Abstract
A mitochondrial inner membrane protein, designated MIX, seems to be essential for cell viability. The deletion of both alleles was not possible, and the deletion of a single allele led to a loss of virulence and aberrant mitochondrial segregation and cell division in Leishmania major. However, the mechanism by which MIX exerts its effect has not been determined. We show here that MIX is also expressed in the mitochondrion of Trypanosoma brucei, and using RNA interference, we found that its loss leads to a phenotype that is similar to that described for Leishmania. The loss of MIX also had a major effect on cytochrome c oxidase activity, on the mitochondrial membrane potential, and on the production of mitochondrial ATP by oxidative phosphorylation. Using a tandem affinity purification tag, we found that MIX is associated with a multiprotein complex that contains subunits of the mitochondrial cytochrome c oxidase complex (respiratory complex IV), the composition of which was characterized in detail. The specific function of MIX is unknown, but it appears to be important for the function of complex IV and for mitochondrial segregation and cell division in T. brucei.
Trypanosomatids are unicellular flagellates of major medical and veterinary significance. They cause serious diseases in humans, such as sleeping sickness (Trypanosoma brucei), Chagas' disease (Trypanosoma cruzi), and leishmaniasis (Leishmania spp.). They undergo a complex life cycle as they alternate between a mammalian host and blood-feeding insect vectors. A striking feature of T. brucei is its ability to adapt to diverse environments encountered throughout the stages of its life cycle (18, 30). In the mammalian host, the bloodstream form (BF) of the T. brucei mitochondrion lacks a complete respiratory chain, and energy production occurs via glycolysis (7). In contrast, in the procyclic form (PF) vector stage of the parasite, the mitochondrion has many cristae, a complete cytochrome-mediated respiratory chain, and Krebs cycle enzymes (3). T. cruzi and Leishmania species undergo different but less characterized metabolic adaptations.
The mitochondrial (mt) respiratory pathway, carried out by complexes I, II, III, IV, and V, generates ATP by oxidative phosphorylation. Furthermore, a membrane potential is created across the mt membrane by this process in PF cells and by ATP hydrolysis in BF cells. This potential is absolutely required for protein import into the mitochondrion. The trypanosomatid cytochrome c (cyt c) oxidase complex (complex IV), like in other eukaryotes, is a multicomponent complex composed of three large mitochondrially encoded subunits and up to 10 smaller nuclear-encoded subunits (11, 33). However, it has several unique features: 8 of 10 nuclear-encoded subunits have no apparent homologues outside the Kinetoplastida (32); mRNAs of two of three mitochondrially encoded subunits are posttranscriptionally edited by the insertion and deletion of uridine residues via a process called RNA editing (34). Two nuclear-encoded subunits (COXIV and COXVI) are bifunctional, also being part of the Leishmania tropica tRNA import complex (20), and the expression of T. brucei nuclear-encoded subunits is developmentally regulated, being absent in BF cells and induced upon the transformation of these cells into the PF (19, 25).
Trypanosomatid cells contain only a single large mitochondrion per cell that contains uniquely structured mt DNA that is called kinetoplast DNA (15). The division of the kinetoplast is tightly coordinated with the division of the flagellar basal body, the flagellum, the nucleus, and the cell itself (9). Recently, a protein unique to the Kinetoplastida that has a role that affects cell morphology, mt segregation, and virulence in Leishmania major has been identified and designated as mt protein X (MIX) (38).
In this study, we characterized the T. brucei MIX protein using RNA interference (RNAi) and tandem affinity purification (TAP) tag approaches. The repression of MIX expression in T. brucei PF cells led to slowed cell growth, abnormalities in the number and distribution of nuclei and kinetoplasts, and reduced cyt c oxidase activity, ATP production by oxidative phosphorylation, and mt membrane potential. Mass spectrometric analysis of the tandem-affinity-purified MIX complex showed that it is associated with subunits of the cyt c oxidase complex. The tandem-affinity-purified cyt c oxidase complex, by tagging its subunits, confirmed the association with MIX and was also used to determined its composition.
MATERIALS AND METHODS
Construction of plasmids.
To create the vectors for the inducible expression of C-terminally tagged proteins, the open reading frames were PCR amplified from T. brucei strain 427 genomic DNA using oligonucleotides TAPMIX-Fw (5′-TGATCAAGCTTGGATTGAATGCTACGT-3′), TAPMIX-Rev (5-ACTCCGAGATCTGGTGTGCGTCG-3′), TAPCOXIV-Fw (CACAAGCTTATGTTTGCTCGCCGCTC), TAPCOXIV-Rev (CACGGATCCAATCTTGTTTGAGAG), TAPCOXV-Fw (CACAAGCTTATGAAGCGCTTTGTC), TAPCOXV-Rev (CACAGATCTGTTACTGATTTTG), TAPTb2320-Fw (CACAAGCTTATGTTACGCAAATCTTC), TAPTb2320-Rev (CACAAGCTTATGTTACGCAAATCTTC), TAPTb1900-Fw (CACAAGCTTATGAAACGAACAGCT), and TAPTb1900-Rev (CACAGATCTAATAAGTCGCTTTGAC) (restriction sites are underlined).
The PCR products were cloned into the pGEM-T Easy vector (Promega), digested with BamHI or BglII and HindIII enzymes, and ligated into vector pLEW79-MHT (12, 23). The TAPMIX PCR product was inserted directly into pLEW79-MHT via HindIII and BglII sites. The constructs were verified by sequencing. Genes used in this study were TAPMIX-Tb927.5.3040 (hypothetical protein [HP]), TAPCOXIV-Tb927.1.4100 (subunit trCOXIV), TAPCOXV-Tb09.160.1820 (subunit COXV), TAPTb2320-Tb10.70.2320 (HP), and TAPTb1900-Tb09.211.1900 (HP).
To create the RNAi construct for MIX repression, the MIX gene was amplified by PCR with the oligonucleotides 5′-AACGTGCTCGAGATGCTACGTCG-3′ (forward) and 5′-TGCACTAAGCTTTTAGGTGTGCGTCG-3′ (reverse) and cloned into plasmid pZJM (39) using XhoI and HindIII restriction sites (underlined).
Cell growth, transfection, and induction.
T. brucei PF cells of strain 29.13, transgenic for T7 RNA polymerase and the tetracycline (Tet) repressor, were grown in vitro at 27°C in SDM-79 medium containing hemin (7.5 mg/ml) and 10% fetal bovine serum. The TAP plasmids and RNAi plasmid were linearized with NotI enzyme and transfected into the cell line as described previously (40). The synthesis of double-stranded RNAi was induced by the addition of Tet at a 1-μg/ml concentration. The cells were counted using the Z2 cell counter (Beckman Coulter Inc.), and growth curves were generated for clonal cell lines over a period of 13 days. In TAP-tagged cell lines, the expression of tagged protein was induced by 100 ng/ml of Tet.
Northern blot analysis.
Total RNA was isolated from 1 × 108 exponentially growing noninduced and RNAi-induced cells by extraction with Trizol reagent (Invitrogen) according to instructions provided by the manufacturer. The whole open reading frame of the MIX gene was labeled using a High Prime DNA labeling kit (Roche) with [α-32P]dCTP and was used as a probe. Hybridization was carried out using standard procedures (16). Signal was visualized using Molecular Dynamics PhosphorImager screens, and autoradiograms were analyzed by densitometry.
Staining of parasite nuclei and kinetoplasts.
Log-phase noninduced and RNAi-induced cells were pelleted by centrifugation, washed, fixed with 4% formaldehyde, and treated with DAPI (4′,6′-diamidino-2-phenylindole) to visualize DNA. Phase-contrast images of the cells and their fluorescence were captured with a Nikon fluorescence microscope equipped with a camera and the appropriate filters.
