Abstract
The cell wall of the human pathogen Candida glabrata governs initial host-pathogen interactions that underlie the establishment of fungal infections. With the aim of identifying species-specific features that may directly relate to its virulence, we have investigated the cell wall of C. glabrata using a multidisciplinary approach that combines microscopy imaging, biochemical studies, bioinformatics, and tandem mass spectrometry. Electron microscopy revealed a bilayered wall structure in which the outer layer is packed with mannoproteins. Biochemical studies showed that C. glabrata walls incorporate 50% more protein than Saccharomyces cerevisiae walls and, consistent with this, have a higher mannose/glucose ratio. Evidence is presented that C. glabrata walls contain glycosylphosphatidylinositol (GPI) proteins, covalently bound to the wall 1,6-β-glucan, as well as proteins linked through a mild-alkali-sensitive linkage to 1,3-β-glucan. A comprehensive genome-wide in silico inspection showed that in comparison to other fungi, C. glabrata contains an exceptionally large number, 67, of genes encoding adhesin-like GPI proteins. Phylogenetically these adhesin-like proteins form different clusters, one of which is the lectin-like EPA family. Mass spectrometric analysis identified 23 cell wall proteins, including 4 novel adhesin-like proteins, Awp1/2/3/4, and Epa6, which is involved in adherence to human epithelia and biofilm formation. Importantly, the presence of adhesin-like proteins in the wall depended on the growth stage and on the genetic background used, and this was reflected in alterations in adhesion capacity and cell surface hydrophobicity. We propose that the large repertoire of adhesin(-like) genes of C. glabrata contributes to its adaptability and virulence.
In the adult human population, Candida glabrata is the second most frequent cause of mucosal and disseminated candidiasis after Candida albicans (50). In a survey by the nationwide German laboratory network MykolabNet-D, 19.1% of all Candida isolates from primarily sterile body sites were identified as C. glabrata (3). The high degree of resistance against azoles in clinical C. glabrata strains makes it more complicated for clinicians to develop an adequate therapeutic strategy for their patients. Thus, there is a clear need for the identification of fungal structures that might serve as new targets for antifungal drug development or that promote the design of improved non-culture-based diagnostic tests.
In C. albicans, morphological switching between a budding yeast form, pseudohyphae, and an invasive filamentous form is regarded as an important virulence factor (55). Although the haploid yeast C. glabrata may develop pseudohyphae under specific in vitro conditions (10), formation of pseudo- or true hyphae during colonization or tissue infection has not been observed. Morphologically and phylogenetically, C. glabrata is much more closely related to the nonpathogenic yeast Saccharomyces cerevisiae than to C. albicans or other medically relevant Candida species in the CTG clade. Since the cell walls of pathogenic fungi form the first point of contact with the human host, they are extremely challenging structures to explore (52, 65). Since candidiasis is caused by a heterogeneous group of biologically distinct fungi, it seems especially important to identify species-specific differences or genus-specific consistencies in cell walls of human-pathogenic Candida spp. The multitude of data concerning the cell wall of S. cerevisiae and recent advances in the genomic toolbox for the pathogen C. glabrata offer unique opportunities to successfully exploit the structure of the cell wall in this species and improve our understanding of the relationship between cell wall structure and virulence.
The fungal cell wall is an essential organelle that shapes the cell, provides physical strength, and limits permeability, thereby retaining periplasmic proteins and protecting fungi from hostile degrading enzymes in the environment. The cell wall also plays an important role in crucial host-fungus interactions that facilitate the establishment of human mycoses. For instance, cell wall components mediate tissue adhesion and invasion, provide protection against host defense reactions, are involved in biofilm formation, trigger the host immune response, and may also confer resistance to antifungal drugs (15, 17, 41, 45, 48, 62, 69).
In S. cerevisiae, it was shown that the structure of the cell wall is highly dynamic during the cell cycle and depends on environmental conditions (31, 34). Several signaling pathways have been identified that regulate the expression of cell wall proteins in response to nutrient limitation, stress, or other signals. Additional adaptive properties are provided through an enormous genetic variability of cell wall proteins, e.g., through subtelomeric epigenetic switching or recombination of intragenic tandem repeats within cell wall genes (17, 43, 63, 64). The effect of variability of tandem repeats on functional diversity of cell surface proteins has been shown for the Flo family in S. cerevisiae, members of which mediate yeast flocculation and adherence to abiotic surfaces, such as agar and plastic (23, 61, 64). Similar mechanisms and a highly adaptive nature of the cell wall may help human-pathogenic Candida spp. to adhere to and thrive in different host niches of the human body. A limited number of fungal cell wall proteins (CWPs) have already been shown or suggested to be instrumental in adhesion to human tissues (e.g., Als proteins, Eap1, and Hwp1 in C. albicans and Epa1 in C. glabrata) (8, 25, 35, 59) and in other infection-related processes, such as invasion, biofilm formation, iron acquisition, scavenging of reactive molecules, and proteolytic cleavage (2, 26, 37, 51, 68). Differential expression of the ALS gene family offers C. albicans optimal ability to adhere to various host sites and medical devices, giving rise to different forms of human infections (24, 25). In C. glabrata, silent adhesin-encoding genes of the Epa family can be induced by nicotinic acid limitation, resulting in the incorporation of Epa proteins in the wall during urinary tract infection (17). Therefore, understanding the cell wall architecture and proteome of C. glabrata is crucial to get more insight into the fungal pathogenesis of this unique organism.
In this study, we have performed detailed biochemical, bioinformatic, and proteomic investigations to improve our understanding of the cell wall of C. glabrata in relation to pathogenesis. The cell wall organization appears similar to that of the closely related nonpathogenic yeast S. cerevisiae; however, C. glabrata walls contain more mannoprotein. The cell wall mannoproteins can be divided into two groups, the largest group being glycosylphosphatidylinositol (GPI)-modified proteins that are covalently bound to the wall 1,6-β-glucan. Previously we reported that C. glabrata contains 106 putative GPI proteins, about 50% of which have features of adhesin-like CWPs (67). We now systematically searched the genome for adhesin-like proteins and classified them into different subgroups. Using a direct cell wall “′shaving” approach and tandem mass spectrometry, we identified 23 covalently bound CWPs, and we showed that the incorporation of adhesin-like wall proteins is dependent on the growth phase and on the strain background. The availability of a large repertoire of differentially regulated adhesin genes may provide C. glabrata ample opportunities to adhere to and form biofilms on a large variety of surfaces and to thrive under many different environmental growth conditions.
MATERIALS AND METHODS
Strains and cell culture.
The C. glabrata strains used in this study were ATCC 90876 (isolated from blood) and ATCC 2001 (from feces). Biochemical analyses were performed with ATCC 90876 grown at 30°C in liquid yeast extract-peptone-dextrose (YEPD) (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto peptone, 2% [wt/vol] glucose) or in synthetic complete (SC) medium, containing 2% (wt/vol) glucose, 1.1% (wt/vol) Casamino Acids (Difco), 0.8% (wt/vol) yeast nitrogen base with ammonium sulfate without amino acids (Becton Dickinson), 110 μg/ml l-leucine (Sigma), 55 μg/ml l-tyrosine (Sigma), 55 μg/ml l-tryptophan (Sigma), and 55 μg/ml adenine sulfate (Fluka), unless otherwise stated. Growth experiments at different pHs were carried out in SC medium buffered with 100 mM HEPES-NaOH. Cultures were inoculated at a starting optical density (OD) of ∼0.1 with cells from fresh overnight cultures (in YEPD) and harvested at log phase (OD = ∼2) or after 24 h (stationary phase). Strain ATCC 2001, from which the genome sequence is derived, was included in cell wall proteomic experiments. S. cerevisiae (strain BY4741) and C. albicans (CAF2-1) were used for comparison of the cell wall composition.
Microscopic methods. (i) Transmission electron microscopy.
C. glabrata was grown at 37°C to an OD at 600 nm (OD600) of 0.9 in liquid YEPD medium. Cells were harvested and fixed in 3% glutaraldehyde for 3 h. The samples were postfixed in 1% OsO4 at 4°C. After several washing and dehydration steps, the samples were embedded in araldite. Ultrathin sections (70 to 74 nm) were cut using an ultramicrotome (Ultracut) and were contrasted with lead citrate and examined using a Zeiss EM 10 transmission electron microscope operating at 60 kV, at magnifications between 1,000- and 50,000-fold.
(ii) Fluorescence microscopy of CFW-stained cells.
Cells were grown at 37°C in liquid YEPD medium to an OD600 of 2, washed extensively, and incubated with calcofluor white (CFW) (100 μg/ml in phosphate-buffered saline) for 15 min. After unbound CFW was removed by washing with phosphate-buffered saline, cells were examined using a Leica DMR fluorescence microscope.
Cell surface hydrophobicity and in vitro adhesion capacity.
The hydrophobicity of the cell surface of C. glabrata was determined by measuring the relative distribution of yeast cells in a two-phase system consisting of an aqueous phase and the organic solvent octane. Briefly, liquid YEPD was inoculated with C. glabrata cells at a starting OD600 of 0.08 and subsequently cultured at 30°C. Cells were harvested during exponential growth (OD600 of ∼2) or after 24 h, when all glucose was consumed. Harvested cells were washed extensively with distilled water. Two ml of the cell suspension, adjusted to an OD600 of 0.7, was transferred to a glass tube containing 2 ml octane (Sigma Aldrich) and mixed for 1 min by gentle vortexing. After separation of the phases, the aqueous phase was carefully transferred to a cuvette and the OD600 was measured. Each value represents an average of three independent biological replicates for each strain, with five measurements per individual cell culture.
For testing of in vitro adhesion to a plastic surface, 25 μl of liquid YEPD cultures of C. glabrata were grown at 30°C as droplets in polystyrene petri dishes (Greiner). Dehydration was prevented by spotting additional water droplets. The cultures were started with cells grown to an OD600 of 2. After 3 days, nonadherent cells were washed away with water. Staining of adherent cells was carried out with 0.1% crystal violet for 10 min at 25°C (22). For quantification of adhesion, cells were grown in YEPD to exponential phase, transferred to fresh growth medium (OD600 = 2), and incubated (50 μl/well) in polystyrene microtiter plates (Greiner) at 30°C. Adhesion was allowed to take place for 1 day. Nonadherent cells were removed by washing with water, and adherent cells were stained with crystal violet. Quantification of adherent cells was achieved by measuring the OD595 after solubilizing cell-bound crystal violet using a solution of 50% ethanol and 1% sodium dodecyl sulfate (SDS). Given values represent the averages of three independent biological replicates, each performed in duplicate.
