Abstract
In 2001, a bioterrorism attack involving Bacillus anthracis spore-laced letters resulted in 22 cases of inhalation anthrax, with five fatalities. This incident identified gaps in our health care system and precipitated a renewed interest in identifying both therapeutics and rapid diagnostic assays. To address those gaps, well-characterized animal models that resemble the human disease are needed. In addition, a rapid assay for a reliable diagnostic marker is key to the success of these efforts. In this study, we exposed African green monkeys to B. anthracis spores; examined clinical signs and physiological parameters, including fever, heart rate, complete blood count, and bacteremia; and evaluated the PCR assay and electrochemiluminescence (ECL) immunoassay for the biomarkers protective antigen and capsule. The results demonstrated that although there were neither objective clinical nor physiological signs that consistently identified either infection or the onset of clinical anthrax disease, the African green monkey is a suitable animal model exhibiting a disease course similar to that observed in the rhesus model and humans. We also demonstrated that detection of the biomarkers protective antigen and capsule correlated with bacterial loads in the blood of these nonhuman primates. The ECL immunoassay described here is simple and sensitive enough to provide results in one to two hours, making this assay a viable option for use in the diagnosis of anthrax, leading to timely initiation of treatment, which is a key component of B. anthracis therapeutic development.
Bacillus anthracis is a gram-positive, nonmotile, spore-forming bacterium that is the etiological agent of anthrax. Bacilli that are exposed to free oxygen form spores that are resistant to extremes of temperature and desiccation and can remain viable for years in the environment. Spores are approximately 1 to 10 μm and when inhaled are deposited deep in the lung. Infection begins when the spores germinate into vegetative cells.
The capsule and two exotoxins, lethal toxin (LT) and edema toxin, are believed to be primarily responsible for the symptoms and pathogenesis of B. anthracis infection. The two toxins are synthesized from different genes but share a protein, protective antigen (PA), that is responsible for the cell-binding component for each (7, 41). Toxic activity is expressed only when PA is combined with lethal factor (LF), forming LT, or edema factor (EF), forming edema toxin. EF and LF have enzymatic functions but require PA to achieve their biological effect (34). LF is a metalloprotease which cleaves the amino terminus of MAP kinase kinases 1 and 2 and thereby inhibits the intracellular MAP kinase pathway (9). EF acts as a calmodulin-dependent adenylate cyclase that induces edema (2). The toxin components are encoded on the pX01 plasmid, while the capsule is encoded on the pX02 plasmid (21, 38). The anthrax toxin system has been studied extensively (7) and linked to many stages of the disease, from clinical symptoms, such as edema, fever, and malaise, to the cellular release of bacilli, which leads to systemic disease and ultimately death (17, 39).
There is a known correlation between bacteremia and PA concentration (13, 32), and, in vitro, production of PA by the vegetative organism peaks during the shift from exponential to stationary replication phase (49). In addition, production of the components of the LT by an isolate has proven to be the most reliable means of identifying B. anthracis strains (35). For these reasons, PA is a logical target for diagnostic assays.
The capsule, composed of poly-d-glutamic acid, is also a virulence factor present during an anthrax infection. By virtue of its negative charge, the capsule is thought to inhibit phagocytosis of the vegetative cells by macrophages (40). Since the capsule is required for virulence, it may also be a good target for diagnostic assays (21, 52).
Inhaled spores are highly infectious, and there is a high mortality rate associated with the ensuing respiratory illness; therefore, B. anthracis is considered a serious bioterrorism threat. Within 1 to 6 days, depending on the inhaled dose, nondescript clinical symptoms of the disease are seen, such as fatigue, muscle pain, and fever, which usually last 2 to 3 days (26). Once clinical symptoms are apparent, mortality is nearly 100% in untreated cases; however, with early diagnosis and proper treatment, the prognosis is improved (1, 5, 14, 15, 22). In the United States, a bioterrorism attack in 2001 resulted in 22 cases (five were fatal) from letters sent through the U.S. Postal Service (30). Rapid diagnosis was critical to initiation of treatment and survival in these cases.
Historically, diagnosis of B. anthracis infection has been made through classical culture and microscopic techniques. These techniques require at least 24 h, longer if B. anthracis-specific fluorescent antibodies are not available to confirm identity. They also require experienced personnel working in a well-equipped laboratory. Reliable, simpler, and faster methods for detection and identification of the organism are needed. In fact, in order to achieve agent identification with a high level of confidence, an integrated approach to diagnostics, combining both immunological and nucleic acid analysis, may be the best course of action. Over the last decade, methods based on antibody capture of antigens (immunoassays) and nucleic acid-based detection have been developed and are rapidly being assimilated into laboratories. Immunoassays based on the detection of PA are available. Colorimetric enzyme-linked immunosorbent assays (ELISAs) claim detection of approximately 10 ng/ml from blood or serum samples (32). However, these assays can be time-consuming and labor-intensive due to the need for a vigorous washing and numerous separate reagent addition steps. The electrochemiluminescence (ECL) assay using BioVeris's M-Series instruments has proven to be superior to conventional immunoassays such as the ELISA or time-resolved fluorescent assays for the detection of a number of different agents (6, 50). ECL assays have proven to be simple (one step), robust (shelf life, ∼2 years), reproducible (coefficients of variation of <15%), rapid (18 min/test sample), and 10 to 1,000 times more sensitive than ELISAs using the same antibodies. For the first time, immunoassays that can potentially achieve sensitivity levels seen with many nucleic acid-based assays are available. Thus, a PA and a capsule ECL assay were developed in our laboratory. A number of PCR assays are also available, including those used in the U.S. Public Health System's Laboratory Response Network as well as those developed by commercial companies, such as Roche, Idaho Technology, and Cepheid (6). Like the Laboratory Response Network assays, most of these assays target three distinct loci, one chromosomal and one on each of the two plasmids (25). When coupled to rapidly evolving sample processing methods for removing inhibitors of PCR, these assays have been shown to be sensitive and specific, providing a means of detection from culture-negative samples even after initiation of antimicrobial therapy (25).
This bacterium, and the disease anthrax, has been studied for decades. Based on vaccine studies, the rabbit and the rhesus monkey (Macaca mulatta) are considered the most appropriate models at this time (3, 16, 20, 28, 29). Although alternatives have been characterized (3, 16, 20, 36, 53), appropriate animal models for evaluating postexposure prophylactic or therapeutic efficacy for inhalation anthrax are still needed. The Food and Drug Administration's animal rule (21 CFR 314 [12a]), which requires pivotal studies in well-characterized animal models with pathophysiology that resembles the disease in humans, dictates that these models be developed to secure licensure of the new medical countermeasures to inhalation anthrax.
The African green monkey (AGM) is being successfully used as a predictive model for several human diseases (4, 10, 11, 27, 33, 42). The AGM is an Old World monkey that has been classified into four subspecies, based not only on phenotypic differences but also on geographical distribution. Our research is focused on characterizing Chlorocebus aethiops, which has been primarily isolated on St. Kitts Island in the West Indies since the 17th century (43). It is thought that C. aethiops was introduced to the island during the slave trade and that 300 years of isolation from the parent African species has caused divergence that allows the St. Kitts AGM to be free from many of the diseases endemic to other Old World monkeys (43).
We have studied the pathology of inhalation anthrax in the AGM and shown that it is similar to the rhesus model as well as to what is known to occur in humans (51). In this study, we exposed AGMs to B. anthracis spores, monitored clinical signs and physiological endpoints, including fever, heart rate, complete blood count (CBC), and bacteremia, and also determined the utility of PCR and immunoassays for B. anthracis PA and capsule. In this model, we identified PA as an appropriate biomarker of infection in addition to establishing a rapid assay platform. The results of this assay correlate with bacteremia and PCR results while permitting medical intervention at an appropriate time point.
MATERIALS AND METHODS
Animals.
Five female and four male AGMs, ranging in weight from 3.2 to 6.1 kg, were commercially obtained from the St. Kitts colony. These were maintained and fed according to facility standard operating procedures and were provided water ad libitum. Research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations related to animals and experiments involving animals and adhered to principles stated in the Guide for the Care and Use of Laboratory Animals (41a). The U.S. Army Medical Research Institute of Infectious Diseases is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Before inclusion in the study, AGMs were determined to be clinically healthy, acclimated to the facilities, and identified by tattoo.
Implantation of telemetry devices.
A radiotelemetry device (Data Sciences International, St. Paul, MN) used to monitor body temperature, heart rate, and blood pressure was surgically implanted into AGMs at least 7 days before exposure. The telemetry body was placed in a pocket between the external and internal abdominal oblique muscles.
Telemetry data analysis.
