Abstract
Chitin is an essential component of fungal cell walls, where it forms a crystalline scaffold, and chitooligosaccharides derived from it are signaling molecules recognized by the hosts of pathogenic fungi. Oomycetes are cellulosic fungus-like microorganisms which most often lack chitin in their cell walls. Here we present the first study of the cell wall of the oomycete Aphanomyces euteiches, a major parasite of legume plants. Biochemical analyses demonstrated the presence of ca. 10% N-acetyl-d-glucosamine (GlcNAc) in the cell wall. Further characterization of the GlcNAc-containing material revealed that it corresponds to noncrystalline chitosaccharides associated with glucans, rather than to chitin per se. Two putative chitin synthase (CHS) genes were identified by data mining of an A. euteiches expressed sequence tag collection and Southern blot analysis, and full-length cDNA sequences of both genes were obtained. Phylogeny analysis indicated that oomycete CHS diversification occurred before the divergence of the major oomycete lineages. Remarkably, lectin labeling showed that the Aphanomyces euteiches chitosaccharides are exposed at the cell wall surface, and study of the effect of the CHS inhibitor nikkomycin Z demonstrated that they are involved in cell wall function. These data open new perspectives for the development of antioomycete drugs and further studies of the molecular mechanisms involved in the recognition of pathogenic oomycetes by the host plants.
The microbial cell wall is involved in growth, development, signaling, and interaction with the environment. It represents an essential structure conferring osmotic protection and maintaining cell shape. In pathogenic microorganisms, the cell wall is a reservoir of molecules involved in host recognition, adhesion, and colonization. It also contains major antigens that elicit innate immunity in animals and plants, and it is a target of host defense responses (43, 58, 82). Analysis of its structure and composition allows the identification of specific components that are attractive targets for the design of antimicrobial compounds (35). A common constituent of cell walls is chitin, a crystalline polymer of β-1,4-linked N-acetyl-d-glucosamine (GlcNAc), which is frequently partially deacetylated, and usually associated with other polysaccharides and proteins. Chitin is found only in eukaryotic organisms, usually as a shell or cuticle component of crustaceans, insects, molluscs, and nematodes, and as an essential cell wall component of fungi. It also occurs in marine sponges and algae (28, 29) and in various protists, such as amoebae, ciliates, diatoms, and chrysoflagellates (41). Chitin is thus widely distributed and is the second most abundant polysaccharide in nature after cellulose, with at least 10 gigatons synthesized and degraded each year in the biosphere. However, it is absent from vertebrates and higher plants and therefore represents a target for the development of antimicrobial compounds aimed at controlling human or crop diseases caused by fungi (53, 55, 75).
The oomycetes are a group of diploid eukaryotes that include the most numerous, most important, and earliest known water molds (24). Oomycetes were classified for many years within the kingdom Fungi because of their similar ecological and morphological traits, but phylogenetic studies have shown that they are closer to diatoms, chromophyte algae, and other heterokont protists of the Stramenopile kingdom (5). They include numerous pathogenic species, such as the major salmonid parasite Saprolegnia parasitica (85) and the causal agent of potato late blight, Phytophthora infestans (44). They share some common infection strategies with fungi (40, 49) and apicomplexans (11, 38). A feature usually mentioned when distinguishing oomycetes from true fungi is the presence of cellulose and the absence of chitin in their cell wall (40). This assumption stems from early studies mainly focused on the Phytophthora cell wall (6, 7) and was recently reinforced by the demonstration that cellulose synthesis is required for normal appressorium formation in Phytophthora infestans (37). However, the cell wall of the closely related hyphochytridiomycetes contains both cellulose and chitin, and chitin was unambiguously detected by biophysical analyses in some oomycete species of the Leptomitale and Saprolegniale orders (2, 12). In Saprolegnia monoica, a chitin synthase (CHS) activity was characterized in detail, and two CHS genes were identified, among which one was fully sequenced (50, 54). In the Peronosporale and Pythiale orders, partial putative CHS sequences were identified in Plasmopara viticola and Phytophthora capsici (54, 87). However, there is only scarce biochemical evidence of the occurrence of chitin in these organisms (25, 26). Lectin labeling and/or cytochemical analyses gave negative results in Phytophthora parasitica (16) and Phytophthora cinnamomi (3) but were positive in Plasmopara viticola (87) and Pythium ultimum (16). In these latter cases, however, the presence of the polymer in its typical crystalline form was not shown.
Within the oomycetes, the Saprolegniale genus Aphanomyces comprises plant and animal pathogens found in both terrestrial and aquatic habitats. Aphanomyces euteiches Drechs. causes seedling and root rot diseases on many legumes and is the most serious pathogen of pea in several countries (34). Neither effective chemicals nor resistant cultivars are available to control the disease in pea. While a huge genomic research effort has been devoted to Phytophthora, Aphanomyces has received little attention. Recently, a database of 19,000 A. euteiches expressed sequence tags (ESTs) assembled in ca. 8,000 unigenes was created (52), representing a first step toward the identification of Aphanomyces pathogenicity effectors (27). Because of the importance of the cell wall in microbial fitness and interaction with hosts, we have undertaken the characterization of the A. euteiches cell wall. This report shows that it contains noncrystalline chitosaccharides. It presents evidence for the first time that chitosaccharides, in the absence of crystalline chitin, might be involved in cell wall integrity of a filamentous microorganism. In addition, it shows that the Aphanomyces chitosaccharides are exposed at the cell wall surface. This opens new perspectives for the development of antioomycete drugs and further studies of the molecular mechanisms involved in the recognition of pathogenic oomycetes by the host plants.
MATERIALS AND METHODS
Strains, media, and culture conditions.
A. euteiches strain ATCC 201684 is a Danish pea isolate kindly provided by F. Krajinski (Hannover University, Germany). Unless otherwise stated, all experiments were performed with this strain. Ae5 and RB84 are pea isolates, and Ae106 is an alfalfa isolate, all kindly provided by B. Tivoli (INRA, Rennes, France). The Phytophthora parasitica (syn. Phytophthora nicotianae) strain used is a tobacco isolate of race 0 (kindly provided by J. P. Helgeson, University of Wisconsin). P. parasitica was maintained by routine subculture every 2 months on clarified V-8 agar medium (5% [vol/vol]; pH 5) incubated in the dark at 24°C, and zoospore suspensions were prepared as previously described (33). A. euteiches was maintained on 1.7% corn meal agar (CMA) (Sigma-Aldrich) by routine subculture each month in the dark at 24°C. For long-term storage, mycelium explants were kept under sterile source water (Volvic, France) at 15°C in the dark. Zoospores were produced by a modified version of the method of Deacon and Saxena (22), with all steps performed at 24°C: 20 mycelium explants from 10- to 15-day-old CMA plates were inoculated into 50 ml of 2% Bacto peptone and 0.5% glucose and incubated 3 days in the dark, then the medium was decanted, and the mycelial mats were washed once with 30 ml of source water and resuspended in 30 ml of source water. After 2 h of incubation in the dark, the water was drained and replaced by fresh source water. This procedure was repeated 2 h later, and the mycelium was incubated overnight in source water in the dark. The released zoospores were counted using a hemocytometer and diluted with distilled water to the concentration required for the various experiments.
For biomass production, 50,000 A. euteiches or 33,000 P. parasitica zoospores were inoculated into 100 ml of 1% glucose and 0.5% yeast extract (GY medium) in Roux flasks. After a period of growth of 3 days (A. euteiches) or 11 days (P. parasitica) in the dark at 24°C, the mycelium was harvested by filtration on a sintered glass filter, washed with cold distilled water, frozen, and stored at −80°C. For each culture, a sample of fresh mycelium was weighed before and after drying at 50°C during 24 h, a procedure that allowed estimation of the total dry weight of the harvested mycelium.
To measure the effect of nikkomycin Z (NZ) on A. euteiches, GY medium aliquots containing various concentrations of NZ were distributed in the wells of a 96-well microtiter plate (100 μl per well) and subsequently inoculated with 5,000 zoospores per well. In some experiments, inositol or sorbitol was added to the medium at a final concentration of 100 or 200 mM. Three replicates were prepared for each condition. The plates were incubated in the dark at 24°C for 3 days, and the hyphal density was determined by measuring the turbidity at 595 nm. Alternatively, in order to obtain sufficient biomass either to extract RNA for gene expression analyses or to prepare cell walls for measuring their GlcNAc content (see below), 5 ml of GY medium in small petri dishes were inoculated with 250,000 zoospores in the presence or absence of either 50 μM NZ alone (gene expression analyses) or of 100 μM NZ plus 200 mM sorbitol (GlcNAc measurements).
For plant inoculation, axenically grown Medicago truncatula Gaertn seedlings of the F83005.5 accession (obtained from J. M. Prosperi, INRA, Montpellier, France) were prepared as follows: the seeds were surface sterilized by incubation in H2SO4 for 7 min, followed by four washings in sterile deionized water and a 1-hour imbibition step in sterile deionized water. The seeds were sown onto 2% water agar and incubated 2 days at 24°C in the dark for germination. The seedlings were then transferred onto a synthetic M agar medium (9), and the plates were incubated vertically, resulting in growth of the rootlets on the surface of the medium, under a 16-hour illumination period at 22°C and an 8-hour night period at 20°C. The primary root of 2-week-old seedlings was then inoculated by three 3-μl drops of a 105 cell·ml−1 zoospore suspension that were deposited onto the root apex and the middle part and the upper part of the piliferous region, respectively.
Cell wall preparation.
For detailed biochemical analyses, cell walls were prepared by grinding the equivalent of 2 g (dry weight) of mycelium in liquid nitrogen to a fine powder using a mortar and pestle and resuspending the powder in 20 ml of 10 mM HEPES-KOH (pH 7.2), 0.2% sodium deoxycholate, 1 mM phenylmethanesulfonyl fluoride, and 2 mM sodium metabisulfite. After 1-h incubation at room temperature with constant stirring, the sample was centrifuged at 2,900 × g for 20 min at room temperature. All further centrifugation steps were performed under the same conditions. The pellet was resuspended in the same volume of buffer, incubated for 30 min, and recentrifuged. This procedure was repeated twice, then the pellet was washed three times by centrifugation and resuspension in ultrahigh-quality water. Lipids were eliminated by two extractions with 20 ml methanol-chloroform (2/1 [vol/vol]) and one extraction with 20 ml acetone. The resulting residue, referred to as crude cell walls, was lyophilized. Alternatively, the cell walls were isolated by the method of Aronson and Bertke (2) which uses 5 g·l−1 KOH dissolved in methanol-water (4/1 [vol/vol]), a solution referred to hereafter as KOH-MeOH. The ground mycelium was incubated for 10 min in KOH-MeOH (1 ml·g−1 [wet weight] of mycelium) at 100°C. After the sample was allowed to cool, it was centrifuged at 1,000 × g for 10 min at 15°C. The resulting pellet was further extracted twice following the same procedure, and then it was repeatedly washed with ultrahigh-quality water until the pH of the supernatant reached neutrality. The residue was finally washed twice with acetone and dried at 50°C and is referred to hereafter as KOH-MeOH cell walls.
For routine determination of the GlcNAc content of the cell wall, cell wall samples were prepared from small amounts of mycelium (less than 0.4 g [fresh weight]) by a quick procedure: the mycelium was ground in liquid nitrogen and resuspended in 5 ml deionized water. After centrifugation at 2,000 × g for 10 min at 4°C, the pellet was resuspended in the same volume of water and recentrifuged. Subsequent steps performed at room temperature involved one wash in MeOH-chloroform (1/1 [vol/vol]), two washes in acetone, and an overnight incubation at 50°C to dry the pellet. The whole pellet was then hydrolyzed for GlcNAc measurement as described below.
Biochemical analysis of the cell walls.
