Abstract
Sigma B (σB) is an alternative sigma factor that controls the transcriptional response to stress in Listeria monocytogenes and is also known to play a role in the virulence of this human pathogen. In the present study we investigated the impact of a sigB deletion on the proteome of L. monocytogenes grown in a chemically defined medium both in the presence and in the absence of osmotic stress (0.5 M NaCl). Two new phenotypes associated with the sigB deletion were identified using this medium. (i) Unexpectedly, the strain with the ΔsigB deletion was found to grow faster than the parent strain in the growth medium, but only when 0.5 M NaCl was present. This phenomenon was independent of the carbon source provided in the medium. (ii) The ΔsigB mutant was found to have unusual Gram staining properties compared to the parent, suggesting that σB contributes to the maintenance of an intact cell wall. A proteomic analysis was performed by two-dimensional gel electrophoresis, using cells growing in the exponential and stationary phases. Overall, 11 proteins were found to be differentially expressed in the wild type and the ΔsigB mutant; 10 of these proteins were expressed at lower levels in the mutant, and 1 was overexpressed in the mutant. All 11 proteins were identified by tandem mass spectrometry, and putative functions were assigned based on homology to proteins from other bacteria. Five proteins had putative functions related to carbon utilization (Lmo0539, Lmo0783, Lmo0913, Lmo1830, and Lmo2696), while three proteins were similar to proteins whose functions are unknown but that are known to be stress inducible (Lmo0796, Lmo2391, and Lmo2748). To gain further insight into the role of σB in L. monocytogenes, we deleted the genes encoding four of the proteins, lmo0796, lmo0913, lmo2391, and lmo2748. Phenotypic characterization of the mutants revealed that Lmo2748 plays a role in osmotolerance, while Lmo0796, Lmo0913, and Lmo2391 were all implicated in acid stress tolerance to various degrees. Invasion assays performed with Caco-2 cells indicated that none of the four genes was required for mammalian cell invasion. Microscopic analysis suggested that loss of Lmo2748 might contribute to the cell wall defect observed in the ΔsigB mutant. Overall, this study highlighted two new phenotypes associated with the loss of σB. It also demonstrated clear roles for σB in both osmotic and low-pH stress tolerance and identified specific components of the σB regulon that contribute to the responses observed.
Listeria monocytogenes is a gram-positive bacterium that has a remarkable ability to grow and survive in very diverse environments. It is almost ubiquitous in nature and can readily be isolated from soil, water, and decaying vegetation. It has been found associated with a wide range of different food products (21, 30, 39, 48), which can be problematic since it causes life-threatening illness in humans, particularly individuals who are immunocompromised. It can grow at temperatures as low as 0°C (17, 57) and can survive in the presence of wide ranges of salt concentrations (17) and pH values (56). In animals L. monocytogenes is a facultative intracellular pathogen that is capable of invading and growing within epithelial cells and macrophages. It can also grow and persist within the gall bladder (28), displaying a remarkable tolerance for bile salts.
The tolerance of L. monocytogenes to a variety of harsh environmental conditions is at least partly attributed to genes under the control of the alternative sigma factor, sigma B (σB). This sigma factor is conserved in closely related gram-positive bacteria, where it coordinates the transcription of genes required for the general stress response (11, 12, 14, 26, 32, 43, 44, 50, 60). Mutants of L. monocytogenes that lack the sigB gene have a variety of stress-sensitive phenotypes. They are sensitive to low pH (24, 55, 59), osmotic stress (5, 25, 55), bile salts (8), bacteriocins (7), and high hydrostatic pressure (58). They survive poorly during prolonged carbon limitation (16, 23, 29), and they grow poorly at low temperatures (5, 6). In addition, mutants lacking sigB have a reduced ability to invade epithelial cells, a finding that is explained by the low levels of internalin expression in such mutants (35, 38). While some of the stress-sensitive phenotypes can be partially explained at the molecular level, it is clear that full elucidation of the σB regulon is required to understand all of the phenotypes observed in sigB mutant strains.
A number of studies have sought to identify components of the σB regulon in L. monocytogenes, either using gene array technology (34, 45) or using proteomic approaches (1, 58). Collectively, these studies have identified about 150 L. monocytogenes genes that are expressed in a σB-dependent manner. The functional categories represented by these genes include stress tolerance, carbon metabolism, transport, cell envelope-related functions, and virulence. In each of these studies L. monocytogenes was cultured in brain heart infusion (BHI) medium, a complex growth medium with an undefined composition. In our recent proteomic study we found that σB-dependent differences in expression could not be detected in the exponential phase of growth in BHI medium, suggesting that cells experience little stress under these growth conditions. This medium also does not allow clear analysis of osmotolerance since it potentially contains an undefined mixture of different compatible solutes. Moreover, the complexity of this growth medium makes it impossible to study the utilization of individual nutrients by wild-type or sigB mutant strains. For these reasons in the present study we used a chemically defined growth medium (DM) to compare protein expression in wild-type and ΔsigB strains of L. monocytogenes.
Overall, this study highlighted two new phenotypes associated with the loss of σB. Mutants lacking σB have a rapid-growth phenotype in DM with added NaCl, and they also exhibit an unusual pattern of Gram staining. The present study also revealed clear roles for σB in both osmotic and low-pH stress tolerance and identified specific components of the σB regulon that contribute to these characteristics.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. All the L. monocytogenes strains were cultured either in BHI broth or in the DM described previously (2) supplemented with either 0.04% (wt/vol) glucose, 0.4% (wt/vol) glucose, 0.4% (wt/vol) mannose, 0.4% (wt/vol) fructose, 0.4% (wt/vol) trehalose, or 0.4% (wt/vol) cellobiose in the presence or absence of 0.5 M NaCl with continuous shaking at 37°C. L. monocytogenes strains containing the shuttle vector pKSV7 or derivatives of this vector were cultured in BHI broth supplemented with 12.5 μg ml−1 chloramphenicol. Escherichia coli strains were routinely grown in Luria-Bertani broth (1% [wt/vol] tryptone peptone, 1% [wt/vol] NaCl, 0.5% [wt/vol] yeast extract) with continuous shaking at 37°C. E. coli strains containing the shuttle vector pKSV7 or derivatives of this vector were cultured in Luria-Bertani broth containing 50 μg ml−1 ampicillin. When required, growth was monitored using 96-well plates. The plates were incubated at 37°C in a 96-well plate reader (Genios, Tecan) and shaken every 30 min for 30 s. Optical densities at 595 nm (OD595) were automatically recorded in triplicate for each well for 48 h. All cultures were inoculated to obtain a starting OD600 of ∼0.05 using 16-h overnight cultures as inocula. Two-dimensional gel electrophoresis (2-DGE) experiments were carried out with cells grown in DM either to the exponential phase (OD600, ∼0.6) or following 8 h of growth to stationary phase.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| E. coli strain TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| L. monocytogenes strains | ||
| 10403S | Serotype 1/2a | S. Foster |
| ΔsigB | 10403S derivative with sigB deleted | K. Boor |
| Δlmo0796 | 10403S derivative with lmo0796 deleted | This study |
| Δlmo0913 | 10403S derivative with lmo0913 deleted | This study |
| Δlmo2391 | 10403S derivative with lmo2391 deleted | This study |
| Δlmo2748 | 10403S derivative with lmo2748 deleted | This study |
| Plasmids | ||
| pKSV7 | Temperature-sensitive replication, chloramphenicol resistance | 52 |
| pKAK0913 | pKSV7 containing an insert with the lmo0913 gene | This study |
| pKAK2391 | pKSV7 containing an insert with the lmo2391 gene | This study |
| pKAK2748 | pKSV7 containing an insert with the lmo2748 gene | This study |
| pFA1 | pKSV7 containing Δlmo0796 DNA deletion cassette | This study |
| pFA2 | pKSV7 containing Δlmo0913 DNA deletion cassette | This study |
| pFA4 | pKSV7 containing Δlmo2391 DNA deletion cassette | This study |
| pFA5 | pKSV7 containing Δlmo2748 DNA deletion cassette | This study |
2-DGE.
