Abstract
A number of Micromonospora strains isolated from the water column, sediment, and cellulose baits placed in freshwater lakes were shown to be able to degrade cellulose in lake water without any addition of nutrients. A selective isolation method was also developed to demonstrate that CFU arose from both spores and hyphae that inhabit the lake environment. Gyrase B gene sequencing performed on the isolates identified a number of new centers of variation within Micromonospora, but the most actively cellulolytic strains were recovered in a single cluster that equated with the type species of the genus, M. chalcea.
Cellulose, the most abundant form of fixed carbon in the biosphere, is insoluble and its efficient degradation is largely restricted to those specific groups of microorganisms that are able to produce multiple cellulases (14). Many microorganisms can attack amorphous cellulose to some degree, but relatively few taxa can completely degrade highly crystalline cellulose, of which the best example is cotton. In terrestrial environments, cellulose tends to be highly lignified and more difficult to degrade, and both fungi (14) and actinomycetes (15) can obtain access to cellulose in woody tissue due to their hyphal growth form. In contrast, little is known about the contributions of different groups of microorganisms to cellulose degradation in aquatic ecosystems.
Micromonosporas are a group of actinomycetes that are usually present in large numbers in soil but are well adapted to water dispersal, as evidenced by the absence of aerial hyphae and production of hydrophilic spores. They can be readily recovered from aquatic environments, especially lake sediments (4), but it is important that their presence is not equated with activity, for micromonosporas produce enormous numbers of spores that can accumulate and remain viable in lakes. Although they grow slowly and can be easily missed or outcompeted in culture-based surveys (27), the ability to degrade complex polysaccharides such as cellulose and chitin is regarded as common among the micromonosporas (12). Against this background, the study reported here concentrates on determining the presence, growth, diversity, and cellulolytic ability of Micromonospora strains colonizing cellulose baits placed in freshwater lakes.
Micromonosporas were recovered from two lakes of contrasting trophic status in the English Lake District: Esthwaite Water and Priest Pot. The former is 15 m in depth, 113 ha in area, and eutrophic (average chlorophyll a concentration, 54 μg liter−1) (5). Priest Pot is 3.5 m deep, 1 ha in area, and is hypereutrophic (average chlorophyll a concentration, 300 μg liter−1) (3). Stratification leading to the formation of an anaerobic zone occurs in the summer months in both lakes but is more persistent in Priest Pot, which is sheltered and has steep oxygen and temperature gradients. Cotton was dewaxed by repeated chloroform-ethanol extraction (29) in a Soxhlet apparatus (Quickfit; SciLabware, Stone, England) prior to use as baits for the recovery of cellulolytic microorganisms from the lakes. The baits (0.5 g) were placed in nylon mesh bags, tethered to ropes at different depths, and recovered at intervals of up to 3 months. Water was also sampled at two depths from both lakes using a Niskin bottle, and loose surface sediment samples were taken with a Jenkins sediment corer (21).
Micromonospora strains were isolated by vigorous physical shaking of the recovered baits in 10 ml phosphate-buffered saline (Sigma) followed by heat treatment of the suspension at 65°C for 10 min prior to plating in triplicate on M3 agar (17, 22). Sediment samples (20 ml) were serially diluted in sterile buffer before the heat treatment and plating on M3, while lake water samples (20 to 40 ml) were membrane filtered (0.2 μm pore size; Millipore) after heat treatment, and the membranes placed face down on M3 agar plates, incubated at 30°C and removed after 24 h. The M3 plates were incubated for a further 2 weeks, Micromonospora colonies were enumerated, and representative morphotypes were isolated and subcultured. Micromonospora colonies were identified by their appearance (small orange, red, brown or black colonies, finely branching substrate hyphae of <0.5 μm in diameter, and few or no aerial hyphae) verified by direct observation of colonies under a light microscope fitted with a 32× long-working-distance objective lens (Leitz, Wetzlar, Germany). A total of 72 isolates were obtained in this way, and these were routinely cultivated on oatmeal agar (24) plus 0.2% yeast extract with incubation at 30°C. Cultures were stored as suspensions of hyphae and spores in 12.5% (vol/vol) glycerol at −80°C.