Total nitrogen was measured in the cell walls by a colorimetric assay adapted from the method of Strauch (76) using the Nessler reagent (Sigma-Aldrich) and (NH4)2SO4 as a standard. The protein content of the cell walls was calculated as the difference between the amount of total nitrogen and the amount of nitrogen present in hexosamines. The hexosamine content was measured after hydrolysis in 6 M HCl (2 ml·mg−1 [dry weight] of cell walls) at 120°C for 1 h and analysis of the neutralized hydrolysate by high-performance anion-exchange chromatography coupled to pulsed amperometric detection (HPAEC-PAD), using a high-performance liquid chromatography Dionex series 4500i device equipped with an analytical Carbo Pac PA1 column (4 by 250 mm) and a gold electrode for detection (Dionex, Sunnyvale, CA). Elution was achieved at room temperature with 15 mM NaOH, and detection was performed in the presence of 300 mM NaOH. The eluted compounds were identified and quantified using commercial standards (Sigma-Aldrich). Quantification of the other monosaccharides from the cell walls was performed by HPAEC-PAD analysis under the same conditions after hydrolysis with 2 M trifluoroacetic acid (2 ml·mg−1 [dry weight] of cell walls) at 120°C for 1 h.
Enzymatic hydrolysis of the cell walls.
The crude cell walls (5 to 10 mg [dry weight]) were incubated overnight with either 0.5 unit of chitinase from Serratia marcescens (catalog no. C7809; Sigma-Aldrich) in 1 ml of 50 mM potassium phosphate (pH 6.8) at 30°C or with 0.5 unit of chitosanase from Streptomyces griseus (catalog no. C9830; Sigma-Aldrich) in 1 ml of 50 mM sodium acetate (pH 5) at 37°C. A control was performed by incubating the cell walls in the absence of hydrolytic enzyme.
For glucanase treatments of KOH-MeOH cell walls, the absence of contaminating chitinase activity in the various glucanases was verified by incubating the enzymes in the presence of colloidal chitin (67) and checking for the absence of hexosamine in the supernatant by HPAEC-PAD analysis after acid hydrolysis (6 M HCl) as described above.
In the first series of experiments, the enzymatic treatment of KOH-MeOH cell walls with a mixture of glucanases was a two-step procedure. In the first step, the cell walls (65 mg) were first incubated in 5 ml of 100 mM sodium acetate (pH 5.2) at 37°C for 24 h in the presence of Westase (Takara) [0.6 unit of (1→ 6)-β-glucanase and 0.08 unit of (1→ 3)-β-glucanase per mg (dry weight) of cell walls] and laminarinase [0.8 unit of (1→ 3)-β-glucanase per mg (dry weight) of cell walls] purified from elicited Medicago truncatula plants (C. Lafitte, unpublished data). Hydrolysis was stopped by incubation at 100°C for 5 min, and the cell wall residue was collected by centrifugation at 3,000 × g for 10 min at 6°C. In the second step, the cell wall residue was treated with 0.3 unit·mg−1 cell wall (dry weight) of cellulase from Trichoderma longibrachiatum (Megazyme International, Libios, France) in 10 ml of 100 mM sodium acetate (pH 5.2) at 37°C for 24 h. Hydrolysis was stopped, and the residual cell walls were pelleted as described above. The supernatants and cell wall pellets obtained after each step were pooled, and their hexosamine content was determined by HPAEC-PAD analysis after acid hydrolysis (6 M HCl) as described above.
In the second series of experiments, the sequential treatment of KOH-MeOH cell walls with single or multiple enzymes coupled to chromatography analyses was as follows: the cell walls (300 mg) were first incubated in 25 ml of 100 mM sodium acetate (pH 4) at 37°C for 16 h in the presence of laminarinase (0.7 unit·mg−1 [dry weight] of cell walls). Hydrolysis was stopped, and the residual cell walls were pelleted as described above and then incubated in 25 ml of 100 mM sodium acetate (pH 5.2) at 37°C for 16 h in the presence of Westase [0.5 unit of (1→ 6)-β-glucanase and 0.07 unit of (1→ 3)-β-glucanase per mg (dry weight) of cell walls]. Hydrolysis was stopped, and the residual cell walls were pelleted as described above and eventually further treated with 0.3 unit·mg−1 cell wall (dry weight) of cellulase (Megazyme) in 25 ml of 100 mM sodium acetate (pH 5.2) at 37°C for 16 h. The hexosamine content of the supernatants and cell wall pellets obtained after each step was determined by HPAEC-PAD analysis after acid hydrolysis (6 M HCl) as described above. The supernatants from the (1→ 3)- and (1→ 6)-β-glucanase treatments were lyophilized and analyzed separately by size exclusion chromatography on a Bio-Gel P-60 fine column (Bio-Rad) in 100 mM ammonium formate (pH 6.8). The elution profile was determined by measuring the glucose (Glc) and GlcNAc concentrations of the fractions by a colorimetric assay using the anthrone reagent (81) and by HPAEC-PAD analysis after acid hydrolysis, respectively. The fractions of interest were lyophilized, resulting in a sample of 38 mg (dry weight) that was further treated with by a mixture of laminarinase (5 units·mg−1 sample [dry weight]), Westase [4 units of (1→ 6)-β-glucanase and 0.5 unit of (1→ 3)-β-glucanase per mg sample (dry weight)], and cellulase (1.4 units·mg−1 sample [dry weight]) in 6 ml of 100 mM sodium acetate (pH 5.2) at 37°C for 16 h. The hydrolysate was then analyzed by size exclusion chromatography as described above.
Biophysical analysis of the cell walls.
For 13C cross polarization/magic angle spinning (CP/MAS) nuclear magnetic resonance (NMR) measurements, KOH-MeOH cell walls were analyzed either in a dried state or after soaking in water. The spectral data of these samples were compared with those of a standard specimen of purified α-chitin from crab shell. This specimen and the cell wall samples were inserted individually into tightly sealed 4-mm BL type ZrO2 rotors. 13C CP/MAS NMR spectra were recorded with a Bruker Avance spectrometer equipped with a 4-mm BL type probe and operated at 100 MHz. The spectra were acquired at room temperature with a 80-kHz proton dipolar decoupling field, matched CP fields of 80 kHz, a proton 90° pulse of 2.5 μs, and MAS at a spinning speed of 12 kHz. The CP transfer was achieved using a ramped amplitude sequence (RAMP-CP) for an optimized total time of 2 ms. The sweep width was of 50,000 Hz to avoid baseline distortion with 2,994 time domain points, and the Fourier transformation was achieved without apodization over 8,000 points. The repetition time was 4 s, and an average number of 20,000 scans were acquired for each spectrum. The 13C chemical shifts were determined relative to the carbon chemical shift of the carbonyl signal of glycine at 176.03 ppm.
Microscopy techniques.
Staining with a fluorescein isothiocyanate-wheat germ agglutinin (FITC-WGA) conjugate (Vector Laboratories, Burlingame, CA) of P. parasitica or A. euteiches was performed on mycelium grown for 2 days on a cellophane membrane placed on top of GY agar medium. Lectin labeling was also performed on Aphanomyces structures (cysts, germinated cysts, and mycelium) formed in GY broth distributed in the wells of 96-well microtiter plates and inoculated with zoospores as described above. For labeling of A. euteiches structures formed in the presence of the host plant, infected roots were either directly cut longitudinally with a razor blade or first embedded in 5% agar (agar with a low melting point; Sigma-Aldrich) and transversally cut with a Leica VT1000S vibratome to obtain 200-μm sections. Lectin labeling was performed under reduced light in phosphate-buffered saline (PBS) solution at pH 7.2; the samples were equilibrated in PBS for 15 min, then incubated for 15 min in 50 μg·ml−1 FITC-WGA, and finally washed three times for 5 min in PBS. Autofluorescence controls were prepared following the same procedure omitting the FITC-WGA conjugate. The specificity of the labeling was checked by preincubating the conjugate with a chitin hydrolysate. The latter was prepared from crab shell chitin (Sigma-Aldrich) by solubilization in 12 M HCl (20 ml·g−1 of crab shell chitin) at −20°C followed by 1 h at 40°C and concentration in a Büchi rotavapor until near dryness. The solubilized fraction was recovered for lectin labeling. Observation was performed with an inverted light microscope (Leitz DMIRBE, Leica, Germany) equipped with a color CoolView charge-coupled-device camera (Photonic Science, United Kingdom) connected to Image Acquisition ProPlus software (Media Cybernetics, Silver Spring, MD). A Leitz filter set I3 was used for epifluorescence analysis.
For the quantification of lectin labeling by image analysis, GY broth containing 200 mM sorbitol distributed in the wells of microtiter plates was inoculated with Aphanomyces zoospores as described above. After 3 days of growth, the mycelium was labeled with a dilute solution of FITC-WGA (12.5 μg·ml−1) in 200 mM sorbitol, then washed in 200 mM sorbitol, and carefully transferred onto microscopy slides in a drop of 200 mM sorbitol. The profile of fluorescence intensity across an hypha was obtained by collecting along a line perpendicular to the hyphal axis the gray value (from 0 to 255) of each individual pixel. For each sample, an average intensity profile was then calculated from four different hyphae. As individual hyphae exhibited different widths, the distance from wall to wall of the average profile was adjusted to the shorter distance obtained for individual hyphae. In this way, only some pixel values in the middle of the larger hyphae were discarded, and all the other pixels were unchanged and were taken into account for the calculation of the average profile.
Molecular techniques.
For Southern blot analysis, A. euteiches genomic DNA was extracted from the mycelium in the presence of cetyltrimethylammonium bromide after a procedure adapted from Dellaporta et al. (23). Genomic DNA samples (100 μg) digested with restriction enzymes were electrophoresed in a 1% (wt/vol) agarose gel. After alkaline denaturation, the DNA fragments were transferred to a positive nylon membrane (Hybond N+; Amersham) and subjected to Southern hybridization (69). The probes were PCR fragments obtained after amplification of plasmid DNA templates prepared from cDNA library clones (52) using primers SP6 and qCHS1F1 (5′-TCGCTGTTGTGTATGCTATTATTGTT) for the A. euteiches CHS1 (AeCHS1) gene and primers T7 and CHS2R1 (5′-ATTGAGCTGGATAGCAAC) for the AeCHS2 gene. The PCR fragments were purified using the Wizard SV gel and PCR cleanup system (Promega) according to the recommendations of the supplier and radiolabeled with [α-32P]dCTP by random priming using the RadPrime DNA labeling system (Invitrogen). Autoradiography of the membranes was performed using a PhosphorImager system.
For real-time quantitative PCR (RT-qPCR) analysis of A. euteiches gene expression and for rapid amplification of cDNA ends and PCR (RACE-PCR) targeting AeCHS1 and AeCHS2 transcripts, total RNA was extracted and purified from ca. 50 mg of A. euteiches mycelium using the RNeasy plant mini kit (Qiagen) coupled to an on-column DNase I (RNase-free) digestion. The absence of contaminating genomic DNA in the RNA preparations was checked by PCR. The RACE-PCR procedure involved reverse transcription of 5.5 μg total RNA using the Superscript III first-strand cDNA synthesis kit (Invitrogen) following the recommendations of the supplier, and 5′ cDNA sequences were obtained using the GeneRacer kit for full-length, RNA ligase-mediated RACE (RLM-RACE, Invitrogen). This technology takes advantage of the 5′ capping of only full-length mRNAs to allow the preparation of only full-length cDNA templates for PCR amplification. The reverse primers used for the first and second (nested) PCR amplification steps were, respectively, 5′-AAGTCGACCCAGACTACAACTTGCTC and 5′-CTTATGGTTCTTCAACGCTTTTTGCAAC for AeCHS1 and 5′-GCTCAATGGCTCTTTCTTTGCTGGT and 5′-CGATGCACTATGTAAAAGATGCCATTG for AeCHS2. The PCR products were cloned using the TOPO TA cloning kit for sequencing (Invitrogen) following the recommendations of the supplier. Their sequence was determined at the Genopole of Toulouse using the ABI PRISM dye terminator cycle sequencing ready reaction kit with ADN polymerase (FS Perkin Elmer).