Cell wall isolation and determination of the protein and carbohydrate content.
The procedure for cell wall isolation has been described in detail by de Groot et al. (13). Briefly, cells were fully disintegrated with 0.25- to 0.50-mm glass beads (Emergo BV) in the presence of a protease inhibitor cocktail (Sigma) using a Bio-Savant Fast Prep 120 machine (Qbiogene). To remove noncovalently linked proteins and intracellular contaminants, isolated cell walls were washed extensively with 1 M NaCl and twice extracted with 2% SDS, 150 mM NaCl, 100 mM Na-EDTA, 100 mM β-mercaptoethanol, and 50 mM Tris-HCl, pH 7.8, for 5 min at 100°C. SDS-treated walls were washed with water, freeze-dried, and stored at −20°C until use.
Protein and chitin contents in the cell wall were determined following the protocols described by Kapteyn et al. (28) using bovine serum albumin and glucosamine as standards, respectively. For determination of the total glucan and mannan content, the polysaccharides in cell wall preparations were hydrolyzed to monosaccharides using sulfuric acid (11) and analyzed by high-performance liquid chromatography analysis as described previously (48). For determination of alkali-resistant 1,6-β- and 1,3-β-glucans, cell walls (about 4 mg [dry weight]) were hydrolyzed by incubating them three times in succession in 1 ml 3% (wt/vol) NaOH at 75°C for 1 h (36). Alkali-resistant glucans were solubilized by subsequent incubations with recombinant endo-1,6-β-glucanases (Prozyme) and endo-1,3-glucanases (Quantazyme, Quantum Biotechnology) as described previously (48). Supernatants containing either 1,6-β-glucan or 1,3-β-glucan were analyzed with the phenol-sulfuric acid assay using glucose as a reference (18).
CWP extraction and fractionation.
Cell wall protein fractions were obtained with various chemical and enzymatic treatments, as detailed elsewhere (13, 32). To solubilize all covalently bound cell wall proteins, SDS-treated walls were digested with Quantazyme, which hydrolyzes the glucan backbone of the cell wall. Specific solubilization of GPI proteins was achieved in two ways. (i) Freeze-dried walls were extracted for 3 h at 0°C with undiluted hydrogen fluoride (HF)-pyridine (Sigma-Aldrich), which under these conditions preferentially cleaves phosphodiester bonds. (ii) Cell walls were incubated with endo-1,6-β-glucanase to hydrolyze the 1,6-β-glucan chains. Pir proteins and other mild-alkali-sensitive linkage CWPs (ASL-CWPs) were released by incubating cell walls with 30 mM NaOH at 4°C for 17 h. In those cases where we performed successive extractions on cell wall material, dialysis against H2O was carried out between the extraction steps.
Solubilized CWP fractions were separated by SDS-PAGE using a linear gradient of 2.2% to 20%. Proteins were stained with silver following the manufacturer's protocol (Bio-Rad) except that an oxidation with 50 mM periodic acid and 100 mM sodium acetate (pH 4.5) for 30 min was included after the first fixing step to enhance reactivity of the silver reagent toward the glycan parts of the mannoproteins. For Western blot analysis, the fractionated proteins were transferred to polyvinylidene difluoride membranes (Millipore). Membranes were probed with peroxidase-labeled concanavalin A (ConA) to visualize mannosylated proteins, as described elsewhere (32). 1,6-β-Glucan moieties attached to CWPs were visualized using polyclonal antiserum against 1,6-β-glucan (39). We also used cross-reacting polyclonal antisera raised against S. cerevisiae CWPs Pir2 (54), Gas1 (46), and Cwp1 (57). The absence of cytosolic proteins in our SDS-treated wall samples was verified using a cross-reacting antiserum against the abundant enzyme glyceraldehyde-3-phosphate dehydrogenase of S. cerevisiae (results not shown) (16). Secondary antibodies were goat-anti-rabbit immunoglobulin G-peroxidase conjugates (Pierce). Blots were developed using ECL reagents (Amersham). The detailed procedure for immunoblot analysis can be found in reference 29.
Mass spectrometric analysis of CWPs.
A detailed protocol for reduction, S alkylation, and subsequent proteolytic digestion of isolated cell walls with sequencing-grade trypsin (Roche) or with endoprotease Glu-C (Sigma) is described by Yin et al. (72). Released CWPs in the mild-alkali fraction were prepared for liquid chromatography-tandem mass spectrometry (LC/MS/MS) analysis as described previously (13). After digestion, peptides were desalted and collected on Omix C18 pipette tips (Varian), washed with 0.1% formic acid, and eluted with 60% acetonitrile-0.1% formic acid. Proteolytic digests derived from 30 μg of freeze-dried cell walls were fractionated on a 150-mm by 75-μm (length × inner diameter) reversed-phase capillary column (PepMap C18; Dionex) using an Ultimate nano-LC system (Dionex). The peptides were separated over a period of 30 min with a two-step linear gradient of 0 to 50% acetonitrile plus 0.1% formic acid, and the outflow at 300 nl/min was directly electrosprayed into the Q-Tof1 (Waters) operating in data-dependent MS and MS/MS mode. The Masslynx software (Waters) was instructed to select ions ranging from m/z 350 to 1,500 with a charge state of at least 2+ above a base peak intensity (BPI) ion count threshold of 40 for collision-induced fragmentation using argon as the collision gas. The resulting MS/MS spectra were processed with the Biolynx tool of the Masslynx software program, which generated a peak list (.pkl file) with all precursor and product ions calculated to the corresponding MH+ charge state. Each LC/MS/MS run was repeated at least twice, thereby excluding abundant ions from previous runs.
Database searching and protein identification.
Different versions of release 2 of the C. glabrata ATCC 2001 genome sequence have been released into the public domain by the Génolevures consortium (http://cbi.labri.fr/Genolevures/) (56) in 2004 and 2006. In the 2006 file, many incomplete (“pseudogenes”) open reading frames (ORFs) from the 2004 version, in particular those lacking a start codon, have been removed, and consequently these ORFs also have not been entered into the NCBI protein database. In silico analysis of adhesin-like GPI proteins (see below) and preliminary analysis of our MS/MS data indicated that this has led to erroneous omission of expressed C. glabrata genes. Therefore, for optimal analysis of our MS/MS data, we created an in-house C. glabrata proteome database by combining the two genome files. Also included were N-terminally truncated (mature) versions of putative cell wall proteins, the used endoproteases trypsin and Glu-C, and keratin, a common contaminant in protein samples. Altogether, this in-house database contains 10,642 protein sequences.
The peak lists (.pkl file) generated with the Masslynx software were submitted to MASCOT (version 2.05, Matrix Science) using the in-house database. The search parameters were as follows: a fixed modification of carbamidomethyl for cysteine, variable modifications of oxidized methionine and N/Q deamidation, semiTrypsin with the allowance of one missed cleavage, peptide and MS/MS tolerance of ±0.2 Da, protein mass unrestricted, and peptide charge state of +1. Probability-based Mascot scores were used to evaluate protein identifications (http://www.matrixscience.com/), and for these settings the threshold confidence score for P values of <0.05 was >42. All cell wall protein identifications are based on multiple peptide matches, of which at least one peptide fulfills the criteria described above. Additional peptide matches with lower scores were added to the list of identified peptides only after manual verification of MS/MS spectra in the raw data using the Masslynx software tools. Inclusion of “semitryptic” peptides (P < 0.05) increased the coverage of identified proteins. Mascot searching with the total NCBI protein database (MSDB 20060831; 3,239,079 sequences), intact or scrambled (decoy), gave a false-positive rate of 0%.
In silico analysis of adhesin-like proteins.
In silico identification of adhesin-like proteins and classification into subgroups were performed using the following approach. Most adhesin-like GPI proteins, defined as large S/T-rich proteins with internal tandem repeats, were identified in a genomic screen for GPI proteins by Weig et al. (67). Additional (fragments of) adhesin-like proteins were identified by the following techniques: (i) searching the C. glabrata 2004 proteome file for ORFs containing the VSHITT motif, which is adhesin specific and conserved in Awp2 and Awp4, using an in-house Perl script, (ii) BLAST analysis using identified adhesin-like proteins as a query, and (iii) analysis of ORFs in telomeric regions for adhesin-like properties. BLAST analysis against the C. glabrata genome was performed using a local BLAST tool, obtained from EMBOSS (http://emboss.sourceforge.net/). Closely linked consecutive ORF fragments on the same DNA strand, as indicated by the NCBI genome browser tool, and with the same BLAST result are considered to be parts of the same protein, taking into account that signal peptides for secretion and for GPI anchoring occur only on N and C termini, respectively. DNA sequences adjacent to (assembled) ORFs that lack N- or C-terminal signal peptides were analyzed for the presence of such features using the genome sequence at NCBI. For prediction of N-terminal signal peptides, we used SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/); C-terminal GPI anchor peptides were identified using big-PI (http://mendel.imp.ac.at/gpi/fungi_server.html) or a complementary algorithm as described previously (67). The phylogenetic tree of adhesin-like proteins was calculated using the ClustalX software program, and the resulting tree was plotted using the NJplot software program. The tree is based on the N-terminal 300-amino-acid high-complexity regions of the proteins, which are likely to comprise the functional domains as described for the Epa family (74). Low-complexity and repeat regions were excluded from this analysis.
RESULTS
Ultrastructure and composition of the cell wall in C. glabrata.
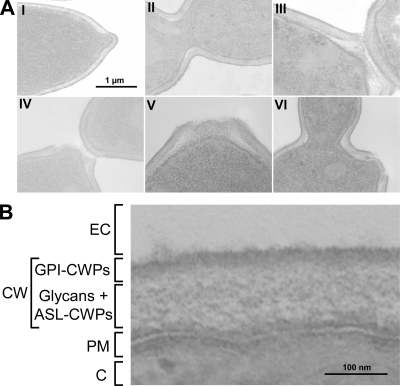
Electron microscopy of C. glabrata cells showed that they are surrounded by a 100- to 200-nm-thick wall (Fig. 1). The inner part of the wall is semitransparent and is surrounded by a more electron-dense layer. The structure of the cell wall in the neck region is modified during the cell cycle, in particular during cytokinesis (Fig. 1A). Fluorescence microscopy with CFW showed intense staining of septa, bud necks, and bud scars, indicating abundant chitin at these loci (data not shown). A similar organization has been described for several other fungi, including the closely related baker's yeast S. cerevisiae (33).
