Abstract
The yeast (Saccharomyces cerevisiae) Snf7 family consists of six highly charged, coiled-coil-forming proteins involved in multivesicular body (MVB) formation. Although all proteins perform a common function at late endosomes, individual mutants also show distinct phenotypes. This suggests that Snf7 homologues have additional functions separate from their role in MVB formation. In this report, we explored the molecular basis for the sucrose-nonfermenting phenotype of snf7 mutants. Our Northern blotting experiments provide evidence that Snf7 is involved in the regulation of SUC2 transcription. The Snf7-dependent regulation of SUC2 transcription does not appear to involve the transcription factor Mig1, since Mig1 phosphorylation after glucose derepression was not affected in a Δsnf7 mutant. Instead, we show that Snf7 influences SUC2 expression by regulating the level of the transcription factor Nrg1. Snf7 exerts its effects on Nrg1 levels through activation of the transcription factor Rim101, which is part of the yeast alkaline response pathway (“Rim101 pathway”). This is supported by the findings that deletion of RIM101 or overexpression of NRG1 from the ADH1 promoter leads to the same SUC2 expression level as deletion of SNF7. In addition, deletion of other components of the Rim101 pathway, like RIM13 and RIM20, led to the same growth phenotype on raffinose media as deletion of SNF7. Furthermore, Snf7 turned out to be dispensable for SUC2 expression in an NRG1 deletion background. Thus, the effects of Snf7 on SUC2 expression can be completely accounted for by its effect on Nrg1 levels.
The class E vacuolar protein sorting (Vps) proteins are involved in the formation of vesicles that bud into the interior of late endosomes forming a multivesicular body (MVB) (18). Endocytic cargo is incorporated into these vesicles. When the MVB fuses with the vacuole, the vesicles are released into the vacuolar lumen and proteins contained in the vesicles are degraded. Class E proteins have been grouped into three protein complexes, ESCRT-I, -II and -III, associated with the endosomal membrane (22). It is thought that these complexes act consecutively in cargo recruitment and MVB vesicle formation. The ESCRT-III complex consists of four proteins: Did4/Vps2, Snf7/Vps32, Vps20, and Vps24/Did3. The ESCRT-III proteins are part of a protein family of highly charged coiled-coil-forming proteins with six members in yeasts (1, 6, 8). The two members of this family (Did2/Vps46, Mos10/Vps60) that have not been mapped to ESCRT-III are class E proteins as well and are also required for late endosome function.
Here, we focus on the Snf7 protein. Originally, this protein was identified in a screen for mutants defective for growth on sucrose as a carbon source (snf represents sucrose nonfermenting) (15). The enzyme invertase encoded by the SUC2 gene is required for sucrose utilization. SUC2 is expressed under low-glucose conditions and repressed by high glucose. Most of the snf mutants isolated in the screen turned out to be defective for invertase derepression under low-glucose conditions. Two classes of Snf proteins could be distinguished: proteins involved in glucose signaling (Snf1 and -4) (7) and proteins that are part of the Swi-Snf chromatin-remodeling complex (Snf2, -5, -6, and -11) (29). Two additional Snf proteins, Snf7 and Snf8/Vps22 (a component of ESCRT-II), are found among the collection of class E Vps functions. How the snf phenotype is related to the endosomal function of these proteins is unclear.
By genome-wide screening (mostly two-hybrid screens), several proteins interacting with Snf7 have been identified. One interaction partner is the class E Vps protein Bro1/Vps31, which appears to act late in the MVB pathway, probably after or at the level of ESCRT-III (17, 19). Bro1 seems to coordinate the deubiquitination of endocytic cargo proteins by recruiting the deubiquitinating enzyme Doa4 to endosomes (13). Another interaction partner is the Bro1 homologue Rim20. Rim20 is one of the components of the Rim101 pathway that is involved in the transcriptional response to alkaline pH (30). Despite the homology to Bro1, Rim20 does not appear to have an endosomal function. In addition to Rim20, a further component of the Rim101 pathway, Rim13, a calpain-like cysteine protease, interacts with Snf7. Further interaction partners are Vps20, which forms a subcomplex of ESCRT-III with Snf7, and Vps4, an AAA ATPase involved in disassembly of ESCRT complexes at endosomes (3).
The six proteins of the Snf7 family are all required for late endosome function, yet mutants of individual members of this family have distinct phenotypes (8). This points to additional nonendosomal functions for these proteins. We analyzed the role of Snf7 in expression of the glucose-repressed SUC2 gene. We found that Snf7 affects SUC2 transcription at a step after Mig1 inactivation and mediates its effects on SUC2 via the Rim101 pathway.
MATERIALS AND METHODS
Yeast strains.
The yeast (Saccharomyces cerevisiae) strains used are listed in Table 1. Deletion strains and strains carrying tagged gene variants were derived from the wild-type strain, JD52. They were constructed by one-step gene replacement with PCR-generated cassettes (12). The deletions and insertions were verified by PCR.
TABLE 1.
