Abstract
Assessment of antibody responses to Epstein-Barr virus (EBV) antigens has been used to assist in nasopharyngeal carcinoma (NPC) diagnosis by several methods. In this study, we evaluated an in-house Luminex multianalyte profiling (xMAP) technology and commercial enzyme-linked immunosorbent assay (ELISA) kits for serological examination of EBV-specific antibody responses in 135 NPC patients and 130 healthy controls. Four EBV biomarkers were measured: immunoglobulin A (IgA) against viral capsid antigen (VCA), EBV nuclear antigen 1 (EBNA1), diffused early antigen (EA-D), and IgG against EA-D. The sensitivities and specificities of the four markers ranged between 71.5 and 90% for xMAP assays and 80 and 92% for ELISA. Logistic regression analysis revealed that the combined markers in the xMAP assay had overall sensitivity and specificity values of 82% and 92%, respectively. The correlation coefficient (r) values for the xMAP assay and ELISA were lowest for IgA-VCA (0.468) and highest for IgA-EBNA1 (0.846); for IgA-EA-D and IgG-EA-D, the r values were 0.719 and 0.798, respectively. The concordances of the two methods for NPC discrimination were good (79 to 88%). Our results suggest that both the xMAP assay and ELISA are satisfactory for EBV antibody evaluation when multiple antigens are included.
Nasopharyngeal carcinoma (NPC) is a squamous-cell carcinoma that arises in the epithelium of the nasopharynx (34). NPC is rare worldwide, with an incidence lower than 1 per 100,000 persons per year in Western countries. However, it has a high incidence in south China, especially in the Cantonese region around Guangzhou, where the incidence is ∼30 to 80 per 100,000 persons per year (26, 31). NPC is usually poorly differentiated or undifferentiated but has a relatively high sensitivity to radiation therapy (31). Therefore, more than 70% of NPC patients treated by radiotherapy and chemotherapy at early stages have a 5-year survival rate (24). Unfortunately, most NPC patients are at advanced stages when first diagnosed due to a lack of an efficient method for earlier diagnosis of NPC. In order to increase the NPC survival rate, it is urgent and important that an efficient method for screening of NPC be developed.
It is well documented that Epstein-Barr virus (EBV) infection is associated with NPC. First, NPC patients typically have higher titers of immunoglobulin A (IgA) and IgG against lytic antigens of EBV than healthy EBV carriers (14, 16). Second, elevated EBV antibody levels can precede clinical onset of NPC by 1 to 5 years (3, 18). Third, there are fluctuations of EBV antibody levels after NPC therapy (35). Thus, serological testing for EBV could be useful for diagnosis and prognosis of NPC. Currently, titers of IgA antibody against the EBV viral capsid antigen (VCA) and the diffuse early antigens (EA-D) are regularly tested in many clinical centers (7, 15-17). Furthermore, many serological markers of EBV infection, including VCA, EA, EBV nuclear antigen 1 (EBNA1), Zta, and DNase, have also been developed in recent years (3, 5, 13, 14).
Both IgA-VCA and IgA-EA-D serology assays are being tested in most laboratories in south China by immunoenzymatic assays with slides, using EBV-infected cell lines as a target (7, 18). However, this method is only semiquantitative and is difficult to standardize. As an alternative approach, the enzyme-linked immunosorbent assay (ELISA) technique is easy to automate and is more suitable for mass screening. But the current ELISA for EBV serology can examine only one or two antigens. One technology, called Luminex multianalyte profiling (xMAP), based on flow cytometry analysis of microbeads, has been developed recently. The beads are internally color coded with two fluorescent dyes, and each bead is covalently coupled with any specific molecule, such as an antigen or antibody, and has a unique ratio of these dyes to represent a detection signal. By use of the relative fluorescence intensity (FI) levels detected by R-phycoerythrin-labeled detection antibodies, the antigen-antibody reactions occurring on the bead surface are quantitated (10, 23). Furthermore, more than 100 distinct reactions can be carried out simultaneously on the various beads in one tube, in which the individual bead is identified by a Luminex 100 or 200 instrument (11, 12, 21). Obviously, the xMAP assay requires a smaller sample volume, fewer procedure steps, and less total reaction time than traditional ELISA (27, 29). Moreover, the xMAP technology is highly reproducible because each result is the mean for ∼100 readings (28). Therefore, xMAP assays have been applied in allergy testing, cancer marker detection, diagnosis of infectious diseases, and cytokine quantification, etc. (6, 12, 30).