Proteins were extracted by sonication and subsequently separated by 2-DGE using a modified version of the O'Farrell method (42), as previously described (1). Briefly, the first dimension consisted of isoelectric focusing using 11-cm IPG strips with linear pH gradients (pH 4 to 7; Amersham). The second-dimension gels contained 12% acrylamide and were run in pairs along with molecular mass markers with a range of 10 to 225 kDa (Broad Range protein molecular markers supplied by Promega). Gels were stained overnight in GelCode Blue staining reagent (Pierce) and then destained in deionized, distilled water for several hours. For each growth condition and strain investigated six gels were run, representing samples extracted from two independent cultures and three technical replicates. Gels were analyzed with PDQuest-Advanced software, version 8.0 (Bio-Rad), and data were normalized using the local regression model. Protein expression differences greater than twofold that were obtained for all six pairs of replicates were considered significant. The proteins were identified using a combination of tryptic digestion and matrix-assisted laser desorption ionization-time of flight mass spectrometry, as previously described (10).
Construction of in-frame deletion mutants and complementation.
We constructed four individual in-frame deletion mutants of L. monocytogenes using the splicing by overlap extension PCR technique, followed by allelic replacement (33). This procedure relies on two recombination events: homologous recombination into the L. monocytogenes chromosome of a truncated copy of the gene of interest carried on the suicide shuttle vector pKSV7 (52), followed by a second recombination event leading to the loss of the wild-type copy of the gene of interest along with the suicide vector. Nonpolar lmo0796, lmo0913, lmo2391, and lmo2748 mutants with 492-, 1,407-, 516-, and 399-bp internal deletions, respectively, were created. Primers were designed for each gene (Table 2) to amplify two fragments (AB and CD) of similar size flanking the portion of the gene to be deleted. The amplified flanking regions were mixed at a 1:1 ratio and reamplified using outer flanking primers A and D (Table 2). The resulting AD fragments were digested with EcoRI and cloned into pKSV7. Each plasmid carrying the truncated copy of the gene of interest was transformed into E. coli TOP10 competent cells (Invitrogen), purified, and subsequently electroporated into L. monocytogenes 10403S. Transformants were selected by growth for 48 h at 30°C on BHI agar plates containing chloramphenicol (12.5 μg ml−1). Subsequently, plasmid integration was forced by growing the organisms at a nonpermissive temperature (42°C) on BHI agar plates in the presence of chloramphenicol. Plasmid excision was achieved by continuous passage of cells growing at 30°C in BHI medium containing no antibiotics with shaking and spreading at intervals onto BHI agar plates. Replica plating on BHI agar plates with and without 12.5 μg ml−1 chloramphenicol allowed screening for vector loss. Homologous recombination was confirmed by PCR using outside primers (for and rev primers [Table 2]) with chloramphenicol-sensitive colonies.
TABLE 2.
Primers used in this study
| Primera | Sequence (5′ to 3′) |
|---|---|
| lmo0796 A | CGGAATTCCCTAAACTCGCTGCATTTTG |
| lmo0796 B | CCATTTTTCTACTGTCATTGTTTCATTCCTCC |
| lmo0796 C | ATGACAGTAGAAAAATGGCAAATCGAAGCAAGTAAATAATTG |
| lmo0796 D | CGGAATTCCTGTCACTAAAAAAGCTGC |
| lmo0796 for | GCAGTAGAGCGTTTTGATAAG |
| lmo0796 rev | CGACTTTTTTAATTAGCCCG |
| lmo0913 A | CGGAATTCGGAGCAGTTTTTGTTAGCC |
| lmo0913 B | TGCTGTTTCTTTAATACTCAAAAATACCACTCC |
| lmo0913 C | AGTATTAAAGAAACAGCAATCCAAGTGAAATTC |
| lmo0913 D | CGGAATTCAAGTGTGCCATAAGTTGC |
| lmo0913 for | GGTTACTATTCATGGGCAG |
| lmo0913 rev | CATCCTGTTATCCCTCC |
| lmo0913-compF | AGGGATCCGGAGCAGTTTTTGTTAGCC |
| lmo0913-compR | CGGGATCCAAGTGTGCCATAAGTTGC |
| lmo2391 A | CCGAATTCCCGAAACGCTAGAGATTG |
| lmo2391 B | TTGCTCGGCTTTTCTAACCATTGCTCGGAC |
| lmo2391 C | GTTAGAAAAGCCGAGCAATAATAAAAAGCGATGAGC |
| lmo2391 D | CGGAATTCTTTGTGGTGGAGTATGTGG |
| lmo2391 for | CTATCGCGATTAAAACACC |
| lmo2391 rev | GGAAACAGAGGAATGGC |
| lmo2748 A | CGGAATTCTTGCTGGTACCACTTTTG |
| lmo2748 B | TTCATTTCTCATCTCAAACTCCTTCC |
| lmo2748 C | TTTGAGATGAGAAATGAAATTGGTTAAAGATTTGC |
| lmo2748 D | CCGAATTCCTCCTCCTTTATTTTTTTCC |
| lmo2748 for | GCTGTGAATGGGTTTGG |
| lmo2748 rev | CTCGTCAATTTCGCGTAG |
| M13 for | TGTAAAACGACGGCCAG |
| M13 rev | CAGGAAACAGCTATGAC |
Most of the primers were designed based on the previously published sequences of L. monocytogenes EGD from the Listilist website (http://genolist.pasteur.fr/listilist/); the exceptions were the M13 for and M13 rev primers, which were directed against the vector pKSV7 and were purchased from Invitrogen. The A, B, C, and D primers were used to generate the deletion cassettes.
Complementation studies were carried out by transforming each of the deletion mutants with a vector carrying the wild-type version of the deleted gene. In short, each gene was amplified by PCR, resulting in an amplicon containing the wild-type version of the gene, including its native promoter region. The corresponding A and D primers used for construction of the deletion cassettes were also used for amplification of the lmo0796, lmo2391, and lmo2748 genes for cloning, while primers lmo0913-compF and lmo0913-compR were used for cloning lmo0913 (Table 2). Subsequently, each of the amplicons carrying the lmo0913 (2,196 bp), lmo2391 (1,511 bp), or lmo2748 (1,165 bp) gene was cloned into pCR-XL-TOPO (Invitrogen Life Technologies) and then subcloned in pKSV7, resulting in plasmids pKAK0913, pKAK2391, and pKAK2748, respectively (Table 1); it was not possible to clone the lmo0796 gene amplicon (1,278 bp) into either pCR-XL-TOPO or pKSV7. Cloning in pKSV7 was performed by digestion of the vector and the amplicons with EcoRI for lmo0796 and lmo2391 and with BamHI for lmo0913 and lmo2748, followed by ligation with T4 ligase. The newly constructed plasmids were electroporated into E. coli One-Shot TOP10 electrocompetent cells purchased from Invitrogen Life Technologies (200 Ω, 25 μF, and 2.4 kV for 0.2-cm cuvettes) and subsequently into both the L. monocytogenes wild-type strain and the corresponding mutant strains (400 Ω, 25 μF, and 2 kV for 0.2-cm cuvettes). All strains were also transformed with the empty vector pKSV7 as a control. The resulting transformants were used for the complementation studies.