Yeast strains used in this study
| Straina | Genotype | Source |
|---|---|---|
| JD52 | MATahis3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 | J. Dohmen, Cologne, Germany |
| JD116 | Δdoa4::LEU2 | J. Dohmen, Cologne, Germany |
| RKY1511 | Δvps4::HIS3 | This study |
| RKY1654 | Δmos10::HIS3 | This study |
| RKY1728 | Δdid2::HIS3 | This study |
| RKY1730 | Δvps24::HIS3 | This study |
| RKY1732 | Δdid4::HIS3 | This study |
| RKY1828 | Δdid4::kanMX Δvps24::HIS3 | This study |
| RKY1829 | Δsnf7::HIS3 Δvps20::kanMX | This study |
| RKY1835 | Δdid2::HIS3 Δmos10::kanMX | This study |
| RKY1852 | Δsnf7::TRP1 | This study |
| RKY1853 | Δvps20::TRP1 | This study |
| RKY1876 | Δvps27::kanMX | This study |
| RKY1922 | Δbro1::kanMX | This study |
| RKY2058 | MIG1::13 myc-HIS3 | This study |
| RKY2100 | Δrim20::kanMX | This study |
| RKY2102 | Δrim13::kanMX | This study |
| RKY2104 | Δsnf7::kanMX MIG1::13 myc-HIS3 | This study |
| RKY2175 | Δrim101::HIS3 | This study |
| RKY2176 | NRG1::3HA-HIS3 | This study |
| RKY2180 | Δsnf7::TRP1 NRG1::3HA-HIS3 | This study |
| RKY2206 | Δnrg1::HIS3 | This study |
| RKY2207 | Δsnf7::TRP1 Δnrg1::HIS3 | This study |
| RKY2208 | ADH1p-3HA::NRG1::HIS3 | This study |
| RKY2284 | Δrim13 NRG1::3HA-TRP1 | This study |
| RKY2285 | Δrim20 NRG1::3HA-TRP1 | This study |
| RKY2286 | Δrim101 NRG1::3HA-TRP1 | This study |
All strains were derived from JD52.
Phosphatase treatment.
Cells were grown overnight to exponential phase (optical density at 600 nm [OD600] of <0.8 [5 × 107/ml]) in YPD (yeast extract, peptone, dextrose) medium. Cells at an OD600 of 4 were pelleted and heated at 95°C for 5 min. After resuspension in 100 μl sodium dodecyl sulfate (SDS) gel sample buffer (2% SDS, 10% glycerol, 60 mM Tris-Cl [pH 6.8], 0.1% bromphenol blue, 20 mM dithiothreitol), the cells were broken by agitation with glass beads for 5 min at 4°C and then centrifuged for 5 min at 500 × g to remove cell debris. The supernatant was diluted 1:5 with 60 mM Tris-Cl (pH 6.8). An aliquot corresponding to an OD600 of 2 of cells was incubated with 5 U calf intestine phosphatase for 4 h at 37°C. The control samples without phosphatase were treated in the same way. After addition of 1 volume of 2× SDS gel sample buffer, the samples were heated again for 5 min at 95°C and then loaded onto the gel.
Preparation of cell extracts for Western blotting.
Cells were grown overnight to exponential phase (OD600 of <0.8 [5 × 107/ml]) in YPD medium. Cells at an OD600 of 4 were pelleted, washed in cold 10 mM NaN3, and then resuspended in 100 μl cold lysis buffer (50 mM HEPES, 0.3 M sorbitol, 10 mM NaN3, pH 7.5, plus protease inhibitor cocktail). Cells were broken by agitation with glass beads for 5 min at 4°C. After addition of 1 volume of 2× SDS gel sample buffer, proteins were solubilized at 50°C for 15 min. Cell debris was removed by a 13,000 × g spin for 2 min in a tabletop centrifuge. An aliquot corresponding to an OD600 of cells of 0.2 was loaded onto a SDS-polyacrylamide gel electrophoresis gel and examined by Western blotting.
Northern blotting.
For RNA isolation, cells were grown overnight to exponential phase (OD600 of <0.8 [5 × 107/ml]) in YPD medium. Cells at an OD600 of 4 were harvested and resuspended in 200 μl extraction buffer (0.5 M NaCl, 0.2 M Tris-Cl [pH 7.6], 10 mM EDTA, 1% SDS). After addition of 400 mg glass beads and 200 μl of a 1:1 mixture of phenol-chloroform, the samples were vortexed for 2.5 min. Then 300 μl extraction buffer and 300 μl phenol-chloroform were added and the samples were vortexed again for 1 min. After 5 min of centrifugation in a tabletop centrifuge at full speed, the aqueous phase was recovered and reextracted with 1 volume of phenol-chloroform. The aqueous phase was mixed with 2 volumes of ethanol and spun for 10 min at full speed. The pellet was washed two times with 3 M Na-acetate (pH 6) to remove DNA, washed with 1 ml 70% ethanol, and finally resuspended in 100 μl sterile H2O. The RNA was separated on a formaldehyde agarose gel, blotted to a GeneScreen Plus membrane (Perkin Elmer, Boston, MA), and hybridized to specific DNA probes according to the instructions of the manufacturer. Autoradiograms were scanned, and the signal intensities were quantified with the program ImageJ (http://rsbweb.nih.gov/ij/). SUC2 signals were normalized to the actin signal.
