Abstract
To study how Listeria monocytogenes survives and grows in ultrahigh-temperature-processed (UHT) skim milk, microarray technology was used to monitor the gene expression profiles of strain F2365 in UHT skim milk. Total RNA was isolated from strain F2365 in UHT skim milk after 24 h of growth at 4°C, labeled with fluorescent dyes, and hybridized to “custom-made” commercial oligonucleotide (35-mers) microarray chips containing the whole genome of L. monocytogenes strain F2365. Compared to L. monocytogenes grown in brain heart infusion (BHI) broth for 24 h at 4°C, 26 genes were upregulated (more-than-twofold increase) in UHT skim milk, whereas 14 genes were downregulated (less-than-twofold decrease). The upregulated genes included genes encoding transport and binding proteins, transcriptional regulators, proteins in amino acid biosynthesis and energy metabolism, protein synthesis, cell division, and hypothetical proteins. The downregulated genes included genes that encode transport and binding proteins, protein synthesis, cellular processes, cell envelope, energy metabolism, a transcriptional regulator, and an unknown protein. The gene expression changes determined by microarray assays were confirmed by real-time reverse transcriptase PCR analyses. Furthermore, cells grown in UHT skim milk displayed the same sensitivity to hydrogen peroxide as cells grown in BHI, demonstrating that the elevated levels of expression of genes encoding manganese transporter complexes in UHT skim milk did not result in changes in the oxidative stress sensitivity. To our knowledge, this report represents a novel study of global transcriptional gene expression profiling of L. monocytogenes in a liquid food.
Listeria monocytogenes, a gram-positive bacterium, is of major concern to the food industry. This bacterium is pathogenic to both humans and animals, particularly susceptible individuals such as pregnant women, newborns, people over 65 years old, and immunocompromised patients. L. monocytogenes is widely distributed in the environment, including soil and food. Well-documented outbreaks of listeriosis have been associated with the consumption of contaminated food products including milk, soft cheese, and ready-to-eat meats (10). Because L. monocytogenes can survive in foods under harsh conditions, including high acidity and low temperature, it is very difficult to eliminate this pathogen from foods and/or food processing plants. The psychrotrophic nature of L. monocytogenes allows it to grow at refrigeration temperatures (9, 35); therefore, refrigerated storage is no guarantee of protection against the growth of L. monocytogenes in foods. Thus, special precaution should be taken to prevent the contamination of refrigerated/ready-to-eat foods by this microorganism.
The ability of L. monocytogenes to contaminate milk and milk products presents a major concern to food safety and public health (18). The consumption of certain types of cheese, such as Mexican-style cheese made from raw milk, has been linked to both sporadic cases and outbreaks of listeriosis (6, 20, 23). Physiological studies revealed that L. monocytogenes was able to survive and grow at 4°C in milk (25, 33, 35). However, the molecular mechanism of its growth remains unclear. Therefore, there is a great need for an understanding of how L. monocytogenes grows in milk to subsequently eliminate this pathogen.
The availability of whole-genome sequence information for several food-borne pathogens, including L. monocytogenes, has paved the way for a global analysis of microbial adaptation to various environments (1). For example, both genomic and proteomic approaches were used to study the behavior of lactic bacteria in milk (12, 39). Genes of Escherichia coli O157:H7 that are involved in high-pressure resistance have also been identified using the tools of genomics (24). Recently, microarrays were used to study the responses of L. monocytogenes and Bacillus subtilis to cold stress, and a number of genes that are differentially expressed at low temperatures were identified (5, 7). However, genomic studies of food-borne pathogens in a real food matrix are generally lacking.
The primary goal of this paper was to investigate the gene expression profile(s) of L. monocytogenes strain F2365 during incubation in refrigerated ultrahigh-temperature-processed (UHT) skim milk using whole-genome microarray technology. Our objective was to identify genes whose expression patterns were altered in UHT skim milk. Such studies can provide baseline data that are useful for an understanding of the adaptations of this pathogen in foods that pose special risks, such as milk and dairy products.
MATERIALS AND METHODS
Bacterial strains and culture conditions in skim milk.
L. monocytogenes strain F2365 was used since its genome is fully sequenced and annotated (28) and since it was isolated from Mexican-style soft cheese that was implicated in an outbreak of listeriosis in California in 1985 (20). Strain F2365 was streaked onto a brain heart infusion (BHI) (catalog no. 53286; Sigma-Aldrich, St. Louis, MO) agar plate from a glycerol stock culture (stored at −80°C) followed by incubation at 37°C overnight. A single colony was picked from the plate, inoculated into 5 ml of BHI broth, and grown at 37°C with agitation at 250 rpm overnight. A 5-ml aliquot of this culture grown overnight was used to inoculate 95 ml of BHI broth. After growth to mid-log phase (optical density at 600 nm of 0.4 to 0.6), 10 ml of the suspension was placed into Spectra/Por Biotech polyvinylidene difluoride dialysis tubing (Spectrum Laboratories Inc., Rancho Dominguez, CA) with a molecular mass cutoff of 1,000 kDa. The relatively high molecular mass cutoff (1,000 kDa) of the dialysis tubing keeps the bacteria inside the tube while allowing the milk components such as proteins and sugars to pass through. The dialysis tubings were placed into either 1 liter of BHI broth or 1 liter of UHT skim milk (Parmalat grade A UHT fat-free milk; Farmland Dairies, Willington, NJ), with gentle agitation. After incubation at 4°C for 24 h, bacterial cells were collected for RNA isolation. Two independent growth experiments in UHT skim milk and BHI broth were performed to ensure reproducibility. Bacterial cells were plated before and after the treatments to obtain the CFU/ml. L. monocytogenes in UHT skim milk grew after 24 h at 4°C from 1.3 × 107 CFU/ml to 2.2 × 107 CFU/ml. UHT skim milk was chosen to eliminate the potential interference of milk fat in bacterial RNA isolation. One hundred microliters of milk was plated onto BHI agar plates, and there was no growth at 37°C after 2 days, indicating that no other microorganisms were present in UHT skim milk.
RNA isolation, microarray chip design, hybridization, and data analysis.