To perform RT-qPCR, total RNA (600 ng) was subjected to reverse transcription using the Superscript II first-strand cDNA synthesis kit (Invitrogen) following the recommendations of the supplier. The 25-μl quantitative PCR mixtures were prepared in 96-well reaction plates using the Sybr green master mix (Applied Biosystems) and the following primers at a final concentration of 100 nM: qCHS1F2 (5′-GTCCGTTCTACGGCAACATGT) and qCHS1R2 (5′-CGTACGTGCCAAGAGCTCAA) for AeCHS1 sequence amplification, qCHS2F2 (5′-GAAGCAGATAAACAAAAGCAAAGC) and qCHS2R2 (5′-TTGCGAAGTAGAGCCATGCA) for AeCHS2 sequence amplification, and qTUBF1 (5′-CGGCTCTGGTTTGGGTAGTTT) and qTUBR1 (5′-AACCGAGCTTGCTCTTGCG) for A. euteiches α-tubulin sequence amplification. The PCRs were performed in an ABI PRISM 7900 HT device, and the data were analyzed using the SDS 2.0 software (Applied Biosystems). The identity of the amplicons was verified by thermal dissociation curves and resequencing. The cDNA targets were quantified using standard curves established with plasmid DNA serial dilutions.
Bioinformatic and phylogenetic analyses.
Local or global alignments of CHS amino acid sequences were performed at http://www.ebi.ac.uk/emboss/align/ using the Smith-Waterman algorithm (73) or Needleman-Wunsch algorithm (57). ClustalW sequence alignments (79) and TopPred prediction of transmembrane domains (19) were performed at http://bioweb.pasteur.fr/. TMHMM transmembrane domain prediction (47) and signal peptide prediction (10) were performed at http://www.cbs.dtu.dk/services/TMHMM-2.0/ and http://www.cbs.dtu.dk/services/SignalP/, respectively. For phylogenetic analyses, the 7.0.4 version (31 May 2005) of the BioEdit software (39) was used to assemble an initial database of oomycete chitin synthases. The AeCHS1 and AeCHS2 sequences were used as probes to perform BLASTp similarity searches (1) against the nonredundant protein database at the National Center for Biotechnology (NCBI) (http://www.ncbi.nlm.nih.gov/) and tBLASTn similarity searches against the Phytophthora genome project databases at the U.S. Department of Energy (DOE) Joint Genome Institute (http://www.jgi.doe.gov/) and the Broad Institute (http://www.broad.mit.edu/). The identified Phytophthora sojae and P. infestans CHS sequences corresponded to ESTs found at the Oomycete Genomics Database (OGD) (http://www.oomycete.org/ogd/), Phytophthora Functional Genomics Database (PFGD) (http://www.pfgd.org/), or Phytophthora Soybean EST database (PhESTDB 1.0) (http://phytophthora.vbi.vt.edu/EST/) (31, 80). A series of consensus sequences for the aligned oomycete CHSs, spanning from 50 to 90% similarity, was built using the software Consensus (http://www.bork.embl.de/). The 50% consensus sequence was then used as a probe to search against the same databases as described above, but no additional hits were retrieved. The resulting oomycete data set consisted of CHS sequences that were referred to as CHS1 or CHS2, according to their grouping in the phylogenetic and molecular evolutionary analysis that was conducted using MEGA version 3 (48). CHS sequences retrieved from genome project databases are the following: from the JGI website, Phytophthora ramorum CHS1 and CHS2 correspond to Phyra1_1:77229 and Phyra1_1:84041, and P. sojae CHS1 and CHS2 correspond to Physo1_1:143614 and Physo1_1:15524; from the Broad Institute website, P. infestans CHS corresponds to PITG_02050.1. Saprolegnia monoica CHS2 and Emericella nidulans CHSB were found at the NCBI database under the accession numbers P48017 and Q00757, respectively.
Nucleotide sequence accession numbers.
The AeCHS1 and AeCHS2 cDNA sequences were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/) under the accession numbers EU522489 and EU447431, respectively.
RESULTS
Biochemical characterization of Aphanomyces euteiches cell walls.
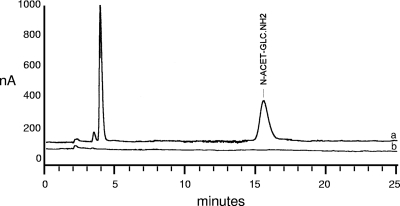
The composition of a crude cell wall preparation from A. euteiches was first determined and compared to a similar cell wall preparation from P. parasitica. Proteins were present in similar proportions in both organisms, as was glucose, which accounted for more than 70% of the cell wall weight (dry weight) (Table 1). The sugar composition of the Phytophthora cell wall was similar to that reported by Bartnicki-García (7) and showed the presence of mannose, whereas the Aphanomyces cell wall contained no detectable amount of mannose. An even more salient feature was the higher proportion of glucosamine (10.7%) in Aphanomyces than in Phytophthora (∼0.5%). This was observed not only for the strain under study (strain ATCC 201684) but also for three other strains (strains Ae5, Ae106, and RB84) in which the proportion of glucosamine ranged between 8 and 17%. Such a high content in glucosamine in cell wall hydrolysates usually accounts for the presence of chitin or chitosan. In order to clarify this, the cell walls were incubated with a chitosanase or with a chitinase, and the products of hydrolysis were analyzed by HPAEC-PAD. No glucosamine was released upon chitosanase treatment, indicating the absence of chitosan. This result was confirmed by the observations that no chitosan could be extracted from the cell walls by a specific chemical extraction method (59) and that the eosin Y stain for chitosan (4) did not label the Aphanomyces hyphae (data not shown). Thus, the glucosamine measured in cell wall hydrolysates originated from GlcNAc, which was deacetylated by the hydrolysis procedure (6 M HCl). Indeed, treatment of the cell walls with a chitinase released GlcNAc monomers (Fig. 1). This was observed not only for the A. euteiches strain ATCC 201684 but also for the three other strains, namely, Ae5, Ae106, and RB84. In contrast, no GlcNAc was released from Phytophthora cell walls by the chitinase treatment (data not shown), which is consistent with a small amount of hexosamines in these cell walls. In conclusion, our data strongly suggest the presence of chitosaccharides in the Aphanomyces cell wall, which are either chito(oligo)saccharides in a noncrystalline form or crystalline chitin.
TABLE 1.
Composition of Aphanomyces and Phytophthora cell walls
| Compounds | Composition of cell wall hydrolysatesa in:
|
|
|---|---|---|
| A. euteiches | P. parasitica | |
| Proteinsb | 158 ± 52 | 124 ± 16 |
| Glucosec | 714 ± 83 | 839 ± 112 |
| Mannosec | NDd | 17 ± 12 |
| Glucosaminec | 107 ± 16 | 5.2 ± 3.6 |
The data are means ± SD from four independent experiments and are expressed as μg·mg−1 (dry weight) of crude cell walls.
Protein content was calculated from the amount of total nitrogen corrected for the amount of nitrogen brought about by glucosamine.
Neutral sugars and glucosamine were quantified by HPAEC-PAD after acid hydrolysis as described in Materials and Methods. Note that N-acetyl-d-glucosamine is converted to glucosamine by the hydrolysis procedure.
ND, not detected.
FIG. 1.
Chromatographic measurement of N-acetyl-d-glucosamine (N-ACET-GLC.NH2) released by chitinase treatment of Aphanomyces euteiches cell walls. (a) HPAEC-PAD profile of a hydrolysate obtained after treatment of crude cell walls with the Serratia marcescens chitinase for 2 h at 30°C. The retention time of 15.7 min was identical to that of a GlcNAc standard run under the same conditions (not shown). (b) Profile of a control hydrolysate obtained under the same conditions but in the absence of chitinase. For better visualization, this profile has been slightly moved downwards relative to the vertical axis.
Characterization of Aphanomyces euteiches cell wall chitosaccharides.
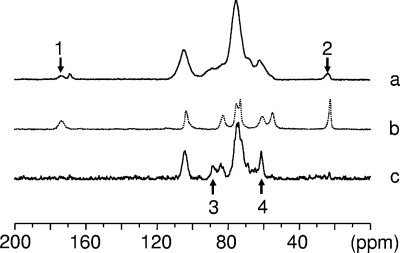
To investigate the nature of the Aphanomyces chitosaccharides, a purified cell wall preparation referred to as KOH-MeOH cell walls was used. This preparation did not contain proteins, and the monosaccharide analysis yielded only glucose and glucosamine, the latter accounting for 98 ± 13 μg·mg−1 of cell walls (means ± standard deviations [SD] from four independent experiments), a proportion similar to that of crude cell walls (Table 1). However, X-ray diffraction analysis of KOH-MeOH cell walls did not show the presence of crystalline chitin, and no chitin could be purified from this starting material when a relevant procedure described by Aronson and Bertke (2) was followed (data not shown). This suggested that the GlcNAc fraction of the Aphanomyces cell wall consists of noncrystalline components. This interpretation was corroborated by the 13C solid-state NMR spectroscopy analysis (Fig. 2): spectrum a, which corresponds to cell walls analyzed under dry conditions, shows two small resonances at 174 and 23.6 ppm (arrows 1 and 2) that are also observed when crystalline crab shell chitin is analyzed either under dry or wet conditions (spectrum b). These resonances can be ascribed to the C=O and the CH3 moieties of N-acetyl-d-glucosamine (77). Similar resonances were also observed in Aspergillus niger and ascribed to chitinaceous compounds in interaction with glucans by CP-MAS NMR (42). Quantitative analysis indicated that the corresponding GlcNAc material accounted for ca. 10% of the cell wall sample, confirming the aforementioned results. Remarkably, when the same cell wall sample was analyzed under wet conditions, the GlcNAc resonances disappeared (Fig. 2, spectrum c). This disappearance indicated that unlike crystalline chitin, which is insensitive to hydration, the GlcNAc component of the cell wall corresponds to a noncrystalline material that becomes mobile under the influence of hydration and that was therefore not observed under our spectral acquisition conditions. Resonance signals at 88.7 and 61.0 ppm were also observed (spectrum c, arrows 3 and 4), suggesting the presence of cellulose and (1→ 3)-β-d-glucan (68). Whereas further investigations are needed to confirm this, the occurrence of these polymers would be consistent with the glucose content of the cell wall and the general finding that oomycetes have a β-glucan/cellulose cell wall type (6).
FIG. 2.
13C CP/MAS NMR spectroscopy analysis of Aphanomyces euteiches cell walls. (a) KOH-MeOH-treated cell walls analyzed under dry conditions; (b) crystalline standard of poly(1→4)-β-d-N-acetylglucosamine (crab α-chitin); (c) as in panel a, but under wet conditions. Arrows 1 and 2 correspond to the small resonances identified as the C=O (at 174 ppm) and the CH3 (at 23.6 ppm) moieties of N-acetyl-d-glucosamine. Arrows 3 and 4 correspond to the resonances of the C-4 of cellulose and of the C-6 of (1→ 3)-β-D-glucan, respectively.