Although commercial xMAP assay kits for testing IgG and IgM against EBV have been developed to evaluate EBV infection status (22), NPC patients usually have higher IgA but not IgM responses to EBV antigens (16). In this study, we established in-house xMAP assays for four EBV antibody markers for NPC diagnosis (IgAs against VCA, EBNA1, and EA-D and IgG against EA-D) and analyzed the correlation and concordance between this new assay and the commercial ELISA.
MATERIALS AND METHODS
Patients and controls.
One hundred thirty-five patients with newly diagnosed and pathologically confirmed NPC were collected from Sun Yat-Sen University Cancer Center. The stages of disease progression were classified according to the 1996 Union International Cancer Control classification. The NPC case group, including 1 patient with stage I NPC, 20 with stage II NPC, 75 with stage III NPC, and 39 with stage IV NPC, had 97 males and 38 females, with an age range of 23 to 77 (mean, 45.8 ± 11.3) years. One hundred thirty healthy volunteers were collected as normal controls, including 66 males and 64 females, with an age range of 18 to 75 (mean, 40.1 ± 14.6) years. Informally written consent was obtained from all participants.
xMAP analysis. (i) Coupling of proteins to beads.
Coupling of recombinant EBV antigens VCA-gp125, EA-D, and EBNA1 (Biodesign, Saco, ME) to the carboxylated beads (Luminex Corp., Austin, TX) was done by previously reported protocols (30). Briefly, 2.5 × 106 beads were washed with activation buffer (10 mM NaH2PO4, pH 6.3), resuspended in 80 μl of activation buffer, and sonicated for 10 s. Both 10 μl of N-hydroxysulfosuccinimide (Pierce, Rockford, IL) solution and 10 μl of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (Pierce, Rockford, IL) solution were diluted to 50 mg/ml in activation buffer and added to the bead suspension. After being mixed, the beads were incubated with rotation for 20 min at room temperature (RT) in the dark. The activated beads were subsequently washed twice with coupling buffer (10 mM NaH2PO4, 150 mM NaCl, pH 7.4) and then incubated with 500 μl of azide-free protein solution (diluted to 250 μg/ml in coupling buffer) for 2 h. The beads were washed thrice with washing buffer (1× phosphate-buffered saline [PBS], 0.05% Tween 20) and resuspended in 100 μl of blocking/storage buffer (1× PBS, 1% bovine serum albumin). Finally, the beads were counted with a hemocytometer, adjusted to a concentration of 1.25 × 106 beads/ml with storage buffer, and protected from light at 4°C.
(ii) Serum sample test.
The conjugated beads were diluted with storage buffer at 1,000 beads in 50 μl per reaction well and then added to the 96-well filtration system (Millipore, Billerica, MA). Sera diluted to 1:21 in storage buffer (20 μl/well) were added and incubated with the beads for 30 min at RT in the dark. After three washes, 150 μl of R-phycoerythrin-conjugated goat anti-human IgA or IgG (1:200 in PBS; SouthernBiotech, Birmingham, AL) was added to each reaction well and incubated for 30 min. The detection analysis was performed by the Luminex 100 multianalytic system (Bio-Rad, Hercules, CA). All tests were carried out in duplicate.
ELISA.
We performed VCA-IgA, EBNA1-IgA, EA-IgA, and EA-IgG immunoassays by using commercial EBV ELISA kits (IBL, Hamburg, Germany), following the recommended protocols. Briefly, microtiter plates were incubated with sera diluted to 1:100 in diluent buffer (100 μl/well) for 1 h at RT in the dark. After three washes, 100 μl of enzyme conjugate was added and incubated for 30 min at RT in the dark. After three washes, 100 μl of TMB (3,4,5-trimethoxybenzaldehyde) substrate solution was added to each well and incubated for 20 min at RT in the dark. The reaction was stopped by adding 100 μl/well of TMB stop solution, and optical density at 450 nm (OD450) was determined by an immunoanalyzer (Thermo Electron Co., Waltham, MA). Each individual serum sample was tested in duplicate. Standard sera were included in each plate.
