Abstract
Fungal glycosylphosphatidylinositol (GPI)-anchored proteins localize to the plasma membrane (PM), cell wall (CW), or both. To study signals that regulate PM versus CW targeting in Candida albicans, we (i) fused the N and/or C termini of the GPI CW protein Hwp1p and the GPI PM protein Ecm331p to green fluorescent protein (GFP) and (ii) expressed and localized the resulting fusions. Forty-seven amino acids from the C terminus of Hwp1p were sufficient to target GFP to the CW, and 66 amino acids from the C terminus of Ecm331p were sufficient to target GFP to the PM. Truncation and mutagenesis studies showed that G390 was the ω cleavage site in Ecm331p. Domain exchange and mutagenesis studies showed that (i) the 5 amino acids immediately N-terminal to the ω sites (the ω − 5 to ω − 1 amino acids) played key roles in targeting to the PM or CW; (ii) KK and FE residues at positions ω − 1 and ω − 2, respectively, targeted to the PM and CW; and (iii) a loss of I at position ω − 5 increased PM retention. Small fluorescent reporters can be used to study the peptide signals that regulate PM versus CW targeting of GPI proteins and may be useful for identifying proteins that interact with key targeting signals.
Eukaryotic cells transport glycosylphosphatidylinositol (GPI)-anchored proteins to the plasma membrane (PM) via the protein secretion pathway (32). In fungi and other walled eukaryotes, some GPI-anchored proteins are retained on the PM, whereas others are released from the PM by cleaving them within the GPI anchors (10, 26, 28), after which the remnant proteins are linked to β-1,6-glucans (2, 24, 28) and transported to the cell wall (CW), where they are covalently cross-linked to β-1,3-glucan networks(25). The means by which fungal GPI-anchored proteins are differentially targeted to the PM or CW are complex and poorly understood. It is known, however, that several types of peptide signals on the GPI-anchored proteins play key roles in cell surface targeting. For example, GPI attachment signals near the C terminus are required for targeting of GPI-anchored proteins to the PM (37, 39). These signals consist of a conserved ω domain (the ω or GPI attachment site and the ω + 1 and ω + 2 sites), a spacer domain consisting of 5 to 12 amino acids, and a hydrophobic tail consisting of 11 to 15 amino acids at the extreme C terminus.
Studies of Saccharomyces cerevisiae indicate that fungi utilize several types of signals to target GPI-anchored proteins to the cell surface. One type is located immediately N-terminal to the ω sites (the ω-minus regions) near the C termini of many fungal GPI-anchored proteins. For example, the basic dipeptides KK and KR located in the ω − 1 to ω − 5 regions of several GPI-anchored S. cerevisiae proteins (e.g., Gas1p, Ecm33p, and Yps1p) appear to function as PM retention signals (4, 12). In contrast, V, I, or L residues at the ω − 4 or ω − 5 position and Y, N, or V residues at the ω − 2 position appear to target S. cerevisiae GPI-anchored proteins (e.g., Sed1p and Fit1p) to the CW (18, 19). Moreover, studies of fungal GPI-anchored CW proteins (GPI-CWPs) demonstrate that the central Ser/Thr-rich repeats in these proteins (i) are necessary for proper localization of some GPI-CWPs to the outer CW (13) and (ii) can override KK-type PM retention signals and thereby target GPI-anchored proteins to the CW (14). Lastly, some GPI-anchored PM proteins (GPI-PMPs) also target partially to the CW (9), suggesting that they carry uncharacterized CW-targeting signals (14).
The genome of the pathogenic fungus Candida albicans encodes more than 100 predicted GPI-anchored proteins (7, 27), and biochemical studies have shown that GPI-anchored proteins account for 30% of the organism's CW (23). In C. albicans, GPI-anchored proteins play key roles in several virulence-associated processes, including adhesion to mammalian cells (5, 20, 34), switching between yeast and hyphal morphologies (3, 15), biofilm formation (17), CW biosynthesis (11), stress tolerance (31, 33), degradation of host tissues (1, 16), interaction with the host immune system (30), and virulence in mice (3, 6, 15, 34). The means by which C. albicans transports GPI-anchored proteins from the PM to the CW and the signals that regulate this process have not been studied. One reason is that most of the C. albicans GPI-anchored proteins that have been studied to date are large and heavily glycosylated CWPs whose targeting signals would be difficult to analyze using traditional biochemical methods (22, 35). Also, the apparent abilities of some targeting signals to override others (14) substantially complicates the analysis of specific targeting signals. For these reasons, it is likely that convenient GPI-anchored reporters that differentially target to the PM or the CW might be very useful both for studying the peptide signals that regulate PM versus CW targeting and for identifying and characterizing the effector proteins that interact with these signals.
In an earlier study, we fused a C. albicans-adapted green fluorescent protein (GFP) to short peptides from the N and C termini of the C. albicans GPI-CWP Hwp1p, an outer CW adhesin of hyphal-phase C. albicans cells that can form covalent linkages with surface components of mammalian cells (34, 35). We then used the resulting fusion proteins (i) to assess the abilities of the N- and the C-terminal signal peptides from Hwp1p to target the GFP reporter to the cell surface, (ii) to identify the ω site in Hwp1p, and (iii) to assess the effects of specific amino acid substitutions on cell surface targeting (29). Some advantages of this approach over traditional biochemical cell fractionation methods are the small sizes of the fluorescent reporters, the relatively high levels at which they are expressed in C. albicans, and the fact that targeting of these reporters to destinations near the cell surface can be assessed by fluorescence microscopy. Also, the effects of specific mutations in C-terminal signal peptides can be assessed without interference by interacting or overriding signals located in other parts of the protein of interest. In this study, we used similar approaches to show that the C. albicans GPI-anchored protein Ecm331p targets mostly to the PM, to identify the protein's ω cleavage site, to identify the domains that target the protein to the PM, and to analyze peptide signals in Ecm331p or Hwp1p that differentially target reporters to the PM or the CW.
MATERIALS AND METHODS
Strains and media.
C. albicans CAI4 (Δura3::imm434/Δura3::imm434) (from William Fonzi, Georgetown University) were cultured in YPD (1% yeast extract, 2% peptone, and 2% glucose) or minimal glucose (0.67% yeast nitrogen base without amino acids, 2% glucose). Germ tubes were obtained by inoculating cells incubated overnight in minimal glucose at 30°C into Lee's medium at an optical density at 600 nm of 0.3 and then by shaking them at 100 rpm at 34°C for 3.5 to 4 h (29).
Plasmid construction.
All C. albicans plasmids used in this study were derived from pHwp1.GFP.Hwp1c, which was called pHwp1.Sig.GFP.GPI in an earlier study (29). pEcm331.GFP.Hwp1c was constructed by replacing the nucleotides encoding the 47 N-terminal amino acids from Hwp1p in pHwp1.GFP.Hwp1c with a PacI- and SpeI-digested PCR product encoding 50 amino acids from the N terminus of Ecm331p (which was generated from C. albicans genomic DNA with Pfu polymerase and primers Ecm331.Sig5 and Ecm331.Sig3 [Table 1 lists all oligonucleotides used in this study]). pHwp1.GFP.Ecm331c and pEcm331.GFP.Ecm331c were constructed by replacing the nucleotides encoding the 53 C-terminal amino acids from Hwp1p in pHwp1.GFP.Hwp1c and in pEcm331.GFP.Hwp1c with a BamHI- and SmaI-digested PCR product encoding the 66 C-terminal amino acids from Ecm331p (which was generated from C. albicans genomic DNA with primers Ecm331C66.5 and Ecm331SspC).
TABLE 1.