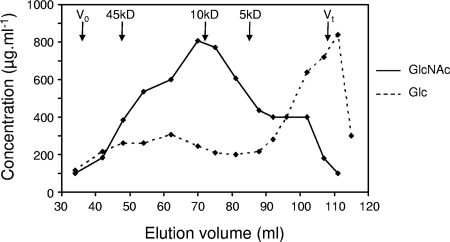
In order to confirm the conclusions drawn from biophysical analyses and to further characterize the noncrystalline chitosaccharides, a biochemical approach based on the use of specific glucanases was developed. First, cell wall preparations were hydrolyzed with a mixture of (1→ 3)-, (1→ 6)-, and (1→ 4)-β-glucanases. This led to the solubilization of virtually all the GlcNAc material together with the other glucans, leaving a residue containing only 1.4% ± 0.1% of the GlcNAc present in the starting material (means ± SD from two independent experiments). This result indicated that the A. euteiches chitosaccharides are soluble and noncrystalline, therefore supporting the conclusion drawn from the biophysical analyses. Second, cell wall preparations were submitted to sequential treatments with glucanases of various specificities, and the GlcNAc content of the resulting fractions was measured in order to obtain insight into the potential linkages between the chitosaccharides and glucans. Treatment with (1→ 3)-β-glucanases led to the solubilization of 14.2% ± 3.3% of the GlcNAc present in the starting material, and subsequent treatment with Westase, a (1→ 6)-β-glucanase preparation with minor (1→ 3)-β-glucanase activity, further solubilized 55.3% ± 12.4% of the GlcNAc present in the starting material (means ± SD from two independent experiments). When the resulting cell wall pellet from one experiment was further treated with a (1→ 4)-β-glucanase, 16.7% of the GlcNAc present in the starting material was recovered in the supernatant and 7.8% remained in the pellet. These data suggested that most of the A. euteiches chitosaccharides are associated with (1→ 6)-β-glucans. In order to further characterize these chitosaccharides, the supernatants from the (1→ 3)-β-glucanase and the (1→ 6)-β-glucanase digestion steps were separately analyzed by size exclusion chromatography on Bio-Gel P-60, and the GlcNAc/Glc ratio from the eluted fractions was determined. This ratio varied between 10 and 40% for the high-molecular-weight material eluted before the bulk of low-molecular-weight oligoglucosides released by the glucanases (data not shown). This suggested that, despite the glucanase treatments, the chitosaccharides were still associated with glucans. In order to further purify them, the corresponding fractions were pooled, treated again with a concentrate mixture of (1→ 3)-, (1→ 6)-, and (1→ 4)-β-glucanases, and the resulting hydrolysate was rechromatographed on a Bio-Gel P-60 column. The elution profile presented in Fig. 3 shows that the GlcNAC-containing material was mostly enriched in a fraction with a GlcNAc/Glc ratio of 80% which was eluted near a 10-kDa dextran standard. Most of the chitosaccharides had an apparent molecular weight ranging from ca. 4,000 to 45,000 and were eluted in fractions containing similar amounts of glucose. This profile shows that the chitosaccharides are soluble high-molecular-weight compounds and suggests that their solubility is due to a branched structure involving glucose residues.
FIG. 3.
Chromatography of Aphanomyces euteiches chitosaccharides on a Bio-Gel P-60 column. The chitosaccharides were prepared by multistep digestion of KOH-MeOH cell walls with (1→ 3)-, (1→ 4)-, and (1→ 6)-β-glucanases. V0, void volume, determined with dextran 2000; Vt, total volume of the column. The positions of peaks of dextran standards of the respective molecular masses are shown by vertical arrows. The GlcNAc and Glc concentrations of the fractions were determined by colorimetry using the anthrone reagent and HPAEC-PAD analysis after acid hydrolysis, respectively.
In conclusion, our data demonstrate that, in contrast to Saprolegnia, Aphanomyces does not contain detectable amounts of crystalline chitin in its cell wall but contains soluble noncrystalline chitosaccharides associated with other cell wall polysaccharides. This conclusion is consistent with the fact that our attempts to measure the chitin synthase activity of Aphanomyces, following a standard assay based on the quantification of insoluble GlcNAc polymeric products (12), were not successful.
Lectin labeling of Aphanomyces euteiches cell wall chitosaccharides.
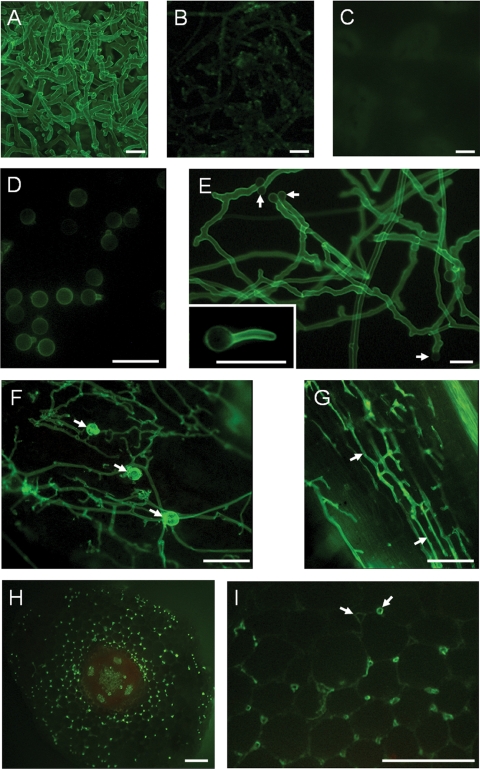
Wheat germ agglutinin is a lectin that specifically interacts with chitosaccharides. It has been widely used to label chitin in fungal cell walls (16, 18, 70, 84). In order to obtain additional evidence for the occurrence of chitosaccharides in the cell wall of Aphanomyces, labeling with a FITC-WGA conjugate was analyzed by epifluorescence microscopy (Fig. 4). The hyphal external surface of Aphanomyces was strongly labeled (Fig. 4A), and this labeling was reduced to a great extent upon preincubation of the conjugate with a chitin hydrolysate (Fig. 4B). In contrast, the mycelium of Phytophthora used as a control hardly showed any detectable labeling (Fig. 4C). This result is consistent with the very small amount of glucosamine in Phytophthora cell wall hydrolysates (Table 1) and with a previous report (16) in which no labeling of Phytophthora was observed in the presence of a WGA-ovomucoid-gold complex. Further experiments were conducted to investigate the distribution and abundance of surface chitosaccharides at various stages of the life cycle of Aphanomyces and to evaluate FITC-WGA labeling as a tool to detect the microorganism during its interaction with the host plant (Fig. 4D to I). Encysted zoospores (cysts) were labeled (Fig. 4D), but the intensity of labeling of individual cells was weak and highly variable. Germ tubes and resulting hyphae were labeled more intensely than the cysts (Fig. 4E), which reflects a higher accessibility or abundance of the chitosaccharides. This is consistent with an early study in the crayfish parasite Aphanomyces astaci, which showed that cyst and germ tube cell walls were of a different structure (61). Hyphae running on the surface of Medicago truncatula host roots were readily detected by FITC-WGA labeling, as well as oospores, which were intensely labeled (Fig. 4F). Attempts to directly label Aphanomyces structures within the roots were unsuccessful because the conjugate did not diffuse inside the tissues, but fresh tissue sectioning allowed detection of the pathogen within the tissues. Longitudinal sections showed hyphae preferentially growing parallel to the root axis, with some abrupt turns suggesting that they were actually growing between the host cells (Fig. 4G). This intercellular growth pattern was confirmed by observing transverse sections (Fig. 4H and I). The weak labeling of the plant cell wall allowed visualization of the boundaries of plant cells and clear differentiation between the plant cells and the intensely labeled Aphanomyces transversally cut hyphae. The latter were mostly localized in the intercellular spaces between three adjacent plant cells (Fig. 4I). In summary, labeling with FITC-WGA allows the study of the A. euteiches infectious process and suggests the presence of surface chitosaccharides in all its life stages presenting a cell wall.
FIG. 4.
Wheat germ agglutinin labeling of Aphanomyces euteiches. Samples were incubated with a FITC-WGA conjugate and analyzed by epifluorescence microscopy. (A) A. euteiches mycelium; (B) as in panel A, but the FITC-WGA conjugate was preincubated with a chitin hydrolysate; (C) control with P. parasitica mycelium; (D) cysts and germinating cysts of A. euteiches obtained in vitro; (E) hyphae arising from in vitro-germinated cysts (arrows), with an insert showing that the cyst surface is much less labeled than the hyphal surface; (F) hyphae and oospores (arrows) produced on the surfaces of infected Medicago truncatula roots; (G to I) hyphae growing within an infected root cut longitudinally (G) or transversally (H and I) before lectin labeling. The arrows in panel G show hyphae growing along the axis of the root, and the arrows in panel I show intercellular spaces in the root cortex either free (left arrow) or colonized (right arrow) by A. euteiches hyphae. Bars = 25 μm (A to E) or 100 μm (F to I).
Characterization of Aphanomyces euteiches CHS genes.
The presence of chitosaccharides in the Aphanomyces cell wall is expected to result from the expression of CHS-related genes. CHSs are membrane-associated processive glycosyltransferases, i.e., enzymes that catalyze multiple transfers of GlcNAc residues from UDP-GlcNAc onto the nonreducing end of growing chitin chains. They belong to glycosyltransferase family 2 (15). Sequence similarity analyses lead to the classification of fungal CHSs into seven classes, with most of them being distributed into two divisions: class I, II, and III enzymes are grouped into division 1, and class IV, V, and VI enzymes are grouped into division 2, whereas class VII fungal CHSs and animal CHSs form distinct groups (17, 65). The A. euteiches EST database (52) contains three gene entries (Ae_1AL1952, Ae_1AL4892, and Ae_6AL6991) with similarity to CHS genes (E values of <1e−25). Complete sequencing of the two strands of cDNA clones led to the assumption that Ae_1AL1952 and Ae_6AL6991 correspond to partial cDNAs of the same gene, referred to as AeCHS1, whereas Ae_1AL4892 corresponds to a partial cDNA of a different gene, referred to as AeCHS2. A Southern blot analysis was performed in order to check the presence of these putative AeCHS genes (Fig. 5). Sequence analysis predicted that KpnI and ScaI would cleave once within the AeCHS1 and AeCHS2 probe fragments, respectively. This is consistent with the presence of two hybridizing fragments in the corresponding lanes (Fig. 5, AeCHS1, lane 3, and AeCHS2, lane 1, respectively), whereas the other enzymes whose sites were absent in the probe sequences resulted in unique hybridizing fragments. None of the hybridizing fragments were revealed by both probes. These data confirm the occurrence of two different single-copy genes.
FIG. 5.
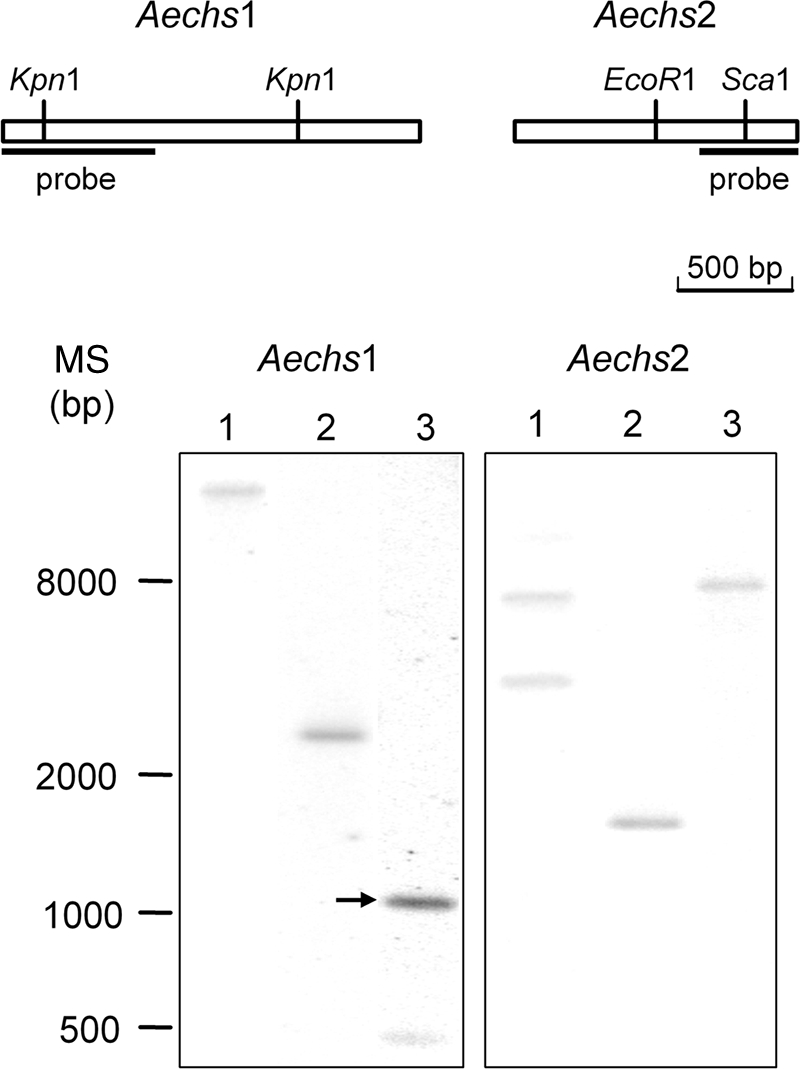
Southern hybridization analysis of Aphanomyces euteiches chitin synthase genes. Genomic DNA (100 μg) digested with either ScaI (lane 1), EcoRI (lane 2), or KpnI (lane 3) was analyzed on a 1% agarose gel, transferred to a nylon membrane, and hybridized with a radiolabeled AeCHS1 probe (Aechs1) or AeCHS2 probe (Aechs2). The arrow shows a KpnI restriction fragment hybridizing with the AeCHS1 that was predicted from sequence analysis to generate a 1,080-bp fragment. The positions of molecular size standards (MS) (in base pairs) are shown to the left of the gels.
