Abstract
The HAP1 (CYP1) gene product of Saccharomyces cerevisiae is known to regulate the transcription of many genes in response to oxygen availability. This response varies according to yeast species, probably reflecting the specific nature of their oxidative metabolism. It is suspected that a difference in the interaction of Hap1p with its target genes may explain some of the species-related variation in oxygen responses. As opposed to the fermentative S. cerevisiae, Kluyveromyces lactis is an aerobic yeast species which shows different oxygen responses. We examined the role of the HAP1-equivalent gene (KlHAP1) in K. lactis. KlHap1p showed a number of sequence features and some gene targets (such as KlCYC1) in common with its S. cerevisiae counterpart, and KlHAP1 was capable of complementing the hap1 mutation. However, the KlHAP1 disruptant showed temperature-sensitive growth on glucose, especially at low glucose concentrations. At normal temperature, 28°C, the mutant grew well, the colony size being even greater than that of the wild type. The most striking observation was that KlHap1p repressed the expression of the major glucose transporter gene RAG1 and reduced the glucose uptake rate. This suggested an involvement of KlHap1p in the regulation of glycolytic flux through the glucose transport system. The ΔKlhap1 mutant showed an increased ability to produce ethanol during aerobic growth, indicating a possible transformation of its physiological property to Crabtree positivity or partial Crabtree positivity. Dual roles of KlHap1p in activating respiration and repressing fermentation may be seen as a basis of the Crabtree-negative physiology of K. lactis.
In both prokaryotes and eukaryotes, the transcription of many genes is controlled by oxygen availability. This regulation has been extensively studied in Saccharomyces cerevisiae. However, this species is rather exceptional among yeasts because of its extreme fermentation-oriented physiology. Other species that have comparable sets of genes in their genomes (45) conspicuously differ from S. cerevisiae in their mode of carbon metabolism in general and in their response to oxygen in particular. The difference may reflect evolutionary variation of the regulatory networks that characterize each species. In the present work, an aerobic species, Kluyveromyces lactis, was chosen for a comparative study of oxygen-linked regulation. The genomic sequence of this yeast is entirely known (EMBL accession numbers CR3821121 to CR382126), and its carbon metabolism is well documented (7, 8, 51).
In S. cerevisiae, the transcriptional activator Hap1p is involved in cell response to oxygen via heme (25). A number of genes appear to be targets of this regulator, but the range and mode of these interactions are only partially known. In S. cerevisiae, oxygen-regulated genes are of two kinds: the “aerobic genes,” which are activated under aerobic conditions, and the “hypoxic genes,” which are fully expressed only under anoxic or hypoxic conditions and repressed by oxygen. This regulation occurs at the level of transcription. Indeed, several transcription factors responding to oxygen have been identified. They include (i) Rox1p and Mot3p, which repress the transcription of the hypoxic genes under aerobic conditions; (ii) conversely, Mga2p, which activates the transcription of the hypoxic genes under hypoxia; and (iii) Hap1p, which activates aerobic genes (26, 29, 32, 46, 53, 56). The activities of Rox1p and Hap1p are in turn controlled by heme. In most organisms, the role of heme is thought to be central in the oxygen-sensing mechanism. It has been suggested that heme synthesis is regulated by oxygen concentration at three steps of its synthetic pathway (25): coproporphyrinogen III oxidase (Hem13p), protoporphyrin oxidase (Hem14p), and ferrochelatase (Hem15p). Heme control of Hap1p appears to be direct and stringent. In S. cerevisiae, the target genes of Hap1p include CYC1 (encoding iso-1-cytochrome c), CYC7 (encoding iso-2-cytochrome c), CYT1 (encoding cytochrome c1), CTT1 (encoding catalase), and YHB1 (encoding a flavohemoprotein), as well as the transcriptional repressor-encoding genes ROX1 and MOT3 (31). Thus, Hap1p plays a pivotal role in the molecular events underlying the oxygen response. Gene microarray-assisted surveying has suggested that Hap1p potentially regulates at least 24 genes in S. cerevisiae (46).
S. cerevisiae and K. lactis show quite different responses to oxygen. S. cerevisiae, known as a Crabtree-positive species, ferments sugars even in the presence of oxygen, while K. lactis, which is Crabtree negative, metabolizes sugars preferentially through the respiratory circuit (51). A reason for this difference is that in S. cerevisiae, glucose repression is a determinant regulatory device that strongly represses mitochondriogenesis and related oxygen-regulated processes. The mechanisms that link glucose repression to the expression of respiratory genes are, however, unclear in K. lactis. For example the Hap2/3/4/5p complex is known to be involved in this regulation in S. cerevisiae (21), but this connection is often absent or uncertain in K. lactis (1, 6, 7, 9, 37). Such data prompted us to examine in K. lactis the role of the supposed major regulator Hap1p.
MATERIALS AND METHODS
Strains and culture media.
Yeast strains are listed in Table 1. Yeast cells were routinely grown in a complete medium containing 1% Bacto yeast extract, 2% Bacto peptone (YP), and 2% glucose. The minimal medium contained 0.67% yeast nitrogen base (Difco) without amino acids (YNB) and a carbon source as specified. For auxotrophic requirements, relevant amino acids were added at the concentration of 40 μg ml−1 and adenine or uracil was added at 20 μg ml−1. The Δhem1 and Δhem1 Δhap1 mutants were grown in media supplemented with 30 μg ml−1 of 5-aminolevulinic acid (δ-ALA) (for heme-sufficient conditions) or with 30 μg ml−1 of ergosterol and 0.2% Tween 80 (for heme-deficient conditions). Tween 80 (polyoxyethylenesorbitan monooleate) is used as a source of unsaturated fatty acids (3, 4). For hypoxic cultures, the complete medium supplemented with ergosterol and Tween 80 was flushed with nitrogen for 1 h without stirring and then the flasks were closed during culture. Note that K. lactis cannot grow under strict anoxia but grows under hypoxic conditions to a limited degree.
TABLE 1.
Yeast strains
| Yeast strain | Genotype | Reference or source |
|---|---|---|
| K. lactis | ||
| PM6-7A | MATauraA1-1 adeT-600 | 12 |
| 2359/152 | MATametA1-1 | H. Fukuhara, Institut Curie |
| MW270-7B | MATauraA1-1 leu2 metA1-1 | M. Wésolowski-Louvel, University of Lyon 1 |
| MW270-7B/Δklhap1 | MW270-7B Klhap1::KanMX | This work |
| S. cerevisiae | ||
| 334-ΔHAP1 | MATapep4-3 prb1-1122 leu2-3,2-112 ura3-1 gal1 hap1::LEU2 | This work |
| W303-1A | MATaade2-1 leu2-3 ura3-1 trp1-1 his3-11,3-15 can1-100 | R. Rothstein, Columbia University |
| W303-ΔHEM1 | W303-1A hem1::LEU2 | This work |
| W303-ΔHAP1 | W303-1A hap1::ADE2 | This work |
| W303-ΔHEM1-ΔHAP1 | W303-1A hem1::LEU2 hap1::ADE2 | This work |
| W303-ΔHAP1(K) | W303-1A hap1::KanMX | This work |
Cloning of KlHAP1.