Micromonospora numbers recovered from cellulose baits varied between 2 × 103 and 1.2 × 104 CFU per bait (0.5 g cellulose). The numbers of micromonosporas recovered from the lake water itself were in the range of 1 × 103 to 1.3 × 104 CFU liter−1, and those from sediment were in the range of 1.5 × 105 to 8.4 × 105 CFU g−1 wet weight. The numbers and distributions of micromonosporas were similar in each of the two lakes. In order to obtain evidence for the growth of micromonosporas in the lakes, a differential isolation method was developed based on that used by Wakisaka et al. (26), in which the antibiotic tunicamycin is incorporated in the M3 isolation agar at 10 μg ml−1 to inhibit the growth of gram-positive bacteria other than micromonosporas. This procedure enabled their isolation without the use of heat treatment, and by comparing CFU obtained, the relative numbers of micromonosporas from hyphal fragments and spores could be determined in any sample. Extracts were obtained from cellulose baits as described above, and 4.5 ml was added to 0.5 ml of 0.1 M NaOH on ice, neutralized with 0.5 ml of 0.1 M HCl after 5 min. This reduces the number of gram-negative bacteria (26). A 150-μl aliquot was plated on M3 agar containing 10 μg ml−1 tunicamycin. Heat treatment (65°C for 10 min) was used to destroy hyphae in the remaining aliquot (2 ml), 150 μl of which was also spread on M3 agar plates containing tunicamycin, as were 10−1 dilutions. The plates were incubated at 30°C for 15 days before identifying and counting the Micromonospora colonies. Sediment samples were diluted 10-fold in phosphate-buffered saline and processed as described above, and undiluted water column samples were filtered through 0.2-μm-pore-diameter membranes, which were placed face down on tunicamycin-containing M3 agar and removed after 24 h, after which the plates were returned to the incubator.
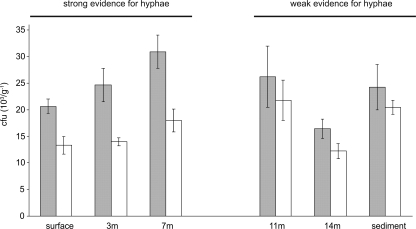
Strong evidence for the presence of Micromonospora hyphal fragments was obtained. The data in Fig. 1 show that the proportion of Micromonospora colony counts from hyphal fragments was higher in the baits closer to the surface and decreased with depth. Although there was evidence for hyphae in the baits suspended deep in the water column and at the sediment, the differences between total CFU numbers and those from Micromonospora spores were not statistically significant. Likewise, there was weak evidence for the presence of hyphae in surface waters themselves and in the sediment. In the deepest water column samples (7 and 14 m), micromonosporas were present only as spores (data not shown). This method is not ideal for quantifying absolute numbers of vegetative hyphae in the baits, but it certainly enables the conclusion that (i) micromonosporas will grow in lakes when particulate organic matter is available for attachment under aerobic conditions and (ii) the organisms are not simply present as spores washed in from the surrounding soil.
FIG. 1.
Presence of hyphae in cellulose baits placed in Esthwaite Water. Gray and white columns represent numbers of micromonosporas recovered from the baits (103 CFU per g cellulose bait) before and after heat treatment, respectively. A decrease in CFU numbers after heat treatment is indicative of the presence of hyphae in the samples, which can be observed with strong confidence in the surface, 3-m, and 7-m samples. Baits placed deeper in the water column and sediment also showed evidence for the presence of hyphae, but this was not statistically significant.