Body temperature, heart rate, and blood pressure were recorded every 30 min by the DataQuest A.R.T.3.1 system (Data Sciences International). Monitoring began 7 to 10 days preexposure to develop a baseline training period to fit an autoregressive integrated moving average model. Forecasted values for the postexposure times were based on the training model extrapolated forward in time. Residual changes postexposure were determined by subtracting the predicted value from the actual value recorded for each point. For body temperature, residual changes greater than three standard deviations above the training period values were used to compute fever duration (number of hours of significant temperature elevation), fever hours (sum of the significant temperature elevations), and average fever elevation (fever hours divided by fever duration in hours).
Implantation of indwelling catheters.
AGMs were implanted with indwelling central venous catheters to facilitate frequent phlebotomy sampling. Central venous lines were inserted in the internal jugular vein and tunneled beneath the skin in the subcutaneous tissue to exit through the skin in the upper back. The AGMs wore nonrestraining jackets to prevent access to the exit site. Catheters were attached to a tether system and maintained using a heparin-saline solution (19).
Spore preparation.
B. anthracis (Ames strain) spores were produced in flask cultures of Leighton and Doi medium. Spores were harvested by centrifugation, washed in sterile water for injection, and purified on gradients of 60% Hypaque-76. Spores were stored until use at 4°C in 1% phenol. Before aerosolization, phenol was removed, and spores were resuspended in water for injection and heat shocked at 60°C for 45 min.
Aerosol exposure.
AGMs were exposed to B. anthracis spores (Ames) in a head-only chamber contained in a class III biological safety cabinet maintained under negative pressure. AGMs were exposed for 10 min to an aerosol created by a 3-jet collision nebulizer (BGI, Inc., Waltham, MA) and controlled by the automated bioaerosol exposure system (23). Whole-body plethysmography (Buxco Research Systems, Wilmington, NC) was performed on each animal to obtain the minute volume, immediately before the exposure, as previously described (18, 29). AGMs were anesthetized by an intramuscular injection of a ketamine-acepromazine-xylazine mixture before plethysmography and aerosol exposure. Aerosol concentration of B. anthracis was determined by constant sampling of the chamber using an all-glass impinger (AGI; Ace Glass, Vineland, NJ) containing sterile water as the collection medium. Spore concentrations were determined by plating on tryptic soy agar. The presented dose (in CFU) was determined using the respiratory minute volume determined by the plethysmograph. Presented dose was calculated by multiplying the total volume of experimental atmosphere inhaled by the aerosol concentration. The disease course was compared in animals receiving 4.6 × 105 ± 2.5 × 105 CFU (low-dose group) and those receiving 1.5 × 107 ± 7.2 × 106 CFU (high-dose group). The median lethal dose in 50% of the population (LD50) in our laboratory has been determined to be 1.1 × 104 CFU (95% confidence limits: 2.9 × 103, 8.1 × 104).
Clinical observation.
AGMs were observed at least twice daily, and appearance, clinical signs, natural behavior and activity, provoked behavior, and food intake were recorded. Descriptive criteria for each category were as follows: appearance included coat and grooming, as well as condition and clarity of eyes and nose; clinical signs included breathing rate and pattern; natural behavior included peer interaction, vocalization, and alertness; provoked behavior included a response when one approached the cage or rattled a toy, versus no response. Each category was ranked on an ascending numerical scale from 0 to 3, with 0 being normal in each category. A total clinical score was determined by adding the categories. When a clinical score was 5 to 8, the frequency of daily observations was increased to carefully monitor the progression of disease and welfare of the nonhuman primate.
CBCs.
Blood (250 to 500 μl) was collected every 12 h in an EDTA tube and processed on a Beckman Coulter hematology analyzer.
Bacteremia.
Whole-blood samples were collected in Wampole isolator tubes (Inverness Medical, Princeton, NJ), every 12 h beginning 48 h postexposure, until the AGM became positive by culture. Blood was cultured on tryptic soy agar by the spread plate method (0.1 ml/plate, three plates/time point). Plates were incubated for 18 to 24 h at 37°C. Colonies were counted and reported approximately 24 h postsampling.
ECL assay for PA and capsule.
Three PA-specific monoclonal antibodies (MAb) were used as a capture antibody mix, and a polyclonal rabbit serum made against PA was used as a detector antibody to measure PA (37). A single capsule-specific MAb was used as both capture and detector antibody for the capsule assay (12). While use of a single MAb on both sides of an assay is not ideal, the existence of multiple copies of the epitope on the capsule surface, demonstration of a dose response, and the specificity of the assay for its target (not reactive to PA, LF, EF, or spore coat; data not shown) confirmed that the optimized capsule ECL assay performed as expected. Purified antibodies were labeled with biotin or ruthenium using standard coupling methods. Biotinylated antibodies were prebound to streptavidin-coated paramagnetic beads (2.8-μm diameter; M-280; Dynal Corp, Lake Success, NY). Concentrations of labeled antibodies were then optimized in a 15-min assay by using standard checkerboard titrations (31). The optimized PA assay was then lyophilized into single-use assay tubes. The capsule assay was not lyophilized, with reagents remaining in a wet format and mixed just before conducting the assay. An M1R instrument (BioVeris, Gaithersburg, MD) was used to run the assays. Data were analyzed using custom-designed Excel spreadsheets. Whole blood from the AGMs as well as four negative controls and two levels of positive controls (whole blood spiked with either PA or capsule) were added to the appropriate assay tubes. All samples (50 μl) were tested in duplicate. An additional 50 μl of 10 mM phosphate-buffered saline with 0.3% Tween-20 (PBS-T) was then added to each of the lyophilized PA assay tubes. All assay tubes were then mixed by shaking on a platform shaker for 15 min at room temperature. Finally, 300 μl of PBS-T was added to each tube, and tubes were then read using the M1R analyzer. Negative or uninfected blood served as matrix controls and was used to determine assay cutoffs. Samples were considered positive if the ECL signal was ≥1.2 times the average of the negative matrix controls. This assay cutoff value was possible due to the reproducibility of ECL assays, as evidenced by the low coefficient of variation (<15%) seen on this platform. Positive controls must reach a threshold value in order for the assay to be valid. Differences in ECL signals of the various instruments were negated by determining the signal-to-noise ratio (S/N value) of each sample, calculated by dividing the sample ECL value by the average ECL value of the negative matrix controls. The limit of detection (LOD) of the PA ECL assay from blood samples was 125 pg of purified PA per 50-μl reaction, or 2.5 ng/ml. The LOD of the capsule assay from whole blood was 100 pg of Cu+-purified capsule per reaction or 2 ng/ml. The LOD for each assay was determined by adding recombinant PA or purified capsule to whole blood and then assaying, to ensure that there were no inhibitory components in whole blood (method development, unpublished data). Recombinant-PA reactivity was also shown to be identical to that of native PA.
DNA purification and PCR analysis.
DNA was extracted from AGM whole blood by using a QIAamp DNA minikit (Qiagen) as previously described (8). Positive extraction controls at a concentration of 1 × 103 CFU/ml as well as negative extraction controls prepared from control nonhuman primate blood were included after being subjected to the same extraction protocol as the samples to ensure extraction efficiency. All nucleic acid was analyzed by real-time PCR with a Roche LightCycler. Five μl of DNA per 20-μl total reaction volume was tested with both PA (plasmid pX01; forward primer, TTCAAGTTGTACTGGACCGATTCTC; reverse primer, TCCATCATTGTCACGGTCTGG; probe, CCGTAGGTCCAGCACTTGTACTTCGCTT) and capsule B (plasmid pX02; forward primer, CAGATAATGCATCGCTTGCTTTAG; reverse primer, GGATGAGCATTCAACATACCACG; probe, CAGAGGCTCTTGGGATTGATGAGGAAACA) gene-specific primers and TaqMan probes (5). In addition, an internal positive control assay was run on all samples to monitor the presence of PCR inhibitors remaining in the purified DNA eluates (24). For all PCR assays, a total of 45 cycles were completed per run. Data analysis was automatically performed by the instrument and reported as a cycle threshold (CT) value. Sample curves were also reported for each sample, and the CT value was evaluated in conjunction with this curve to determine if the CT value was interpreted correctly by the instrument software. The LOD of the PA and capsule B PCR assays was 50 fg of genomic DNA per 5 μl of reaction mixture, which corresponds to 9 genome equivalents based on total genome size (6).
Statistical analysis.
SAS version 9.1.3 (SAS Institute, Cary, NC) was used. Due to small sample sizes, Wilcoxon rank-sum tests were used to compare CBC raw values and percentage change from baseline between groups at each time point. Step-down Bonferroni corrections were used for multiple comparisons. Bacteremia load and ECL PA S/N values were correlated using Spearman rank-order correlation. Dose and time to death (TTD) were correlated using Pearson product-moment correlation. Times to bacteremia were also compared using a Wilcoxon test. Kaplan-Meier survival analysis was used to calculate survival curves and median TTD. Log-rank tests were used to compare resulting survival curves. Sensitivity and specificity of the various assays were determined by comparing the assay results to the presence or absence of bacteria in the blood. Sensitivity measured the ability of the assay to identify bacteremic samples, while specificity measured the ability of the assay to identify nonbacteremic samples.