FIG. 1.
The cell wall of C. glabrata has a bilayered structure. (A) The budding process and cytokinesis in C. glabrata visualized by transmission electron microscopy. I, formation of a new bud; II, isotropic bud growth and chitin deposition at the inner part of the wall in the mother/bud neck; III and IV, septum formation and cell separation; V, reinforcement of the bud scar; VI, formation of a new bud proximal to a previous budding event. (B) Close-up of a lateral wall fragment. Based on extrapolation using data from the well-studied yeast S. cerevisiae, the electron-dense outer layer of the cell wall is expected to predominantly consist of GPI-modified proteins (GPI-CWPs), which are covalently bound to 1,6-β-glucan, whereas the more transparent inner layer consists of a glycan network interspersed with ASL-CWPs or at least Pir proteins. This view is fully consistent with the data presented in this paper. CW, cell wall; EC, extracellular environment; PM, plasma membrane; C, cytosol.
Cell walls in S. cerevisiae and C. albicans are composed of 1,3-β-glucan, 1,6-β-glucan, chitin, and mannoproteins, all covalently attached to each other (12, 31). The abundance and ratios of the above-mentioned molecules in cell walls of exponentially growing cells of C. glabrata were investigated and compared to those of S. cerevisiae and C. albicans (Table 1). Dried walls of C. glabrata ATCC 90876 accounted for 19% of the dried total cell mass, implying that cells invest a substantial amount of metabolic energy in wall biosynthesis during growth. Notably, C. glabrata walls contain 6% protein, which is about 50% more than S. cerevisiae and C. albicans (31, 48). Consistent with this, the amount of mannan, presumably present as O and N glycosylation on CWPs (31), was also significantly larger in C. glabrata walls. Consequently, the relative level of total glucan is lower and the mannose/glucose ratio is higher in C. glabrata. The amount of alkali-insoluble glucans, relative to both total wall mass and total glucan, is lower than those in C. albicans and S. cerevisiae, suggesting that there are fewer cross-links between glucan and chitin in the cell wall of C. glabrata. Chitin is a minor wall component in all three species, accounting for 1.1 to 1.3% of the wall mass in log-phase cells of C. glabrata (Table 1). Since chitin is mostly concentrated in the septum, bud neck, and bud scar areas, little chitin will be incorporated in lateral walls. We conclude that the cell wall of C. glabrata contains the same basic components as those in S. cerevisiae and C. albicans; however, the contribution of individual components, with an increased level of mannoproteins, and the extent of cross-links between the components are different.
TABLE 1.
Cell wall composition of C. glabrata in comparison to those of S. cerevisiae and C. albicans
| Species | Cell wall contenta
|
||||||
|---|---|---|---|---|---|---|---|
| Protein (%) | Chitin (%) | Man (%) | Glu (%) | M/G | Alkali-insoluble glucan (%)b
|
||
| 1,6-β | 1,3-β | ||||||
| C. glabrata | 6.4 ± 0.1 | 1.2 ± 0.1 | 43.8 ± 0.5 | 54.0 ± 0.2 | 0.81 | 4.2 ± 0.1 | 16.7 ± 1.7 |
| S. cerevisiae | 4.0 ± 0.1 | 1.4 ± 0.2 | 34.2 ± 1.6 | 60.3 ± 2.5 | 0.57 | 7.1 ± 0.2 | 26.8 ± 0.9 |
| C. albicans | 3.5 ± 0.2 | 4.2 ± 0.1 | 26.6 ± 2.3 | 64.0 ± 4.9 | 0.42 | 10.6 ± 0.6 | 26.2 ± 1.1 |
Values are from exponentially growing cells (in YEPD), are the means and standard deviations for two independent samples measured in duplicate, and are expressed as percentages of freeze-dried cell walls. Man, mannose; Glu, glucose; M/G, ratio of mannose to glucose.
Enzymatically released with 1,6-β-glucanase or 1,3-β-glucanase.
C. glabrata cell wall protein incorporation.
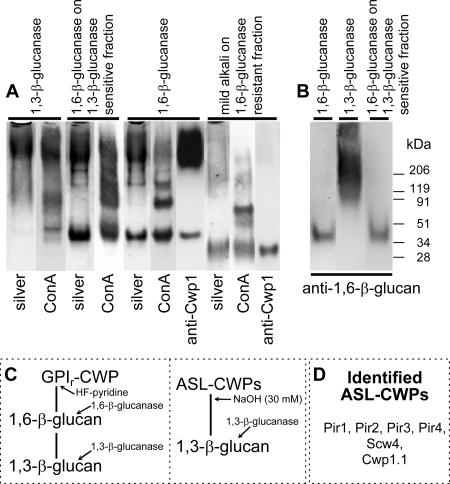
Transmission electron microscopy in combination with cell wall composition analysis indicated that by analogy to S. cerevisiae, the outer layer of the wall of C. glabrata is mainly comprised of mannoproteins. These proteins are expected to play key roles in adhesion and biofilm formation and other host-pathogen interactions that mediate fungal virulence. We therefore proceeded to examine the cell wall architecture of C. glabrata with a focus on identifying covalently bound mannoproteins and resolving the bonds to the carbohydrate network of the wall. First, SDS-treated walls were digested with recombinant endo-1,3-β-glucanase, and the released fraction was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Silver staining and probing of blots with the lectin ConA both revealed high-molecular-mass smears (Fig. 2A), indicating that the glucanase had released heavily glycosylated wall mannoproteins. In C. albicans and S. cerevisiae, the majority of the CWPs are attached to the wall carbohydrate network through a covalent bond between a sugar remnant of their GPI anchor and 1,6-β-glucan. Digestion of cell walls and 1,3-β-glucanase-released wall material with recombinant 1,6-β-glucanase indicated that this is also the case in C. glabrata (Fig. 2A). Silver staining showed an abundant protein band of approximately 45 kDa, as well as additional bands and a smear of high molecular mass. ConA revealed four major protein bands, of 45 kDa, 85 kDa, 150 kDa, and more than 200 kDa, well distinguishable in the 1,6-β-glucanase digest. Immunoblot analysis using anti-1,6-β-glucan antibodies further showed that the 1,3-β-glucanase- and the 1,6-β-glucanase-extracted protein(s) contains 1,6-β-glucan moieties (Fig. 2B). We conclude that the cell wall of C. glabrata contains mannoproteins that are linked to 1,3-β-glucan via 1,6-β-glucan, as would be expected for GPI-modified CWPs (Fig. 2C). Consistent with these findings, HF-pyridine extraction released multiple mannoproteins from C. glabrata (67), similar to results with the 1,6-β-glucanase treatment. Thus, the visualized proteins released by both methods most likely are GPI-modified mannoproteins, linked through a phosphodiester bridge to 1,6-β-glucan (Fig. 2C). The most abundant protein of 45 kDa in mass in the 1,6-β-glucanase extract (Fig. 2A) and the 37-kDa band in the HF-pyridine extract (67) both react with antibodies raised against the abundant GPI-modified CWP Cwp1 in S. cerevisiae (ScCwp1). The difference in masses of the ScCwp1 homologs in the two fractions can be explained by 1,6-β-glucan remnants and/or phosphomannan groups which were removed from the protein by the HF-pyridine treatment.
FIG. 2.
CWP-polysaccharide complexes in the cell wall of C. glabrata. (A and B) SDS-PAGE analysis of cell wall material extracted as indicated from isolated walls of C. glabrata grown in SC medium. Gels were analyzed by silver staining or by blotting and probing with the lectin ConA or anti-ScCwp1 antiserum (A) or with anti-1,6-β-glucan antiserum (B). (C) Identified cross-links between proteins and β-glucans in the cell wall of C. glabrata. The extraction methods used to solubilize CWPs are indicated. (D) Cell wall proteins identified in mild-alkali extracts by LC/MS/MS.
A minor group of fungal CWPs is covalently connected to 1,3-β-glucan via a mild-alkali-sensitive linkage(s) (Fig. 2C, ASL-CWPs); this includes Pir (Proteins with internal repeats) proteins (15). Multiple ASL-CWPs are also present in C. glabrata, and among them is at least one Pir protein of 75 kDa in mass (67). Incorporation of Pir proteins is governed by the formation of an ester linkage between a glutamine residue in the repeat sequence and cell wall 1,3-β-glucan. Thus, Pir proteins with multiple repeats may cross-link different 1,3-β-glucan chains, thereby strengthening the cell wall. Pattern searching with the consensus Pir repeat sequence, Q-[IV]-X-D-G-Q-[IVP]-Q (Prosite format) showed that besides the four Pir proteins in C. glabrata, six predicted GPI proteins (the three Cwp1 homologs and three Srp1/Tip1 family members) contain (a single copy of) the Pir repeat. They may therefore be bound, and thus cross-link, 1,3-β-glucan and 1,6-β-glucan chains, as has been shown previously for Cwp1 in S. cerevisiae (28). Mild-alkali treatment of the 1,6-β-glucanase-resistant insoluble wall fraction released a high-molecular-mass smear and a major band of approximately 30 kDa (Fig. 2A), which is also visualized by probing with ConA and antibodies against ScCwp1. These results indicate that at least an abundant homolog of ScCwp1 in C. glabrata has the potential to cross-link 1,3-β-glucan and 1,6-β-glucan in the cell wall.
Identification of covalently bound CWPs in C. glabrata.
In previous studies, we have identified covalently linked CWPs in C. albicans and S. cerevisiae. A comparative analysis with C. glabrata therefore might reveal the following: (i) common proteins needed for fungal cell wall biosynthesis and (ii) pathogen-specific proteins that may be important virulence factors. SDS-treated walls from C. glabrata ATCC 90876 cells, grown in YEPD to mid-log phase (similar conditions to those used for C. albicans and S. cerevisiae), were directly incubated with the endoprotease trypsin (which cleaves after K or R, except when followed by P) to obtain peptide fragments. LC/MS/MS analysis resulted in the unambiguous identification of 18 CWPs (Table 2) (see mass spectrometric details in Table S1 in the supplemental material). Repeating the same analysis with the protease Glu-C (which cleaves after D or E) did not identify extra proteins, since only 11 peptides were obtained for proteins already identified with trypsin. All 18 identified C. glabrata proteins possess predicted signal peptides for secretion (analyzed using the SignalP 3.0 server) and are orthologs of identified CWPs in S. cerevisiae (Table 2) (72). This suggests that the proteins we have identified under these conditions have general roles in cell wall biosynthesis rather than pathogenesis-related functions.