Statistical analysis.
The results were analyzed using the statistics software program SPSS (version 16.0). The unpaired t test was used to compare the mean values from the NPC and healthy groups; one-way analysis of variance was used to compare the mean FI values of EBV antibody markers among a population with different ages and cancer stages; receiver operating characteristic (ROC) curve analysis was employed to assess the diagnostic values. Logistic regression was used to create a diagnostic model of NPC. Linear correlations between the xMAP assay and ELISA were analyzed with Spearman's method.
RESULTS
Examination of EBV serology by the xMAP assay.
The antibody levels of IgA-VCA gp125, IgA-EBNA1, IgA-EA-D, and IgG-EA-D were detected by xMAP assays with 135 NPC patients and 130 healthy controls. The mean FI values for NPC patients and healthy controls are summarized in Table 1, showing that levels for all four antibodies were significantly higher in NPC patients than in controls in the xMAP assays (P < 0.0001).
TABLE 1.
Analysis of anti-EBV antibody values for 135 NPC patients and 130 healthy controls
| Anti-EBV antibody | FI (mean ± SEM) for xMAP assay
|
OD (mean ± SEM) for ELISA
|
||
|---|---|---|---|---|
| NPC patients | Healthy controls | NPC patients | Healthy controls | |
| IgA-VCA | 2,193 ± 232 | 657 ± 77 | 0.991 ± 0.064 | 0.145 ± 0.012 |
| IgA-EBNA1 | 9,380 ± 389 | 4,674 ± 356 | 0.380 ± 0.020 | 0.125 ± 0.016 |
| IgA-EA-D | 2,931 ± 317 | 426 ± 64 | 1.111 ± 0.070 | 0.161 ± 0.014 |
| IgG-EA-D | 7,948 ± 545 | 784 ± 170 | 1.201 ± 0.080 | 0.211 ± 0.012 |
To evaluate the accuracy of our xMAP-based assays for discrimination between NPC cases and healthy controls, we performed ROC analysis to define the optimal cutoff values and examine the sensitivity and specificity for NPC diagnosis. The corresponding diagnostic values for serological markers of EBV infection as determined by the xMAP technology are shown in Table 2. The data demonstrated that IgG-EA-D had the highest area under the ROC curve (AUC), with sensitivity and specificity values of 87.4% and 90%, respectively, whereas IgA-VCA gp125 had the lowest AUC, with sensitivity and specificity values of 76.3% and 71.5%, respectively. However, analysis of variance showed no significant differences among these four EBV biomarkers (P > 0.05) when NPC patients were further classified separately by cancer stage, gender, or age.
TABLE 2.
Diagnostic characteristics of EBV IgA-VCA, IgA-EBNA1, IgA-EA, and IgG-EA for the xMAP technology and ELISA for NPC patients (n = 135) and healthy controls (n = 130)
| Assay and EBV marker | AUC (95% CI) | Cutoff | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| xMAP | ||||
| IgA-VCA | 0.785 (0.730-0.841) | 700 (FI) | 76.3 | 71.5 |
| IgA-EBNA1 | 0.812 (0.760-0.864) | 6,000 (FI) | 74.1 | 75.4 |
| IgA-EA-D | 0.882 (0.843-0.922) | 500 (FI) | 79.3 | 81.8 |
| IgG-EA-D | 0.943 (0.916-0.970) | 1,000 (FI) | 87.4 | 90.0 |
| ELISA | ||||
| IgA-VCA | 0.946 (0.919-0.972) | 0.20 (OD) | 91.9 | 83.8 |
| IgA-EBNA1 | 0.863 (0.817-0.909) | 0.15 (OD) | 80.0 | 80.0 |
| IgA-EA-D | 0.960 (0.938-0.981) | 0.25 (OD) | 92.6 | 90.8 |
| IgG-EA-D | 0.926 (0.897-0.956) | 0.30 (OD) | 85.2 | 82.3 |
Assessment of combined EBV markers for NPC diagnosis.