Oligonucleotide primers
| Primer no. | Primer name | Sequence |
|---|---|---|
| 1 | ECM331C66.5 | 5′ CCTGACGGATCCGTTTGTACTCATCCAGCTAATCCTTC 3′ |
| 2 | ECM331SspC | 5′ GTCGGTCCCGGGCTAAATTAATGCCAATAAAACACCCAAGGC 3′ |
| 3 | ECM331sig5 | 5′ GGCCTGATTAATTAAATGCAAATTAAGTCATTTCTTTTACCAATAGTC 3′ |
| 4 | ECM331sig3 | 5′ GCCTGAACTAGTATCTAAAGTAGAGCATGCATTTAATTGGGA 3′ |
| 5 | EcmC20NoQ5 | 5′ GACGGATCCGTATTGGTTGTTCCTGGTATGGTATTG 3′ |
| 6 | EcmC28NoQ5 | 5′ GACGGATCCTCATCTAAAAAAGGTGCTAGTAATGTATTGG 3′ |
| 7 | EcmC34NoQ5 | 5′ GACGGATCCAGCTCAAATTCTAGTAGCTCATCTAAAAAAGG 3′ |
| 8 | EcmC40NoQ5 | 5′ GACGGATCCTCTTCAGATGGTTCATCAAGCTC 3′ |
| 9 | 3xFlagGFP | 5′ GTCACTAGTGATTATAAAGATCATGATGGTGATTATAAAGATCATGATATTGATTATAAAGATGATGATGATAAAATGTCTAAAGGTGAAGAATTATTCACTGG 3′ |
| 10 | ECM66-25/Hwp122.3 | 5′ TGACCCGGGTTAGATCAAGAATGCAGCAATACCAATAATAGCAGCACCGAAAGTCAATCTCATGTTGTTACCAGCACCTTTTTTAGATGAGCTACTAGAATTTGAGCTTGATGA 3′ |
| 11 | HWP47-23/ECM24.5 | 5′ AGCGGATCCACTACATTAGAATCAGTTCCACTCATGCAACCATCTGCCAATTACTCAAGTGTCGCTCCTATTTCTACATTTGAAGGTGCTAGTAATGTATTGGTTGTTCCTGGT 3′ |
| 12a | ECM331.wT4 | 5′ TGACCCGGGCTAAATTAATGCCAATAAAACACCCAAGGCTGTAGTCAATACCATACCAGGAACAACCAATACATTACTAGCAGTTTTTTTAGATGAGCTACTAGAATTTGAGCT 3′ |
| 13b | ECM.SSSSS3 | 5′ TGACCCGGGCTAAATTAATGCCAATAAAACACCCAAGGCTGTAGTCAATACCATACCAGGAACAACCAATACATTACTAGCACCAGATGAAGATGAGCTACTAGAATTTGAGCT 3′ |
| 14b | HWP.SSSKK3 | 5′ TGACCCGGGTTAGATCAAGAATGCAGCAATACCAATAATAGCAGCACCGAAAGTCAATCTCATGTTGTTACCAGCACCTTTTTTAGATGAACTAGGAGCGACACTTGAGTAATTGGC 3′ |
Codon GGT at G390 was changed to AGA at G390R, CAA at S392Q, GAT at G390D, TCA at G390S, and GGT at S392G.
Oligonucleotides used in the mutagenesis of the ω-minus region are the same, except for the sequences encoding 1 to 5 amino acids that were mutagenized.
A three-Flag tag was fused to the N terminus of GFP by amplifying the GFP gene in pHwp1.GFP.Hwp1c with primers 3xFlagGFP and GFP3 and by replacing the GFP gene in pHwp1.GFP.Hwp1c and pHwp1.GFP.Ecm331c with the three-Flag-tagged GFP, which yielded pHwp1.FlagGFP.Hwp1c and pHwp1.FlagGFP.Ecm331c.
pHwp1.GFP.Ecm331FL and pHwp1.GFP.Hwp1FL, respectively, were constructed by replacing the part of pHwp1.GFP.Hwp1C that encodes the C terminus of Hwp1p with BamHI- and SmaI-digested PCR products encoding all of Hwp1p except for its 53 N-terminal amino acids or all of Ecm331p except for its 25 N-terminal amino acids.
To truncate the C terminus of Ecm331p, PCR products encoding the C-terminal 40, 34, 28, and 20 amino acids of Ecm331p were (i) amplified from pHwp1.FlagGFP.Ecm331c with primers ECM331SspC and either Ecm331GPI405, Ecm331GPI345, Ecm331GPI285, or Ecm331GPI205 and (ii) inserted into BamHI- and SmaI-digested pHwp1.FlagGFP.Ecm331 in place of the fragment encoding the 66 C-terminal amino acids from Ecm331p.
To introduce specific amino acid substitutions into the C terminus of Ecm331p, primers Ecm331C66.5 and either ECM331.ωT4 or its derivatives encoding G390T, G390R, S392Q, G390D, G390S, and S392G substitutions (Table 1) were used to amplify from pHwp1.FlagGFP.Ecm331 DNA products encoding the wild-type C-terminal 66 amino acids of Ecm331p or derivatives encoding the listed mutations. These BamHI- and SmaI-digested fragments were used to replace the corresponding fragment in pHwp1.FlagGFP.Ecm331c, which yielded pHwp1.FlagGFP.Ecm331G390T, pHwp1.FlagGFP.Ecm331G390R, pHwp1.FlagGFP.Ecm331S392Q, pHwp1.FlagGFP.Ecm331G390D, pHwp1.FlagGFP.Ecm331G390S, and pHwp1.FlagGFP.Ecm331S392G.
To generate a C-terminal sequence that contained the ω-minus region of Hwp1p and the ω-plus region of Ecm331, primers HWP47-23/ECM24.5 and ECM331SspC were used to amplify from Hwp1/FlagGFP/Hwp1c the ω-minus region of Hwp1p. This BamHI-SmaI PCR fragment was used to replace the BamHI-SmaI fragment in pHwp1.FlagGFP.Hwp1c, which yielded pHwp1.FlagGFP.Hwp1ω-.Ecm331ω+. Similarly, to generate a C-terminal sequence that contained the ω-minus region of Ecm331p and the ω-plus region of Hwp1p, primers ECM331C66.5 and Ecm66-25/Hwp22.3 were used to amplify from Hwp1/FlagGFP/Ecm331c the ω-minus region of Ecm331p. This BamHI-SmaI PCR fragment was used to replace the BamHI-SmaI fragment in pHwp1.FlagGFP.Ecm331c, which yielded Hwp1.FlagGFP.Ecm331ω-.Hwp1ω+.
To generate additional mutations in the ω-minus region of Ecm331p, the amino acids in the ω − 1 to ω − 5 region of Ecm331p were mutagenized by PCR using primers ECM331C66.5 and Ecm.SSSSS3 or derivatives thereof (Table 1). Similarly, the amino acids in the ω − 1 to ω − 5 region of Hwp1p were mutagenized by PCR using primers Hwp1GPI5 and Hwp1GPI.SSSKK3 or derivatives thereof. Each PCR product was ligated into the BamHI and SmaI sites in pHwp1.FlagGFP.Hwp1 to replace the DNA encoding the C-terminal 47 amino acids.
The accuracy of all DNA constructs used in this study was verified by DNA sequencing.
Expression and detection of fluorescent proteins.
Plasmids were introduced into C. albicans CAI4 by the lithium acetate method, and URA+ colonies were selected on minimal glucose plates. Germ tubes were induced by harvesting yeast phase cells from overnight cultures in minimal glucose medium, transferring them to Lee's medium, and incubating them at 33°C for 4 h (29). The cells were fixed in 2% paraformaldehyde in 1× phosphate-buffered saline (PBS), and they were examined for green fluorescence by fluorescence microscopy (Axiophot; Zeiss, Germany) with 490-nm excitation and 525-nm emission filters (29).
Indirect immunofluorescence staining was as described by Hoyer et al. (21). Briefly, cells from an overnight culture were incubated in RPMI 1640 (Gibco BRL) at a density of 5 × 106 cells/ml in a plastic culture chamber. This culture was incubated at 33°C and 120 rpm for 3 h, followed by a thorough wash with PBS. The cells were blocked with 200 μl of antibody incubation buffer (1.5% normal goat serum and 2% bovine serum albumin in PBS) and incubated at room temperature for 1 hour. Then, 200 μl of anti-Flag primary antibody (Sigma) was added to each chamber. After 1 hour of incubation, the cells were washed with PBS. The Alexa Fluor 594-conjugated secondary antibody (Invitrogen) was added, and the cells were incubated for 1 h. The cells were washed thoroughly with PBS and examined under a fluorescence microscope equipped with a Texas red filter.
Cellular fractionation.