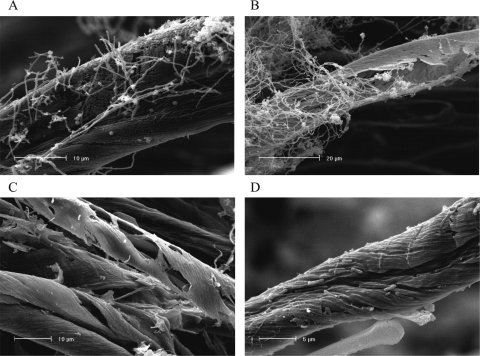
Scanning electron microscopy (SEM) was used to observe the extent of physical breakdown of cellulose by micromonosporas in lake water. A selection of 17 strains were incubated at 30°C in 20-ml universal bottles containing 4 ml of sterile lake water with dewaxed cotton (0.2 g), the degradation of which was examined by SEM. Samples were dehydrated in ethanol, critical point dried, and coated with 60% Au/Pd in a Polaron E5100 sputter coater. Degraded cotton samples were examined using a Phillips XL 30 scanning electron microscope with an accelerating voltage of 10 to 15 kV. SEM revealed different levels of degradation; colonized cellulose strands sometimes only showed superficial degradation (Fig. 2A). Often, however, the strands were penetrated and degraded from the interior, leaving an outside “shell” almost intact (Fig. 2B). Scanning electron micrographs of baits recovered from the lakes themselves revealed two main degradation patterns: hollow, similar to that observed above with Micromonospora pure cultures (Fig. 2C); and in the form of pits in the fiber surface, which appeared to be due to erosion by unicellular bacteria (Fig. 2D).
FIG. 2.
Scanning electron micrographs of cellulose degradation patterns produced by Micromonospora strains (A and B) and by natural microbial populations in the lakes Priest Pot(C) and Esthwaite Water (D).
Endoglucanase production was determined by growing Micromonospora isolates in minimal salts medium (16) supplemented with 1% fibrous filter paper powder (Whatman, Maidstone, United Kingdom) and then measuring reducing sugars released from carboxymethylcellulose incubated with culture supernatants. The enzyme assay procedure was modified from that described previously (16). One milliliter of culture supernatant, diluted appropriately in 0.1 M potassium phosphate buffer (pH 7), was added to 2 ml of 5% (wt/vol) low-viscosity carboxymethylcellulose (Sigma) dissolved in the same buffer and incubated for 1 h at 50°C with shaking at 170 rpm. Substrate and enzyme controls were used for every sample. Reducing sugars were determined by the dinitrosalicylic acid method (19). Enzyme activity was expressed as units, each unit corresponding to 1 μmol of glucose equivalents released per minute. Protein was measured in the culture supernatant and biomass extracts using the Bradford method (2). For the determination of biomass, the culture was centrifuged and cell pellets were resuspended in 1 M NaOH and boiled for 20 min. Cell debris was removed by centrifugation prior to determination of the protein concentration. In order to assess levels of activity on crystalline cellulose, the strains were incubated in test tubes containing filter paper strips and M3 plus 0.4% yeast extract to a depth of ca. 20% of the tube. Degradation was assessed by visual examination after 1 month at 30°C.
The best eight strains included the type strain of Micromonospora chalcea and produced very high levels of endoglucanase (240 to 800 U liter−1), and all extensively degraded filter paper. When incubated in minimal salts medium supplemented with 0.4% yeast extract for 6 weeks at 30°C, these strains almost completely solubilized the cellulose baits (ca. 0.2 g). The levels of enzyme activity produced per unit of biomass (cell pellet protein) indicated that these highly cellulolytic strains were not simply those with the highest growth yields. A second group of 17 isolates produced intermediate levels of endoglucanase activity (42 to 169 U liter−1), but only two of these strains degraded filter paper to any great extent, with the majority producing little or no evidence of degradation. The remaining 48 isolates were poor producers of endoglucanase activity, and none of these strains degraded filter paper to a significant extent.