Since EBV serological spectra are different individually, evaluation of NPC status by EBV serology is not reliable when based on any single antibody. Therefore, we attempted to combine these EBV biomarkers for better diagnosis of NPC. As shown in Table 3, a combination of any two EBV markers is more efficient for discriminating NPC patients from healthy populations, and with a combination of all four biomarkers, only 1.5% of the NPC patients were negative for all and none of healthy controls was positive for all. If more than two markers with FI values higher than the cutoff were selected as the positive standard for NPC diagnosis, the sensitivity could reach 95.8%.
TABLE 3.
Distribution of results for combinations of two EBV biomarkers for NPC patients and healthy controls
| EBV marker and result | No. of sera for indicated antibody, treatment group, and EBV marker
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IgA-EBNA1
|
IgA-EA-D
|
IgG-EA-D
|
||||||||||
| NPC
|
Healthy
|
NPC
|
Healthy
|
NPC
|
Healthy
|
|||||||
| Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | |
| IgA-VCA | ||||||||||||
| Positive | 80 | 23 | 15 | 21 | 87 | 16 | 15 | 21 | 92 | 26 | 1 | 35 |
| Negative | 20 | 12 | 18 | 76 | 21 | 11 | 12 | 82 | 11 | 6 | 12 | 82 |
| IgA-EBNA1 | ||||||||||||
| Positive | 86 | 14 | 9 | 24 | 87 | 31 | 3 | 30 | ||||
| Negative | 22 | 13 | 14 | 83 | 13 | 4 | 10 | 87 | ||||
| IgA-EA-D | ||||||||||||
| Positive | 95 | 23 | 4 | 19 | ||||||||
| Negative | 13 | 4 | 9 | 98 | ||||||||
Subsequently, we performed multivariate analysis to identify the best combination of EBV biomarkers for serological diagnosis of NPC. In the logistic regression model, the sensitivity and specificity were 81.5% and 92.3%, respectively, when all four EBV markers were combined, showing a good diagnostic panel with satisfactory accuracy for NPC detection.
Comparison of the xMAP assay and ELISA for detection of EBV antibodies.
For comparison, all serum samples were detected by ELISA (see the table in the supplemental material). The mean ODs for IgA-VCA, IgA-EBNA1, IgA-EA-D, and IgG-EA-D are shown in Table 1. The mean OD values for all EBV biomarkers were significantly different in the two group populations. It appeared that, considered separately (except for IgG-EA-D), the evaluations of IgA-VCA, IgA-EBNA1, and IgA-EA-D by ELISA were better than those by the xMAP assay for NPC diagnosis (Table 2). In addition, among the four ELISAs, the one for IgA-EA-D had the highest AUC (0.960) while that for IgA-EBNA1 had the lowest AUC (0.863).
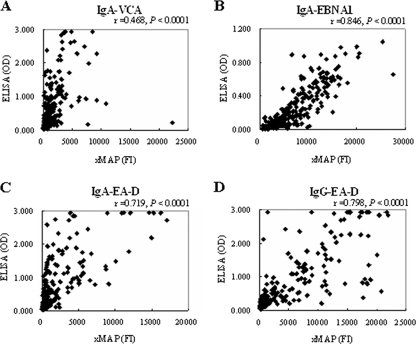
The correlation between the xMAP assay and ELISA results for IgA-VCA was relatively low in the total population (r = 0.468), whereas that for IgA-EBNA1 was high (r = 0.846). The correlation coefficient values for the two methods were 0.719 and 0.798 for IgA-EA-D and IgG-EA-D, respectively (Fig. 1). All correlations were highly significant (P < 0.0001). Moreover, the concordances between the xMAP assay and ELISA were assessed for different EBV markers in NPC diagnosis. As shown in Table 4, there was strong concordance between the two methods for each test evaluated. The IgA-VCA assay has the lowest value (79%), and both the IgA-EBNA1 and the IgG-EA-D assays have the highest (88%).