The procedures for cellular fractionations were described previously (29). Briefly, C. albicans germ tube cells were broken by vortexing them with glass beads in lysis buffer (50 mM Tris-HCl, pH 7.4, 5 mM EDTA, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, and proteinase inhibitors Σ). The cell lysate and the CW pellets were obtained by centrifugation at 10,000 × g for 10 min. The CW pellets were heated twice to 100°C for 10 min each time in 2% sodium dodecyl sulfate (SDS) and 2 mM β-mercaptoethanol and washed five times with water. The washed pellets were treated with (i) laminarinase (Sigma), (ii) β-1,6-glucanase from Trichoderma (Glyko), (iii) Quantazyme ylg (Qbiogene), and/or Westase (Takara) for 4 h or overnight according to the manufacturer's instructions. The supernatants obtained by centrifugation at 12,000 × g for 5 min were subjected to SDS-polyacrylamide gel electrophoresis (PAGE). Monoclonal antibodies to GFP (Zymed Laboratories, Inc.) or to Flag tag (Sigma) were used to detect the GFP fusion proteins on Western blots by enhanced chemiluminescence (Amersham). All fractionation experiments included as negative controls C. albicans cells that had been transformed only with the insertless vector.
Phase separation was conducted as described previously (29). Cell lysates were extracted with Triton X-114 at a final concentration of 1.4%. The mixture was incubated on ice for 40 min. The sample was centrifuged at 10,000 × g for 5 min at 4°C, the supernatant was transferred into a new tube, and the solution was incubated at 33°C for 3 min. After centrifugation at 10,000 × g for 20 s at room temperature, the solution was separated into an aqueous phase and a detergent phase. The detergent phase was extracted twice more with the lysis buffer. The samples from both phases were separated by SDS-PAGE, transferred to nitrocellulose, and probed with monoclonal antibodies to GFP or the Flag epitope.
Lysates from germ tube cells expressing the Hwp1/GFP/Ecm331FL fusion protein were incubated with phosphatidylinositol-phospholipase C (PI-PLC) (Sigma) for 4 h under conditions described by Staab et al. (35). The digestion product was separated into the detergent and aqueous phases with Triton X-114. The detergent phase was washed twice with PBS containing protease inhibitors. The fresh detergent was added five times to the aqueous phase to separate the potential contamination. The resulting detergent and aqueous phases were loaded on SDS-PAGE and detected by Western blotting with anti-Flag antibody.
Western blotting and quantitation of GFP fusion proteins.
Crude lysates, SDS extracts of the CW, and glucanase digests of CW pellets of the C. albicans transformants were loaded on SDS-PAGE and transferred to the nitrocellulose filters. The GFP fusion proteins on the blot were detected with a primary anti-Flag antibody (Sigma) and a secondary goat anti-mouse immunoglobulin G. For each Western blot, protein bands on two X-ray films exposed for different durations were quantitated by photodensitometry. The relative amount of each protein of interest in the CW (CW%) was calculated using the formula CW% = (densityCW × volCW)/[(densityCW × volCW) + (densityLYS × volLYS)] × 100, where “density” is the density reading per μl of sample, “vol” is the sample volume in μl, “LYS” is the proteins from the crude lysate, and “CW” is the proteins released from the CW by β-glucanases. For each fusion construct, data from three Western blots were averaged and analyzed.
RESULTS
The C terminus from Ecm331p targets fluorescent reporters to the PM.
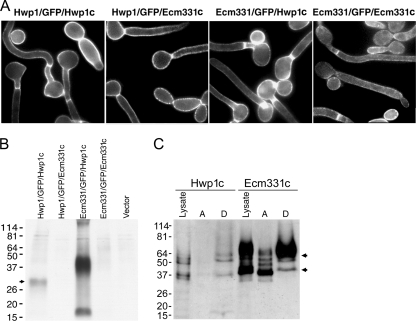
A search of the C. albicans genome database revealed a 1,242-bp intronless open reading frame, the 414-amino acid deduced product of which (Ecm331p) is homologous to the S. cerevisiae Sps2 family of the GPI-anchored proteins (which localize primarily on the PM [4]) and to the C. albicans GPI-anchored protein Ecm33p. C. albicans Ecm33p has been demonstrated in the CW by mass spectrometry (8), but the presence of a KK dipeptide immediately N terminal to the predicted ω site near the protein's C terminus suggests that it also targets to the PM. We therefore predicted that Ecm331p would target primarily to the PM, and we tested this prediction by localizing fusion proteins consisting of (i) the N terminus from Hwp1p, GFP, and the 47 C-terminal amino acids from Hwp1p (Hwp1/GFP/Hwp1c) (29); (ii) the N terminus from Hwp1p, GFP, and the 66 C-terminal amino acids from Ecm331p (Hwp1/GFP/Ecm331c); (iii) the N terminus from Ecm331p, GFP, and the C terminus from Hwp1p (Ecm331/GFP/Hwp1c); or (iv) the N terminus from Ecm331p, GFP, and the C terminus from Ecm331p (Ecm331/GFP/Ecm331c). We found that C. albicans cells expressing each of these four fusion proteins were fluorescent at the cell periphery (Fig. 1A), as would be expected for GPI-anchored proteins that target to the PM or the CW.
FIG. 1.
Targeting of GFP reporters by the C terminus of Hwp1p or Ecm331p. (A) C. albicans CAI4 cells expressing the Hwp1/GFP/Hwp1c, Hwp1/GFP/Ecm331c, Ecm331/GFP/Hwp1c, or Ecm331/GFP/Ecm331c fusion protein were fluorescent at or near the cell surface. (B) Western blots of laminarinase-digested CW probed with antibodies to GFP showed that the Hwp1/GFP/Hwp1c and Ecm331/GFP/Hwp1c fusions targeted mostly to the CW, whereas the Hwp1/GFP/Ecm331c and the Ecm331/GFP/Ecm331c fusions did not. (C) Triton X-114 extraction of the whole-cell lysates into aqueous (lanes A) and detergent (lanes D) fractions showed that both fusion proteins were present in the membrane-rich detergent phases. The arrows in B and C indicate the expected monomeric and dimeric forms of the proteins of interest, and molecular sizes in kDa are shown at the left. Hwp1c and Ecm331c represent Hwp1/FlagGFP/Hwp1c and Hwp1/FlagGFP/Ecm331c, respectively.
To determine if the marker proteins targeted to the PM or the CW, we digested CW from the transformants of interest with β-glucanase, and we probed the supernatants with monoclonal antibodies to GFP. CW from the transformants expressing the Hwp1/GFP/Hwp1c and the Ecm331/GFP/Hwp1c fusions contained abundant amounts of immunoreactive GFP, whereas CW from the transformants expressing the Ecm331/GFP/Ecm331c and the Hwp1/GFP/Ecm331c fusions did not (Fig. 1B). To determine if the C terminus from Ecm331p targeted fluorescent reporters to the membrane fractions instead of the CW, we inserted a three-Flag epitope tag immediately N terminal to the GFP moiety in the Hwp1/GFP/Ecm331c and Hwp1/GFP/Hwp1c fusion proteins, and we probed Western blots of fractionated C. albicans transformants with monoclonal anti-Flag antibodies. When lysates of the C. albicans transformants of interest were extracted with Triton X-114, the membrane-rich detergent phase contained abundant amounts of the Hwp1/FlagGFP/Ecm331c fusion protein and much smaller amounts of the Hwp1/FlagGFP/Hwp1c fusion protein (Fig. 1C).
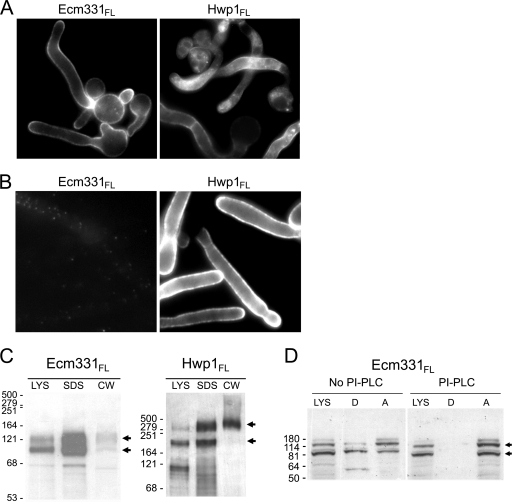
PM versus CW targeting of reporters containing full-length forms of Ecm331p and Hwp1p.