Hap1p of S. cerevisiae has a typical zinc finger motif, CTICRKRKVKC (15, 40). According to this sequence, oligonucleotides were synthesized that contained mixed degenerate codons (Table 2, oligonucleotide 1). The mixed nucleotides were 5′ labeled with [γ-32P]ATP and used as a hybridization probe to search the HAP1 ortholog in the K. lactis genome. Genomic DNA (strain PM6-7A) was digested with EcoRI and HindIII and electrophoresed. Southern blot hybridization gave an unambiguous signal with an EcoRI-HindIII fragment of 3.6 kb and an EcoRI fragment of 7.1 kb (data not shown). Therefore, the DNA fragments of the 3.6-kb size range were collected and inserted into pUC19 vector to construct a partial library. Colony hybridization detected a positive clone (named pBW-1). Sequencing of its 3.6-kb insert revealed a partial open reading frame orthologous to the N terminus of S. cerevisiae HAP1 (ScHAP1). To clone the sequence downstream of this partial open reading frame, inverse PCR (39) was applied with a self-circularization mixture of EcoRI-digested genomic DNA of K. lactis (Table 2, oligonucleotides 2 and 3). The PCR product was cloned into pCR-Script Amp SK(+) vector. The resulting plasmid (named pBW-2) contained a 3.8-kb DNA fragment overlapping the 3.6-kb insert of pBW-1. Colony hybridization and Southern blot hybridization were carried out at 45°C in a solution containing 0.75 M NaCl, 0.075 M trisodium citrate, 5× Denhardt's mixture, and 0.5% sodium dodecyl sulfate. Denatured herring sperm DNA was included at the concentration of 100 μg ml−1.
TABLE 2.
List of oligonucleotides
| Purpose and oligonucleotide no. | Target | Sequence (5′→3′) |
|---|---|---|
| KlHAP1 cloning probe (containing degenerate codons) | ||
| 1 | TG(C/T)ACCAT(C/T)TGTCG(A/C/G/T)AA(AG)AG(A/G)AA(A/G)GT(C/T)AA(A/G)TGT | |
| Inverse PCR for KlHAP1 | ||
| 2 | TGGATTGGCTATAATGG | |
| 3 | ACCACACAAAGTTTTCC | |
| KlHAP1 disruption | ||
| 4 | CGGGATCCGAGAGAATATCGCTAATGCC | |
| 5 | GGGGTACCAGTTCAAAAAATATCCACCTC | |
| 6 | CGGGATCCGTTCATTGTTACTTAGATCG | |
| 7 | GGGGTACCGGCTGCACTTGCTAAGGG | |
| Northern hybridization probes | ||
| 8 | KlHAP1 | ATGAGCTCTATTACCTCACCTGGAACAG |
| 9 | GGTTGGAGTTCCCTCGCGAAGCATTGGACA | |
| 10 | KlCYC1 | TGCCAGCTCCATACAAGAAG |
| 11 | ACAAGCCTTCAACATGTAAG | |
| 12 | KlCYT1 | AGATCATTTTCTACTGCCGC |
| 13 | TCATTTCTTTGGTGGGTTGA | |
| 14 | KlQCR2 | TTACAATTTGCCCAGCAAAC |
| 15 | TTCATCAGCGAAAGGTAGCTT | |
| 16 | KlHMG1 | GGCTAAACATCCAATTCACG |
| 17 | AAGGACCAATAACACCAACCG | |
| 18 | KlERG11 | CGAAATCAGAATCGGTGGTT |
| 19 | GTCCATTCAATTTCAGATGGC | |
| 20 | KlHEM13 | TCCATTCGACTCACCAACTG |
| 21 | TTCAAACCCATTCAACAGGG | |
| 22 | KlYHB1 | ATGCTGTCCGTTAATACGAA |
| 23 | TCTCAAGGCGTCATATTCGT | |
| 24 | RAG1 | TAGTACGAGTGCTGGAAGTGG |
| 25 | TGTCGTCATGTTGCAAAGCA | |
| 26 | HGT1 | AAATTGGCTTTTGCTACGTGA |
| 27 | AATTGGAGTTGCTTGCTGAG | |
| 28 | KlPOX1 | CAGTCTACGTATAATGCCAAG |
| 29 | TTAGTTGCCAGCAAGTCTGGA | |
| 30 | KlEEB1 | TGTTATGCATGGTTTGGCTG |
| 31 | GCATCAAAATATTGGGCAACC | |
| 32 | KlACT1 | GGTCGCTGCTTTAGTTATTG |
| 33 | ACACTTCAAGTGAACGATGG | |
| Sc-UASCYC1 (Hap1p binding site in CYC1 promoter) | ||
| 34 | ScCYC1(UAS1B-342) | CTCTTTGGCCGGGGTTTACGGACGATGACCG |
| 35 | ScCYC1(UAS1B-313) | CGGTCATCGTCCGTAAACCCCGGCCAAAGAG |
| Kl-UASCYC1 (Hap1p binding site in KlCYC1 promoter) | ||
| 36 | KlCYC1(UAS1B-276) | CTATCATTTCGGGAACATCGGTCAAGACAC |
| 37 | KlCYC1(UAS1B-305) | GTGTCTTGACCGATGTTCCCGAAATGATAG |
Disruption of KlHAP1.
To construct the Klhap1 null mutant, the “split-marker recombination” procedure was used as described previously (17). The DNA fragments upstream and downstream of the KlHAP1 open reading frame were PCR amplified (Table 2, primers 4 and 5 and primers 6 and 7) and cloned into pKA and pAN vectors, respectively. The resulting plasmids were transformed into K. lactis strain MW270-7B. The expected structure of integration was confirmed by Southern hybridization: the entire open reading frame of KlHAP1, exactly from the initiation codon ATG to the stop codon TGA, has been deleted and replaced by the KanMX selection marker to give the ΔKlhap1 mutant.
Northern blot analysis.
Total RNA was isolated (42), fractionated on an agarose-formaldehyde denaturing gel, and immobilized onto a Hybond-N+ membrane (Amersham). Hybridization was performed at 65°C in a buffer containing 7% sodium dodecyl sulfate, 0.5 M sodium phosphate buffer, pH 7.2, and 10 mM EDTA. Probes were synthesized by PCR (Table 2, oligonucleotides 8 to 33) and labeled with 32P using the Ready to Go DNA labeling kit (Pharmacia).
The intensity of hybridization bands was quantified with the program ImageQuant 5.2 and normalized for RNA loading against signal of KlACT1.
Protein-DNA binding assay.
Formation of the Hap1p-DNA complex was assayed by a gel retardation procedure according to the method of Fytlovich et al. (20). Double-stranded synthetic oligonucleotides Sc-UASCYC1 and Kl-UASCYC1 (Table 2, oligonucleotides 34 and 35 and oligonucleotides 36 and 37, respectively) were labeled with 32P and incubated with cell extracts (see below) in 20 μl of incubation buffer (6% glycerol, 20 mM HEPES buffer, pH 8, 50 mM KCl, 5 mM MgCl2, 10 μM ZnCl2, and 0.5 to 5 μg of denatured salmon sperm DNA as a nonspecific competitor). Binding reactions were carried out at 4°C for 15 min, and the reaction mixtures were loaded onto a 4% polyacrylamide gel in 0.5× TBE buffer, pH 8.3 (45 mM Tris base, 45 mM H3BO3, 1.2 mM EDTA, 2% glycerol). After electrophoresis at 4°C, the gel was transferred to the surface of Whatman 3MM paper, dried, and autoradiographed. The cell extracts were prepared as follows. S. cerevisiae strain 334-ΔHAP1 was transformed with the plasmid pDP-Kl-HAP1 (carrying KlHAP1 fused to the inducible promoter GAL10-CYC1). To prepare control extracts, the empty vector pDP8-10 was also used for transformation. The transformants were grown in liquid minimal medium containing 2% glucose as carbon source. At the cell density of A600 = 1.5, KlHAP1 expression was induced by the addition of galactose (final concentration, 2%). After 8 h, cells were harvested by centrifugation and washed with extraction buffer (20 mM Tris-HCl, pH 7.5, 0.1 mM EDTA, 1 mM dithiothreitol, 20% glycerol, and 1 mM phenylmethylsulfonyl fluoride). The cells suspended in the extraction buffer (1 ml for 1 g of cells) were disrupted in a Carver press. The broken cells were centrifuged for 30 min at 27,000 × g, and the supernatant fraction was collected and stored at −80°C until use.
Measurement of glucose uptake rate.
