Abstract
Photobiological hydrogen production using microalgae is being developed into a promising clean fuel stream for the future. In this study, microarray analyses were used to obtain global expression profiles of mRNA abundance in the green alga Chlamydomonas reinhardtii at different time points before the onset and during the course of sulfur-depleted hydrogen production. These studies were followed by real-time quantitative reverse transcription-PCR and protein analyses. The present work provides new insights into photosynthesis, sulfur acquisition strategies, and carbon metabolism-related gene expression during sulfur-induced hydrogen production. A general trend toward repression of transcripts encoding photosynthetic genes was observed. In contrast to all other LHCBM genes, the abundance of the LHCBM9 transcript (encoding a major light-harvesting polypeptide) and its protein was strongly elevated throughout the experiment. This suggests a major remodeling of the photosystem II light-harvesting complex as well as an important function of LHCBM9 under sulfur starvation and photobiological hydrogen production. This paper presents the first global transcriptional analysis of C. reinhardtii before, during, and after photobiological hydrogen production under sulfur deprivation.
The development of new systems to produce zero CO2 emission fuels for the future is of major importance to combat the effects of climate change. Consequently, biofuel production using conventional crops has increased substantially in recent years with the aim of reducing our CO2 footprint. A downside of this approach has been the increased competition between food and fuel production. Microalgae offer a promising alternative and likely a higher efficiency route (6) to the production of a wide range of biofuels, as they can be cultivated in PBRs sited on nonarable land in which nutrient, light, and temperature levels can be carefully regulated.
In 2000, Melis and coworkers (30) reported a two-phase microalgal photobiological H2 production process. This process consists of an aerobic and an anaerobic phase and is summarized by the following two reactions: aerobic phase = H2O → 2H+ + 2e− + 1/2O2; anaerobic phase = 2H+ + 2e− → H2.
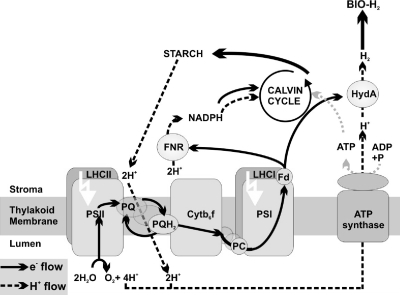
Photosystem II (PSII) drives the first stage of the process by splitting H2O into protons (H+), electrons (e−), and O2. Normally, the photosynthetic light reactions and the Calvin cycle produce carbohydrates that fuel mitochondrial respiration and cell growth (Fig. 1). However, under anaerobic conditions, mitochondrial oxidative phosphorylation is largely inhibited. Under these conditions, some organisms (e.g., Chlamydomonas reinhardtii) reroute the energy stored in carbohydrates to a chloroplast hydrogenase, likely using an NAD(P)H-plastoquinone (PQ) e− transfer mechanism (29), to facilitate ATP production via photophosphorylation (Fig. 1). Thus, hydrogenase essentially acts as an H+/e− release valve by recombining H+ (from the medium) and e− (from reduced ferredoxin) to produce H2 gas that is excreted from the cell (30). C. reinhardtii and potentially other green algae could therefore provide the basis for solar-driven biological hydrogen production. The combustion of the evolved H2 yields only H2O and thereby completes the clean energy cycle.
FIG. 1.
Photobiological hydrogen production in Chlamydomonas reinhardtii. Under aerobic conditions, electrons derived from the water-splitting reaction of PSII are passed along the photosynthetic ETC via PQ, cytochrome b6f (Cyt b6f), PSI, and ferrodoxin (Fd) before being used in the production of NADPH and starch, which can be used for mitochondrial respiration. H+ released into the thylakoid lumen by PSII and the PQ/PQH2 cycle (H+ flow; dashed lines), generates an H+ gradient, which drives ATP production via ATP synthase. Under anaerobic conditions, mitochondrial respiration is not functional, H+/e− is stored in starch, and NADPH is used by hydrogenase for bio-H2 production.
During the aerobic phase, the microalgae can be grown either photoautotrophically or photoheterotrophically to increase the cell density of the culture. The H+ and e− extracted from water by PSII (and under heterotrophic growth conditions, potentially from added exogenous substrates) during this phase are stored in a range of metabolic products, including starch and protein, the latter reported to be an important potential H+ and e− source for H2 production (30).
To induce H2 production, Melis and coworkers (30) depleted the cultures of S to inhibit the repair of the methionine-containing PSII reaction center protein (D1) after photodamage. When PSII functions below normal capacity, the culture turns anaerobic. Anaerobiosis is a strict requirement for algal photobiological H2 production, since O2 blocks the activity of hydrogenases by binding to the reaction center of the already assembled enzyme, preventing it from catalyzing the reduction of protons (12, 15). Oxygen also competes with hydrogenase as an electron acceptor, making hydrogen production even more oxygen sensitive (25). Consequently, Melis and coworkers (30) included acetate in the medium to maintain a high level of respiration during the early stage of the S depletion phase and so assist in the consumption of residual O2. Melis and coworkers concluded that “acetate is consumed by respiration for as long as there is O2 in the culture medium (0 to 30 h)” for wild-type cells, but that “it does not contribute significantly to the source of electrons in the H2-production process (30 to 120 h)” (30).
Although Chlamydomonas reinhardtii cultures can also evolve H2 under dark and anaerobic conditions, the yield is very much lower than that observed in illuminated anaerobic cultures (16, 20). The photobiological hydrogen production process is reported to source H+ and e− directly from the water-splitting reaction or via an indirect route in which they are first stored in starch/protein (22, 30). The possibility that some H+ and e− are derived from acetate must also be considered. The precise contribution from the water-splitting reaction in PSII, from starch degradation and from acetate, remains a debated issue. PSII has been suggested to be the main source of electrons for hydrogen production, since hydrogen production was reduced to 20% when the PSII inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) was added (13, 22). In addition, mutants defective in performing water photolysis or starch accumulation were also reported to have reduced H2 production capacities (38, 46). Furthermore, mutant strains that have increased starch reserves (stm6) or the ability to take up externally supplied glucose via an introduced glucose transporter (stm6glc4) have shown significant improvements in H2 production (8, 24). The degree of starch contribution to increased H2 production in stm6 cannot be easily quantified because stm6 is also inhibited in cyclic electron flow around photosystem I (PSI) (arguably reducing electron competition with the hydrogenases) as well as its respiratory metabolism. The matter is further complicated by the fact that starch degradation not only provides H+ and e− for photobiological H2 production but is also thought to be able to contribute to the establishment of anaerobiosis, which is the requirement for high hydrogenase activity. A recent study using NAD(P)H dehydrogenase inhibitors strongly suggested that a NAD(P)H dehydrogenase is involved in this pathway (34).
Given the metabolic complexity of inducing and producing H2 under S-deprived conditions, further studies are required. Here, detailed transcriptional analyses of wild-type C. reinhardtii cultures sampled at different time points during the aerobic and anaerobic phase of the photobiological H2 production process are presented to provide new insights into the complex interplay between these biochemical pathways.
MATERIALS AND METHODS
Strains and growth conditions.
Chlamydomonas reinhardtii wild-type strain CC124 was obtained from the Chlamydomonas Genetics Center. The alga was cultured in Tris-acetate-phosphate (TAP) medium (pH 7.3) (23) in sterilized conical 2-liter Erlenmeyer flasks under continuous white fluorescent illumination at 200 μmol photons m−2 s−1 on an orbital shaker at 80 rpm. For the microarray experiment, 3 liters of CC124 culture was inoculated from the same starter culture which was first grown to mid-logarithmic growth phase (an optical density at 750 nm [OD750] of 1.5 and a chlorophyll concentration of 30 mg/liter). For the control sample, 100 ml of culture from each flask was harvested from the cultures described above by centrifugation at 3,000 × g for 1.5 min at 4°C. The resulting cell pellets were immediately frozen in liquid nitrogen and stored at −80°C for later RNA extractions. The remaining cultures were harvested for S deprivation and H2 production. The cultures were washed three times to remove residual sulfur by centrifugation at 3,000 × g for 5 min at 4°C before resuspending the pellets in 100 ml of sulfur-free TAP medium. The washed cells were pelleted again under the same centrifugation conditions, resuspended, and then pooled together in a total of 2.7 liters of sulfur-free TAP medium, resulting in a sulfur-free suspension with an OD750 of 1.2 and a chlorophyll concentration of ∼22.8 mg/liter. The suspension was poured into four custom-built 650-ml photobioreactors (PBRs) for H2 production and sample collection at later time points. The OD of the cultures stayed relatively constant after S deprivation, and no significant growth was observed. This experimental design allows direct comparisons between the samples taken after S depletion (time points 1 to 6 [T1 to T6]; see details described below) and the sample just prior to S deprivation (T0).
H2 production conditions.
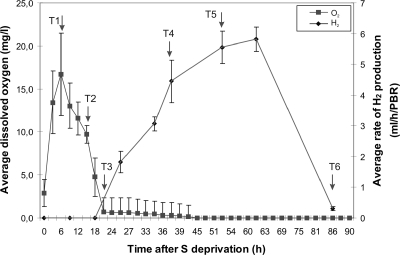
The PBRs were tightly sealed and connected to custom-built gas collection cylinders placed on corner positions of a six-spot HP magnetic stirrer and illuminated from opposite sides of the stirrer unit with white fluorescent light. In all cases, the light intensity measured at the surfaces of the PBRs ranged from 250 μE m−2 s−1 (on the darker side of the two PBRs) to 450 μE m−2 s−1 (on the side closer to the light source). The cultures were stirred continuously with magnetic bars at 150 rpm. The temperature, pH, and dissolved O2 (DO) levels in the PBRs were recorded every 5 min by a D130 data logger (Consort, Belgium). The volume of gas produced by the cultures was constantly monitored once the DO level reached zero. The hydrogen gas content was analyzed as described previously (24). The samples were collected from the PBRs through a gas-tight septum attached to a sidearm by a syringe to ensure no O2 was introduced into the system. Cells were immediately centrifuged (3,000 × g for 90 s at 4°C), and pellets were flash-frozen in liquid nitrogen and stored at −80°C before RNA isolations. When the gas volume in the collection cylinders was lower than 20 ml, nitrogen was used to flush the gas-collecting cylinders before sample collections to avoid diluting the cultures with the incoming distilled water used to submerge the cylinders' outlet tubes. Samples were taken at six time points (T1 to T6). These correspond to the peak DO level (“peak oxygen” production; T1), the DO level at approximately half of its peak (“mid-oxygen” production; T2), the DO level at zero (“zero oxygen” production; T3), the hydrogen production rate at approximately half its peak (“mid-hydrogen” production; T4), the H2 production rate at its maximum (“peak hydrogen” production; T5), and the H2 production rate at near zero (“zero hydrogen” production; T6) (Fig. 2). The exact times for the T1 to T6 sampling were 6 h, 16 h, 21 h, 37 h, 52 h, and 86 h after the start of S deprivation, respectively (Fig. 2). The volume of culture collected was 20 ml each for T1, T2, T3, T4, and T5 and 50 ml for T6. The control/reference sample taken just prior to S deprivation allowed direct comparison between samples at different time points while minimizing the volume of culture taken from the PBRs.
FIG. 2.
Changes in the DO level and rate of H2 production during the course of S deprivation-induced photobiological hydrogen production. Standard error values are shown from three independent PBRs; arrows show sampling time points.
RNA preparation.
All samples were collected directly from the active PBRs using a syringe through an airtight sampling port. Samples were immediately transferred to 50-ml Falcon tubes and centrifuged at 3,000 × g for 90 s, and the supernatant was poured off and then transferred to liquid nitrogen. This sampling process was generally completed within 5 min; it was designed to minimize the impact on the active PBRs and limit changes within the RNA. Frozen cells were stored at −80°C before RNA extraction. RNA was extracted following the centrifugation protocol for plant tissues using the SV total RNA isolation system (Promega) without sample grinding. Purified RNA from each sample was eluted in 100 μl of Milli-Q H2O. RNA was quantified using a NanoDrop ND-1000 spectrophotometer. RNA integrity was checked by gel electrophoresis and quantitative reverse transcription-PCR (qRT-PCR). All the control RNA samples were pooled together. Purified RNA was further purified by using the RNeasy MinElute cleanup kit (Qiagen) before preparation of microarray probes. The RNA extracted from each time point was used for both microarray and qRT-PCR analyses.