RESULTS
Survival of AGMs after aerosol exposure to B. anthracis.
Nine AGMs were exposed in two cohorts to aerosols containing B. anthracis spores at a range of presented doses between 2.1 × 105 and 1.9 × 107 CFU to gain an understanding of the disease course in AGMs. These doses were chosen based on preliminary studies (E. Leffel, unpublished observations) and were expected to give a gradation of responses (Table 1). Four out of five AGMs in the low-dose group succumbed to disease 1 to 5 days after becoming bacteremic. The survivor did not become bacteremic. All four AGMs in the high-dose group succumbed to inhalation anthrax 1 to 3 days after becoming bacteremic.
TABLE 1.
Summary of B. anthracis aerosol-exposed AGMs' disease course
| Animal ID | Body wt (kg) | Gender | No. of inhaled CFU | No. of days to death | Bacteremia (day post-exposure) | Fever |
|---|---|---|---|---|---|---|
| V302 | 3.4 | F | 210,000 | Survived | No | No |
| V461 | 3.4 | F | 210,000 | 3 | Yes (2) | No |
| 05072 | 5.5 | M | 520,000 | 7.5 | Yes (2) | Yes |
| 05065 | 6.1 | M | 630,000 | 11 | Yes (9) | Yes |
| W247 | 4.6 | M | 750,000 | 12.5 | Yes (10) | Yes |
| V540 | 3.2 | F | 11,200,000 | 5 | Yes (3) | Unknowna |
| V547 | 3.4 | F | 12,800,000 | 6 | Yes (3) | Unknowna |
| V537 | 3.2 | F | 15,900,000 | 3 | Yes (2) | No |
| W164 | 5.5 | M | 18,900,000 | 3.5 | Yes (2) | No |
Unknown due to disruption of telemetry device.
Clinical observations.
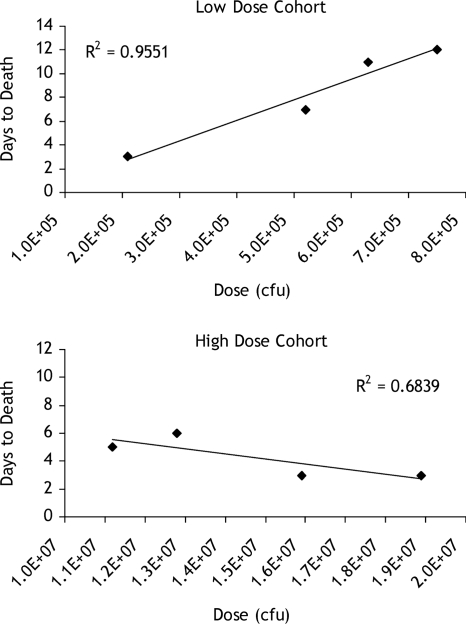
There was a positive relationship (r = 0.98, P = 0.0470) between presented dose of B. anthracis spores and survival time in the low-dose but not in the high-dose group (r = −0.82, P = 0.18) (Fig. 1). The median survival time in the low-dose cohort (11 days) was significantly longer than the median survival time in the high-dose cohort (4 days) (P = 0.0276). Over the course of the infections, some AGMs exhibited decreases in food intake, but this did not correlate with presence of biomarkers, total clinical scores, or mortality. Breathing patterns and respiration rates rarely changed from normal. A change in provoked behavior was the single clinical observation that most indicated a declining state of health: however, the increase in the score was considerably different in only one animal in the high-dose cohort. Because clinical observations are very individualized and abnormal signs do not present long before death, these subjective observations are not useful predictors of infection or state of pathogenesis (Tables 2 and 3).
FIG. 1.
Relationship between dose and survival of B. anthracis aerosol-exposed AGMs.
TABLE 2.
Correlation of bacteremia and ECL and PCR assay results with clinical score in the low-dose AGM cohorta
| Monkey | Dose, CFU/ml of blood (LD50) | Time
|
Bacteremia (CFU/ml) | ECL result (S/N value)
|
PCR result (CT)b
|
Clinical score | |||
|---|---|---|---|---|---|---|---|---|---|
| Day | Hour | PA | Capsule | PA | Capsule | ||||
| V461 | 2.1E + 05 (19) | −7 | NT | 0.94 | 0.99 | BLD | BLD | NT | |
| 0 | 7 | 0 | 0.91 | 0.98 | BLD | BLD | NT | ||
| 1 | 24 | 0 | 0.82 | 0.89 | BLD | BLD | NT | ||
| 36 | 0 | 0.94 | 0.89 | BLD | BLD | NT | |||
| 2 | 48 | 235 | 3.24 | 1.09 | 33.86 | 37.73 | 0 | ||
| 60 | >1,000 | 92.67 | 4.09 | 30.59 | 31.92 | 0 | |||
| 3 | 72c | NT | NT | NT | NT | NT | NT | ||
| 05072 | 5.2E + 05 (47) | −7 | NT | 0.97 | 1.10 | BLD | BLD | NT | |
| 0 | 7 | 0 | 0.96 | 0.98 | BLD | BLD | NT | ||
| 1 | 24 | 0 | 0.97 | 1.01 | BLD | BLD | NT | ||
| 36 | 0 | 1.02 | 1.13 | BLD | 33.79* | NT | |||
| 2 | 48 | 40 | 1.33 | 1.62 | BLD | BLD | 0 | ||
| 60 | 95 | NT | NT | 33.03 | BLD | 0 | |||
| 3 | 72 | 165 | 11.77 | 3.02 | 33.90 | 35.97 | 0 | ||
| 84 | NT | NT | NT | NT | NT | NT | |||
| 4 | 96 | 155 | 7.61 | 5.48 | 29.54 | 30.88 | 2 | ||
| 108 | NT | NT | NT | NT | NT | 2 | |||
| 5 | 120 | 60 | 8.57 | 6.99 | 28.16 | 29.23 | 2 | ||
| 132 | NT | NT | NT | NT | NT | 2 | |||
| 6 | 144 | 860 | 13.12 | 3.57 | 28.14 | 29.57 | 1 | ||
| 156 | NT | NT | NT | NT | NT | 0 | |||
| 7 | 168 | 205 | 26.10 | 7.79 | 27.47 | 28.86 | 0 | ||
| 180c | >1,000 | NT | NT | 26.78 | 27.80 | NT | |||
| 05065 | 6.3E + 05 (57) | −7 | NT | 0.94 | 0.99 | BLD | BLD | NT | |
| 0 | 7 | 0 | 0.96 | 1.05 | BLD | BLD | NT | ||
| 1 | 24 | 0 | 0.92 | 1.06 | BLD | BLD | NT | ||
| 36 | 0 | NT | NT | BLD | BLD | NT | |||
| 2 | 48 | 0 | 0.91 | 1.05 | BLD | BLD | 0 | ||
| 60 | 0 | NT | NT | BLD | BLD | 0 | |||
| 3 | 72 | 0 | 0.92 | 1.07 | BLD | BLD | 0 | ||
| 84 | 0 | NT | NT | BLD | BLD | 0 | |||
| 4 | 96 | 0 | 0.95 | 1.05 | BLD | BLD | 0 | ||
| 108 | 0 | 1.00 | 1.00 | BLD | BLD | 0 | |||
| 5 | 120 | 0 | 0.97 | 1.07 | BLD | BLD | 0 | ||
| 132 | 0 | NT | NT | BLD | BLD | NT | |||
| 6 | 144 | 0 | 0.96 | 1.07 | BLD | BLD | 2 | ||
| 156 | 0 | NT | NT | BLD | BLD | 2 | |||
| 7 | 168 | 0 | 0.90 | 1.00 | BLD | BLD | 0 | ||
| 180 | NT | NT | NT | NT | NT | 0 | |||
| 8 | 192 | 0 | 0.98 | 0.99 | BLD | BLD | 0 | ||
| 9 | 216 | >1,000 | 185.78 | 1.72 | 25.47 | 25.92 | 0 | ||
| 10 | 240 | >1,000 | 109.90 | 12.12 | 29.84 | 30.57 | 0 | ||
| 11 | 264c | NT | NT | NT | NT | NT | NT | ||