SDS-PAGE and Western blot analysis.
The protein samples were fractionated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and detected by Sypro Ruby (Molecular Probes) staining overnight. For Western analysis, the proteins were blotted onto a polyvinylidene difluoride membrane and probed with anti-His monoclonal antibody (MAb) (1:2,000; Invitrogen); polyclonal rabbit antibodies against the L. major MIX protein (1:500), T. brucei cyt c1 (1:500) (kindly provided by S. L. Hajduk), cyt c oxidase subunit 6 (COXVI) (1:500), mt RNA binding protein 2 (MRP2) (1:1,000), Leishmania tarentolae cyt c oxidase subunit IV (trCOXIV) (1:1,000) (kindly provided by J. Lukeš), and phosphoglycerate kinase (1:5,000) (kindly provided by M. Parsons); and monoclonal mouse antibody 33 against the Hsp70 protein (1:5,000) (24) and the trypanosome alternative oxidase (1:100) (kindly provided by M. Chaudhuri), and proteins were visualized using the ECL system (Roche).
cyt c oxidase and cyt c reductase assays.
The mt vesicles from 5 × 108 cells were isolated by hypotonic lysis as described elsewhere previously (10) and lysed with 1% dodecyl maltoside. cyt c reductase (complex III) activity was measured as described previously (29). cyt c oxidase (complex IV) activity was measured as described previously (42). In parallel, the activity of cyt c oxidase was detected by an in-gel assay following the electrophoresis of mt lysates (100 μg of protein per lane) on a 3 to 15% blue native (BN) PAGE gel as described previously (42).
ATP production assay.
ATP production was measured as described previously (1). Briefly, a crude mt preparation from the RNAi knockdown cell line was obtained by digitonin extraction (36), ATP production was induced by 5 mM indicated substrates (succinate, pyruvate, and α-ketoglutarate), and 67 μM ADP was added. Inhibitors were preincubated with mitochondria on ice for 10 min and used at the following concentrations: 6.7 mM malonate and 33 μg/ml atractyloside. The concentration of ATP was determined by a luminometer using the CLS I ATP bioluminescence assay kit (Roche Applied Science).
Analysis of mt membrane potential by flow cytometry.
A 1-ml portion of mid-log-phase cells was incubated in the presence of 250 nM Rh123 (Molecular Probes) for 30 min at 27°C, harvested (1,300 × g for 10 min), and washed with 1 ml CytoMix (25 mM HEPES [pH 7.6], 120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4-KH2PO4 [pH 7.6], 2 mM EDTA, 5 mM MgCl2, 6 mM glucose). Cells were resuspended in 0.5 ml CytoMix and analyzed for green fluorescence in a Beckman Counter Epics XL-MCL flow cytometer.
Digitonin fractionation.
The crude mt fraction isolated by hypotonic lysis (10) was treated with 320 μM digitonin followed by centrifugation at 3,450 × g for 3 min to produce the outer membrane/intermembrane space-enriched fraction and mitoplasts. To separate the inner membrane fragments from soluble matrix components, mitoplasts were sonicated, followed by centrifugation at 7,000 × g for 5 min.
Immunocytochemistry.
The subcellular localization of the expressed tagged proteins within the cell was determined by immunofluorescence assay using anti-myc MAb (Invitrogen) as described previously (43).
TAP of tagged complexes.
The TAP protocol was adapted from methods described previously (23, 26-28). We purified the tagged complexes from 110 to 410 cells by three complementary methods (see Fig. S1 in the supplemental material). All three methods were applied for the purification of MIX and TAPCOXV complexes. Methods 1 and 2 were used to purify TAPCOXIV, TAPTb2320, and TAPTb1900 complexes. Briefly, in method 1, the harvested cells were lysed by 1% Triton X-100, and the tagged complexes were isolated by immunoglobulin G (IgG) affinity chromatography. The bound complexes were eluted by tobacco etch virus protease cleavage and fractionated on a 10 to 30% glycerol gradient by centrifugation for 5 h at 38,000 rpm at 4°C in a Sorvall SW-40 rotor (28). The sedimentation profiles of the tagged complexes were monitored by Western analyses using anti-His6 MAb. Peak reactive fractions were pooled and further purified by calmodulin affinity chromatography. In method 2, the tagged complexes were purified from cells lysed with 0.25% NP-40 and cleared by low-speed centrifugation, and the supernatant was further lysed with 1.25% NP-40 and cleared by high-speed centrifugation (40,000 rpm at 4°C in a Sorvall SW-55 rotor for 40 min). The tagged complexes were isolated by sequential binding to IgG and calmodulin affinity columns. This method was adapted from a protocol described previously (8). In method 3, a crude mt fraction was isolated by hypotonic lysis as described elsewhere previously (10) and lysed with 1% dodecyl maltoside for 30 min on ice. The tagged complexes were isolated from cleared supernatant by sequential binding to IgG and calmodulin affinity columns.
Mass spectrometry analysis.
We prepared and analyzed the samples by gel-based and gel-free approaches. In the first method, 30 μl of the sample was separated by SDS-PAGE, and protein bands were visualized by Sypro Ruby staining (Molecular Probes). All visible protein bands were excised from the gel and digested with trypsin in gel as described elsewhere previously (22). In method 2, elution fraction 2 from the calmodulin affinity column was precipitated with acetone, and the samples were further denatured with 8 M urea-1 mM dithiothreitol and treated overnight with 100 ng of trypsin. The resulting peptides were purified using RPC-18 magnetic beads (Dynabeads). Peptides were identified using a Thermo Electron LTQ linear ion trap mass spectrometer. The collision-induced dissociation spectra were compared to the T. brucei protein database downloaded from GeneDB using TurboSequest software, and protein matches were determined using PeptideProphet and ProteinProphet software (13, 21).
RESULTS
Inhibition of MIX gene expression leads to growth defects and defects in kinetoplast position and cell division.
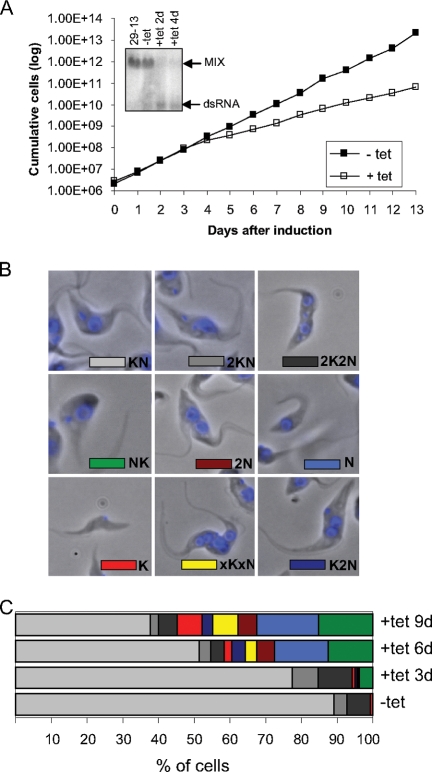
The expression of the MIX gene was repressed by RNAi induction. Northern analysis showed that in RNAi-induced cells, MIX mRNA was virtually eliminated after 2 days of induction (Fig. 1A). The RNAi-induced cells grew slowly compared to noninduced cells, and the reduced growth rate was apparent by day 4 (Fig. 1A). Based on the growth curves, RNAi-induced cells were collected on days 3, 6, and 9 following induction for all subsequent biochemical analyses.
FIG. 1.