Acid resistance.
L. monocytogenes strains were grown for 16 h in BHI broth containing 0.5 M NaCl (BHIS) with shaking at 37°C. Two 1-ml aliquots were centrifuged at 13,000 × g for 2 min. For each culture, one of the cell pellets was resuspended in 1 ml of fresh BHIS in order to determine the initial count for the bacterial population. The other cell pellet was resuspended in 1 ml of fresh BHIS previously acidified to pH 2.5 with HCl. Assay tubes were incubated at room temperature, and 20-μl aliquots were removed at regular intervals, serially diluted, and plated in triplicate on BHI agar plates, which were incubated at 37°C for 48 h before enumeration of CFU.
Caco-2 invasion assays.
The gentamicin protection assay was performed with the wild-type strain and the mutants derived from it, as described previously by Elsinghorst (20), with minor modifications. Two days before the invasion assays were performed, 1.5 × 105 Caco-2 human colon adenocarcinoma cells (European Collection of Cell Cultures number 86010202) were seeded in 24-well plates in Dulbecco's modified Eagle's medium containing 2 mM glutamine, 1% (wt/vol) nonessential amino acids, and 20% (vol/vol) fetal bovine serum supplemented with 100 U ml−1 penicillin/streptomycin (Sigma). Thirty minutes before coincubation, the medium in each well was replaced with prewarmed fresh medium without antibiotics. The OD600 of stationary-phase bacterial cultures grown in BHI broth overnight at 37°C were determined, all cultures were washed twice with sterile phosphate-buffered saline (PBS), and the concentrations were adjusted to obtain similar OD600 values. We previously confirmed that there was a good correlation between OD600 and the number of cells for each strain, as assessed by comparing numbers of CFU and OD600 values. Coincubation was performed with approximately 108 CFU of stationary-phase bacteria for 45 min at 37°C. Subsequently, Caco-2 cells were washed twice with PBS and suspended in Dulbecco's modified Eagle's medium containing 150 mg liter−1 gentamicin. After 45 min of incubation at 37°C, cells were washed twice with sterile PBS and lysed with 2 ml Triton X-100 (1% vol/vol) in PBS. Following incubation for 5 min at 37°C, cell lysates were serially diluted and plated on BHI agar to determine the number of intracellular bacteria. The invasion efficiency (expressed as a percentage) was calculated by using the ratio of the number of bacterial cells that survived the gentamicin assay to the total number of bacterial cells added initially to each well. A statistical analysis was performed using the Mann-Whitney nonparametric test, which was appropriate for the number of observations conducted (8 < n < 20). P values less than 0.05 were considered statistically significant.
Gram staining.
Gram staining of L. monocytogenes strains was performed with colonies from BHI agar plates. Single colonies were suspended in sterilized distilled water, and a drop of a suspension was placed on a glass microscope slide and fixed by heating. The heat-fixed cells were flooded with crystal violet for 1 min. Then an iodine solution was added for 3 min, and decolorization with alcohol was performed for 20 s, which was followed by counterstaining by addition of safranin for 2 min. Stained bacterial cells were visualized with a Nikon Eclipse E600 microscope, and the images were captured using Q Capture Pro software, version 5.1 (QImaging).
RESULTS
L. monocytogenes ΔsigB mutant grows faster than the wild type in DMS.
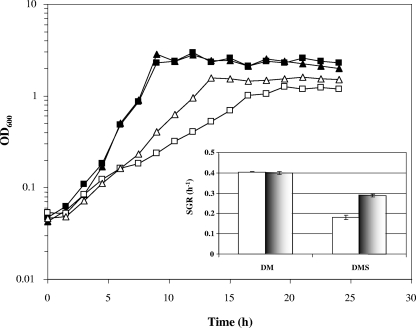
The growth of L. monocytogenes 10403S and the growth of the corresponding ΔsigB mutant were monitored in DM supplemented with 0.4% (wt/vol) glucose both in the presence of 0.5 M NaCl (DMS) and in the absence of 0.5 M NaCl (DM). In DM the two strains exhibited very similar patterns of growth, and during exponential phase they grew with a doubling time of approximately 100 min (Fig. 1). The presence of 0.5 M NaCl caused both strains to grow more slowly, but surprisingly, the sigB deletion mutant was found to grow significantly faster than the wild-type strain. The growth rate of the ΔsigB mutant under these conditions was approximately 60% higher than that of the parent strain (Fig. 1, inset). This result was quite unexpected since σB is known to play a positive role in osmoregulation, at least in media containing compatible solutes. The presence of the ΔsigB allele in mutant cultures was confirmed by performing PCRs with samples taken from each culture (data not shown).
FIG. 1.
Rapid-growth phenotype of an L. monocytogenes ΔsigB mutant in the presence of 0.5 M NaCl. L. monocytogenes wild-type strain 10403S (squares) and the ΔsigB mutant (triangles) were grown in defined media supplemented with 0.4% (wt/vol) glucose at 37°C with shaking in the absence (DM) (filled symbols) and in the presence (DMS) (open symbols) of 0.5 M NaCl. Representative growth curves for the conditions investigated are shown; all experiments were performed in triplicate. The inset shows the specific growth rates (SGR) during exponential growth derived from the curves for the wild type (open bars) and the ΔsigB mutant (shaded bars) in DM and DMS. The error bars indicate the standard deviations from the means of triplicate measurements.
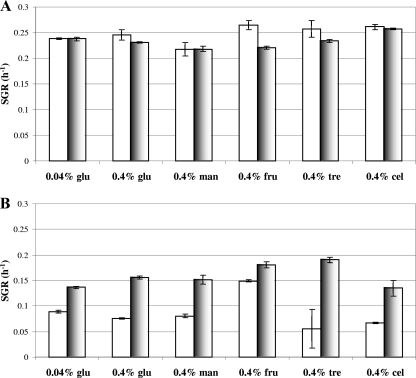
We investigated the possibility that the higher growth rate of the ΔsigB mutant in DMS might have been due to an ability of this strain to utilize glucose more efficiently than the parent strain. If this phenomenon was specific for glucose, then it should not have been observed if alternative carbon sources were provided in the growth media. Therefore, the growth rates of the two strains were compared in DM and DMS without glucose and supplemented with either mannose, fructose, trehalose, or cellobiose. These growth experiments were performed in a temperature-controlled 96-well plate reader, where the growth rate of L. monocytogenes is typically about 50% of that observed in conical flasks, presumably due to differences in culture aeration. Under these growth conditions with 0.4% (wt/vol) glucose, the wild-type strain and ΔsigB mutant were again found to grow at the same rate in DM (Fig. 2A), while the rapid-growth phenotype of the ΔsigB mutant in DMS was confirmed (Fig. 2B). The rapid-growth phenotype of the ΔsigB mutant was also observed after addition of 0.5 M NaCl when the growth medium was supplemented with either 0.4% (wt/vol) fructose, 0.4% (wt/vol) mannose, 0.4% (wt/vol) trehalose, 0.4% (wt/vol) cellobiose, or a limiting concentration of glucose (0.04%, wt/vol) (Fig. 2B). The effect was less dramatic for fructose than for the other carbon sources tested. In all cases, in the absence of salt (Fig. 2A), the wild type and the sigB deletion strain grew at similar rates. In each case addition of salt to the medium resulted in both strains growing at a lower rate, and the decrease was more dramatic for the wild-type strain (Fig. 2). These data indicate that the rapid-growth phenotype of the ΔsigB mutant in DMS is not dependent on the carbon source provided in the medium.
FIG. 2.