The cells were grown in YP medium supplemented with 2% glucose and harvested at an optical density at 600 nm (OD600) of 1 to 1.5. The cells were washed twice with ice-cold 100 mM potassium phosphate buffer, pH 6.5, and suspended in the same buffer, at a concentration of about 80 mg (dry weight) ml−1. To start the uptake reaction, 100 μl of cell suspension was added with 100 μl of a buffered (pH 6.5) aqueous solution of 200 mM [14C]glucose (603 dpm/nmol). Aliquots of 48 μl were taken at different time intervals (7, 15, 30, and 60 s), uptake was stopped by dilution with 10 ml of ice-cold water, and the aliquots were filtered through a glass microfiber disc and washed twice with 5 ml ice-cold water. The blank in each experiment was determined by adding labeled substrate simultaneously with the cold water. The filters were placed in scintillation vials, and 5 ml of Ultima Gold (Packard) was added; the retained radioactivity was determined in a liquid scintillation counter.
Glucose concentration and ethanol concentration determination.
Yeast cells were harvested by centrifugation after a 36-h precultivation at 28°C in YP medium containing 2% glucose, reinoculated into 50 ml YP medium containing 5% glucose in a flask to an OD600 of 3.0, and then incubated at 28°C on a shaker (220 rpm). Aliquots (0.5 to 1.0 ml) were taken from the cultures at appropriate time intervals for analysis of cell OD600, glucose consumption, and ethanol production. Glucose levels in medium were determined by use of dinitrosalicylic acid reagent as described previously (38). Ethanol levels in culture supernatant were assayed enzymatically using a procedure with NAD-dependent alcohol dehydrogenase (Sigma) based on the work of Kagi and Vallee (27).
Nucleotide sequence accession number.
The nucleotide sequence of KlHAP1 has been assigned GenBank accession number AY648979.
RESULTS
KlHAP1 complements the hap1 mutation of S. cerevisiae.
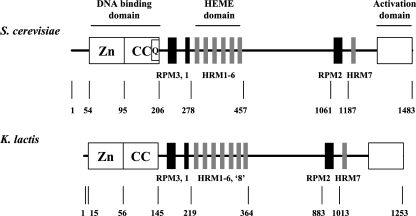
Compared with Hap1p, the distinct functional domains, including the Zn(II)2Cys6 DNA binding domain, the dimerization domain, repression modules, the heme-responsive motif, and the activation domain, are generally well conserved in KlHap1p (Fig. 1). We therefore examined whether this K. lactis gene could complement the S. cerevisiae hap1 mutation.
FIG. 1.
Comparison of functional modules between S. cerevisiae Hap1p and K. lactis KlHap1p. Both proteins are schematically represented and composed of a typical Zn(II)2Cys6 binuclear cluster (box Zn) followed by a coiled-coil dimerization domain (box CC), three repression modules (RPM; indicated with solid bars), several heme-responsive modules (HRM; indicated with stippled bars; seven in S. cerevisiae and eight in K. lactis), and an activation domain (box at C terminus). The stretch of 12 glutamine residues (box Q) present in the C-terminal part of the dimerization domain in Hap1p is absent in KlHap1p. Numbers indicate amino acid residue positions in the proteins. The distances are not in proportion.
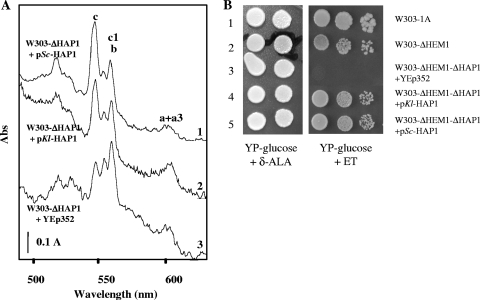
In S. cerevisiae, one of the best-known genes under the control of Hap1p is CYC1, encoding iso-1-cytochrome c. In aerobiosis, the amount of cytochrome c in the Δhap1 strain (W303-ΔHAP1) was reduced to one-half of the wild-type level (Fig. 2A, spectrum 3 versus spectrum 1). The KlHAP1 gene was placed behind the promoter of the ScHAP1 gene and introduced into the multicopy vector YEp352, to give the pKl-HAP1 plasmid. The use of KlHAP1's own promoter was avoided in this case, because (i) the promoter regions of KlHAP1 and ScHAP1 had little similarity of sequence and (ii) it had previously been reported that the promoter of KlHAP4 could not function in S. cerevisiae (6). The pKl-HAP1 plasmid was then transformed into the Δhap1 strain. In the transformant, the amount of cytochrome c was increased twofold (Fig. 2A, spectrum 2), an effect similar to that obtained by transformation with ScHAP1. Thus, KlHap1p can replace ScHap1p as a regulator of transcription of the CYC1 gene in the S. cerevisiae host.
FIG. 2.
Complementation of S. cerevisiae Δhap1 mutants by KlHAP1. (A) Increase of cytochrome c synthesis by introduction of KlHAP1 into the Δhap1 mutant of S. cerevisiae. KlHAP1 (placed under the control of the S. cerevisiae HAP1 promoter) was inserted into the multicopy vector YEp352, resulting in the plasmid pKl-HAP1. The plasmid pSc-HAP1 containing the S. cerevisiae HAP1 gene was used as a positive control. The Δhap1 mutant W303-ΔHAP1 was transformed with pSc-HAP1 (curve 1), pKl-HAP1 (curve 2), or the empty vector YEp352 (curve 3). All strains were grown at 28°C for 2 days on plates of complete medium containing 2% glucose. Cytochrome spectra were determined at liquid nitrogen temperature in a Cary 400 spectrophotometer according to the protocol of Claisse et al. (13), and three resulting curves were placed at different heights in panel A so that they could be compared with each other. The absorption peak at 550 nm corresponds to cytochrome c. (B) KlHap1p complements the defective phenotype of the S. cerevisiae Δhap1 Δhem1 strain in the presence of ergosterol and Tween 80. Line 1, W303-1A (HEM1 HAP1); line 2, W303-ΔHEM1 (hem1 HAP1); line 3, W303-ΔHEM1-ΔHAP1 (hem1 hap1) transformed with the empty vector YEp352; line 4, W303-ΔHEM1-ΔHAP1 with pKl-HAP1; line 5, W303-ΔHEM1-ΔHAP1 with pSc-HAP1. A serial dilution of cultures was deposited on YP-2% glucose medium supplemented with 30 μg ml−1 ergosterol and 0.2% Tween 80 (right panel, YP-glucose + ET, heme-deficient condition), as well as on the medium supplemented with 30 μg ml−1 δ-ALA as control (left panel, YP-glucose + δ-ALA, heme-sufficient condition). Cells were grown at 28°C for 3 days.
According to published reports, the Δhap1 mutants of S. cerevisiae do not show a modified growth phenotype in aerobiosis under heme-sufficient conditions. Chantrel et al. (10) have found that Hap1p was essential for S. cerevisiae to grow under anaerobiosis or under heme-deficient conditions. Deletion of the HAP1 gene in the S. cerevisiae Δhem1 strain (HEM1 encodes the enzyme of the first step of heme biosynthesis) resulted in an inability to grow even in the presence of ergosterol and Tween 80 (Fig. 2B, line 3). Introduction of KlHAP1 into this Δhem1 Δhap1 double mutant restored limited growth on this medium (Fig. 2B, line 4) to a level similar to that of the Δhem1 single mutant (Fig. 2B, line 2). Complementation by KlHAP1 was as efficient as that by the S. cerevisiae HAP1 gene (Fig. 2B, line 5). These experiments showed that KlHap1p, similarly to the S. cerevisiae protein, has a role essential for growth under hypoxic or heme-deficient conditions when being introduced into S. cerevisiae (20, 49).
KlHap1p binds to the consensus cis element CGGN6CGG of the CYC1 and KlCYC1 promoters.