TABLE 2.
Covalently bound C. glabrata CWPs identified by LC/MS/MSa
| Category and protein name | ORF no. | Properties and proposed functionb | MS/MS result
|
Conserved functional domain(s)d | Closest S. cerevisiae homolog (SGD namee) | Closest C. albicans homolog (CGD namee) | Reference for C. glabrata protein name | |
|---|---|---|---|---|---|---|---|---|
| Sequence coverage (%) | No. of peptides identifiedc | |||||||
| Carbohydrate-active enzymes | ||||||||
| Crh1 | CAGL0G09449g | SP, GPI, 452 aa. GH16 transglycosidase, involved in chitin incorporation | 22 | 11 | GH16: 34-240 | Crh1f | Crh11f | 67 |
| Utr2 | CAGL0C02211g | SP, GPI, 481 aa. GH16 transglycosidase, involved in chitin incorporation | 6 | 2 | CBM 18: 23-64; GH16: 93-303 | Utr2f | Utr2f | 67 |
| Gas1 | CAGL0G00286g | SP, GPI, 559 aa. GH72 transglycosidase, elongation of 1,3-β-glucan | 8 | 1 (+ 2) | GH72: 25-329; X8: 377-459 | Gas1f | Phr2,f Phr1f | 66 |
| Gas2 | CAGL0M13849g | SP, GPI, 565 aa. GH72 transglycosidase, elongation of 1,3-β-glucan | 10 | 2 (+ 2) | GH72: 26-330; X8: 378-460 | Gas1f | Phr2,f Phr1f | 66 |
| Gas4 | CAGL0F03883g | SP, GPI, 480 aa. GH72 transglycosidase, elongation of 1,3-β-glucan | 12 | 3 | GH 2: 20-350 | Gas3f | Phr3, Pga4f | |
| Gas5 | CAGL0F01287g | SP, GPI, 523 aa. GH72 transglycosidase, elongation of 1,3-β-glucan | 24 | 10 | GH72: 27-331 | Gas5f | Pga4f | |
| Scw4 | CAGL0G00308g | SP, no GPI, 374 aa. GH17 transglycosidase, modification of 1,3-β-glucan | 28 | 8 | GH17: 118-372 | Scw4f | MP65/Scw1f | |
| Other enzymatic activity | ||||||||
| Plb2 | CAGL0J11748g | SP, GPI, 695 aa. phospholipase | 10 | 6 | PLAc: 33-550 | Plb2f | Plb3 | |
| Nonenzymatic CWPs | ||||||||
| Cwp1.1 | CAGL0F07601g | SP, GPI, 218 aa. structural mannoprotein | 53 | 13 (+ 26) | None | Cwp1f | None | 67 |
| Cwp1.2 | CAGL0F07579g | SP, GPI, 212 aa. structural mannoprotein | 55 | 4 (+ 26) | None | Cwp1f | none | 67 |
| Ssr1 | CAGL0H06413g | SP, GPI, 212 aa. contains CFEM domain | 27 | 6 | CFEM: 22-81 | Ccw14f | Ssr1f | |
| Tir1 | CAGL0F01463g | SP, GPI, 221 aa. mannoprotein of the Srp1p/Tip1p family | 12 | 2 | None | Tir1f | None | |
| Pir1 | CAGL0I06204g | SP, no GPI, 349 aa. conserved 4-cysteine domain | 21 | 1 (+ 9) | 4× Cys: 252-349 | Pir1-4f | Pir1f | 67 |
| Pir2 | CAGL0I06182g | SP, no GPI, 340 aa. conserved 4-cysteine domain | 26 | 2 (+ 10) | 4× Cys: 243-340 | Pir1-4f | Pir1f | 67 |
| Pir3 | CAGL0M08492g | SP, no GPI, 335 aa. conserved 4-cysteine domain | 20 | 11 (+ 2) | 4× Cys: 238-335 | Pir1-4f | Pir1f | 67 |
| Pir4 | CAGL0I06160g | SP, no GPI, 233 aa. conserved 4-cysteine domain | 43 | 11 | 4× Cys: 136-233 | Pir1-4f | Pir1f | 67 |
| Unknown proteins | ||||||||
| Ecm33 | CAGL0M01826g | SP, GPI, 421 aa. unknown role in cell wall biosynthesis | 18 | 7 | Unknown | Ecm33f, Pst1f | Ecm33f, Ecm331 | |
| Pst1 | CAGL0E04620g | SP, GPI, 429 aa. unknown role in cell wall biosynthesis | 6 | 3 | Unknown | Ecm33f, Pst1f | Ecm33f, Ecm331 | |
| Adhesin-like wall proteinsh | ||||||||
| Awp1 | CAGL0J02508g | SP, GPI, 870 aa. putative adhesin | 6 | 4 | Unknown | Awa1, Hpf1, Hpf1′ | None | |
| Awp2 | CAGL0K00110g | SP, GPI, 832 aa. putative adhesin | 8 | 5 | Unknown | Awa1, Hpf1, Hpf1′ | Iff family, Hyr1 | |
| Awp3 | CAGL0J11902g- CAGL0J11924g | SP, GPI, unknown size putative adhesin | 6g | 2 | Unknown | None | None | |
| Awp4 | CAGL0J11990-CAGL0J12056g | SP, GPI, unknown size putative adhesin | 11 + 13g | 2 (+ 3) | Unknown | None | Iff family, Hyr1 | |
| Epa6 | CAGL0C00110g | SP, GPI, 715 aa. adhesin | 15 | 5 (+ 1) | PA14: 139-247 | Flocculins | None | 4 |
For mass spectrometric details, see Table S1 in the supplemental material.
Predicted signal peptides for secretion (SP) (http://www.cbs.dtu.dk/services/SignalP/) and C-terminal signatures for GPI anchoring (http://mendel.imp.ac.at/gpi/fungi_server.html) are indicated; see also reference 67. Conserved functional domains were identified using CDD v2.03 (http://www.ncbi.nlm.nih.gov/structure/cdd/cdd.shtml), CAZy (http://www.cazy.org/), and reference 15. GH, glycoside hydrolase; aa, amino acids.
Numbers in parentheses indicate nonunique peptides.
Identified as outlined in footnote b.
SGD, Saccharomyces Genome Database (http://www.yeastgenome.org/); CGD, Candida Genome Database (http://www.candidagenome.org/).
Sequence coverage of identified ORF fragments.
Not identified in ATCC 90876 cells that were grown in YEPD to mid-log phase.
Gas1, Gas2, Gas4, Gas5, Crh1, Utr2, and Scw4 specify putative carbohydrate-active enzymes (9) that may modify cell wall polysaccharides and thus are implicated in building and remodeling of the cell wall glycan network during growth. Ecm33 and Pst1 are homologs of a family of four GPI proteins (the Sps2 family) in S. cerevisiae. This family, judged from studies of the ascomycetes S. cerevisiae, C. albicans, and Aspergillus fumigatus, has a crucial rule in establishing and maintaining proper cell wall integrity (6, 38, 49). Identification of Plb2 is consistent with the presence of its ortholog in cell walls of S. cerevisiae (72). Cwp1.1, Cwp1.2, Ssr1, and Tir1 are relatively small CWPs that are unlikely to have enzymatic functions and may serve as cross-linking or coat-forming wall proteins. Finally, we identified four Pir proteins (Pir1 to -4) in log-phase ATCC 90876 cells. The fifth Pir protein homolog that lacks Pir repeat sequences (67) was not detected, consistent with the observation that the repeats are responsible for the formation of covalent links between Pir proteins and 1,3-β-glucan (5, 19).
Thirteen of the eighteen identified CWPs are predicted GPI proteins (67). The five non-GPI proteins are the four Pir proteins and Scw4, which lacks obvious Pir repeat sequences. Crucial steps in our cell wall isolation procedure are extractions with hot solutions containing SDS and reducing agents. We therefore are confident that the identified Scw4 molecules are covalently bound to the cell wall, similar to the case with Scw4 and Scw10 in S. cerevisiae and MP65/Scw1 in C. albicans (13, 72). LC/MS/MS analysis of protein pools extracted with NaOH (Fig. 2D) (see Table S1 in the supplemental material) confirmed that Scw4 can be extracted from cell walls with mild alkali. In the NaOH extract, we also identified Pir proteins and Cwp1.1. The latter confirms our in silico and immunoblot analyses, indicating that it can be linked through an alkali-sensitive bond to 1,3-β-glucan in addition to being coupled to 1,6-β-glucan in a GPI-dependent manner.
The genome of C. glabrata harbors a large diversity of adhesin-like proteins.