Invertase assay.
Yeast cells grown overnight to the exponential phase (OD600 of <0.8 [5 × 107/ml]) in YPD plus 5% glucose at 25°C were harvested, washed in H2O, and resuspended in YPD plus 0.1% glucose. At time intervals after the shift to low glucose, cell extracts were prepared and invertase activity was determined. For determination of invertase activity, cells at an OD600 of 1 were harvested, washed twice in 10 mM NaN3, and resuspended in 100 μl 0.1 M Na-acetate (pH 5). After breakage of the cells by glass beading for 5 min, 150 μl 0.1 M Na-acetate (pH 5) was added and cell debris was removed by centrifugation at 500 × g for 5 min. Five microliters of the supernatant was mixed with 100 μl 0.1 M Na-acetate (pH 5) and 25 μl of freshly prepared 0.5 M sucrose and incubated for 20 min at 37°C. The reaction was stopped by the addition of 150 μl 0.2 M K2HPO4 and heating at 95°C for 3 min. After cooling on ice, 1 ml reaction mix (2.5 mg/ml glucose oxidase, 0.5 mg/ml peroxidase, 0.3 mg/ml o-dianisidine, 0.1 M K phosphate [pH 7]) was added. After incubation at 37°C for 30 min, the reaction was stopped by the addition of 6 M HCl and the A540 was measured in a spectrophotometer.
RESULTS
Deletion mutants of SNF7 family members show distinct phenotypes.
The Snf7 family proteins perform a function at late endosomes, probably as part of a larger protein complex (1, 2, 8). The phenotypes of SNF7 family member mutants, however, suggested that these proteins carry out additional functions. To investigate this more systematically, deletion mutants of all six SNF7 family members were examined for their growth phenotypes under conditions that activate different cell signaling pathways (Fig. 1). Cells were incubated at elevated temperature (37°C) to test for a role in heat shock signaling. To see whether the Snf7 homologues are involved in glucose derepression, glucose-grown cells were spotted onto plates containing raffinose as the sole carbon source. Cells that are defective in glucose derepression, as suggested previously for snf7 mutants, cannot grow on raffinose and on other carbon sources whose utilization is prevented by the presence of high concentrations of glucose in the growth medium. Also, cells were spotted onto plates containing the cell-wall-disturbing agent Congo red and onto plates containing caffeine. Both Congo red and caffeine activate the Slt2-mitogen-activated protein (MAP) kinase cell integrity pathway (23).
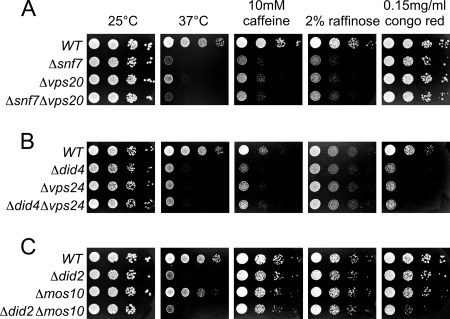
FIG. 1.
Growth phenotypes of SNF7 family deletion mutants. Tenfold serial dilutions of overnight cultures of different yeast strains grown in YPD medium were spotted onto YPD plates containing 10 mM caffeine, 2% raffinose as the sole carbon source (plus 2 μg/ml antimycin), or 0.15 mg/ml Congo red, as indicated. Plates were incubated at 25°C, except for one set of plates that were incubated at 37°C. The following strains were used (from top to bottom): (A) JD52 (wild type [WT]), RKY1852 (Δsnf7), RKY1853 (Δvps20), RKY1829 (Δsnf7 Δvps20); (B) JD52 (WT), RKY1732 (Δdid4), RKY1730 (Δvps24), and RKY1828 (Δdid4 Δvps24); and (C) JD52 (WT), RKY1728 (Δdid2), RKY1654 (Δmos10), and RKY1835 (Δdid2 Δmos10).
Deletion mutants of the four Snf7 homologues that were assigned to the ESCRT-III complex (Did4, Snf7, Vps20, and Vps24) resembled each other in that they were all sensitive to high temperature (37°C) and 10 mM caffeine. However, in addition, these mutants also showed distinct growth phenotypes: the Δsnf7 and Δvps20 mutants grew poorly on raffinose but were unaffected by Congo red (Fig. 1A), whereas the opposite was true for the Δdid4 and Δvps24 mutants (Fig. 1B). A completely different pattern of growth phenotypes was observed for the deletion mutants of the other two Snf7 homologues that have not been mapped to the ESCRT-III complex. While the Δmos10 strain grew like the wild type under all conditions tested, the only phenotype observed for the Δdid2 strain was temperature sensitivity.