The results summarized above established that 66 C-terminal amino acids from Ecm331p targeted fluorescent reporters mostly to the PM and that 47 C-terminal amino acids from Hwp1p targeted the same reporter mostly to the CW, but it was not known if the PM versus CW targeting by these short C-terminal peptides accurately reflected the behavior of the corresponding full-length proteins. Therefore, we fused the N terminus from Hwp1p, a three-Flag tag, and GFP to (i) all of Ecm331p except for its 25 N-terminal amino acids and (ii) all of Hwp1p except for its 53 N-terminal amino acids. Both of the resulting fusion proteins (Hwp1/FlagGFP/Ecm331FL and Hwp1/FlagGFP/Hwp1FL) targeted to the cell periphery (Fig. 2A), but only the Hwp1/FlagGFP/Hwp1FL fusion protein was recognized on the surfaces of intact C. albicans cells by antibodies to the Flag epitope (Fig. 2B). Moreover, digestion of purified CW with β-glucanase released only a small amount of the Hwp1/FlagGFP/Ecm331FL reporter compared to a much larger amount of the Hwp1/FlagGFP/Hwp1FL reporter (Fig. 2C). Lastly, to determine if the Hwp1/FlagGFP/Ecm331FL reporter was attached to membranes via GPI anchors, we extracted PI-PLC-treated and untreated cell lysates with Triton X-114 and probed Western blots of the aqueous and detergent phases with anti-Flag antibodies. There were abundant amounts of the reporter in the membrane-rich detergent fraction of the non-PI-PLC-treated lysates, but the reporter was not detectable in the detergent fraction of the PI-PLC-treated lysates (Fig. 2D).
FIG. 2.
Targeting of GFP reporters by full-length forms of Ecm331p and Hwp1p. C. albicans cells expressing the Hwp1/FlagGFP reporter fused to all of Ecm331p or Hwp1p except for the N-terminal signal peptides (Hwp1/FlagGFP/Ecm331FL or Hwp1/FlagGFP/Hwp1FL, respectively) were fluorescent at the cell surface (A), but only the Hwp1/FlagGFP/Hwp1FL reporter was recognized on the surfaces of intact C. albicans cells by antibodies to the Flag epitope (B). (C) The cells expressing each of these proteins were fractionated into lysate (LYS), noncovalently linked CW (SDS), and glucanase-released CW fractions. When Western blots of fractionated cells were probed with antibodies to the Flag epitope, abundant amounts of the Hwp1/FlagGFP/Hwp1FL fusion targeted to the CW, whereas the Hwp1/FlagGFP/Ecm331FL fusion did not. (D) The Hwp1/FlagGFP/Ecm331FL reporter was present in the detergent fraction of Triton X-114 extracts of non-PI-PLC-treated whole-cell lysates, but not in PI-PLC-treated lysates. The arrows indicate the major bands of the GFP fusion proteins, and molecular sizes in kDa are shown at the left.
Identification of the ω site of Ecm331p.
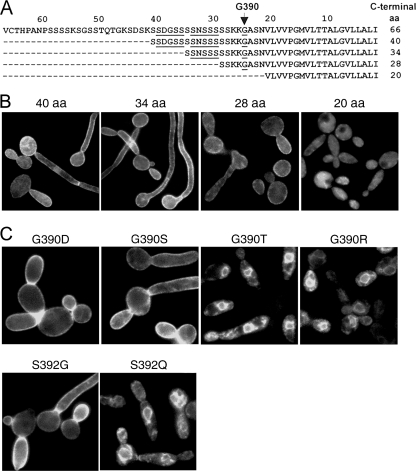
In S. cerevisiae, the amino acids immediately N terminal to the ω cleavage site (the ω-minus region) play a key role in targeting GPI-anchored proteins to the PM or CW. To determine if the ω-minus regions of C. albicans GPI-anchored proteins would have similar effects, it was first necessary to know the ω sites in the proteins of interest. In an earlier study, we showed that G613 was the ω site in Hwp1p and that the rules for predicting ω sites in GPI-anchored proteins from S. cerevisiae and other eukaryotes generally apply in C. albicans (29). The ω rules indicated that there were 11 potential ω sites within 40 amino acids of the C terminus of Ecm331p (38), so we sought to narrow the possibilities by fusing 40, 34, 28, or 20 amino acids from the C terminus of Ecm331p to a reporter consisting of 53 amino acids from the N terminus of Hwp1p, a three-Flag epitope, and GFP (Fig. 3A). When the resulting fusions were expressed in C. albicans using the TEF2-regulated plasmid pYM6, we found that 40, 34, or 28 C-terminal amino acids from Ecm331p were sufficient to target GFP to the cell periphery, whereas 20 C-terminal amino acids were not (Fig. 3B).
FIG. 3.
Identification of the ω site in Ecm331p. Sixty-six, 40, 34, or 28 amino acids (aa) from the C terminus of Ecm331p (A) (potential ω sites are underlined) targeted fluorescent reporters to the cell periphery, but 20 C-terminal amino acids did not (B). (C) That G390 and S392 were the ω and ω + 2 sites, respectively, in Ecm331p was verified by showing that amino acid substitutions permitted by the ω rules (G390D, G390S, and S392D) did not interfere with cell surface targeting, whereas substitutions not permitted by the ω rules (G390T, G390R, and S392Q) caused accumulation of fluorescent reporters in intracellular endoplasmic reticulum-like structures.
Since the only predicted ω site between 20 and 28 amino acids of the C terminus of Ecm331p was G390, these results suggested that G390 was the ω amino acid in Ecm331p. However, the GPI Fungal Prediction Server (http://mendel.imp.ac.at/gpi/cgi-bin/gpi_pred_fungi.cgi) predicted that the ω site in Ecm331p was either S384 or S385, and the targeting behavior we observed might also occur if G390 could serve as an alternative ω site after the true ω amino acid (either S384 or S385) had been deleted. Therefore, we directly tested the hypothesis that G390 is the ω site in Ecm331p by using site-directed mutagenesis (i) to change G390 in the C terminus of Ecm331p into the permitted ω amino acid D or S or the nonpermitted ω amino acid T or R and (ii) to change the predicted ω + 2 amino acid S392 into the permitted ω + 2 amino acid G or the nonpermitted ω + 2 amino acid Q. When the resulting reporters were expressed in C. albicans, those carrying the permitted substitutions G390D, G390S, and S392G all targeted to the cell periphery, whereas those carrying the nonpermitted substitutions G390T, G390R, and S392Q all accumulated within endoplasmic reticulum-like structures in the cytoplasm (Fig. 3C). Lastly, since fluorescent reporters containing only short C-terminal peptides from Ecm331p might behave differently from the corresponding full-length proteins, we also tested the effects on targeting of introducing the same amino acid substitutions into the full-length Hwp1/FlagGFP/Ecm331FL reporter. In all cases, full-length and truncated reporters bearing the same amino acid substitutions localized to the same cellular destinations (data not shown). Since the above-mentioned studies established that G390 was both necessary and sufficient to target fluorescent reporters containing either 66 amino acids from the C terminus of Ecm331p or all of Ecm331p except for its extreme N terminus to the cell periphery, it was clear that G390 was the ω amino acid in Ecm331p.
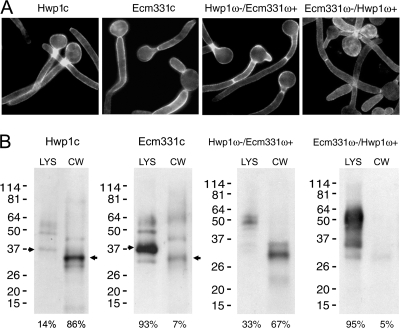
Analysis of the ω-plus and ω-minus regions of Ecm331p and Hwp1p.
