Abstract
PCR screening of the shellfish-borne pathogen Vibrio vulnificus revealed csrA-negative strains, and these strains formed increased biofilm compared to csrA-positive strains. Complementation in trans with csrA resulted in reduced biofilm formation, similar to that by csrA+ strains. Our results provide evidence that csrA inhibits biofilm formation in V. vulnificus.
The opportunistic human pathogen Vibrio vulnificus is an inhabitant of marine environments worldwide (2, 6, 12, 14, 20). It is responsible for 95% of all seafood-related fatalities in the United States, with its most serious disease (primary septicemia) associated with the consumption of raw or undercooked oysters (15). Clinical and environmental strains of this bacterium can be sorted through rapid PCR screening of a gene (vcg) that has been correlated with virulence in this pathogen. Strains which possess the sequence associated with clinical isolates (vcgC) are designated C type, while strains containing the sequence associated almost exclusively with environmental isolates (vcgE) are designated E type (17). Both of these genotypes have been isolated from the environment, and yet their distribution among environmental sources varies. For example, we found E-type strains to be overwhelmingly predominant in oysters compared to C-type strains, indicating a differential ability for strains with this genotype to colonize oyster tissue (19). Attachment to oysters likely involves the formation of biofilms, and several surface structures have been shown to be involved in biofilm formation by this organism. However, of these structures, only capsular polysaccharide (CPS) has also been shown to be differentially expressed between the C and E genotypes. The absence of surface capsule has been linked to increased biofilm formation in V. vulnificus, and E-type strains demonstrate an increased transition to a phenotype with reduced capsule (CPS) expression (5, 8).
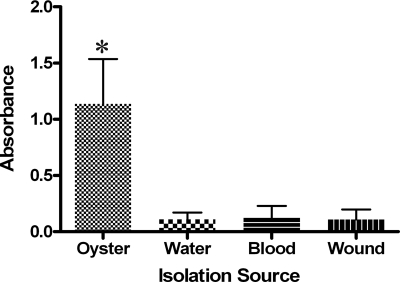
Given these indications that E-type isolates may form increased biofilm compared with C-type isolates, the two genotypes were compared in their abilities to form biofilms by using a microtiter plate method. Briefly, overnight cultures of V. vulnificus were inoculated at a 1:100 dilution into 99 μl of Luria broth (LB) contained in 96-well microtiter plates (Costar). Cells were grown statically at 30°C for 48 h. Following incubation, the wells were emptied and then washed three times with 150 μl of one-half-strength artificial seawater. Two hundred microliters of 1% crystal violet was added to the washed wells and incubated at room temperature for 15 min. The crystal violet was removed, and the wells were washed three times with 200 μl of one-half-strength artificial seawater. The remaining dye was solubilized in 200 μl of 95% ethanol, and the absorbance values were measured using a uQuant Microplate spectrophotometer (Biotek). Results from this assay showed that upon screening of 18 C-type and 16 E-type strains, there was no significant difference in the level of biofilm formation between the genotypes (data not shown). However, when these strains were grouped by isolation source, oyster isolates demonstrated a significantly (P = 0.021) higher level of biofilm formation than did strains from all other sources of isolation (Fig. 1).
FIG. 1.
Biofilm formation by V. vulnificus isolates based on their source of isolation. Those strains (n = 9) isolated from oysters demonstrated a significantly (asterisk; P = 0.021) higher level of biofilm formation than did strains isolated from other sources (nine water, nine blood, and nine wound isolates).
The global regulatory protein CsrA has been shown to influence biofilm formation by Escherichia coli by binding to mRNA and affecting translation (7, 11). Microarray analysis comparing clinical and environmental strains of V. vulnificus found csrA to be missing in many environmental isolates (G. Vora, personal communication). Therefore, strains previously analyzed for biofilm formation were screened for the presence of csrA using PCR. BLAST comparison of E. coli K-12 CsrA (accession no. AAC75738) with the two published V. vulnificus genomes (CMCP6, accession no. NC_004459, and YJ016, accession no. NC_005139) was used to locate csrA in V. vulnificus, and results showed 96% amino acid identity and 78% nucleotide identity between E. coli and both V. vulnificus strains. Alignment of the V. vulnificus nucleotide sequences (YJ016 [accession no. VV2801] and CMCP6 [accession no. VV1_1595]) revealed 100% nucleotide identity for csrA and 99% nucleotide identity for the surrounding regions (1 kb upstream and downstream).