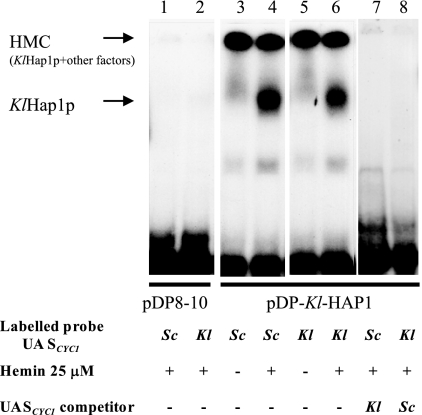
In S. cerevisiae, Hap1p binds to the cis element CGGN6CGG of the target gene promoters (23, 34), although this may not be the only mode of interaction (24). Both Sc-UASCYC1 and Kl-UASCYC1 contain the CGGN6CGG motif. The physical interaction of KlHap1p with this sequence was shown by a gel retardation experiment (Fig. 3). To do this experiment, we overproduced KlHap1p by introducing a multicopy KlHAP1-carrying plasmid into the S. cerevisiae Δhap1 strain (334-ΔHAP1) under the control of a galactose-inducible promoter. Labeled oligonucleotides Sc-UASCYC1 and Kl-UASCYC were used for binding assays. We found that (i) in the presence of hemin, the formation of a KlHap1p-DNA complex was observed (Fig. 3, lanes 4 and 6), and (ii) in the absence of hemin, a diffuse weak band with slower migration was formed (Fig. 3, lanes 3 and 5). Clearly heme was important for in vitro binding of KlHap1p to the cis element of the cytochrome gene. At the same time, a high-molecular-weight complex appeared on the top of the gel (Fig. 3, lanes 3 to 6), as previously reported (20, 54). All these data confirmed that KlHap1p could bind DNA by the same mechanism as that used by Hap1p.
FIG. 3.
Binding of KlHap1p to the upstream activation sequence element CCGN6CCG in the promoters of CYC1 and KlCYC1. Cell extracts were prepared from (i) S. cerevisiae strain 334-ΔHAP1 transformed with the plasmid pDP-Kl-HAP1, which contained the KlHAP1 gene fused to the GAL10-CYC1 promoter of S. cerevisiae (lanes 3 to 8), and (ii) the same strain transformed with the empty vector pDP8-10 as a control (lanes 1 and 2). The cell extracts were incubated with 32P-labeled probes (Sc-UASCYC1, lanes 1, 3, 4, and 7; Kl-UASCYC1, lanes 2, 5, 6, and 8), either in the absence (lanes 3 and 5) or in the presence (lanes 1, 2, 4, 6, 7, and 8) of 25 μM hemin. One hundred nanograms of unlabeled Kl-UASCYC1 or Sc-UASCYC1 was added as a competitor (lanes 7 and 8, respectively). Other details are specified in Materials and Methods.
Expression of KlHAP1 in K. lactis.
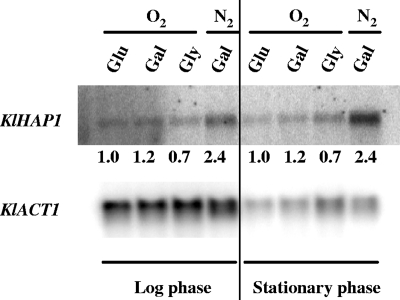
Figure 4 shows the transcript level of KlHAP1 under several growth conditions. The cells grown aerobically on glucose, galactose, or glycerol did not show any significant difference in the transcript level. By contrast, the transcription was markedly increased (2.4-fold) under hypoxia, at both exponential and stationary phases of growth. This response to hypoxia is similar to that known for ScHAP1 (20, 49).
FIG. 4.
Expression of KlHAP1 under various growth conditions. The K. lactis reference strain 2359/152 was grown under aerobic conditions (O2) in YP medium containing 2% glucose (Glu), 2% galactose (Gal), or 2% glycerol (Gly) or under hypoxic conditions (N2) in the Gal medium (galactose was used to avoid possible glucose repression). Total RNA was extracted at exponential and stationary phases of growth. The KlHAP1 probe was prepared by PCR amplification (primers are shown in Table 2). KlACT1 was used as a loading control. Signal quantitation and normalization were carried out as described in Materials and Methods, and the value 1.0 in each panel indicates the reference of normalized intensity with which other signals were compared.
Genes under the control of KlHap1p.
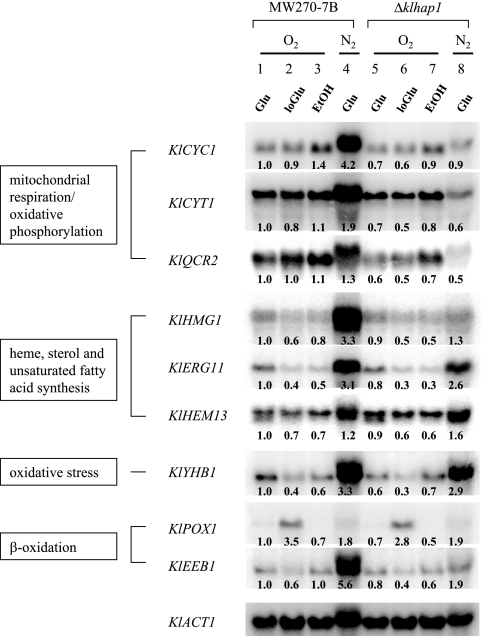
In order to identify the target genes of KlHap1p, we constructed a strain with a disrupted Klhap1 gene (see Materials and Methods). The transcript levels of several K. lactis genes, which are orthologous to the known target genes of Hap1p in S. cerevisiae (46), were compared between the wild-type and ΔKlhap1 strains under different growth conditions. Results are presented in Fig. 5.
FIG. 5.
Regulation by KlHap1p of the transcription of target genes in K. lactis. The wild-type strain MW270-7B and its ΔKlhap1 isogenic mutant were grown in YP medium containing 2% glucose (Glu; lanes 1 and 5), 0.02% glucose (loGlu; lanes 2 and 6), or 2% ethanol (EtOH; lanes 3 and 7) under aerobic conditions (O2). Under hypoxic conditions (N2; lanes 4 and 8), the YP-2% glucose medium was supplemented with ergosterol and Tween 80 and flushed with nitrogen gas. Total RNA was extracted at the exponential phase of growth. Gene probes were obtained by PCR amplification (primers are listed in Table 2). KlACT1 was used as a loading control. Signal quantitation and normalization were carried out as described in Materials and Methods, and the value 1.0 in each panel indicates the reference of normalized intensity with which other signals were compared.
KlCYC1, KlCYT1, and KlQCR2 genes encode the unique cytochrome c, the cytochrome c1, and subunit II of ubiquinol cytochrome c reductase of K. lactis, respectively (9, 11, 22). As with their S. cerevisiae counterparts, which are regulated by Hap1p (16, 43, 46), the expression of these three K. lactis genes was markedly reduced (>2.5-fold) by the Klhap1 mutation when oxygen was limited.
KlHMG1 is an ortholog of S. cerevisiae HMG1 encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase. The S. cerevisiae gene is known to be under the control of Hap1p (46, 47). The K. lactis ortholog was also activated (2.5-fold) by KlHap1p under hypoxic conditions.
ERG11 of S. cerevisiae encoding lanosterol 14-α-demethylase (cytochrome P450) is controlled by Hap1p (48). By contrast, the expression of its K. lactis ortholog was not affected by Klhap1 mutation under any of the conditions tested.
HEM13 of S. cerevisiae encodes coproporphyrinogen III oxidase. Its counterpart KlHEM13 has previously been reported to be subject to heme- and oxygen-dependent negative regulation (23), as is the case for the S. cerevisiae gene (2, 49). Our experiment indicated that KlHap1p had such a repressive effect on the KlHEM13 expression, but to a limited degree (1.4-fold) under oxygen-limited conditions.