Since the ω amino acid in Hwp1p was previously known to be G613 (29), it was now possible to examine the roles in PM versus CW targeting of specific regions within the C termini of Ecm331p and Hwp1p. First, the Hwp1/FlagGFP reporter was fused to chimeric C termini consisting of (i) the ω-minus region (25 amino acids N terminal to the ω site) from Hwp1p, the ω amino acid G, and the ω-plus region (23 amino acids C terminal to the ω site) from Ecm331p or (ii) the ω-minus region (42 amino acids) from Ecm331p, the ω amino acid G, and the ω-plus region (21 amino acids) from Hwp1p. C. albicans transformants expressing both chimeric fusions were fluorescent at the cell periphery (Fig. 4A). When Western blots of whole-cell lysates and of CW digested with β-glucanases were probed with anti-Flag antibodies, the 47 C-terminal amino acids from Hwp1p and the chimeric C terminus consisting of the ω-minus region from Hwp1p and the ω-plus region from Ecm331p targeted most of the GFP-Flag reporter molecule to the CW. In contrast, the reporters containing the 66 C-terminal amino acids from Ecm331p or the chimeric C terminus consisting of the ω-minus domain from Ecm331p and the ω-plus region from Hwp1p targeted mostly to the whole-cell lysate and not significantly to the CW (Fig. 4B).
FIG. 4.
Effects of ω-plus and ω-minus region domain exchanges. (A) The C termini from Hwp1p, Ecm331p, a chimeric C terminus consisting of the ω-minus region from Hwp1p and the ω-plus region of Ecm331p (Hwp1ω−/Ecm331ω+), and a chimeric C terminus consisting of the ω-minus region from Ecm331p and the ω-plus region from Hwp1p (Ecm331ω−/ Hwp1ω+) all targeted the Hwp1/FlagGFP reporter to the cell periphery. (B) The C terminus from Hwp1p and the chimeric molecule consisting of the ω-minus region from Hwp1p and the ω-plus region of Ecm331p targeted the reporter mostly to the CW, and the C terminus from Ecm331p and the chimeric molecule consisting of the ω-minus region from Ecm331p and the ω-plus region from Hwp1p targeted the reporter mostly to the cell lysate (LYS). The expected molecular masses of monomeric Hwp1/FlagGFP/Hwp1 were 36 kDa in cell lysates and 31 kDa in the CW, and the expected molecular masses of monomeric Hwp1/FlagGFP/Ecm331 were 38 kDa in cell lysates and 33 kDa in the CW.
Effects of selected amino acid substitutions on PM versus CW targeting.
Since the domain exchange experiments indicated that the ω-minus regions of Hwp1p and Ecm331p played key roles in PM versus CW targeting, we next examined the effects of a series of amino acid substitutions in these proteins' ω-minus regions. We focused on the 5 amino acids immediately N-terminal to the ω site (positions ω − 1 to ω − 5) because residues in these positions are important in targeting S. cerevisiae GPI-anchored proteins to the PM or CW (12, 19, 36). C. albicans transformants expressing each of the ω-minus mutations listed in Table 2 were brightly fluorescent at the cell periphery (data not shown), but amino acid substitutions at positions ω − 1 to ω − 5 in both proteins' C-terminal signal peptides had substantial effects on PM versus CW targeting. For example, when SSSKK from positions ω − 1 to ω − 5 in Ecm331p were replaced with ISTFE (ω − 1 to ω − 5 from Hwp1p), SSSFE (ω − 3 to ω − 5 from Ecm331p and ω − 1 and ω − 2 from Hwp1p), ISSYS (ω − 1 to ω − 5 from a consensus S. cerevisiae CW targeting signal) (36), or VSVEA (predicted ω − 1 to ω − 5 from the C. albicans GPI-CWP Rbt5p), CW targeting increased from 7% to 22 to 39% (Table 2). Conversely, when ISTFE from positions ω − 1 to ω − 5 of Hwp1p were replaced with SSSKK from Ecm331p or with SSSSS or SSSLL, CW targeting decreased from 86% to 3%, 13%, or 23%, respectively (Table 2).
TABLE 2.
Effects of amino acid substitutions on PM versus CW targetinga
| Mutagenesis | Sequence | Relative distribution (%)
|
||
|---|---|---|---|---|
| LYS | CW | SD | ||
| Ecm331p | VCTHPANPSSSSKSGSSTQTGKSDSKSSDGSSSSNSSSSSKKGASNVLVVPGMVLTTALGVLLALI | 93 | 7 | ±7.9 |
| --------------------------------------GAGNNMRLTFGAAIIGIAAFLI | 95 | 5 | ±4.0 | |
| ----------------------------------ISTFE------------------------ | 77 | 23 | ±0.1 | |
| ----------------------------------SSSSS------------------------ | 96 | 4 | ±1.5 | |
| ----------------------------------SSSFE------------------------ | 61 | 39 | ±2.1 | |
| ----------------------------------SSSLL------------------------ | 89 | 11 | ±1.4 | |
| ----------------------------------ISSSS------------------------ | 90 | 10 | ±2.5 | |
| ----------------------------------ISSYS------------------------ | 78 | 22 | ±0.1 | |
| ----------------------------------VSVEA------------------------ | 77 | 23 | ±2.3 | |
| Hwp1p | TTLESVPLMQPSANYSSVAPISTFEGAGNNMRLTFGAAIIGIAAFLI | 14 | 86 | ±7.2 |
| -----------------------GASNVLVVPGMVLTTALGVLLALI | 33 | 67 | ±1.7 | |
| -------------------SSSKK---------------------- | 97 | 3 | ±0.1 | |
| -------------------SSSSS---------------------- | 87 | 13 | ±3.1 | |
| -------------------SSSLL---------------------- | 77 | 23 | ±2.3 | |
| -------------------ISTKK---------------------- | 81 | 19 | ±1.6 | |
| -------------------STSS---------------------- | 77 | 23 | ±1.6 | |
| -------------------SSTFE---------------------- | 61 | 39 | ±4.2 | |
| -------------------ISSYS--------------------- | 34 | 66 | ±0.5 | |
| -------------------VSVEA---------------------- | 59 | 41 | ±11.7 | |
The effects on PM versus CW targeting of a series of domain exchanges and amino acid substitutions in the C termini of Ecm331p and Hwp1p are shown (the ω sites are underlined; dashes indicate amino acids identical to those above; LYS, lysate; SD, standard deviation; Relative distributions were determined in at least three experiments).
We also examined the effects on PM versus CW targeting of specific amino acids in the ω − 1 to ω − 5 regions of Ecm331p and Hwp1p. For example, basic dipeptides, such as KK, in the ω − 1 and ω − 2 positions of GPI-anchored proteins appear to function as PM retention signals in S. cerevisiae (4, 11), and the ω − 1 and ω − 2 amino acids in Ecm331p are also KK. Therefore, we examined whether basic amino acids in positions ω − 1 and ω − 2 were necessary and/or sufficient to retain GPI-anchored proteins on the PM of C. albicans. We found that replacing the KK at positions ω − 1 and ω − 2 of Ecm331p with the neutral dipeptides SS or LL did not significantly decrease membrane targeting (Table 2). However, when the FE dipeptide at positions ω − 1 and ω − 2 of Hwp1p were replaced with KK or SS, membrane targeting increased from 14% to 81% or 77%, respectively. Lastly, since I or L residues at position ω − 4 or ω − 5 have been reported to target S. cerevisiae GPI-anchored proteins to the CW (18, 19), we replaced the I at position ω − 5 in Hwp1p with S and found that this substitution decreased CW targeting from 86% to 39% (Table 2).
DISCUSSION
To understand the signals that regulate targeting of GPI-anchored proteins in C. albicans, we compared the abilities of peptides derived from Ecm331p and Hwp1p to target fluorescent reporters to the PM or the CW. To identify a model GPI-PMP, we searched the C. albicans genome for open reading frames whose deduced protein products were homologous to known GPI-PMPs from other organisms. We predicted from its deduced peptide sequence that Ecm331p was a GPI-PMP, and several lines of evidence supported this prediction. First, we found that either 66 amino acids from its C terminus or all of Ecm331p except for its 25 N-terminal amino acids targeted a fluorescent reporter to the peripheries of C. albicans cells. Second, antibodies that recognized a full-length GPI-CWP reporter (Hwp1/FlagGFP/Hwp1FL) on the surfaces of intact C. albicans cells did not recognize the Hwp1/FlagGFP/Ecm331FL reporter. Third, analyses of subcellular fractions demonstrated much larger amounts of both the full-length and truncated Ecm331p reporters in whole-cell lysates than in β-glucanase digests of purified CW. Lastly, the Hwp1/FlagGFP/Ecm331FL and the Hwp1/FlagGFP/Ecm331c reporters could both be extracted from whole-cell lysates by Triton X-114, and treatment of whole-cell lysates with an enzyme that cleaves within GPI anchors (PI-PLC) eliminated the Hwp1/FlagGFP/Ecm331FL reporter from the membrane-rich Triton X-114 fraction.