Total RNA was isolated using a RiboPure-Bacteria kit (catalog number 1925; Ambion, Austin, TX) according to the manufacturer's instructions, with the following modification: RNA samples were incubated for 2.5 h at 37°C for DNase I treatment. The concentration and purity of RNA were evaluated using an Agilent (Wilmington, DE) 2100 bioanalyzer, and absorbance readings at 260 nm and 280 nm were performed using the Nanodrop (Wilmington, DE) ND100 UV-Vis spectrophotometer.
A whole-genome microarray was constructed to include 35-mer oligonucleotides representing the 2,847 open reading frames identified based on the annotated genome for L. monocytogenes strain F2365 (GenBank accession number AE017262) (28). For each open reading frame, two unique probes were selected to be specific as judged by pairwise BLASTN (3). The probes were designed to have similar annealing stabilities, that being a melting temperature of 72°C, as judged by a nearest-neighbor thermodynamic model (2). Probes that had significant secondary structure (melting temperature of >45°C), significant repeat structure, and/or %GC content outside of the range of 35% to 65% were rejected. Each probe was custom synthesized in duplicate by Combimatrix (Mukilteo, WA).
To save on the cost of microarray chips, the balanced block design (8) with dual-labeled microarrays was used in this study. Dye swap experiments were performed to eliminate the dye bias caused by Alexa 555 and Alexa 647. Two biological (two independent RNA sources) and two technical (same RNA samples divided into two aliquots) replicates were included to ensure accurate measurements. Ten micrograms of total RNA was reverse transcribed into cDNA and labeled with Alexa Fluor dyes (either Alexa Fluor 555 or Alexa Fluor 647) using Superscript Reverse Transcriptase III (Invitrogen Inc., Carlsbad, CA). The fluorescence incorporation into the cDNA was measured using the Nanodrop spectrophotometer. Equal amounts (50 to 100 pmol) of Alexa Fluor 555- and 647-labeled probes were mixed and used for microarray hybridization. All samples were hybridized twice with one experiment (chip 1) using Alexa Fluor 555 to label the cDNA from milk and Alexa Fluor 647 to label the cDNA from BHI medium, and in the reciprocal experiment (chip 2), Alexa Fluor 647 was used to label the cDNA from milk and Alexa Fluor 555 was used to label the cDNA from BHI medium. The expression ratio of a particular gene was calculated as follows: [chip 1 (Alexa Fluor 555/647) + chip 2 (Alexa Fluor 647/555)]/2. Each experiment was performed in duplicate. Microarray hybridization and washing were performed according to the CustomArray 12K microarray protocols provided by Combimatrix. The microarray slide was scanned at 5-μm resolutions by the scanArray ExpressHT microarray scanner (Packard Bioscience, Biochip Technologies, Billerica, MA). The intensity of the signal was quantified by Microarray Imager software provided by Combimatrix. The microarray raw data can be found elsewhere (see the supplemental material).
Microarray data were analyzed using the software package BRB-ArrayTools (version 3.4), developed by the Biometric Research Branch of the U.S. National Cancer Institute (http://linus.nci.nih.gov/BRB-ArrayTools.html) according to the instructions provided and by using statistical analysis (SAS) (36). For BRB-ArrayTools analysis, the lowest 5% of the signals were used as background. The Lowess method was used for data normalization. A minimum threshold of a twofold change in gene expression and a P value of <0.01 were used as the cutoff values. For SAS analysis, the data from the two independent experiments were transformed by taking the logarithm (base 2) of the intensity. For each experiment, the data were tested for departures from normality. The values were then analyzed using a two-stage analysis of variance approach (41) that incorporates a “normalization” model and a “gene” model. Fitting of the models was performed using the mixed procedure of the SAS software system. The genes that exceeded a P value cutoff of <0.05 (experiment-wise error rate using the Bonferroni method to account for multiple comparisons) were selected as possible sites of up- or downregulated genes.
Hydrogen peroxide sensitivity assays (disk diffusion assays).
L. monocytogenes strain F2365 was tested for its sensitivity to hydrogen peroxide as described previously (42), with the following modifications: a 5-ml stationary-phase culture of strain F2365 grown overnight was diluted into 75 ml of BHI medium and grown at 37°C until the optical density at 600 nm reached approximately 0.4. Next, 10 ml of the freshly grown culture was placed into Spectra/Por Biotech polyvinylidene difluoride dialysis tubings (molecular mass cutoff of 1,000 kDa; Spectrum Laboratories Inc., Rancho Dominguez, CA) that were then separately placed into 1 liter each BHI and UHT skim milk (Parmalat). The BHI- and UHT skim milk-containing dialysis tubings were placed onto a Stovall (Greensboro, NC) Belly Dancer shaker at speed setting 2 and placed at 4°C for 18 h. The dialysates were harvested and diluted 1:50 in BHI broth, and 150 μl was spread onto BHI agar plates to generate a lawn of cells. A 5-μl portion of sterile water containing 125 μg, 250 μg, 500 μg, 750 μg, or 1,000 μg hydrogen peroxide (Fisher Scientific, Fairlawn, NJ) was added to 6-mm filter disks and placed onto the surface of the BHI and UHT skim milk dialysate spread plates. After overnight incubation at 37°C, the diameters of the zones of inhibition were measured in millimeters. All experiments were performed in duplicate with two replicates each time.
Primer design, cDNA synthesis, and real-time PCR analysis.
Primers were designed using Primer3 (v.0.4.0) software (http://frodo.wi.mit.edu/) and selected based on the gene sequence of upregulated and downregulated genes from the microarray data. The specificity of the primer sequences was further determined using the NCBI BLASTN program against the nonredundant database, and analyses revealed that the primer sequences showed 100% homology only to L. monocytogenes strain F2365 (GenBank accession number AE017262). Primers were synthesized and purchased from IDT (www.idtdna.com). Primers from the 16S rRNA, gyrase B, and spoG housekeeping genes were tested as internal controls. Of these three housekeeping genes, spoG displayed the most consistent threshold cycle (CT) values and was chosen as the internal control gene for quantification. The same RNA samples that were used for microarray experiments were used for real-time PCR assays. The synthesis of cDNA was performed using SuperScript III First-Strand Synthesis SuperMix (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Two hundred fifty nanograms of DNase I-treated RNA was used for each cDNA synthesis reaction. Reactions without reverse transcriptase were used as negative controls. cDNA synthesis was performed using an Applied Biosystems GeneAmpPCR System 9600 apparatus.