ΔsigB mutant exhibits greater specific growth rates than the wild-type strain in DM supplemented with different carbon sources in the presence of 0.5 M NaCl. Specific growth rates (SGR) of L. monocytogenes 10403S (open bars) and the corresponding sigB deletion mutant (shaded bars) in defined media supplemented with 0.04 or 0.4% (wt/vol) glucose (glu), 0.4% (wt/vol) mannose (man), 0.4% (wt/vol) fructose (fru), 0.4% (wt/vol) trehalose (tre), or 0.4% (wt/vol) cellobiose (cel) were determined. Cells were grown at 37°C in 96-well plates in the absence (A) and presence (B) of 0.5 M NaCl. The errors bars indicate the standard deviations from the means of triplicate measurements.
σB-Dependent protein expression.
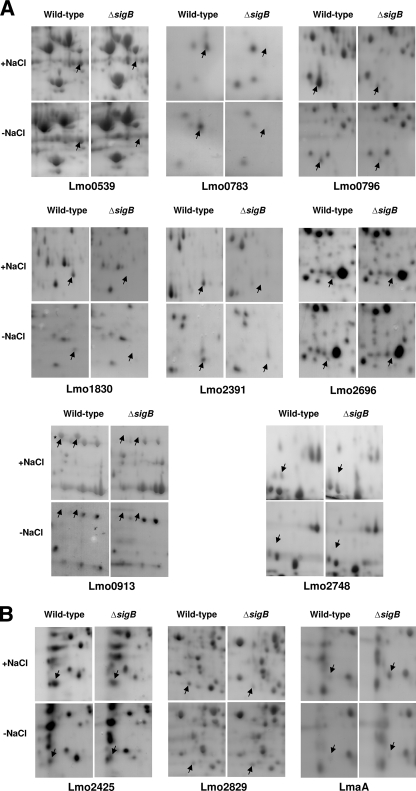
We performed a proteomic comparison of L. monocytogenes wild-type strain 10403S and the corresponding ΔsigB mutant. Protein expression in both DM and DMS was investigated in both the exponential and stationary phases of growth. Overall, 11 proteins were found to be differentially expressed in the ΔsigB background under all conditions tested (Fig. 3 and Table 3). The expression pattern of each of these 11 proteins is summarized in Table 3. Ten proteins were found to be under positive control of σB, while the level of expression of one protein, LmaA, was higher in the ΔsigB mutant than in the wild-type strain. Eight proteins (Lmo0539, Lmo0783, Lmo0796, Lmo1830, Lmo2391, Lmo2696, Lmo0913, and Lmo2748) (Fig. 3A) were always expressed in a σB-dependent manner when they were detectable. The levels of two of these proteins, Lmo0796 and Lmo0539, were lower in the ΔsigB mutant strain, while the other six proteins were undetectable in extracts prepared from the ΔsigB mutant but were clearly detectable in extracts of the wild-type strain (Fig. 3A).
FIG. 3.
Eleven proteins were found to be expressed in a σB-dependent manner. The gel images show representative sections of 2-DGE profiles for proteins extracted from either L. monocytogenes wild-type strain 10403S or ΔsigB cells grown to exponential phase or stationary phase in DM. Proteins were extracted from cells growing either in the presence (+NaCl) or absence (−NaCl) of 0.5 M NaCl. The arrows indicate the locations of the proteins showing altered expression patterns in the ΔsigB background. (A) Proteins showing σB-dependent expression in stationary phase (the same proteins showed σB-dependent expression in exponential phase [data not shown]). The asterisk indicates the Lmo0913 protein referred to as Lmo0913a. (B) Proteins showing σB-dependent expression under specific conditions. Extracts of exponential-phase cells were used for Lmo2425, while extracts of stationary-phase cells were used for Lmo2829 and LmaA.
TABLE 3.
Proteins identified in extracts of DM and DMS cultures
| Protein | Homologous protein (% identity/% similarity)a | Microorganism with homologous protein | Mol wt, kDa (predicted/observed) | pI (predicted/observed) | % Coverage | % Matched peptides | Score (%)b | Expression in DMc
|
Expression in DMSc
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Exponential phase | Stationary phase | Exponential phase | Stationary phase | ||||||||
| Lmo0539 | Tagatose-1,6-diphosphate aldolase (55/76) | Staphylococcus aureus | 37.8/39.0 | 4.9/4.8 | 51 | 71 | 1,024 | x | x | x | x |
| Lmo0783 | Phosphotransferase system comp-ponent IIB (72/82) | Escherichia coli | 17.9/17.0 | 6.2/6.6 | 39 | 57 | 265 | x | x | x | x |
| Lmo0796 | YceI (44/61) | Escherichia coli | 19.3/19.0 | 4.6/4.5 | 47 | 69 | 484 | x | x | x | x |
| Lmo1830 | Short-chain dehydrogenase (47/67) | Pseudomonas aeruginosa | 20.9/22.0 | 5.9/6.2 | 26 | 31 | 150 | x | x | x | x |
| Lmo2391 | YhfK (42/61) | Bacillus subtilis | 22.7/25.0 | 6.0/6.7 | 62 | 81 | 607 | x | x | x | x |
| Lmo2696 | Dihydroxyacetone kinase (50/68) | Lactococcus lactis | 21.5/20.0 | 5.1/5.1 | 44 | 36 | 385 | x | x | x | x |
| Lmo0913 | Succinate semialdehyde dehydrogenase (48/67) | Bacillus subtilis | 53.2/55.0 | 5.8/6.4 | 50 | 64 | 977 | x | − | x | x |
| Lmo0913a | Succinate semialdehyde dehydrogenase (48/67) | Bacillus subtilis | 53.2/55.0 | 5.8/6.3 | 87 | 92 | 579 | x | − | x | x |
| Lmo2425 | Glycine cleavage system protein H (51/76) | Escherichia coli | 13.8/17.0 | 3.9/4.0 | 20 | 25 | 105 | = | = | x | = |
| Lmo2748 | YdaG (42/68) | Bacillus subtilis | 15.7/15.0 | 4.6/4.3 | 31 | 33 | 102 | − | − | x | x |
| Lmo2829 | Nitroreductase (56/75) | Clostridium acetobutylicum | 22.2/22.0 | 4.7/4.6 | 55 | 86 | 630 | = | = | x | x |
| LmaA | Antigen A (100/100) | Listeria monocytogenes | 18.0/19.0 | 4.5/4.2 | 24 | 25 | 260 | − | = | − | 1/x |
The identity and similarity values were obtained by performing a protein-protein BLAST search on the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST/).
The score was derived from ion scores that were equal to −10 × log(P), where P is the probability that the observed match is a random event.
x, protein expressed in the ΔsigB strain at a reduced level compared to the wild-type strain; 1/x, protein expressed in the ΔsigB strain at an increased level compared to the wild-type strain; =, protein expressed at similar levels in the ΔsigB and wild-type strains; −, protein not detectable in either strain.
Three other proteins (Lmo2425, Lmo2829, and LmaA) (Fig. 3B and Table 3) were found to be expressed in a σB-dependent manner only under specific growth conditions. Lmo2425 was under σB control only in the presence of salt in exponential phase (Fig. 3B). Under all other conditions this protein was detectable but was not found to be differentially expressed (data not shown). Lmo2829 was expressed in a σB-dependent manner in both phases of growth, but only in the presence of salt (Fig. 3B; data not shown for exponential phase). In the absence of NaCl, similar levels of Lmo2829 were detected in both genetic backgrounds in stationary phase (Fig. 3B) and in exponential phase (data not shown). LmaA was the only protein that showed increased expression in the ΔsigB mutant compared to the wild type. This protein was detectable only in stationary phase and was found to be differentially expressed only when salt was present in the medium (Fig. 3B).