Preparation of fluorescent microarray probes using the indirect labeling method.
Fluorescent microarray probes were prepared by using the indirect labeling method. Total RNA (30 to 40 μg) was denatured at 70°C for 10 min with 1 μg of oligonucleotide deoxyribosylthymine-12 in a reaction mixture volume of 15.5 μl. Master mix 2 was then added to the ice-chilled reaction mixture, containing 6 μl of 5× SuperScript III reverse transcriptase buffer, 1.5 μl of 0.1 M dithiothreitol, 5 μl of deoxynucleoside triphosphate mix (0.9 mM dTTP, 3 mM dATP, 3 mM dGTP, 3 mM dCTP, and 1.8 mM aminoallyl-dUTP), and 2 μl of SuperScript III reverse transcriptase (200 U/μl; Invitrogen). The 30-μl reaction mixture was incubated at 50°C in a Thermocycler for 3 h and was then stopped by being heated to 95°C for 5 min. RNA was hydrolyzed by the addition of 15 μl of 1 M NaOH and incubation at 70°C for 15 min before the pH was neutralized by the addition of 15 μl of 1 M HCl. The amine-modified cDNA was purified using the Labeled Star purification kit (Qiagen) according to the manufacturer's directions and then concentrated to a 5-μl volume using a vacuum centrifuge. The concentrated cDNA was then labeled with either Alexa Fluor dye 555 or 647 (Invitrogen), 5 μl of cDNA with 3 μl of 0.3 M NaHCO3, and 2 μl of the Alexa Fluor succinimidyl ester dye in dimethyl sulfoxide and was incubated at room temperature for 1 h in the dark. The labeled cDNA from each time point following the initiation of S deprivation was combined with the labeled control cDNA. The labeled sample/control combined cDNA was purified with a Qiagen Labeled Star purification kit. Purified cDNA was recovered in 25 μl of Milli-Q Ultrapure H2O. The incorporation of the fluorescent dyes into cDNA was quantified with a NanoDrop ND-1000 spectrophotometer. A direct labeling method using Alexa Flour 555 5-aminohexylacrylamido-dUTP (aha-dUTP) or Alexa Flour 647 aha-dUTP (Invitrogen) was trialed at T2 (oxygen at approximately half its peak) but was found to be less efficient than the indirect method. The cDNA synthesis method was similar to the indirect labeling method described above but with the following changes. Master mix 2 consisted of the following: 6 μl of 5× buffer, 3 μl of 0.1 M dithiothreitol, 0.5 μl of deoxynucleoside triphosphates (10 mM dTTP, 25 mM dATP, 25 mM dGTP, and 25 mM dCTP), 2 μl of SuperScript II reverse transcriptase (200 U/μl), and 5 μl of 2 mM Alexa Fluor 555 aha-dUTP or Alexa Fluor 647 aha-dUTP. Hydrolysis, purification, and quantification were performed as described above.
Preparation, hybridization, washing, and scanning of the microarrays.
Chlamydomonas microarray slides version 2 (9) were obtained from Arthur Grossman (Stanford, CA). The microarray slides were immobilized, prehybridized, hybridized, and washed as instructed in the accompanying protocol. The slides were scanned at 532 nm and 635 nm in an Axon GenePix 4000B scanner at a 10-μm resolution. Photomultiplier tube voltages were adjusted to minimize the background signal and the number of saturated spots. Images of the fluorescence at 532 nm (for Alexa Fluor 555) and 635 nm (for Alexa Fluor 647) were recorded and analyzed.
Microarray data analysis.
Microarray images from scanned slides were imported into the GenePix Pro 6.0 program (Axon Instruments). Spots were automatically identified using the built-in spot alignment algorithm based on the information of spot positions provided in the GenePix array list file. All spots were visually checked and adjusted (if necessary) manually. Spots with signals distorted by dusts or high local background as well as spots that were marked absent by the GenePix program were not included in the analyses. The spot intensity medians were normalized by GeneSpring GX using the Lowess algorithm. Induction/repression ratios were obtained by dividing the sample's median intensity by the control's median intensity. Spots with signal intensities more than threefold away from the average signal intensity from all valid spots of the same gene at one time point were not included in further analyses. At each time point, for each gene, a paired two-way Student's t test was conducted to determine if a gene showed significant change (either an increase or decrease in its relative transcript compared to the control). The null hypothesis was that the log2 of the unaveraged ratio for a gene was 0 (sample/control ratio = 1): for H0, log2(sample/control) = 0 (sample/control = 1); for H1, log2(sample/control) <> 0 (sample/control <> 1). Further paired one-way t tests were conducted to assess the significance of the relative transcript abundance levels, with the thresholds being twofold changes. For this, the null hypothesis was either that the log2 of the unaveraged ratio for a gene was not higher than 1 or was not lower than −1: for H0, log2(sample/control) ≥ −1 (sample/control ≥ 0.5); for H1, log2(sample/control) < −1 (sample/control < 0.5); and for H0, log2(sample/control) ≤ 1 (sample/control ≤ 2); for H1: log2(sample/control) > 1 (sample/control > 2). The degrees of freedom used to determine the significance of the test equaled the number of spots that gave valid intensities for that gene minus one. In this case, duplicate spots on the same slide were also considered pseudoindependent samples. This approach has been used in other microarray experiments where the number of true independent samples (biological replicates) was limited (51). On the Chlamydomonas microarray slide v2.0, there are duplicate spots for each gene. Therefore, in the data analysis, genes that have less than three valid spots were excluded to ensure that at least two independent biological samples were present in the statistical analysis. A total of 12 microarray slides were used, with two slides for each time point and three slides for T3 and T5. The microarray analysis for T5 (peak H2) was prepared from samples taken from three PBRs in a separate experiment. The culturing conditions and RNA isolation were as described previously. Labeled cDNA was prepared from total RNA using the ChipShot indirect labeling and clean-up system (Promega), following the supplier's protocol. The labeled cDNA was resuspended in DIG EasyHyb Solution (Roche) and hybridized to prehybridized microarray slides in a Tecan HS 4800 hybridization station at 42°C for 2 h and washed as described previously. Slides were scanned with a resolution of 10 μm using the ScanArray 4000 (PerkinElmer), and images were processed using the ImaGene 6.0 software (BioDiscovery). Microarray slides used for T5 analysis are from a slightly different version of the C. reinhardtii version 2 slides. These slides contain two extra rows of spots in each subgrid, giving the well-characterized genes three replica spots on each slide. The layout of this version can be found in the NCBI Gene Expression Omnibus platform GPL6589.
Real-time qRT-PCR.
Real-time qRT-PCR and analysis were carried out as previously described (27, 35). A list of all primers used for qRT-PCR is shown in Table S2 in the supplemental material. Names of the 57 genes used for qRT-PCR (as shown in Fig. 3) encoding for proteins and corresponding to GenBank accession numbers or annotations as reported by Eberhard et al. (9) are as follows: LHCBM1 (AF495473); LHCBM2 (XM_001693935); LHCBM3 (XM_001703647); LHCBM5 (XM_001697474); LHCBM6 (AF495472); LHCBM8 (XM_001695415); LHCBM9 (AF479778); LHCBM11 (CHLRE3.0:scaffold_26: 641372:644769); TBC2, TBC2 translation factor (contig 114.36.1.0); LOX, lipoxygenase (scaffold_11 1392588 1392986 [399 bp]); PETC, cytochrome b6f Rieske subunit (D32003.1); PETG, cytochrome b6f complex subunit 5 (X66250); LHCA, LHCI-4/LHCA8 (XM_001696150); LHCI, LHCA7 (XM_001691907); LHC1-6, LHCA1 (AB122119); LHCB4 (AB051211); PSB1, oxygen-evolving enhancer 1 (OEE1; X13826); PSB3, OEE3 (X13832); FDX, ferredoxin (L10349.1); LHCSR1, chlorophyll a/b binding protein (XM_001696086); CPX1, coproporphyrinogen oxidase (AF133672.1); OXR, putative oxidoreductase (contig 15.82.2.11); HYDA1, hydrogenase 1 (AY055755); HYDA2, hydrogenase 2 (AY090770); NPK, novel protein kinase (U36196); RBP, RNA binding protein (contig 53.14.3.11); GPAT, putative glycerol-3-phosphate acyltransferase (XM_001694926); VFL, variable flagellar number protein (XM_001697461); TPR, tetratricopeptide repeat (TPR) domain-containing protein (contig 81.50.2.11); PFL, pyruvate formate lyase (PFL) (AJ620191); ICL, isocitrate lyase (U18765.1); NAB1, nucleic acid binding protein 1 (AY157846); SDB, scaffold attachment region DNA binding protein (contig 15.105.2.51); VSP3, VSP3 protein (L29029); AOX1, alternative oxidase 1 (AF047832); COX2A, cytochrome c oxidase subunit 2a (XM_001697494); GLPV/PHOA, starch phosphorylase (XM_001700039); AMYA2, alpha amylase 2 (XM_001695962); GBP, GTP binding protein (contig 11.120.1.5); NAD, NADH ubiquinone oxidoreductase (AY347479); Nucleolin (contig 7.42.1.5); HPP3, hypothetical protein 3 (XM_001692849); Nad-MDH, cytoplasmic malate dehydrogenase (AJ250844.1); NADP-MDH, chloroplast NADP-malate dehydrogenase (AJ277281.2); ARS2, arylsulfatase 2 (X52304); ECP88, extracellular protein 88 (AF359252); CAMK4, calcium/calmodulin-dependent protein kinase 4 (contig 25.9.2.51); SRP, hypothetical serine-rich protein (XM_001703235); HPP1, hypothetical protein 1 (XM_001693665); HYDEF, iron hydrogenase assembly protein (AY582739); Unknown 1 (contig 116.19.1.5); Unknown 2 (contig 85.6.1.0); Unknown 4 (contig 8.45.1.0); Unknown 5 (XM_001698756); Unknown 7 (contig 134.13.3.11); Unknown 8 (contig 400.2.1.5); and Unknown 9 (XM_001703534).
FIG. 3.
Real-time qRT-PCR analysis of photosynthetic genes and other genes of interest that were identified by microarray analysis. Shown are averages ± the standard error of induction ratios of 57 genes from three independent biological replicates from six different time points before, during, and after hydrogen production. Full gene names and accession numbers/annotations are provided in Materials and Methods.
Isolation of thylakoid membranes.
For the analysis of differentially expressed LHCBM proteins during sulfur starvation, thylakoid membranes were isolated from a photobiological H2 production experiment identical in design to that described above. All protein isolation procedures were performed under low-light conditions to prevent photodamage to the isolated thylakoid membranes. Cells were harvested by centrifugation (10 min at 2,200 × g for 4°C) and washed in 30 ml of buffer A (25 mM HEPES [pH 7.5], 1 mM MgCl2, 0.3 M sucrose). The washed cells were again pelleted by the same centrifugation procedure and then resuspended in 8 ml of buffer A and broken by two passes through a French press (2,000 lb/in2 at 4°C). The sample volume was then increased to 30 ml with buffer A. Thylakoid membranes were precipitated by centrifugation (45 min at 20,000 × g) at 4°C. The pellet was resuspended in 30 ml of buffer B (5 mM HEPES [pH 7.5], 10 mM EDTA, 0.3 M sucrose), centrifuged (45 min at 4°C and 48,000 × g), and finally resuspended in 2.65 ml of buffer B and 1 ml of buffer C (5 mM HEPES [pH 7.5], 10 mM EDTA, 2.2 M sucrose) to adjust the final sucrose concentration (not including the volume of the pellet) to 1.82 M. The suspension was dispensed into a Beckman SW 32 Ti centrifugation tube overlaid with 3 ml of 1.75 M sucrose solution and a further 5-ml layer of buffer D (5 mM HEPES, [pH 7.5], 0.5 M sucrose) before centrifugation (60 min at 100,000 × g and 4°C). The centrifuged thylakoid membrane sample formed a dense green band at the position of the sucrose cushion step. Upon harvesting, the sample was diluted with 5 volumes of buffer E (20 mM MES [morpholineethanesulfonic acid], pH 6.3, 5 mM MgCl2, 15 mM NaCl, 10% [vol/vol] glycerol) to facilitate pelleting of the purified thylakoid membranes upon centrifugation (20 min at 40,000 × g for 4°C). The thylakoid membranes were then resuspended in a minimal volume (∼1 ml) of MMNB buffer (25 mM MES, pH 6.0, 5 mM MgCl2, 10 mM NaCl, 2 M betaine) and flash-frozen in liquid N2 prior to storage at −80°C.