The PCR was performed in a 96-well plate using a Bio-Rad (Hercules, CA) iQ5 real-time PCR system in a 25-μl total volume and contained 2.5 μl of 10× PCR buffer (without MgCl2), 1.5 μl 50 mM MgCl2, 0.5 μl 10 mM deoxynucleoside triphosphate mix, 0.75 μl recombinant Taq DNA polymerase (Invitrogen, Carlsbad, CA), 0.3 μl 20× EvaGreen (Biotium, Hayward, CA), 1.25 μl of each primer at 10 μM, 0.5 μl of cDNA, and nuclease-free water (Ambion). Amplification involved an initial denaturation step at 95°C, followed by 40 cycles of 95°C for 15 s, 59°C for 25 s, and 72°C for 30 s. Fluorescence data were collected at the 59°C annealing step. The final step was a melt curve of 81 cycles with a temperature range of 55°C to 95°C for 30 s and with an increase set point temperature after cycle 2 by 0.5°C. Results were visualized using the iQ5, v. 2.0, software package provided with the thermocycler. All quantitative reverse transcriptase real-time PCR (qRT-PCR) experiments were performed with three replicates in a 96-well plate. To evaluate genomic DNA contamination in the cDNA samples, “no amplification controls” (a minus-reverse transcriptase control) were included. In addition, three “no-template controls” containing all of the reverse transcriptase PCR reagents except the RNA template were included in each reaction plate. To determine relative gene expression, the CT value of the internal control gene (spoG) was subtracted from the BHI and milk samples. The ΔCCT, ΔΔCT, and the 2−fx values were calculated as described previously (32).
Microarray data accession number.
The microarray data have been deposited into the Gene Expression Omnibus database under accession number GPL6489 (www.ncbi.nlm.nih.gov/geo).
RESULTS
To identify genes that are differentially expressed in UHT skim milk, RNA isolated from L. monocytogenes strain F2365 cells incubated in UHT skim milk held at 4°C for 24 h was labeled and subjected to microarray experiments. The total RNA isolated from strain F2365 held under otherwise similar conditions in BHI broth was used as a comparison. A minimum threshold of a twofold change in gene expression with a P value of <0.01 was used as the cutoff value. All of the genes identified by microarray analysis that were differentially expressed in UHT skim milk were confirmed by qRT-PCR (see Tables 1 and 2 for primer sequences). Only genes that were up- and downregulated by both microarray and qRT-PCR assays are presented here.
TABLE 1.
Oligonucleotides used for real-time PCR to evaluate upregulated genes in L. monocytogenes strain F2365
| Gene | Forward primer sequence | Reverse primer sequence | Amplicon size (bp) |
|---|---|---|---|
| lmo0153 | GACAAAGCAAATGGTTCAGG | AAATAGCGGTGCGTAAACAG | 144 |
| lmo0154 | TGCGAAAGGTCTATCCAAAG | AAAGCGCCTGTAATGACTTG | 114 |
| lmo0234 | GCTACCAAGCGATTTTAGCAAT | AGCTCTTCCGCTTTAGCAAT | 136 |
| lmo0243 | TGAATATGGTGCGGTTCTTT | CTGCCTCTAACGCATCAAAT | 122 |
| lmo0703 | GGGGTAGTGAAAATGGGAAA | TGCAACGTAACCCATGACTA | 120 |
| lmo0899 | GTTTCGAGAAATGCGACAGT | GTGGACCAAAATCTGTTTCG | 148 |
| lmo0914 | ATTGCTGATTTCATCGGTGT | CATCCGAATCAGCTTCAATC | 109 |
| lmo0963 | AAGGCGACAACGTAACAAAC | ATTCTAATGGCGCTTTTCCT | 102 |
| lmo0974 | GAAGCAAGGTCCGTATGAGA | AGGCACCAACACCAATTAGA | 128 |
| lmo0975 | GAAAAACGTCGCAATTCAGT | GAGATTTCAGTTCGCGTTGT | 100 |
| lmo0977 | CAAGTGCTTGGAATTGGAAC | AGCTTCACGAGTGGAATCTG | 108 |
| lmo1365 | CCGATTCATCCACTTTATGC | GGCCTCATGTTCTGCTTTTA | 102 |
| lmo1443 | TACCAGCGACATTATGCTGA | AGAAGCGATTCGTTTTTCCT | 128 |
| lmo1520 | AGCTAGATGCAAGGATTTCG | GGATCACAAACGAAGAAGGA | 141 |
| lmo1622 | CGGCTTTGCTTTGTTTAGAA | GGTGTAGCCAAAGGAACAAA | 115 |
| lmo1875 | TTGTCTGGATCCGCTTTTAC | AGGCAAGACGTCTGAAACAG | 105 |
| lmo1876 | GACCATATGCTTCTGCCATC | CGGATATGTGGATGACGATT | 114 |
| lmo1877 | GTTCCGAGCAACACCATATC | TGGCTGACAAACCACTTACA | 112 |
| lmo1996 | TCCATTTTCCTTGTCATGCT | AAAATAGGATAGCCGCAACA | 103 |
| lmo2063 | TCACTCACCAAAGCTCTTCC | TCATGCGCTTCAGTTAAACA | 123 |
| lmo2064 | TCCAGTTAAGGCTTGTTTCG | ACCTGTTATCGCTCAAATCG | 109 |
| lmo2190 | TTAGCGTCGCCAGTTTTATC | GGTGACGCAAAAGACAAGTT | 110 |
| lmo2541 | TTGGCTTGGACGAGTAATTC | CCGGGAATCGTGTTTAGATA | 126 |
| lmo2459 | TGCGTTTTGAGTGAAGACAA | GTCGATGGAACACTTGGAAC | 105 |
| lmo2460 | CAACATCACGAATTTCACCA | TCCGGTCGTGATTTATTTGT | 130 |
| lmo2484 | ATCTCTTTCTGCCCCTTTGT | TGCTACGTGCAGAAGAAACA | 134 |
TABLE 2.