Deletion of σB-dependent genes lmo0796, lmo0913, lmo2391, and lmo2748.
We attempted to obtain new insights into the σB regulon by constructing deletion mutants with mutations in targeted σB-dependent genes. Five genes were selected for this deletion analysis, lmo0796, lmo0913, lmo1830, lmo2391, and lmo2748. These genes were selected because they were σB dependent under every condition that we tested and they are also known to be σB dependent when cells are grown in BHI medium (or BHIS) to stationary phase (1). In addition, we previously confirmed that all of these genes are transcribed in a σB-dependent manner by reverse transcription-PCR (1). Indeed, for three of them (lmo0796, lmo1830, and lmo2391) σB promoters have been mapped (1). Deletions were constructed using the splicing by overlap extension PCR method (33), resulting in in-frame deletion of 492, 1,407, 516, and 399 bp for lmo0796, lmo0913, lmo2391, and lmo2748, respectively. The presence of the correct deletion constructs in the chromosome of L. monocytogenes was confirmed by PCR analysis and by DNA sequencing (data not shown). Several attempts to delete the lmo1830 gene were unsuccessful. Heterozygous recombinants (lmo1830+ and Δlmo1830) were obtained, but repeated attempts to obtain Δlmo1830 secondary recombinants failed to yield the required mutant.
To test the possibility that reduced expression of lmo0796, lmo0913, lmo2391, or lmo2748 might contribute to the rapid-growth phenotype of the ΔsigB mutant, we measured the growth of each deletion mutant in DM and in DMS. The mutants all grew at the same rate and reached the same final optical densities as the wild-type strain, and only the ΔsigB mutant grew faster in DMS (data not shown). This result suggested that none of these four σB-regulated genes is responsible for the higher growth rate observed for the ΔsigB strain in DMS. It also showed that these genes are not essential for growth in a chemically defined medium.
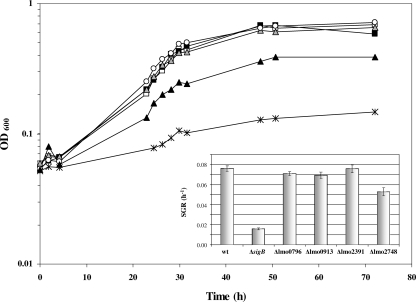
Growth of the ΔsigB and Δlmo2748 mutants in complex media is impaired in the presence of 1.75 M NaCl.
To further characterize the Δlmo0796, Δlmo0913, Δlmo2391, and Δlmo2748 mutants, we investigated whether any of them displayed reduced osmotolerance in complex growth media. In BHI broth with an NaCl concentration of 1 M or less, there were no detectable differences in the growth rate between the wild type and either the ΔsigB mutant or the four newly constructed deletion mutants (data not shown). However, when the organisms were grown in BHI broth supplemented with 1.75 M NaCl, the growth of the ΔsigB mutant was clearly impaired compared to the growth of the wild-type strain (0.016 h−1 versus 0.076 h−1) (Fig. 4). The Δlmo0796, Δlmo0913, and Δlmo2391 mutants did not show any growth defect in the presence of 1.75 M NaCl. However, the Δlmo2748 mutant was reproducibly found to grow at a lower rate than the parent strain (0.053 h−1 versus 0.076 h−1) under these conditions (Fig. 4). These data suggest that the loss of Lmo2748 expression in the ΔsigB mutant may contribute to the osmotic sensitivity of this strain.
FIG. 4.
Δlmo2748 and ΔsigB mutants have a slow-growth phenotype in BHI medium supplemented with 1.75 M NaCl. L. monocytogenes wild-type strain 10403S (black squares), Δlmo0796 mutant (open squares), Δlmo0913 mutant (gray triangles), Δlmo2391 mutant (open circles), Δlmo2748 mutant (black triangles), and ΔsigB mutant (asterisks) cells were grown in BHI medium supplemented with 1.75 M NaCl at 37°C with shaking. Representative growth curves for the conditions investigated are shown; all experiments were performed in triplicate. The inset shows the specific growth rates (SGR) derived from the curves. The error bars indicate the standard deviations from the means of triplicate measurements. wt, wild type.
Lmo0796, Lmo0913, and Lmo2391 contribute to acid tolerance.
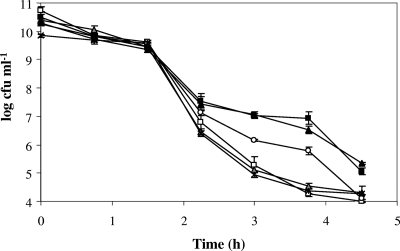
The ΔsigB mutant had a clear disadvantage for survival under acidic conditions (pH 2.5) in BHI medium (Fig. 5), a result that confirmed previous findings (24, 55, 59). A 4-log reduction in the size of the ΔsigB population after 2.25 h was observed, compared with a 3-log reduction in the size of the wild-type population. After 3.75 h of incubation at pH 2.5 in BHI medium, the size of the ΔsigB mutant population had decreased by a total of 6 logs, while the size of the wild-type population had decreased by only 3 logs. The Δlmo0796 and Δlmo0913 mutants both displayed an acid-sensitive profile that was very similar to that of the ΔsigB mutant (Fig. 5). The level of survival of the Δlmo2391 strain was reduced compared to that of the wild-type strain, but the difference was not as marked as it was in the case of the ΔsigB strain; after 3 h of incubation at pH 2.5, the number of Δlmo2391 mutant cells was reduced by 4 logs, whereas the number of wild-type cells was reduced by only 3 logs. In contrast, the Δlmo2748 mutant did not show any defect in survival under acid conditions (Fig. 5). These data suggest that lmo0796, lmo0913, and lmo2391 (albeit to a lesser extent) contribute to acid tolerance in L. monocytogenes and that decreased levels of the corresponding proteins in the ΔsigB mutant might contribute to the acid sensitivity of this strain.
FIG. 5.
Δlmo0796, Δlmo0913, Δlmo2391, and ΔsigB mutants have a survival defect in BHIS at pH 2.5. L. monocytogenes wild-type strain 10403S (black squares), Δlmo0796 mutant (open squares), Δlmo0913 mutant (gray triangles), Δlmo2391 mutant (open circles), Δlmo2748 mutant (black triangles), and ΔsigB mutant (asterisks) cells were grown to stationary phase in BHIS at 37°C with shaking and were subsequently subjected to pH 2.5 in fresh BHIS at room temperature. Survival under the acid conditions was monitored in triplicate, and the errors bars indicate the standard deviations from the means.
Genetic complementation of deletion mutants.
In order to confirm the roles of the lmo0796, lmo0913, lmo2391, and lmo2748 genes in the newly identified phenotypes associated with the deletions, attempts were made to genetically complement each mutant. Three of the genes, lmo0913, lmo2391, and lmo2748, were successfully amplified by PCR and cloned into the shuttle vector pKSV7, generating pKAK0913, pKAK2391, and pKAK2748, respectively. These genes were cloned together with their native promoter and regulatory elements. It was not possible to clone the lmo0796 gene despite repeated attempts, suggesting that the presence of multiple copies of this gene may be deleterious to cell growth. It was therefore not possible to complement the acid-sensitive phenotype associated with the Δlmo0796 deletion. When plasmid pKAK0913 or pKAK2391 was present, there was a significant increase in survival under acid conditions (28- and 310-fold, respectively) (data not shown) compared with the survival of the corresponding deletion strains carrying the empty cloning vector, pKSV7, confirming the role of the lmo0913 and lmo2391 genes in acid tolerance. When plasmid pKAK0913 was present, there was a smaller increase in acid tolerance than when plasmid pKAK2391 was present, but this might be attributed to the presence of a missense mutation in the cloned allele of the lmo0913 gene. Sequencing of the clone revealed the presence of a missense mutation (A to C at position 697), which is predicted to change codon 232 from Thr to Ala in the translated protein, a change that could conceivably influence the activity of the protein. Complementing the salt-sensitive phenotype of the Δlmo2748 mutant proved to be difficult because the presence of the empty vector was found to have a significant effect on the growth rate when a high NaCl level (1.6 M) was present in the growth medium. However, it was observed that the wild-type strain carrying pKAK2748 grew significantly faster (0.026 ± 0.003 h−1) than an isogenic strain carrying the empty vector (0.012 ± 0.001 h−1) when 1.6 M NaCl was present in the growth medium, suggesting that the lmo2748 product may confer some protection against osmotic stress.