The transcribed AeCHS1 and AeCHS2 sequences were extended by 476 and 1,321 nucleotides, respectively, by 5′RACE-PCR. Virtual translation of the available 2,585 nucleotides of AeCHS1 and 2,506 nucleotides of AeCHS2 resulted in polypeptide sequences of 840 and 802 amino acids, respectively. The best hit for both sequences after BLASTp analysis was chitin synthase CHS2 from S. monoica (referred to hereafter as SmCHS2) (54). As the SignalP algorithm (10) did not predict the presence of a signal peptide in either of the AeCHS sequences, it might be questioned whether they are full-length proteins. However, they are likely complete because (i) their sizes fall within the range of 738 to 1,131 amino acid residues recorded for division 1 fungal CHSs (65) and (ii) the RACE-PCR technology that was used allows only the amplification of fragments corresponding to full-length cDNAs (see Materials and Methods). In addition, there is a stop codon shortly upstream of the putative start codon of the AeCHS2 cDNA. Finally, both A. euteiches CHS (AeCHS) sequences show all features of a functional CHS enzyme as described below.
Analysis of SmCHS2, AeCHS1, and AeCHS2 amino acid sequences is presented in Fig. 6, where conserved CHS motifs (motifs a to h) (17, 54, 65) are indicated. Both A. euteiches CHSs show putative transmembrane domains with similar localizations to those found in SmCHS2, except at the C terminus where an additional putative transmembrane domain is predicted in AeCHS enzymes (Fig. 6A). This similarity suggests that the three proteins have the same topology in the region between motifs a and h. Both AeCHS sequences contain the complete CHS signature found in SmCHS2 (motifs a to h), including motifs or residues presumably involved in UDP-GlcNAc binding (motifs a, b, and c), catalysis (motif f), and enzyme processivity (motifs g and h). Motif g in both AeCHS enzymes is QRRRW, a sequence most often found in fungal CHS, from which the S. monoica enzyme diverges significantly by the substitution of the first arginine by an alanine residue (Fig. 6B). Motifs a, c, and f match the consensus sequences of division 1 CHSs, which are T(M/Y)YNE, DXGT, and LAEDRIL, respectively (17). In addition, the sequence-spanning motifs d and e strongly resemble the consensus sequence NPLV(A/Y)XQNFEYKXSNILDK(P/T)XESX(F/M)GX(I/V)(S/T)VLP(G/A)A(F/L)(S/C)AYR found in division 1 CHSs, and the dipeptide LN following motif g is also typical of division 1 CHSs (65) (Fig. 6B). These observations suggest a phylogenic relatedness between division 1 fungal CHSs and oomycete CHSs, as already noticed by Ruiz-Herrera et al. (65).
FIG. 6.
Alignment of Aphanomyces euteiches and Saprolegnia monoica chitin synthase sequences. (A) Schematic structure of the AeCHS1, AeCHS2, and SmCHS2 deduced amino acid sequences. Conserved chitin synthase motifs are shown by open circles (motifs a to h [see below]), and predicted transmembrane domains are indicated by black boxes. The boundaries of the sequences aligned in panel B are shown by dotted lines. aa, amino acids. (B) Alignment of the AeCHS1, AeCHS2, and SmCHS2 regions encompassing the complete CHS signature. Chitin synthase motifs (T/P)XYXE (motif a), D (motif b), DX(G/D)(T/C) (motif c), QXXEY (motif d), LP(G/A) (motif e), L(A/G)EDRXL (motif f), Q(R/G)RRW (motif g), and (S/T)WG (motif h) are shown by open boxes, and amino acid residues of the glycosyltransferase family 2 signature are in bold type. Gaps introduced to maximize alignment of the sequences are indicated by dashes.
Phylogenetic analysis of oomycete chitin synthase genes.
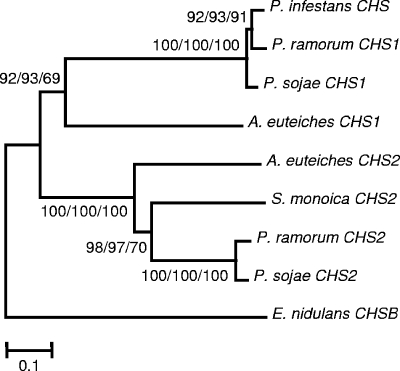
Pairwise global alignments between the AeCHS1, AeCHS2, and SmCHS2 sequences suggested that AeCHS2 and SmCHS2 are actually more similar to each other (with 55.2% similarity) than they are to AeCHS1 (with 50.4% and 42.4% similarities, respectively). This observation is reminiscent of the finding that orthologous CHSs are more similar than paralogous CHS in organisms of the kingdom Fungi. In these organisms, it has been recognized that the CHS phylogeny does not reflect the kingdom phylogeny, a phenomenon that is explained by assuming that the diversification of CHS genes into more than five classes occurred before the diversification of the fungal phylum itself (65). The oomycete orders are distributed into two major lineages, the Pythiales and Peronosporales on the one hand and the Saprolegniales, Leptomitales, and Rhipidiales on the other hand (63). Phytophthora belongs to the first lineage, whereas Aphanomyces belongs to the second. We took advantage of the progress in Phytophthora genomics since the last exhaustive study of Ruiz-Herrera et al. (65) to construct a dendrogram of oomycete CHSs and check whether the CHS phylogeny reflects the oomycete classification. We looked for sequences in databases from whole-genome sequencing projects of Phytophthora species (described by Tyler et al. [83] for P. sojae and P. ramorum and at the Broad Institute website for P. infestans) and verified, by searching for corresponding ESTs in dedicated databases (see Materials and Methods for the URLs), that the identified Phytophthora sojae and Phytophthora infestans genes are actually expressed and not pseudogenes. These genes were called CHS1 or CHS2 according to their position in the resulting dendrogram (Fig. 7). This dendrogram shows that oomycete CHSs form two clusters of so-called CHS1 and CHS2 enzymes. Phytophthora CHS2 are more related to Saprolegnia and Aphanomyces CHS2 than to Phytophthora CHS1, thereby suggesting that oomycete CHSs originate from a common ancient ancestor gene that duplicated before the divergence of the Saprolegniale and Peronosporale lineages.
FIG. 7.
Phylogeny of oomycete chitin synthases. The A. euteiches CHS amino acid sequences were aligned with other oomycete CHS sequences, such as Phytophthora and Saprolegnia CHS sequences. The phylogenetic tree was constructed with the Mega version 3.1 software using minimum evolution (ME), neighbor-joining (NJ), and maximum parsimony (MP) methods (Poisson correction for multiple amino acid substitutions; 1,000 random bootstrap replicates). All three methods produced similar topologies; this figure shows the ME tree. Numbers on each branch represent bootstrap values of ME/NJ/MP analyses. The Emericella nidulans CHSB sequence was employed as an outgroup taxon.
Biological roles of the Aphanomyces euteiches cell wall chitosaccharides.
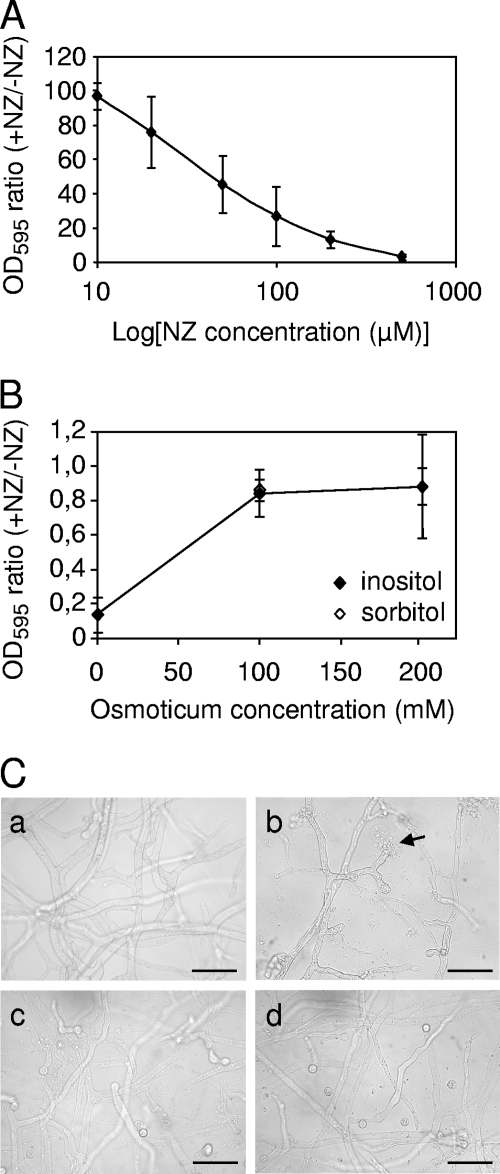
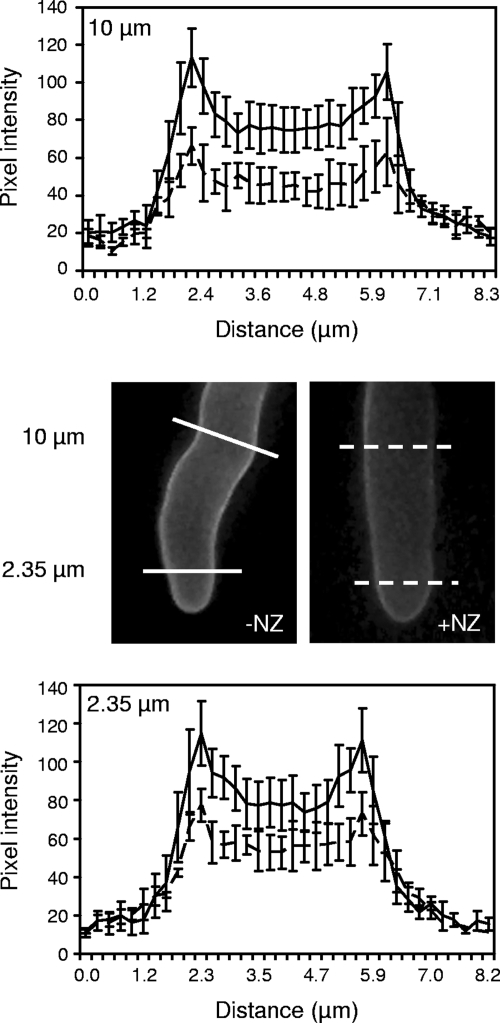
To obtain insight into the biological roles of Aphanomyces chitosaccharides, the microorganism was grown in the presence of nikkomycin Z, a competitive inhibitor of CHS enzymes (66) (Fig. 8). This resulted in a marked growth inhibition, which reached ca. 50% in the presence of 50 μM NZ (Fig. 8A). This 50% inhibitory concentration is comparable to the concentrations that are required to inhibit the true fungi that are sensitive to NZ, such as Candida albicans which shows an 50% inhibitory concentration of ca. 25 μM NZ (45). It should be noted that sensitivity to NZ varies greatly between fungal species (32, 36, 78). In Aphanomyces, growth in the presence of 100 μM NZ led to hyphal tip bursting and cytoplasmic leakage (Fig. 8C, panel b). This suggested that cell wall synthesis at the hyphal apex was affected by the CHS inhibitor and that the resulting cell wall structure could not afford efficient osmotic protection. To confirm that NZ was directly affecting cell wall integrity, we searched for osmoprotective compounds in order to check whether they could reverse the effect of NZ. Polyols, such as inositol or sorbitol, were reported not to be used as carbon sources by A. euteiches (62) and therefore were good candidates as osmoprotectants during growth. The microorganism was thus incubated in the presence of 100 μM NZ and with various concentrations of inositol or sorbitol. Both inositol and sorbitol at concentrations in the range from 100 to 200 mM protected Aphanomyces from the adverse effect of NZ, restoring growth to nearly control levels and abolishing hyphal tip bursting (Fig. 8B and C, panels c and d). In addition, when the microorganism was grown in GY medium supplemented with 200 mM sorbitol, the intensity of WGA labeling of the cell wall was significantly lower in the presence of 100 μM NZ than in the absence of the drug, showing a reduction between one-third and one-half depending on the distance to the hyphal tip (Fig. 9). Quantification by HPAEC-PAD analysis of cell wall hydrolysates indicated that the GlcNAc content of the cell wall of mycelium grown in the presence of 100 μM NZ was reduced by 33% ± 12% compared to the control grown in the absence of the drug (mean ± SD from three independent experiments). This demonstrates that the difference in in vivo WGA labeling intensity reflects an actual difference in chitosaccharide content and not in chitosaccharide accessibility to the lectin. Taken together, these data strongly suggest that nikkomycin Z affects cell wall integrity by inhibiting a CHS activity that is needed for the synthesis of chitosaccharides conferring the mechanical strength of the cell wall.