Interestingly, two each of the ESCRT-III mutants (Δsnf7/Δvps20 and Δdid4/Δvps24) showed exactly the same pattern of growth phenotypes. This is in line with earlier observations that Snf7/Vps20 and Did4/Vps24 form subcomplexes of the ESCRT-III complex (2). The pairwise identity of growth phenotypes suggests that the two proteins of each subcomplex perform a common function. If this notion is correct, the double-deletion mutant of one pair should show the same phenotype as the single mutants. This is what we observed. In contrast, the Δdid2 Δmos10 double mutant showed a synthetic growth phenotype not observed with the single mutants, i.e., sensitivity to Congo red. This suggests that these two proteins are not part of a common subcomplex but instead perform parallel functions.
Role of Snf7 in SUC2 expression.
The phenotypes of the Snf7 family deletion mutants suggest that these proteins perform additional functions unrelated to their role in MVB formation. To explore this nonendosomal function in more detail, we focused on the role of Snf7 in invertase expression. Invertase is required for the utilization of carbon sources like sucrose and raffinose. Its expression from the SUC2 gene is under glucose control: SUC2 is expressed on low-glucose media and repressed on high-glucose media. Snf7 is somehow involved in high-level expression of SUC2 (27, 28). However, how Snf7 participates in SUC2 expression has not been thoroughly investigated so far.
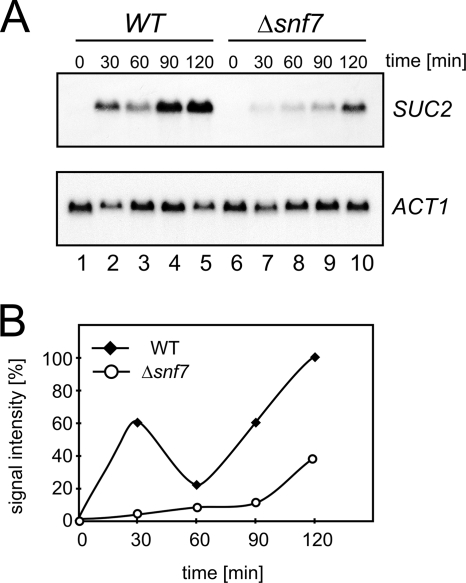
Several possibilities are conceivable: Snf7 could directly affect transcription of the SUC2 gene, or it could affect later steps that lead to expression of invertase activity (like nuclear export of mRNA, translation, or transport and turnover of invertase). To see whether Snf7 acts at the transcriptional or posttranscriptional level, Northern blot experiments were performed. Cells were pregrown under repressing conditions (5% glucose) and were then shifted to inducing medium (0.1% glucose). At time intervals after a shift to low-glucose medium, RNA was prepared and examined by Northern blotting for the amount of SUC2 mRNA. In the wild-type strain, an increase in the amount of SUC2 mRNA was observed during the time course of the experiment (2 h) (Fig. 2A). In the Δsnf7 strain, SUC2 mRNA also increased but to a much lower level. The amount of actin mRNA (ACT1) was not affected by the glucose shift, and the amounts were the same in both strains. This experiment shows that Snf7 affects transcript levels and that it is required for full derepression of the SUC2 gene after the shift to low-glucose medium.
FIG. 2.
Effect of Δsnf7 on invertase expression. (A) Northern blotting. JD52 (wild type [WT], lanes 1 to 5) and RKY1852 (Δsnf7, lanes 6 to 10), pregrown in YPD medium (plus 5% glucose), were transferred to low-glucose medium (0.1%). At time zero, RNA was prepared at time intervals after the shift to low glucose (as indicated) and examined for the amounts of SUC2 (upper panel) and ACT1 mRNA (lower panel) by Northern blotting with specific DNA probes. (B) Quantification of the Northern blot signals. Closed diamonds, JD52 (WT); open circles, RKY1852 (Δsnf7). The SUC2 signals were normalized to the ACT1 signals.
Quantification of the Northern blot signals revealed that the SUC2 expression profile was biphasic (Fig. 2B). After an initial increase, SUC2 transcript levels drop and then rise again. A similar observation has been described before (5). It appears that the SNF7 deletion mainly affects the initial burst of SUC2 transcriptional activity, since the amount of SUC2 transcript at later time points was comparable to that of the wild type. The reason for this behavior is at present unclear.
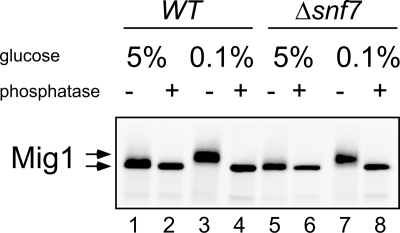
Under high-glucose conditions, expression of the SUC2 gene is prevented by the Mig1 repressor, which binds to the SUC2 promoter (14). When the glucose concentration drops to low levels, the Snf1 kinase is activated, which in turn inactivates the Mig1 repressor through phosphorylation (26). Snf7 could interfere at several steps with this signaling cascade. To see whether the glucose-signaling cascade is intact in the Δsnf7 mutant, we examined whether Mig1 is still phosphorylated after the shift to low-glucose medium. For detection, Mig1 was marked with a 13-Myc tag. After the shift to low glucose, more slowly migrating Mig1 species were observed on Western blots that were condensed to a sharp band that migrated slightly faster by phosphatase treatment (Fig. 3). Both wild-type and Δsnf7 strains showed the same phosphorylation pattern. This suggests that the glucose-signaling cascade resulting in phosphorylation of Mig1 is still intact in the Δsnf7 mutant. Thus, Snf7 appears to be involved in a step independent of Mig1 inactivation.