The gyrase B gene was chosen as a phylogenetic marker for the isolates, as the 16S rRNA gene sequence does not resolve species structure in the genus Micromonospora (11), which was confirmed by our 16S rRNA gene sequence data for 20 of the isolates. Prior to sequencing, restriction fragment length polymorphism analysis of the amplified gyrB gene fragments (1.18 kbp) was performed in order to group the isolates and to select a subset of strains for sequence determination. The strains were grown in yeast extract-malt extract broth (10 g glucose, 3 g yeast extract, 3 g malt extract, 5 g peptone) at 30°C with shaking for 10 days. The biomass was harvested by centrifugation, and total DNA was extracted using a DNA extraction kit (Mo Bio UltraClean). PCR amplification of the gyrB gene was performed using gyrB universal bacterial primers UP1TL and UP2rTL (11). The amplification products from 67 of the 72 Micromonospora isolates were concentrated and partially purified by ethanol-sodium acetate precipitation (23) and incubated with BstNI restriction enzyme overnight at 60°C. The products were run on a 2% (wt/vol) agarose gel containing ethidium bromide (0.5 μg ml−1) for 2 h and examined using a transilluminator. This enabled recognition of seven distinguishable restriction fragment length polymorphism patterns, and multiple examples of each were sequenced.
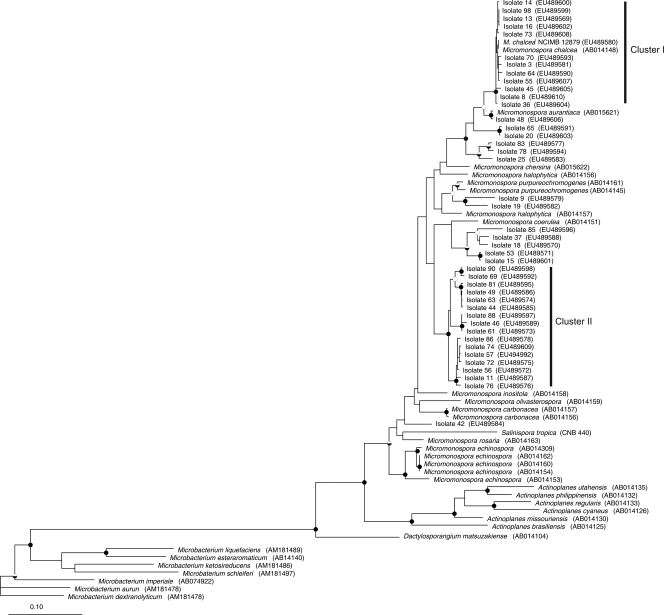
The amplification products were gel extracted using a QIAprep spin Miniprep kit and cloned using the Promega pGEM-T Easy vector plasmid cloning kit. The plasmids with inserts were extracted using the Eppendorf Perfectprep plasmid isolation kit, and insert sequences were determined by GATC Biotech (Constance, Germany). Forward and reverse gyrB gene sequences of 42 Micromonospora strains and that of the type strain of M. chalcea NCIMB 12879 were assembled together into contigs (1,180 bp) using the Pregap4 and Gap4 programs of the Staden package (25), and accuracy of the assembled traces was manually checked. Sequences in GenBank that were closely related to experimental sequences were identified using BlastN (1), and these sequences were aligned using ARB software (beta v. 2003-08-22) (13). The positions corresponding to primer sequences, gap columns, and hypervariable bases were removed, leaving a final alignment of 1,151 bp covering the region 314 to 1486 of the gyrB sequence of Escherichia coli K-12. The optimized alignment was exported, and a maximum likelihood tree was constructed using PhyML (v. 2.4.4) (6), with 100 bootstrap samplings. Phylip DNAPARS was run in ARB in order to calculate a maximum parsimony tree with vertical gap compression, random sequence order, and 100 bootstrap samplings. Tree topologies and bootstrap support for branching were compared between the two trees and found to be similar, and the maximum likelihood tree is presented in Fig. 3. All of those isolates that exhibited high cellulolytic activity were recovered in cluster I, as well as the two gyrB M. chalcea sequences: one from the type strain cultured in the laboratory and the other downloaded from the GenBank database. A second major cluster (II) containing 16 strains was delineated, but it did not include any named species from the database and may represent a novel center of variation within the genus. Other strains were related to Micromonspora coerulea and Micromonospora purpureochromogenes, whereas isolates 48, 65, 20, 83, 78, and 25 branched close to but not within the Micromonospora aurantiaca and M. chalcea clusters. Micromonospora strain 42 was not closely associated with any named species or any other lake isolate.