FIG. 1.
Correlation between the xMAP and ELISA assays for serological detection of EBV in NPC patients (n = 135) and healthy controls (n = 130). xMAP FI versus ELISA OD values for IgA-VCA (A), for IgA-EBNA1 (B), for IgA-EA-D (C), and for IgG-EA-D (D). Positive correlations were observed for all four EBV biomarkers.
TABLE 4.
Comparison of the xMAP assay and ELISA for NPC diagnosis by serological detection of EBV
| EBV marker and result for xMAP assay | No. of sera for ELISA
|
Concordance (%) | |
|---|---|---|---|
| Positive | Negative | ||
| IgA-VCA | 79 | ||
| Positive | 114 | 25 | |
| Negative | 30 | 96 | |
| IgA-EBNA1 | 88 | ||
| Positive | 119 | 14 | |
| Negative | 19 | 113 | |
| IgA-EA-D | 82 | ||
| Positive | 110 | 21 | |
| Negative | 26 | 108 | |
| IgG-EA-D | 88 | ||
| Positive | 119 | 12 | |
| Negative | 20 | 114 | |
DISCUSSION
To reveal the individual diverse EBV antigen recognition patterns, ELISA is currently utilized, but it has obvious practical issues, such as its limitation to one or two antigens per reaction and its concentration range of 2 to 3 logs. In this report, we developed xMAP assays for examining multiple EBV antigens based on the Luminex platform, which needs only two parallel plates to measure levels of IgA and IgG. For example, evaluations of the four EBV antigens used in this report can be achieved with only two sets of reagents and serum samples. Moreover, the xMAP assay has a wider dynamic range (4 to 5 logs) with fluorescence than ELISA with OD (8, 20). In addition, both the xMAP assay and ELISA showed good concordances for most EBV biomarkers for discriminating NPC patients from healthy populations, although there were some discrepancies between the two methods for each individual marker tested, based upon the correlation analysis. Therefore, the xMAP assay may replace ELISA for some EBV biomarkers.
The discrepancies between the xMAP and ELISA methods could be attributed to differences in antigen coating, reaction principles, and procedure (29). Our results showed that ELISA was better for anti-EBV IgA and that the xMAP assay was better for IgG-EA-D for distinguishing NPC patients from healthy populations, although the reasons remain unclear. In any immunoassay, specific recognition of high affinity between an antigen and its antibody contributes to both specificity and sensitivity. In fact, in our initial experiments, we performed both multiple and single xMAP assays for measuring EBV biomarkers, showing that both assays were consistent. Thus, we speculate that the antigens can be specifically detected by individual antibodies with minor cross-reactions. In contrast, some reports showed that the FI values of reporter antibodies were influenced by multiple xMAP assays, which might be due to the cross-reactions (8, 12).
The EBV VCA antigen consists of several proteins, such as gp125, p18, and p23 (32). The VCA gp125 and EA-D used in our xMAP assay were recombinant protein fragments derived from Escherichia coli; however, the VCA and EA-D used in other general serological detections of EBV were protein mixtures. Thus, the low correlation between the results for our xMAP analysis and the commercial ELISA kit for IgA-VCA might be due to the different sources of antigens. This notion is also supported by the fact that there is a high correlation coefficient for IgA-EBNA1 due to the use of similar components in both methods. Overall, the correlation coefficients for the xMAP assay and ELISA ranged from 0.7 to 0.9, consistent with previous studies (22, 29). Therefore, if the proper components of EBV antigens, such as EA-D and EBNA1, were selected, both xMAP and ELISA assays could have relatively high sensitivity, specificity, and AUC for EBV serodiagnosis of NPC.