FIG. 3.
Mig1 phosphorylation. RKY2058 (wild type [WT], lanes 1 to 4) and RKY2104 (Δsnf7, lanes 5 to 8) expressing a 13-Myc-tagged Mig1 variant from the chromosomal copy of the gene were pregrown in YPD medium with 5% glucose. Half of the cells were transferred to low-glucose medium (0.1%) for 1 h, while the other half were further incubated with 5% glucose. Equal amounts of cell extracts were examined for Mig1 phosphorylation by Western blotting with anti-myc antibodies. +, samples treated with phosphatase (even lanes); −, untreated samples (odd lanes).
Effect of class E vps mutants on SUC2 expression.
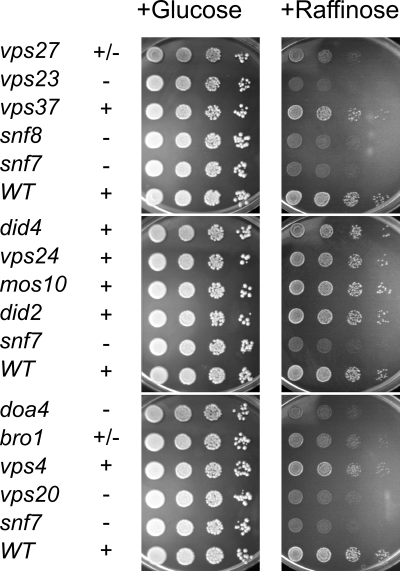
We were interested to see whether the function of Snf7 in SUC2 expression is tied to a functional Vps pathway. Therefore, several class E vps mutants were tested for growth on raffinose plates (Fig. 4). At least one member each of the different ESCRT complexes was included in the analysis (22). Lack of growth on raffinose was observed for vps23 (ESCRT-I), snf8 (ESCRT-II), snf7, and vps20 (ESCRT-III) and doa4. A partial growth defect was seen for Δvps27 (ESCRT-0) and bro1. Basically, the data indicate that the ESCRT complexes functioning upstream of Snf7 are required for growth on raffinose medium. Among the ESCRT-III proteins, only Vps20, which forms a subcomplex with Snf7 (2), is required for growth on raffinose, while the other members of the complex are dispensable. Downstream functions, like bro1 and vps4, also do not seem to be crucial for SUC2 expression. However, some exceptions from this scheme were noted: Vps37, another subunit of ESCRT-I, was not required for growth on raffinose and Vps27, a component of ESCRT-0, was only partially required. On the contrary, the deubiquitinating enzyme Doa4, which is recruited to late endosomes by Bro1 and which is involved in deubiquitination of endocytic cargo proteins (1, 13, 16), appears to be essential for invertase expression. Since the deubiquitinating activity of Doa4 at endosomes is dependent on Bro1 and since deletion of BRO1 has only a moderate effect on raffinose growth, the Δdoa4 effect is probably unrelated to the endosomal function of Doa4.
FIG. 4.
Growth of “class E” mutant strains on raffinose plates. Tenfold serial dilutions of overnight cultures of different yeast strains grown in YPD medium were spotted onto YPD plates containing 2% glucose (left panels) or 2% raffinose (right panels) as the sole carbon source. Plates were incubated at 25°C for 4 days. The yeast mutants are derived from the wild-type (WT) strain JD52 and carry deletion mutations of the genes indicated. +, ±, and −, respectively, indicate growth, some growth, and no growth on raffinose plates.
Regulation of SUC2 expression by the Rim101 pathway.
A similar dependence on ESCRT functions as observed for SUC2 expression has been reported for the alkaline response pathway (the Rim101 pathway). In response to alkaline pH, the transcription factor Rim101 is activated by proteolytic cleavage (11). Snf7 plays a central role in this processing event by recruiting the processing factors Rim13 and Rim20 (30). Because of the similar requirements for upstream ESCRT functions (31), we examined whether SUC2 is also a target of the Rim101 pathway by testing rim mutants for growth on raffinose plates (Fig. 5A). Indeed, we found that rim13, rim20, and rim101 deletion mutants are unable to grow on raffinose. This suggests that SUC2 is regulated by the Rim101 pathway.
FIG. 5.
Growth phenotypes of Rim101 pathway mutants. Tenfold serial dilutions of overnight cultures of different yeast strains grown in YPD medium were spotted onto YPD plates containing 2% glucose (left panels) or 2% raffinose (right panels) as the sole carbon source (A) or onto a normal YPD plate (left panel) and a YPD plate containing 0.3 M LiCl (right panel) (B). Plates were incubated at 25°C for 4 days. The yeast mutants were derived from the wild-type (WT) strain JD52 and carry deletion mutations of the genes indicated. NRG1-3HA represents expression of epitope-tagged Nrg1, and ADH1p-NRG1 represents overexpression of NRG1 from the ADH1 promoter. +, ±, and −, respectively, indicate growth, some growth, and no growth on raffinose plates.