Oligonucleotides used for real-time PCR to evaluate downregulated genes in L. monocytogenes strain F2365
| Gene | Forward primer sequence | Reverse primer sequence | Amplicon size (bp) |
|---|---|---|---|
| lmo0037 | TTTCCTCATCCCAAATACCA | TTGCGGCGCTTATAAAGTAG | 107 |
| lmo0172 | AGGGTTATGCCCGAAAATAC | ATATTTTCCCGGTCCTTCTG | 149 |
| lmo0516 | GTACGTCCAAAGCGAAGAAA | GTGAGATGCGGTGTGACATA | 111 |
| lmo0633 | CAACTACACCAGCGACAAAA | ATTACTCCCGCGATTATCTG | 103 |
| lmo0886 | TCTGGATTAGGGTGGATTGA | CCATCGTTAGCGAGAAAAGA | 142 |
| lmo1264 | TATTTTTCGCCTTTTGCATC | TGCTTACATTCCTTGCTTCC | 139 |
| lmo1265 | TGGAGTCAGACACAGCAGAA | GCAGCCTCGTACGGTAATAA | 114 |
| lmo1352 | CTGAATGCGGTGATCGTAA | GTTACACGGCGTAATCTTGG | 100 |
| lmo1381 | ACATTTCAGCGCTATCCAAG | TTTGAACGTTAGCTGCTTGA | 110 |
| lmo1392 | CATTAATACCCCGGTTAAAACA | GAGTTTTCTTCCGCATGACT | 101 |
| lmo1808 | TTCATTACCGCTTGGTTAGC | CGTAAAAGGCGGAACAGTAA | 114 |
| lmo1814 | AGTTTCGCACGACGTACTTT | GCGAAACTTTCACTGTTCGT | 124 |
| lmo1824 | GCACCATTATGCATCCATTT | GGCCGTTCAATCGAAACTAT | 101 |
| lmo1844 | AATCACGTTCCGGTAACAAA | TTAGGTTTGCCGTTAACCAG | 103 |
| lmo1946 | TTACGCTTCCAATCCAACAT | TTTGGTTCTGCTGAGTTTCC | 132 |
| lmo2041 | AACGCTTTGACCTTCGTCTA | AGTGAAAAAGGCTTCGGTTT | 111 |
| lmo2078 | TTGCTACATCTTTGCCATCA | CAAAAGCCAAAAAGCGTAAG | 141 |
| lmo2182 | TACCGGACTTATCGGTTTTG | TAAGAACGTTTACGGCGTTT | 102 |
| lmo2209 | GCACCTGTTATTGTGCCAGT | ACCAGCGATAGTCGGAATAA | 108 |
| lmo2521 | TGGATACTCGTTGCCATCTT | CATCCTGAGTACCGTCCAGT | 117 |
| lmo2631 | CCAAAGGCCTGTAGCAGTAA | ACTGTGAGGCAAGGTGAAAG | 129 |
| lmo2846 | TCTTCCTTTACGACGACGAC | TCAACCAAGTAAACGTAAAAGAAA | 103 |
Upregulated genes of L. monocytogenes strain F2365 in UHT skim milk.
The 26 upregulated genes grouped into the following categories: genes encoding transport and binding, transcriptional regulators, proteins in amino acid biosynthesis and energy metabolism, protein synthesis, toxin production and resistance, cell division, and hypothetical proteins (Table 3).
TABLE 3.
L. monocytogenes strain F2365 genes upregulated in skim milk at 4°C as identified by microarraya and real-time PCR analysis
| Category and gene | Functionb | Fold changec
|
|
|---|---|---|---|
| Microarrayd | qRT-PCRe | ||
| Gene encoding proteins involved in transcription | |||
| lmo0153 | Oligopeptide ABC transporter; oligopeptide-binding protein | 4.9 | 163.1 |
| lmo0154 | Oligopeptide ABC transporter; permease protein | 4.7 | 14.0 |
| lmo1443 | Transporter; NRAMP family | 4.4 | 49.1 |
| lmo1875 | ABC transporter; manganese-binding protein | 16.7 | 435.5 |
| lmo1876 | Manganese ABC transporter; permease protein | 11.5 | 290.0 |
| lmo1877 | Manganese ABC transporter; ATP-binding protein | 12.3 | 71.8 |
| Gene encoding proteins involved in transcription | |||
| lmo0914 | RNA polymerase δB factor | 2.1 | 3.8 |
| Gene encoding proteins involved in amino acid biosynthesis | |||
| lmo0234 | Cysteine synthase A | 3.7 | 14.8 |
| Gene encoding proteins involved in energy metabolism | |||
| lmo1365 | Glycine cleavage system T protein | 3.7 | 10.5 |
| Gene encoding proteins involved in central intermediary metabolism | |||
| lmo0977 | Glucosamine-6-phosphate isomerase | 3.2 | 3.3 |
| Gene encoding proteins involved in protein synthesis | |||
| lmo2484 | Ribosomal subunit interface protein | 2.4 | 5.4 |
| Gene encoding proteins involved in toxin production and resistance | |||
| lmo0963 | Peroxide resistance protein Dpr | 2.6 | 8.0 |
| Gene encoding proteins involved in cell division | |||
| lmo2064 | Cell division protein FtsZ | 2.7 | 5.7 |
| Genes encoding hypothetical or unknown proteins | |||
| lmo0243 | ATP:guanido phosphotransferase family protein | 2.3 | 2.4 |
| lmo0703 | Hypothetical protein | 4.0 | 2.7 |
| lmo0899 | LysM domain protein | 4.2 | 57.2 |
| lmo0974 | Conserved hypothetical protein | 3.1 | 50.4 |
| lmo0975 | Conserved hypothetical protein | 2.9 | 34.2 |
| lmo1520 | Conserved hypothetical protein | 2.5 | 4.6 |
| lmo1622 | Conserved hypothetical protein | 2.2 | 6.9 |
| lmo1996 | Conserved hypothetical protein | 3.4 | 22.7 |
| lmo2063 | Conserved hypothetical protein | 2.6 | 3.3 |
| lmo2190 | Conserved hypothetical protein | 2.5 | 7.7 |
| lmo2459 | PspC domain protein | 4.0 | 31.6 |
| lmo2460 | Conserved hypothetical protein | 4.3 | 26.7 |
| lmo2541 | Conserved hypothetical protein | 3.2 | 31.0 |
Only the genes that met the stringent criteria for being upregulated in milk (i.e., change of more than twofold; P < 0.01) are listed here.