The flexibility of the xMAP technology as well as its throughput ability gives it broad applications, such as cytokine and chemokine analysis, antibody screening, HLA typing, pathogen detection, and single-nucleotide polymorphism or mutation analysis (1, 4, 8, 9, 19, 25, 36). Our results showed that the xMAP assay is suitable for EBV antibody detection when proper biomarkers are used and provides valid serological profiles of EBV in NPC patients. Thus, the xMAP assay is better for serological detection of EBV titers for clinical diagnosis of NPC than other methods currently used in clinical centers. In addition, the xMAP technology may evaluate the antibodies in serum, plasma, and other body fluids, such as saliva or DNA or RNA from plasma or cancer tissues.
Supplementary Material
Acknowledgments
This study was supported by the 863 and 973 projects of the Ministry of Science and Technology of China, the Scientific and Technologic Project of Guangzhou City (2007Z-E4021), and the China Postdoctoral Science Foundation (20070410862).
We thank Tie Bang Kang for carefully reading the manuscript. We thank all the patients and healthy volunteers who participated in this study.
Footnotes
Published ahead of print on 3 September 2008.
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1.Bovers, M., M. R. Diaz, F. Hagen, L. Spanjaard, B. Duim, C. E. Visser, H. L. Hoogveld, J. Scharringa, I. M. Hoepelman, J. W. Fell, and T. Boekhout. 2007. Identification of genotypically diverse Cryptococcus neoformans and Cryptococcus gattii isolates by Luminex xMAP technology. J. Clin. Microbiol. 45:1874-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reference deleted.
- 3.Chien, Y. C., J. Y. Chen, M. Y. Liu, H. I. Yang, M. M. Hsu, C. J. Chen, and C. S. Yang. 2001. Serologic markers of Epstein-Barr virus infection and nasopharyngeal carcinoma in Taiwanese men. N. Engl. J. Med. 345:1877-1882. [DOI] [PubMed] [Google Scholar]
- 4.Dalva, K., and M. Beksac. 2007. HLA typing with sequence-specific oligonucleotide primed PCR (PCR-SSO) and use of the Luminex technology. Methods Mol. Med. 134:61-69. [DOI] [PubMed] [Google Scholar]
- 5.Dardari, R., W. Hinderer, D. Lang, A. Benider, B. El Gueddari, I. Joab, A. Benslimane, and M. Khyatti. 2001. Antibody responses to recombinant Epstein-Barr virus antigens in nasopharyngeal carcinoma patients: complementary test of ZEBRA protein and early antigens p54 and p138. J. Clin. Microbiol. 39:3164-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jager, W., H. te Velthuis, B. J. Prakken, W. Kuis, and G. T. Rijkers. 2003. Simultaneous detection of 15 human cytokines in a single sample of stimulated peripheral blood mononuclear cells. Clin. Diagn. Lab. Immunol. 10:133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, H., Y. Zeng, Y. Lei, Z. Zhao, P. Wang, B. Li, Z. Pi, B. Tan, Y. Zheng, W. Pan, et al. 1995. Serological survey of nasopharyngeal carcinoma in 21 cities of south China. Chin. Med. J. (England) 108:300-303. [PubMed] [Google Scholar]
- 8.Dernfalk, J., K. Persson Waller, and A. Johannisson. 2007. The xMAP technique can be used for detection of the inflammatory cytokines IL-1beta, IL-6 and TNF-alpha in bovine samples. Vet. Immunol. Immunopathol. 118:40-49. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar, S. A., C. A. Vander Zee, K. G. Oliver, K. L. Karem, and J. W. Jacobson. 2003. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the Luminex LabMAP system. J. Microbiol. Methods 53:245-252. [DOI] [PubMed] [Google Scholar]