Several other phenotypes have been described for rim mutants, like cold sensitivity, sensitivity to Na+ or Li+ ions (30), defects in sporulation (24), and haploid-invasive growth (11). Indeed, we could show that the Δsnf7 mutant shows the same lithium sensitivity as the rim mutants (Fig. 5B).
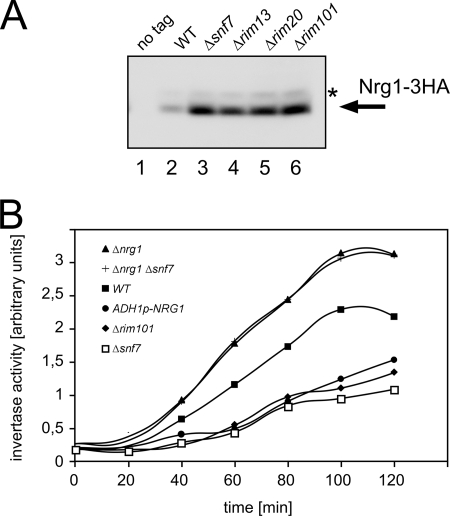
How does Rim101 regulate SUC2 expression? Rim101 could either bind directly to the SUC2 promoter, or it could regulate the expression of a transcription factor that binds to the SUC2 promoter. Since there was no evidence for a direct role of Rim101 at the SUC2 promoter, we looked for Rim101 targets that could be involved in SUC2 regulation (9). The Rim101 target Nrg1 has been implicated before in SUC2 regulation (32). We therefore examined the role of Nrg1 in Snf7-dependent regulation of SUC2 expression. Nrg1 acts as a SUC2 repressor, while Rim101 in turn represses NRG1. Thus, Rim101 should have a positive effect on SUC2 transcription by lowering Nrg1 levels. In turn, Nrg1 levels should be higher in a Δsnf7 mutant, where Rim101 activation is prevented. Indeed, we could show that the level of Nrg1 is much higher in the Δsnf7 mutant than in the wild type (Fig. 6A). Also, a similar elevated Nrg1 expression level was observed in the rim mutants.
FIG. 6.
Effect of Nrg1 on SUC2 expression. (A) Expression levels of Nrg1-3HA in several yeast strains were determined by Western blotting with anti-HA antibodies. Lane 1, JD52 (wild type [WT], no tag), lane 2, RKY2176 (SNF7); lane 3, RKY2180 (Δsnf7); lane 4, RKY2284 (Δrim13); lane 5, RKY2285 (Δrim20); lane 6, RKY2286 (Δrim101). The arrow points to the main Nrg1-3HA band; the phosphorylated form is marked with an asterisk. (B) Yeast cells pregrown in YPD medium with 5% glucose were shifted to low-glucose medium (0.1%). At time intervals after the shift to low glucose, invertase activity was determined. Strains: closed squares, JD52 (WT); open squares, RKY1852 (Δsnf7); closed diamonds, RKY2175 (Δrim101); closed triangles, RKY2206 (Δnrg1); crosses, RKY2207 (Δnrg1 Δsnf7); and closed circles, RKY2208 (ADH1p-NRG1).
To test whether altered Nrg1 levels could explain the effect of Δsnf7 on SUC2 expression, induction of invertase activity after the shift to low-glucose medium was measured in various mutants (Fig. 6B). Compared to the wild type, the invertase activity in the Δsnf7 strain after 2 h of derepression was much lower (about 30% of the wild-type level). Exactly the same level of activity was reached in a Δrim101 mutant and upon overexpression of NRG1 from the ADH1 promoter. This is consistent with the view that Δsnf7 exerts its effect on SUC2 by increased Nrg1 levels. Also, invertase activity was clearly higher in a Δnrg1 strain than in the wild type. Most interestingly, invertase activity was no longer dependent on Snf7 in the Δnrg1 background. Thus, Snf7 exlusively acts on SUC2 by affecting Nrg1 levels. As expected, altered Nrg1 levels affected the growth on raffinose plates (Fig. 5A). While the Δnrg1 mutant grew like the wild type, the NRG1-overexpressing strain had a growth defect similar to that of the Δsnf7 mutant.
DISCUSSION
In this report, we explore the molecular basis for the sucrose-nonfermenting phenotype of snf7 mutants. We demonstrate that the ESCRT-III protein Snf7 mediates high-level expression of the SUC2 gene via the Rim101 pathway.
Our Northern blotting experiments provide evidence that Snf7 is involved in the regulation of SUC2 transcript levels. One potential target for Snf7 could be glucose signaling at the SUC2 promoter. Upon glucose limitation, the Mig1 repressor, which represses SUC2 expression on high glucose, is inactivated by phosphorylation (26). This Mig1 phosphorylation, however, was not affected in the Δsnf7 mutant. Thus, Snf7 appears to affect events at the SUC2 promoter independent of Mig1. This notion is also supported by earlier findings that galactose utilization, which is also under Mig1 control, is not affected in snf7 mutants (28).