Pigment measurements.
Pigments were extracted from C. reinhardtii cells in 80% acetone, and the insoluble fraction was precipitated by centrifugation (1 min at 17,000 × g) before measuring the chlorophyll concentration and the chlorophyll a/b ratio according to Arnon (2).
SDS gel electrophoresis.
Polypeptide fractions from the sucrose density gradient were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 10% Tris-tricine gel. The stacking gel was made of 5% acrylamide (Bio-Rad), 0.7 M Tris, pH 8.45, 0.07% SDS, 0.11% AMPS (ammonium persulfate), 3.8 μl TEMED (N,N,N′,N′-tetramethylethylenediamine). The resolving gel was made of 10% acrylamide (Bio-Rad), 0.9 M Tris, pH 8.45, 0.09% SDS, 12.5% glycerol, 0.05% AMPS, 3.8 μl TEMED. Samples from sucrose gradient fractions were concentrated from 2 ml to ∼30 μl with Vivaspin 500 μl. Pigments were extracted from the protein samples with acetone in excess, dried with N2 gas, and resuspended in H2O. Solubilization was conducted in a buffer containing 0.2 M Tris, pH 6.8, 5 mM EDTA, 1 M sucrose, 0.4% dithiothreitol, 3.1% SDS, 0.8% glycerol, and bromophenol blue for approximately 15 min. Gel lanes were loaded with an equal amount of chlorophyll (10 μg). Gels were stained with 0.1% Coomassie blue R250 and G250 for protein visualization.
Identification of proteins by MS, MALDI-TOF, and MALDI-TOF-TOF.
All bands from protein gels were excised and digested with trypsin (Promega) at 4 μg/ml. The standard preparation of the calibration mixture for matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) contained des-Arg-bradykinin (1.0 pmol/liter), angiotensin I (2.0 pmol/liter), Glu-fibrino-peptide B (1.3 pmol/liter), adrenocorticotropin (ACTH) 1-17 clip (2.0 pmol/liter), ACTH 18-39 clip (1.5 pmol/liter), and ACTH 7-38 clip (3.0 pmol/liter). A 10-fold dilution of the standard calibration mixture was used for calibration of the MALDI plates on which the tryptic digest samples were spotted. The sample matrix used was αCHCA (α-cyano-4-hydroxycinnamic acid) at a concentration of (5 mg/ml) in 50% acetonitrile in 5 mM ammonium phosphate and 1% formic acid (aqueous). Samples were analyzed using a 4700 Proteomics analyzer MALDI-tandem TOF (MALDI-TOF-TOF) (Applied Biosystems, CA). The peptides were resuspended directly with 5 mg/ml of αCHCA in 60% acotinase/0.1% formic acid (aqueous) onto a MALDI target plate. All mass spectrometry (MS) spectra were recorded in the positive reflector mode at a laser energy setting of 4800. For MS data, 1,000 shots were accumulated for each spectrum using an MS-positive ion reflectron mode acquisition method. All tandem MS (MS-MS) data from the TOF-TOF were acquired using the default positive ion, 1 kV collision energy, reflectron mode, MS-MS method at a laser energy of 5500. MS-MS data acquisition was performed in a four-step process. First, MS spectra were recorded from each of the six calibration spots, and the default calibration parameters of the instrument and the plate model for that plate were updated. Second, MS spectra were recorded for all sample spots on the plate. Each spectrum was generated by accumulating the data from 1,000 laser shots, using the newly updated default calibration settings. Third, the TOF MS spectra were analyzed using the Peak Picker software supplied with the instrument. The 10 most abundant spectral peaks that met the threshold (a signal/noise ratio of >20) criteria and were not on the exclusion list were included in the acquisition list for the TOF-TOF, MS-MS portion of the experiment. The threshold criteria were set as follows: a mass range of 850 to 4,000 Da, a minimum cluster area of 500, a minimum signal/noise ratio of 20, and a maximum number of MS-MS spectra per spot of 10. A mass filter excluding matrix cluster ions and trypsin autolysis peaks was applied. An XML file was generated that contains the list of the precursor masses selected for MS-MS. Database searching of noninterpreted TOF-MS and TOF-TOF MS-MS data was carried out using the Mascot search engine (Matrix Science) (44) and the MSDB 20040630 containing 1,501,893 sequences and 480,537,669 residues with the Viridiplantae taxonomy. Proteins were identified from the MS-MS data with confidence of at least 95%. Confidence was based on the number of peptides found in MS and the ion score from MS-MS.
Microarray data accession numbers.
The complete microarray data set has been submitted to Gene Expression Omnibus and can be accessed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=prszzawmwoakcty&acc=GSE9165/.
RESULTS
Global expression profiling in Chlamydomonas reinhardtii before, during, and after hydrogen production.
Cultures of C. reinhardtii (CC124) were grown as independent biological replicates and induced for hydrogen production by sulfur deprivation. Samples for RNA extraction were taken from the PBRs at T1 to T6 corresponding to 6 h, 16 h, 21 h, 37 h, 52 h, and 86 h after the initiation of S depletion. The first three time points correspond with different DO levels in the PBRs, referred as peak oxygen, mid-oxygen, and zero oxygen production rates (Fig. 2). The last three time points represent mid-hydrogen, peak hydrogen, and zero hydrogen production rates (Fig. 2). Quality control of all RNA samples was carried out by gel electrophoresis and real-time qRT-PCR to ensure that the RNA samples did not show any signs of degradation. Only a limited amount of quality RNA could be obtained from T6 samples due to cell death at this late time point. Consequently, microarray hybridizations were only conducted on samples collected during the first five time points (T1 to T5). All samples were compared to the control taken immediately prior to sulfur deprivation (T0). Stringent analyses of the data identified 166 genes that showed at least a twofold change in one or more of the five time points consistently across technical (duplicate spots on the array) and biological replicates of the experiment and that had shown statistical significance (Table 1). Based on their closest match to GenBank entries, genes were assigned to different groups according to their putative functions. These include photosynthesis (22 genes), sulfur metabolism (8 genes), carbon metabolism (4 genes), proteolysis (5 genes), amino acid synthesis (4 genes), transcription and translation (10 genes), redox cycling (3 genes), other pathways and processes (27 genes), and a large number of genes with unknown functions (83 genes).
TABLE 1.
Induction and repression ratios of 166 C. reinhardtii genes that showed at least a twofold change in transcript abundance in one or more time points (T1 to T5) before and during hydrogen productiona
| Gene function and GenBank accession no. or contig (no. of genes) | GAL file ID | Gene annotation description (closest match) | Peak O2 | Mid O2 | Zero O2 | Mid H2 | Peak H2 |
|---|---|---|---|---|---|---|---|
| Photosynthesis (22) | |||||||
| AB122117.1 | 257.A | LhcI-4 gene for light-harvesting chlorophyll a/b protein of PSI | 0.39 | 0.20 | 0.19 | 0.22 | NA |
| AY171231 | 262.A | Light-harvesting complex I protein | 0.36 | NA | 0.29 | 0.25 | NA |
| X15166 | 250.A | PsaK mRNA for 8.4-kDa subunit of PSI | 0.31 | NA | 0.28 | 0.32 | NA |
| XM_001691032 | 8094.D | PSI reaction center subunit XI, chloroplast precursor | 0.47 | NA | NA | 0.45 | NA |
| AB050007 | 217.A | CP26 | NA | NA | NA | 0.43 | NA |
| AF244524 | 158.A | Chlorophyll a/b binding protein | NA | NA | 0.20 | 0.56 | NA |
| AB051211 | 159.A | Light-harvesting chlorophyll a/b binding protein Lhcb4 | 0.52 | 0.50 | 0.27 | 0.14 | 0.46 |
| AF170026.1 | 426.A | PSII reaction center W protein, chloroplast precursor | NA | NA | 0.50 | 0.34 | 0.50 |
| XM_001701652 | 4465.C | 4.1-kDa PSII subunit (PSBX) | NA | NA | 0.30 | 0.34 | NA |
| J05524 | 463.A | Plastocyanin, chloroplast precursor | 0.27 | NA | 0.11 | 0.22 | 0.29 |
| X04472.1 | 45.A | RbcS2 for ribulose bisphosphate carboxylase/oxygenase | NA | NA | 0.12 | 0.13 | NA |
| D32003 | 155.A | Cytochrome B6-F complex iron-sulfur subunit, chloroplast precursor | 0.53 | NA | 0.30 | 0.36 | NA |
| X92488 | 84.A | Cytochrome B6-F complex 4-kDa subunit, chloroplast precursor | NA | NA | NA | 0.90 | 0.27 |
| L10349 | 243.A | Ferredoxin, chloroplast precursor | 0.21 | 0.44 | 0.11 | 0.12 | 0.27 |
| X13826 | 54.A | Oxygen-evolving enhancer protein 1, chloroplast precursor | 0.56 | 0.77 | 0.42 | 0.42 | NA |
| M15187 | 96.A | Oxygen-evolving enhancer protein 2, chloroplast precursor | 0.44 | NA | NA | 0.31 | NA |
| X13832 | 83.A | Oxygen-evolving enhancer protein 3, chloroplast precursor | NA | NA | 0.39 | 0.39 | NA |
| M36123 | 352.A | Phosphoribulokinase, chloroplast precursor (PRK) | NA | NA | 1.68 | 3.37 | NA |
| AF107303 | 458.A | Open reading frame of intron 2 from chloroplast psbA gene | NA | NA | 1.48 | 1.91 | 2.84 |
| XM_001696073 | 251.A | Chlorophyll a/b binding protein homolog LHCSR1 precursor | 1.93 | 1.30 | NA | 4.55 | 3.07 |
| XM_001696086 | 8770.D | Chlorophyll a/b binding protein homolog LHCSR3 precursor | 4.89 | NA | NA | 1.29 | NA |
| AF479778 | 350.A | Major light-harvesting complex II protein m9 (LHCBM9) | NA | 11.26 | 17.41 | 109.03 | 22.30 |
| Sulfur assimilation (8) | |||||||