Gene functions are based on annotations provided by TIGR (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi).
Change indicates the transcript ratios between L. monocytogenes F2365 grown in milk and BHI medium at 4°C as determined by microarray and real-time PCR.
Numbers are average values from four independent experiments.
Numbers are average values from three independent experiments.
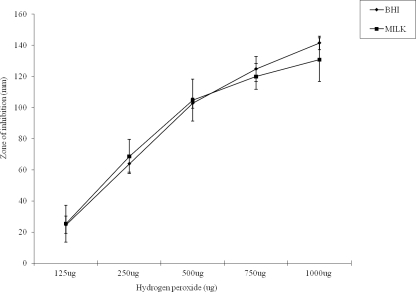
Of the six transporters whose transcript levels were upregulated, lmo1875 (ABC transporter; manganese-binding protein), lmo1876 (manganese ABC transporter; permease protein), and lmo1877 (manganese ABC transporter; ATP-binding protein) showed a >10-fold induction in the microarray assay and a >70-fold induction by real-time PCR assays (Table 3). These gene complexes are involved in manganese transport (31). In addition, lmo1443 (transporter; NRAMP) that encodes another class of manganese transporters was also upregulated by 4.4-fold on the microarray and by 49.1-fold via qRT-PCR. Manganese is involved in a number of cellular functions such as virulence and oxidative stress (31). Furthermore, lmo0963 (peroxide resistance protein Dpr) was also upregulated. The product of lmo0963 is an iron-binding protein that is responsible for oxygen tolerance in lactic acid bacteria (13). To test whether the induction of these genes in UHT skim milk resulted in any alteration in response to oxidative stress, disk diffusion assays measuring the tolerance of strain F2365 to hydrogen peroxide were performed. As shown in Fig. 1, the diameters of inhibition zones increased with increased levels of hydrogen peroxide, and cells in UHT skim milk were inhibited to the same extent as cells in BHI broth (Fig. 1). These data suggest that the elevated levels of genes encoding manganese transporter complexes in strain F2365 cells in UHT skim milk did not result in changes in the oxidative stress sensitivity. In addition, lmo0153 (oligopeptide ABC transporter; oligopeptide-binding protein) and lmo0154 (oligopeptide ABC transporter; permease protein) were upregulated about fivefold in microarray assays and 163- and 14-fold in real-time PCR assays (Table 3). These data suggest that the transporters may play important roles in the survival and growth of L. monocytogenes strain F2365 in UHT skim milk.
FIG. 1.
Susceptibility of L. monocytogenes F2365 to hydrogen peroxide in milk (rectangles) and BHI growth medium (diamonds) by disk diffusion assays. The sensitivity of L. monocytogenes F2365 to hydrogen peroxide was expressed as the diameter (mm) of the inhibition zone. Sterile 6-mm disks were saturated with different amounts of hydrogen peroxide (from 125 μg to 1,000 μg) and applied to BHI agar plates containing L. monocytogenes F2365 from milk and BHI medium. The diameters of zones of inhibition were measured after overnight growth at 37°C. Data presented here are means for four replicates ± standard deviations.
lmo0914 (RNA polymerase δB factor) regulates both virulence and stress responses (17) and was also moderately upregulated in UHT skim milk (Table 3). The fact that both lmo0914 and lmo0963 (encoding the peroxide resistance protein Dpr) were induced is consistent with the findings described previously by Polidoro et al. (34), who found that the transcription of dpr is regulated by δB. The expression of lmo0234 (cysteine synthase A) and that of lmo1365 (glycine cleavage system T protein), which are involved in amino acid biosynthesis and energy metabolism, were also induced 3.7-fold by microarray and over 10-fold by qRT-PCR assay. Likewise, lmo0977 (glucosamine-6-phosphate isomerase), which is involved in bacterial cell wall synthesis and glycolysis (19), was also upregulated. Consistent with our genomic data, a proteomic study showed that the glucosamine-6-phosphate isomerase protein was also upregulated in the growth of Lactococcus lactis in milk (12). lmo2064 encodes FtsZ, which is an abundant protein essential for cell division in bacteria (22), and was also induced at the transcriptional level (Table 3). Most of the genes that were upregulated were related to bacterial growth, which is consistent with data from previous physiological studies of the growth of L. monocytogenes in milk (25, 35). Finally, 13 genes encoding hypothetical proteins were also upregulated in UHT skim milk (Table 3), indicating that these genes might be related to the growth of L. monocytogenes in UHT skim milk.
Downregulated genes of L. monocytogenes strain F2365 in UHT skim milk.
Of the 30 downregulated genes identified by microarray assay, 14 were also downregulated by real-time PCR assay. There was a 47% correlation between microarray data and real-time PCR assay data. The lower correlation between the microarray and real-time PCR for downregulated genes may be due to the increased variability observed in low-intensity array spots, i.e., downregulated genes. Alternatively, the lower correlation between the microarray and real-time PCR assay may also be due to the effects of the greater variability associated with decreased reaction efficiencies found in real-time PCR measurements at later cycles, where the expression of genes with low expression levels becomes detected (26). The 14 downregulated genes in UHT skim milk held at 4°C included genes that encoded a cold-shock protein, a membrane protein, ribosomal proteins, a hypothetical protein, a protein involved in energy metabolism, a transporter, and a transcriptional regulator (Table 4). The transcription of lmo1264, which encodes a putative transporter, was significantly decreased (less than eightfold) in UHT skim milk. The substrate for this putative transporter remains uncharacterized in L. monocytogenes. lmo2209 encodes a putative membrane protein that was downregulated in UHT skim milk (less than twofold). The function of the putative membrane protein is unknown. lmo1381, encoding a 68-amino-acid cold shock domain family protein (CspA), was downregulated moderately (less than twofold). CspA is the major cold shock protein in E. coli that is dramatically induced after cold shock (43). The induction of the CspA protein was also caused by a nutritional upshift (44). The switching of the cells of strain F2365 from BHI broth to UHT skim milk could be considered a nutritional downshift; therefore, the downregulation of this gene in UHT skim milk is expected. The CspA protein also functions as a translational enhancer (27). Since eight genes encoding ribosomal proteins were downregulated in UHT skim milk (Table 4), the protein translation rate would be reduced; therefore, the downregulation of cspA genes is not surprising. The expression of lmo1265, which encodes a putative transcriptional regulator, was significantly decreased (less than fourfold), indicating that this gene may relate to the growth of L. monocytogenes in UHT skim milk. The potential function of lmo1265 remains uncharacterized.