YHB1 encodes a flavohemoglobin that could play a role against the effects of oxidative stress (55). Previous reports showed that the expression of YHB1 was high under oxygen-replete conditions and low under hypoxia (14, 55). In contrast to that, KlYHB1 expression was increased under hypoxic conditions. Furthermore, it appeared that KlHap1p had little effect on the expression of KlYHB1.
Interestingly, the overall amino acid sequence of KlHap1p has some similarity with those of Oaf1p and Pip2p of S. cerevisiae. Oaf1p and Pip2p are key transcription factors when the genes encoding peroxisomal proteins are activated in the presence of a fatty acid such as oleate (28). This sequence similarity raises the possibility that KlHap1p also has a regulatory role in fatty acid metabolism. Oaf1p has been reported to regulate POX1 and EEB1 (YPL095C) genes in S. cerevisiae (28). Sequences orthologous to these genes have been detected in the K. lactis genome. We found that the K. lactis ortholog of EEB1/EHT1 (but not that of POX1) was indeed regulated by KlHap1p (Fig. 5). EEB1 and EHT1 encode acyl coenzymeA:ethanol O-acyltransferase in S. cerevisiae (41). Thus, KlHap1p probably has a role in medium-chain fatty acid ethyl ester biosynthesis in K. lactis, the culture of which smells fruity and is easily distinguishable from that of S. cerevisiae.
The phenotype of the ΔKlhap1 mutant suggests important roles of KlHAP1 in glucose metabolism of K. lactis.
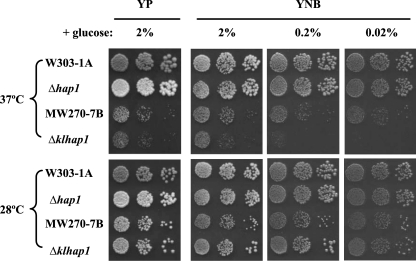
To further investigate the KlHAP1 function in K. lactis, we examined the phenotype of the ΔKlhap1 mutant grown under different conditions. The S. cerevisiae hap1 mutant was also examined for comparison.
The S. cerevisiae hap1 mutant did not present any modified phenotype in the culture conditions shown in Fig. 6. In contrast to the S. cerevisiae mutant, the ΔKlhap1 mutant showed an increased sensitivity to the temperature 37°C in glucose media, in particular at lower concentrations of glucose (upper panels in Fig. 6). At 28°C in media with higher levels of glucose, a careful observation of individual colonies led to a finding that the colonies formed by the ΔKlhap1 mutant were markedly larger than those of the wild type (lower panels in Fig. 6). Such differences between Klhap1 and hap1 mutants in glucose media might suggest that KlHAP1 could have a particular role in K. lactis, one so far unknown for HAP1 of S. cerevisiae.
FIG. 6.
Phenotype of the ΔKlhap1 mutant. The strains, including the S. cerevisiae wild-type strain W303-1A and its hap1 isogenic mutant and the K. lactis wild-type strain MW270-7B and its ΔKlhap1 isogenic mutant, were grown aerobically to stationary phase at 28°C in YP-2% glucose medium. The cells were serially diluted in 0.15 M NaCl and dropped onto complete medium (YP) or minimal medium (YNB) containing different concentrations of glucose as specified and then grown at 28°C (lower panels) or 37°C (upper panels) for 2 to 4 days. The cells grown at 28°C for 2 days in YP-2% glucose medium (the lower leftmost panel) were used as a reference for cell amount loaded.
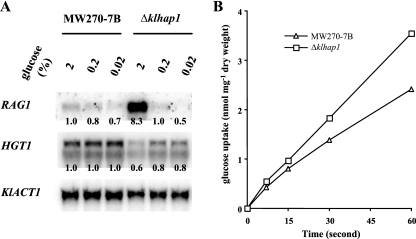
KlHAP1 negatively controls the transcription of RAG1.
When yeast grows on sugars under hypoxia or under conditions limiting respiration, the fermentation requires a much higher flow of substrate than does the respiratory mode of growth. Sugar uptake often becomes the limiting step of fermentation. In this sense, oxygen availability may possibly modify the gene activities involved in sugar uptake. K. lactis has two glucose permease genes, RAG1 (for low-affinity transport; Km of about 20 to 50 mM) (12, 52) and HGT1 (for high-affinity transport; Km of about 1 mM) (5). It has been reported that the colonies of the strain having the rag1 deletion were smaller than those of the wild type in 2% glucose medium (52). Therefore, we examined whether KlHap1p could influence the expression of the two glucose transporter genes. The results of Northern hybridization clearly revealed that at a high glucose concentration, RAG1 transcript level was significantly higher in the ΔKlhap1 mutant than in the wild type while full expression of HGT1, in contrast, seemed to be dependent on KlHap1p (Fig. 7A). These effects were also confirmed in minimal medium (data not shown). It appeared therefore that KlHap1p negatively regulated RAG1 transcription specifically at a high glucose concentration.
FIG. 7.
KlHap1p controls the transcription of the glucose transporter genes (A) and glucose uptake rate (B) in K. lactis. (A) The wild-type strain MW270-7B and its ΔKlhap1 isogenic mutant were grown aerobically at 28°C in YP medium containing different concentrations of glucose (2% [≈110 mM], 0.2% [≈11 mM], and 0.02% [≈1.1 mM]) as specified. Total RNA was extracted at the exponential phase of growth. Gene probes were obtained by PCR amplification (primers are listed in Table 2). KlACT1 was used as a loading control. Signal quantitation and normalization were carried out as described in Materials and Methods, and the value 1.0 in each panel indicates the reference of normalized intensity with which other signals were compared. Note that HGT1 has two transcripts of different sizes whose precise nature is not known (5). (B) The K. lactis wild-type strain MW270-7B and its ΔKlhap1 isogenic mutant were grown at 28°C in YP-2% glucose. Cells were suspended in phosphate buffer, and the rate of uptake of [14C]glucose (100 mM) was measured as described in Materials and Methods. The values are means of two independent experiments. In no case was the variation higher than 15% of the mean.
KlHAP1 represses glucose uptake in K. lactis.
The finding that KlHAP1 controls the expression of glucose transporter genes in K. lactis raised the question of whether such regulation might have a physiological significance in glucose transport. We then determined the rate of glucose uptake in vivo. When a low concentration of glucose (10 mM) was used in the uptake assay system, no radical changes were observed in the ΔKlhap1 mutant with respect to the wild type (data not shown). However, when a high concentration of glucose (100 mM) was used, we found a 47% increase in the overall rate of glucose uptake compared to that of the wild type (Fig. 7B). It should be noted that the observed glucose uptake was due to both RAG1 and HGT1 products. Since HGT1 transcription was lowered in the mutant under high-glucose conditions (Fig. 7A), the increase of glucose uptake due to the RAG1 product was thought to be even higher than 47%.
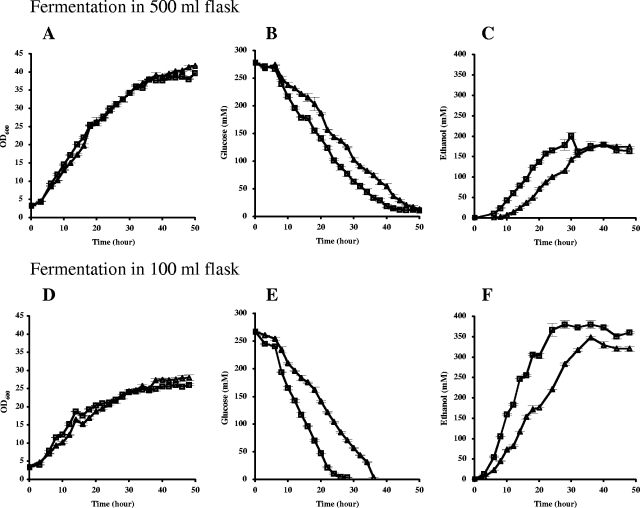
The ΔKlhap1 mutant has an increased rate of ethanol production.