| W247 | 7.5E + 05 (68) | −7 | NT | 0.91 | 0.94 | BLD | BLD | NT | |
| 0 | 7 | 0 | 0.95 | 0.98 | BLD | BLD | NT | ||
| 1 | 24 | 0 | 0.95 | 0.97 | BLD | BLD | NT | ||
| 36 | 0 | NT | NT | BLD | BLD | NT | |||
| 2 | 48 | 0 | 0.97 | 0.94 | BLD | BLD | 0 | ||
| 60 | 0 | NT | NT | BLD | BLD | 0 | |||
| 3 | 72 | 0 | 0.81 | 0.98 | BLD | BLD | 1 | ||
| 84 | 0 | NT | NT | BLD | 19.48* | NT | |||
| 4 | 96 | 0 | 0.99 | 0.94 | BLD | BLD | 2 | ||
| 108 | 0 | 0.89 | 0.94 | BLD | 26.32* | 2 | |||
| 5 | 120 | 0 | 0.88 | 0.97 | BLD | BLD | 0 | ||
| 132 | 0 | NT | NT | BLD | BLD | 0 | |||
| 6 | 144 | 0 | 0.93 | 0.94 | BLD | BLD | 0 | ||
| 156 | 0 | NT | NT | BLD | BLD | 0 | |||
| 7 | 168 | 0 | 0.98 | 0.93 | BLD | BLD | 0 | ||
| 180 | NT | NT | NT | NT | NT | 0 | |||
| 8 | 192 | 0 | 0.87 | 0.93 | BLD | BLD | 0 | ||
| 9 | 216 | 0 | 1.02 | 0.98 | 36.09 | BLD | 0 | ||
| 10 | 240 | 575 | 11.93 | 3.66 | 30.74 | 32.05 | 0 | ||
| 11 | 264 | 160 | 22.86 | 5.79 | 27.21 | 27.65 | 2 | ||
| 12 | 288 | >1,000 | 78.06 | 3.40 | 23.95 | 25.03 | 3 | ||
| 300c | >1,000 | 134.57 | 1.11 | BLD | BLD | NT | |||
| V302 | 2.1E + 05 (19) | −7 | NT | 0.95 | 0.93 | BLD | BLD | NT | |
| 0 | 7 | 0 | 0.96 | 1.04 | BLD | BLD | NT | ||
| 1 | 24 | 0 | 0.99 | 0.98 | BLD | BLD | NT | ||
| 36 | 0 | NT | NT | BLD | BLD | NT | |||
| 2 | 48 | 0 | 0.95 | 1.04 | BLD | BLD | 0 | ||
| 60 | 0 | NT | NT | 35.93* | BLD | 0 | |||
| 3 | 72 | 0 | 1.03 | 0.97 | 34.55* | BLD | 0 | ||
| 84 | 0 | 0.99 | 0.96 | 34.82* | BLD | NT | |||
| 4 | 96 | 0 | 1.55 | 1.05 | 32.86* | BLD | 0 | ||
| 108 | 0 | 0.95 | 1.00 | BLD | BLD | 0 | |||
| 5 | 120 | 0 | 1.03 | 0.95 | BLD | BLD | 0 | ||
| 132 | 0 | NT | NT | 15.58* | BLD | 0 | |||
| 6 | 144 | 0 | 0.94 | 0.98 | BLD | BLD | 0 | ||
| 156 | 0 | NT | NT | BLD | BLD | 0 | |||
| 7 | 168 | 0 | 0.93 | 0.92 | BLD | BLD | 0 | ||
| 180 | NT | NT | NT | NT | NT | 0 | |||
| 8 | 192 | 0 | 0.95 | 0.94 | BLD | BLD | 0 | ||
| 9 | 216 | 0 | 0.98 | 0.97 | BLD | BLD | 0 | ||
| 10 | 240 | 0 | 0.98 | 0.96 | BLD | BLD | 0 | ||
| 11 | 264 | 0 | 0.96 | 0.96 | BLD | BLD | 0 | ||
| 12 | 288 | 0 | 0.98 | 0.93 | BLD | BLD | 0 | ||
| 300 | 0 | 1.00 | 0.99 | 18.73 | 19.81 | 0 | |||
| 13 | 312 | 0 | 0.97 | 0.95 | BLD | BLD | NT | ||
| 324 | 0 | NT | NT | NT | NT | NT | |||
| 14 | 336 | 0 | 0.98 | NT | NT | NT | NT | ||
Values in boldface are positive. BLD, below LOD; NT, not tested.
For PCR results, both markers must be present for a sample to be considered positive. Asterisks indicate samples which had a positive CT value but did not exhibit an expected response curve and thus are considered negative.
Time of death of the animal.
TABLE 3.
Correlation of bacteremia and ECL and PCR assay results with clinical score in the high-dose AGM cohorta
| Monkey | Dose, CFU/ml of blood (LD50) | Time
|
Bacteremia (CFU/ml) | ECL result (S/N value)
|
PCR result (CT)b
|
Clinical score | |||
|---|---|---|---|---|---|---|---|---|---|
| Day | Hour | PA | Capsule | PA | Capsule | ||||
| V537 | 1.6E + 07 (1,455) | −2 | −48 | 0 | 0.99 | 1.08 | BLD | BLD | NT |
| 0 | 7 | 0 | 0.98 | BLD | BLD | BLD | NT | ||
| 1 | 24 | 0 | 1.00 | 1.17 | BLD | BLD | NT | ||
| 36 | 0 | 1.15 | 1.18 | 33.92 | BLD | NT | |||
| 2 | 48 | 940 | 67.76 | 43.19 | 28.70 | 28.85 | 0 | ||
| 60 | NT | NT | NT | NT | NT | NT | |||
| 3 | 72 | >1,000 | NT | NT | NT | NT | NT | ||
| 84c | NT | NT | NT | NT | NT | 0 | |||
| W164 | 1.9E + 07 (1,727) | −2 | −48 | 0 | 0.96 | 0.90 | BLD | BLD | NT |
| 0 | 7 | 0 | 0.97 | NT | BLD | BLD | NT | ||
| 1 | 24 | 0 | 0.96 | 0.95 | 36.68 | BLD | NT | ||
| 36 | 0 | 1.04 | NT | BLD | BLD | NT | |||
| 2 | 48 | >1,000 | 11.39 | 9.24 | 29.86 | 30.17 | 1 | ||
| 60 | 900 | 50.75 | 26.96 | 28.69 | 29.63 | 1 | |||
| 3 | 72 | >1,000 | 68.43 | 20.43 | 27.45 | 27.50 | 3 | ||
| 84c | >1,000 | 70.54 | 1.19 | 11.74 | 12.64 | NT | |||
| V540 | 1.1E + 07 (1,000) | −2 | −48 | 0 | 1.09 | 1.10 | BLD | BLD | NT |
| 0 | 7 | 0 | 1.00 | NT | BLD | BLD | NT | ||
| 1 | 24 | 0 | 1.04 | NT | BLD | BLD | NT | ||
| 36 | 0 | 1.04 | NT | BLD | BLD | NT | |||
| 2 | 48 | 0 | 1.11 | 1.05 | BLD | BLD | 0 | ||
| 60 | 0 | 1.12 | NT | BLD | BLD | 0 | |||
| 3 | 72 | 0 | 1.27 | 1.08 | BLD | BLD | 1 | ||
| 84 | 260 | 2.92 | 2.48 | 36.10 | 35.91 | 0 | |||
| 4 | 96 | >1,000 | 223.83 | 1.82 | 23.90 | 22.73 | 0 | ||
| 5 | 120c | NT | NT | NT | NT | NT | NT | ||
| V547 | 1.3E + 07 (1,182) | −2 | −48 | 0 | 0.98 | 0.92 | BLD | BLD | NT |
| 0 | 7 | 0 | 0.99 | NT | BLD | BLD | NT | ||
| 1 | 24 | 0 | 0.99 | NT | BLD | BLD | NT | ||
| 36 | 0 | 1.01 | NT | BLD | BLD | NT | |||
| 2 | 48 | 0 | 1.02 | 0.87 | BLD | BLD | 0 | ||
| 60 | NT | NT | NT | NT | NT | 0 | |||
| 3 | 72 | 0 | 1.18 | 0.93 | 35.99 | BLD | 2 | ||
| 84 | 60 | 3.62 | 4.23 | 34.44 | 37.49 | 0 | |||
| 4 | 96 | 820 | 25.62 | 15.24 | 32.30 | 33.32 | 0 | ||
| 5 | 120 | 10 | NT | NT | NT | NT | NT | ||
| 6 | 144c | 144 | NT | NT | NT | NT | NT | ||
Values in boldface are positive. BLD, below the LOD; NT, not tested.
For PCR results, both markers must be present for a sample to be considered positive.
Time of death of the animal.
Telemetry.