| X52304 | 73.A | Arylsulfatase 2 precursor (ARS2) | 15.39 | NA | 7.62 | 18.10 | 5.63 |
| XM_001697170 | 7778.D | Sulfate transporter (SULTR2) | 3.61 | 2.90 | 2.68 | 3.16 | NA |
| XM_001699252 | 8577.D | Sulfite reductase | NA | NA | NA | 0.93 | 3.96 |
| X16179.1 | 382.A | Arylsulfatase 2 precursor | NA | NA | NA | NA | 7.25 |
| U57088 | 468.A | Probable sulfate adenylyl transferase ATS1 | NA | NA | NA | 2.86 | 2.22 |
| AF359251 | 432.A | Extracellular polypeptide Ecp76 | 2.19 | NA | NA | 4.27 | NA |
| 199.8.2.31 | 7885.D | Extracellular polypeptide Ecp88 | 2.96 | 1.31 | 2.75 | 2.15 | NA |
| AF359252 | 167.A | Extracellular polypeptide Ecp88 | 3.25 | NA | 2.40 | 18.90 | 2.33 |
| Carbon metabolism (4) | |||||||
| AF394513 | 337.A | 6-Phosphogluconate dehydrogenase | 2.65 | NA | 1.66 | 2.73 | NA |
| U18765 | 40.A | Probable isocitrate lyase | 0.14 | 0.11 | 0.17 | 0.14 | NA |
| X66410 | 254.A | Pyruvate formate lyase | NA | NA | 3.54 | 1.82 | NA |
| U42979.1 | 252.A | Malate dehydrogenase, sodium acetate induced | 0.64 | NA | NA | 0.42 | 0.6 |
| Proteolysis (5) | |||||||
| X60826 | 67.A | UBI3 mRNA for ubiquitin fusion protein | NA | NA | 1.47 | 3.18 | NA |
| XM_001696123 | 3750.C | 26S proteasome regulatory complex, lid subcomplex, non-ATPase subunit RPN3 (subunit 3) (PSD3) (PSMD3) | NA | 3.63 | 2.77 | 0.76 | NA |
| 72.37.1.5 | 7281.C | F15M7.13, ubiquitin carboxyl-terminal hydrolase, Arabidopsis | NA | 4.01 | NA | NA | NA |
| AY062935 | 348.A | Ubiquitin-conjugating enzyme E2 isoform | 2.98 | 2.09 | NA | 6.00 | NA |
| XM_001695251 | 4464.C | Putative serine carboxypeptidase precursor | NA | NA | NA | 2.48 | |
| Amino acid synthesis (4) | |||||||
| U36197 | 135.A | 5-Methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | NA | NA | 0.38 | 0.29 | NA |
| AF078693 | 447.A | Putative O-acetylserine lyase precursor | NA | 1.89 | 3.00 | 2.89 | 10.93 |
| U46207 | 112.A | Glutamine synthetase, cytosolic isozyme | NA | NA | 0.37 | 0.56 | NA |
| XM_001695007 | 9339.E | Aspartate-semialdehyde dehydrogenase (ASSD1) | NA | 0.80 | 0.74 | 0.40 | 1.17 |
| Transcription and translation (10) | |||||||
| XM_001696696 | 5869.C | PRP3, pre-mRNA processing factor 3 | NA | NA | NA | 2.53 | NA |
| XM_001691400 | 9120.E | Eukaryotic initiation factor 4A-10-like protein | NA | NA | NA | 2.84 | 1.45 |
| XM_001691255 | 4469.C | SF3A3 splicing factor 3a, subunit 3 (SPL3) | NA | NA | 2.78 | 4.11 | NA |
| AY337612 | 4147.C | Chloroplast 30S ribosomal protein S11 (rps11) | NA | NA | NA | NA | 2.49 |
| X95313 | 110.A | 60S ribosomal protein L11 | NA | NA | NA | 2.55 | 0.67 |
| 2019.1.1.5 | 8710.D | 60S ribosomal protein L22 [Drosophila melanogaster], 54.2% identity | NA | 5.31 | NA | 0.56 | NA |
| XM_001696564 | 819.C | 60S ribosomal protein L37A | 0.55 | NA | NA | 0.33 | 0.82 |
| XM_001692618 | 3219.C | 60S ribosomal protein L13 | NA | NA | NA | 0.43 | 0.53 |
| XM_001689821 | 9565.E | 60S ribosomal protein L6 | NA | NA | NA | 0.35 | NA |
| AY177616 | 312.A | 41-kDa ribosome-associated protein precursor | 0.60 | NA | 0.42 | 0.48 | 0.7 |
| Redox cycling (3) | |||||||
| XM_001693558 | 9401.E | Similar to vanadium chloroperoxidase | 2.72 | 1.28 | NA | 3.22 | 7.71 |
| 15.82.2.11 | 9635.E | Putative oxidoreductase [Oryza sativa subsp. japonica], 82.3% identity | 0.67 | NA | 3.11 | 1.90 | NA |
| X78822 | 17.A | Thioredoxin H-type | NA | NA | NA | 0.46 | NA |
| Other pathways and processes (27) | |||||||
| U36196 | 161.A | Protein kinase, 48K | 0.45 | NA | NA | 0.46 | NA |
| XM_001691558 | 4567.C | WNK protein kinase (WNK1) | NA | NA | 2.86 | 1.37 | 0.74 |
| XM_001702027 | 516.B | CDK inhibitor kinase (WEE1) | NA | NA | NA | 3.98 | NA |
| XM_001692562 | 506.B | Rhodanese domain phosphatase (RDP3) | 5.99 | NA | 2.96 | 1.94 | 2.32 |
| XM_001696853 | 645.C | G protein-coupled seven transmembrane receptor | NA | 3.31 | NA | 0.64 | NA |
| XM_001703235 | 9249.E | PWR motif protein (PWR1) | NA | 3.15 | 2.85 | 6.72 | NA |
| XM_001703306 | 3414.C | Selenium binding protein (SBD1) | 5.17 | NA | 2.48 | 2.39 | 4.54 |
| XM_001698919 | 4207.C | Conserved protein similar to EMP70/nonaspanin | NA | 3.51 | NA | 0.55 | NA |
| AF399653.1 | 389.A | Ezy2 | NA | 2.91 | NA | 0.60 | NA |
| L29029 | 186.A | VSP-3 | NA | 3.33 | NA | 0.39 | 0.58 |
| U31975 | 188.A | Alanine aminotransferase | NA | 3.46 | NA | 0.67 | NA |
| XM_001694402 | 9383.E | Hydroxylamine reductase, hybrid cluster protein HCP4 | 2.18 | 3.42 | NA | 1.32 | NA |
| XM_001690300 | 6995.C | NOP58 structural component of C/D snoRNPs | NA | 4.38 | NA | 0.46 | NA |
| XM_001703004 | 8375.D | Type II NADH dehydrogenase (NDA7) | 2.42 | NA | NA | NA | NA |
| XM_001702995 | 8352.D | Dehydrogenase with pyrroloquinoline quinone as a cofactor | NA | NA | 3.84 | 1.34 | NA |
| XM_001690491 | 4875.C | Endoplasmic reticulum-located HSP110/SSE-like protein | NA | NA | 2.67 | 1.46 | NA |
| XM_001701229 | 2968.C | Flagellar-associated protein | NA | NA | 3.04 | NA | NA |
| AF133672 | 311.A | Coproporphyrinogen III oxidase precursor | NA | NA | NA | 3.53 | 2.40 |
| XM_001696259 | 2161.C | Mu1-adaptin (AP1M1) | NA | NA | 1.85 | 2.44 | NA |
| XM_001693906 | 1657.C | 1-Deoxy-d-xylulose 5-phosphate reductoisomerase (DXR1) | NA | NA | NA | 2.98 | NA |
| X76117 | 212.A | Zygote-specific protein | 0.40 | 0.55 | 0.46 | 0.55 | NA |
| XM_001698620 | 9223.E | THI4 regulatory protein | 0.38 | 0.37 | NA | 0.28 | NA |
| XM_001693730 | 166.A | Acyl carrier protein (ACP2) | 0.25 | NA | 0.47 | 0.44 | NA |
| AF295371 | 63.A | Chlamyopsin 2 | 0.53 | NA | 0.48 | 0.28 | NA |
| XM_001690256 | 9591.E | Cell wall protein pherophorin C2 (PHC2) | NA | NA | 0.25 | 0.36 | NA |
| XM_001697461 | 6272.C | Flagellar-associated protein (FAP218) | NA | NA | NA | 0.43 | 1.58 |
| XM_001697963 | 4463.C | Cell wall protein pherophorin C3 (PHC3) | NA | NA | 0.52 | 0.35 | NA |
| Unknown (83) | |||||||
| XM_001690783 | 1650.C | Low-CO2-inducible protein (LCI30) | NA | 2.31 | NA | 1.35 | 0.39 |
| AF015661 | 477.A | Low-CO2-inducible protein (LCI3) | 0.71 | NA | NA | 0.35 | 0.20 |
| 141.1.1.0 | 5077.C | Phenylcoumaran benzylic ether reductase homolog TH6 | NA | NA | NA | NA | 6.82 |
| 25.9.2.51 | 6103.C | CAMK4, calcium/calmodulin-dependent kinase IV [Homo sapiens], 62.2% identity | NA | NA | 3.21 | NA | 0.68 |
| 44.35.1.5 | 6796.C | Fibroin heavy chain precursor [Bombyx mori], 6.8% identity | NA | NA | 3.75 | NA | NA |
| 688.1.1.0 | 9070.E | Large tegument protein [Herpes simplex virus], 10.7% identity | 2.49 | NA | NA | 1.19 | NA |
| 13.36.2.11 | 7462.C | SRP40, suppressor protein SRP40 [Saccharomyces cerevisiae], 68.7% identity | NA | 4.62 | NA | 0.56 | NA |
| 26.39.1.0 | 2318.C | EBNA-1 nuclear protein [Human herpesvirus 4], 27.0% identity | NA | NA | NA | 0.38 | NA |
| X96877 | 483.A | Hypothetical luminal protein precursor, chloroplast | 0.57 | 0.54 | 0.34 | 0.40 | 0.72 |
| XM_001695499 | 1422.C | Predicted protein (SSA15) with TPR, ANK, and zf-MYND domains | NA | NA | 3.72 | 1.38 | NA |
| XM_001700193 | 668.C | Predicted protein CAP_ED, effector domain of CAP transcription factor | NA | NA | 2.56 | 1.44 | 0.63 |
| 3.43.1.0 | 4962.C | Putative glucan synthase [Arabidopsis thaliana], 72.8% identity | NA | NA | 2.76 | 1.49 | NA |
| 16.97.1.5 | 6593.C | Y25C1A.3 protein [Caenorhabditis elegans], 57.9% identity | NA | NA | 3.96 | 1.68 | NA |
| 121.14.1.5 | 6312.C | Vegetative cell wall protein gp1 precursor [C. reinhardtii], 30% identity | NA | NA | NA | 2.27 | NA |
| XM_001694590 | 3153.C | Hypothetical protein | NA | 2.30 | NA | NA | NA |
| 111.18.4.11 | 724.C | Cullin 1B (Nicotiana tabacum), 99.7% identity | NA | 2.87 | NA | NA | NA |
| 7.42.1.5 | 3362.C | Nucleolin (protein C23) [Mesocricetus auratus], 67.8% identity | NA | 6.30 | NA | 0.14 | 0.50 |
| XM_001697504 | 9998.E | Hypothetical protein | NA | NA | 0.49 | 0.39 | 0.33 |
| 152.14.1.5 | 1904.C | CreA protein (Shigella flexneri), 55.4% identity | NA | NA | 2.74 | 1.29 | NA |
| 58.41.1.5 | 1097.C | Fibroin heavy chain precursor (Fib-H) (H-fibroin) [B. mori], 10.5% identity | NA | NA | 2.20 | 0.89 | |
| 576.2.1.0 | 9678.E | 0.61 | NA | 0.52 | 0.33 | 1.81 | |
| 356.1.3.11 | 9243.E | Hypothetical protein | 0.56 | NA | 0.52 | 0.12 | NA |
| 46.52.2.11 | 2626.C | CG2839 protein | NA | 3.73 | 0.86 | 0.79 | NA |
| XM_001689556 | 3709.C | Hypothetical protein, homology to Phytophthora infestans CRN1 | NA | NA | 1.35 | 2.16 | NA |
| XM_001696388 | 9338.E | Hypothetical protein | NA | NA | 1.39 | 2.23 | 1.57 |
| 116.19.1.5 | 8373.D | C_1160006 [158018:161045] | 2.53 | 2.55 | 1.56 | 0.44 | NA |
| XM_001700674 | 4539.C | Hypothetical protein 45% identity to A. thaliana UDP glucosyl transferase | NA | NA | 1.62 | 2.68 | NA |
| XM_001694976 | 6697.C | Hypothetical protein | NA | NA | 2.14 | 3.64 | 2.48 |
| Contig 85.6.1.0 | 2154.C | 0.85 | 6.61 | 2.42 | 0.10 | NA | |
| XM_001693665 | 9725.E | CAS_like, clavaminic acid synthetase-like | 5.31 | NA | 2.64 | 3.94 | 3.02 |
| XM_001689728 | 5855.C | Hypothetical protein with ring finger domain | NA | 3.79 | 2.39 | 1.31 | NA |
| 1.8.3.11 | 8427.D | HC_10364 [926397:929841] | 1.73 | NA | 5.22 | 4.16 | NA |
| 400.2.1.5 | 7404.C | NA | NA | 0.47 | 0.69 | NA | |
| 8.83.1.5 | 4854.C | Tax_Id=9606 splice isoform 1 of Q15149 plectin 1 | NA | NA | 2.39 | 0.70 | NA |