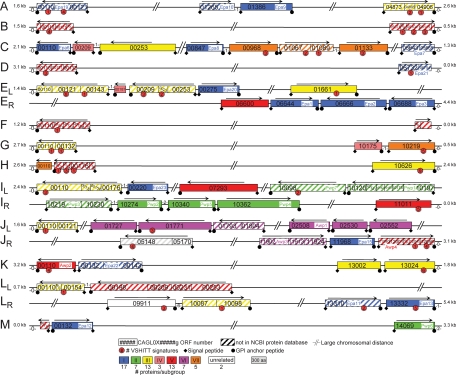
Genome-wide predictions of GPI proteins using the second assembly of the C. glabrata genome, released in 2004, revealed an impressive number of large modular proteins with a putative effector domain in the N-terminal part followed by a low-complexity region with a high S/T content and internal tandem repeats, which is typical of cell wall proteins with adhesive properties (67). Careful reexamination of putative adhesins in the C. glabrata proteome files, detailed in Materials and Methods, revealed a total of 67 adhesin-like proteins (Table 3). Importantly, 44 of these are located in subtelomeric regions, and at both ends of all chromosomes, adhesin-like sequences are present (Fig. 3). Comparison of the N-terminal domains specifying the putative ligand-binding parts showed that these proteins can be divided into multiple subgroups (Fig. 4; Table 3). The largest subgroup is the Epa family of lectin-like adhesins, described by the Cormack laboratory (4), of which we found 17 proteins in strain ATCC 2001. The N-terminal parts of the putative adhesins in cluster II share with Epa proteins and with flocculins in S. cerevisiae the presence of a conserved PA14 domain, named after an anthrax protective antigen, suggesting a functional relationship with respect to ligand binding (14, 74). The proteins in cluster II were therefore tentatively designated Pwp1 to Pwp7 (PA14-containing wall protein). The interrelationship with and between other clusters is poor in the N-terminal domains. The N-terminal parts of the clustered CAGL0C00209g and CAGL0G10175g show weak similarity with Dan1/2/3/4 (delayed anaerobic) and the seripauperin (Pau) multiprotein family in S. cerevisiae. Like many of the adhesin-like proteins in C. glabrata, PAU and DAN genes are mostly located in subtelomeric regions. Interestingly, PAU and DAN genes are repressed under aerobic conditions, and this silencing is alleviated when the oxygen availability is low (53). Inspection of the low-complexity regions in the C-terminal half of adhesin-like proteins in different subgroups revealed 46-amino-acid repeats (named Awp2 repeats in Table 3), including conserved TTVVT and VSHITT sequences. Pattern searching with the VSHITT sequence revealed that it seems specific for and is present in the C-terminal half of at least 31 adhesin-like proteins, across most of the subgroups, including the Epa family (Table 3; Fig. 3).
TABLE 3.
The genome of C. glabrata strain ATCC 2001 harbors 67 genes putatively encoding adhesin-like proteins
| Subgroupa | No. of proteins | No. of proteins with subtelomeric localization | Presence of PA14 domain | No. of proteins with Awp2 repeats | No. of proteins present in NCBI protein databaseb |
|---|---|---|---|---|---|
| I, Epa family (blue) | 17 | 14 | Yes | 4 | 11 |
| II, Pwp family (green) | 7 | 1 | Yes | 1 | 4 |
| III (yellow) | 13 | 11 | No | 11 | 5 |
| IV (pink) | 3 | 3 | No | 0 | 3 |
| V, includes Awp2 and -4 (red) | 13 | 12 | No | 8 | 4 |
| VI, includes Awp1 and -3 (purple) | 7 | 1 | No | 1 | 5 |
| VII (orange) | 5 | 2 | No | 4 | 4 |
| Others (white) | 2 | 0 | No | 2 | 1 |
| Total | 67 | 44 | 31 | 37 |
Colors corresponding to Fig. 3 are given in parentheses.
As of 21 August 2008.
FIG. 3.
Genomic organization for adhesin-like proteins encoded in the genome of C. glabrata ATCC 2001. GPI-modified adhesin-like proteins were primarily identified by a genome-wide in silico analysis, as described previously (29). Additional adhesin-like proteins were found by pattern searching using the conserved VSHITT motif, by BLAST analysis, and by analysis of telomeric regions, where most of the adhesin-like proteins are located. Chromosomes and ORFs are numbered following Génolevures' systematic ORF numbering. Adjacent ORF fragments belonging to a single gene, as also indicated by the NCBI genome browser, are connected. Unannotated ORF fragments identified by BLASTX and containing N- or C-terminal signal peptides were connected to CAGL0B00110g, CAGL0B05115g, and Epa11. ORF sizes are to scale, but distances between ORFs are not. Colors indicate subfamilies I to VII, sharing homology in the N-terminal putative ligand-binding parts, as presented in Fig. 4. CAGL0L09911g and CAGL0J05170g (white) are unrelated outgroups in Fig. 4. Numbers of proteins in each subgroup are indicated. For CAGL0H00110g (group VII, orange) and CAGL0E00187g (group IV, pink), only C-terminal parts of the proteins were identified; their classification is therefore based on BLASTP analysis of these regions. Numbers of nonadhesive ORFs separating adhesive-like proteins and telomeres and distances of terminal adhesive-like proteins to telomeres are indicated. Arrows indicate directions of transcription.
FIG. 4.
Multiple subfamilies of adhesin-like proteins exist in C. glabrata. A neighbor-joining phylogenetic tree, with bootstrap values added (1,000 bootstraps performed), of adhesin-like wall proteins based on the putative functional domains (the 300 N-terminal amino acids or fewer in cases where the N-terminal ORF fragment is shorter) of the ORFs is shown. S/T-rich low-complexity regions within the first 300 amino acids of CAGL0C00209g and CAGL0G10175 were excluded from this analysis. CAGL0L09911g, which together with CAGL0J05170g is least related to other adhesin-like proteins, is plotted as an outgroup. The distantly related S. cerevisiae Flo1 and Dan1 and C. albicans Hyr1 are included for comparison. CAGL0H00110g (group VII) and CAGL0E00187g (group IV) were excluded from this analysis since their N-terminal parts are unidentified; see Fig. 3. The scale bar indicates phylogenetic distances, in number of amino acid substitutions per position.
Due to general difficulties in sequencing and the assembly of telomere regions, many of the adhesin-like proteins appear to be artificially broken into multiple adjacent ORF fragments and have been annotated as pseudogenes. Consequently, in the later (2006) version of the C. glabrata proteome file, these fragments have been deleted, causing many (putative) adhesins, including six Epa proteins, to be overlooked in the NCBI protein database (Table 3; Fig. 3). Consistent with their proposed role as adhesins, the absence of consecutive basic amino acid residues in the sequences immediately preceding the GPI attachment sites (67) suggests that these proteins will reside in the cell wall. However, the proteomic analysis on ATCC 90876 log-phase cells did not result in identification of any adhesin-like proteins. This raised the question of whether this was due to allelic variations or differential regulation of protein expression or whether we failed to identify adhesins due to technical limitations of our methodology.
Strain- and growth phase-dependent in vitro adhesion capacity.
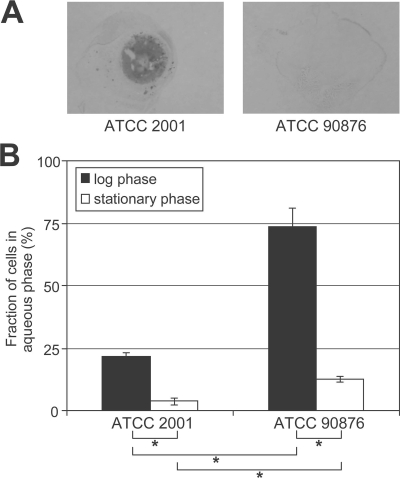
Frequently, large allelic variations occur in the structure and expression of fungal cell wall adhesin genes (43, 63, 64). Independent isolates may express and incorporate different proteins, with direct consequences for cell surface properties, such as the adhesion capacity. To determine whether the adhesion capacity of C. glabrata depends on the genetic strain background, strain ATCC 90876 was compared with the sequenced strain ATCC 2001. Adhesion to plastic in a liquid environment in vitro was tested by growing both strains as droplets on polystyrene plates. After removal of nonadherent cells by gentle washing, adherent cells were stained with crystal violet. As shown in Fig. 5A, ATCC 2001 adhered strongly to the plastic surface, whereas ATCC 90876 cells were entirely washed away. Quantification of cell-bound crystal violet using a similar assay with polystyrene microtiter plates confirmed the superior adhesion capacity of ATCC 2001 compared to that of ATCC 90876 (OD595 = 0.32 ± 0.02 versus 0.10 ± 0.01 [Student's t test, P < 0.05]).
FIG. 5.
Strain- and growth phase-dependent cell surface hydrophobicity and in vitro adhesion. (A) In vitro adhesion of ATCC 2001 and ATCC 90876 was analyzed by growing cells in 25-μl spots on polystyrene plates for 3 days at 30°C. Nonadherent cells were washed away with water, and remaining adherent cells were stained with crystal violet. (B) Surface hydrophobicity of exponentially growing or stationary-phase cells of C. glabrata strains ATCC 2001 and ATCC 90876 was measured using a two-phase assay. Surface hydrophobicity can be inferred by subtraction of the fraction of cells measured in the aqueous phase. Significant differences (P values of <0.05, Student's t test) are indicated with asterisks.
The structure of the cell wall is highly dynamic (Fig. 1), and the incorporation of fungal CWPs is tightly controlled during the cell cycle (31) but also is dictated by the environmental conditions, the growth phase, and the morphology of the cells. For instance, induction of filamentation in pleomorphic C. albicans is accompanied by alterations in the cell wall composition and the expression of certain cell wall genes, including the hyphal adhesins Als3 and Hwp1 (40, 42). In C. glabrata, the expression of Epa adhesins is tightly regulated (4), and we observed by lectin and immunoblot analysis of different CWP fractions that changes in environmental parameters, e.g., growth temperature and pH (data not shown), lead to alterations in wall protein incorporation. Stress experienced during stationary phase has been shown to increase the expression of several CWPs in S. cerevisiae, including the flocculin Flo5 (20, 31). The incorporation of such flocculins in the cell wall determines the surface hydrophobicity and adhesion properties (23, 27). Development of biofilms on abiotic surfaces (e.g., plastic catheters and dentures) and colonization/infection of human epithelia by Candida are preceded by adhesion, and this whole process takes place over longer periods of time. Stationary cells of C. glabrata appear more adherent to plastic than exponentially growing cells (26). We therefore speculated that prolonged culturing of C. glabrata would affect the incorporation of adhesins into the cell wall, thereby influencing the surface properties. As indicated in Fig. 5B, both ATCC 2001 and ATCC 90876 cells show a significant increase in surface hydrophobicity when they entered the stationary phase. As expected from our adhesion tests, the surface hydrophobicity of strain ATCC 2001 is higher than that of ATCC 90876 both during exponential growth and in stationary phase.
Identification of adhesins in the cell wall of C. glabrata.