We concluded from these results that (i) either the C-terminal signal peptide from Emc331p or all of Ecm331p except for its extreme N terminus targeted GFP fusions mostly to the PM and (ii) either the C-terminal signal peptide of Hwp1p or all of Hwp1p except for its extreme N terminus targeted GFP mostly to the CW. It should be noted that the differential PM versus CW targeting we observed was incomplete. That small amounts of reporters with C termini from Hwp1p were present in the membrane fractions was not surprising, because GPI-CWPs are attached to the PM prior to their removal and transport to the CW. However, small amounts of the Hwp1/GFP/Ecm331c and the Hwp1/GFP/Ecm331FL reporters were also found in CW fractions. We do not believe that this result was caused by contamination of CW fractions with membrane proteins because our results were consistent with the earlier observations that small amounts of S. cerevisiae GPI-PMPs are also present in the CW (9, 12, 40). Taken together, these earlier studies and the results of the present study support the view that all GPI-PMPs also target to the CW, at least to some extent (9). Nevertheless, since we found that fluorescent reporters with short C-terminal signal peptides from Ecm331p targeted mostly to the PM and that fluorescent reporters with short C-terminal signal peptides from Hwp1p targeted mostly to the CW, we concluded that reporters of this type can be used to study the PM versus CW targeting signals in C. albicans GPI-anchored proteins.
By constructing and analyzing a series of deletion, domain exchange, and amino acid substitution mutants, we found that (i) the ω site in Ecm331p was G390, (ii) the PM versus CW targeting signals were in the ω-minus regions of Ecm331p and Hwp1p, and (iii) the ω − 1 to ω − 5 amino acids in both proteins of interest played key roles in PM versus CW targeting. Since earlier studies showed that the ω rules from S. cerevisiae and other eukaryotes generally applied in C. albicans Hwp1p (29) and since the amino acids immediately N terminal to the ω sites play key roles in targeting of other fungal GPI-anchored proteins to the PM or CW (12, 19, 36), most of these results were expected. However, some properties of the PM versus CW targeting signals in Ecm331p and Hwp1p were new or unexpected.
First, the observation that G390 was both necessary and sufficient to target fluorescent reporters containing either 66 amino acids from the C terminus of Ecm331p or all of Ecm331p except for its extreme N terminus to the cell periphery showed that ω sites predicted by commonly used algorithms are not necessarily correct. For example, the GPI Fungal Prediction Server (http://mendel.imp.ac.at/gpi/cgi-bin/gpi_pred_fungi.cgi) predicted incorrectly that the ω amino acid in Ecm331 was either S384 or S385. Thus, when the ω rules suggest multiple possibilities or when identification of the ω site is critical, the deletion and mutagenesis strategy used in this study and an earlier study of Hwp1p (29) can be used to test the hypothesis that a specific amino acid is the ω site in a fungal GPI-anchored protein.
A second unexpected finding was the observation that replacing only 2 to 5 amino acids in the C terminus of Hwp1p with the corresponding amino acids from Ecm331p had such marked effects on PM versus CW targeting. For example, replacing the ISTFE pentapeptide from positions ω − 5 to ω − 1 from Hwp1p with the corresponding SSSKK pentapeptide from Ecm331p reduced CW targeting from 86% to 3%. Similarly, replacing only the FE dipeptide from positions ω − 2 and ω − 1 of Hwp1p with the corresponding KK dipeptide from Ecm331p reduced CW targeting from 86% to 19%. These results support earlier proposals that basic amino acids at positions ω − 2 and ω − 1 function as PM retention signals in S. cerevisiae (4, 12), but we also found that SSSSS at positions ω − 5 to ω − 1 and SS at positions ω − 2 and ω − 1 were almost as effective at retaining the reporters on the PM as SSSKK and KK. Thus, basic amino acids at positions ω − 2 and ω − 1 were sufficient, but not necessary, to retain GPI-anchored proteins on the PM of C. albicans.
Replacing the amino acids between positions ω − 5 and ω − 1 in the C terminus of Ecm331p with the corresponding amino acids from Hwp1p also increased CW targeting, but the magnitude of this effect was less than was observed with substitutions in the opposite direction. For example, when the SSSKK pentapeptide or the KK dipeptide from Ecm331p were replaced with ISTFE or FE from Hwp1p, CW targeting increased from 7% to 23% and 39%, respectively. These results were similar to those observed by Terashima et al. (36), who showed that replacing the ω − 1 to ω − 5 residues in the S. cerevisiae GPI-PMP Ecm33p with the corresponding amino acids from S. cerevisiae GPI-CWPs increased CW targeting from 10% to 40%. We concluded that the 5 amino acids at positions ω − 1 to ω − 5, the 2 amino acids at positions ω − 1 and ω − 2, and also the single amino acid at position ω − 5 all play key roles in regulating whether a C. albicans GPI-anchored protein is retained in the PM or is transported to and incorporated into the CW. However, since the effects of inserting several identical ω − 1 to ω − 5 pentapeptides into the C termini of Hwp1p or Ecm331p varied considerably in magnitude (Table 2), it is also clear that portions of the C-terminal signal domains other than positions ω − 1 to ω − 5 also contribute significantly to targeting of GPI-anchored-proteins to the PM or CW.
Another new observation was that introducing the dipeptide FE from positions ω − 2 and ω − 1 of Hwp1p into the C terminus of Ecm331p substantially increased CW targeting (from 7% to 39%). Since the FE dipeptide is not present in the analogous position in any S. cerevisiae GPI-CWP, we searched the C-terminal signaling domains of all known or predicted C. albicans GPI-anchored proteins (7, 27) for similar sequences. Sixteen predicted C. albicans GPI-anchored proteins had an F at position ω − 2 (or ω − 3) and an acidic amino acid E at position ω − 1 (or ω − 2) (e.g., Hwp1p, Rbt1p, and Cht2p), and another four proteins had an F at position ω − 2 (or ω − 3) and an acidic amino acid D at position ω − 1 (or ω − 2) (e.g., Als3p and Als4p). Since 12 of these 20 proteins are known GPI-CWPs, it is likely that the dipeptide FE or FD in positions ω − 2 and ω − 1 functions as a previously unrecognized CW targeting signal in C. albicans.
The results from mutagenesis and cellular localization from the present study and those of Hamada et al. (18, 19) in S. cerevisiae suggest (i) that I, V, or L residues at positions ω − 3 to ω − 8 or FE, FD, Y, or N residues at positions ω − 1 to ω − 3 are CW targeting signals and (ii) that K or R residues at positions ω − 1 to ω − 7 or KK or SS dipeptides at positions ω − 1 and ω − 2 are PM retention signals. When we examined the 45 C-terminal deduced amino acids of all 104 predicted C. albicans GPI-anchored proteins (7, 27), we found CW targeting signals in 64 proteins, PM targeting signals in 22 proteins, CW and PM targeting signals in 9 proteins, and neither type of signal in 9 proteins. To assess the accuracy of predictions based on the putative signals listed above, we examined the C termini of 27 C. albicans GPI-anchored proteins that have been localized experimentally. Table 3 shows that 25 of these deduced GPI-anchored proteins have been demonstrated in the CW, one (Dfg5p) has been demonstrated in both the PM and the CW, and one (Ecm331p from the present study) targeted mostly to the PM but also to the CW. Nineteen of these 27 proteins had CW targeting signals, and all of these have been demonstrated in the CW. Moreover, Ecm331p was correctly predicted to target mostly to the PM, and Dfg5p was correctly predicted to target to both the CW and the PM. In contrast, the predicted GPI-PMPs Ecm33p, Pga4p, Phr1p, and Rbr1p have all been demonstrated in the CW; whether and to what extent these proteins also target to the PM has not been examined experimentally. The above analyses provide some support for the CW versus PM targeting signals we have proposed, but firm conclusions about the roles these specific signals play in targeting will require more direct experimental evidence.
TABLE 3.