FIG. 3.
Maximum likelihood tree constructed with gyrB gene sequences from Micromonospora isolates, named strains, and other members of the Micromonosporaceae. The tree topology generated by both maximum likelihood and maximum parsimony methods was strongly supported by bootstrap values which were similar for both methods. The bootstrap values are represented by the circles in the branch nodes: filled circles, bootstrap values of >95% for both methods; half-filled circles, bootstrap values of >95% by only one method; open circles, bootstrap values of 75 to 95% for both methods. The tree was rooted with Microbacterium spp. as the outgroup; sequence accession numbers are in parentheses at the end of each branch. The scale represents 0.1 substitution per nucleotide position.
There are few studies on the ecology of micromonosporas in lakes to which the data presented here can be compared. Most of the information on the occurrence of Micromonospora spp. in aquatic habitats was published over 20 years ago (4, 10, 12). The presence of micromonosporas along with other actinomycetes and their interaction with fungi in submerged macrophytes and decomposing leaf matter have been investigated in some more recent studies (18, 27, 28), whereas others have focused on the diversity and isolation of members of the Micromonosporaceae from different habitats such as marine sediments and Antarctic rocks (7-9, 20). None of these studies have addressed the potential role of micromonosporas as degraders of particulate organic matter in the aquatic environment, and there have been no previous attempts to relate function to species identity.
The data presented here show that micromonosporas are part of the indigenous cellulolytic community in freshwater lakes. The importance of micromonosporas in cellulose degradation in lakes relative to other aerobic microorganisms remains to be determined. Recognition that micromonosporas can be truly aquatic actinomycetes of freshwater lakes may be added to the relatively recent classification of certain members of the Micromonosporaceae as marine microorganisms and the suggestion that they are the first actinomycetes to be identified as such (20).
Acknowledgments
We acknowledge Paul Loughnane for technical support; Cornelis Veltkamp for performing SEM analysis; Roger Pickup at the Centre of Ecology and Hydrology, Lancaster; and the Freshwater Biological Association, Ambleside, for their support with the fieldwork and access to the sampling sites.
This study was supported by a University of Liverpool studentship to A.B.M. and was partially funded by the Natural Environment Research Council of the United Kingdom.