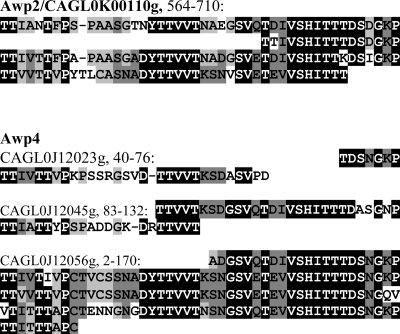
The observed strain- and growth phase-dependent differences in surface hydrophobicity and adhesiveness prompted us to further analyze the differential incorporation of cell wall adhesins in C. glabrata on a proteomic level. First, ATCC 90876 was grown to stationary phase (24 h in YEPD, OD600 of ∼25, glucose depleted) at both 30°C and 37°C to achieve an increase in surface hydrophobicity (see above). For LC/MS/MS identification of the cell wall proteome, we applied the direct “cell wall shaving” method using trypsin as an endoprotease (71). In addition to the proteins found in exponentially growing cells, this analysis resulted at both temperatures in the identification of one GPI protein with adhesin-like features, tentatively named adhesin-like wall protein 1 (Awp1) (Table 2). Second, since we found clear differences in the surface characteristics of ATCC 90876 and ATCC 2001, we compared the cell wall proteomes of these two clinical isolates using the same differential growth conditions. LC/MS/MS analysis of ATCC 2001 cells resulted in the identification of four more adhesin-like proteins. CAGL0C00110g is the closest homolog of Epa6 (sequenced from strain BG2) (4). The other three proteins are uncharacterized and were termed Awp2, Awp3, and Awp4 (Table 2) (see Table S1 in the supplemental material for mass spectrometric details). In the most recent C. glabrata genome assembly, Awp3 appears to be mistakenly dispersed into three overlapping fragments, of which our MS analysis identified the N-terminal fragment. This fragment is not S/T rich, and therefore it will have a relatively low level of glycosylation, as would be expected for a GPI-modified adhesin with an N-terminal ligand-binding domain (7). Similarly, we identified the first two of five fragments of Awp4. Between the three middle fragments, ORF numbers CAGL0J12001g, CAGL0J12023g and CAGL0J12045, unannotated gaps of 234 and 554 bp, respectively, exist in the current genome sequence. BLAST analysis of the DNA sequences in the gaps revealed homology with other putative adhesins, supporting our view that these ORF fragments and gaps are indeed parts of a single gene. Based on homology in N-terminal regions, Awp3 and Awp1 belong to the same subgroup, all of which reside on chromosome J (Fig. 3). Awp2 and Awp4 belong to another subgroup, which is spread over several chromosomes, as is also the case for the Epa family. Both Awp2 and -4 contain multiple Awp2 repeats (Fig. 6).
FIG. 6.
Awp2 and Awp4 have adhesin-specific intragenic tandem repeats. Multiple alignment of tandem repeats present in both Awp2 and Awp4 is shown. Awp2 is encoded by CAGL0K00110g. Awp4 is dispersed into five ORF fragments, CAGL0J11990g, CAGL0J12001g, CAGL0J12023g, CAGL0J12045g, and CAGL0J12056g. The two N-terminal parts (not shown) were identified by LC/MS/MS. The three C-terminal ORF fragments contain (parts of) repeat units. Numbers indicate the aligned regions of the ORFs (fragments).
Awp4 and Epa6 were identified only in stationary-phase cells and Awp3 only in log-phase cells. Awp1 was not identified in strain ATCC 2001. Obvious temperature-dependent changes in incorporation of adhesin-like proteins were not detected. We have to stress that the used methodology provides a qualitative inventory rather than precise quantitative data. Nevertheless, careful examination of raw data from MS survey spectra points to quantitative differences in incorporation of adhesin-like proteins, dependent on strain background and growth phase. This may have direct consequences for surface properties such as adhesion capacity and biofilm formation.
DISCUSSION
Fungal cell walls are essential organelles and are composed of molecules that are largely absent in mammals. They thus are excellent targets for development of antifungal drugs and diagnostic tools. Being the interface between fungal pathogens and the human host, cell wall components such as glucan and N- and O-mannan represent important pathogen-associated molecular patterns recognized by the innate immune system (21, 44, 45). Moreover, the cell wall proteome defines surface properties, such as biofilm formation and adhesiveness to (host) cells and abiotic medical devices. Therefore, the aim of our study was to identify species-specific differences in the glycan and protein composition and in the cross-links between these components. The major constituents of the cell wall of C. glabrata are the polysaccharides 1,3-β-glucan, 1,6-β-glucan, and chitin and mannoproteins. The mannoprotein content appeared to be 50% higher than that of baker's yeast and C. albicans, concomitant with a reduced glucan content (Table 1). The relatively low levels of alkali-insoluble glucans suggests that fewer cross-links may exist between glucan and chitin. The cell walls obtained from C. glabrata (this study) and S. cerevisiae (1) growing exponentially in rich medium both constitute about 20% of the cell's dry weight. This would imply that the glucan network is thinner in C. glabrata and the outer layer of the cell wall is more densely packed with mannoproteins. Thus, β-glucans in the cell wall of C. glabrata may be more effectively masked from host immune recognition by the receptor dectin-1 than is the case for C. albicans (69). Conceivably, the host's innate immune system is well equipped to monitor subtle species-specific differences in fungal pathogen-associated molecular patterns to readily build an appropriate response.
Transmission electron microscopy revealed that cell walls of C. glabrata have a dynamic bilayered structure, consisting of an electron-dense outer layer that surrounds a semitransparent inner layer (Fig. 1). This is typical for ascomycetous yeasts (33). Permanganate staining (47) and protease treatment (73) of S. cerevisiae cells have indicated that the outer layer primarily contains (manno)proteins whereas the inner layer is mainly composed of carbohydrates. Our biochemical experiments with protein incorporation indicated that C. glabrata contains covalently bound CWPs that can be divided into two separate groups. The majority are GPI-modified proteins, whereas the second group is directly bound to 1,3-β-glucan via an ASL. Immunostaining of S. cerevisiae and C. albicans cells with specific antisera against individual CWPs has shown that GPI proteins localize predominantly to the outer layer of the wall. On the other hand, Pir proteins, belonging to the group ASL-CWPs, are found throughout the wall (for a review, see reference 33). In C. glabrata walls, we found four Pir proteins, which explains the semitransparency of the inner layer and is consistent with the hypothesis that they can cross-link different 1,3-β-glucan molecules via their Pir-specific repeats (19). Scw4, another ASL-CWP, lacks such repeats, which suggests that covalent incorporation of ASL proteins may depend on various mechanisms.
Many of the CWPs previously identified in ascomycetous yeasts are ubiquitous and have a role in cell wall construction, maintenance, and remodeling and presumably also in biofilm formation. Others are species specific and may dictate virulence-related properties, such as adhesion (to host tissues or medical devices) or counteractivity toward host defense responses. Cell wall proteomic studies of C. albicans have identified some important proteins that are present only in (some) CTG-clade species and might directly relate to Candida virulence (Table 2). In particular, under nonlimiting growth conditions, C. albicans cell walls contained two adhesins, Als1 and 4 (13). Furthermore, growth in a vagina-simulative medium induced two other adhesins, Als3 and Hwp1, as well as proteins involved in iron acquisition (58). To identify possible pathogenesis-related CWPs in C. glabrata, covalently attached proteins were identified from SDS-treated walls using our “wall shaving” method followed by LC/MS/MS. Large functional similarity was found between the set of CWPs in log-phase C. glabrata cells and those previously identified in C. albicans and S. cerevisiae (Table 2), which is consistent with the idea that many of them have a role in cell wall biosynthesis. For instance, all three organisms contain multiple homologous carbohydrate-active enzymes, which may use cell wall components as their substrates, and small structural or coat-forming proteins in their wall (13, 72). As deduced from SDS-PAGE and mass spectrometric analysis and in agreement with codon adaptation index values (67), the nonenzymatic proteins Cwp1.1 and Cwp1.2 are by far the most abundant CWPs in C. glabrata. In view of the relatively high level of total mannoprotein, this implies an important role for Cwp1.1 and Cwp1.2 in cell wall organization, probably by contributing to such surface characteristics as permeability and (negative) charge. In addition, by being linked to 1,3-β-glucan through alkali-labile ester linkages and to 1,6-β-glucan through GPI modification, Cwp1.1 and Cwp1.2 may cross-link different glucan chains, thereby significantly contributing to cell wall strengthening. Noteworthy in this respect is the anti-Cwp1 reactive material of high molecular mass in the 1,6-β-glucanase extract (Fig. 2A), which also suggests a more complex incorporation of Cwp1 molecules.
Bioinformatic analysis showed that the genome of C. glabrata harbors a large set of putative GPI-modified adhesins. This is supported by the identification of Epa6 and the novel putative adhesins Awp1/2/3/4 by mass spectrometry and by confirmation of the expression of their corresponding genes by reverse transcription-PCR (E. Kraneveld, personal communication). However, none of the putative adhesins was identified in exponentially growing cells of ATCC 90876. Development of candidiasis is often preceded by the formation of a biofilm on mucosal tissues or medical devices. Candida cells in biofilms grow and adhere, and they then age if they are retained within the biofilm matrix. Therefore, in the human body, adherent C. glabrata cells probably survive in a semistationary phase for most of their lifetime. Differential expression of several cell wall genes was observed in S. cerevisiae during the postdiauxic and stationary phases (20). Moreover, in the stationary phase, mutant strains lacking selected covalently bound nonenzymatic CWPs showed significantly decreased viability, leading to up to 25% of dead cells in the culture (60). These results indicate that the functions and/or incorporation of certain CWPs is related to the stationary growth phase. Furthermore, large allelic variations may occur for adhesin genes, as has been observed, for instance, in C. albicans and S. cerevisiae (24, 63). The two C. glabrata strains used in this study show different surface hydrophobicities and in vitro adhesion capacities, with ATCC 2001 being more hydrophobic and adherent to plastic than ATCC 90876. Also, both strains showed a significant increase in surface hydrophobicity when the cells were grown from the logarithmic phase to the stationary phase. Strikingly, the observed differences in hydrophobicity seem to coincide with incorporation of different adhesins, as indicated by mass spectrometric analysis of cell wall proteins under the same conditions. Of the five identified adhesin-like proteins, Awp2, Awp3, Awp4, and Epa6 were found only in ATCC 2001. Awp1, on the other hand, was identified only in ATCC 90876. In addition, identification of Awp1, Awp4, and Epa6 in stationary cells only and Awp3 in log-phase cells only further affirmed that incorporation of certain (putative) adhesins is indeed growth phase dependent. Although it awaits experimental confirmation, it is tempting to speculate that the reduced adhesion capacity of log-phase cells as observed for ATCC 2001 might be related to the dissemination of yeast cells from biofilms.