The effect of the Klhap1 mutation on RAG1 expression and glucose transport led us to compare the time course of ethanol accumulation between the wild-type MW270-7B and the ΔKlhap1 mutant. Two S. cerevisiae strains, the wild-type W303-1A and an isogenic hap1 mutant (disrupted by KanMX to avoid influence on fermentation resulting from auxotrophy), were examined in parallel for each experiment.
Oxygen limitation has been regarded as the primary environmental factor in triggering alcoholic fermentation in K. lactis (30). We therefore carried out the fermentation experiments in smaller flasks (50 ml medium in a 100-ml flask) simulating an oxygen-limiting condition (lower panels in Fig. 8). In conditions where the two strains grew at similar rates (Fig. 8D), the ΔKlhap1 mutant consumed glucose much faster than did the wild type (a difference of about 8 to 10 h [Fig. 8E]), in agreement with the results of glucose uptake assays. Consequently, the ΔKlhap1 mutant produced ethanol more rapidly than did the wild type (Fig. 8F).
FIG. 8.
Kinetics of ethanol production of the K. lactis wild-type strain MW270-7B (▵) and its ΔKlhap1 isogenic mutant (□). Cells were grown at 28°C in 50 ml YP-5% glucose contained in 500-ml flasks (upper panels) or 100-ml flasks (lower panels). Aliquots were taken for analysis of cell OD600 (A and D), glucose consumption (B and E), and ethanol production (C and F). Other details are described in Materials and Methods. The values are means of analysis for two flask cultures. Bars indicate the up and down variations.
As described above, the expression of KlHAP1 responded to oxygen availability. We therefore performed the fermentation experiments under normoxia (50 ml medium in a 500-ml flask [upper panels in Fig. 8]), which is usually used as an aerobic condition in routine manipulation. Compared to growth under the oxygen-limiting condition, both the wild type and the ΔKlhap1 mutant grew to a higher final optical density (OD600 of ≈40 [Fig. 8A] versus OD600 of ≈25 under the oxygen-limiting condition) and consumed glucose much more slowly (Fig. 8B), showing a strong Pasteur effect (33, 44). Even in this case, the difference in glucose consumption still existed between the wild type and the ΔKlhap1 mutant (a difference of about 6 h [Fig. 8B]). A similar increase in ethanol production was found in the ΔKlhap1 mutant, but at a lower ethanol concentration (Fig. 8C). Although cultivation in a shake-flask is not regarded as a strictly aerobic condition, we noticed that the K. lactis wild type did not produce ethanol until after 8 h of incubation. Meanwhile, 10 mM ethanol was already detected in the 6-h culture of the ΔKlhap1 mutant.
However, such differences were not observed between the S. cerevisiae strains under the growth conditions that we tested (data not shown), indicating differences in roles between KlHAP1 in K. lactis and HAP1 in S. cerevisiae which may relate to the distinctness of their glucose metabolisms.
DISCUSSION
The spectrum of target genes regulated by KlHAP1 and its mode of regulation in K. lactis could deviate from those of HAP1 in S. cerevisiae.
The sequence of KlHap1p contains the equivalents of all the functional domains described for the S. cerevisiae counterpart. Therefore, the functional similarity of KlHap1p to ScHap1p can be anticipated from the sequence features. Complementation of the Schap1 mutation by the KlHAP1 gene showed that KlHap1p was an activator in aerobiosis and could also restore growth in heme-depleted conditions. The capacity of heterospecific binding to common cis elements indicated that the Hap1p proteins of the two species share a common specific interaction with these target sites. Our results, together with previous reports (18, 23), show that the basic regulatory scheme “O2 → heme → Hap1p → some respiratory genes (as for example CYC1),” known in S. cerevisiae, also applies to K. lactis. In this sense, the two yeasts thus share a common mechanism of Hap1p-mediated transcriptional regulation in response to oxygen availability, despite the fact that the two species have a number of important metabolic differences.
However, the action of KlHap1p seems to diverge essentially by the nature of the target genes. The comparisons are summarized in Table 3. Among the structural orthologs of the Hap1p-regulated genes of S. cerevisiae, some K. lactis genes seem to escape from KlHap1p regulation. KlERG11 is in this category, and the putative binding site is not detected in the promoters of KlERG11. For many genes, we observed the regulation only under hypoxic conditions. In aerobiosis, they are less regulated by KlHAP1, an observation also reported in a recent paper (35). On the other hand, some other genes such as KlEEB1 (KlEHT1), RAG1, and HGT1, orthologs of which have not been reported as Hap1p-regulated targets in S. cerevisiae, are in turn under the control of KlHap1p in K. lactis.
TABLE 3.
Target genes of Hap1p: comparison between S. cerevisiae and K. lactisa
| Target gene |
S. cerevisiae
|
K. lactis
|
||
|---|---|---|---|---|
| CGGN6CGG sites in the promoterb | Regulation by ScHap1pc | CGGN6CGG sites in the promoterb | Regulation by KlHap1pc | |
| CYC1 | −349 CGGAAAGATCGG −360 | + | −285 CGGGAACATCGG −296 | + |
| −337 CGGGGTTTACGG −326 | ||||
| CYC7 | −612 GGGACTCCCCGG −623 | + | ||
| CYT1 | −527 CGGATTTCTTGG −516 | + | Not detected | + |
| −494 CGGAAATACCGG −505 | ||||
| QCR2 | −472 CGGGATTCAAGG −461 | + | −917 CGGGGATCAGGG −906 | + |
| −509 GGGTTTTAGCGG −520 | ||||
| HMG1 | −620 CGGGGTAAAAGG −631 | + | Not detected | + |
| ERG11 | −789 CGGGACAGGCGG −800 | + | Not detected | n |
| −731 GGGAATTACCGG −720 | ||||
| −642 CGGCTTAGTCGG −653 | ||||
| HEM13 | −778 CGGATCTTGCGG −767 | − | −782 TGGCATACACGG −771 | − |
| −358 CGGCTCCGCGGG −347 | −259 CGGAGACATAGG −270 | |||
| YHB1 | −469 TGGATCCTCCGG −458 | + | −595 CGGAGCTACCGG −606 | n |
| RAG1 (K. lactis) | HXTs? | ? | Not detectedd | − |
| HGT1 (K. lactis) | HXTs? | ? | −531 CGGAATTCTGGG −542 | + |
| EEB1 | −713 CGGCCACAATGG −724 | −240 CGGATCCTTCGG −229 | + | |
| −677 GGGTGCCTTCGG −666 | ? | |||
The phenotypes of the Δhap1 mutants for each strain were as follows: S. cerevisiae, no obvious change in growth phenotype; K. lactis, growth defect in low-glucose medium at 37°C and increased colony size in high-glucose medium at 28°C.
A search for putative Hap1p binding site CGGN6CGG (in two orientations) was carried out over a 1-kb region upstream of each gene by using the program at http://rulai.cshl.edu/, one mismatch allowed (C→A, G, T). The numbers refer to the distance from the motif to the ATG initiation codon. Boldface characters indicate nucleotides which match the CGG triplets of the consensus sequence.
Symbols: +, activated; −, repressed; n, little affected; ?, unknown or unclear.
The RAG1 gene has two CGGN5CGG sequences (one nucleotide shorter than the consensus sequence) at −629 and −952.
Transcription of CYC1 and KlCYC1 is regulated by KlHap1p through its binding to the upstream cis element CGGN6CGG of their promoters. However, this motif is not always found in other genes that are apparently regulated by KlHap1p. Such was the case for the genes RAG1, KlCYT1, and KlHMG1 in our data (Table 3). KlHap1p may indirectly act on these genes through intermediary regulators. It is also possible that the sequence of this motif can deviate from the consensus without losing the binding specificity, as previously suggested for the case of the S. cerevisiae d-lactate dehydrogenase gene DLD1 (36).
KlHap1p may be involved in the Crabtree effect.
