. Caister Academic Press, Wymondham, United Kingdom.
- 9.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 10.Hosaka, T., S. Ui, T. Ohtsuki, A. Mimura, M. Ohkuma, and T. Kudo. 2001. Characterization of the NADH-linked acetylacetoin reductase/2,3-butanediol dehydrogenase gene from Bacillus cereus YUF-4. J. Biosci. Bioeng. 91:539-544. [DOI] [PubMed] [Google Scholar]
- 11.Hubbert, M. K. 1956. Drilling and production practice, p. 7-25. American Petroleum Institute, New York, NY.
- 12.Jörnvall, H., O. Danielsson, H. Eklund, L. Helmqvist, J.-O. Höög, X. Parés, and J. Shafqat. 1993. Enzyme and isozyme developments within the medium-chain alcohol dehydrogenase family, p. 533-544. In H. Weiner, D. W. Crabb, and T. G. Flynn (ed.), Enzymology and molecular biology of carbonyl metabolism, vol. 4. Plenum Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 13.Killefer, D. H. 1927. A microbe in international affairs. Sci. Am. 137:30-32. [Google Scholar]
- 14.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertaro, P. Bessieres, A. Bolotin, S. Borchert, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 15.López, J., B. Thoms, and P. Fortnagel. 1973. Mutants of Bacillus subtilis blocked in acetoin reductase. Eur. J. Biochem. 40:479-483. [DOI] [PubMed] [Google Scholar]
- 16.Mekjian, K. R., E. M. Bryan, B. W. Beall, and C. P. Moran, Jr. 1999. Regulation of hexuronate utilization in Bacillus subtilis. J. Bacteriol. 181:426-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer, D., V. Schlensog, and A. Böck. 1995. Identification of the transcriptional activator controlling the butanediol fermentation pathway in Klebsiella terrigena. J. Bacteriol. 177:5261-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. M. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Nakano, M. M., and F. M. Hulett. 1997. Adaptation of Bacillus subtilis to oxygen limitation. FEMS Microbiol. Lett. 157:1-7. [DOI] [PubMed] [Google Scholar]
- 20.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52:165-190. [DOI] [PubMed] [Google Scholar]
- 21.Nakano, M. M., and P. Zuber. 2002. Anaerobiosis, p. 393-404. In A. L. Soneneshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 22.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. J. Wiley and Sons, New York, NY.
- 23.Otero, J. M., G. Panagiotou, and L. Olsson. 2007. Fueling industrial biotechnology growth with bioethanol. Adv. Biochem. Eng. Biotechnol. 108:1-40. [DOI] [PubMed] [Google Scholar]
- 24.Palsson, B. O., S. Fathi-Afshar, D. F. Rudd, and E. N. Lightfoot. 1981. Biomass as a source of chemical feedstocks: an economic evaluation. Science 213:513-517. [DOI] [PubMed] [Google Scholar]
- 25.Patel, M. A., M. S. Ou, R. Harbrucker, H. C. Aldrich, M. L. Buszko, L. O. Ingram, and K. T. Shanmugam. 2006. Isolation and characterization of acid-tolerant, thermophilic bacteria for effective fermentation of biomass-derived sugars to lactic acid. Appl. Environ. Microbiol. 72:3228-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel, M. A., M. S. Ou, L. O. Ingram, and K. T. Shanmugam. 2005. Simultaneous saccharification and co-fermentation of crystalline cellulose and sugar cane bagasse hemicellulose hydrolysate to lactate by a thermotolerant acidophilic Bacillus sp. Biotechnol. Prog. 21:1453-1460. [DOI] [PubMed] [Google Scholar]
- 27.Persson, B., J. S. Zigler, Jr., and H. Jörnvall. 1994. A super-family of medium-chain dehydrogenases/reductases (MDR). Eur. J. Biochem. 226:15-22. [DOI] [PubMed] [Google Scholar]
- 28.Renna, M. C., N. Najimudin, L. R. Winik, and S. A. Zahler. 1993. Regulation of the alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J. Bacteriol. 175:3863-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricca, E., A. O. Henriques, and S. M. Cutting (ed.). 2004. Bacterial spore formers: probiotics and emerging applications. Horizon Bioscience, Wymondham, United Kingdom.
- 30.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Sonenshein, A. L., J. A. Hoch, and R. Losick (ed.) 1993. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, DC.
- 32.Sonenshein, A. L., J. A. Hoch, and R. Losick (ed.). 2002. Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 33.St. John, F. J., J. D. Rice, and J. F. Preston. 2006. Paenibacillus sp. strain JDR-2 and XynA1: a novel system for methylglucuronoxylan utilization. Appl. Environ. Microbiol. 72:1496-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Underwood, S. A., S. Zhou, T. B. Causey, L. P. Yomano, K. T. Shanmugam, and L. O. Ingram. 2002. Genetic changes to optimize carbon partitioning between ethanol and biosynthesis in ethanologenic Escherichia coli. Appl. Environ. Microbiol. 68:6263-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voges, O., and B. Proskauer. 1898. Beitrag zur Ernährungsphysiologie und zur Differentialdiagnose der hämorrhagischen Septicämie. Z. Hyg. Infekt. 28:20-32. [Google Scholar]
- 36.Voloch, M., N. B. Jansen, M. R. Ladisch, G. T. Tsao, R. Narayan, and V. W. Rodwell. 1985. 2,3-Butanediol, p. 933-947. In M.-Y. Murray, C. L. Cooney, and A. E. Humphrey (ed.), Comprehensive biotechnology. Pergamon Press, New York, NY.
- 37.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]