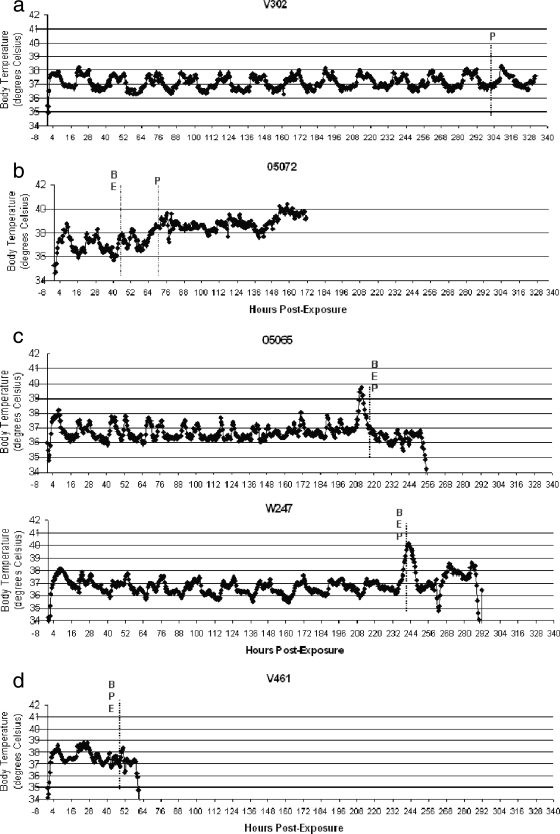
In the low-dose cohort, the survivor maintained a normal biorhythm throughout the study (Fig. 2a). One AGM appeared to develop a low-grade fever approximately 24 h after testing positive for bacteremia (Fig. 2b). Two AGMs exhibited a fever spike at approximately the time of bacteremia (Fig. 2c). One AGM did not develop fever even though it became bacteremic (Fig. 2d).
FIG. 2.
Time course of temperature data in B. anthracis aerosol-exposed AGMs and illustration of time points positive for clinical assays. (a) Survivor; (b) AGM with low-grade fever 24 h after becoming bacteremic and antigenemic; (c) AGM with fever spike preceding bacteremia and antigenemia; (d) AGM with absence of fever in the presence of bacteremia and antigenemia. B, bacteremia (the first time point at which B. anthracis was detected in blood by culture); E, antigenemia (the first time point at which the PA was detected in blood by the ECL assay); P, antigenemia (the first time point in which both PA and capsule markers were detected in blood by the PCR assay).
In the high-dose cohort, collection of data was disrupted, due to unknown technical failure of the system, at approximately 80 h postexposure. Two animals died in this window, without developing signs of fever at the time of bacteremia. The other two AGMs became bacteremic during the window in which telemetry was disrupted, so conclusions could not be formed.
An increase in heart rate was seen with the spike in body temperature but began to decrease as the animal became moribund. Little or no change was seen in systolic or diastolic blood pressure postexposure until animals were moribund, and a profound drop in blood pressure was then observed (data not shown).
CBCs.
CBCs were compared between the high-dose and low-dose cohorts for the first 3 days postexposure. No significant differences in raw CBCs or percentage change from baseline were found at any of the time points for any of the 18 tests (data not shown).
Bacteremia.
The data obtained in this study indicated that bacteremia occurred 2 to 3 days after AGMs were exposed to high doses of aerosolized spores (time to bacteremia, 2.5 ± 0.3 days [mean ± standard error of the mean]). Death in this cohort occurred 1 to 3 days after the detection of bacteremia. In the low-dose cohort, there was a greater range: onset of bacteremia occurred 2 to 10 days postexposure and death 3 to 12 days postexposure, with one survivor (time to bacteremia, 5.8 ± 2.1 days) (Table 1).
ECL assays for PA and capsule.
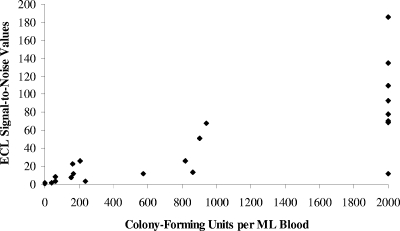
The ECL assays were easy to perform and very reproducible, with a coefficient of variation of less than 15%. In this study, results from each of these assays were obtained in approximately 35 min, providing “real-time” information about the AGM. Detection of PA by the ECL assay correlated with plate culture results at every time point, with the exception of one instance in the surviving AGM. Detection of capsule by the ECL assay was a bit more variable, and in some cases, capsule was not detected even when PA was present at very high levels (Tables 2 and 3). The PA ECL assay was 100% sensitive (23/23) and 97% specific (64/66) relative to bacteremia data. In contrast, the capsule ECL assay was 91.3% sensitive (21/22) and 100% specific (56/56). If both assay results were required to be positive before a sample was considered positive, the sensitivity and specificity would be the same as the capsule results, 91.3% and 100%, respectively. Bacterial load was positively correlated with ECL PA S/N values (r = 0.75, P < 0.0001) (Fig. 3).
FIG. 3.
Comparison of ECL PA detection results to bacterial load in the blood of B. anthracis aerosol-exposed AGMs (87 samples). For graphing purposes, values that were greater than 1,000 CFU/ml were assigned the value of 2,000.
Relative concentrations of PA and capsule could be determined if a standard curve was included with test samples. Using TableCurve 2D (Systat Inc., Richmond, CA) and a transition curve fit, a standard curve was generated and used to quantitate concentrations of test samples. This type of analysis also required a number of dilutions of each unknown to be tested to ensure that the S/N value was within the linear range of the assay. For both PA and capsule, the linear range was between 1 and 1,000 ng/ml (data not shown). An example of quantitative PA and capsule results is shown in Table 4. Quantitation was complicated by the fact that these assays exhibited a prozone at concentrations above 1,000 ng/ml.
TABLE 4.
Example of ECL data from AGMs aerosol exposed to high doses of B. anthracisa
| Monkey | Dose, CFU/ml of blood (LD50) | Time
|
Bacteremia (CFU/ml) | ECL data for:
|
||||
|---|---|---|---|---|---|---|---|---|
| Day | Hour | PA
|
Capsule
|
|||||
| S/N value | Concn (ng/ml) | S/N value | Concn (ng/ml) | |||||
| V537 | 1.6E + 07 (1,455) | −2 | −48 | 0 | 0.99 | BLD | 1.08 | BLD |
| 0 | 7 | 0 | 0.98 | BLD | NT | BLD | ||
| 1 | 24 | 0 | 1.00 | BLD | 1.17 | BLD | ||
| 36 | 0 | 1.15 | BLD | 1.17 | BLD | |||
| 2 | 48 | 940 | 67.76 | 54 | 43.19 | >1,000 | ||
| 60 | NT | NT | NT | NT | NT | |||
| 3 | 72 | >1,000 | NT | NT | NT | NT | ||
| 84 | NT | NT | NT | NT | NT | |||
| W164 | 1.9E + 07 (1,727) | −2 | −48 | 0 | 0.96 | BLD | 0.90 | BLD |
| 0 | 7 | 0 | 0.97 | BLD | NT | NT | ||
| 1 | 24 | 0 | 0.96 | BLD | 0.95 | BLD | ||
| 36 | 0 | 1.04 | BLD | NT | NT | |||
| 2 | 48 | >1,000 | 11.39 | 8 | 9.24 | 312 | ||
| 60 | 900 | 50.75 | 40 | 26.96 | >1,000 | |||
| 3 | 72 | >1,000 | 68.43 | 55 | 20.43 | >1,000 | ||
| 84 | >1,000 | 70.54 | 57 | 1.19 | Prozone | |||
| V540 | 1.1E + 07 (1,000) | −2 | −48 | 0 | 1.09 | BLD | 1.10 | BLD |
| 0 | 7 | 0 | 1.00 | BLD | NT | NT | ||
| 1 | 24 | 0 | 1.04 | BLD | NT | NT | ||
| 36 | 0 | 1.04 | BLD | NT | NT | |||
| 2 | 48 | 0 | 1.11 | BLD | 1.05 | BLD | ||
| 60 | 0 | 1.12 | BLD | NT | NT | |||
| 3 | 72 | 0 | 1.27 | BLQ | 1.08 | BLD | ||
| 84 | 260 | 2.92 | 1 | 2.48 | 88 | |||
| 4 | 96 | >1,000 | 223.83 | 202 | 1.82 | >1,000 | ||
| 5 | 120 | NT | NT | NT | NT | NT | ||
| V547 | 1.3E + 07 (1,182) | −2 | −48 | 0 | 0.98 | BLD | 0.92 | BLD |
| 0 | 7 | 0 | 0.99 | BLD | NT | NT | ||
| 1 | 24 | 0 | 0.99 | BLD | NT | NT | ||
| 36 | 0 | 1.01 | BLD | NT | NT | |||
| 2 | 48 | 0 | 1.02 | BLD | 0.87 | BLD | ||
| 60 | NT | NT | NT | NT | NT | |||
| 3 | 72 | 0 | 1.18 | BLD | 0.93 | BLD | ||
| 84 | 60 | 3.62 | 2 | 4.23 | 114 | |||
| 4 | 96 | 820 | 25.62 | 40 | 15.24 | >1,000 | ||
| 5 | 120 | 10 | NT | NT | NT | NT | ||
| 6 | 144 | 144 | NT | NT | NT | NT | ||
Values in boldface are positive. BLD, below the LOD; NT, not tested; BLQ, below the limit of quantitation.