RNAi of MIX inhibits cell growth, and cells display abnormalities in the number and distribution of nuclei and kinetoplasts. (A) Growth effect. The numbers of noninduced cells (filled squares) and RNAi-induced cells (open squares) were plotted logarithmically as the product of cell density and total dilution. The levels of MIX mRNA were analyzed by blotting 10 μg of total RNA extracted from the 29-13 cell line, noninduced (−tet) cells, and induced (+tet) cells (days 2 and 4 after induction of RNAi). The positions of the targeted mRNA and the double-stranded RNA (dsRNA) synthesized following induction are indicated on the right. (B) DAPI staining was used to visualize various nucleus/kinetoplast (NK) phenotypes. (Top) Normal cells in G1/S and G2/M phases and undergoing cytokinesis. (Middle) Abnormal position of kinetoplast in the cell and cells without kinetoplast (dyskinetoplast cells). (Bottom) Example of a zoid (cell without nucleus) and abnormal cells with more than two nuclei. (C) Quantification of the results based on the number of nuclei and kinetoplasts in more than 200 cells per each time point.
The effect on cell division and kinetoplast segregation was monitored by staining of the nuclear (N) and mt DNA (kinetoplast [K]) with DAPI and visualization by fluorescent microscopy. The cells in G1 and S phases would have one nucleus and one kinetoplast (1N1K). Since mt division precedes nuclear division, cells in G2/M phase have one nucleus and two kinetoplasts (1N2K), and cells undergoing cytokinesis have two nuclei and two kinetoplasts (2N2K). In the control MIX cell line (noninduced cells), 89% of the cells were found to have a normal-sized single nucleus and a single kinetoplast (1N1K); the rest of the cells (11%) were either 1N2K or 2N2K. However, in RNAi-induced cells, a reduction in the proportion of cells in G1/S phase (1N1K) and a proportional increase in cells with an aberrant nuclear and kinetoplast phenotype were observed. Examples of these abnormalities are shown in Fig. 1B. These include cells in which the position of the kinetoplast in relation to the nucleus was inverted as well as cells that, in addition to this inversion, also had more kinetoplasts or more nuclei than normal (phenotypes 1K1N, KNK, and KNNK). There were also cells lacking kinetoplast(s) (phenotypes N and 2N), cells lacking nuclei (phenotype K), and larger cells with multiple nuclei and multiple kinetoplasts (phenotype xNxK). The percentage of cells harboring these effects increased over time, 6% at day 3, 42% at day 6, and 55% at day 9 (Fig. 1C). Cells with inverted kinetoplasts and cells lacking the kinetoplast were the most common phenotype, constituting about 17% and 15% of the total number of cells, respectively.
ATP production by oxidative phosphorylation but not by substrate phosphorylation is reduced following MIX repression.
The variable phenotype described above may not be directly attributed to MIX alone but rather to the disruption of more extensive cellular processes such as mt metabolism, since MIX localizes to the inner mt membrane (38). A defect in the membrane may affect functions of respiratory complexes, which also localize to the inner membrane. Previous studies have shown that the inactivation of trypanosome respiratory complex II leads to the reduction of ATP produced by oxidative phosphorylation (4). Thus, using an ATP production assay (1), we investigated whether MIX affects the stability and activity of the respiratory pathway.
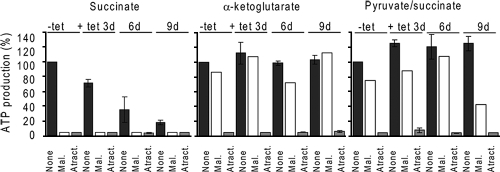
The procyclic-stage mt ATP is produced via three different pathways that can be assayed in the isolated intact mitochondria (1). Succinate is the main substrate for one of the pathways, oxidative phosphorylation. In the second pathway, α-ketoglutarate induces ATP production by substrate-level phosphorylation occurring in the citric acid cycle. Finally, pyruvate and succinate induce ATP production by substrate-level phosphorylation occurring during the acetate-succinate coenzyme A (CoA) transferase/succinyl-CoA synthetase cycle. All three forms of ATP production are sensitive to atractyloside treatment, which prevents mt import of the added ADP.
The results presented in Fig. 2 show that the repression of the MIX protein selectively reduces ATP production by oxidative phosphorylation by succinate but does not interfere with either of the two forms of ATP production by substrate phosphorylation. The ATP produced by oxidative phosphorylation was decreased by ∼30% at day 3 and by ∼65% and ∼82% at days 6 and 9, respectively. The residual ATP synthesis (around 5%) was insensitive to malonate, a competitive inhibitor of the respiratory pathway (succinate dehydrogenase). ATP produced by the acetate-succinate CoA transferase/succinyl-CoA synthetase cycle was slightly increased, by ∼20%. Thus, the repression of the MIX protein has a specific effect on the mt respiratory pathway.
FIG. 2.
ATP production by oxidative phosphorylation is severely affected in mitochondria of MIX RNAi cells. ATP production was measured in digitonin-extracted mitochondria. The three ATP-producing pathways were triggered by the addition of ADP plus one of the following substrates: succinate, α-ketoglutarate, and pyruvate-succinate. ATP production in mitochondria isolated from noninduced cells and tested without any additions (None) was set to 100%. All other values in each panel are means expressed as percentages of this sample. Malonate (Mal.) was used to inhibit ATP production by oxidative phosphorylation, and atractyloside (Atract.) was used to inhibit the import of ADP into mitochondria.
Silencing of MIX affects the steady-state abundance of two complex IV subunits and also specifically reduces cyt c oxidase activity.
Since ATP production by oxidative phosphorylation depends on the intact function of the respiratory complexes, we investigated the possibility that the activity and/or stability of complexes III and IV is impaired in MIX RNAi-induced cells. Western analysis with specific antibodies showed that the MIX protein is depleted substantially in RNAi-induced cells and correlated with a reduction in the amounts of trCOXIV and COXVI subunits of the cyt c oxidase complex, whereas the abundance of the complex III cytochrome c1 protein remained unchanged (Fig. 3). The level of the alternative oxidase protein was slightly increased by day 9. The upper band visible in the Western blot is thought to be the improperly solubilized dimeric form (6).
FIG. 3.
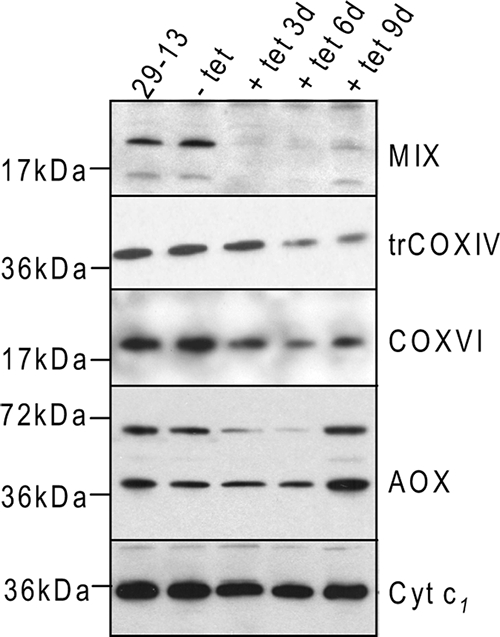
Effect of MIX RNAi on the steady-state abundance of several mt proteins. The steady-state abundance of the examined proteins was analyzed by Western blotting in mt extracts from the 29-13 cell line, noninduced (−tet) cells, and induced (+tet) cells. Each lane was loaded with 7.5 μg of protein, and blots were analyzed using polyclonal antibody against MIX, trCOXIV, COXVI, cyt c1, and alternative oxidase (AOX).
The specific activity of the cyt c oxidase complex was measured in noninduced and RNAi-induced cells. While the specific activities in different mt lysates of the noninduced cells fluctuated between 1.9 mU/mg and 1.1 mU/mg, the levels obtained for MIX RNAi-induced cells showed a substantial decrease in cyt c oxidase activity down to 0.25 mU/mg, which represents an 85% reduction in cyt c oxidase activity (Table 1). In order to determine if MIX RNAi affects the cyt c oxidase activity specifically, we simultaneously measured the activity of the cyt c reductase complex. The absence of the MIX protein had no influence on the activity of this complex (Table 1).