TABLE 4.
L. monocytogenes strain F2365 genes downregulated in skim milk at 4°C as identified by microarraya and real-time PCR analysis
| Category and gene | Functionb | Fold changec
|
|
|---|---|---|---|
| Microarrayd | qRT-PCRe | ||
| Genes encoding proteins involved in protein synthesis | |||
| lmo1352 | Ribosomal protein L3 | −5.0 | −3.3 |
| lmo1808 | Ribosomal protein L20 | −2.4 | −3.4 |
| lmo1814 | Ribosomal protein L19 | −3.1 | −2.2 |
| lmo1824 | Ribosomal protein S16 | −2.8 | −3.2 |
| lmo1844 | Ribosomal protein L28 | −6.5 | −3.7 |
| lmo2078 | Ribosomal protein L32 | −5.3 | −5.7 |
| lmo2521 | Ribosomal protein L31 | −3.5 | −2.4 |
| lmo2846 | Ribosomal protein L34 | −3.4 | −2.8 |
| Gene encoding proteins involved in cellular processes | |||
| lmo1381 | Cold shock domain family protein | −2.4 | −2.2 |
| Gene encoding proteins involved in regulatory function | |||
| lmo1265 | Putative transcriptional regulator | −4.0 | −4.7 |
| Gene encoding proteins involved in cell envelope | |||
| lmo2209 | Putative membrane protein | −2.4 | −2.5 |
| Gene encoding proteins involved in transport and binding | |||
| lmo1264 | Putative transporter | −8.3 | −16.3 |
| Gene encoding proteins involved in energy metabolism | |||
| lmo1946 | Formate acetyltransferase | −4.8 | −3.5 |
| Gene encoding hypothetical or unknown proteins | |||
| lmo2182 | Hypothetical protein | −3.5 | −2.1 |
Only the genes that met the stringent criteria for being downregulated in milk (i.e., change of twofold; P < 0.01) are listed here.
Gene functions are based on annotations provided by TIGR (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi).
Change indicates the transcript ratios between L. monocytogenes F2365 grown in milk and BHI medium at 4°C as determined by microarray and real-time PCR; negative values indicate transcript levels that are lower in milk than in BHI broth (e.g., a value of −3.2 indicates a 3.2-fold-lower transcript level in milk than in BHI medium).
Numbers are average values from four independent experiments.
Numbers are average values from three independent experiments.
DISCUSSION
The major components of milk are proteins, sugars (mainly lactose), vitamins, and minerals. Milk contains epithelial and white blood cells (somatic cells), and the somatic cell count is commonly used to measure milk quality (30). Skim milk provides an ideal growth medium for some bacteria, such as lactic acid bacteria (37), since it is a good source of all principal nutrients including carbon, nitrogen, and microminerals. Milk proteins are comprised predominantly of casein and whey proteins. In addition, small amounts of peptides are also present in milk (38). However, milk may not be the most favorable growth medium for L. monocytogenes, since it is not able to directly use the proteins and lactose in milk (25) as nitrogen and carbon sources. The peptides present in skim milk provide an essential nitrogen source for the growth of L. monocytogenes since the concentration of free amino acids in milk is low and unbalanced (16).
Our data demonstrated that the levels of expression of both lmo0153 (oligopeptide ABC transporter; oligopeptide-binding protein) and lmo0154 (oligopeptide ABC transporter; permease protein) were elevated significantly in microarray and real-time PCR assays (Table 3). Consistent with our findings, these two genes were also induced during the growth of Lactobacillus helveticus in milk (39). Given the fact that these two genes are both required for bacterial growth at low temperatures and favor the intracellular survival of L. monocytogenes in macrophages (4), these genes are potential targets for gene knockout mutations for studying the survival of L. monocytogenes in milk. Although L. monocytogenes is unable to hydrolyze milk proteins since it does not have proteolysis activity (25), it is able to use oligopeptides (40). Since the growth of L. monocytogenes in milk is dependent upon the transport of peptides followed by their intracellular hydrolysis, the oligopeptide transporters must be activated to utilize the peptides present in the milk. The oligonucleotide transport system may be crucial to supply essential amino acids for growth. The elevated level of an oligopeptide transport system may result in the growth of L. monocytogenes in milk, but further research is needed for confirmation.
In our study, genes involved in manganese transport, including lmo1875 (ABC transporter; manganese-binding protein), lmo1876 (manganese ABC transporter; permease protein), and lmo1877 (manganese ABC transporter; ATP-binding protein), showed over a 10-fold induction in the microarray assay and over a 70-fold induction in real-time PCR assays. This result is not surprising given that milk contains manganese (21). Manganese transporters have been shown to be related to oxidative stress (31). For example, the deletion of the putative Mn(II) ABC transporter (MntA) in Bacillus anthracis resulted in increased sensitivity to oxidative stress (11). However, our data demonstrated that the elevated levels of manganese transporters in milk did not result in changes in the sensitivity of L. monocytogenes to hydrogen peroxide under the conditions of our study.
Our study demonstrated that lmo0914 (RNA polymerase δB factor) was upregulated 2.1- and 3.8-fold by microarray and real-time PCR assays, respectively. This gene regulates both virulence and stress responses (17). Other virulence genes were not appreciably altered in UHT skim milk under our experimental conditions. The reasons for this are unclear at this time. Studies with other strains would be useful in this regard, since F2365 is a member of a specific clonal group and may also have certain atypical virulence-associated characteristics. This strain carried a nonsense mutation in inlB, a key virulence gene, and was reported to be less virulent in human Caco-2 cells than were other L. monocytogenes serotype 4b strains (29). In another study, a different strain of L. monocytogenes isolated from milk also showed reduced virulence in mice and chicken embryos compared to reference strains such as 10403S and EGD (15).