FIG. 8.
Effect of nikkomycin Z on Aphanomyces euteiches in vitro growth. (A and B) Effect of NZ on mycelium growth in the absence (A) or presence (B) of osmoprotectants. A nutritive medium was inoculated with zoospores in the presence (+NZ) or absence (−NZ) of various concentrations of NZ (A) or of 100 μM NZ and 0, 100, or 200 mM inositol or sorbitol (B). The hyphal density was determined after 3 days of incubation as described in Materials and Methods. The ratios between the absorbances of the mycelium treated with NZ and not treated with NZ are reported. Data are means ± SD (error bars) from three independent experiments. OD595, optical density at 595 nm. (C) Microscopy observation of A. euteiches treated with NZ in the presence or absence of osmoprotectants. The assay was performed as above in the absence (a) or presence (b) of 100 μM NZ alone or in combination with 200 mM inositol (c) or sorbitol (d). The arrow shows a hyphal tip bursting. Bars = 50 μm.
FIG. 9.
Effect of nikkomycin Z on wheat germ agglutinin labeling of Aphanomyces euteiches. A nutrient medium containing 200 mM sorbitol was inoculated with zoospores in the presence (+NZ) or absence (−NZ) of 100 μM NZ. After 3 days of growth, the samples were incubated with a FITC-WGA conjugate and analyzed by epifluorescence microscopy. The pixel intensity of fluorescence was scanned along lines perpendicular to hyphal axes at a distance of either 10 μm (top panel) or 2.35 μm (bottom panel) from hyphal tips. Note the decrease of fluorescence intensity in the NZ-treated samples compared to the untreated ones. Within the hyphae (i.e., between the peaks corresponding to the cell wall), the fluorescence intensity does not return to background level due to signal originating from out-of-focus planes. The experiment was repeated twice independently, and the data are means ± standard errors (error bars) of scans performed on four different hyphae from a representative experiment.
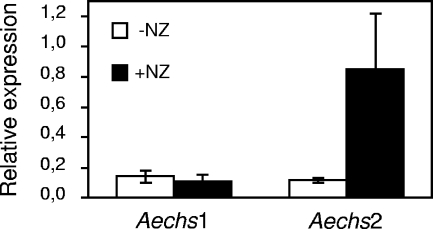
In true fungi, inhibition of the synthesis of a cell wall polysaccharide often induces compensatory mechanisms, such as the elevated synthesis of the targeted polysaccharide or of other polysaccharides that participate in resistance or tolerance to the inhibitor (see, for example, references 20 and 56). To check whether Aphanomyces is able to respond to NZ treatment by an elevated CHS gene expression, we measured by RT-qPCR the expression of the AeCHS1 and AeCHS2 genes in the mycelium grown in the presence or absence of 50 μM NZ. Figure 10 illustrates that the two AeCHS genes were expressed at similar levels in the untreated mycelium. AeCHS1 expression was not modified in the presence of NZ, whereas AeCHS2 showed an eightfold induction under this condition. Such an induction confirms that NZ affects the cell wall dynamics and suggests that this stress induces a compensatory response, including the induction of the AeCHS2 gene.
FIG. 10.
Effect of nikkomycin Z on Aphanomyces euteiches chitin synthase gene expression. Expression of the AeCHS1 and AeCHS2 genes in the absence (−NZ) or presence (+NZ) of 50 μM NZ. AeCHS1 and AeCHS2 mRNA copy numbers were determined by RT-qPCR and standardized by the mRNA copy numbers of an α-tubulin gene. The means ± SD (error bars) of the resulting gene expression ratios calculated from three independent experiments are shown.
DISCUSSION
This work represents the first study of the cell wall of the Saprolegniale Aphanomyces euteiches. Biochemical analysis indicated a strong predominance of glucose, in agreement with the data obtained in other oomycetes, which most often have a β-glucan/cellulose cell wall type (2, 6, 8, 64). However, whereas oomycetes usually contain minute quantities of hexosamines in their cell walls, this study showed that a significant proportion of GlcNAc is present in Aphanomyces. Biophysical analyses suggested that the GlcNAc-containing material of the Aphanomyces cell wall is not crystalline chitin but that it corresponds to noncrystalline chitosaccharides instead. The detrimental effect of the chitin synthase inhibitor nikkomycin Z showed that these components are necessary for cell wall integrity. Thus, it can be hypothesized that they might participate in cell wall scaffolding by being linked to other cell wall polysaccharides. In Saccharomyces cerevisiae, for example, chitin occurs in various proportions depending on the physiological state and on its subcellular localization, as a free polymer or as a polymer bound to (1→ 3)-β-glucan or (1→ 6)-β-glucan chains (13, 46). Remarkably, it was observed that linkage of chitin to (1→ 6)-β-glucan greatly increases its solubility compared to free chitin (13). Here the use of specific glucanases allowed us to show that most of the A. euteiches chitosaccharides are associated with (1→ 6)-β-glucans, and size exclusion chromatography analysis of enzymatic hydrolysis products showed that GlcNAc-enriched compounds were distributed around the molecular weight value of 10,000. The fact that these chitosaccharides are soluble and the presence of glucose in the corresponding fractions suggest that they correspond to GlcNAc polymers branched with glucose oligomers. The major difference between A. euteiches and S. cerevisiae cell wall GlcNAc polymers might then be summarized as the absence of free chitin in A. euteiches. Further studies are necessary to confirm this interpretation and fully characterize the A. euteiches chitosaccharides.
Comparison of the cell walls of A. euteiches and S. monoica reveals similarities but also striking differences. Both organisms have a glucose-rich cell wall that in addition contains GlcNAc (12), and WGA labeling showed in both organisms that the GlcNAc components are not present only in the inner cell wall layers, in contrast to what is usually observed in true fungi (16, 70, 84). Leal-Morales et al. (50) mentioned that the GlcNAc-containing material was uniformly distributed throughout the mycelial walls of Saprolegnia, a feature that is likely to occur also in Aphanomyces, as the proportion of GlcNAc in KOH-MeOH cell walls, where the amorphous superficial layers have been solubilized by treatment with hot KOH-MeOH, was the same as in crude cell walls. In Saprolegnia, however, the GlcNAc components are true crystalline chitin that accounts for only 0.7% of the cell wall and is not essential for hyphal growth (12), whereas in Aphanomyces these components are noncrystalline chitosaccharides that account for ca. 10% of the cell wall and are required for hyphal growth. These differences suggest that there is substantial variability in cell wall structure, and thus biology, within a single phylum, such as the Saprolegniales.
At the molecular level, our study led to the identification of two new putative chitin synthase sequences of a Saprolegniale species, corresponding to two putative CHS genes that are expressed in A. euteiches. Two CHS genes were also detected in both S. monoica and Plasmopara viticola (54, 87), and additional putative CHS sequences from a few oomycete species can be found in the NCBI database. With the exception of the SmCHS2 sequence, they are all very short. Careful examination of the available Phytophthora genome sequences revealed the presence of two genes in P. sojae and P. ramorum, whereas only one gene was found in P. infestans. The corresponding full-length protein sequences were included in a data set together with the SmCHS2, AeCHS1, and AeCHS2 sequences to draw a phylogenetic tree of oomycete CHSs. Evolutionary distances at the constructed dendrogram suggest that oomycete enzymes originate from an ancient common ancestor gene that underwent duplication before the major oomycete lineages diverged, with subsequent loss of one paralog in P. infestans. The various other putative CHS fragments that have been characterized thus far are likely to share this ancestry (data not shown), except for the Plasmopara viticola CHS2 (PvCHS2) fragment which is not related to the other oomycete CHS sequences (87). This discrepancy suggests that oomycete CHSs might be more diverse than expected. More sequence information is needed to draw a more comprehensive scheme of oomycete CHS evolution, and the significance of the presence of CHS genes in organisms reported not to contain chitin (e.g., Phytophthora) should be further investigated.
True fungi have at least three CHS genes, a redundancy that corresponds to different patterns of expression and complementary biological roles. In Saccharomyces cerevisiae, chs1 encodes a repair enzyme, whereas chs2 is involved in the synthesis of the central layer of the primary septum and chs3 forms the chitin ring during budding (14, 71). There is evidence for differential expression of CHS genes in Plasmopara viticola, where PvCHS1 was shown to be constitutively expressed, whereas PvCHS2 is specifically transcribed in sporangiophores and sporangia (87). In S. monoica, weak expression of both CHS genes was observed in the mycelium, but their regulation was not investigated. For A. euteiches, we observed similar expression of both genes in the mycelium, but only one responded to NZ treatment, thus suggesting that they do not share the same biological role. Their expression pattern during development of Aphanomyces in vitro and in planta is worthy of investigation. In addition, reverse genetics studies should help decipher their respective role in cell wall synthesis and function and the possible occurrence of a cell wall salvage pathway. Procedures for such studies, based on gene silencing or RNA interference after protoplast or Agrobacterium-mediated transformation, have been established in some Phytophthora species (44) but are not routine (33) and are not yet established in Aphanomyces.