Invasion is not impaired by the Δlmo0796, Δlmo0913, Δlmo2391, and Δlmo2748 mutations.
The invasiveness of the deletion strains was investigated using the Caco-2 human epithelial cell line. The wild-type bacteria were found to invade Caco-2 cells with an invasion efficiency of approximately 0.075%, while the ΔsigB mutant had a significantly lower invasion efficiency (0.025%) (Table 4), which was consistent with previous reports (35, 36). None of the four deletion mutants showed a decrease in invasion compared to the wild-type bacteria, suggesting that none of the genes is required for efficient host cell invasion. The Δlmo0913 mutant was found to invade Caco-2 cells at a higher rate than the wild-type strain, and the difference appeared to be significant (Table 4). The reason for this difference is not clear at present.
TABLE 4.
Invasion of Caco-2 epithelial cells
| Strain | % Invasiona
|
P valueb | |
|---|---|---|---|
| Mean | SD | ||
| Wild type | 0.073 | 0.031 | |
| ΔsigB | 0.025 | 0.022 | 0.0004* |
| Δlmo2391 | 0.180 | 0.123 | 0.0691 |
| Δlmo2748 | 0.122 | 0.071 | 0.1072 |
| Δlmo0913 | 0.562 | 0.597 | 0.0001* |
| Δlmo0796 | 0.150 | 0.093 | 0.0650 |
Invasion assays with Caco-2 cells were performed at least eight times for each strain.
P values were calculated using a Mann-Whitney nonparametric test by comparing each mutant to the wild-type parent. Values considered statistically significant (P < 0.05) are indicated by an asterisk.
ΔsigB and Δlmo2748 mutants have an altered pattern of Gram staining.
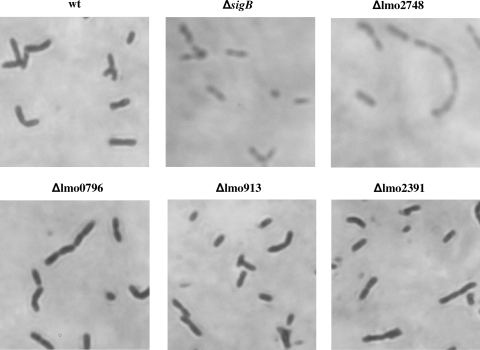
In order to investigate if the deletion mutants had a defect in cell morphology, microscopic observation following Gram staining was performed. The Δlmo0796, Δlmo0913, and Δlmo2391 strains appeared to be identical to the wild type (Fig. 6). They were all found to be dark purple, as expected for gram-positive bacteria, while both the ΔsigB and Δlmo2748 mutant strains stained red (Fig. 6). Similar results were obtained regardless of whether the microscope slides were prepared directly from colonies growing on BHI agar or from liquid cultures grown in DMS or BHIS (data not shown). These results suggest that σB might contribute to the integrity of the cell wall and that the absence of Lmo2748 expression in the ΔsigB mutant background could be involved in this phenotype.
FIG. 6.
Unusual pattern of Gram staining of the Δlmo2748 and ΔsigB mutants. L. monocytogenes wild-type strain 10403S (wt), Δlmo0796 mutant, Δlmo0913 mutant, Δlmo2391 mutant, Δlmo2748 mutant, and ΔsigB mutant cells were observed with the microscope after Gram staining.
DISCUSSION
In this study we investigated the role of σB in L. monocytogenes cells growing in a chemically defined medium (DM), using a combination of proteomic and genetic approaches. Overall, 11 proteins were found to be differentially expressed in the ΔsigB background compared to the parent. Eight of these proteins were σB dependent under all conditions under which they were detectable (Fig. 3A). The remaining three proteins (Lmo2425, Lmo2829, and LmaA) showed σB-dependent expression only under specific conditions. For example, Lmo2829 was found to be σB dependent only when NaCl was present in the growth medium. In the absence of NaCl this protein was expressed at detectable levels, but the sigB deletion had no effect on its expression level. Seven of the proteins identified here (Lmo0539, Lmo0796, Lmo1830, Lmo2391, Lmo2696, Lmo0913, and Lmo2748) were previously described as proteins that were under σB control when L. monocytogenes was grown in complex medium (1). Four of these proteins have putative functions related to carbon utilization (Lmo0539, Lmo0913, Lmo1830, and Lmo2696), while three are similar to proteins that have unknown functions but are known to be stress inducible (Lmo0796, Lmo2391, and Lmo2748). Thus, this study identified four new members of the σB regulon in L. monocytogenes, namely, Lmo0783, Lmo2425, Lmo2829, and LmaA. Lmo0783 is related to component IIB of the mannose-specific phosphotransferase system (Table 3). Despite the reduced levels of this protein in the ΔsigB background, no effect on the ability of the Lmo0783 mutant to grow was observed when mannose was the sole carbon source in the medium (Fig. 2A). However, there are at least two other genes in the L. monocytogenes genome encoding proteins that also have similarity to component IIB of this transport system (Lmo0096 and Lmo2002), suggesting that there may be functional redundancy that allows mannose transport. Lmo2425 is similar to a subunit of the glycine cleavage system, protein H (Table 3), which is involved in balancing the requirements of the cells for glycine and single carbon units. Lmo2829 is related to nitroreductase, which may have a role in membrane bioenergetics. LmaA is the only protein that has been found to be induced in the ΔsigB mutant compared to the parent, suggesting that σB has an indirect negative effect on its expression. LmaA is a surface-expressed antigen that induces cell-mediated responses in mice (27). Moreover, lmaA appears to be absent in nonpathogenic species of Listeria and in some L. monocytogenes strains that are attenuated for virulence (47).
Surprisingly, the loss of σB was found to result in a higher growth rate in DM in the presence of 0.5 M NaCl (Fig. 1), regardless of the carbon source in the medium (Fig. 2B). This result was particularly unexpected since σB is known to play a central role in osmoregulation (5, 25, 55). Indeed, the present study revealed that σB is required for osmotolerance in a complex medium with 1.75 M NaCl added (Fig. 4). Neither the proteomic data nor the genetic analysis performed here provided any obvious insight into this unusual phenotype. However, this phenotype is not without precedent in other bacteria; Bacillus subtilis mutants lacking σB have a selective growth advantage in glucose-limited continuous cultures (49). A similar phenomenon has been described for E. coli, continuous glucose-limited cultures of which are known to accumulate rpoS mutations that confer a growth advantage (40). The nature of the nutrient limitation in E. coli was not the determining factor in selecting for the emergence of these mutants (37, 40). For E. coli these observations have been explained by proposing that sigma factors compete for an RNA polymerase core enzyme whose availability is limited (22). In this model σs provides protection against adverse conditions to the bacterium, but at the cost of diverting resources away from growth-related functions; in the absence of σs faster growth can therefore be achieved (40). Nyström (41) suggested that environmental conditions can influence the trade-off between stress resistance and growth by altering the availability of RNA polymerase and competing sigma factors. Further study is necessary to determine if the same phenomenon exists in L. monocytogenes.