| XM_001698598 | 8873.D | Hypothetical protein | NA | NA | 2.85 | 1.10 | NA |
| XM_001699124 | 5272.C | Hypothetical protein | NA | NA | 2.63 | 1.11 | NA |
| 117.7.1.0 | 3343.C | NA | NA | 3.64 | 1.25 | NA | |
| 30.47.1.5 | 3534.C | Y25C1A.3 protein | NA | NA | 3.26 | 1.32 | NA |
| 205.4.1.5 | 1902.C | NA | NA | 4.02 | 1.41 | 1.40 | |
| XM_001691394 | 4410.C | Hypothetical protein | NA | NA | 3.46 | 1.73 | 1.33 |
| 103.28.2.31 | 8178.D | Hypothetical protein | NA | 6.32 | NA | 0.47 | NA |
| 667.1.1.0 | 4037.C | NA | 4.04 | NA | 0.51 | NA | |
| 17.108.1.0 | 6106.C | C_1740018 [20231:20880] | NA | 4.90 | NA | 0.61 | NA |
| XM_001699240 | 1640.C | TPR repeat protein | NA | 4.17 | NA | 0.78 | NA |
| 26.58.1.5 | 7656.D | NA | 4.40 | NA | 0.81 | NA | |
| 26.55.1.5 | 4842.C | NA | 6.04 | NA | 0.12 | NA | |
| 3.101.2.11 | 6615.C | MGC33630, hypothetical protein MGC33630 | NA | 4.03 | NA | 0.38 | NA |
| XM_001698756 | 5644.C | Hypothetical protein | NA | 3.91 | NA | 0.33 | NA |
| 8.45.1.0 | 2919.C | NA | 4.64 | NA | 0.20 | NA | |
| XM_001692849 | 4703.C | Hypothetical protein, 26% identity to DNA-dependent RNA polymerase E′ (ISS) [Ostreococcus tauri] | NA | 3.88 | NA | 0.45 | 0.74 |
| XM_001695799 | 2172.C | Hypothetical protein | 1.76 | NA | NA | 2.64 | 1.37 |
| XM_001692679 | 1212.C | Hypothetical protein | NA | NA | NA | 2.18 | NA |
| 1.59.2.11 | 3917.C | NA | NA | NA | 2.70 | 1.28 | |
| 10.23.2.11 | 4728.C | NA | NA | NA | 2.68 | NA | |
| 12.107.1.5 | 8247.D | Tax_Id=10090 Ensembl_locations:None RAD54-like | NA | NA | NA | 0.42 | NA |
| XM_001702460 | 8328.D | 44% identity to molecular cochaperone STI1 (ISS) [O. tauri] | NA | NA | NA | 2.24 | NA |
| XM_001694296 | 5751.C | 45% identity to PBCV-1 prolyl 4-hydroxylase (ISS) [O. tauri] | NA | NA | NA | 0.17 | NA |
| 145.2.2.11 | 5700.C | NA | NA | NA | 2.40 | 1.38 | |
| XM_001695405 | 4344.C | 37% identity to PEX22 (peroxin 22), protein binding [A. thaliana] | NA | NA | NA | 2.51 | NA |
| XM_001702135 | 1572.C | Hypothetical protein | NA | NA | NA | 2.18 | NA |
| XM_001691387 | 2256.C | Tax_Id=9606 | NA | NA | NA | 2.33 | NA |
| XM_001701095 | 5542.C | LETM1-like protein | NA | NA | NA | 3.31 | 1.40 |
| XM_001699679 | 2712.C | CG10555 protein | NA | NA | NA | 3.12 | NA |
| XM_001702888 | 6018.C | CG1517 protein | NA | NA | NA | 2.24 | NA |
| 53.15.3.11 | 8244.D | NA | NA | NA | 2.16 | NA | |
| 59.52.1.0 | 1456.C | Tax_Id=10090, Ensembl_locations:7-33233606 | NA | NA | NA | 0.47 | NA |
| 6.97.1.0 | 9930.E | C_60084 [691340:694926] | NA | NA | NA | 3.75 | 1.74 |
| XM_001703534 | 4240.C | Hypothetical protein, 31% identity to ERD4-related membrane protein | NA | NA | NA | 0.38 | NA |
| 93.32.2.31 | 3081.C | NA | NA | NA | 2.20 | NA | |
| 117.16.2.11 | 9185.E | C_1170030 [93583:99018] | NA | NA | 3.75 | 1.51 | 0.77 |
| XM_001696892 | 5915.C | SPRY superfamily | NA | NA | 2.46 | 1.56 | NA |
| XM_001696875 | 8243.D | Hypothetical protein | NA | NA | 2.43 | 1.62 | NA |
| 69.8.2.31 | 632.C | Hypothetical protein | NA | NA | 4.76 | 1.63 | NA |
| 3.168.1.0 | 5052.C | Hypothetical 38.7-kDa protein, 65.6% identity | NA | NA | 2.51 | 1.65 | NA |
| XM_001701276 | 9373.E | Hypothetical protein | NA | NA | 6.36 | 5.95 | 6.53 |
| XM_001701229 | 2968.C | Flagellar-associated protein [Chlamydomonas reinhardtii] | NA | NA | 3.04 | NA | NA |
| 5.123.2.31 | 5749.C | 2.94 | 1.21 | NA | NA | NA | |
| 329.4.1.5 | 4269.C | C_3290004 [33926:36211] | NA | 1.63 | NA | 0.33 | NA |
| XM_001699578 | 4127.C | Hypothetical protein | NA | 1.77 | NA | 2.16 | 1.17 |
| XM_001699290 | 5393.C | 0.79 | NA | NA | 2.03 | 3.39 | |
| 341.1.1.5 | 6015.C | NA | NA | 1.22 | 2.08 | 3.40 | |
| XM_001703283 | 2353.C | 31% identity to 2OG-Fe(II) oxygenase [Frankia sp. strain EAN1pec] | 1.92 | 2.79 | NA | NA | NA |
| 81.50.2.11 | 9274.E | Hypothetical protein containing the TPR domain | NA | NA | 1.48 | 2.70 | 2.44 |
Shown are averages obtained from microarray data using two to three independent biological replicates. Ratios of at least twofold induction or repression that passed all stringent criteria for data analysis, including Student's t test (P < 0.05), are bold. Student's t test was carried out on normalized microarray signals against control signals to establish significant differences. A complete list, including results for all genes used for microarray analysis, is shown in Table S1 in the supplemental material. NA, data points that did not pass the background cutoff or other stringent analysis criteria; GAL file ID, GenePix array list file identification number.
Photosynthesis genes.
The majority of transcripts encoding proteins involved in photosynthesis declined soon after S deprivation and were consistently suppressed throughout the experiment (Table 1, photosynthesis). These included transcripts encoding a ferredoxin; cytochrome b6f; plastocyanin; an oxygen-evolving enhancer (OEE1/PSB1) 1, 2, and 3; subunits of PSI and PSII; and light-harvesting complex proteins (LHCA, LHCB4, and LHCB5). Surprisingly, there was an increase in transcript abundance of the genes encoding the major light-harvesting complex 9 (LHCBM9) and two different stress-related chlorophyll a/b binding proteins (LHCSR1 and LHCSR3) (39, 40). In addition, the transcripts of PRK encoding phosphoribulokinase and the open reading frame containing psbA intron 2 increased more than twofold during mid-hydrogen production. Together with the increase of LHCBM9 and LHCSR transcripts, this result indicates that photosynthetic activity and its regulation did not undergo a simple decrease but underwent a more complex adjustment during photobiological hydrogen production (PBHP) (see Discussion).
Sulfur assimilation and carbon metabolism genes.
As expected, the microarray data showed strong and consistent responses to S deprivation for genes involved in S assimilation. Expression was strongly induced at all four time points for genes encoding arylsulfatase (ARS2) and the sulfate transporter (SULTR2). Genes encoding sulfite reductase and extracellular proteins ECP76 and two different ECP88 proteins were induced in at least one time point. It was apparent that major changes also occurred in the carbon metabolic pathways after the initiation of S depletion. There was a marked and consistent decrease in the transcript levels of genes encoding isocitrate lyase and malate dehydrogenase, two enzymes in the glyoxylate cycle and the citric acid cycle (Table 1, carbon metabolism). On the other hand, transcript levels of the genes encoding 6-phosphogluconase dehydrogenase (an enzyme in the pentose phosphate pathway) and PFL (belonging to the fermentative PFL pathway) increased during the H2 production phase.
Proteolysis and amino acid synthesis.
There were also indications of increased protein degradations in C. reinhardtii cells during PBHP. Genes involved in protein degradation that showed an increase in transcript abundance encode a 26S proteasome subunit S3, a putative serine carboxypeptidase, a putative ubiquitin, an E2 ubiquitin-conjugating enzyme and a ubiquitin carboxyl-terminal hydrolase (Table 1, proteolysis). Furthermore, transcript levels of genes involved in amino acid synthesis, such as those encoding methionine synthase and aspartate β-semialdehyde dehydrogenase, were consistently decreased (two- to threefold), especially at mid-H2 production (T4). This is consistent with a previous report by Melis et al. (30), demonstrating that significant protein degradation occurred during H2 production. The only exception was the induction of a gene encoding the putative cysteine-synthesizing O-acetylserine lyase precursor which could also be classed to the sulfur assimilation category.
Genes involved in transcription, translation, and regulation.
Among the more than twofold differentially expressed genes, 10 were classified as having a putative role in transcription or translation. Four of these were upregulated during the hydrogen production phase, including the genes encoding pre-mRNA processing factor 3 (PRP3), the eukaryotic initiation factor 4A-10 (eIF4A-10), splicing factor 3a subunit 3 (SP3a3), and the chloroplast 30S ribosomal protein L11 (rps11). The genes encoding the remaining ribosomal proteins were mostly repressed throughout the course of the experiment. Other genes also potentially involved in regulation or signal transduction include, for example, genes encoding protein kinases (WNK1 and WEE1), phosphatase (RDP3), or the G protein-coupled transmembrane receptor (Table 1, other pathways and processes).
Genes involved in other pathways and processes and unspecified genes with unknown functions.
A total of 27 genes were identified with differential expression during hydrogen production that are likely to be involved in other general cell maintenance, regulation, and developmental processes not specified above (Table 1). Interestingly, these include a relatively large number of genes (11) that were induced at early time points (T1 or T2). A large number of genes (83 genes) could not be assigned to any putative function. Although their sequences were used for BLAST searches to check against GenBank entries (3), none returned any significant match to any other gene or protein with known functions.
Expression profiles using real-time qRT-PCR.