Two sets of primers were developed for PCR screening. The first primer set localized to a conserved region within csrA (csrAup, 5′-GCGTAGGCGAAACACTGA-3′, and csrAdwn, 5′-CGTTGCCTTTCTCAGCC-3′), while the second pair of primers were found in conserved regions upstream and downstream of the gene (csrAF2, 5′-GTCAGCCTCTATCATTCAGAG-3′, and csrAR1, 5′-GGATAATAGCCTCGTAGCTA-3′, respectively). Cycling conditions for both primer pairs were optimized using a gradient thermal cycler (Techne), and the following parameters were used for amplification: 94°C for 3 min followed by 30 cycles of 94°C for 45 s, 58°C for 45 s, and 72°C for 45 s with a final extension at 72°C for 2 min. All strains were initially screened with csrAup and csrAdwn. Strains yielding a negative PCR result were then screened with the second csrA primer set and with primers for vvhA (19) to verify that the isolates were V. vulnificus and the DNA was amplifiable. Strains which yielded negative results for both csrA primer sets and a positive result with vvhA primers were designated csrA negative. Results showed that only 33.3% of the oyster isolates previously examined for biofilm formation were positive for csrA (henceforth designated csrA+), while 66.7% of water isolates and 100% of both blood and wound isolates were csrA+. These data suggested that csrA may be absent with greater frequency in oyster isolates than in strains from other environmental sources. However, a more extensive screening of additional strains revealed that 72.3% of oyster isolates (n = 47), 72.7% of water isolates (n = 32), and 100% of blood and wound isolates (n = 18 and 9, respectively) were positive for csrA, indicating that there is no correlation between isolation source and the presence of csrA.
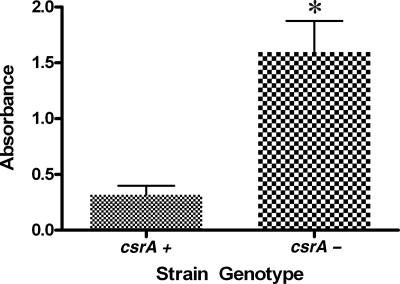
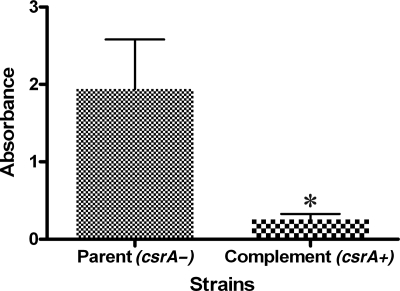
The oyster isolates which formed high levels of biofilm were predominately csrA negative, suggesting that csrA may be involved in biofilm formation by V. vulnificus. To investigate this hypothesis, biofilm formation by csrA+ (n = 17) and that by csrA-negative (n = 12) strains isolated from both clinical and environmental sources were compared. Results showed that strains without csrA produced significantly (P ≤ 0.001) more biofilm than did strains possessing the gene, indicating that the presence of csrA negatively influences biofilm formation by V. vulnificus (Fig. 2). To confirm this hypothesis, a crsA-negative strain was complemented in trans with csrA. Primers csrAF2 and csrAR1 (located 28 bp upstream and 49 bp downstream of csrA, respectively) were used to amplify the gene in strain V. vulnificus C7184/K2 by using the PCR conditions described above, and the resulting product was cloned into the TOPO TA cloning vector as described by the manufacturer (Invitrogen). The presence of csrA in the resulting construct (pTCSRA) was confirmed by PCR using csrAup and csrAdwn primers (data not shown). pTCSRA and the broad-host-range vector pRK404 were digested with EcoRI per the manufacturer's instructions (Promega), and the digests were gel purified and cleaned using the Ultra-Clean Gel Spin kit (Mo-Bio). Digested pRK404 and csrA were then ligated overnight at 4°C using T4 DNA ligase per the manufacturer's instructions (Promega) to yield pRKCSRA, which was then cloned into chemically competent E. coli S17-λ-pir. pRKCSRA was conjugated into the csrA-negative V. vulnificus strain NRO 102-418 through filter mating as previously described (21). Complementation was confirmed by PCR using csrAup and csrAdwn, and the complement was then evaluated for biofilm formation. Results showed that complementation in trans of csrA significantly (P ≤ 0.005) reduced biofilm formation compared to the parent strain, and levels were similar to those of csrA+ isolates (Fig. 3). These results strongly suggest that csrA is an inhibitor of biofilm formation by V. vulnificus.