KlHap1p repressed the expression of the major glucose transporter gene RAG1 and overall glucose transport at high glucose concentrations in K. lactis. Since the rate of sugar uptake is a major determinant of the glycolytic flux for fermentation (19) and the regulation of glucose uptake is crucial for sensitivity to glucose repression in K. lactis (50), KlHap1p might have a role in negatively controlling the glycolytic flux by reducing sugar uptake, while it positively regulates the expression of some respiratory genes in K. lactis. The logic of this dual regulation may be that it would help maximize the respiratory pathway and minimize fermentation in this Crabtree-negative species. A possible role of Hap1p in the transcription of sugar transporter genes has not been reported so far for any organism. It is still unclear how KlHap1p regulates the transcription of the glucose transporter gene.
Acknowledgments
We thank Tiziana Lodi (University of Parma) for useful advice and help in glucose uptake experiments and Lydie Belliard for her assistance in several experiments.
This work received support from the Institut Fédératif de Recherche Génomes-IFR115 and the European Union contract QLK3-CT-2000-00174. F.M.L.P. received a sabbatical fellowship from CNPq-Brazil. Z.-A.F. is a recipient of a fellowship from the French-Chinese joint doctoral program (French Embassy in China).
Footnotes
Published ahead of print on 19 September 2008.
REFERENCES
- 1.Alberti, A., P. Goffrini, I. Ferrero, and T. Lodi. 2000. Cloning and characterization of the lactate-specific inducible gene KlCYB2, encoding the cytochrome b(2) of Kluyveromyces lactis. Yeast 16657-665. [DOI] [PubMed] [Google Scholar]
- 2.Amillet, J. M., N. Buisson, and R. Labbe-Bois. 1995. Positive and negative elements involved in the differential regulation by heme and oxygen of the HEM13 gene (coproporphyrinogen oxidase) in Saccharomyces cerevisiae. Curr. Genet. 28503-511. [DOI] [PubMed] [Google Scholar]
- 3.Andreasen, A. A., and T. J. Stier. 1953. Anaerobic nutrition of Saccharomyces cerevisiae. I. Ergosterol requirement for growth in a defined medium. J. Cell. Physiol. 4123-36. [DOI] [PubMed] [Google Scholar]
- 4.Andreasen, A. A., and T. J. Stier. 1954. Anaerobic nutrition of Saccharomyces cerevisiae. II. Unsaturated fatty acid requirement for growth in a defined medium. J. Cell. Physiol. 43271-281. [DOI] [PubMed] [Google Scholar]
- 5.Billard, P., S. Menart, J. Blaisonneau, M. Bolotin-Fukuhara, H. Fukuhara, and M. Wesolowski-Louvel. 1996. Glucose uptake in Kluyveromyces lactis: role of the HGT1 gene in glucose transport. J. Bacteriol. 1785860-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourgarel, D., C. C. Nguyen, and M. Bolotin-Fukuhara. 1999. HAP4, the glucose-repressed regulated subunit of the HAP transcriptional complex involved in the fermentation-respiration shift, has a functional homologue in the respiratory yeast Kluyveromyces lactis. Mol. Microbiol. 311205-1215. [DOI] [PubMed] [Google Scholar]
- 7.Breunig, K. D., M. Bolotin-Fukuhara, M. M. Bianchi, D. Bourgarel, C. Falcone, I. I. Ferrero, L. Frontali, P. Goffrini, J. J. Krijger, C. Mazzoni, C. Milkowski, H. Y. Steensma, M. Wesolowski-Louvel, and A. M. Zeeman. 2000. Regulation of primary carbon metabolism in Kluyveromyces lactis. Enzyme Microb. Technol. 26771-780. [DOI] [PubMed] [Google Scholar]
- 8.Breunig, K. D., and H. Y. Steensma. 2003. Kluyveromyces lactis: genetics, physiology and application. Top. Curr. Genet. 2171-206. [Google Scholar]
- 9.Brons, J. F., A. A. Dryla, E. B. Pluger, T. M. Vinkenvleugel, N. C. Hornig, L. A. Grivell, and J. Blom. 2001. Carbon source-dependent transcriptional regulation of the QCR8 gene in Kluyveromyces lactis. Identification of cis-acting regions and trans-acting factors in the KlQCR8 upstream region. Curr. Genet. 39311-318. [DOI] [PubMed] [Google Scholar]
- 10.Chantrel, Y., M. Gaisne, C. Lions, and J. Verdiere. 1998. The transcriptional regulator Hap1p (Cyp1p) is essential for anaerobic or heme-deficient growth of Saccharomyces cerevisiae: genetic and molecular characterization of an extragenic suppressor that encodes a WD repeat protein. Genetics 148559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, X. J., and G. D. Clark-Walker. 1993. Mutations in MGI genes convert Kluyveromyces lactis into a petite-positive yeast. Genetics 133517-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, X. J., M. Wesolowski-Louvel, and H. Fukuhara. 1992. Glucose transport in the yeast Kluyveromyces lactis. II. Transcriptional regulation of the glucose transporter gene RAG1. Mol. Gen. Genet. 23397-105. [DOI] [PubMed] [Google Scholar]
- 13.Claisse, M. L., G. A. Pere-Aubert, L. P. Clavilier, and P. P. Slonimski. 1970. Method for the determination of cytochrome concentrations in whole yeast cells. Eur. J. Biochem. 16430-438. (In French.) [DOI] [PubMed] [Google Scholar]
- 14.Crawford, M. J., D. R. Sherman, and D. E. Goldberg. 1995. Regulation of Saccharomyces cerevisiae flavohemoglobin gene expression. J. Biol. Chem. 2706991-6996. [DOI] [PubMed] [Google Scholar]
- 15.Creusot, F., J. Verdiere, M. Gaisne, and P. P. Slonimski. 1988. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J. Mol. Biol. 204263-276. [DOI] [PubMed] [Google Scholar]
- 16.Dorsman, J. C., and L. A. Grivell. 1990. Expression of the gene encoding subunit II of yeast QH2: cytochrome c oxidoreductase is regulated by multiple factors. Curr. Genet. 17459-464. [DOI] [PubMed] [Google Scholar]
- 17.Fairhead, C., B. Llorente, F. Denis, M. Soler, and B. Dujon. 1996. New vectors for combinatorial deletions in yeast chromosomes and for gap-repair cloning using ‘split-marker’ recombination. Yeast 121439-1457. [DOI] [PubMed] [Google Scholar]
- 18.Freire-Picos, M. A., C. P. Hollenberg, K. D. Breunig, and M. E. Cerdan. 1995. Regulation of cytochrome c expression in the aerobic respiratory yeast Kluyveromyces lactis. FEBS Lett. 36039-42. [DOI] [PubMed] [Google Scholar]
- 19.Fukuhara, H. 2003. The Kluyver effect revisited. FEMS Yeast Res. 3327-331. [DOI] [PubMed] [Google Scholar]
- 20.Fytlovich, S., M. Gervais, C. Agrimonti, and B. Guiard. 1993. Evidence for an interaction between the CYP1(HAP1) activator and a cellular factor during heme-dependent transcriptional regulation in the yeast Saccharomyces cerevisiae. EMBO J. 121209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gbelska, Y., K. Horvathova, Q. J. van der Aart, B. J. Zonneveld, H. Y. Steensma, and J. Subik. 1996. Isolation and molecular analysis of the gene for cytochrome c1 from Kluyveromyces lactis. Curr. Genet. 30145-150. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Dominguez, M., M. A. Freire-Picos, E. Ramil, B. Guiard, and M. E. Cerdan. 2000. Heme-mediated transcriptional control in Kluyveromyces lactis. Curr. Genet. 38171-177. [DOI] [PubMed] [Google Scholar]