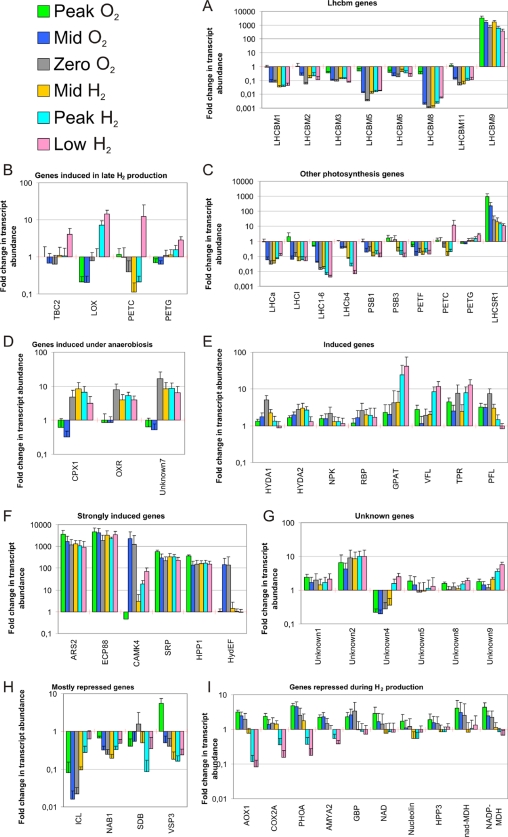
Real-time qRT-PCR was carried out on cDNA from all C. reinhardtii CC124 samples (three independent biological replicates) and using all six time points (T1 to T6). Among the 166 genes that were found by microarray analysis to be significantly (P < 0.05) differentially expressed by more than twofold, 30 were chosen for further expression profiling by qRT-PCR using specific primers. In addition, another 27 genes of particular interest were subjected to further qRT-PCR analysis (Fig. 3). These genes were grouped according to their functions (e.g., photosynthesis) or sorted by similar expression patterns. As shown in Fig. 3A and C and in agreement with the microarray data, most photosynthesis genes analyzed by qRT-PCR were consistently downregulated, with the exception of LHCSR1 (encoding a light-induced chlorophyll a/b binding protein), LHCBM9 (both strongly upregulated in all time points), and the gene encoding the chloroplast Rieske Fe-S precursor protein (PETC; T6 only). The upregulation of LHCBM9 was further investigated (see below). Other repressed genes include isocitrate lyase (ICL), light-harvesting gene translation suppressor (NAB1), and the scaffold attachment region DNA binding protein (SDB), as shown in Fig. 3H. Genes that were notably induced include, as expected, the hydrogenase-encoding genes HYDA1, HYDA2, and HYDEF as well as genes encoding a putative glycerol-3-phosphate acyltransferase (GPAT) involved in glycerol lipid biosynthesis, the first step in phospholipid biosynthesis. Also induced were TPR, PFL, the gene encoding the variable flagellar number protein VFL, and an unknown gene termed “Unknown 2” (Fig. 3E to G). Apart from LHCBM9, strong induction (up to several thousand-fold) throughout the time course experiment was found for the genes encoding arylsulfatase ARS2, the extracellular 88-kDa polypeptide ECP88, and calcium-dependent calmodulin kinase CAMK4 (not in T1), suggesting that these genes may play important roles during S starvation and PBHP (Fig. 3F). For the same genes, the microarray results display the same induction trend although at a much lower magnitude (Table 1, sulfur assimilation). A gene termed “Unknown 7” and genes encoding the coproporphyrinogen III oxidase precursor CPX1 and a putative NADPH oxidoreductase (OXR) showed a strong induction from T3 onwards when anaerobiosis started (Fig. 3D), making them potential targets for hydrogen production improvements. Genes that were differentially regulated in an antagonistic fashion during the early S starvation (T1 and T2) and the late-hydrogen-production phase (T5 and T6) include a lipoxygenase (LOX), cytochrome b6f complex small subunit PETG and PETC as well as an unknown gene (“Unknown 4”); these were generally repressed in T1 to T4 and then induced in T5 and/or T6 (Fig. 3B and G). The opposite expression pattern (early induction followed by repression during H2 production) was observed for a range of genes, notably those encoding alternative oxidase 1 (AOX1), cytochrome c oxidase subunit II (COX2A), glycogen/starch phosphorylase (GLPV), and the gene encoding alpha amylase 2 (AMYA2) (repressed during hydrogen production but induced in earlier time points) (Fig. 3I). Modification of these genes, together with genes listed under transcription and translation in Table 1, including regulatory genes, may also be suitable for further improvements of PBHP.
PSII light-harvesting complex is remodeled during S deprivation and PBHP.
Our microarray analyses showed a strong increase of LHCBM9 transcripts before (T2 and T3) and during PBHP with induction ratios up to 109-fold (mid-H2 production; T4). To further investigate this, we carried out detailed expression profiling of LHCBM-encoding genes, followed by biochemical analyses of LHCBM isoforms. The Lhc family of light-harvesting proteins in C. reinhardtii, which capture the light required for normal photosynthesis and PBHP, consists of 11 highly conserved proteins (LHCBM1 to LHCBM6, -8, -9, and -11; LHCBM4 and LHCBM5) that are predominantly associated with PSII (10), and nine proteins (LHCA1 to LHCA9) that are more closely associated with PSI (43). Although the LHC genes and proteins share a high degree of sequence similarity, using specific primers designed in the 5′ untranslated region, qRT-PCR allowed us to distinguish individual members of this gene family (35). Results obtained by qRT-PCR confirmed that LHCBM9 was massively induced (up to 3,000-fold) immediately after S deprivation (T1) and then decreased but maintained high levels along the course of PBHP (Fig. 3A). While LHCBM9 was highly induced throughout the experiment, there was strong suppression of the other genes encoding LHCBM isoforms (Fig. 3A). Interestingly, under normal conditions, LHCBM9 transcript abundance was very low (∼0.25% of that of LHCBM1). However, during S deprivation, its abundance rose to as high as twice the combined abundance of all the other LHCBM-encoding genes under normal conditions (data not shown).
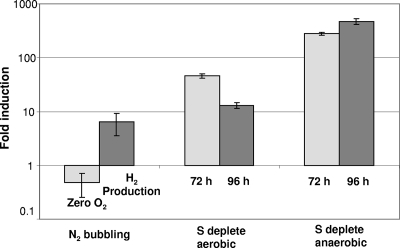
To test whether remodeling of the antenna system of PSII was linked not only to S deprivation but also to anaerobiosis or PBHP, we induced PBHP solely by anaerobiosis using N2 bubbling (“microoxic H2 production”) rather than by S starvation. In addition, we carried out S deprivation under aerobic and anaerobic conditions. In this experiment, transcriptional regulation of LHCBM9 in cells of C. reinhardtii wild-type strain CC124 was analyzed under the following three conditions: (i) anaerobicity was induced by purging the culture with N2; (ii) cells were deprived of sulfur but compressed air was bubbled to keep the culture oxygenated; and (iii) cells were deprived of sulfur, the culture became anaerobic, and H2 production was induced. As shown in Fig. 4, LHCBM9 induction (sixfold) was also observed during PBHP, following anaerobiosis independently of S starvation. It was also about 45-fold induced at 72 h following aerobic sulfur deprivation and up to 470-fold during anaerobic sulfur deprivation. This suggests that LHCBM9 regulation depends on both sulfur deprivation and the microoxic H2 production process. Although LHCBM9 transcript levels increased during anaerobic H2 production conditions induced by N2 bubbling, S deprivation had a more marked effect. The strongest upregulation was observed during anaerobic conditions induced by S depletion at all time points analyzed (Fig. 3A and 4). This result therefore suggests that although LHCBM9 is upregulated during H2 production, its induction is regulated mainly by sulfur depletion. In an independent time course experiment under aerobic conditions, LHCBM9 showed initial induction at 12 h, as expected, as a response to sulfur deprivation; however, this was followed by a steady decrease in transcript abundance until 96 h (data not shown).
FIG. 4.
Induction ratios of LHCBM9 in C. reinhardtii CC124 following anaerobiosis by N2 bubbling in comparison to sulfur depletion under aerobic or anaerobic conditions. N2 bubbling, transcripts measured at zero O2 production and under anaerobic H2 production conditions; S deplete aerobic, S-depleted cultures bubbled with air to maintain aerobic conditions; S deplete anaerobic, anaerobiosis (microoxic conditions) induced by S depletion. Transcript levels were determined by qRT-PCR and compared to T0 from two to three independent biological replicates (shown are averages ± the standard error; hours show time after S depletion).
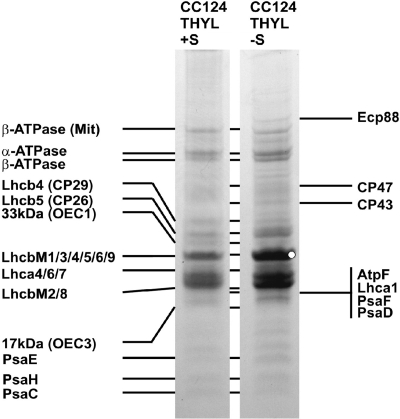
To directly analyze the protein composition of the C. reinhardtii Lhc family of light-harvesting proteins under normal and under S starvation-induced PBHP conditions, we isolated thylakoid membrane fractions (same samples as those shown in Fig. 4) and subjected the thylakoid proteins to one-dimensional SDS-PAGE. As shown in Fig. 5, a difference in intensity could be observed for proteins around 27 kDa. To further identify the proteins, bands corresponding to C. reinhardtii before/after S thylakoid membrane proteins were excised, digested with trypsin, and analyzed by MS-MS. The proteins that were identified with a confidence greater than 95% are listed in Fig. 5, most notably LHCBM9 and ECP88, which were identified only in thylakoid fractions after S induction. This confirms the marked upregulation of expression for both genes observed in the microarray and qRT-PCR studies. Although the homology between LHCBM proteins is very high (66 to 95% amino acid sequence identity, with an average of 77% (43), the differentiation between LHCBM9 and other LHCBM proteins was possible thanks to a specific precursor ion which is unique to LHCBM9, which has an observed mass of 2,540.25 Da. Among the SDS-PAGE-resolved proteins, LHCBM isoforms were by far the most dominant, followed by α and β subunits of ATP synthase.
FIG. 5.
Identification of C. reinhardtii CC124 thylakoid proteins before (+ S) and after (− S) induction of photobiological hydrogen production by sulfur deprivation. Shown are protein bands with corresponding names after size separation by 10% SDS-PAGE. All proteins were identified in both fractions (indicated by the connecting lines between both lanes) except for LhcbM9 and Ecp88, which were identified only under conditions of S deprivation. Note that LhcbM9 contributed to the band labeled with the white dot. Proteins were identified by MS-MS or by selective extraction from thylakoid membranes (17-kDa and 33-kDa extrinsic proteins).
DISCUSSION
Following microarray experiments and stringent data analysis, we were able to confidently measure the regulation of 7,124 transcripts from C. reinhardtii CC124 during sulfur deprivation at five time points: peak O2 production, mid-O2 production (consumption), zero O2 production, mid-H2 production, and peak H2 production. Among the characterized genes, 166 showed more than a twofold change in transcript abundance. Out of these, 18 genes were confirmed by qRT-PCR, 4 were not differentially regulated, and 8 did not confirm the microarray results (Fig. 3; Table 1; see Table S1 in the supplemental material). Different results obtained by both methods are likely to be caused by weak signals and cross-hybridization events to other genes, a common problem with microarray hybridizations that are carried out at relatively low temperatures (5, 14, 18, 23, 41), suggesting that further confirmations by qRT-PCR may be required for certain genes of interest. Nevertheless, our microarray data from three biological replicates that passed all stringent criteria for data analysis, including Student's t test, will provide a useful platform for gene discovery in C. reinhardtii during PBHP. The Blast search homology matches in GenBank provide a guide for putative functions of C. reinhardtii genes (listed in this paper after their closest match), and the following paragraphs discuss the possible roles of genes during PBHP, keeping in mind that further experimentation may be required to determine the exact functions.
Expression patterns in C. reinhardtii cells during initial phases of PBHP reveal commonalities to responses to sulfur stress.
A large portion of the expression signature presented here shows overlaps to results from S starvation experiments (51), suggesting that S starvation presents a main factor for gene expression changes in our experiment. PBHP in C. reinhardtii using sulfur depletion is a complex process. From the start of the experiment to peak H2 production, C. reinhardtii cells undergo two major physiological changes, S starvation and then anaerobiosis. In the first two phases (oxygen evolution phase and oxygen consumption phase), both the cell's structure and its metabolic system must adapt to the lack of sulfur supply. Consequently, the cell has to conserve S by minimizing usage and maximizing scavenging efforts. Such adaptation has been studied in detail (37, 42, 51). Evidence at the transcript level and some evidence at the protein level have shown that during this phase, major cellular activities such as photosynthesis and protein synthesis decrease, while a significant increase in the abundance of transcripts encoding stress-responsive proteins was observed. A steep decline in photosynthetic activity at 20 h after sulfur deprivation was confirmed by pulse amplitude modulation measurements in four independent PBRs (data not shown). The following three phases (anaerobic phase, hydrogen production phase, and termination phase) have not been analyzed for transcriptional changes until now. During these phases, in addition to S starvation, cells also suffer a lack of oxygen. As a result, carbon metabolism shifts from respiration to fermentation and reduced hydrogen is released as H2 gas to circumvent NAD(P)+ reduction (17). Therefore, a large number of sulfur starvation and anoxia-responsive genes mask the process of hydrogen production. The results showed that photosynthetic activities were reduced under S starvation. With a few exceptions, the transcript level of most photosynthesis genes decreased significantly throughout the first 40 h (Fig. 3A and C). Previous studies showed that the concentration of functional PSII (QA), cytochrome b6f, and PSI (P700) as well as the chlorophyll concentration in cultures undergoing PBHP decreased steadily after the initiation of S deprivation (30). In contrast to those of most photosynthesis genes, transcripts of C. reinhardtii sulfur assimilation genes increased significantly during PBHP in response to the lack of sulfur in the medium to increase the capacity for S scavenging (37, 42, 51). All transcripts encoding sulfur assimilation genes presented in this study have been described as sulfur acclimation-specific genes (37). Our data show that cells undergoing PBHP also display a downregulation of genes required for amino acid synthesis and an upregulation of genes involved in protein degradation, as expected for cells that do not multiply but maintain only their basic functions for survival. This is in agreement with Melis et al. (30), who reported that under PBHP conditions, the protein concentration of the culture decreased steadily over time.