FIG. 2.
Biofilm formation abilities of csrA+ (n = 17) and csrA-negative (n = 12) strains. Strains which lacked csrA (csrA−) displayed significantly (asterisk; P ≤ 0.001) higher biofilm formation than did strains which possessed csrA.
FIG. 3.
Complementation in trans with pRKCSRA resulted in a significant (asterisk; P ≤ 0.005) reduction in biofilm formation compared to that by the csrA-negative (csrA−) parent strain.
V. vulnificus is a natural inhabitant of oysters, and many studies have investigated physical properties which contribute to its attachment to and colonization of such shellfish. Several surface structures (including CPS, extracellular polysaccharide, pili, and flagellum) are involved in biofilm formation by this organism (3, 4, 8, 9, 16). However, the genetic regulators responsible for their expression during biofilm formation have yet to be elucidated. Recently, the global carbon storage regulator CsrA has been identified as a major posttranscriptional regulator of biofilm formation by E. coli (7). This protein often binds to mRNA transcripts and promotes their decay, thereby impacting gene expression at the translational level (1).
In this study biofilm formation by isolates of V. vulnificus was examined, and results indicated that strains lacking csrA formed significantly (P ≤ 0.001) higher levels of biofilm than did csrA+ strains, indicating that csrA is an inhibitor of biofilm formation by this species. This observation was confirmed through complementation of a csrA-negative strain, which resulted in a significant (P ≤ 0.005) reduction in biofilm formation compared to that by the parent strain. In E. coli, biofilm formation is inhibited by CsrA through downregulation of both glycogen synthesis and production of a polysaccharide adhesion which is required for biofilm formation to occur (7, 18). Conversely, in V. vulnificus CPS expression has been reported to reduce biofilm formation (8). Our studies demonstrated that csrA negatively regulates biofilm formation (as is seen in E. coli), suggesting that either (i) csrA is not a regulator of group I polysaccharide synthesis or (ii) there are other factors in addition to the presence of CPS required for biofilm formation. Recently, however, several studies have reported that capsule expression does not play a role in biofilm formation by V. vulnificus, throwing the role of CPS in biofilm formation into question (3, 10, 13). Together, these findings suggest that csrA may be an important regulator in colonization by V. vulnificus and highlight the need for more investigation into the role of this protein in polysaccharide production and biofilm formation.
Acknowledgments
These studies were funded, in part, by USDA grant CSREES 2007-35201-18381.
Footnotes
Published ahead of print on 26 September 2008.