The well-described Epa1 protein was not identified under the conditions used in this study, which is in agreement with the observation that it is present in cell walls during very early growth stages but is removed later on through proteolytic digestion by aspartic proteases (30). Four of the adhesin-like proteins identified in our study are localized close to telomeres (Fig. 3). Subtelomeric localization has already been shown to cause EPA genes to be normally repressed under nonlimiting growth conditions (4). EPA6 and also EPA1 and EPA7 become transcriptionally activated as a result of nicotinic acid limitation and may therefore be relevant for establishment of infections under conditions that occur in the urinary tract (17). Epa6 has also been shown to play an important role during biofilm formation (26). Transcriptional regulation of a large repertoire of different adhesins may therefore help C. glabrata to adapt to specific environmental circumstances imposed by different host niches by stimulating cell-cell adhesion or biofilm formation. In the genome of C. glabrata, we detected at least 7 subgroups comprising 67 putative CWPs with adhesin-like characteristics. Most of these are located in subtelomeric regions. Seventeen of these proteins can be allocated to the Epa family, which constitutes the largest subgroup of adhesins in C. glabrata. Also interesting in this respect is a subfamily of adhesins in C. glabrata with similarity to the multiprotein family of Dan and Pau proteins. In S. cerevisiae, subtelomerically located PAU and DAN genes are repressed under aerobic conditions and by heme and are induced under anaerobic conditions (53). A similar regulation in C. glabrata might, for instance, lead to upregulation of a specific set of adhesin-like proteins during infection or biofilm formation, which both represent conditions with low oxygen availability. Such a specific regulation would also explain why many of the putative GPI-modified adhesive-like proteins are not identified in the limited set of growth conditions tested in our studies. Together with current developments in quantitative proteomics (70), this work therefore opens new lines of investigation where changes in the cell wall proteome composition can be related to different clinical strain backgrounds and medically relevant growth or host conditions. Furthermore, detailed analysis of the newly identified Awp1 to -4 proteins should elucidate the precise functional role of these adhesins and their contribution to fungal virulence.
In conclusion, this study shows unique and distinctive features of the cell wall network in the human pathogen C. glabrata, which may contribute to its virulence. In particular, we discovered a large family of novel adhesin-like wall proteins. Differential incorporation into the wall, as shown for some of the proteins, probably governs the ability to adhere and form biofilms on various host surfaces.
Supplementary Material
Acknowledgments
We thank P. Schmidt, W. Brück, and B. Maruschak for excellent assistance in electron microscopy, B. Brandt for writing Perl scripts, B. Granger for sharing experience with the adhesion assays, and B. Cormack for valuable discussions of Epa nomenclature. Antisera used in this paper were generously provided by H. Shimoi, M. Makarow, H. Riezman, and D. Gozalbo.
This work was supported by a DFG grant (WE 3537/1-2) to M.W. and an EU grant (STREP FungWall, contract LSHB-CT-2004-511952) to F.M.K.
Footnotes
Published ahead of print on 19 September 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Aguilar-Uscanga, B., and J. M. Francois. 2003. A study of the yeast cell wall composition and structure in response to growth conditions and mode of cultivation. Lett. Appl. Microbiol. 37268-274. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht, A., A. Felk, I. Pichova, J. Naglik, M. Schaller, P. De Groot, D. MacCallum, F. C. Odds, W. Schäfer, F. Klis, M. Monod, and B. Hube. 2006. Glycosylphosphatidylinositol-anchored proteinases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281688-694. [DOI] [PubMed] [Google Scholar]
- 3.Borg-von Zepelin, M., L. Kunz, R. Ruchel, U. Reichard, M. Weig, and U. Gross. 2007. Epidemiology and antifungal susceptibilities of Candida spp. to six antifungal agents: results from a surveillance study on fungaemia in Germany from July 2004 to August 2005. J. Antimicrob. Chemother. 60424-428. [DOI] [PubMed] [Google Scholar]
- 4.Castaño, I., S. J. Pan, M. Zupancic, C. Hennequin, B. Dujon, and B. P. Cormack. 2005. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol. Microbiol. 551246-1258. [DOI] [PubMed] [Google Scholar]
- 5.Castillo, L., A. I. Martinez, A. Garcera, M. V. Elorza, E. Valentin, and R. Sentandreu. 2003. Functional analysis of the cysteine residues and the repetitive sequence of Saccharomyces cerevisiae Pir4/Cis3: the repetitive sequence is needed for binding to the cell wall β-1,3-glucan. Yeast 20973-983. [DOI] [PubMed] [Google Scholar]
- 6.Chabane, S., J. Sarfati, O. Ibrahim-Granet, C. Du, C. Schmidt, I. Mouyna, M. C. Prevost, R. Calderone, and J. P. Latgé. 2006. Glycosylphosphatidylinositol-anchored Ecm33p influences conidial cell wall biosynthesis in Aspergillus fumigatus. Appl. Environ. Microbiol. 723259-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, M. H., Z. M. Shen, S. Bobin, P. C. Kahn, and P. N. Lipke. 1995. Structure of Saccharomyces cerevisiae α-agglutinin. Evidence for a yeast cell wall protein with multiple immunoglobulin-like domains with atypical disulfides. J. Biol. Chem. 27026168-26177. [DOI] [PubMed] [Google Scholar]
- 8.Cormack, B. P., N. Ghori, and S. Falkow. 1999. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 285578-582. [DOI] [PubMed] [Google Scholar]
- 9.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 10.Csank, C., and K. Haynes. 2000. Candida glabrata displays pseudohyphal growth. FEMS Microbiol. Lett. 189115-120. [DOI] [PubMed] [Google Scholar]
- 11.Dallies, N., J. Francois, and V. Paquet. 1998. A new method for quantitative determination of polysaccharides in the yeast cell wall. Application to the cell wall defective mutants of Saccharomyces cerevisiae. Yeast 141297-1306. [DOI] [PubMed] [Google Scholar]
- 12.De Groot, P. W. J., B. W. Brandt, and F. M. Klis. 2007. Cell wall biology of Candida, p. 293-325. In C. d'Enfert and B. Hube (ed.), Candida comparative and functional genomics. Caister Academic Press, Norfolk, United Kingdom.
- 13.De Groot, P. W. J., A. D. de Boer, J. Cunningham, H. L. Dekker, L. de Jong, K. J. Hellingwerf, C. de Koster, and F. M. Klis. 2004. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell 3955-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Groot, P. W. J., and F. M. Klis. 2008. The conserved PA14 domain of cell wall-associated fungal adhesins governs their glycan binding specificity. Mol. Microbiol. 68535-537. [DOI] [PubMed] [Google Scholar]
- 15.De Groot, P. W. J., A. F. Ram, and F. M. Klis. 2005. Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet. Biol. 42657-675. [DOI] [PubMed] [Google Scholar]
- 16.Delgado, M. L., J. E. O'Connor, I. Azorin, J. Renau-Piqueras, M. L. Gil, and D. Gozalbo. 2001. The glyceraldehyde-3-phosphate dehydrogenase polypeptides encoded by the Saccharomyces cerevisiae TDH1, TDH2, and TDH3 genes are also cell wall proteins. Microbiology 147411-417. [DOI] [PubMed] [Google Scholar]
- 17.Domergue, R., I. Castaño, A. De las Peñas, M. Zupancic, V. Lockatell, J. R. Hebel, D. Johnson, and B. P. Cormack. 2005. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science 308866-870. [DOI] [PubMed] [Google Scholar]
- 18.Dubois, M., K. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28350-356. [Google Scholar]
- 19.Ecker, M., R. Deutzmann, L. Lehle, V. Mrša, and W. Tanner. 2006. PIR-proteins of Saccharomyces cerevisiae are attached to β-1,3-glucan by a new protein-carbohydrate linkage. J. Biol. Chem. 28111523-11529. [DOI] [PubMed] [Google Scholar]
- 20.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 114241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gow, N. A. R., M. G. Netea, C. A. Munro, G. Ferwerda, S. Bates, H. M. Mora-Montes, L. Walker, T. Jansen, L. Jacobs, V. Tsoni, G. D. Brown, F. C. Odds, J. W. M. Van der Meer, A. J. P. Brown, and B. J. Kullberg. 2007. Immune recognition of Candida albicans β-glucan by dectin-1. J. Infect. Dis. 1961565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Granger, B. L., M. L. Flenniken, D. A. Davis, A. P. Mitchell, and J. E. Cutler. 2005. Yeast wall protein 1 of Candida albicans. Microbiology 1511631-1644. [DOI] [PubMed] [Google Scholar]
- 23.Guo, B., C. A. Styles, Q. Feng, and G. R. Fink. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 9712158-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoyer, L. L. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9176-180. [DOI] [PubMed] [Google Scholar]
- 25.Hoyer, L. L., C. B. Green, S. H. Oh, and X. Zhao. 2008. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med. Mycol. 461-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iraqui, I., S. Garcia-Sanchez, S. Aubert, F. Dromer, J.-M. Ghigo, C. d'Enfert, and G. Janbon. 2005. The Yak1p kinase controls expression of adhesins and biofilm formation in Candida glabrata in a Sir4p-dependent pathway. Mol. Microbiol. 551259-1271. [DOI] [PubMed] [Google Scholar]
- 27.Ishigami, M., Y. Nakagawa, M. Hayakawa, and Y. Iimura. 2006. FLO11 is the primary factor in flor formation caused by cell surface hydrophobicity in wild-type flor yeast. Biosci. Biotechnol. Biochem. 70660-666. [DOI] [PubMed] [Google Scholar]
- 28.Kapteyn, J. C., B. Ter Riet, E. Vink, S. Blad, H. De Nobel, H. Van den Ende, and F. M. Klis. 2001. Low external pH induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 39469-479. [DOI] [PubMed] [Google Scholar]
- 29.Kapteyn, J. C., P. Van Egmond, E. Sievi, H. Van den Ende, M. Makarow, and F. M. Klis. 1999. The contribution of the O-glycosylated protein Pir2p/Hsp150 to the construction of the yeast cell wall in wild-type cells and β1,6-glucan-deficient mutants. Mol. Microbiol. 311835-1844. [DOI] [PubMed] [Google Scholar]