- 24.Ha, N., K. Hellauer, and B. Turcotte. 1996. Mutations in target DNA elements of yeast HAP1 modulate its transcriptional activity without affecting DNA binding. Nucleic Acids Res. 241453-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hon, T., A. Dodd, R. Dirmeier, N. Gorman, P. R. Sinclair, L. Zhang, and R. O. Poyton. 2003. A mechanism of oxygen sensing in yeast. Multiple oxygen-responsive steps in the heme biosynthetic pathway affect Hap1 activity. J. Biol. Chem. 27850771-50780. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, Y., M. J. Vasconcelles, S. Wretzel, A. Light, L. Gilooly, K. McDaid, C. S. Oh, C. E. Martin, and M. A. Goldberg. 2002. Mga2p processing by hypoxia and unsaturated fatty acids in Saccharomyces cerevisiae: impact on LORE-dependent gene expression. Eukaryot. Cell 1481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kagi, J. H., and B. L. Vallee. 1960. The role of zinc in alcohol dehydrogenase. V. The effect of metal-binding agents on the structure of the yeast alcohol dehydrogenase molecule. J. Biol. Chem. 2353188-3192. [PubMed] [Google Scholar]
- 28.Karpichev, I. V., and G. M. Small. 1998. Global regulatory functions of Oaf1p and Pip2p (Oaf2p), transcription factors that regulate genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 186560-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kastaniotis, A. J., T. A. Mennella, C. Konrad, A. M. Torres, and R. S. Zitomer. 2000. Roles of transcription factor Mot3 and chromatin in repression of the hypoxic gene ANB1 in yeast. Mol. Cell. Biol. 207088-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiers, J., A. M. Zeeman, M. Luttik, C. Thiele, J. I. Castrillo, H. Y. Steensma, J. P. van Dijken, and J. T. Pronk. 1998. Regulation of alcoholic fermentation in batch and chemostat cultures of Kluyveromyces lactis CBS 2359. Yeast 14459-469. [DOI] [PubMed] [Google Scholar]
- 31.Kwast, K. E., P. V. Burke, and R. O. Poyton. 1998. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J. Exp. Biol. 2011177-1195. [DOI] [PubMed] [Google Scholar]
- 32.Kwast, K. E., L. C. Lai, N. Menda, D. T. James III, S. Aref, and P. V. Burke. 2002. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol. 184250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagunas, R. 1986. Misconceptions about the energy metabolism of Saccharomyces cerevisiae. Yeast 2221-228. [DOI] [PubMed] [Google Scholar]
- 34.Lalonde, B., B. Arcangioli, and L. Guarente. 1986. A single Saccharomyces cerevisiae upstream activation site (UAS1) has two distinct regions essential for its activity. Mol. Cell. Biol. 64690-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamas-Maceiras, M., L. Nunez, E. Rodriguez-Belmonte, M. I. Gonzalez-Siso, and M. E. Cerdan. 2007. Functional characterization of KlHAP1: a model to foresee different mechanisms of transcriptional regulation by Hap1p in yeasts. Gene 40596-107. [DOI] [PubMed] [Google Scholar]
- 36.Lodi, T., A. Alberti, B. Guiard, and I. Ferrero. 1999. Regulation of the Saccharomyces cerevisiae DLD1 gene encoding the mitochondrial protein D-lactate ferricytochrome c oxidoreductase by HAP1 and HAP2/3/4/5. Mol. Gen. Genet. 262623-632. [DOI] [PubMed] [Google Scholar]
- 37.Lodi, T., M. Saliola, C. Donnini, and P. Goffrini. 2001. Three target genes for the transcriptional activator Cat8p of Kluyveromyces lactis: acetyl coenzyme A synthetase genes KlACS1 and KlACS2 and lactate permease gene KlJEN1. J. Bacteriol. 1835257-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31426-428. [Google Scholar]
- 39.Ochman, H., J. W. Ajioka, D. Garza, and D. L. Hartl. 1990. Inverse polymerase chain reaction. Bio/Technology 8759-760. [DOI] [PubMed] [Google Scholar]
- 40.Pfeifer, K., B. Arcangioli, and L. Guarente. 1987. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell 499-18. [DOI] [PubMed] [Google Scholar]
- 41.Saerens, S. M., K. J. Verstrepen, S. D. Van Laere, A. R. Voet, P. Van Dijck, F. R. Delvaux, and J. M. Thevelein. 2006. The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium-chain fatty acid ethyl ester synthesis and hydrolysis capacity. J. Biol. Chem. 2814446-4456. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 183091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider, J. C., and L. Guarente. 1991. Regulation of the yeast CYT1 gene encoding cytochrome c1 by HAP1 and HAP2/3/4. Mol. Cell. Biol. 114934-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snoek, I. S. I., and H. Y. Steensma. 2007. Factors involved in anaerobic growth of Saccharomyces cerevisiae. Yeast 241-10. [DOI] [PubMed] [Google Scholar]
- 45.Souciet, J., M. Aigle, F. Artiguenave, G. Blandin, M. Bolotin-Fukuhara, E. Bon, P. Brottier, S. Casaregola, J. de Montigny, B. Dujon, P. Durrens, C. Gaillardin, A. Lepingle, B. Llorente, A. Malpertuy, C. Neuveglise, O. Ozier-Kalogeropoulos, S. Potier, W. Saurin, F. Tekaia, C. Toffano-Nioche, M. Wesolowski-Louvel, P. Wincker, and J. Weissenbach. 2000. Genomic exploration of the hemiascomycetous yeasts. 1. A set of yeast species for molecular evolution studies. FEBS Lett. 4873-12. [DOI] [PubMed] [Google Scholar]
- 46.Ter Linde, J. J., and H. Y. Steensma. 2002. A microarray-assisted screen for potential Hap1 and Rox1 target genes in Saccharomyces cerevisiae. Yeast 19825-840. [DOI] [PubMed] [Google Scholar]
- 47.Thorsness, M., W. Schafer, L. D'Ari, and J. Rine. 1989. Positive and negative transcriptional control by heme of genes encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase in Saccharomyces cerevisiae. Mol. Cell. Biol. 95702-5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turi, T. G., and J. C. Loper. 1992. Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14 alpha-demethylase (ERG11). J. Biol. Chem. 2672046-2056. [PubMed] [Google Scholar]
- 49.Verdiere, J., M. Gaisne, and R. Labbe-Bois. 1991. CYP1 (HAP1) is a determinant effector of alternative expression of heme-dependent transcribed genes in yeast. Mol. Gen. Genet. 228300-306. [DOI] [PubMed] [Google Scholar]
- 50.Weirich, J., P. Goffrini, P. Kuger, I. Ferrero, and K. D. Breunig. 1997. Influence of mutations in hexose-transporter genes on glucose repression in Kluyveromyces lactis. Eur. J. Biochem. 249248-257. [DOI] [PubMed] [Google Scholar]
- 51.Wésolowski-Louvel, M., K. D. Breunig, and H. Fukuhara. 1996. Kluyveromyces lactis, p. 139-201. In K. Wolf (ed.), Nonconventional yeasts in biotechnology. Springer-Verlag, Berlin, Germany.
- 52.Wésolowski-Louvel, M., P. Goffrini, I. Ferrero, and H. Fukuhara. 1992. Glucose transport in the yeast Kluyveromyces lactis. I. Properties of an inducible low-affinity glucose transporter gene. Mol. Gen. Genet. 23389-96. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, L., and A. Hach. 1999. Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cell. Mol. Life Sci. 56415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, L., A. Hach, and C. Wang. 1998. Molecular mechanism governing heme signaling in yeast: a higher-order complex mediates heme regulation of the transcriptional activator HAP1. Mol. Cell. Biol. 183819-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao, X. J., D. Raitt, P. V. Burke, A. S. Clewell, K. E. Kwast, and R. O. Poyton. 1996. Function and expression of flavohemoglobin in Saccharomyces cerevisiae. Evidence for a role in the oxidative stress response. J. Biol. Chem. 27125131-25138. [DOI] [PubMed] [Google Scholar]
- 56.Zitomer, R. S., and C. V. Lowry. 1992. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]