From our data, we conclude that Snf7 influences SUC2 expression by regulating the level of Nrg1, a transcription factor already implicated in SUC2 regulation. NRG1 transcription is negatively controlled by the transcription factor Rim101 (9). This transcription factor in turn is activated by proteolytic cleavage in response to alkaline pH (11). Snf7 appears to be an integral part of the complex involved in Rim101 cleavage consisting of the Bro1 homologue Rim20 and the calpain-like protease Rim13 (30). The effect of Snf7 on SUC2 expression can be completely accounted for by its effect on Nrg1 levels. This notion is supported by the following findings: (i) deletion of RIM101 or overexpression of NRG1 from the ADH1 promoter leads to the same SUC2 expression level as deletion of SNF7 and (ii) Snf7 is dispensable for SUC2 expression in an NRG1 deletion background. This implies that the RIM101 pathway is fully active at the pH of our growth media used for the invertase assays. Indeed, we found that Rim101 is completely processed under these conditions (data not shown).
SUC2 shows a biphasic transcription profile after derepression (5). After an initial burst of transcription, transcript levels drop and then rise again. In the Δsnf7 mutant, the initial burst of transcription seems to be missing. Since the only role of Snf7 in SUC2 expression is to modulate Nrg1 levels, the release from Nrg1 repression appears to be defective in the Δsnf7 mutant. How Nrg1 repression is released after the shift to low glucose is not known, but apparently inactivation is counteracted by high Nrg1 levels. It has been reported that Nrg1 becomes phosphorylated by protein kinase CK2 after exposure to various stress conditions, among them the shift to low glucose (4). The consequences of this modification, however, remained unclear. We could also detect phosphorylation of Nrg1 (Fig. 6A), but in our hands, there was no significant change in phosphorylation upon shift to low glucose or in the Δsnf7 mutant compared to the wild type (data not shown). Thus, how exactly Nrg1 repression is released after the shift to low glucose remains elusive. In any case, at later time points after derepression, Nrg1 does not act any longer as a repressor irrespective of the amounts of Nrg1 present.
It is not immediately obvious why the SUC2 gene, which is mainly controlled by the carbon source, should be under the control of the alkaline response pathway. One possible explanation could be that carbon source utilization is somehow connected to the pH of the growth medium. Indeed, it has been reported that growth on high glucose concentrations lowers the pH, while growth on a poor carbon source, like acetate, results in a pH increase (11). Thus, active growth, independent from the carbon source used, would exert a negative effect on SUC2 expression via lowered pH and a less active RIM101 pathway.
Alternatively, the RIM101 pathway could have other roles in gene expression beyond pH control. In fact, the association between the RIM101 pathway and pH control in Saccharomyces cerevisiae is not as tight as in Aspergillus nidulans, where this system was initially discovered (9). By macroarray analysis, alkaline-induced genes in S. cerevisiae were identified (10). Only some of these genes turned out to be regulated by Rim101. In addition, some Rim101 target genes were still pH regulated in the presence of the constitutively active Rim101-531 mutant protein. This suggests that there are additional mechanisms of pH control in S. cerevisiae. Furthermore, rim mutants show many phenotypes, like cold sensitivity, sensitivity to Na+ or Li+ ions (30), defects in sporulation (24), and haploid-invasive growth (11), which may not be directly linked to pH control.
SUC2 is not the only yeast gene that is controlled by both Mig1 (and/or the homologous protein Mig2) and by Nrg1 (and/or the homologous protein Nrg2). For instance, ENA1, coding for the Na+-ATPase, is similarly regulated by Nrg1 and Mig2 (and additional factors) (20). In addition, a database query in the YEASTRACT database (25) revealed 40 genes whose expression is affected by both Mig1/2 and Nrg1/2. Thus, for a certain set of genes, it appears to be beneficial to link carbon source control via Mig1/2 and stress signaling via Nrg1/2.
Proteolytic activation of Rim101 is connected to a functional ESCRT pathway (31). The current view is that the protein complexes of the ESCRT pathway are assembled at the endosomal membrane in a fixed order and that each complex assists in recruitment of the following complex (22). In line with previous findings, we found that the components placed “upstream” of the Snf7/Vps20 subcomplex in this assembly pathway are required for optimal growth on raffinose (i.e., full SUC2 expression), while the “downstream” components appear to be dispensable. From this, it can be concluded that the main function of the ESCRT pathway, with respect to Rim101 processing, is to bring Snf7/Vps20 to the membrane. Membrane association appears to be a prerequisite for assembly of an active Snf7/Rim20/Rim13 processing complex.
However, there are some conflicts with published reports as to the contribution of some of the “class E” functions (Vps27, Vps37, Bro1, and Doa4) to Rim101 activation (21, 31). Basically, our results are in agreement with the study by Rothfels et al. (21). We find that Vps37 is not required for growth on raffinose, while Doa4 is required and Bro1 is partially required. It is unclear why some upstream components of the ESCRT pathway (like Vps37 and maybe Vps27) should not be required for Rim101 activation. However, it is conceivable that the requirements for Rim101 activation may not be as stringent as the requirements for the proper functioning of the ESCRT pathway.