TABLE 1.
Functional assays for cyt c oxidase and cyt c reductase activities
| RNAi | Mean cyt c oxidase activity (mU mg−1) ± SDa | % cyt c oxidase activity | Mean cyt c reductase activity (mU mg−1) ± SDa | % cyt c reductase activity |
|---|---|---|---|---|
| MIX noninduced | 1.56 ± 0.42 | 100 | 1.05 ± 0.39 | 100 |
| MIX induced at 3 days | 0.466 ± 0.16 | 29.8 | 1.39 ± 0.29 | 132 |
| MIX induced at 6 days | 0.24 ± 0.14 | 15.3 | 0.996 ± 0.5 | 94.8 |
| MIX induced at 9 days | 0.25 ± 0.2 | 16 | 1.08 ± 0.6 | 102.8 |
All activities were measured in mt lysate prepared from at least three independent RNAi induction experiments. One unit of activity catalyzes the reduction or oxidation of 1μmol of cyt c per minute. Specific activity is calculated as U per mg of mt proteins. Mean values and standard deviations are shown.
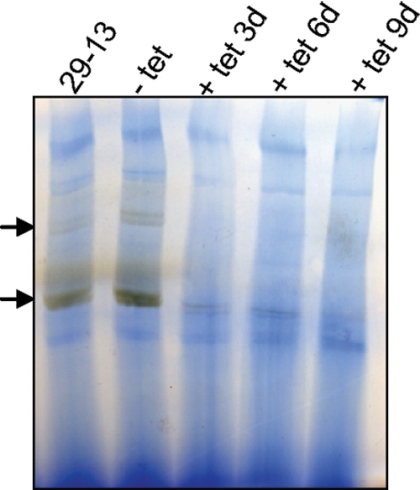
The results were further confirmed by a parallel approach, where we used a BN gel-based assay for the detection of multisubunit complex IV (10, 42). The mt lysates from RNAi-induced and noninduced cells were fractionated on a BN gel, and complex IV activity was detected in gel by histochemical staining (Fig. 4). In agreement with data from the spectrophotometric assay, we consistently observed a very strong decrease in complex IV activity in RNAi-induced MIX mt lysates (Fig. 4).
FIG. 4.
Effect of MIX RNAi on the in-gel activity of the cyt c oxidase complex. Extracts of mt vesicles from trypanosomes (MIX RNAi induced for 3, 6, and 9 days) were fractionated in a BN-PAGE gel followed by staining specific for cyt c oxidase activity. Arrows point to bands visualized by the specific activity of cyt c oxidase.
MIX protein depletion causes a reduction in mt membrane potential.
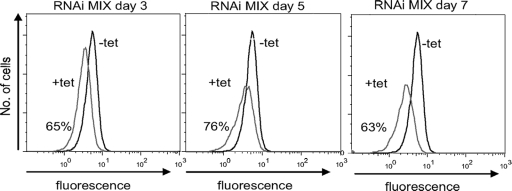
mt membrane potential depends on the intact function of respiratory complexes and is indispensable for the import of mt proteins. Membrane potential was measured every 24 h for 7 days after RNAi induction. Analysis of Rh123 staining revealed a continuous decrease in fluorescence intensity, which is indicative of reduced mt membrane potential (Fig. 5). At day 3 after MIX RNAi induction, before the growth phenotype became obvious, membrane potential was impaired down to 65% of its level in noninduced parasites. At days 5 and 7, the membrane potential was decreased down to 76% and 63%, respectively.
FIG. 5.
Depletion of MIX protein decreases mt membrane potential. mt membrane potential was measured in noninduced cells (−tet) and RNAi-induced cells (+tet) every 24 h after induction for 7 days. The graphs are shown for days 3, 5, and 7 after RNAi induction. The fluorescence distribution was plotted as a frequency histogram.
MIX protein copurifies with several subunits of cyt c oxidase complex.
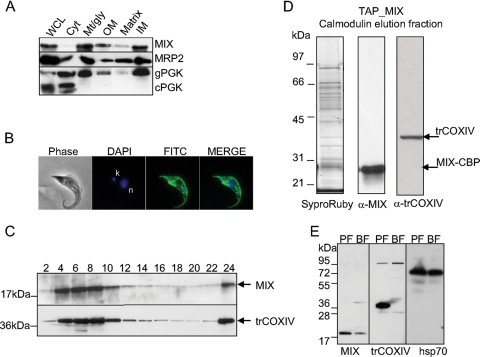
To determine if the association of the MIX protein with the cyt c oxidase complex is not only functional but also structural, we tagged, affinity purified, and analyzed the MIX-containing protein complex by mass spectrometry. We verified that MIX is localized to mitochondria by Western and immunofluorescence analyses. Western analysis of subcellular fractionations showed that the MIX protein was present in the mitochondrion-rich fraction and primarily in the inner membrane fraction, whereas no signal was observed in the cytosol (Fig. 6A). Using anti-myc MAb in immunofluorescence experiments, we showed that the tagged MIX protein was evenly distributed throughout the reticulated mitochondrion (Fig. 6B).
FIG. 6.
Expression, localization, and purification of the TAP-tagged MIX complex. (A) Subcellular fractionation and Western blotting using anti-MIX antibody indicates that MIX is localized to the mt inner membrane. The 25-kDa mt MRP2 protein and 56-kDa glycosomal phosphoglycerate kinase (gPGK) and 45-kDa cytosolic phosphoglycerate kinase (cPGK) isozymes were used as marker proteins. WCL, whole-cell lysate; Cyt, cytosolic fraction; Mt/gly, mt and glycosomal fraction; OM, outer membrane fraction; IM, inner membrane fraction. (B) Immunolocalization of tagged MIX protein shows an even distribution in the reticulated mitochondrion of procyclic T. brucei. Phase-contrast light microscopy shows procyclic T. brucei cells, DAPI staining is specific to DNA contents (n, nucleus; k, kinetoplast), and fluorescein isothiocyanate (FITC)-conjugated secondary antibody shows mt localization of the target protein. (C) Sedimentation analysis of MIX and the trCOXIV subunit of the cyt c oxidase complex. The cleared lysate of hypotonically purified mitochondria was loaded onto a 10 to 30% glycerol gradient and fractionated. The even fractions were screened for the presence of MIX and trCOXIV proteins using specific polyclonal antibodies against these proteins. (D) The TAP-tagged MIX complex was separated on a 10 to 14% polyacrylamide Tris-glycine gel and stained with Sypro Ruby. Positions of bait protein (MIX-CBP) and trCOXIV were determined by immunoblot analysis. The sizes of the protein markers are indicated on the left. (E) MIX is expressed throughout the Trypanosoma life cycle, with reduced expression in the BF. Western analysis using anti-MIX, anti-trCOXIV, and anti-Hsp70 as a loading control was carried out on lysates from the PF and BF.
To test if the MIX protein is associated with a larger protein mass, the sedimentation of the MIX protein on a glycerol gradient on which mt lysate was fractionated was determined by probing the even fraction using anti-MIX polyclonal antibody (38). We also probed these fractions with anti-trCOXIV antibody, and the results showed that these proteins cosediment together in a glycerol gradient, peaking in fractions 6 and 8, and a strong signal was also observed in the last fraction. This may be due to their association with pieces of membranes, cell particles, or kinetoplast DNAs, which sediment at the bottom of the tube following centrifugation (Fig. 6C). This result indicates that MIX is associated with a multiprotein complex, possibly with cyt c oxidase based on cosedimentation and the results obtained from the functional studies described above.