This study also identified two other novel phenotypes associated with the loss of σB. Strains lacking σB are known to be defective for the uptake of compatible solutes under osmotic stress conditions (25, 34, 54). This is explained by the known role of σB in the transcription of the opuC and gbu operons, which are involved in carnitine and betaine uptake, respectively (13, 15, 25, 54). Despite this, no obvious growth defect has been reported for sigB mutants growing in complex medium in the presence of additional NaCl. Here we show that at a very high salt concentration (1.75 M NaCl) the growth of the ΔsigB mutant was extremely poor compared to the growth of the parent strain (Fig. 4). Microscopic examination of the ΔsigB mutant after Gram staining suggested that σB may contribute to the maintenance of cell wall integrity (Fig. 6). In a recent study we found that the expression of DapE is under control of σB (1). This enzyme is involved in biosynthesis of diaminopimelate, a precursor that is required for assembly of the peptidoglycan cell wall, and its absence could contribute to the staining defect seen in the ΔsigB mutant.
Of the 11 proteins identified by proteomics, 5 were targeted for further genetic analysis (Lmo0796, Lmo0913, Lmo1830, Lmo2391, and Lmo2748). Four deletion mutants were successfully constructed; however, despite repeated attempts we were unable to generate an lmo1830 deletion mutant. This might indicate that lmo1830 is an essential gene, at least under the conditions used during mutant construction. Using the deletion mutants, the contributions of lmo0796, lmo0913, lmo2391, and lmo2748 to osmotolerance were investigated using complex medium with a high salt concentration (1.75 M NaCl). As discussed above, the ΔsigB mutant grows very poorly under these conditions. It is clear that lmo2748 contributes to this phenotype since the growth of the Δlmo2748 strain was slower than the growth of the wild-type strain under these conditions (Fig. 4). This apparent function of Lmo2748 correlates well with the finding that this protein is expressed only in the presence of NaCl both in DM and in BHI medium (1). Interestingly, a gene encoding a homologue of Lmo2748 in B. subtilis, designated ydaG, is known to be expressed in a σB-dependent manner and is also induced in response to osmotic shock (43). Although the function of Lmo2748 remains unknown at present, this protein may be involved in maintaining the integrity of the cell wall since, like the ΔsigB mutant, the Δlmo2748 strain has an unusual color after Gram staining (Fig. 6). Motif searching revealed that Lmo2748 possesses a potential pyridoxamine 5′-phosphate oxidase motif, which might indicate that it is involved in pyridoxal phosphate biosynthesis. Pyridoxal phosphate is a cofactor for many enzymatic reactions in the cell, and it is possible that one or more of these reactions could account for the phenotypes observed for the Δlmo2748 mutant.
σB is known to contribute to acid tolerance in L. monocytogenes (24, 55, 58, 59). Three of the deletion mutants tested here were found to have acid-sensitive phenotypes; two were as sensitive as the ΔsigB mutant (Δlmo0913 and Δlmo0796), while one (Δlmo2391) had an intermediate phenotype (Fig. 5). Interestingly, both lmo0796 and lmo2391 have been shown to be induced in exponentially growing L. monocytogenes cells exposed to pH 5.0 (46). Lmo0796 has a conserved potential lipid/isoprenoid binding motif and is homologous (44% identity and 61% similarity) to YceI from E. coli, which is a periplasmic protein that is induced in response to basic pH stress but whose function remains unknown (53). Lmo2391 is homologous to YhfK (46% identity and 61% similarity) from B. subtilis, a general stress protein that is induced in response to osmotic shock (31), heat shock, and ethanol stress (40).
Lmo0913 is homologous (46% identity and 61% similarity) to succinate semialdehyde dehydrogenase from B. subtilis, which is proposed to be involved in the metabolism of γ-aminobutyrate (GABA) to succinate (19). GABA is the decarboxylated product of glutamate, and its production plays a central role in acid tolerance in L. monocytogenes (18, 19). During the response to low pH, intracellular GABA is thought to be exchanged via an antiporter for extracellular glutamate, which is then decarboxylated to produce another GABA molecule, thereby establishing a cycle (4, 51). It might be that an inability to metabolize intracellular GABA increases the susceptibility of cells to low pH, perhaps through the known toxic effects of this metabolite (3, 9). Although the mutation in none of the four mutants tested reduced the ability of L. monocytogenes to invade human epithelial cells (Table 4), the Δlmo0913 mutant appeared to be significantly more invasive (seven- to eightfold) than the parent strain. The underlying reason for this difference is not clear at present.
In summary, this study identified two new phenotypes associated with the loss of σB in L. monocytogenes. It demonstrated clear roles for σB in both osmotic and low-pH stress tolerance and identified specific components of the σB regulon that contribute to these responses.
Acknowledgments
We are grateful to members of the Bacterial Stress Response Group at National University of Ireland—Galway, Galway, Ireland, for useful discussions and for providing critical comments on the manuscript. We thank Cyril Carroll for providing unrestricted access to the 2-DGE apparatus and also Gavin Collins for his assistance with microscopy. We are grateful to Eoin Cosgrave for help with some preliminary experiments.
This work was supported by National Institutes of Health award RO1-AI052151-01A1 (to K.J.B.), by the Science Foundation Ireland Research Frontiers Programme, and by a Framework 6 Marie Curie Transfer of Knowledge grant.
Footnotes
Published ahead of print on 19 September 2008.