- 10.duPont, N. C., K. Wang, P. D. Wadhwa, J. F. Culhane, and E. L. Nelson. 2005. Validation and comparison of luminex multiplex cytokine analysis kits with ELISA: determinations of a panel of nine cytokines in clinical sample culture supernatants. J. Reprod. Immunol. 66:175-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Earley, M. C., R. F. Vogt, Jr., H. M. Shapiro, F. F. Mandy, K. L. Kellar, R. Bellisario, K. A. Pass, G. E. Marti, C. C. Stewart, and W. H. Hannon. 2002. Report from a workshop on multianalyte microsphere assays. Cytometry 50:239-242. [DOI] [PubMed] [Google Scholar]
- 12.Elshal, M. F., and J. P. McCoy. 2006. Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA. Methods 38:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fachiroh, J., D. K. Paramita, B. Hariwiyanto, A. Harijadi, H. L. Dahlia, S. R. Indrasari, H. Kusumo, Y. S. Zeng, T. Schouten, S. Mubarika, and J. M. Middeldorp. 2006. Single-assay combination of Epstein-Barr Virus (EBV) EBNA1- and viral capsid antigen-p18-derived synthetic peptides for measuring anti-EBV immunoglobulin G (IgG) and IgA antibody levels in sera from nasopharyngeal carcinoma patients: options for field screening. J. Clin. Microbiol. 44:1459-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fachiroh, J., T. Schouten, B. Hariwiyanto, D. K. Paramita, A. Harijadi, S. M. Haryana, M. H. Ng, and J. M. Middeldorp. 2004. Molecular diversity of Epstein-Barr virus IgG and IgA antibody responses in nasopharyngeal carcinoma: a comparison of Indonesian, Chinese, and European subjects. J. Infect. Dis. 190:53-62. [DOI] [PubMed] [Google Scholar]
- 15.Hadar, T., M. Rahima, E. Kahan, J. Sidi, E. Rakowsky, B. Sarov, and I. Sarov. 1986. Significance of specific Epstein-Barr virus IgA and elevated IgG antibodies to viral capsid antigens in nasopharyngeal carcinoma patients. J. Med. Virol. 20:329-339. [DOI] [PubMed] [Google Scholar]
- 16.Henle, G., and W. Henle. 1976. Epstein-Barr virus-specific IgA serum antibodies as an outstanding feature of nasopharyngeal carcinoma. Int. J. Cancer 17:1-7. [DOI] [PubMed] [Google Scholar]
- 17.Ho, S., P. Teo, W. H. Kwan, P. Choi, J. Tjong, and P. J. Johnson. 1998. Staging and IgA VCA titre in patients with nasopharyngeal carcinoma: changes over a 12-year period. Oral Oncol. 34:491-495. [DOI] [PubMed] [Google Scholar]
- 18.Ji, M. F., D. K. Wang, Y. L. Yu, Y. Q. Guo, J. S. Liang, W. M. Cheng, Y. S. Zong, K. H. Chan, S. P. Ng, W. I. Wei, D. T. Chua, J. S. Sham, and M. H. Ng. 2007. Sustained elevation of Epstein-Barr virus antibody levels preceding clinical onset of nasopharyngeal carcinoma. Br. J. Cancer 96:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johannisson, A., R. Jonasson, J. Dernfalk, and M. Jensen-Waern. 2006. Simultaneous detection of porcine proinflammatory cytokines using multiplex flow cytometry by the xMAP technology. Cytometry A 69:391-395. [DOI] [PubMed] [Google Scholar]
- 20.Kellar, K. L., and M. A. Iannone. 2002. Multiplexed microsphere-based flow cytometric assays. Exp. Hematol. 30:1227-1237. [DOI] [PubMed] [Google Scholar]
- 21.Kettman, J. R., T. Davies, D. Chandler, K. G. Oliver, and R. J. Fulton. 1998. Classification and properties of 64 multiplexed microsphere sets. Cytometry 33:234-243. [PubMed] [Google Scholar]