Footnotes
Published ahead of print on 26 September 2008.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Casper, P., S. C. Maberly, G. H. Hall, and B. J. Finlay. 2000. Fluxes of methane and carbon dioxide from a small productive lake to the atmosphere. Biogeochemistry 49:1-19. [Google Scholar]
- 4.Cross, T. 1981. Aquatic actinomycetes—a critical survey of the occurrence, growth and role of actinomycetes in aquatic habitats. J. Appl. Bacteriol. 50:397-423. [DOI] [PubMed] [Google Scholar]
- 5.George, D. G., J. F. Talling, and E. Rigg. 2000. Factors influencing the temporal coherence of five lakes in the English Lake District. Freshw. Biol. 43:449-461. [Google Scholar]
- 6.Guindon, S., F. Lethiec, P. Duroux, and O. Gascuel. 2005. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557-W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch, P., U. Mevs, R. M. Kroppenstedt, P. Schumann, and E. Stackebrandt. 2004. Cryptoendolithic actinomycetes from antarctic sandstone rock samples: Micromonospora endolithica sp nov and two isolates related to Micromonospora coerulea Jensen 1932. Syst. Appl. Microbiol. 27:166-174. [DOI] [PubMed] [Google Scholar]
- 8.Jensen, P. R., E. Gontang, C. Mafnas, T. J. Mincer, and W. Fenical. 2005. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ. Microbiol. 7:1039-1048. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, C.-L., and L.-H. Xu. 1996. Diversity of aquatic actinomycetes in lakes of the Middle Plateau, Yunnan, China. Appl. Environ. Microbiol. 62:249-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnston, D. W., and T. Cross. 1976. Actinomycetes in lake muds—dormant spores or metabolically active mycelium. Freshw. Biol. 6:465-470. [Google Scholar]
- 11.Kasai, H., T. Tamura, and S. Harayama. 2000. Intrageneric relationships among Micromonospora species deduced from gyrB-based phylogeny and DNA relatedness. Int. J. Syst. Evol. Microbiol. 50:127-134. [DOI] [PubMed] [Google Scholar]
- 12.Kawamoto, I. 1984. Genus Micromonospora, p. 2442-2450. In S. T. Williams, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 4. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 13.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarthy, A. J. 1987. Lignocellulose-degrading actinomycetes. FEMS Microbiol. Rev. 46:145-163. [Google Scholar]
- 16.McCarthy, A. J., E. Peace, and P. Broda. 1985. Studies on the extracellular xylanase activity of some thermophilic actinomycetes. Appl. Microbiol. Biotechnol. 21:238-244. [Google Scholar]
- 17.McCarthy, A. J., and S. T. Williams. 1990. Methods for studying the ecology of actinomycetes. Methods Microbiol. 22:533-563. [Google Scholar]
- 18.Mille-Lindblom, C., and L. J. Tranvik. 2003. Antagonism between bacteria and fungi on decomposing aquatic plant litter. Microb. Ecol. 45:173-182. [DOI] [PubMed] [Google Scholar]
- 19.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 20.Mincer, T. J., P. R. Jensen, C. A. Kauffman, and W. Fenical. 2002. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 68:5005-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohnstad, F. R., and J. G. Jones. 1982. The Jenkins surface mud sampler. User manual. Occasional publication 15. Freshwater Biological Association, Windermere Laboratories, Cumbria, United Kingdom.
- 22.Rowbotham, T. J., and T. Cross. 1977. Ecology of Rhodococcus coprophilus and associated actinomycetes in fresh-water and agricultural habitats. J. Gen. Microbiol. 100:231-240. [Google Scholar]
- 23.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed., p. A8.12-A8.14. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Shirling, E. B., and D. Gottlieb. 1966. Methods for the characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16:313-340. [Google Scholar]
- 25.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233-241. [DOI] [PubMed] [Google Scholar]
- 26.Wakisaka, Y., Y. Kawamura, Y. Yasuda, K. Koizumi, and Y. Nishimoto. 1982. A selective isolation procedure for Micromonospora. J. Antibiot. 35:822-836. [DOI] [PubMed] [Google Scholar]
- 27.Wohl, D. L., and J. V. McArthur. 1998. Actinomycete-flora associated with submersed freshwater macrophytes. FEMS Microbiol. Ecol. 26:135-140. [Google Scholar]
- 28.Wohl, D. L., and J. V. McArthur. 2001. Aquatic actinomycete-fungal interactions and their effects on organic matter decomposition: a microcosm study. Microb. Ecol. 42:446-457. [DOI] [PubMed] [Google Scholar]
- 29.Wood, T. M. 1988. Preparation of crystalline, amorphous, and dyed cellulase substrates. Methods Enzymol. 160:19-25. [Google Scholar]