REFERENCES
- 1.Abram, F., S. Wan-Lin, M. Wiedmann, K. J. Boor, P. Coote, C. Botting, K. A. G. Karatzas, and C. P. O'Byrne. 2008. Proteomic analyses of a Listeria monocytogenes mutant lacking σB identify new components of the σB regulon and highlight a role for σB in the utilization of glycerol. Appl. Environ. Microbiol. 74:594-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amezaga, M. R., I. Davidson, D. McLaggan, A. Verheul, T. Abee, and I. R. Booth. 1995. The role of peptide metabolism in the growth of Listeria monocytogenes ATCC 23074 at high osmolarity. Microbiology 141:41-49. [DOI] [PubMed] [Google Scholar]
- 3.Arst, H. J. 1976. Integrator gene in Aspergillus nidulans. Nature 262:231-234. [DOI] [PubMed] [Google Scholar]
- 4.Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147:173-180. [DOI] [PubMed] [Google Scholar]
- 5.Becker, L. A., M. S. Cetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, L. A., S. N. Evans, R. W. Hutkins, and A. K. Benson. 2000. Role of σB in adaptation of Listeria monocytogenes to growth at low temperature. J. Bacteriol. 182:7083-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Begley, M., C. Hill, and R. P. Ross. 2006. Tolerance of Listeria monocytogenes to cell envelope-acting antimicrobial agents is dependent on sigB. Appl. Environ. Microbiol. 72:2231-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begley, M., R. D. Sleator, C. G. Gahan, and C. Hill. 2005. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 73:894-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belitsky, B. R., and A. L. Sonenshein. 2002. GabR, a member of a novel protein family, regulates the utilization of gamma-aminobutyrate in Bacillus subtilis. Mol. Microbiol. 45:569-583. [DOI] [PubMed] [Google Scholar]
- 10.Birch, R. M., C. P. O'Byrne, I. R. Booth, and P. Cash. 2003. Enrichment of Escherichia coli proteins by column chromatography on reactive dye columns. Proteomics 3:764-776. [DOI] [PubMed] [Google Scholar]
- 11.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the sigma B transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cetin, M. S., C. Zhang, R. W. Hutkins, and A. K. Benson. 2004. Regulation of transcription of compatible solute transporters by the general stress sigma factor, σB, in Listeria monocytogenes. J. Bacteriol. 186:794-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan, Y. C., K. J. Boor, and M. Wiedmann. 2007. σB-dependent and σB-independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth. Appl. Environ. Microbiol. 73:6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaturongakul, S., and K. J. Boor. 2004. RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 70:5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole, M. B., M. V. Jones, and C. Holyoak. 1990. The effect of pH, salt concentration and temperature on the survival and growth of Listeria monocytogenes. J. Appl. Bacteriol. 69:63-72. [DOI] [PubMed] [Google Scholar]
- 18.Cotter, P. D., C. G. M. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 19.Cotter, P. D., S. Ryan, C. G. Gahan, and C. Hill. 2005. Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 71:2832-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elsinghorst, E. A. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol. 236:405-420. [DOI] [PubMed] [Google Scholar]
- 21.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farewell, A., K. Kvint, and T. Nystrom. 1998. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 29:1039-1051. [DOI] [PubMed] [Google Scholar]
- 23.Ferreira, A., C. P. O'Byrne, and K. J. Boor. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferreira, A., D. Sue, C. P. O'Byrne, and K. J. Boor. 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fraser, K. R., D. Sue, M. Wiedmann, K. Boor, and C. P. O’Byrne. 2003. Role of σB in regulating the compatible solute uptake systems of Listeria monocytogenes: osmotic induction of opuC is σB dependent. Appl. Environ. Microbiol. 69:2015-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gertz, S., S. Engelmann, R. Schmid, A.-K. Ziebandt, K. Tischer, C. Scharf, J. Hacker, and M. Hecker. 2000. Characterization of the σB regulon in Staphylococcus aureus. J. Bacteriol. 182:6983-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gohmann, S., M. Leimeister-Wachter, E. Schiltz, W. Goebel, and T. Chakraborty. 1990. Characterization of a Listeria monocytogenes-specific protein capable of inducing delayed hypersensitivity in Listeria-immune mice. Mol. Microbiol. 4:1091-1099. [DOI] [PubMed] [Google Scholar]
- 28.Hardy, J., K. P. Francis, M. DeBoer, P. Chu, K. Gibbs, and C. H. Contag. 2004. Extracellular replication of Listeria monocytogenes in the murine gall bladder. Science 303:851-853. [DOI] [PubMed] [Google Scholar]
- 29.Herbert, K. C., and S. J. Foster. 2001. Starvation survival in Listeria monocytogenes: characterization of the response and the role of known and novel components. Microbiology 147:2275-2284. [DOI] [PubMed] [Google Scholar]
- 30.Hof, H. 2003. History and epidemiology of listeriosis. FEMS Immunol. Med. Microbiol. 35:199-202. [DOI] [PubMed] [Google Scholar]
- 31.Hoffmann, T., A. Schutz, M. Brosius, A. Volker, U. Volker, and E. Bremer. 2002. High-salinity-induced iron limitation in Bacillus subtilis. J. Bacteriol. 184:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoper, D., U. Volker, and M. Hecker. 2005. Comprehensive characterization of the contribution of individual sigB-dependent general stress genes to stress resistance of Bacillus subtilis. J. Bacteriol. 187:2810-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horton, R., Z. Cai, S. Ho, and L. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 34.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, H., K. J. Boor, and H. Marquis. 2004. Listeria monocytogenes σB contributes to invasion of human intestinal epithelial cells. Infect. Immun. 72:7374-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, H., H. Marquis, and K. J. Boor. 2005. σB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 151:3215-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King, T., S. Seeto, and T. Ferenci. 2006. Genotype-by-environment interactions influencing the emergence of rpoS mutations in Escherichia coli populations. Genetics 172:2071-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGann, P., M. Wiedmann, and K. J. Boor. 2007. The alternative sigma factor σB and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins. Appl. Environ. Microbiol. 73:2919-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLauchlin, J. 1996. The relationship between Listeria and listeriosis. Food Control 7:187-193. [Google Scholar]
- 40.Notley-McRobb, L., T. King, and T. Ferenci. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184:806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nyström, T. 2004. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol. Microbiol. 54:855-862. [DOI] [PubMed] [Google Scholar]
- 42.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 43.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 45.Raengpradub, S., M. Wiedmann, and K. J. Boor. 2008. Comparative analysis of the σB-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol. 74:158-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Satorhelyi, P. 2005. Microarray-analyse der pH-stressantwort von Listeria monocytogenes und Corynebacterium glutamicum. Ph.D. thesis. Technische Universitat Munchen, Munich, Germany.
- 47.Schaferkordt, S., and T. Chakraborty. 1997. Identification, cloning, and characterization of the ima operon, whose gene products are unique to Listeria monocytogenes. J. Bacteriol. 179:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlech, W. F., III, P. M. Lavigne, R. A. Bortolussi, A. C. Allen, E. V. Haldane, A. J. Wort, A. W. Hightower, S. E. Johnson, S. H. King, E. S. Nicholls, and C. V. Broome. 1983. Epidemic listeriosis-evidence for transmission by food. N. Engl. J. Med. 308:203-206. [DOI] [PubMed] [Google Scholar]
- 49.Schweder, T., A. Kolyschkow, U. Volker, and M. Hecker. 1999. Analysis of the expression and function of the σB-dependent general stress regulon of Bacillus subtilis during slow growth. Arch. Microbiol. 171:439-443. [DOI] [PubMed] [Google Scholar]
- 50.Singh, V. K., J. L. Schmidt, R. K. Jayaswal, and B. J. Wilkinson. 2003. Impact of sigB mutation on Staphylococcus aureus oxacillin and vancomycin resistance varies with parental background and method of assessment. Int. J. Antimicrob. Agents 21:256-261. [DOI] [PubMed] [Google Scholar]
- 51.Small, P. L. C., and S. R. Waterman. 1998. Acid stress, anaerobiosis and gadCB: lessons from Lactococcus lactis and Escherichia coli. Trends Microbiol. 6:214-216. [DOI] [PubMed] [Google Scholar]
- 52.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 53.Stancik, L. M., D. Stancik, M. B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sue, D., K. J. Boor, and M. Wiedmann. 2003. σB-dependent expression patterns of compatible solute transporter genes opuCA and lmo1421 and the conjugated bile salt hydrolase gene bsh in Listeria monocytogenes. Microbiology 149:3247-3256. [DOI] [PubMed] [Google Scholar]
- 55.Sue, D., D. Fink, M. Wiedmann, and K. J. Boor. 2004. σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150:3843-3855. [DOI] [PubMed] [Google Scholar]
- 56.Vasseur, C., L. Baverel, M. Hebraud, and J. Labadie. 1999. Effect of osmotic, alkaline, acid or thermal stresses on the growth and inhibition of Listeria monocytogenes. J. Appl. Microbiol. 86:469-476. [DOI] [PubMed] [Google Scholar]
- 57.Walker, S. J., P. Archer, and J. G. Banks. 1990. Growth of Listeria monocytogenes at refrigeration temperatures. J. Appl. Bacteriol. 68:157-162. [DOI] [PubMed] [Google Scholar]
- 58.Wemekamp-Kamphuis, H. H., J. A. Wouters, P. P. de Leeuw, T. Hain, T. Chakraborty, and T. Abee. 2004. Identification of sigma factor σB-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 70:3457-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, S., H. de Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]