- 22.Klutts, J. S., R. S. Liao, W. M. Dunne, Jr., and A. M. Gronowski. 2004. Evaluation of a multiplexed bead assay for assessment of Epstein-Barr virus immunologic status. J. Clin. Microbiol. 42:4996-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kofoed, K., U. V. Schneider, T. Scheel, O. Andersen, and J. Eugen-Olsen. 2006. Development and validation of a multiplex add-on assay for sepsis biomarkers using xMAP technology. Clin. Chem. 52:1284-1293. [DOI] [PubMed] [Google Scholar]
- 24.Lin, J. C., J. S. Jan, C. Y. Hsu, W. M. Liang, R. S. Jiang, and W. Y. Wang. 2003. Phase III study of concurrent chemoradiotherapy versus radiotherapy alone for advanced nasopharyngeal carcinoma: positive effect on overall and progression-free survival. J. Clin. Oncol. 21:631-637. [DOI] [PubMed] [Google Scholar]
- 25.Linkov, F., A. Lisovich, Z. Yurkovetsky, A. Marrangoni, L. Velikokhatnaya, B. Nolen, M. Winans, W. Bigbee, J. Siegfried, A. Lokshin, and R. L. Ferris. 2007. Early detection of head and neck cancer: development of a novel screening tool using multiplexed immunobead-based biomarker profiling. Cancer Epidemiol. Biomarkers Prev. 16:102-107. [DOI] [PubMed] [Google Scholar]
- 26.Lo, K. W., K. F. To, and D. P. Huang. 2004. Focus on nasopharyngeal carcinoma. Cancer Cell 5:423-428. [DOI] [PubMed] [Google Scholar]
- 27.Olsson, A., H. Vanderstichele, N. Andreasen, G. De Meyer, A. Wallin, B. Holmberg, L. Rosengren, E. Vanmechelen, and K. Blennow. 2005. Simultaneous measurement of beta-amyloid(1-42), total tau, and phosphorylated tau (Thr181) in cerebrospinal fluid by the xMAP technology. Clin. Chem. 51:336-345. [DOI] [PubMed] [Google Scholar]
- 28.Prabhakar, U., E. Eirikis, and H. M. Davis. 2002. Simultaneous quantification of proinflammatory cytokines in human plasma using the LabMAP assay. J. Immunol. Methods 260:207-218. [DOI] [PubMed] [Google Scholar]
- 29.Reijn, T. S., M. O. Rikkert, W. J. van Geel, D. de Jong, and M. M. Verbeek. 2007. Diagnostic accuracy of ELISA and xMAP technology for analysis of amyloid beta(42) and tau proteins. Clin. Chem. 53:859-865. [DOI] [PubMed] [Google Scholar]
- 30.Skogstrand, K., P. Thorsen, B. Norgaard-Pedersen, D. E. Schendel, L. C. Sorensen, and D. M. Hougaard. 2005. Simultaneous measurement of 25 inflammatory markers and neurotrophins in neonatal dried blood spots by immunoassay with xMAP technology. Clin. Chem. 51:1854-1866. [DOI] [PubMed] [Google Scholar]
- 31.Spano, J. P., P. Busson, D. Atlan, J. Bourhis, J. P. Pignon, C. Esteban, and J. P. Armand. 2003. Nasopharyngeal carcinomas: an update. Eur. J. Cancer 39:2121-2135. [DOI] [PubMed] [Google Scholar]
- 32.Tarbouriech, N., M. Buisson, T. Geoui, S. Daenke, S. Cusack, and W. P. Burmeister. 2006. Structural genomics of the Epstein-Barr virus. Acta Crystallogr D 62:1276-1285. [DOI] [PubMed] [Google Scholar]
- 33.Reference deleted.
- 34.Wei, W. I., and J. S. Sham. 2005. Nasopharyngeal carcinoma. Lancet 365:2041-2054. [DOI] [PubMed] [Google Scholar]
- 35.Yip, T. T., R. K. Ngan, W. H. Lau, Y. F. Poon, I. Joab, C. Cochet, and A. K. Cheng. 1994. A possible prognostic role of immunoglobulin-G antibody against recombinant Epstein-Barr virus BZLF-1 transactivator protein ZEBRA in patients with nasopharyngeal carcinoma. Cancer 74:2414-2424. [DOI] [PubMed] [Google Scholar]
- 36.Yurkovetsky, Z., S. Ta'asan, S. Skates, A. Rand, A. Lomakin, F. Linkov, A. Marrangoni, L. Velikokhatnaya, M. Winans, E. Gorelik, G. L. Maxwell, K. Lu, and A. Lokshin. 2007. Development of multimarker panel for early detection of endometrial cancer. High diagnostic power of prolactin. Gynecol. Oncol. 107:58-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.