- 30.Kaur, R., B. Ma, and B. P. Cormack. 2007. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc. Natl. Acad. Sci. USA 1047628-7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klis, F. M., A. Boorsma, and P. W. J. De Groot. 2006. Cell wall construction in Saccharomyces cerevisiae. Yeast 23185-202. [DOI] [PubMed] [Google Scholar]
- 32.Klis, F. M., P. De Groot, and S. Brul. 2007. Identification, characterization, and phenotypic analysis of covalently linked cell wall proteins, p. 281-301. In I. Stansfield and M. J. R. Stark (ed.), Yeast gene analysis. Methods in microbiology, vol. 36, 2nd ed. Academic Press, London, United Kingdom. [Google Scholar]
- 33.Klis, F. M., A. F. J. Ram, and P. W. J. De Groot. 2007. A molecular and genomic view of the fungal cell wall, p. 95-117. In R. J. Howard and N. A. R. Gow (ed.), The Mycota, vol. 8. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 34.Lesage, G., and H. Bussey. 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70317-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, F., and S. P. Palecek. 2003. EAP1, a Candida albicans gene involved in binding human epithelial cells. Eukaryot. Cell 21266-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magnelli, P., J. F. Cipollo, and C. Abeijon. 2002. A refined method for the determination of Saccharomyces cerevisiae cell wall composition and β-1,6-glucan fine structure. Anal. Biochem. 301136-150. [DOI] [PubMed] [Google Scholar]
- 37.Martchenko, M., A. M. Alarco, D. Harcus, and M. Whiteway. 2004. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell 15456-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Lopez, R., L. Monteoliva, R. Diez-Orejas, C. Nombela, and C. Gil. 2004. The GPI-anchored protein CaEcm33p is required for cell wall integrity, morphogenesis and virulence in Candida albicans. Microbiology 1503341-3354. [DOI] [PubMed] [Google Scholar]
- 39.Montijn, R. C., J. van Rinsum, F. A. van Schagen, and F. M. Klis. 1994. Glucomannoproteins in the cell wall of Saccharomyces cerevisiae contain a novel type of carbohydrate side chain. J. Biol. Chem. 26919338-19342. [PubMed] [Google Scholar]
- 40.Munro, C. A., D. A. Schofield, G. W. Gooday, and N. A. R. Gow. 1998. Regulation of chitin synthesis during dimorphic growth of Candida albicans. Microbiology 144391-401. [DOI] [PubMed] [Google Scholar]
- 41.Naglik, J. R., F. Fostira, J. Ruprai, J. F. Staab, S. J. Challacombe, and P. Sundstrom. 2006. Candida albicans HWP1 gene expression and host antibody responses in colonization and disease. J. Med. Microbiol. 551323-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A. P. Bouin, C. W. Sensen, H. Hogues, M. van het Hoog, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 133452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nather, K., and C. A. Munro. 2008. Generating cell surface diversity in Candida albicans and other fungal pathogens. FEMS Microbiol. Lett. 285137-145. [DOI] [PubMed] [Google Scholar]
- 44.Netea, M. G., G. D. Brown, B. J. Kullberg, and N. A. Gow. 2008. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 667-78. [DOI] [PubMed] [Google Scholar]
- 45.Netea, M. G., N. A. R. Gow, C. A. Munro, S. Bates, C. Collins, G. Ferwerda, R. P. Hobson, G. Bertram, H. B. Hughes, T. Jansen, L. Jacobs, E. T. Buurman, K. Gijzen, D. L. Williams, R. Torensma, A. McKinnon, D. M. MacCallum, F. C. Odds, J. W. M. Van der Meer, A. J. P. Brown, and B. J. Kullberg. 2006. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J. Clin. Investig. 1161642-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nuoffer, C., P. Jeno, A. Conzelmann, and H. Riezman. 1991. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae Gas1 protein to the plasma membrane. Mol. Cell. Biol. 1127-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osumi, M. 1998. The ultrastructure of yeast: cell wall structure and formation. Micron 29207-233. [DOI] [PubMed] [Google Scholar]
- 48.Pardini, G., P. W. J. De Groot, A. T. Coste, M. Karababa, F. M. Klis, C. G. de Koster, and D. Sanglard. 2006. The CRH family coding for cell wall glycosylphosphatidylinositol proteins with a predicted transglycosidase domain affects cell wall organization and virulence of Candida albicans. J. Biol. Chem. 28140399-40411. [DOI] [PubMed] [Google Scholar]
- 49.Pardo, M., L. Monteoliva, P. Vazquez, R. Martinez, G. Molero, C. Nombela, and C. Gil. 2004. PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology 1504157-4170. [DOI] [PubMed] [Google Scholar]
- 50.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phan, Q. T., C. L. Myers, Y. Fu, D. C. Sheppard, M. R. Yeaman, W. H. Welch, A. S. Ibrahim, J. E. Edwards, Jr., and S. G. Filler. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pitarch, A., A. Jimenez, C. Nombela, and C. Gil. 2006. Decoding serological response to Candida cell wall immunome into novel diagnostic, prognostic, and therapeutic candidates for systemic candidiasis by proteomic and bioinformatic analyses. Mol. Cell. Proteomics 579-96. [DOI] [PubMed] [Google Scholar]
- 53.Rachidi, N., M. J. Martinez, P. Barre, and B. Blondin. 2000. Saccharomyces cerevisiae PAU genes are induced by anaerobiosis. Mol. Microbiol. 351421-1430. [DOI] [PubMed] [Google Scholar]
- 54.Russo, P., N. Kalkkinen, H. Sareneva, J. Paakkola, and M. Makarow. 1992. A heat shock gene from Saccharomyces cerevisiae encoding a secretory glycoprotein. Proc. Natl. Acad. Sci. USA 893671-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saville, S. P., A. L. Lazzell, A. P. Bryant, A. Fretzen, A. Monreal, E. O. Solberg, C. Monteagudo, J. L. Lopez-Ribot, and G. T. Milne. 2006. Inhibition of filamentation can be used to treat disseminated candidiasis. Antimicrob. Agents Chemother. 503312-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sherman, D., P. Durrens, F. Iragne, E. Beyne, M. Nikolski, and J.-L. Souciet. 2006. Génolevures complete genomes provide data and tools for comparative genomics of hemiascomycetous yeasts. Nucleic Acids Res. 34D432-D435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimoi, H., Y. Iimura, and T. Obata. 1995. Molecular cloning of CWP1: a gene encoding a Saccharomyces cerevisiae cell wall protein solubilized with Rarobacter faecitabidus protease I. J. Biochem. 118302-311. [DOI] [PubMed] [Google Scholar]
- 58.Sosinska, G. J., P. W. J. De Groot, M. J. Teixeira de Mattos, H. L. Dekker, C. G. De Koster, K. J. Hellingwerf, and F. M. Klis. 2008. Hypoxic conditions and iron restriction affect the cell wall proteome of Candida albicans grown under vagina-simulative conditions. Microbiology 154510-520. [DOI] [PubMed] [Google Scholar]
- 59.Sundstrom, P. 2002. Adhesion in Candida spp. Cell. Microbiol. 4461-469. [DOI] [PubMed] [Google Scholar]
- 60.Teparic, R., I. Stuparevic, and V. Mrša. 2004. Increased mortality of Saccharomyces cerevisiae cell wall protein mutants. Microbiology 1503145-3150. [DOI] [PubMed] [Google Scholar]
- 61.Teunissen, A. W., and H. Y. Steensma. 1995. Review: the dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 111001-1013. [DOI] [PubMed] [Google Scholar]
- 62.Tumbarello, M., B. Posteraro, E. M. Trecarichi, B. Fiori, M. Rossi, R. Porta, K. de Gaetano Donati, M. La Sorda, T. Spanu, G. Fadda, R. Cauda, and M. Sanguinetti. 2007. Biofilm production by Candida species and inadequate antifungal therapy as predictors of mortality for patients with candidemia. J. Clin. Microbiol. 451843-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verstrepen, K. J., A. Jansen, F. Lewitter, and G. R. Fink. 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37986-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verstrepen, K. J., and F. M. Klis. 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 605-15. [DOI] [PubMed] [Google Scholar]
- 65.Weig, M., and A. J. Brown. 2007. Genomics and the development of new diagnostics and anti-Candida drugs. Trends Microbiol. 15310-317. [DOI] [PubMed] [Google Scholar]
- 66.Weig, M., K. Haynes, T. R. Rogers, O. Kurzai, M. Frosch, and F. A. Muhlschlegel. 2001. A GAS-like gene family in the pathogenic fungus Candida glabrata. Microbiology 1472007-2019. [DOI] [PubMed] [Google Scholar]
- 67.Weig, M., L. Jansch, U. Gross, C. G. De Koster, F. M. Klis, and P. W. J. De Groot. 2004. Systematic identification in silico of covalently bound cell wall proteins and analysis of protein-polysaccharide linkages of the human pathogen Candida glabrata. Microbiology 1503129-3144. [DOI] [PubMed] [Google Scholar]
- 68.Weissman, Z., and D. Kornitzer. 2004. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol. Microbiol. 531209-1220. [DOI] [PubMed] [Google Scholar]
- 69.Wheeler, R. T., and G. R. Fink. 2006. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yin, Q. Y., P. W. J. de Groot, L. de Jong, F. M. Klis, and C. G. De Koster. 2007. Mass spectrometric quantitation of covalently bound cell wall proteins in Saccharomyces cerevisiae. FEMS Yeast Res. 7887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin, Q. Y., P. W. J. de Groot, C. G. de Koster, and F. M. Klis. 2008. Mass spectrometry-based proteomics of fungal wall glycoproteins. Trends Microbiol. 1620-26. [DOI] [PubMed] [Google Scholar]
- 72.Yin, Q. Y., P. W. J. de Groot, H. L. Dekker, L. de Jong, F. M. Klis, and C. G. de Koster. 2005. Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls: identification of proteins covalently attached via glycosylphosphatidylinositol remnants or mild alkali-sensitive linkages. J. Biol. Chem. 28020894-20901. [DOI] [PubMed] [Google Scholar]
- 73.Zlotnik, H., M. P. Fernandez, B. Bowers, and E. Cabib. 1984. Saccharomyces cerevisiae mannoproteins form an external cell wall layer that determines wall porosity. J. Bacteriol. 1591018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zupancic, M. L., M. Frieman, D. Smith, R. A. Alvarez, R. D. Cummings, and B. P. Cormack. 2008. Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Mol. Microbiol. 68547-559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.