C-terminal 45 amino acids of 27 C. albicans GPI-anchored proteins
| Protein | ORF19a position | C-terminal 45 amino acidsb | Localizationc
|
|
|---|---|---|---|---|
| Expt | Predict | |||
| Als1 | 13163 | TLSQQVTSSSPSTNTFIASTYDGSGSIIQHSTWLYGLITLLSLFI | CW | CW |
| Als3 | 1816 | QTTLSQQMTSSLVSLHMLTTFDGSGSVIQHSTWLCGLITLLSLFI | CW | CW |
| Als4 | 4555 | PTTLSQQTTSSLISTPLASTFDGSGSIVQHSGWLYVLLTAISIFF | CW | CW |
| Als5 | 5736 | TTLIQQVATSSYNQPLITTYAGSSSATKHPSWLLKFISVALFFFL | CW | CW |
| Als6 | 7414 | TTLIQQVATSSYNQPLITTYAGSSSATKHPSWLLKFISVALFFFL | CW | CW |
| Als7 | 7400 | VSTTVTEQYDTSTYTPASLLVSDNSGSVSKYSLWMMAFYMLFGLF | CW | CW |
| Als9 | 5742 | QTTLSQSLISSSTKTVIASTYDGSGSVIKLHSWFYGLVTIFFLFI | CW | CW |
| Hwp1 | 1321 | LESVPLMQPSANYSSVAPISTFEGAGNNMRLTFGAAIIGIAAFLI | CW | CW |
| Rbt1 | 1327 | ESVPAIQPSANSSYTIASVSSFEGAGNNMRLTYGAAIIGLAAFLI | CW | CW |
| Eap1 | 1401 | VTTSTIASSSETSVPPAQVSTFEGSGSALKKPYYGLAVAALVYFM | CW | CW |
| Cht2 | 3895 | YPTSVASNGTNTTVPVFTFEGGAAVANSLNSVWFPVPFLLAAFAF | CW | CW |
| Rhd3 | 5305 | SGVAQAASSSAGPAQASVSNFEGAAGQNKLSYGVGMAAVVAGLVM | CW | CW |
| Ywp1 | 3618 | TISQTTVAKASGSGKAAISTFEGAAAASAGASVLALALIPLAYFI | CW | CW |
| Hwp2 | 3380 | VESISVAVSSAAQSSIAAISSYEGTGNNMKLSFGVVIAGVAAFAI | CW | CW |
| Hyr1 | 4975 | SMPSNTTDSSSSVPTIDTNENGSSIVTGGKSILFGLIVSMVVLFM | CW | CW |
| Rbt5 | 5636 | AEESSVAQSSSSAADVASVSVEAANAGNMPAVAIGGVIAAVAALF | CW | CW |
| Csa1 | 7114 | KETGVSQATVAANTHSVAIANMANTKFASTMSLLVASFVFVGLFI | CW | CW |
| Ssr1 | 7030 | SKTSTTAAASSSESTTATGVLTQSEGSAAKVGLGALVGLVGAVLL | CW | CW |
| Crh11 | 2706 | TGDAAPSSSASEKPSVSTTENNGAVSVAKTTSLFGFVALIGFLFV | CW | CW |
| Ecm331 | 4255 | KSDSKSSDGSSSSNSSSSSKKGASNVLVVPGMVLTTALGVLLALI | PM | PM |
| Ecm33 | 3010 | SETGSDSSSSASGSSSSSKKGGAASNNGKLASVVAAFAAVGVALF | CW | PM |
| Pga4 | 4035 | PSQTSQVSSSSATSANSTSSKKNDAAVEGAGFLSVIALAAGIALL | CW | PM |
| Phr1 | 3829 | SSSSSTSSGSSSSSGVKATQQMSMVKLVSIITIVTAFVGGMSVVF | CW | PM |
| Rbr1 | 535 | SAAKSGASSATGGSSAAKSGSSSGAGFAPVAGAGSLAAIAGLLLL | CW | PM |
| Dfg5 | 2075 | SAGTNSEDNANKNELTITGKDKAGAGVLTAIVLAVILGGAIWMIF | CW+PM | CW+PM |
| Rbr2 | 532 | ASAEKSSGSSASASSTAGGSSSKGGVSELVAPVGAVVGALAVALM | CW | ND |
| Sod4 | 2062 | SSSSKNSTNGSSGSSTSASQGSGAGRAEISGFLAAGIAGVVAALI | CW | ND |
ORF, open reading frame.
Each protein's known or most likely ω site is underlined and in boldface. CW and PM targeting signals are in boldface italics.
In summary, we have shown that short C-terminal peptides from the GPI-anchored proteins Hwp1p and Ecm331p efficiently target fluorescent reporters to the CW or PM of C. albicans and that the 5 amino acids immediately N-terminal to the ω cleavage sites in these two proteins play key roles in PM versus CW targeting. Although we found that regulation of PM versus CW targeting was similar in C. albicans and S. cerevisiae, we observed several new or unexpected results. One of these was that replacing as few as 2 amino acids in the ω-minus region of Hwp1p abolished CW targeting almost completely. Since very little is known about the processes by which some fungal GPI-anchored proteins are removed from the PM and then transported to and incorporated into the CW, the availability of small fluorescent reporters that target almost completely to either the PM or the CW while differing by as few as 2 amino acids may facilitate future efforts to identify interacting proteins that directly mediate key steps in PM versus CW targeting.
Acknowledgments
We thank Qiuming Gong and Zhengfeng Zhou for assistance with the indirect fluorescent-antibody staining, Janet Staab for valuable discussions, Gerald Kayingo for assistance in C. albicans transformations, Brendan Cormack (Johns Hopkins University) for the C. albicans-optimized GFP, William Fonzi (Georgetown University) for C. albicans CAI4, Xiaoming Hong (Apex Medical Associates) for generous support, and Fred Gorelick for assistance with fluorescence microscopy.
This work was supported by the U.S. Department of Veterans Affairs and by grants R01 AI-47442 and R01 AI-64085 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 22 August 2008.
REFERENCES
- 1.Albrecht, A., A. Felk, I. Pichova, J. R. Naglik, M. Schaller, P. de Groot, D. Maccallum, F. C. Odds, W. Schafer, F. Klis, M. Monod, and B. Hube. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281688-694. [DOI] [PubMed] [Google Scholar]
- 2.Benachour, A., G. Sipos, I. Flury, F. Reggiori, E. Canivenc-Gansel, C. Vionnet, A. Conzelmann, and M. Benghezal. 1999. Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J. Biol. Chem. 27415251-15261. [DOI] [PubMed] [Google Scholar]
- 3.Braun, B. R., W. S. Head, M. X. Wang, and A. D. Johnson. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 15631-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caro L.H., H. Tettelin, J. H. Vossen, A. F. Ram, H. van den Ende, and F. M. Klis. 1997. In silico identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 131477-1489. [DOI] [PubMed] [Google Scholar]
- 5.Castano, I., A. De Las Penas, and B. P. Cormack. 2006. Function and regulation of adhesion gene families in Saccharomyces cerevisiae, Candida albicans, and Candida glabrata, p. 163-175. In J. Heitman, S. G. Filler, J. E. Edwards, Jr., and A. P. Mitchell (ed.), Molecular principles of fungal pathogenesis. ASM Press, Washington, DC.