Changes in carbon metabolism.
From the beginning of S deprivation, carbon metabolism in C. reinhardtii changed markedly. Our results indicate a strong suppression of the glyoxylate and Calvin cycles. Both cycles are coordinately regulated at the enzyme level to balance the energy-yielding metabolism and glucogenesis from acetate. In one glyoxylate cycle, one succinate and one NADH are produced for every two acetyl coenzyme A (acetyl-CoA) and one NAD+. When enough energy is produced through the glycolytic and citric acid cycles, acetate is funneled into the glyoxylate cycle for the production of glucose, amino acids, or nucleotides. Although reducing power is generated as NADH, probably the process is not favorable due to the lack of energy for synthesizing acetyl-CoA and due to the decreased demand for succinate under S-depleted conditions. It is expected that both cycles are downregulated during PBHP, since energy supply from respiration is very limited toward the hydrogen production phase of PBHP (29). In contrast, the pentose phosphate pathway was induced. Such changes are expected because the glyoxylate cycle functions only when the cell has enough energy from glycolysis and respiration, which is not the case under microoxic or anaerobic conditions due to S starvation. Although acetate is ubiquitous in the S-deprived culture, acetyl-CoA is not readily available for the glyoxylate cycle. The following conversion of acetate to acetyl-CoA catalyzed by acetyl-CoA synthase enzyme requires ATP: acetate + ATP + CoA → acetyl-CoA + ADP.
The pentose phosphate pathway produces two NADPH, one CO2, and one ribose-5-phosphate from one glucose-6-phosphate. The five-carbon sugar can be used for nucleotide synthesis or recycled into glucose-6-phosphate. In the latter case, the pentose phosphate can provide protection from oxidative damage to the cell by recycling the five-carbon-sugar product ribose-5-phosphate. This generates primarily NADPH, whose oxidative damage protection role is rather well known (e.g., in erythrocytes). S starvation studies have shown that cells suffer from oxidative stress under these conditions (51). Moreover, the NADPH produced from the pentose phosphate pathway potentially can feed electrons into the chloroplast electron transport chain (ETC) via one of the putative NAD(P)H dehydrogenases for hydrogen production and therefore maintains ATP production (34). An increase in transcript abundance of phosphogluconate dehydrogenase was also reported in S-deprived culture (51). The increase in the pentose phosphate pathway indicates that C. reinhardtii cells already suffer from redox stress at the beginning of S starvation. Although no microarray data were available for the alternative oxidase genes, qRT-PCR data showed a substantial increase in the transcript abundance of AOX1 at peak and mid-O2 production levels before gradually decreasing to below that of the control level at mid-H2 and zero H2 production, indicating that C. reinhardtii cells were under redox stress during the initial phases of the experiment (Fig. 3I). Zhang et al. (51) reported that AOX1 was repressed after 24 h of S deprivation (their first point of measurement), which is earlier than that in our experiment. This may be attributed to slightly different conditions used in our experimental setup that led to more severe stress (gas-tight conditions and sulfur depletion under high-light conditions). During the oxygen evolution phase, the rate of photosynthetic O2 evolution rapidly slows down. At peak O2 levels where sample T1 was harvested, the respiration rate began to exceed that of O2 evolution, causing O2 shortage and redox stress in the photosynthetic ETC (Fig. 2). More evidence came from the increase in the transcript abundance of a gene similar to the Yersinia pestis hydroxylamine reductase (HCP)-encoding gene soon after the initiation of S deprivation (Table 1). HCP has been reported to increase under redox stress (1), and more recently, HCP was shown to be strongly induced shortly after the initiation of dark anaerobiosis in C. reinhardtii (33). There was also evidence that fermentative pathways increased from the start of anaerobiosis. Our data show that the transcript encoding formate acetyltransferase (PFL) increased substantially at T0 O2 and mid-H2 production. At those points, the culture had used up all the DO available for respiration. Consequently, fermentation was the only way to generate ATP at the substrate level, with an additional mechanism being through cyclic electron transport. By catalyzing the conversion of pyruvate to formate, ethanol, and acetate with the production of ATP, PFL helps the cell to generate energy without the requirement of O2. This result agrees with previous studies of the role of the PFL pathway in C. reinhardtii carbon metabolism under anoxic conditions (3, 17).
Surprisingly, our qRT-PCR data showed that transcripts encoding starch degradation enzymes starch phosphorylase (GLPV) and alpha amylase (AMY2) were upregulated very early after S depletion, when starch accumulation patterns are normally observed. Prevailing evidence at the substrate level demonstrated that C. reinhardtii cells accumulate starch in the early phase (20 to 30 h) of both S starvation only and in combination with closed conditions for H2 production (4, 11, 21, 30, 44, 49, 50). Similar results to ours were also observed in at least two other shorter time course studies of sulfur (24 h) and phosphate (48 h) deprivation (32, 51). In these studies, genes encoding starch phosphorylase and alpha amylase were significantly upregulated at the same time as the gene encoding the granule-bound starch synthesizing enzyme starch synthase (STA2). Our data further show that GLPV (PHOA) and AMYA2 were suppressed later during H2 production (Fig. 3I) when starch is consumed (21). While transcriptional regulation may not always be linked to enzymatic regulation, Moseley et al. (32) suggested that starch degradation was required for starch synthesis. Another possible explanation for the spatial separation of simultaneous degradation and synthesis of starch can also be excluded, since starch-degrading activities in C. reinhardtii are located solely in the chloroplast (19, 26), unlike in many higher plants where it also occurs in the cytosol. This may suggest that GLPV and AMYA2 are involved in starch accumulation rather than degradation. Supporting evidence from Yu et al. (48) showed that alpha amylase was not required for transitory starch breakdown in Arabidopsis spp. leaves, while a more recent study of starch phosphorylase postulated that PHOA, which is identical to GLPV, also has starch synthase activities (7).
The trigger for HYDA transcription is not just anaerobiosis.
Although anaerobiosis is a requirement for hydrogenase activity, it is not the only trigger for HYDA transcription. Our qRT-PCR results clearly show that HYDA transcription was induced soon after S starvation, long before anaerobiosis in the medium was established, although anaerobiosis did enhance the transcription of HYDA. After the first 16 h of our experiment (peak O2 and mid-O2 production), the DO levels were at ∼9 mg liter−1, which is far above the induction level for hydrogenases reported by others, although fully aerobic sulfur-deprived culture showed repression of HYDA transcription during the first 24 h (51). Therefore, it is possible that the induction of HYDA1 and HYDA2 was influenced by the effect of higher levels of light in our experiment (250 to 450 μE compared to only 100 μE used by Zhang et al. [51]).
Remodeling of PSII during S-deprived PBHP.
It is not surprising that a large number of differentially expressed genes can be classified as S starvation responsive. This group of genes is either constitutively induced or suppressed throughout the experiment. Belonging to this group are most of the photosynthesis, sulfur assimilation, and acquiring genes and some of the genes involved in carbon metabolism, protein synthesis, and degradation (Table 1; Fig. 3). To this end, our data confirm previous findings about the impacts of S starvation on the activities of photosynthesis, S scavenging, protein degradation, and synthesis in C. reinhardtii (51). Furthermore, we provide strong evidence of the remodeling of PSII-LHC where LHCBM9 and LHCSR-encoding genes were constitutively induced up to thousands-fold in spite of the dramatic repression of genes encoding other LHCBM isoforms. This was also confirmed by our biochemical analyses of LHCBM isoforms (Fig. 5). The induction of different LHCSR isoforms is in agreement with previous studies which suggest that LHCSR proteins protect the cell from photodamage induced by low-CO2 and high-light conditions (10, 39). The massive induction of LHCBM9 against other LHCBM isoforms is more surprising. They share a striking number of nucleotide and polypeptide similarities, but their individual functions are not known. Confirmed by the biochemical analyses of the PSII-LHC, our data suggest that LHCBM9 becomes the dominant LHCBM isoform during Lhc remodeling under S starvation. This could be explained by the fact that LHCBM9 displays the smallest S contents (only four atoms) compared to all other LHCBM proteins that contain either six or seven S atoms each.
Remodeling of light-harvesting complexes, for example via the state transition process, is a common adaption of photosynthetic organisms in response to environmental changes (36), in particular to optimize light energy capture and distribution to PSI and PSII and dissipation of excess energy to minimize photooxidative damage to the PSII reaction centers. For example, under high-light conditions, LHCII proteins can be phosphorylated, causing them to dissociate from PSII and to migrate to PSI where they are functionally integrated. By using pulse amplitude modulation to obtain fluorescence yield measurements, Melis (28) and Wykoff et al. (47) showed that the maximal fluorescence (Fmax) of S-starved cells was only 76% that of unstarved cells, indicating a larger amount of light energy absorbed by LHCII was dissipated as heat in starved cells. However, it is unlikely that LHCM9 replaces other LHCBM polypeptides to increase the energy dissipation capacity, as this can probably be better achieved by reducing the antenna size. Moreover, LHCBM9 was found to be downregulated under CO2-limiting conditions (31), suggesting it might actively contribute to photosynthesis. As LHCBM9 has the least number of sulfur-containing amino acids of all the major light-harvesting proteins of PSII and the relative abundance of LHCBM9 transcript under S starvation was about one to two times the abundance of all other LHCBM-encoding transcripts combined under normal conditions, one could suggest that under S-depleted conditions, other major light-harvesting complex proteins are replaced by LHCBM9 to compensate for the sulfur shortage. According to Turkina et al. (45), LHCBM9 contains two phosphorylation sites, which both become phosphorylated under high-light conditions. From our additional study of the regulation of LHCBM9 transcription by sulfur alone or anaerobiosis, which shows that further activation of LHCBM9 occurred under anaerobic conditions, one could also suggest that LHCBM9 plays a role in state transition and/or in the cyclic electron flow that takes place under anaerobic conditions including light. However, despite of the overall reduction in photosynthesis due to S starvation, part of this system remains functional or even slightly upregulated during the H2 production phase. As indicated by the qRT-PCR and the microarray results, a number of photosynthesis genes were actually upregulated during the course of PBHP (PRK, LHCBM9, LHCSR1, and LHCSR3). Such a selective maintenance indicates that not only the light-harvesting system but the whole photosynthesis system are remodeled in response to S starvation and later to hydrogen production. Photosystems do not simply decline but undergo significant transformations to prepare for hydrogen production. Further studies are required to characterize the unknown target genes and to compare the expression of identified genes at the protein level.
Supplementary Material
Acknowledgments
We thank Elizabeth Harris (Duke University) for providing the C. reinhardtii strain CC-137 and Arthor Grossman (Standford University) for providing the microarrays. We are also grateful to Bernie Degnan for the usage of the GenePix scanner and Rosanne Casu (University of Queensland, Australia) for the help with microarray data analysis and Alun Jones (University of Queensland, Australia) for providing us with advice and material for MS-MS analysis.