REFERENCES
- 1.Baker, C. S., I. Morozov, K. Suzuki, T. Romeo, and P. Babitzke. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 44:1599-1610. [DOI] [PubMed] [Google Scholar]
- 2.Bisharat, N., V. Agmon, R. Finkelstein, R. Raz, G. Ben-Dror, L. Lemer, S. Soboh, R. Colodner, D. N. Cameron, D. L. Wykstra, D. L. Swerdlow, and J. J. Farmer III. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421-1424. [DOI] [PubMed] [Google Scholar]
- 3.Grau, B. L., M. C. Henk, and G. S. Pettis. 2005. High-frequency phase variation of Vibrio vulnificus 1003: isolation and characterization of a rugose phenotypic variant. J. Bacteriol. 187:2519-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grau, B. L., M. C. Henk, K. L. Garrison, B. J. Olivier, R. M. Schulz, K. L. O'Reilly, and G. S. Pettis. 2008. Further characterization of Vibrio vulnificus rugose variants and identification of capsular and rugose exopolysaccharide gene clusters. Infect. Immun. 76:1485-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilton, T., T. Rosche, B. Froelich, B. Smith, and J. Oliver. 2006. Capsular polysaccharide phase variation in Vibrio vulnificus. Appl. Environ. Microbiol. 72:6986-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Høi, L., J. L. Larsen, I. Dalsgaard, and A. Dalsgaard. 1998. Occurrence of Vibrio vulnificus biotypes in Danish marine environments. Appl. Environ. Microbiol. 64:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joseph, L. A., and A. C. Wright. 2004. Expression of Vibrio vulnificus capsular polysaccharide inhibits biofilm formation. J. Bacteriol. 186:889-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, J. H., J. B. Rho, K. J. Park, C. B. Kim, Y. S. Han, S. H. Choi, K. H. Lee, and S. J. Park. 2004. Role of flagellum and motility in pathogenesis of Vibrio vulnificus. Infect. Immun. 72:4905-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, J. H., J. E. Rhee, U. Park, H. M. Ju, B. C. Lee, T. S. Kim, H. S. Jeong, and S. H. Choi. 2007. Identification and functional analysis of Vibrio vulnificus SmcR, a novel global regulator. J. Microbiol. Biotechnol. 17:325-334. [PubMed] [Google Scholar]
- 11.Lucchetti-Miganeh, C., E. Burrowes, C. Baysse, and G. Ermel. 2008. The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology 154:16-29. [DOI] [PubMed] [Google Scholar]
- 12.Mahmud, Z. H., S. B. Neogi, A. Kassu, B. T. M. Huong, I. K. Jahid, M. S. Islam, and F. Ota. 2008. Occurrence, seasonality and genetic diversity of Vibrio vulnificus in coastal seaweeds and water along the Kii Chanel, Japan. FEMS Microbiol. Ecol. 64:209-218. [DOI] [PubMed] [Google Scholar]
- 13.McDougald, D., W. H. Lin, S. A. Rice, and S. Kjelleberg. 2006. The role of quorum sensing and the effect of environmental conditions on biofilm formation by strains of Vibrio vulnificus. Biofouling 22:133-144. [DOI] [PubMed] [Google Scholar]
- 14.Myatt, D. C., and G. H. Davis. 1989. Isolation of medically significant Vibrio species from riverine sources in South East Queensland. Microbios 60:111-123. [PubMed] [Google Scholar]
- 15.Oliver, J. D. 2006. Vibrio vulnificus, p. 349-366. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington, DC.
- 16.Paranjpye, R. N., and M. S. Strom. 2005. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 73:1411-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosche, T. M., Y. Yano, and J. D. Oliver. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol. Immunol. 49:381-389. [DOI] [PubMed] [Google Scholar]
- 18.Wang, X., A. K. Dubey, K. Suzuki, C. S. Baker, P. Babitzke, and T. Romeo. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of biofilm polysaccharide adhesion of Escherichia coli. Mol. Microbiol. 56:1648-1663. [DOI] [PubMed] [Google Scholar]
- 19.Warner, E. B., and J. D. Oliver. 2008. Population structures of two genotypes of Vibrio vulnificus in oysters (Crassostrea virginica) and seawater. Appl. Environ. Microbiol. 74:80-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright, A. C., R. T. Hill, J. A. Johnson, M. Roghman, R. R. Colwell, and J. G. Morris, Jr. 1996. Distribution of Vibrio vulnificus in the Chesapeake Bay. Appl. Environ. Microbiol. 62:717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright, A. C., J. L. Powell, J. B. Kaper, and J. G. Morris, Jr. 2001. Identification of a group 1-like capsular polysaccharide operon in Vibrio vulnificus. Infect. Immun. 69:6893-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]