PCR for PA and capsule.
PCR analysis is more complex and requires that the sample be extracted before being analyzed, resulting in a slower turnaround time. Results from each of these assays were obtained in approximately 2 h. In most cases, detection of PA and capsule by the PCR assay was initially done at the same time. These results correlated with culture data and ECL results; although in three cases they were positive one time point earlier. Similar results were seen regardless of which cohort the sample was obtained from (Tables 2 and 3). The PA PCR assay was 95.8% sensitive (23/24) and 93.7% specific (74/79) relative to bacteremia data. In contrast, the capsule PCR assay was 91.7% sensitive (22/24) and 98.7% specific (78/79). As is usually the case, if both assay results were required to be positive before a sample was considered positive, the sensitivity and specificity would be 91.7% and 93.7%, respectively. It is noteworthy, however, that an additional eight samples (five for PA and three for capsule [Table 2]) would have been considered positive if only the CT value were used.
DISCUSSION
One critical aspect of a robust animal model is how well it extrapolates to humans. We demonstrated that the disease course of inhalation anthrax in the AGM is similar to that observed in humans and rhesus macaques. Examination of the pathological changes in AGMs that succumbed to inhalation of B. anthracis spores showed that the pathogenesis of B. anthracis in AGMs is similar to that in rhesus macaques (51). We also identified a biological marker (PA) for infection that can be detected in a rapid diagnostic assay and confirmed by two other laboratory tests.
Observation of animals after infection, particularly nonhuman primates, is highly subjective and peculiar to the individual animal. Although this is much like what would be expected in humans, and unlike many infectious disease models, the stage of illness cannot be clearly defined based on observations of behavior or anorexia. In a review of human cases, fatigue and/or malaise was a presenting symptom in only 64% of cases (26). While decreased activity and interaction were noted in some AGMs, this is a subjective observation that is accurate only when the researcher is familiar with the individual animals. It cannot be used to predict severity of disease or outcome. Previous observations of the rhesus model, in this laboratory, suggested that clinical signs in the AGM would not be overt enough to suggest onset of disease, and this was indeed the case. However, unlike animals, humans can report their symptoms to their physician.
Respiration was rarely found to change from normal, even when it was assessed only hours before the AGM succumbed to inhalation anthrax. This too seems consistent with the human data, as dyspnea was reported in only 52% of known human cases (26). Another clinical finding that was absent was a cough, reported in 62% of human cases (26). We do not feel that this is a weakness of the model, because a cough is not specific to inhalation anthrax.
In the low-dose group, only two out of five of the animals had a slight fever for a brief time, but the others animals did not show the same pattern. The fever did correlate with positive bacteremia and antigenemia in these two animals, but the fever resolved and the AGMs died from inhalation anthrax days later. In one case, the temperature increased steadily for approximately 2.5 days and then the AGM died, thus exhibiting a pattern different from that seen in the other AGMs. Therefore, fever is not a reliable or reproducible sign of infection in AGMs. Fever is considered a prominent clinical sign in human infection with B. anthracis, and the failure of AGMs to develop a significant fever postexposure was surprising. This was not a flaw in the telemetry system employed, as it has been successfully used to detect and quantify fever responses in a number of other diseases (45-48). It is also not an issue peculiar to AGMs, which have been shown to develop significant fevers after aerosol exposure to Yersinia pestis (44). Fever results similar to those obtained with the AGMs have been observed with B. anthracis-infected rhesus macaques (Leffel, unpublished).
It must also be considered that perhaps expectations of the presence of fever are too loosely based. Close evaluation of the human data revealed that the lack of pattern we found in the AGM is not inconsistent with that seen in human cases. While it was reported that 100% of the 2001 cases included fever as a symptom (30), analysis of the significant elevation, duration, and onset shows a lack of pattern similar to that seen in the AGM data presented here. Historically, 81% of cases have had an “abnormal temperature” response (26), and we would describe this model as displaying an abnormal trend: the response ranged from absence of fever to a disruption in diurnal rhythms to low-grade fever.
Changes in the heart rate or blood pressure failed to indicate a reliable time to treat in either cohort (data not shown). Tachycardia was a presenting clinical sign in 66% of human cases (26), but changes in blood pressure were not identified as being relevant. Analysis of CBCs resulted in no consistent pattern or significant changes which could be used to indicate infection, and this is comparable to human findings.
The TTD for the AGM model is another variable that could be compared to human cases and used to speculate on dose-response relationships. In this study, the low-dose cohort had a median TTD of 11 days (range, 3 to 12.5), and in the high-dose cohort, it was 4 days (range, 3.5 to 6). This was comparable to the 2001 U.S. cases, where the TTD after initiation of symptoms ranged from 5 to 8 days (30). The critical variable is probably not the dose inhaled but how quickly one becomes bacteremic after the exposure. If there is a robust host response that results in spores being quickly cleared by alveolar macrophages, for example, then bacteremia can be prevented and there may be survival (as seen in AGM V302). At the same dose in AGM V461, spores germinated and systemic bacteremia was present in 2 days, suggesting that the immune response of this animal was not as vigorous. This AGM did not succumb to inhalation anthrax until day 3, so if treatment had been started at the onset of bacteremia, recovery might have been possible, substantiating the need for a rapid diagnostic assay for this disease.
The inverted dose-response curve for the low-dose cohort, shown in Fig. 1, is counterintuitive. There are two factors to consider when analyzing this set of data. First, one must consider the confidence limits for the LD50 and recognize that an inhalation challenge dose is not as discrete as doses administered by other routes. Therefore, in the low-dose cohort, we saw a varied response that would probably be seen in a diverse human population. These doses are not vastly different, and the graphical representation illustrates nicely a second point to consider. If the exposure is low enough to allow time for the innate immune system to respond by clearing spores, thereby prolonging germination time, then the TTD may be extended. The advantage, in a model, would be more time to initiate treatment and increase chance of survival.
There were neither clinical nor physiological signs that consistently identified either a positive infection or onset of classical anthrax disease, so it was very important to focus on reliable biological markers and determine how to measure them quickly. In this model, we chose to target a marker in whole blood because it is an easily obtainable sample that does not require anesthesia of the AGM and it extrapolates well to traditional clinical sampling of humans. Certainly B. anthracis could be detected in other samples, such as a nasal swab. However, this would be indicative of exposure and not necessarily a systemic infection, which is what we intended to identify. Traditionally, researchers (and, in rare cases, clinicians) have relied on a 24-h culture to confirm bacteremia after exposure to B. anthracis. In a hospital setting, additional time would be required to confirm the identification of the bacteria. However, this could result in a significant lag between admittance and initiation of appropriate treatment if the cause is B. anthracis; in the 2001 attack, rapid diagnosis was critical to survival (30). We have been interested in identifying a correlate of infection that can be used in a diagnostic setting to rapidly determine onset of disease. In these studies, we used ECL and PCR to test for PA and capsule, based on some of the diagnostic assays used in various hospitals during the 2001 U.S. attacks. Our intent was to down-select the most promising assay for use in development of a therapeutic animal model. In high- and low-dose cohorts, both ECL and PCR assays correlated well with bacteremia levels in the exposed AGMs. In all low-dose nonhuman primates that succumbed to the disease (4/5), all assays for each animal became positive within a 24-h window. In the high-dose cohort, four out of four AGMs that succumbed to inhalation anthrax had positive assay results within a 12-h window. While very few samples yielded less than 100 CFU/ml, a single sample containing only 40 CFU/ml was positive by both PA and capsule ECL but just above the assay cutoff. This same sample was negative (below the LOD) by the PCR assays. However, a sample containing 60 CFU/ml was positive by all four assays, perhaps indicating that the assays as described in this study have LODs (40 to 60 CFU/ml) similar to those of isolation of B. anthracis from blood.
The data presented here demonstrated that the AGM is a suitable animal model for the study of inhalation anthrax and may be suitable as a therapeutic or vaccine model. In particular, the availability of a rapid, reliable, and specific diagnostic marker makes it particularly suitable for postexposure prophylaxis or therapeutic studies. Because the TTD is protracted in the AGM at moderate doses, the model may be adjusted by changing the presented aerosol dose, which theoretically alters the time to bacteremia. This also may make the AGM a good postexposure prophylaxis model because the treatment window may be modified by presented dose of aerosolized spores. A therapeutic model is badly needed to allow testing of new pharmaceuticals under the Food and Drug Administration's animal rule (12a).