Compared to other L. monocytogenes strains, there might be some strain-to-strain variation in the gene response following exposure to UHT skim milk. In addition, one incubation time (24 h) and one temperature (4°C) were evaluated in our study. In future experiments, multiple incubation times and temperatures will need to be included.
With the rapid progress of microbial genome sequencing, more and more bacteria are available for genomic and proteomic studies at the whole-genome level (1). There are some genomic and proteomic studies of the growth of lactic bacteria in milk (12, 39). Lactobacillus sakei genes that are induced in meat fermentation have also been identified (14). Our study also identified a number of genes, including some with unknown function, that were altered in L. monocytogenes strain F2365 held for 24 h at 4°C in UHT skim milk. It should be noted that our findings would be useful only to understand how L. monocytogenes behaves in UHT skim milk but may or may not be extrapolated to raw milk (or even to milk subjected to standard pasteurization). The chemical profiles of UHT skim milk and pasteurized milk may have some important differences that may affect the metabolism of the bacteria; in raw milk, the differences would be pronounced due to the presence of both active enzymes and bioactive peptides as well as the other microflora.
Genomic studies will lead to a more detailed and fundamental understanding of the mechanisms of bacterial survival and growth in food. An understanding of how bacteria survive in different food systems may help food processors develop effective preservation strategies to better manage pathogens in food. For example, since L. monocytogenes was able to grow at low temperatures, cold storage and other processing techniques such as high hydrostatic pressure could be utilized in combination to control this pathogen. Since food systems are complex, with different nutritional components, bacterial pathogens in foods may grow differently. This study can be extended to evaluate the gene expression profiling of different bacterial pathogens in different food matrices. To our knowledge, this paper represents the first report of the genomic study of a food-borne pathogen in a food matrix. This study not only provides new insights into the survival and growth of L. monocytogenes in UHT skim milk but also helps identify target genes for future functional genomic experiments. Most importantly, information from this study may help food processors develop more effective preservation strategies to control cells of L. monocytogenes present in milk due to process failures and/or postprocess contamination.
Supplementary Material
Acknowledgments
We thank John Phillips for statistical analyses, Jeff Call for providing the strains and for helpful comments, Stacy Raleigh for helping with RNA isolations, and John B. Luchansky and James Smith (USDA Agricultural Research Service, Eastern Regional Research Center, Wyndmoor, PA) for helpful discussions.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 19 September 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abee, T., W. van Schaik, and R. J. Siezen. 2004. Impact of genomics on microbial food safety. Trends Biotechnol. 22:653-660. [DOI] [PubMed] [Google Scholar]
- 2.Allawi, H. T., and J. SantaLucia, Jr. 1999. Nearest-neighbor thermodynamics and NMR of DNA sequences with internal AA, CC, GG, and TT mismatches. Biochemistry 38:3468-3477. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borezee, E., E. Pellegrini, and P. Berche. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68:7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budde, I., L. Steil, C. Scharf, U. Völker, and E. Bremer. 2006. Adaptation of Bacillus subtilis to growth at low temperature: a combined transcriptomic and proteomic appraisal. Microbiology 152:831-853. [DOI] [PubMed] [Google Scholar]
- 6.Carrique-Mas, J. J., I. Hökeberg, Y. Andersson, M. Arneborn, W. Tham, M. L. Danielsson-Tham, B. Osterman, M. Leffler, M. Steen, E. Eriksson, G. Hedin, and J. Giesecke. 2003. Febrile gastroenteritis after eating on-farm manufactured fresh cheese—an outbreak of listeriosis? Epidemiol. Infect. 130:79-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, Y. C., S. Raengpradub, K. J. Boor, and M. Wiedmann. 2007. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl. Environ. Microbiol. 73:6484-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobbin, K., J. H. Shih, and R. Simon. 2003. Questions and answers on design of dual-label microarrays for identifying differentially expressed genes. J. Natl. Cancer Inst. 95:1362-1369. [DOI] [PubMed] [Google Scholar]
- 9.Donnelly, C. W., and E. H. Briggs. 1986. Psychrotrophic growth and thermal inactivation of Listeria monocytogenes as a function of milk composition. J. Food Prot. 49:994-998. [DOI] [PubMed] [Google Scholar]
- 10.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gat, O., I. Mendelson, T. Chitlaru, N. Ariel, Z. Altboum, H. Levy, S. Weiss, H. Grosfeld, S. Cohen, and A. Shafferman. 2005. The solute-binding component of a putative Mn(II) ABC transporter (MntA) is a novel Bacillus anthracis virulence determinant. Mol. Microbiol. 58:533-551. [DOI] [PubMed] [Google Scholar]
- 12.Gitton, C., M. Meyrand, J. Wang, C. Caron, A. Trubuil, A. Guillot, and M. Y. Mistou. 2005. Proteomic signature of Lactococcus lactis NCDO763 cultivated in milk. Appl. Environ. Microbiol. 71:7152-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higuchi, M., Y. Yamamoto, and Y. Kamio. 2000. Molecular biology of oxygen tolerance in lactic acid bacteria: functions of NADH oxidases and Dpr in oxidative stress. J. Biosci. Bioeng. 90:484-493. [PubMed] [Google Scholar]