In order to identify associated proteins, the MIX-tagged complex was purified by TAP, and the resulting purified complex was fractionated by SDS-PAGE (Fig. 6D). Western analysis using anti-trCOXIV antibody showed that the affinity-purified MIX complex contains the trCOXIV protein (Fig. 6D). In addition to trCOXIV, mass spectrometry analysis of this protein sample showed the COXV, COXX, and Tb11.0400 proteins, which were previously recognized as the components of the cyt c oxidase complex in trypanosomatids (11, 33). Seven more proteins were also identified in this sample by at least two tryptic peptides (Table 2). Thus, the MIX protein is associated with a multiprotein complex that contains several cyt c oxidase subunits. In summary, our results suggest that at least 11 proteins are associated within the MIX complex, half of which are currently annotated as being hypothetical (Table 2).
TABLE 2.
Proteins found in tagged MIX and cyt c oxidase complexesa
| Group | T. brucei gene | No. of predicted transmembrane domainsb | Molecular mass (kDa) | No. of unique peptides detected withc:
|
Possible function and/or similarity (E value) | ||||
|---|---|---|---|---|---|---|---|---|---|
| TAPMIX | TAPCOXIV | TAPCOXV | TAPTb2320 | TAPTb1900 | |||||
| A | Tb927.3.1890 | NI | 33.2 | √ | √ | √ | √ | ND | Hypothetical protein; Trypj specific |
| Tb927.5.3040e | 1 | 23.1 | √ | 1 | ND | 1 | ND | Hypothetical protein; Tryp specific | |
| Tb09.211.1750 | 1 | 34.3 | √ | √ | √ | √ | 1 | mt carrier protein (1e−100) | |
| Tb09.211.4880 | 1 | 42.7 | √ | √ | √ | √ | ND | Cyclophilin isomerase (6e−09) | |
| Tb11.01.3860 | NI | 18.4 | √ | √ | √ | √ | ND | Hypothetical protein; Tryp specific | |
| Tb11.02.2460 | 1 | 50.4 | √ | √ | √ | √ | ND | Similarity to Homo Sec63 (0.063) | |
| Tb11.03.0110 | 1 | 41.7 | √ | 1 | 1 | ND | ND | DnaJ chaperon (5e−14) | |
| B | Tb927.1.4100d,f | NI | 40.5 | √ | √ | √ | √ | √ | cyt c oxidase subunit IV; Tryp specific |
| Tb09.160.1820d,g | NI | 22.2 | √ | √ | √ | √ | √ | cyt c oxidase subunit V; Tryp specific | |
| Tb11.0400d | 1 | 27.7 | √ | √ | √ | √ | √ | Hypothetical protein; Tryp specific | |
| Tb11.01.4702d | NI | 13.7 | √ | √ | √ | √ | √ | cyt c oxidase subunit X; Tryp specific | |
| Tb10.70.2320d,h | NI | 26 | 1 | √ | √ | √ | √ | Hypothetical protein; Tryp specific | |
| Tb09.211.1900i | 1 | 29.6 | ND | √ | √ | √ | √ | Hypothetical protein; Tryp specific | |
| Tb09.211.4740 | 1 | 44.5 | ND | √ | √ | √ | √ | Saccharomyces Oms1 (4e−19) | |
| Tb10.70.1890 | NI | 51.2 | ND | √ | √ | √ | √ | Hypothetical protein; Tryp specific | |
| Tb927.7.6990 | 1 | 32.7 | ND | √ | √ | √ | √ | Hypothetical protein; Tryp specific | |
| Tb11.01.1900 | 1 | 30.3 | ND | √ | √ | √ | √ | Hypothetical protein; Tryp specific | |
| Tb11.46.0006 | 2 | 50.3 | ND | √ | √ | √ | √ | Hypothetical protein; Tryp specific | |
| C | Tb927.3.1940 | 1 | 49.4 | ND | √ | √ | √ | √ | Hypothetical protein; conserved (3e−06) |
| Tb927.5.2580 | NI | 44.3 | ND | √ | √ | √ | √ | Hypothetical protein; Tryp specific | |
| Tb09.v1.0420 | 1 | 23 | 1 | √ | √ | √ | √ | Hypothetical protein; Tryp specific | |
| Tb09.160.1140 | 1 | 37.5 | 1 | √ | √ | √ | √ | SCO1/SCO2 (5e−45) | |
| Tb10.05.0050 | 1 | 18.5 | ND | √ | √ | √ | √ | Hypothetical protein; Tryp specific | |
| Tb10.70.5120 | NI | 39.0 | ND | √ | √ | √ | √ | Putative malate dehydrogenase (8e−42) | |
Only proteins identified in more than three experiments and by at least two unique peptides are shown. Proteins likely to be contaminants (including several ribosomal proteins, translation factors, heat shock proteins, alpha- and beta-tubulin, and a number of mt proteins) are listed in Table S2 in the supplemental material.
Number of predicted transmembrane domains calculated by TMHMM 2.0 software. NI, no transmembrane domain identified.
√ indicates that the number of unique peptides detected with each of the TAP-tagged constructs is higher than 2. ND, protein has not been detected.
Used as bait with TAPMIX.
Used as bait with TAPCOXIV.
Used as bait with TAPCOXV.
Used as bait with TAPTb2320.
Used as bait with TAPTb1900.
Trypanosomatid-specific protein.
In the affinity-purified MIX complex, we also identified other proteins, which we have operationally classified as contaminants based on their identification in unrelated tagged complexes that we are analyzing in our laboratory or abundant proteins such as tubulins and heat shock proteins, etc., and were not included in the list (see Table S1 in the supplemental material).
In the African trypanosome, the subunits of the cyt c oxidase complex are stage regulated, being expressed in PF and repressed in BF cells in which the mitochondrion does not participate directly in ATP production and a typical mt respiratory chain is absent (19). To examine if MIX is also developmentally regulated, total cell lysates from PF and BF cells were analyzed by Western analysis using specific antibodies against the trCOXIV and MIX proteins. As expected, trCOXIV is present in the PF but not detected in the BF (Fig. 6E). Probing the same samples with MIX antibody revealed that MIX is expressed in BF cells but that it is less abundant than the PF, indicating that MIX may be stage regulated, as are the cyt c oxidase subunits (Fig. 6E).
Composition of cyt c oxidase complex.
To confirm the association of cyt c oxidase subunits with MIX, the cyt c oxidase complex was purified by tagging four different subunits. The mt localization of all tagged proteins was confirmed by immunofluorescence assay, indicating that the tags did not interfere with mt import (see Fig. S2 in the supplemental material). Initially, subunits trCOXIV (TAPCOXIV) and COXV (TAPCOXV) were tagged and analyzed. Pilot liquid chromatography-tandem mass spectrometry analysis of calmodulin elution fractions identified 44 proteins that were present in both TAPCOXIV and TAPCOXV complexes and identified by at least two tryptic peptides. To further validate the association of newly identified proteins with cyt c oxidase subunits, we tagged and analyzed two proteins of unknown function (TAPTb2320 and TAPTb1900). A total of 33 proteins were identified in all four tagged complexes, 17 proteins of which are common contaminants of TAP tag purifications and were excluded from our list (see Table S2 in the supplemental material). The SDS-PAGE protein profile of the cyt c oxidase complex purified by TAPTb2320- and TAPTb1900-tagged proteins showed ∼14 protein bands, and we identified the respective proteins by gel band analysis (Fig. 7). Three of these protein bands corresponded to albumin (66 kDa) and alpha- and beta-tubulin (∼50-kDa bands). The other 11 proteins corresponded to Tb09.211.4740, Tb927.1.4100 (TAPCOXIV), Tb09.211.1900 (TAPTb1900), Tb927.7.6990, Tb11.01.1900, Tb10.70.2320 (TAPTb2320), Tb11.0400, Tb09.160.1820 (TAPCOXV), Tb11.01.4702, Tb10.70.1890, and Tb11.46.0006 (Fig. 7). These 11 proteins were also identified by liquid chromatography-tandem mass spectrometry analysis of the whole mixture of calmodulin elution fractions from all four tagged complexes and are considered to be part of the cyt c complex (Table 2).