Chitin is an essential component of fungal cell walls, but the redundancy of the CHS genes has been invoked as a major difficulty in designing antifungal compounds that efficiently target chitin synthesis (66). On the other hand, it was shown that disruption of only one CHS gene could lead to a drastic reduction in the pathogenicity of plant parasites (53, 74, 86, 88), suggesting that targeting only one enzyme might be sufficient. Apparently, there are fewer CHS genes in oomycetes than in fungi. The fact that chitin synthesis is an essential process in Aphanomyces leads to the conclusion that chitin synthesis should not be underestimated as a target for designing new compounds aiming at controlling plant diseases caused by oomycetes. Preliminary data suggest that, although not conferring lasting protection, the addition of 100 μM NZ to the zoospore inoculum can delay the development of root rot symptoms. This suggests that a higher concentration of NZ would be necessary to potentially protect the plants by foliar application or by incorporation to the soil. This, together with the high cost of this chemical and the fact that chitin occurs in numerous other organisms, including beneficial mycorrhiza or pollinating insects, makes necessary the isolation of more-active and specific compounds before envisaging crop protection against oomycetes by targeting CHSs.
Labeling of Aphanomyces hyphae with a FITC-WGA conjugate has important implications, considering that A. euteiches is a major pathogen of legumes. It shows that the chitosaccharides are exposed on the microorganism's surface, as was already shown in Plasmopara viticola (87). This contrasts markedly with the situation in true fungi where chitin is usually embedded in inner cell wall layers. It is well-known that plants defend themselves by producing a large array of hydrolases targeted to pathogen cell walls, including endo- and exochitinases. Upon pathogen attack, the chitooligosaccharides (COS) that are released by these enzymes are perceived as nonself compounds by the host plant and elicit defense responses (43). COS signaling fits with the concept of microbe (or pathogen)-associated molecular patterns that describes the molecular basis of innate immunity in plants and animals (51, 60). Recently, it was shown that the avirulence protein AVR4 of Cladosporium fulvum exhibits chitin-binding activity and functions as a virulence factor by shielding the fungal chitin and presumably protecting it from degradation and/or perception by the host (84). Another possible escape from chitinase-mediated COS generation and recognition is the conversion of chitin to chitosan as soon as the parasite penetrates the plant (30). In this context, it is striking that A. euteiches exposes chitosaccharides at its surface. As it was shown that the elicitor activity of linear COS in various plants depends on their degree of polymerization (72), we have undertaken the purification of the A. euteiches cell wall GlcNAc-containing material in order to determine its structure and assess its activity. Depending on the perception system operating in Medicago truncatula, it might act as an active elicitor by itself. Alternatively, it could act in association with other cell wall glucans, as one of the most potent elicitors in legumes is a well-characterized hepta-β-glucoside oomycete cell wall fragment (21, 72).
In conclusion, our data demonstrate for the first time in a phytopathogenic oomycete species the presence of significant amounts of exposed cell wall chitosaccharides, which might be perceived by the host plant. In addition, they show the involvement of these chitosaccharides in cell wall integrity, suggesting that the chitin biosynthetic pathway may be a potential target of antioomycete compounds.
Acknowledgments
We thank the Genomics Platform of the Genopole of Toulouse for RT-qPCR and sequencing facilities; Bernard Henrissat (AFMB UMR 6098 CNRS/UI/UII, Marseille, France) for advice on chitin synthase sequence analysis; Alain Jauneau (IFR40-RIO Imaging Platform of Toulouse) and Anne-Gabriel Desseix for help with microscopy analyses and with lectin labeling and nikkomycin Z assays, respectively; Henri Chanzy (CERMAV, Grenoble, France) for cell wall X-ray analyses and helpful discussions; and Nickolas Panopoulos (IMBB, Heraklion, Greece) for revising the manuscript.
Vincent Bulone was supported by the Swedish Centre for Biomimetic Fiber Engineering (Biomime).
Footnotes
Published ahead of print on 19 September 2008.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Aronson, J. M., and C. C. Bertke. 1987. Isolation and analysis of cell walls, p. 175-185. In M. S. Fuller and A. Jaworski (ed.), Zoosporic fungi in teaching and research. Southeastern Publishing Corporation, Athens, GA.
- 3.Bacic, A., M. L. Williams, and A. E. Clarke. 1985. Studies on the cell surface of zoospores and cysts of the fungus Phytophthora cinnamomi: nature of the surface saccharides as determined by quantitative lectin binding studies. J. Histochem. Cytochem. 33384-388. [DOI] [PubMed] [Google Scholar]
- 4.Baker, L. G., C. A. Specht, M. J. Donlin, and J. K. Lodge. 2007. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell 6855-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldauf, S. L. 2003. The deep roots of eukaryotes. Science 3001703-1706. [DOI] [PubMed] [Google Scholar]
- 6.Bartnicki-García, S. 1968. Cell wall chemistry, morphogenesis and taxonomy of fungi. Annu. Rev. Microbiol. 2287-108. [DOI] [PubMed] [Google Scholar]
- 7.Bartnicki-García, S. 1966. Chemistry of hyphal walls of Phytophthora. J. Gen. Microbiol. 4257-69. [DOI] [PubMed] [Google Scholar]
- 8.Bartnicki-García, S., and M. C. Wang. 1983. Biochemical aspects of morphogenesis in Phytophthora, p. 121-137. In P. H. Tsao (ed.), Phytophthora: its biology, taxonomy, ecology, and pathology. APS Press, St. Paul, MN.
- 9.Bécard, G., and J. A. Fortin. 1988. Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol. 108211-218. [DOI] [PubMed] [Google Scholar]
- 10.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 11.Birch, P. R. J., A. P. Rehmany, L. Pritchard, S. Kamoun, and J. L. Beynon. 2006. Trafficking arms: oomycete effectors enter host plant cells. Trends Microbiol. 148-11. [DOI] [PubMed] [Google Scholar]
- 12.Bulone, V., H. Chanzy, L. Gay, V. Girard, and M. Fèvre. 1992. Characterization of chitin and chitin synthase from the cellulosic cell wall fungus Saprolegnia monoica. Exp. Mycol. 168-21. [Google Scholar]
- 13.Cabib, E., and A. Durán. 2005. Synthase III-dependent chitin is bound to different acceptors depending on location on the cell wall of budding yeast. J. Biol. Chem. 2809170-9179. [DOI] [PubMed] [Google Scholar]
- 14.Cabib, E., A. Sburlati, B. Bowers, and S. J. Silverman. 1989. Chitin synthase 1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 1081665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell, J. A., G. J. Davies, V. Bulone, and B. Henrissat. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326929-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chérif, M., N. Benhamou, and R. R. Bélanger. 1992. Occurrence of cellulose and chitin in the hyphal walls of Pythium ultimum: a comparative study with other plant pathogenic fungi. Can. J. Microbiol. 39213-222. [Google Scholar]
- 17.Choquer, M., M. Boccara, I. R. Gonçalves, M. C. Soulié, and A. Vidal-Cros. 2004. Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur. J. Biochem. 2712153-2164. [DOI] [PubMed] [Google Scholar]
- 18.Ciopraga, J., O. Gozia, R. Tudor, L. Brezuica, and R. J. Doyle. 1999. Fusarium sp. growth inhibition by wheat germ agglutinin. Biochim. Biophys. Acta 1428424-432. [DOI] [PubMed] [Google Scholar]
- 19.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10685-686. [DOI] [PubMed] [Google Scholar]
- 20.Cota, J. M., J. L. Grabinski, R. L. Talbert, D. S. Burgess, P. D. Rogers, T. D. Edlind, and N. P. Wiederhold. 2008. Increases in SLT2 expression and chitin content are associated with incomplete killing of Candida glabrata by caspofungin. Antimicrob. Agents Chemother. 521144-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Day, B., and T. Graham. 2007. The plant host pathogen interface: cell wall and membrane dynamics of pathogen-induced responses. Ann. N. Y. Acad. Sci. 1113123-134. [DOI] [PubMed] [Google Scholar]
- 22.Deacon, J.-W., and G. Saxena. 1998. Germination triggers of zoospore cysts of Aphanomyces euteiches and Phytophthora parasitica. Mycol. Res. 10233-41. [Google Scholar]
- 23.Dellaporta, S. L., J. Wood, and J. B. Hicks. 1983. A plant DNA minipreparation version II. Plant Mol. Biol. Rep. 119-21. [Google Scholar]
- 24.Dick, M. W. 2001. Straminipilous fungi. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 25.Dietrich, S. M. 1973. Carbohydrates from the hyphal walls of some oomycetes. Biochim. Biophys. Acta 31395-98. [DOI] [PubMed] [Google Scholar]
- 26.Dietrich, S. M. 1975. Comparative study of hyphal wall components of oomycetes: Saprolegniaceae and Pythiaceae. An. Acad. Brasil. Ciênc. 47155-162. [Google Scholar]
- 27.Dumas, B., A. Bottin, E. Gaulin, and M. T. Esquerré-Tugayé. 2008. Cellulose binding domains: cellulose-associated defense sensing partners? Trends Plant Sci. 13160-164. [DOI] [PubMed] [Google Scholar]
- 28.Ehrlich, H., M. Krautter, T. Hanke, P. Simon, C. Knieb, S. Heinemann, and H. Worch. 2007. First evidence of the presence of chitin in skeletons of marine sponges. Part II. Glass sponges (Hexactinellida: Porifera). J. Exp. Zool. B Mol. Dev. Evol. 308473-483. [DOI] [PubMed] [Google Scholar]
- 29.Ehrlich, H., M. Maldonado, K. D. Spindler, C. Eckert, T. Hanke, R. Born, C. Goebel, P. Simon, S. Heinemann, and H. Worch. 2007. First evidence of chitin as a component of the skeletal fibers of marine sponges. Part I. Verongidae (Demospongia: Porifera). J. Exp. Zool. B Mol. Dev. Evol. 308347-356. [DOI] [PubMed] [Google Scholar]
- 30.El Gueddari, N. E., U. Rauchhaus, B. M. Moerschbacher, and H. B. Deising. 2002. Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 156103-112. [Google Scholar]
- 31.Gajendran, K., M. D. Gonzales, A. Farmer, E. Archuleta, J. Win, M. E. Waugh, and S. Kamoun. 2006. Phytophthora functional genomics database (PFGD): functional genomics of Phytophthora-plant interactions. Nucleic Acids Res. 34D465-D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaughran, J. P., M. H. Lai, D. R. Kirsch, and S. J. Silverman. 1994. Nikkomycin Z is a specific inhibitor of Saccharomyces cerevisiae chitin synthase isozyme Chs3 in vitro and in vivo. J. Bacteriol. 1765857-5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaulin, E., N. Haget, M. Khatib, C. Herbert, M. Rickauer, and A. Bottin. 2007. Transgenic sequences are frequently lost in Phytophthora parasitica transformants without reversion of the transgene-induced silenced state. Can. J. Microbiol. 53152-157. [DOI] [PubMed] [Google Scholar]
- 34.Gaulin, E., C. Jacquet, A. Bottin, and B. Dumas. 2007. Root rot disease of legumes caused by Aphanomyces euteiches. Mol. Plant Pathol. 8539-548. [DOI] [PubMed] [Google Scholar]
- 35.Goldman, R. C., and A. Branstrom. 1999. Targeting cell wall synthesis and assembly in microbes: similarities and contrasts between bacteria and fungi. Curr. Pharm. Des. 5473-501. [PubMed] [Google Scholar]
- 36.Gooday, G. W. 1990. Inhibition of chitin metabolism, p. 61-79. In P. J. Kuhn, A. P. J. Trinci, M. J. Jung, M. W. Goosey, and L. G. Copping (ed.), Biochemistry of cell walls and membranes in fungi. Springer Verlag, Berlin, Germany.