We characterized the natural history of inhalation anthrax in the AGM and were able to confirm infection with a rapid diagnostic assay that identifies the presence of PA or capsule in the blood. We also demonstrated that PCR can detect the genetic material responsible for encoding the same two biomarkers at approximately the time when the animal becomes bacteremic. The ECL assay represents a significant improvement over more traditional immunodiagnostic technologies in terms of assay simplicity, robustness, speed, and sensitivity. These rapid ECL assays will make it possible to initiate treatment within 1 to 2 h after detection of a positive PA or capsule result from blood samples. This assay can be structured to provide S/N values or quantitative results; however, quantitative results require substantially more controls in every assay as well as multiple dilutions of test samples (extending assay time). We established that an assay cutoff of 1.2 correlates with the appearance of bacteremia and there is good correlation between S/N values for PA and the level of bacteremia in the blood of infected AGMs. While detection of capsule looks promising, the capsule MAb is an immunoglobulin M, which is difficult to purify and does not label consistently from lot to lot. Efforts are under way to class-switch this MAb and, until this has been achieved, the current capsule assay will not be further studied. Being able to rapidly predict bacteremia allows treatment to be started in a realistic time frame, thereby mimicking a true threat emergency. While the complexity and additional time requirement of the PCR assays make it less attractive than the simpler ECL technique, the PCR assays described in this study worked well and could easily be used in a confirmatory role or in situations that are not as time sensitive.
Acknowledgments
This research was sponsored by the National Institute of Allergy and Infectious Diseases, Bethesda, MD. Assay development was funded by the Joint Science and Technology Office for Chemical and Biological Defense Program, Defense Threat Reduction Agency.
The views, opinions, and/or findings contained herein are those of the authors and should not be construed as an official Department of Army position, policy, or decision unless so designated by other documentation. The authors do not have a commercial or other association that would pose a conflict of interest.
We are grateful to D. Norwood, P. Hobart, and K. Kenyon for critically reviewing the manuscript, and we thank D. Dyer, S. Coyne, and B. Kearney for their excellent technical assistance.
Editor: S. R. Blanke
Footnotes
Published ahead of print on 13 October 2008.
REFERENCES
- 1.Albrink, W. S. 1961. Pathogenesis of inhalation anthrax. Bacteriol. Rev. 25268-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aroura, N., and S. H. Leppla. 1993. Residues 1-254 of anthrax toxin lethal factor are sufficient to cause cellular uptake of fused polypeptides. J. Biol. Chem. 2683334-3341. [PubMed] [Google Scholar]
- 3.Berdjis, C. C., C. A. Gleiser, H. A. Hartmen, R. W. Kuehne, and W. S. Gochenour. 1962. Pathogenesis of respiratory anthrax in Macaca mulatta. Br. J. Exp. Pathol. 43515-524. [PMC free article] [PubMed] [Google Scholar]
- 4.Binhazim, A. A., S. S. Shin, W. L. Chapman, Jr., and J. Olobo. 1993. Comparative susceptibility of African green monkeys (Cercopithecus aethiops) to experimental infection with Leishmania leishmania donovani and Leishmania leishmania infantum. Lab. Anim. Sci. 4337-47. [PubMed] [Google Scholar]
- 5.Brachman, P. S. 1980. Inhalation anthrax. Ann. N. Y. Acad. Sci. 35383-93. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, D. R., L. J. Hartman, B. M. Loveless, M. S. Frye, M. A. Shipley, D. L. Bridge, M. J. Richards, R. S. Kaplan, J. Garrison, C. D. Baldwin, D. A. Kulesh, and D. A. Norwood. 2006. Detection of biological threat agents by real-time PCR: comparison of assay performance on the R.A.P.I.D., the LightCycler, and the Smart Cycler platforms. Clin. Chem. 52141-145. [DOI] [PubMed] [Google Scholar]
- 7.Collier, R., and J. Young. 2003. Anthrax toxins. Annu. Rev. Cell Dev. Biol. 1945-70. [DOI] [PubMed] [Google Scholar]
- 8.Coyne, S. R., P. D. Craw, D. A. Norwood, and M. P. Ulrich. 2004. Comparative analysis of the Schleicher and Schuell IsoCode Stix DNA isolation device and the Qiagen QIAamp DNA mini kit. J. Clin. Microbiol. 424859-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duesbery, N. S., C. P. Webb, S. H. Leppla, V. M. Gordon, K. R. Klimpel, T. D. Copeland, et al. 1998. Proteolytic inactivation of MAP kinase-kinase by anthrax lethal factor. Science 280734-737. [DOI] [PubMed] [Google Scholar]
- 10.Durbin, A. P., C. J. Cho, W. R. Elkins, L. S. Wyatt, B. Moss, and B. R. Murphy. 1999. Comparison of the immunogenicity and efficacy of a replication-defective vaccinia virus expressing antigens of human parainfluenza virus type 3 (HPIV3) with those of a live attenuated HPIV3 vaccine candidate in rhesus monkeys passively immunized with PIV3 antibodies. J. Infect. Dis. 1791345-1351. [DOI] [PubMed] [Google Scholar]
- 11.Durbin, A. P., W. R. Elkins, and B. R. Murphy. 2000. African green monkeys provide a useful nonhuman primate model for the study of human parainfluenza virus types-1, -2, and -3 infection. Vaccine 182462-2469. [DOI] [PubMed] [Google Scholar]
- 12.Ezzell, J. W., T. G. Abshire, and C. Brown. 1999. Analysis of Bacillus anthracis vegetative cell surface antigens and of serum protease cleavage of protective antigen, p. 43-44. In Proceedings of the International Workshop on Anthrax, 1990. Salisbury Printing Co., Wiltshire, United Kingdom.
- 12a.FDA. CFR title 21, food and drugs—part 314. www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfCFR/CFRSearch.cfm.
- 13.Fish, D. C., and R. E. Lincoln. 1968. In vivo-produced anthrax toxin. J. Bacteriol. 95919-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franz, D. R., P. B. Jahrling, A. M. Friedlander, D. J. McClain, D. L. Hoover, W. R. Bryne, J. A. Pavlin, G. W. Christopher, and E. M. Eitzen, Jr. 1997. Clinical recognition and management of patients exposed to biological warfare agents. JAMA 278399-411. [DOI] [PubMed] [Google Scholar]
- 15.Friedlander, A. M. 2000. Anthrax: clinical features, pathogenesis, and potential biological warfare threat. Curr. Clin. Top. Infect. Dis. 20335-349. [PubMed] [Google Scholar]
- 16.Fritz, D. L., N. K. Jaax, W. B. Lawrence, K. J. Davis, M. L. Pitt, J. W. Ezzell, and A. M. Friedlander. 1995. Pathology of experimental inhalation anthrax in the rhesus monkey. Lab. Investig. 73691-702. [PubMed] [Google Scholar]
- 17.Forino, M., S. Johnson, T. Y. Wong, D. V. Rozanov, A. Y. Savinov, W. L. R. Fattorusso, B. Becattini, A. J. Orry, D. Jung, R. A. Abagyan, J. W. Smith, K. Alibek, R. C. Liddington, A. Y. Stongin, and M. Pellecchia. 2005. Efficient synthetic inhibitors of anthrax lethal factor. Proc. Natl. Acad. Sci. USA 1029499-9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster, C. D., T. C. Hunter, P. H. Gibbs, and E. K. Leffel. 2008. Comparison of whole-body plethysmography in jacketed and non-jacketed African green monkeys (Chlorocebus aethiops). J. Am. Assoc. Lab. Anim. Sci. 4752-55. [PMC free article] [PubMed] [Google Scholar]
- 19.Gamble, C. S., K. O. Jacobsen, E. K. Leffel, and M. L. Pitt. 2007. Use of a low-concentration heparin solution to extend the life of central venous catheters in African green monkeys (Chlorocebus aethiops). J. Am. Assoc. Lab. Anim. Sci. 4658-60. [PubMed] [Google Scholar]
- 20.Gleiser, C. A., C. C. Berdjis, H. A. Hartman, and W. S. Gochenour. 1963. Pathology of experimental respiratory anthrax in Macaca mulatta. Br. J. Exp. Pathol. 44416-426. [PMC free article] [PubMed] [Google Scholar]
- 21.Green, B. D., L. Battisti, T. M. Koehler, C. B. Thorne, and B. E. Ivins. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hail, A. S., C. A. Rossi, G. V. Ludwig, B. E. Ivins, R. F. Tammariello, and E. A. Henchal. 1999. Comparison of noninvasive sampling sites for early detection of Bacillus anthracis spores from rhesus monkeys after aerosol exposure. Mil. Med. 164833-837. [PubMed] [Google Scholar]