FIG. 7.
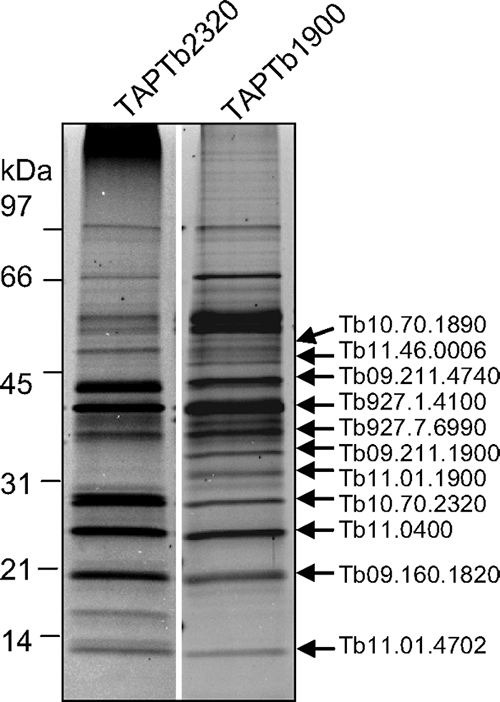
Purification of the cyt c oxidase complex and identification of its protein components. Shown are data for Sypro Ruby staining of purified cyt c oxidase complexes using TAPTb2320 and TAPTb1900 purifications. Arrows on the right point to the proteins, which were identified in individual bands by mass spectrometry. The sizes of the protein markers are indicated on the right.
Additionally, six other proteins (Tb927.3.1940, Tb927.5.2580, Tb09.v1.0420, Tb09.160.1140, Tb10.05.0050, and Tb10.70.5120) (Table 24 that were identified in all four samples by the gel-free method are most likely tightly associated with the cyt c oxidase complex, although their relative concentrations (stoichiometry) may be lower than those of the 11 proteins identified by gel band analysis. Similarly, MIX and most of its associated proteins were identified by gel-free analysis only (Table 2). Some of the MIX complex components were not identified in the TAPTb1900-tagged complex, indicating that the tag in this protein may have affected the association with the MIX complex. Because of the apparent substoichiometry nature of the MIX protein, we infer that it is not a core subunit of the cyt c oxidase complex; however, our results strongly support that it is structurally and functionally associated with the cyt c oxidase complex.
In whole-mixture analyses, we also identified 17 other proteins, which were variably seen in all four tagged complexes, indicating that they may be associated transiently with the cyt c oxidase complex even though our confidence level for the assignment is low (see Table S3 in the supplemental material). We were not successful in the identification of the mitochondrially encoded core subunits I, II, and III, most likely because of their high hydrophobicity and few potential tryptic cleavage sites.
Core subunits and associated proteins of the cyt c oxidase complex.
Using known protein sequences of cyt c oxidase core subunits and assembly factors from Homo sapiens (23 proteins), Saccharomyces cerevisiae (27 proteins), and Arabidopsis thaliana (17 proteins), we searched the T. brucei genome database and identified 13 homologue proteins with various degrees of homology (see Table S4 in the supplemental material). We have not found any homology to core subunits of the cyt c oxidase complex except for subunits COXVI and COXVIII. Conversely, assembly factors seem to be well conserved, and the T. brucei genome contains proteins homologous to Surfeit1, SCO1, SCO2, COX10, COX11, COX15, COX16, COX17, OXA1, COX19, and PET309 (see Table S4 in the supplemental material). Five of these proteins were identified in our pull-down complexes. The SCO1/SCO2 protein has been identified in all four tagged complexes (Table 2), while Surfeit1, COX11, COX15, and PET309 were identified in three and fewer tagged complexes (see Table S3 in the supplemental material). These proteins are not core components of the cyt c oxidase complex, and their association with the core complex is probably transient.
Most of the purified proteins in the cyt c oxidase complex and the MIX-tagged complex have no homology to cyt c oxidase subunits from other organisms and are currently annotated as being HPs in the GeneDB database. To explore the level of homology of these proteins, we performed PSI-BLAST search against the “nr” database and CDD, PFAM, PROSITE, and InterPro domain searches and found various degrees of similarities to other proteins of known functions or motifs for only seven proteins. We have identified the proteins as being cyclophilin isomerase (Tb09.211.4880), mt phosphate carrier protein (Tb09.211.1750), DnaJ chaperone protein (Tb11.03.0110), putative malate dehydrogenase (Tb10.70.5120), conserved bacterial HP with defined DUF477 domain (Tb927.3.1940), and a protein similar to Homo Sec63 (Tb11.02.2460). Also, the homologue of the yeast Oms1 protein (Tb09.211.4740), an inner membrane protein carrying a methyltransferase-like domain and a suppressor of respiratory defects caused by OXA1 mutations (14), was identified. Sixteen of the 24 proteins identified in tagged complexes have no recognizable motifs and/or domains and are unique to kinetoplastids (Table 2).
DISCUSSION
In this study, we showed that the T. brucei MIX protein is both structurally and functionally associated with mt respiratory complex IV. Further compositional analysis showed that the cyt c oxidase complex is unique and highly diverged in trypanosomes. The inhibition of MIX expression in T. brucei resulted in growth defects, indicating that this protein is essential for the viability of PF trypanosomes. The repression of MIX by RNAi resulted in a severe decrease in mt membrane potential and had a major negative effect on ATP production via oxidative phosphorylation and on the activity of the cyt c oxidase complex. This may have led to the observed defects in mt organization and cell division.
Role of MIX.
The MIX protein is unique to kinetoplastids and was initially studied in L. major (38), where the deletion of one allele led to defects in cell morphology and cell growth. The phenotype observed in this study is consistent with that observed for Leishmania. We found that the depletion of MIX by RNAi caused a severe growth defect even though the growth never stopped completely. This may be due to the fact that RNAi did not eliminate transcripts completely, and the residual amount of protein may be sufficient to support growth but at a lower rate. As in Leishmania, we observed a strong accumulation of abnormal configurations of nuclei and kinetoplasts in RNAi-induced cells, reaching 42% and 55% of the total population after 6 and 9 days of RNAi induction, respectively. The most frequently found aberrant kinetoplast/nucleus configurations are characterized by an inverted position of the kinetoplast (whereby cells with a 1K1N configuration have the kinetoplast anterior to the nucleus instead of the expected posterior positioning), by a loss of nucleus (zoids), and by a loss of the kinetoplast (dyskinetoplastids). In trypanosomatids, the division of the kinetoplast is tightly coordinated with the division of the flagellar basal body, the flagellum, and the cell itself (9). Moreover, as the cell goes through its cycle, there is a precisely orchestrated movement of the kinetoplast in relation to the nucleus. The loss of nucleus or kinetoplast is probably the result of ongoing cytokinesis in the cells with misplaced kinetoplasts. The appearance of large cells with the configuration xNxK also may imply that MIX is involved in cytokinesis (9). However, no significant changes were observed in the population of 2K2N cells after RNAi induction, which typically accumulate when cytokinesis is impaired directly (37); thus, the observed phenotype may be due to a secondary effect(s).