- 37.Grenville-Briggs, L. J., V. L. Anderson, J. Fugelstad, A. O. Avrova, J. Bouzenzana, A. Williams, S. Wawra, S. C. Whisson, P. R. Birch, V. Bulone, and P. van West. 2008. Cellulose synthesis in Phytophthora infestans is required for normal appressorium formation and successful infection of potato. Plant Cell 20720-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haldar, K., S. Kamoun, N. L. Hiller, S. Bhattacharje, and C. van Ooij. 2006. Common infection strategies of pathogenic eukaryotes. Nat. Rev. Microbiol. 4922-931. [DOI] [PubMed] [Google Scholar]
- 39.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 40.Hardham, A. R. 2007. Cell biology of plant-oomycete interactions. Cell. Microbiol. 931-39. [DOI] [PubMed] [Google Scholar]
- 41.Herth, W., M. Mulisch, and P. Zugenmaier. 1986. p. 107-120. In R. Muzarelli, C. Jeauniaux, and G.-W. Gooday (ed.), Chitin in nature and technology. Plenum Press, New York, NY.
- 42.Heux, L., J. Brugnerotto, J. Desbrières, M. F. Versali, and M. Rinaudo. 2000. Solid state NMR for determination of degree of acetylation of chitin and chitosan. Biomacromolecules 1746-751. [DOI] [PubMed] [Google Scholar]
- 43.Hückelhoven, R. 2007. Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45101-127. [DOI] [PubMed] [Google Scholar]
- 44.Judelson, H. S. 2007. Genomics of the plant pathogenic oomycete Phytophthora: insights into biology and evolution. Adv. Genet. 5797-141. [DOI] [PubMed] [Google Scholar]
- 45.Kim, M. K., H. S. Park, C. H. Kim, H. M. Park, and W. Choi. 2002. Inhibitory effect of nikkomycin Z on chitin synthases in Candida albicans. Yeast 19341-349. [DOI] [PubMed] [Google Scholar]
- 46.Kollár, R., B. B. Reinhold, E. Petrakova, H. J. Yeh, G. Ashwell, J. Drgonova, J. C. Kapteyn, F. M. Klis, and E. Cabib. 1997. Architecture of the yeast cell wall. β(1→6)-glucan interconnects mannoprotein, β(1→3)-glucan, and chitin. J. Biol. Chem. 27217762-17775. [DOI] [PubMed] [Google Scholar]
- 47.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305567-580. [DOI] [PubMed] [Google Scholar]
- 48.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinformatics 5150-163. [DOI] [PubMed] [Google Scholar]
- 49.Latijnhouwers, M., P. J. de Wit, and F. Govers. 2003. Oomycetes and fungi: similar weaponry to attack plants. Trends Microbiol. 11462-469. [DOI] [PubMed] [Google Scholar]
- 50.Leal-Morales, C. A., L. Gay, M. Fèvre, and S. Bartnicki-García. 1997. The properties and localization of Saprolegnia monoica chitin synthase differ from those of other fungi. Microbiology 1432473-2483. [DOI] [PubMed] [Google Scholar]
- 51.Mackey, D., and A. J. McFall. 2006. MAMPs and MIMPs: proposed classifications for inducers of innate immunity. Mol. Microbiol. 611365-1371. [DOI] [PubMed] [Google Scholar]
- 52.Madoui, M. A., E. Gaulin, C. Mathe, H. San Clemente, A. Couloux, P. Wincker, and B. Dumas. 2007. AphanoDB: a genomic resource for Aphanomyces pathogens. BMC Genomics 8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madrid, M. P., A. Di Pietro, and M. I. Roncero. 2003. Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol. Microbiol. 47257-266. [DOI] [PubMed] [Google Scholar]
- 54.Mort-Bontemps, M., L. Gay, and M. Fèvre. 1997. CHS2, a chitin synthase gene from the oomycete Saprolegnia monoica. Microbiology 1432009-2020. [DOI] [PubMed] [Google Scholar]
- 55.Munro, C. A., and N. A. Gow. 2001. Chitin synthesis in human pathogenic fungi. Med. Mycol. 3941-53. [PubMed] [Google Scholar]
- 56.Munro, C. A., S. Selvaggini, I. de Bruijn, L. Walker, M. D. Lenardon, B. Gerssen, S. Milne, A. J. Brown, and N. A. Gow. 2007. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol. Microbiol. 631399-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Needleman, S. B., and C. D. Wunsch. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48443-453. [DOI] [PubMed] [Google Scholar]
- 58.Netea, M. G., G. D. Brown, B. J. Kullberg, and N. A. Gow. 2008. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 667-78. [DOI] [PubMed] [Google Scholar]
- 59.Niederhofer, A., and B. W. Müller. 2004. A method for direct preparation of chitosan with low molecular weight from fungi. Eur. J. Pharm. Biopharm. 57101-105. [DOI] [PubMed] [Google Scholar]
- 60.Nürnberger, T., F. Brunner, B. Kemmerling, and L. Piater. 2004. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198249-266. [DOI] [PubMed] [Google Scholar]
- 61.Nyhlen, L., and T. Unestam. 1978. Cyst and germ tube wall structure in Aphanomyces astaci, Oomycetes. Can. J. Microbiol. 241296-1299. [DOI] [PubMed] [Google Scholar]
- 62.Papavizas, G. C., and W. A. Ayers. 1974. Aphanomyces species and their root diseases in pea and sugarbeet. A review. USDA technical bulletin 1485. U.S. Department of Agriculture, Washington, DC.
- 63.Riethmueller, A., H. Voglmayr, M. Goeker, M. Weiss, and F. Oberwinkler. 2002. Phylogenetic relationships of the downy mildews (Peronosporales) and related groups based on nuclear large subunit ribosomal DNA sequences. Mycologia 94834-849. [DOI] [PubMed] [Google Scholar]
- 64.Ruiz-Herrera, J. 1992. Fungal cell wall: structure, synthesis, and assembly. CRC Press, Boca Raton, FL.
- 65.Ruiz-Herrera, J., J. M. Gonzalez-Prieto, and R. Ruiz-Medrano. 2002. Evolution and phylogenetic relationships of chitin synthases from yeasts and fungi. FEMS Yeast Res. 1247-256. [DOI] [PubMed] [Google Scholar]
- 66.Ruiz-Herrera, J., and G. San-Blas. 2003. Chitin synthesis as target for antifungal drugs. Curr. Drug Targets Infect. Disord. 377-91. [DOI] [PubMed] [Google Scholar]
- 67.Rupley, J. A. 1964. The hydrolysis of chitin by concentrated hydrochloric acid, and the preparation of low-molecular-weight substrates for lysozyme. Biochim. Biophys. Acta 83245-255. [DOI] [PubMed] [Google Scholar]
- 68.Saito, H., M. Yokoi, and Y. Yoshioka. 1989. Effect of hydration on conformational change or stabilization of (1→3)-β-d-glucans of various chain lengths in the solid state as studied by high resolution solid-state 13C NMR spectroscopy. Macromolecules 223892-3898. [Google Scholar]
- 69.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 70.Schoffelmeer, E. A., F. M. Klis, J. H. Sietsma, and B. J. Cornelissen. 1999. The cell wall of Fusarium oxysporum. Fungal Genet. Biol. 27275-282. [DOI] [PubMed] [Google Scholar]
- 71.Shaw, J. A., P. C. Mol, B. Bowers, S. J. Silverman, M. H. Valdivieso, A. Durán, and E. Cabib. 1991. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 114111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shibuya, N., and E. Minami. 2001. Oligosaccharide signalling for defence responses in plant. Physiol. Mol. Plant Pathol. 59223-233. [Google Scholar]
- 73.Smith, T. F., and M. S. Waterman. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147195-197. [DOI] [PubMed] [Google Scholar]
- 74.Soulié, M. C., C. Perino, A. Piffeteau, M. Choquer, P. Malfatti, A. Cimerman, C. Kunz, M. Boccara, and A. Vidal-Cros. 2006. Botrytis cinerea virulence is drastically reduced after disruption of chitin synthase class III gene (Bcchs3a). Cell. Microbiol. 81310-1321. [DOI] [PubMed] [Google Scholar]
- 75.Soulié, M. C., A. Piffeteau, M. Choquer, M. Boccara, and A. Vidal-Cros. 2003. Disruption of Botrytis cinerea class I chitin synthase gene Bcchs1 results in cell wall weakening and reduced virulence. Fungal Genet. Biol. 4038-46. [DOI] [PubMed] [Google Scholar]
- 76.Strauch, L. 1965. Ultra-mikromethode zur Stickstoff Bestimmung in biologischem Material. Z. Klin. Chem. 5165-167. [Google Scholar]
- 77.Tanner, S. F., H. Chanzy, M. Vincendon, C. Roux, and F. Gaill. 1990. High-resolution solid-state carbon-13 nuclear magnetic resonance study of chitin. Macromolecules 233576-3583. [Google Scholar]
- 78.Tariq, V. N., and P. L. Devlin. 1996. Sensitivity of fungi to nikkomycin Z. Fungal Genet. Biol. 204-11. [DOI] [PubMed] [Google Scholar]
- 79.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Torto-Alalibo, T. A., S. Tripathy, B. M. Smith, F. D. Arredondo, L. Zhou, H. Li, M. C. Chibucos, D. Qutob, M. Gijzen, C. Mao, B. W. Sobral, M. E. Waugh, T. K. Mitchell, R. A. Dean, and B. M. Tyler. 2007. Expressed sequence tags from Phytophthora sojae reveal genes specific to development and infection. Mol. Plant-Microbe Interact. 20781-793. [DOI] [PubMed] [Google Scholar]
- 81.Trevelyan, W. E., and J. S. Harrison. 1952. Studies on yeast metabolism. I. Fractionation and microdetermination of cell carbohydrates. Biochem. J. 50298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tyler, B. M. 2002. Molecular basis of recognition between Phytophthora pathogens and their hosts. Annu. Rev. Phytopathol. 40137-167. [DOI] [PubMed] [Google Scholar]
- 83.Tyler, B. M., S. Tripathy, X. Zhang, P. Dehal, R. H. Jiang, A. Aerts, F. D. Arredondo, L. Baxter, D. Bensasson, J. L. Beynon, J. Chapman, C. M. Damasceno, A. E. Dorrance, D. Dou, A. W. Dickerman, I. L. Dubchak, M. Garbelotto, M. Gijzen, S. G. Gordon, F. Govers, N. J. Grunwald, W. Huang, K. L. Ivors, R. W. Jones, S. Kamoun, K. Krampis, K. H. Lamour, M. K. Lee, W. H. McDonald, M. Medina, H. J. Meijer, E. K. Nordberg, D. J. Maclean, M. D. Ospina-Giraldo, P. F. Morris, V. Phuntumart, N. H. Putnam, S. Rash, J. K. Rose, Y. Sakihama, A. A. Salamov, A. Savidor, C. F. Scheuring, B. M. Smith, B. W. Sobral, A. Terry, T. A. Torto-Alalibo, J. Win, Z. Xu, H. Zhang, I. V. Grigoriev, D. S. Rokhsar, and J. L. Boore. 2006. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 3131261-1266. [DOI] [PubMed] [Google Scholar]
- 84.van den Burg, H. A., S. J. Harrison, M. H. Joosten, J. Vervoort, and P. J. de Wit. 2006. Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant-Microbe Interact. 191420-1430. [DOI] [PubMed] [Google Scholar]
- 85.van West, P. 2006. Saprolegnia parasitica, an oomycete parasite with a fishy appetite: new challenges for an old problem. Mycologist 2099-104. [Google Scholar]
- 86.Weber, I., D. Assmann, E. Thines, and G. Steinberg. 2006. Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell 18225-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Werner, S., U. Steiner, R. Becher, A. Kortekamp, E. Zyprian, and H. B. Deising. 2002. Chitin synthesis during in planta growth and asexual propagation of the cellulosic oomycete and obligate biotrophic grapevine pathogen Plasmopara viticola. FEMS Microbiol. Lett. 208169-173. [DOI] [PubMed] [Google Scholar]
- 88.Werner, S., J. A. Sugui, G. Steinberg, and H. B. Deising. 2007. A chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola. Mol. Plant-Microbe Interact. 201555-1567. [DOI] [PubMed] [Google Scholar]