- 23.Hartings, J. M., and C. J. Roy. 2004. The automated bioaerosol exposure system: preclinical platform development and a respiratory dosimetry application with nonhuman primates. J. Pharmacol. Toxicol. Methods 4939-55. [DOI] [PubMed] [Google Scholar]
- 24.Hartman, L. J., S. R. Coyne, and D. A. Norwood. 2005. Development of a novel internal positive control for Taqman based assays. Mol. Cell. Probes 1951-59. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmaster, A. R., R. F. Meyer, M. P. Bowen, C. K. Marston, R. S. Weyant, G. A. Barnet, J. J. Sejvar, J. A. Jernigan, B. A. Perkins, and T. Popovic. 2002. Evaluation and validation of a real-time polymerase chain reaction assay for rapid identification of Bacillus anthracis. Emerg. Infect. Dis. 81178-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holty, J. E., D. M. Bravata, H. Liu, R. A. Olshen, K. M. McDonald, and D. K. Owens. 2006. Systematic review: a century of inhalational anthrax cases from 1900 to 2005. Ann. Intern. Med. 144270-280. [DOI] [PubMed] [Google Scholar]
- 27.Hurwitz, J. L., K. F. Soike, M. Y. Sangster, A. Portner, R. E. Sealy, D. H. Dawson, and C. Coleclough. 1997. Intranasal Sendai virus vaccine protects African green monkeys from infection with human parainfluenza virus-type one. Vaccine 15533-540. [DOI] [PubMed] [Google Scholar]
- 28.Ivins, B. E., P. F. Fellows, and G. O. Nelson. 1994. Efficacy of a standard human anthrax vaccine against Bacillus anthracis spore challenge in guinea-pigs. Vaccine 12872-874. [DOI] [PubMed] [Google Scholar]
- 29.Ivins, B. E., M. L. Pitt, P. F. Fellows, J. W. Farchaus, G. E. Benner, D. M. Waag, S. F. Little, G. W. Anderson, Jr., P. H. Gibbs, and A. M. Friedlander. 1998. Comparative efficacy of experimental anthrax vaccine candidates against inhalation anthrax in rhesus macaques. Vaccine 161141-1148. [DOI] [PubMed] [Google Scholar]
- 30.Jernigan, D. B., P. L. Raghunathan, B. P. Bell, R. Brechner, E. A. Bresnitz, J. C. Butler, M. Cetron, M. Cohen, T. Doyle, M. Fischer, C. Greene, K. S. Griffith, J. Guarner, J. L. Hadler, J. A. Hayslett, R. Meyer, L. R. Petersen, M. Phillips, R. Pinner, T. Popovic, C. P. Quinn, J. Reefhius, D. Reissman, N. Rosenstein, A. Schuchat, W. J. Shieh, L. Siegal, D. L. Swerdlow, F. C. Tenover, M. Traeger, J. W. Ward, I. Weisfuse, S. Wiersma, K. Yeskey, S. Zaki, D. A. Ashford, B. A. Perkins, S. Ostroff, J. Hughes, D. Fleming, J. P. Koplan, and J. L. Gerberding for the National Anthrax Epidemiologic Investigation Team. 2002. Investigation of bioterrorism-related anthrax, United States, 2001: epidemiologic findings. Emerg. Infect. Dis. 81019-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kijek, T. M., C. A. Rossi, D. Moss, R. W. Parker, and E. A. Henchal. 2000. Rapid and sensitive immunomagnetic-electrochemiluminescent detection of staphylococcal enterotoxin B. J. Immunol. Methods 2369-17. [DOI] [PubMed] [Google Scholar]
- 32.Kobiler, D., S. Weiss, H. Levy, M. Fisher, A. Mechaly, A. Pass, and Z. Altboum. 2006. Protective antigen as a correlative marker for anthrax in animal models. Infect. Immun. 745871-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koech, D. K., J. O. Olobo, A. Wamachi, J. I. Githure, and G. D. Reid. 1992. Cellular responses of vervet monkeys (Cercopithecus aethiops) experimentally infected with Leishmania major. J. Med. Primatol. 21375-376. [PubMed] [Google Scholar]
- 34.Leppla, S. H. 1982. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cAMP concentrations in eukaryotic cells. Proc. Natl. Acad. Sci. USA 793162-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leppla, S. H., B. E. Ivins, and J. W. Ezzell. 1985. Anthrax toxin, p 63-66. In D. Schlessinger (ed.), Microbiology—1985. American Society for Microbiology, Washington, DC.
- 36.Lever, M. S., A. J. Stagg, M. Nelson, P. Pearce, D. J. Stevens, E. A. Scott, A. J. Simpson, and M. J. Fulop. 2008. Experimental respiratory anthrax infection in the common marmoset (Callithrix jacchus). Int. J. Exp. Pathol. 89171-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Little, S. F., S. H. Leppla, and E. Cora. 1988. Production and characterization of monoclonal antibodies to the protective antigen component of Bacillus anthracis toxin. Infect. Immun. 561807-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mikesell, P., B. E. Ivins, J. D. Ristroph, and T. M. Drier. 1983. Evidence for plasmid-mediated toxin production in Bacillus anthracis. Infect. Immun. 39371-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moayeri, M., and S. H. Leppla. 2004. Anthrax toxins in pathogenesis. Curr. Opin. Microbiol. 719-24. [DOI] [PubMed] [Google Scholar]
- 40.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55647-671. [DOI] [PubMed] [Google Scholar]
- 41.Mourez, M. 2004. Anthrax toxins. Rev. Physiol. Biochem. Pharmacol. 152135-164. [DOI] [PubMed] [Google Scholar]
- 41a.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC.
- 42.Olobo, J. O., M. M. Gicheru, and C. O. Anjili. 2001. The African green monkey model for cutaneous and visceral leishmaniasis. Trends Parasitol. 17588-592. [DOI] [PubMed] [Google Scholar]
- 43.Pandrea, I., C. Apetrei, J. Dufour, N. Dillon, J. Barbercheck, M. Metzger, B. Jacquelin, R. Bohm, P. A. Marx, F. Barre-Sinoussi, V. M. Hirsch, M. C. Müller-Trutwin, A. A. Lackner, and R. S. Veazey. 2006. Simian immunodeficiency virus SIVagm.sab infection of Caribbean African green monkeys: a new model for the study of SIV pathogenesis in natural hosts. J. Virol. 804858-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pitt, L. 2004. Nonhuman primates as a model for pneumonic plague. In Public Workshop on Animal Models and Correlates of Protection for Plague Vaccines. http://www.fda.gov/cber/minutes/plague101304t.htm.
- 45.Reed, D. S., C. M. Lind, L. J. Sullivan, W. D. Pratt, and M. D. Parker. 2004. Aerosol infection of cynomolgus macaques with enzootic strains of Venezuelan equine encephalitis viruses. J. Infect. Dis. 1891013-1017. [DOI] [PubMed] [Google Scholar]
- 46.Reed, D. S., C. M. Lind, M. Lackemeyer, L. J. Sullivan, W. Pratt, and M. D. Parker. 2005. Genetically engineered, live attenuated vaccines protect nonhuman primates against aerosol challenge with a virulent IE strain of Venezuelan equine encephalitis virus. Vaccine. 233139-3147. [DOI] [PubMed] [Google Scholar]
- 47.Reed, D. S., T. Larsen, L. J. Sullivan, C. M. Lind, M. G. Lackemeyer, W. D. Pratt, and M. D. Parker. 2005. Aerosol exposure to western equine encephalitis virus causes fever and encephalitis in cynomolgus macaques. J. Infect. Dis. 1921173-1182. [DOI] [PubMed] [Google Scholar]
- 48.Reed, D. S., M. G. Lackemeyer, N. L. Garza, S. Norris, S. Gamble, L. J. Sullivan, C. M. Lind, and J. L. Raymond. 2007. Severe encephalitis in cynomolgus macaques exposed to aerosolized eastern equine encephalitis virus. J. Infect. Dis. 196441-450. [DOI] [PubMed] [Google Scholar]
- 49.Sirard, J. C., M. Mock, and A. Fouet. 1994. The three Bacillus anthracis toxin genes are coordinately regulated by bicarbonate and temperature. J. Bacteriol. 1765188-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, D. R., C. A. Rossi, T. M. Kijek, E. A. Henchal, and G. V. Ludwig. 2001. Comparison of dissociation-enhanced lanthanide fluorescent immunoassays to enzyme-linked immunosorbent assays for detection of staphylococcal enterotoxin B, Yersinia pestis-specific F1 antigen, and Venezuelan equine encephalitis virus. Clin. Diagn. Lab. Immunol. 81070-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Twenhafel, N. A., E. Leffel, and M. L. Pitt. 2007. Pathology of inhalational anthrax infection in the African green monkey. Vet. Pathol. 44716-721. [DOI] [PubMed] [Google Scholar]
- 52.Uchida, I., T. Sekizaki, K. Hashimoto, and N. Terakado. 1985. Association of the encapsulation of Bacillus anthracis with a 60 megadalton plasmid. J. Gen. Microbiol. 131363-367. [DOI] [PubMed] [Google Scholar]
- 53.Vasconcelos, D., R. Barnewall, M. Babin, R. Hunt, J. Estep, C. Nielsen, R. Carnes, and J. Carney. 2003. Pathology of inhalation anthrax in cynomolgus monkeys (Macaca fascicularis). Lab. Investig. 831201-1209. [DOI] [PubMed] [Google Scholar]