This may be supported by the observation in the TrypanoFan project (35) that about 47% of the genes that showed the RNAi phenotype had abnormal DAPI staining. We infer that the changes in kinetoplast/nucleus number and configurations are often an observed general phenotype that may not be a direct effect of a specific protein in most cases.
The phenotype observed in MIX RNAi cells could be a consequence of the interruption of other cellular processes such as mt metabolism or protein import. To investigate this possibility, we examined several typical mt functions such as ATP production, maintenance of mt membrane potential, and the activity of respiratory complexes. The loss of MIX has a major negative effect on ATP production via oxidative phosphorylation and on the activity of the cyt c oxidase complex, with a remarkable decrease in cyt c oxidase activity occurring at day 3 after induction of MIX RNAi. In addition, we found that tagged MIX protein copurifies with at least four known subunits of the cyt c oxidase complex as well as seven proteins that were shown in this study to be present within the same large complex. Thus, MIX is both structurally and functionally associated with respiratory complex IV, and the repression of MIX affects its function, which may be the primary reason for the observed phenotypic defects.
cyt c oxidase composition.
The cyt c oxidase complex has been biochemically purified from L. tarentolae and from Crithidia fasciculata using two-dimensional BN gel electrophoresis (11, 17, 33). In those studies, a total of 10 proteins were identified and were assigned as core subunits (COXIV, COXV, COXVI, COXVII, COXVIII, COXIX, and COXX and homologues of Tb11.0400, Tb10.70.2320, and Tb10.70.0625). The subunit name assignment was done based on the migration sizes of proteins in the SDS-PAGE gel. Here, the cyt c oxidase complex was purified from T. brucei using two-step TAP and four different tagged subunits. By gel band analysis of TAP-tagged complexes, we have identified 11 proteins that appear to be core components of this complex. These include five of the previously described subunits: trCOXIV, Tb10.70.2320, Tb11.0400, COXV, and COXX. The other previously assigned subunits (COXVI, COXVII, COXVIII, and COXIX) were identified by complex mixture analysis but not by gel band analysis, indicating a weaker or transient association with the tagged complexes or a differential composition of the cyt c oxidase complexes between trypanosomatid species. It is also possible that the tag affects the composition of the complex. None of the three mitochondrially encoded subunits were identified, possibly due to the characteristics of these proteins, which include high hydrophobicity, membrane association, nonmigration into SDS-PAGE gels, and few potential trypsin cleavage sites, making their peptide identification by routine mass spectrometry analysis difficult. In summary, if we combine our results with those obtained from studies of Crithidia and Leishmania, it appears that the trypanosomatid cyt c complex has at least 15 core nuclear-encoded subunits.
We carried out homology searches using the sequences of core subunits of cyt c oxidase complexes from human and yeast against the T. brucei protein database, and the results showed that only two of the nuclear-encoded subunits are identifiable in the T. brucei genome. These two subunits, COXVIII (Tb927.4.4620) and COXVI (Tb10.100.0160), have some degree of similarity to human cyt c complex subunits IV and VIb (that correspond to COX5 and COX12 in yeast), respectively. While there is a good conservation of protein sequences for a large set of subunits in human, yeast, and plant (see Table S4 in the supplemental material), in T. brucei, all except two of the designated subunits have no homology outside trypanosomes. Thus, the cyt c oxidase complex appears to be highly diverged in T. brucei.
The association of the cyt c oxidase complex with MIX adds further complexities to the cyt c oxidase complex in trypanosomes. It suggests the possibility that we are dealing with a large complex within the inner mt membrane possessing more than one function or that some of the protein components have dual functions and may be found in different complexes. This is not an unusual circumstance in trypanosomatids, as some of the components of respiratory complexes III, IV, and V have been recently identified as being a part of a complex responsible for tRNA transfer into L. tropica mitochondria (5, 20). Additionally, in the MIX-interacting complex, proteins such as cyclophilin, mt carrier protein, a chaperone, and a protein similar to the human Sec63 protein may be involved in the assembly of the complex or in mt import. The yeast homologue of Sec63 is an essential subunit of the Sec63 complex, which forms a channel competent for protein targeting and import into the endoplasmic reticulum (41). Interestingly, this protein has been found in the mt proteome and is thought to be involved in controlling mt morphology (2, 31).
Summary.
We have found that a unique mt protein, MIX, which is located in the inner mt membrane, is structurally and functionally associated with the cyt c oxidase complex. In addition, we have affinity purified the cyt c oxidase complex and identified its component and associated proteins. The cyt c oxidase complex in trypanosomes is highly diverged compared to those of other eukaryotes and appears to have at least 11 nuclear-encoded core subunits, all except 2 of which are unique to trypanosomes. We have also identified a large group of proteins that are associated with the complex, some of which have characteristics of assembly factors. These results expand our knowledge of the unique mt respiratory machinery in these parasitic protozoa.
Supplementary Material
Acknowledgments
We thank Yuko Ogata and Atashi Anupama for help with mass spectrometry and data analysis. Steve L. Hajduk (University of Georgia, Athens), Minu Chaudhuri (Meharry Medical College, Nashville, TN), Marylin Parsons (Seattle Biomedical Research Institute, Seattle, WA) and Julius Lukeš (Institute of Parasitology, Czech Republic) kindly provided antibodies. We also thank other members of the Stuart laboratory for helpful discussions.
Research was conducted using equipment made possible by the Economic Development Administration-U.S. Department of Commerce and the M. J. Murdock Charitable Trust. This work was supported by NIH grant AI065935 to K.S. and by the Australian National Health and Medical Research Council to E.H.