- 14.Hufner, E., T. Markieton, S. Chaillou, A. M. Crutz-Le Coq, M. Zagorec, and C. Hertel. 2007. Identification of Lactobacillus sakei genes induced during meat fermentation and their role in survival and growth. Appl. Environ. Microbiol. 73:2522-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, L. L., J. J. Xu, N. Chen, J. B. Shuai, and W. H. Fang. 2006. Virulence phenotyping and molecular characterization of a low-pathogenicity isolate of Listeria monocytogenes from cow's milk. Acta Biochim. Biophys. Sin. (Shanghai) 38:262-270. [DOI] [PubMed] [Google Scholar]
- 16.Juillard, V., D. Le Bars, E. R. Kunji, W. N. Konings, J. C. Gripon, and J. Richard. 1995. Oligopeptides are the main source of nitrogen for Lactococcus lactis during growth in milk. Appl. Environ. Microbiol. 61:3024-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiss, R., T. Tirczka, G. Szita, S. Bernáth, and G. Csikó. 2006. Listeria monocytogenes food monitoring data and incidence of human listeriosis in Hungary, 2004. Int. J. Food Microbiol. 112:71-74. [DOI] [PubMed] [Google Scholar]
- 19.Komatsuzawa, H., T. Fujiwara, H. Nishi, S. Yamada, M. Ohara, N. McCallum, B. Berger-Bächi, and M. Sugai. 2004. The gate controlling cell wall synthesis in Staphylococcus aureus. Mol. Microbiol. 53:1221-1231. [DOI] [PubMed] [Google Scholar]
- 20.Linnan, M. J., L. Mascola, X. D. Lou, V. Goulet, S. May, C. Salminen, D. W. Hird, M. L. Yonekura, P. Hayes, R. Weaver, et al. 1988. Epidemic listeriosis associated with Mexican-style cheese. N. Engl. J. Med. 319:823-828. [DOI] [PubMed] [Google Scholar]
- 21.Lonnerdal, B., C. L. Keen, and L. S. Hurley. 1981. Iron, copper, zinc, and manganese in milk. Annu. Rev. Nutr. 1:149-174. [DOI] [PubMed] [Google Scholar]
- 22.Lutkenhaus, J. 1993. FtsZ ring in bacterial cytokinesis. Mol. Microbiol. 9:403-409. [DOI] [PubMed] [Google Scholar]
- 23.MacDonald, P. D., R. E. Whitwam, J. D. Boggs, J. N. MacCormack, K. L. Anderson, J. W. Reardon, J. R. Saah, L. M. Graves, S. B. Hunter, and J. Sobel. 2005. Outbreak of listeriosis among Mexican immigrants as a result of consumption of illicitly produced Mexican-style cheese. Clin. Infect. Dis. 40:677-682. [DOI] [PubMed] [Google Scholar]
- 24.Malone, A. S., Y. K. Chung, and A. E. Yousef. 2006. Genes of Escherichia coli O157:H7 that are involved in high-pressure resistance. Appl. Environ. Microbiol. 72:2661-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall, D. L., and R. H. Schmidt. 1991. Physiological evaluation of stimulated growth of Listeria monocytogenes by Pseudomonas species in milk. Can. J. Microbiol. 37:594-599. [DOI] [PubMed] [Google Scholar]
- 26.Morey, J. S., J. C. Ryan, and F. M. Van Dolah. 2006. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol. Proced. Online 8:175-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakaminami, K., D. T. Karlson, and R. Imai. 2006. Functional conservation of cold shock domains in bacteria and higher plants. Proc. Natl. Acad. Sci. USA 103:10122-10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nightingale, K. K., S. R. Milillo, R. A. Ivy, A. J. Ho, H. F. Oliver, and M. Wiedmann. 2007. Listeria monocytogenes F2365 carries several authentic mutations potentially leading to truncated gene products, including inlB, and demonstrates atypical phenotypic characteristics. J. Food Prot. 70:482-488. [DOI] [PubMed] [Google Scholar]
- 30.Ogola, H., A. Shitandi, and J. Nanua. 2007. Effect of mastitis on raw milk compositional quality. J. Vet. Sci. 8:237-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papp-Wallace, K. M., and M. E. Maguire. 2006. Manganese transport and the role of manganese in virulence. Annu. Rev. Microbiol. 60:187-209. [DOI] [PubMed] [Google Scholar]
- 32.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pine, L., G. B. Malcolm, J. B. Brooks, and M. I. Daneshvar. 1989. Physiological studies on the growth and utilization of sugars by Listeria species. Can. J. Microbiol. 35:245-254. [DOI] [PubMed] [Google Scholar]
- 34.Polidoro, M., D. De Biase, B. Montagnini, L. Guarrera, S. Cavallo, P. Valenti, S. Stefanini, and E. Chiancone. 2002. The expression of the dodecameric ferritin in Listeria spp. is induced by iron limitation and stationary growth phase. Gene 296:121-128. [DOI] [PubMed] [Google Scholar]
- 35.Rosenow, E. M., and E. H. Marth. 1987. Growth of Listeria monocytogenes in skim, whole and chocolate milk, and in whipping cream during incubation at 4, 8, 13, 21 and 35°C. J. Food Prot. 50:452-459. [DOI] [PubMed] [Google Scholar]
- 36.SAS Institute, Inc. 2004. SAS/STAT 9.1 user's guide. SAS Institute, Inc., Cary, NC.
- 37.Savijoki, K., H. Ingmer, and P. Varmanen. 2006. Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 71:394-406. [DOI] [PubMed] [Google Scholar]
- 38.Severin, S., and X. Wenshui. 2005. Milk biologically active components as nutraceuticals: review. Crit. Rev. Food Sci. Nutr. 45:645-656. [DOI] [PubMed] [Google Scholar]
- 39.Smeianov, V. V., P. Wechter, J. R. Broadbent, J. E. Hughes, B. T. Rodríguez, T. K. Christensen, Y. Ardö, and J. L. Steele. 2007. Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium. Appl. Environ. Microbiol. 73:2661-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verheul, A., F. M. Rombouts, and T. Abee. 1998. Utilization of oligopeptides by Listeria monocytogenes Scott A. Appl. Environ. Microbiol. 64:1059-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolfinger, R. D., G. Gibson, E. D. Wolfinger, L. Bennett, H. Hamadeh, P. Bushel, C. Afshari, and R. S. Paules. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8:625-637. [DOI] [PubMed] [Google Scholar]
- 42.Wonderling, L. D., B. J. Wilkinson, and D. O. Bayles. 2004. The htrA (degP) gene of Listeria monocytogenes 10403S is essential for optimal growth under stress conditions. Appl. Environ. Microbiol. 70:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamanaka, K., L. Fang, and M. Inouye. 1998. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol. Microbiol. 27:247-255. [DOI] [PubMed] [Google Scholar]
- 44.Yamanaka, K., and M. Inouye. 2001. Induction of CspA, an E. coli major cold-shock protein, upon nutritional upshift at 37 degrees C. Genes Cells 6:279-290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.