Why Rim101 processing only occurs at the membrane is at present unclear. Perhaps, membrane association is necessary to integrate signaling events with Rim101 processing.
Acknowledgments
We thank Karin Krapka for assistance with some of the experiments.
This work was supported by the DFG grant Ko 963/5-1.
Footnotes
Published ahead of print on 19 September 2008.
REFERENCES
- 1.Amerik, A. Y., J. Nowak, S. Swaminathan, and M. Hochstrasser. 2000. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell 113365-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell 3271-282. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 172982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkey, C. D., and M. Carlson. 2006. A specific catalytic subunit isoform of protein kinase CK2 is required for phosphorylation of the repressor Nrg1 in Saccharomyces cerevisiae. Curr. Genet. 501-10. [DOI] [PubMed] [Google Scholar]
- 5.Geng, F., and B. C. Laurent. 2004. Roles of SWI/SNF and HATs throughout the dynamic transcription of a yeast glucose-repressible gene. EMBO J. 23127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howard, T. L., D. R. Stauffer, C. R. Degnin, and S. M. Hollenberg. 2001. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci. 1142395-2404. [DOI] [PubMed] [Google Scholar]
- 7.Jiang, R., and M. Carlson. 1997. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 172099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kranz, A., A. Kinner, and R. Kölling. 2001. A family of small coiled-coil-forming proteins functioning at the late endosome in yeast. Mol. Biol. Cell 12711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamb, T. M., and A. P. Mitchell. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb, T. M., W. Xu, A. Diamond, and A. P. Mitchell. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 2761850-1856. [DOI] [PubMed] [Google Scholar]
- 11.Li, W., and A. P. Mitchell. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 14563-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 13.Luhtala, N., and G. Odorizzi. 2004. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nehlin, J. O., and H. Ronne. 1990. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 92891-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neigeborn, L., and M. Carlson. 1984. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108845-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikko, E., and B. André. 2007. Evidence for a direct role of the Doa4 deubiquitinating enzyme in protein sorting into the MVB pathway. Traffic 8566-581. [DOI] [PubMed] [Google Scholar]
- 17.Nikko, E., A. M. Marini, and B. André. 2003. Permease recycling and ubiquitination status reveal a particular role for Bro1 in the multivesicular body pathway. J. Biol. Chem. 27850732-50743. [DOI] [PubMed] [Google Scholar]
- 18.Odorizzi, G., M. Babst, and S. D. Emr. 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95847-858. [DOI] [PubMed] [Google Scholar]
- 19.Odorizzi, G., D. J. Katzmann, M. Babst, A. Audhya, and S. D. Emr. 2003. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 1161893-1903. [DOI] [PubMed] [Google Scholar]
- 20.Platara, M., A. Ruiz, R. Serrano, A. Palomino, F. Moreno, and J. Arino. 2006. The transcriptional response of the yeast Na(+)-ATPase ENA1 gene to alkaline stress involves three main signaling pathways. J. Biol. Chem. 28136632-36642. [DOI] [PubMed] [Google Scholar]
- 21.Rothfels, K., J. C. Tanny, E. Molnar, H. Friesen, C. Commisso, and J. Segall. 2005. Components of the ESCRT pathway, DFG16, and YGR122w are required for Rim101 to act as a corepressor with Nrg1 at the negative regulatory element of the DIT1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 256772-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saksena, S., J. Sun, T. Chu, and S. D. Emr. 2007. ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 32561-573. [DOI] [PubMed] [Google Scholar]
- 23.Schmitz, H. P., A. Lorberg, and J. J. Heinisch. 2002. Regulation of yeast protein kinase C activity by interaction with the small GTPase Rho1p through its amino-terminal HR1 domain. Mol. Microbiol. 44829-840. [DOI] [PubMed] [Google Scholar]
- 24.Su, S. S., and A. P. Mitchell. 1993. Identification of functionally related genes that stimulate early meiotic gene expression in yeast. Genetics 13367-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teixeira, M. C., P. Monteiro, P. Jain, S. Tenreiro, A. R. Fernandes, N. P. Mira, M. Alenquer, A. T. Freitas, A. L. Oliveira, and I. Sa-Correia. 2006. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 34D446-D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Treitel, M. A., S. Kuchin, and M. Carlson. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 186273-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu, J., L. G. Vallier, and M. Carlson. 1993. Molecular and genetic analysis of the SNF7 gene in Saccharomyces cerevisiae. Genetics 13517-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallier, L. G., and M. Carlson. 1991. New SNF genes, GAL11 and GRR1 affect SUC2 expression in Saccharomyces cerevisiae. Genetics 129675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 201899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu, W., and A. P. Mitchell. 2001. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 1836917-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu, W., F. J. Smith, Jr., R. Subaran, and A. P. Mitchell. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 155528-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou, H., and F. Winston. 2001. NRG1 is required for glucose repression of the SUC2 and GAL genes of Saccharomyces cerevisiae. BMC Genet. 25. [DOI] [PMC free article] [PubMed] [Google Scholar]