Footnotes
Published ahead of print on 5 September 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Allemann, N., and A. Schneider. 2000. ATP production in isolated mitochondria of procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 11187-94. [DOI] [PubMed] [Google Scholar]
- 2.Altmann, K., and B. Westermann. 2005. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell 165410-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besteiro, S., M. P. Barrett, L. Riviere, and F. Bringaud. 2005. Energy generation in insect stages of Trypanosoma brucei: metabolism in flux. Trends Parasitol. 21185-191. [DOI] [PubMed] [Google Scholar]
- 4.Bochud-Allemann, N., and A. Schneider. 2002. Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J. Biol. Chem. 27732849-32854. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee, S., P. Home, S. Mukherjee, B. Mahata, S. Goswami, G. Dhar, and S. Adhya. 2006. An RNA-binding respiratory component mediates import of type II tRNAs into Leishmania mitochondria. J. Biol. Chem. 28125270-25277. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhuri, M., R. D. Ott, L. Saha, S. Williams, and G. C. Hill. 2005. The trypanosome alternative oxidase exists as a monomer in Trypanosoma brucei mitochondria. Parasitol. Res. 96178-183. [DOI] [PubMed] [Google Scholar]
- 7.Clarkson, A. B., Jr., E. Bienen, G. Pollakis, and R. W. Grady. 1989. Respiration of bloodstream forms of the parasite Trypanosoma brucei brucei is dependent on a plant-like alternative oxidase. J. Biol. Chem. 26417770-17776. [PubMed] [Google Scholar]
- 8.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415141-147. [DOI] [PubMed] [Google Scholar]
- 9.Hammarton, T. C., S. Monnerat, and J. C. Mottram. 2007. Cytokinesis in trypanosomatids. Curr. Opin. Microbiol. 10520-527. [DOI] [PubMed] [Google Scholar]
- 10.Horváth, A., E. Horáková, P. Dunajčíková, Z. Verner, E. Pravdová, I. Šlapetová, L. Cuninková, and J. Lukeš. 2005. Downregulation of the nuclear-encoded subunits of the complexes III and IV disrupts their respective complexes but not complex I in procyclic Trypanosoma brucei. Mol. Microbiol. 58116-130. [DOI] [PubMed] [Google Scholar]
- 11.Horváth, A., T. G. Kingan, and D. A. Maslov. 2000. Detection of the mitochondrially encoded cytochrome c oxidase subunit I in the trypanosomatid protozoan Leishmania tarentolae. Evidence for translation of unedited mRNA in the kinetoplast. J. Biol. Chem. 27517160-17165. [DOI] [PubMed] [Google Scholar]
- 12.Jensen, B. C., C. T. Kifer, D. L. Brekken, A. C. Randall, Q. Wang, B. L. Drees, and M. Parsons. 2006. Characterization of protein kinase CK2 from Trypanosoma brucei. Mol. Biochem. Parasitol. 15128-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller, A., S. Purvine, A. I. Nesvizhskii, S. Stolyar, D. R. Goodlett, and E. Kolker. 2002. Experimental protein mixture for validating tandem mass spectral analysis. Omics 6207-212. [DOI] [PubMed] [Google Scholar]
- 14.Lemaire, C., F. Guibet-Grandmougin, D. Angles, G. Dujardin, and N. Bonnefoy. 2004. A yeast mitochondrial membrane methyltransferase-like protein can compensate for oxa1 mutations. J. Biol. Chem. 27947464-47472. [DOI] [PubMed] [Google Scholar]
- 15.Liu, B., Y. Liu, S. A. Motyka, E. C. Agbo, and P. T. Englund. 2005. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 21363-369. [DOI] [PubMed] [Google Scholar]
- 16.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 17.Maslov, D. A., A. Zíková, I. Kyselová, and J. Lukeš. 2002. A putative novel nuclear-encoded subunit of the cytochrome c oxidase complex in trypanosomatids. Mol. Biochem. Parasitol. 125113-125. [DOI] [PubMed] [Google Scholar]
- 18.Matthews, K. R. 2005. The developmental cell biology of Trypanosoma brucei. J. Cell Sci. 118283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayho, M., K. Fenn, P. Craddy, S. Crosthwaite, and K. Matthews. 2006. Post-transcriptional control of nuclear-encoded cytochrome oxidase subunits in Trypanosoma brucei: evidence for genome-wide conservation of life-cycle stage-specific regulatory elements. Nucleic Acids Res. 345312-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee, S., S. Basu, P. Home, G. Dhar, and S. Adhya. 2007. Necessary and sufficient factors for the import of transfer RNA into the kinetoplast mitochondrion. EMBO Rep. 8589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nesvizhskii, A. I., A. Keller, E. Kolker, and R. Aebersold. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 754646-4658. [DOI] [PubMed] [Google Scholar]
- 22.Panigrahi, A. K., A. Schnaufer, N. Carmean, R. P. Igo, Jr., S. P. Gygi, N. L. Ernst, S. S. Palazzo, D. S. Weston, R. Aebersold, R. Salavati, and K. D. Stuart. 2001. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 216833-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panigrahi, A. K., A. Schnaufer, N. L. Ernst, B. Wang, N. Carmean, R. Salavati, and K. Stuart. 2003. Identification of novel components of Trypanosoma brucei editosomes. RNA 9484-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panigrahi, A. K., A. Zíková, R. A. Dalley, N. Acestor, Y. Ogata, A. Anupama, P. J. Myler, and K. D. Stuart. 2008. Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol. Cell. Proteomics 7534-545. [DOI] [PubMed] [Google Scholar]
- 25.Priest, J. W., and S. L. Hajduk. 1994. Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J. Bioenerg. Biomembr. 26179-192. [DOI] [PubMed] [Google Scholar]
- 26.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24218-229. [DOI] [PubMed] [Google Scholar]
- 27.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 171030-1032. [DOI] [PubMed] [Google Scholar]
- 28.Schnaufer, A., N. Ernst, J. O'Rear, R. Salavati, and K. Stuart. 2003. Separate insertion and deletion sub-complexes of the Trypanosoma brucei RNA editing complex. Mol. Cell 12307-319. [DOI] [PubMed] [Google Scholar]
- 29.Schnaufer, A., S. Sbicego, and B. Blum. 2000. Antimycin A resistance in a mutant Leishmania tarentolae strain is correlated to a point mutation in the mitochondrial apocytochrome b gene. Curr. Genet. 37234-241. [DOI] [PubMed] [Google Scholar]
- 30.Schneider, A. 2001. Unique aspects of mitochondrial biogenesis in trypanosomatids. Int. J. Parasitol. 311403-1415. [DOI] [PubMed] [Google Scholar]
- 31.Sickmann, A., J. Reinders, Y. Wagner, C. Joppich, R. Zahedi, H. E. Meyer, B. Schonfisch, I. Perschil, A. Chacinska, B. Guiard, P. Rehling, N. Pfanner, and C. Meisinger. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA 10013207-13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speijer, D., C. K. D. Breek, A. O. Muijsers, P. X. Groenevelt, H. Dekker, A. De Haan, and R. Benne. 1996. The sequence of a small subunit of cytochrome c oxidase from Crithidia fasciculata which is homologous to mammalian subunit IV. FEBS Lett. 381123-126. [DOI] [PubMed] [Google Scholar]
- 33.Speijer, D., A. O. Muijsers, H. Dekker, A. de Haan, C. K. Breek, S. P. Albracht, and R. Benne. 1996. Purification and characterization of cytochrome c oxidase from the insect trypanosomatid Crithidia fasciculata. Mol. Biochem. Parasitol. 7947-59. [DOI] [PubMed] [Google Scholar]
- 34.Stuart, K. D., A. Schnaufer, N. L. Ernst, and A. K. Panigrahi. 2005. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 3097-105. [DOI] [PubMed] [Google Scholar]
- 35.Subramaniam, C., P. Veazey, S. Redmond, J. Hayes-Sinclair, E. Chambers, M. Carrington, K. Gull, K. Matthews, D. Horn, and M. C. Field. 2006. Chromosome-wide analysis of gene function by RNA interference in the African trypanosome. Eukaryot. Cell 51539-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan, T. H., N. Bochud-Allemann, E. K. Horn, and A. Schneider. 2002. Eukaryotic-type elongator tRNAMet of Trypanosoma brucei becomes formylated after import into mitochondria. Proc. Natl. Acad. Sci. USA 991152-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tu, X., P. Kumar, Z. Li, and C. C. Wang. 2006. An aurora kinase homologue is involved in regulating both mitosis and cytokinesis in Trypanosoma brucei. J. Biol. Chem. 2819677-9687. [DOI] [PubMed] [Google Scholar]
- 38.Uboldi, A. D., F. B. Lueder, P. Walsh, T. Spurck, G. I. McFadden, J. Curtis, V. A. Likic, M. A. Perugini, M. Barson, T. Lithgow, and E. Handman. 2006. A mitochondrial protein affects cell morphology, mitochondrial segregation and virulence in Leishmania. Int. J. Parasitol. 361499-1514. [DOI] [PubMed] [Google Scholar]
- 39.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 27540174-40179. [DOI] [PubMed] [Google Scholar]
- 40.Wirtz, E., S. Leal, C. Ochatt, and G. A. M. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 9989-101. [DOI] [PubMed] [Google Scholar]
- 41.Young, B. P., R. A. Craven, P. J. Reid, M. Willer, and C. J. Stirling. 2001. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 20262-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zíková, A., E. Horáková, M. Jirků, P. Dunajčíková, and J. Lukeš. 2006. The effect of down-regulation of mitochondrial RNA-binding proteins MRP1 and MRP2 on respiratory complexes in procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 14965-73. [DOI] [PubMed] [Google Scholar]
- 43.Zíková, A., A. K. Panigrahi, R. A. Dalley, N. Acestor, A. Anupama, Y. Ogata, P. J. Myler, and K. Stuart. 2008. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol. Cell. Proteomics 71286-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.