- 6.De Bernardis, F., F. A. Muhlschlegel, A. Cassone, and W. A. Fonzi. 1998. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect. Immun. 663317-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Groot, P. W., K. J. Hellingwerf, and F. M. Klis. 2003. Genome-wide identification of fungal GPI proteins. Yeast 20781-796. [DOI] [PubMed] [Google Scholar]
- 8.de Groot, P. W., A. D. de Boer, J. Cunningham, H. L. Dekker, L. de Jong, K. J. Hellingwerf, C. de Koster, and F. M. Klis. 2004. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell 3955-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Sampaio, G., J. P. Bourdineaud, and G. J. Lauquin. 1999. A constitutive role for GPI anchors in Saccharomyces cerevisiae: cell wall targeting. Mol. Microbiol. 34247-256. [DOI] [PubMed] [Google Scholar]
- 10.Fujii, T., H. Shimoi, and Y. Iimura. 1999. Structure of the glucan-binding sugar chain of Tip1p, a cell wall protein of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1427133-144. [DOI] [PubMed] [Google Scholar]
- 11.Fonzi, W. A. 1999. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of beta-1,3- and beta-1,6-glucans. J. Bacteriol. 1817070-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frieman, M. B., and B. P. Cormack. 2003. The ω-site sequence of glycosylphosphatidylinositol-anchored proteins in Saccharomyces cerevisiae can determine distribution between the membrane and the cell wall. Mol. Microbiol. 50883-896. [DOI] [PubMed] [Google Scholar]
- 13.Frieman, M. B., J. M. McCaffery, and B. P. Cormack. 2002. Modular domain structure in the Candida glabrata adhesin Epa1p, a beta1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 46479-492. [DOI] [PubMed] [Google Scholar]
- 14.Frieman, M. B., and B. P. Cormack. 2004. Multiple sequence signals determine the distribution of glycosylphosphatidylinositol proteins between the plasma membrane and cell wall in Saccharomyces cerevisiae. Microbiology 1503105-3114. [DOI] [PubMed] [Google Scholar]
- 15.Fu, Y., A. S. Ibrahim, D. C. Sheppard, Y. C. Chen, S. W. French, J. E. Cutler, S. G. Filler, and J. E. Edwards, Jr. 2002. Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol. Microbiol. 4461-72. [DOI] [PubMed] [Google Scholar]
- 16.Ghannoum, M. A. 2000. Potential role of phospholipases in virulence and fungal pathogenesis. Clin. Microbiol. Rev. 13122-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green, C. B., G. Cheng, J. Chandra, P. Mukherjee, M. A. Ghannoum, and L. L. Hoyer. 2004. RT-PCR detection of Candida albicans ALS gene expression in the reconstituted human epithelium (RHE) model of oral candidiasis and in model biofilms. Microbiology 150267-275. [DOI] [PubMed] [Google Scholar]
- 18.Hamada, K., H. Terashima, M. Arisawa, and K. Kitada. 1998. Amino acid sequence requirement for efficient incorporation of glycosylphosphatidylinositol-associated proteins into the cell wall of Saccharomyces cerevisiae. J. Biol. Chem. 27326946-26953. [DOI] [PubMed] [Google Scholar]
- 19.Hamada, K., H. Terashima, M. Arisawa, N. Yabuki, and K. Kitada. 1999. Amino acid residues in the omega-minus region participate in cellular localization of yeast glycosylphosphatidylinositol-attached proteins. J. Bacteriol. 1813886-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoyer, L. L. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9176-180. [DOI] [PubMed] [Google Scholar]
- 21.Hoyer, L. L., T. L. Payne, and J. E. Hecht. 1998. Identification of Candida albicans ALS2 and ALS4 and localization of Als proteins to the fungal cell surface. J. Bacteriol. 1805334-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imhof, I., I. Flury, C. Vionnet, C. Roubaty, D. Egger, and A. Conzelmann. 2004. Glycosylphosphatidylinositol (GPI) proteins of Saccharomyces cerevisiae contain ethanolamine phosphate groups on the α1,4-linked mannose of the GPI anchor. J. Biol. Chem. 27919614-19627. [DOI] [PubMed] [Google Scholar]
- 23.Kapteyn, J. C., L. L. Hoyer, J. E. Hecht, W. H. Muller, A. Andel, A. J. Verkleij, M. Makarow, H. Van Den Ende, and F. M. Klis. 2000. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol. Microbiol. 35601-611. [DOI] [PubMed] [Google Scholar]
- 24.Kapteyn, J. C., R. C. Montijn, G. J. Dijkgraaf, and F. M. Klis. 1994. Identification of beta-1,6-glucosylated cell wall proteins in yeast and hyphal forms of Candida albicans. Eur. J. Cell Biol. 65402-407. [PubMed] [Google Scholar]
- 25.Klis, F. M., A. Boorsma, and P. W. de Groot. 2006. Cell wall construction in Saccharomyces cerevisiae. Yeast 23185-202. [DOI] [PubMed] [Google Scholar]
- 26.Kollár, R., B. B. Reinhold, E. Petráková, H. J. Yeh, G. Ashwell, J. Drgonová, J. C. Kapteyn, F. M. Klis, and E. Cabib. 1997. Architecture of the yeast cell wall. Beta(1→6)-glucan interconnects mannoprotein, beta(1→)3-glucan, and chitin. J. Biol. Chem. 27217762-17775. [DOI] [PubMed] [Google Scholar]
- 27.Lee, S. A., S. Wormsley, S. Kamoun, K. Joiner, and B. Wong. 2003. An analysis of the Candida albicans genome database for soluble secreted proteins using computer-based prediction algorithms. Yeast 20595-610. [DOI] [PubMed] [Google Scholar]
- 28.Lu, C. F., J. Kurjan, and P. N. Lipke. 1994. A pathway for cell wall anchorage of Saccharomyces cerevisiae alpha-agglutinin. Mol. Cell. Biol. 144825-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao, Y., Z. Zhang, and B. Wong. 2003. Use of green fluorescent protein fusions to analyze the N- and C-terminal signal peptides of GPI-anchored cell wall proteins in Candida albicans. Mol. Microbiol. 501617-1628. [DOI] [PubMed] [Google Scholar]
- 30.Martinez, J. P., M. L. Gil, J. L. Lopez-Ribot, and W. L. Chaffin. 1998. Serologic response to cell wall mannoproteins and proteins of Candida albicans. Clin. Microbiol. Rev. 11121-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Lopez, R., L. Monteoliva, R. Diez-Orejas, C. Nombela, and C. Gil. 2004. The GPI-anchored protein CaEcm33p is required for cell wall integrity, morphogenesis and virulence in Candida albicans. Microbiology 1503341-3354. [DOI] [PubMed] [Google Scholar]
- 32.Mayor, S., and H. Riezman. 2004. Sorting GPI-anchored proteins. Nat. Rev. Mol. Cell Biol. 5110-120. [DOI] [PubMed] [Google Scholar]
- 33.Pardini, G., P. W. de Groot, A. T. Coste, M. Karababa, F. M. Klis, C. G. de Koster, and D. Sanglard. 2006. The CRH family coding for cell wall glycosylphosphatidylinositol proteins with a predicted transglycosidase domain affects cell wall organization and virulence of Candida albicans. J. Biol. Chem. 28140399-40411. [DOI] [PubMed] [Google Scholar]
- 34.Staab, J. F., S. D. Bradway, P. L. Fidel, and P. Sundstrom. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 2831535-1538. [DOI] [PubMed] [Google Scholar]
- 35.Staab, J. F., Y. S. Bahn, C. H. Tai, P. F. Cook, and P. Sundstrom. 2004. Expression of transglutaminase substrate activity on Candida albicans germ tubes through a coiled, disulfide-bonded N-terminal domain of Hwp1 requires C-terminal glycosylphosphatidylinositol modification. J. Biol. Chem. 27940737-40747. [DOI] [PubMed] [Google Scholar]
- 36.Terashima, H., K. Hamada, and K. Kitada. 2003. The localization change of Ybr078w/Ecm33, a yeast GPI-associated protein, from the plasma membrane to the cell wall, affecting the cellular function. FEMS Microbiol. Lett. 218175-180. [DOI] [PubMed] [Google Scholar]
- 37.Tiede, A., I. Bastisch, J. Schubert, P. Orlean, and R. E. Schmidt. 1999. Biosynthesis of glycosyl-phosphatidylinositols in mammals and unicellular microbes. Biol. Chem. 380503-523. [DOI] [PubMed] [Google Scholar]
- 38.Udenfriend, S., and K. Kodukula. 1995. Prediction of omega site in nascent precursor of glycosylphosphatidylinositol protein. Methods Enzymol. 250571-582. [DOI] [PubMed] [Google Scholar]
- 39.Vossen, J. H., W. H. Muller, P. N. Lipke, and F. M. Klis. 1997. Restrictive glycosylphosphatidylinositol anchor synthesis in cwh6/gpi3 yeast cells causes aberrant biogenesis of cell wall proteins. J. Bacteriol. 1792202-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin, Q. Y., P. W. de Groot, L. de Jong, F. M. Klis, and C. G. De Koster. 2007. Mass spectrometric quantitation of covalently bound cell wall proteins in Saccharomyces cerevisiae. FEMS Yeast Res. 7887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]