Footnotes
Published ahead of print on 15 August 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Almeida, C. C., C. V. Romao, P. F. Lindley, M. Teixeira, and L. M. Saraiva. 2006. The role of the hybrid cluster protein in oxidative stress defense. J. Biol. Chem. 28132445-32450. [DOI] [PubMed] [Google Scholar]
- 2.Arnon, D. I. 1949. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 241-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atteia, A., R. van Lis, G. Gelius-Dietrich, A. Adrait, J. Garin, J. Joyard, N. Rolland, and W. Martin. 2006. Pyruvate formate-lyase and a novel route of eukaryotic ATP synthesis in Chlamydomonas mitochondria. J. Biol. Chem. 2819909-9918. [DOI] [PubMed] [Google Scholar]
- 4.Ball, S. G., L. Dirick, A. Decq, J. C. Martiat, and R. F. Matagne. 1990. Physiology of starch storage in the monocellular alga Chlamydomonas reinhardtii. Plant Sci. 661-9. [Google Scholar]
- 5.Chapman, S., P. Schenk, K. Kazan, and J. Manners. 2002. Using biplots to interpret gene expression patterns in plants. Bioinformatics 18202-204. [DOI] [PubMed] [Google Scholar]
- 6.Chisti, Y. 2007. Biodiesel from microalgae. Biotechnol. Adv. 25294-306. [DOI] [PubMed] [Google Scholar]
- 7.Dauvillee, D., V. Chochois, M. Steup, S. Haebel, N. Eckermann, G. Ritte, J. P. Ral, C. Colleoni, G. Hicks, F. Wattebled, P. Deschamps, C. d'Hulst, L. Lienard, L. Cournac, J. L. Putaux, D. Dupeyre, and S. G. Ball. 2006. Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J. 48274-285. [DOI] [PubMed] [Google Scholar]
- 8.Doebbe, A., J. Rupprecht, J. Beckmann, J. H. Mussgnug, A. Hallmann, B. Hankamer, and O. Kruse. 2007. Functional integration of the HUP1 hexose symporter gene into the genome of C. reinhardtii: impacts on biological H2 production. J. Biotechnol. 13127-33. [DOI] [PubMed] [Google Scholar]
- 9.Eberhard, S., M. Jain, C. S. Im, S. Pollock, J. Shrager, Y. A. Lin, A. S. Peek, and A. R. Grossman. 2006. Generation of an oligonucleotide array for analysis of gene expression in Chlamydomonas reinhardtii. Curr. Genet. 49106-124. [DOI] [PubMed] [Google Scholar]
- 10.Elrad, D., and A. R. Grossman. 2004. A genome's-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii. Curr. Genet. 4561-75. [DOI] [PubMed] [Google Scholar]
- 11.Fouchard, S., A. Hemschemeier, A. Caruana, K. Pruvost, J. Legrand, T. Happe, G. Peltier, and L. Cournac. 2005. Autotrophic and mixotrophic hydrogen photoproduction in sulfur-deprived Chlamydomonas cells. Appl. Environ. Microbiol. 716199-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghirardi, M. L., R. K. Togasaki, and M. Seibert. 1997. Oxygen sensitivity of algal H2-production. Appl. Biochem. Biotechnol. 63-65141-151. [DOI] [PubMed] [Google Scholar]
- 13.Ghirardi, M. L., J. P. Zhang, J. W. Lee, T. Flynn, M. Seibert, E. Greenbaum, and A. Melis. 2000. Microalgae: a green source of renewable H2. Trends Biotechnol. 18506-511. [DOI] [PubMed] [Google Scholar]
- 14.Han, T., C. D. Melvin, L. M. Shi, W. S. Branham, C. L. Moland, P. S. Pine, K. L. Thompson, and J. C. Fuscoe. 2006. Improvement in the reproducibility and accuracy of DNA microarray quantification by optimizing hybridization conditions. BMC Bioinformatics 7(Suppl. 2)S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Happe, T., and J. D. Naber. 1993. Isolation, characterization and N-terminal amino-acid sequence of hydrogenase from the green-alga Chlamydomonas reinhardtii. Eur. J. Biochem. 214475-481. [DOI] [PubMed] [Google Scholar]
- 16.Healey, F. P. 1970. Mechanism of hydrogen evolution by Chlamydomonas moewusii. Plant Physiol. 45153-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemschemeier, A., and T. Happe. 2005. The exceptional photofermentative hydrogen metabolism of the green alga Chlamydomonas reinhardtii. Biochem. Soc. Trans. 3339-41. [DOI] [PubMed] [Google Scholar]
- 18.Kazan, K., P. M. Schenk, I. Wilson, and J. M. Manners. 2001. DNA microarrays: new tools in the analysis of plant defence responses. Mol. Plant Pathol. 2177-185. [DOI] [PubMed] [Google Scholar]
- 19.Klein, U. 1986. Compartmentation of glycolysis and of the oxidative pentose-phosphate pathway in Chlamydomonas reinhardtii. Planta 16781-86. [DOI] [PubMed] [Google Scholar]
- 20.Klein, U., and A. Betz. 1978. Fermentative metabolism of hydrogen-evolving Chlamydomonas moewusii. Plant Physiol. 61953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosourov, S., E. Patrusheva, M. L. Ghirardi, M. Seibert, and A. Tsygankov. 2007. A comparison of hydrogen photoproduction by sulfur-deprived Chlamydomonas reinhardtii under different growth conditions. J. Biotechnol. 128776-787. [DOI] [PubMed] [Google Scholar]
- 22.Kosourov, S., M. Seibert, and M. L. Ghirardi. 2003. Effects of extracellular pH on the metabolic pathways in sulfur-deprived, H2-producing Chlamydomonas reinhardtii cultures. Plant Cell Physiol. 44146-155. [DOI] [PubMed] [Google Scholar]
- 23.Kothapalli, R., S. J. Yoder, S. Mane, and T. P. Loughran, Jr. 2002. Microarray results: how accurate are they? BMC Bioinformatics 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruse, O., J. Rupprecht, K. P. Bader, S. Thomas-Hall, P. M. Schenk, G. Finazzi, and B. Hankamer. 2005. Improved photobiological H2 production in engineered green algal cells. J. Biol. Chem. 28034170-34177. [DOI] [PubMed] [Google Scholar]
- 25.Lee, J. W., and E. Greenbaum. 2003. A new oxygen sensitivity and its potential application in photosynthetic H2 production. Appl. Biochem. Biotechnol. 105303-313. [DOI] [PubMed] [Google Scholar]
- 26.Levi, C., and M. Gibbs. 1984. Starch degradation in synchronously grown Chlamydomonas reinhardtii and characterization of the amylase. Plant Physiol. 74459-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGrath, K. C., B. Dombrecht, J. M. Manners, P. M. Schenk, C. I. Edgar, D. J. Maclean, W. R. Scheible, M. K. Udvardi, and K. Kazan. 2005. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139949-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melis, A. 2002. Green alga hydrogen production: progress, challenges and prospects. Int. J. Hydrogen Energy 271217-1228. [Google Scholar]
- 29.Melis, A., and T. Happe. 2001. Hydrogen production. Green algae as a source of energy. Plant Physiol. 127740-748. [PMC free article] [PubMed] [Google Scholar]
- 30.Melis, A., L. P. Zhang, M. Forestier, M. L. Ghirardi, and M. Seibert. 2000. Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 122127-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miura, K., T. Yamano, S. Yoshioka, T. Kohinata, Y. Inoue, F. Taniguchi, E. Asamizu, Y. Nakamura, S. Tabata, K. Yamato, K. Ohyama, and H. Fukuzawa. 2004. Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol. 1351595-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moseley, J. L., C. W. Chang, and A. R. Grossman. 2006. Genome-based approaches to understanding phosphorus deprivation responses and PSR1 control in Chlamydomonas reinhardtii. Eukaryot. Cell 526-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mus, F., A. Dubini, M. Seibert, M. C. Posewitz, and A. R. Grossman. 2007. Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 28225475-25486. [DOI] [PubMed] [Google Scholar]
- 34.Mus, F., L. Cournac, W. Cardettini, A. Caruana, and G. Peltier. 2005. Inhibitor studies on non-photochemical plastoquinone reduction and H2 photoproduction in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1708322-332. [DOI] [PubMed] [Google Scholar]
- 35.Mussgnug, J. H., S. Thomas-Hall, J. Rupprecht, A. Foo, V. Klassen, A. McDowall, P. M. Schenk, O. Kruse, and B. Hankamer. 2007. Engineering photosynthetic light capture: impacts on improved solar energy to biomass conversion. Plant Biotechnol. J. 5802-814. [DOI] [PubMed] [Google Scholar]
- 36.Nield, J., K. Redding, and M. Hippler. 2004. Remodeling of light-harvesting protein complexes in Chlamydomonas in response to environmental changes. Eukaryot. Cell 31370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollock, S. V., W. Pootakham, N. Shibagaki, J. L. Moseley, and A. R. Grossman. 2005. Insights into the acclimation of Chlamydomonas reinhardtii to sulfur deprivation. Photosynthesis Res. 86475-489. [DOI] [PubMed] [Google Scholar]
- 38.Posewitz, M. C., S. L. Smolinski, S. Kanakagiri, A. Melis, M. Seibert, and M. L. Ghirardi. 2004. Hydrogen photoproduction is attenuated by disruption of an isoamylase gene in Chlamydomonas reinhardtii. Plant Cell 162151-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richard, C., H. Ouellet, and M. Guertin. 2000. Characterization of the LI818 polypeptide from the green unicellular alga Chlamydomonas reinhardtii. Plant Mol. Biol. 42303-316. [DOI] [PubMed] [Google Scholar]
- 40.Savard, F., C. Richard, and M. Guertin. 1996. The Chlamydomonas reinhardtii LI818 gene represents a distant relative of the CABI/II genes that is regulated during the cell cycle and in response to illumination. Plant Mol. Biol. 32461-473. [DOI] [PubMed] [Google Scholar]
- 41.Schenk, P. M., K. Kazan, J. M. Manners, J. P. Anderson, R. S. Simpson, I. W. Wilson, S. C. Somerville, and D. J. Maclean. 2003. Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiol. 132999-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi, H., C. E. Braby, and A. R. Grossman. 2001. Sulfur economy and cell wall biosynthesis during sulfur limitation of Chlamydomonas reinhardtii. Plant Physiol. 127665-673. [PMC free article] [PubMed] [Google Scholar]
- 43.Tokutsu, R., H. Teramoto, Y. Takahashi, T. A. Ono, and J. Minagawa. 2004. The light-harvesting complex of photosystem I in Chlamydomonas reinhardtii: protein composition, gene structures and phylogenic implications. Plant. Cell Physiol. 45138-145. [DOI] [PubMed] [Google Scholar]
- 44.Tsygankov, A., S. Kosourov, M. Seibert, and M. L. Ghirardi. 2002. Hydrogen photoproduction under continuous illumination by sulfur-deprived, synchronous Chlamydomonas reinhardtii cultures. Int. J. Hydrogen Energy 271239-1244. [Google Scholar]
- 45.Turkina, M. V., J. Kargul, A. Blanco-Rivero, A. Villarejo, J. Barber, and A. V. Vener. 2006. Environmentally modulated phosphoproteome of photosynthetic membranes in the green alga Chlamydomonas reinhardtii. Mol. Cell. Proteomics 51412-1425. [DOI] [PubMed] [Google Scholar]
- 46.White, A. L., and A. Melis. 2006. Biochemistry of hydrogen metabolism in Chlamydomonas reinhardtii wild type and a Rubisco-less mutant. Int. J. Hydrogen Energy 31455-464. [Google Scholar]
- 47.Wykoff, D. D., J. P. Davies, A. Melis, and A. R. Grossman. 1998. The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117129-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, T. S., S. C. Zeeman, D. Thorneycroft, D. C. Fulton, H. Dunstan, W. L. Lue, B. Hegemann, S. Y. Tung, T. Umemoto, A. Chapple, D. L. Tsai, S. M. Wang, A. M. Smith, J. Chen, and S. M. Smith. 2005. Alpha-amylase is not required for breakdown of transitory starch in Arabidopsis leaves. J. Biol. Chem. 2809773-9779. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, L. P., T. Happe, and A. Melis. 2002. Biochemical and morphological characterization of sulfur-deprived and H2-producing Chlamydomonas reinhardtii (green alga). Planta 214552-561. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, L. P., and A. Melis. 2002. Probing green algal hydrogen production. Philos. Trans. R. Soc. Lond. B 3571499-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, Z. D., J. Shrager, M. Jain, C. W. Chang, O. Vallon, and A. R. Grossman. 2004. Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot. Cell 31331-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.