Abstract
Actinobacillus pleuropneumoniae is the etiological agent of porcine pleuropneumonia, a highly contagious respiratory infection in pigs. AasP, a putative subtilisin-like serine protease autotransporter, has recently been identified in A. pleuropneumoniae. We hypothesized that, similarly to other autotransporters of this type, AasP may undergo autocatalytic cleavage resulting in release of the passenger domain of the protein. Furthermore, AasP may be responsible for cleavage of other A. pleuropneumoniae outer membrane proteins. To address these hypotheses, the aasP gene was cloned and the expressed recombinant AasP protein used to raise monospecific rabbit antiserum. Immunoblot analysis of whole-cell lysates and secreted proteins demonstrated that AasP does not undergo proteolytic cleavage. Immunoblot analysis also confirmed that AasP is universally expressed by A. pleuropneumoniae. Confirmation of the maturation protease function of AasP was obtained through phenotypic analysis of an A. pleuropneumoniae aasP deletion mutant and by functional complementation. Comparison of the secreted proteins of the wild type, an aasP mutant derivative, and an aasP mutant complemented in trans led to the identification of OmlA protein fragments that were present only in the secreted-protein preparations of the wild-type and complemented strains, indicating that AasP is involved in modification of OmlA. This is the first demonstration of a function for any autotransporter protein in Actinobacillus pleuropneumoniae.
The gram-negative bacterium Actinobacillus pleuropneumoniae is the cause of porcine pleuropneumonia, a fatal respiratory disease that contributes to major economic losses in the swine industry (15). The organism is transmitted via respiratory droplets or through direct contact and may result in rapid death or severe pathology characterized by hemorrhagic and necrotic lesions in the lung (8, 26). Swine with partially protective immunity may develop chronic infections resulting in reduced weight gain, poor body condition, and a failure to thrive. These animals can act as subclinical carriers, transmitting the infection to previously unexposed swine (35). A. pleuropneumoniae virulence is thought to be dependent on a variety of putative virulence determinants, including lipopolysaccharide (5, 32), capsule (4), iron binding proteins (2, 33), Apx toxins (17), type 4 fimbriae (7, 40), and flagella (30). Furthermore, proteins involved with anaerobic metabolic activity appear to be required for persistence on porcine lung epithelia (3, 10, 24).
Autotransporters are a large superfamily of outer membrane/secreted proteins of gram-negative bacteria (19). They possess unique structural properties that facilitate their transport across the gram-negative cell envelope to the cell surface. They consist of an amino-terminal signal peptide required for Sec-dependent transport across the inner membrane, a functional N-terminal “passenger domain,” and a C-terminal “β domain,” which facilitates the transfer of the passenger domain to the external surface of the outer membrane. The β domain can either serve to anchor the passenger domain to the surface of the cell or be enzymatically cleaved from the rest of the protein, allowing release of the latter into the extracellular environment (19). Autotransporter proteins have often been associated with virulence functions, including protease activity, toxicity, adhesion, invasion, intracellular motility, and iron scavenging (19).
Recently, Baltes and colleagues described a 104-kDa outer membrane protein, designated AasP, in a serotype 7 strain of A. pleuropneumoniae (1). AasP was identified using a selective-capture-of-transcribed-sequences technique with A. pleuropneumoniae-infected porcine lung tissue (1). Uniquely for an autotransporter protein, under anaerobic conditions AasP expression was found to be regulated by a global anaerobic regulator, HlyX (1). Furthermore, Deslandes et al. used microarray analysis to show that expression of aasP in a serotype 1 strain of A. pleuropneumoniae was upregulated during growth under iron-restricted conditions (14).
Given the strong association of bacterial autotransporter proteins with pathogenesis, we had previously searched the predicted coding sequences of the serotype 1 strain A. pleuropneumoniae 4074 for homologues of well-characterized autotransporters of other gram-negative pathogens and independently identified AasP. The predicted 4074 AasP amino acid sequence is 100% identical to the serotype 7 AasP sequence reported by Baltes et al. (1) and is 58% identical to the sequence for the Ssa1 surface-localized autotransporter of Mannheimia haemolytica (28), 24% identical to that for SphB1 of Bordetella pertussis (13), and 32% identical to that for AspA/NalP from Neisseria meningitidis (36). Like these proteins, AasP is predicted to contain a subtilase proteolytic domain (Gly349 to Gly359), suggesting that AasP may be autocatalytic and, furthermore, may serve as a maturation protease cleaving other A. pleuropneumoniae outer membrane proteins at the cell surface. Here, we report the expression and purification of recombinant AasP and the creation of an A. pleuropneumoniae 4074ΔaasP mutant. We demonstrate that AasP is not autocatalytic but is a maturation protease responsible for the release of fragments of outer membrane lipoprotein A (OmlA) from the cell surface.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli TOP10F′ and BL21(DE3)pLysS (Table 1) were used for the expression of six-histidine-tagged recombinant AasP, encoded by plasmid pRC7 (Table 1). E. coli JM109 was used as a host strain for the construction of the mutagenic and complementation plasmids, pRC6 and pNJO85, respectively. E. coli S17 λ pir was used as the donor strain for conjugative transfer of pNJO85 to A. pleuropneumoniae. E. coli strains were grown in Luria-Bertani (LB) broth supplemented with ampicillin (100 μg ml−1), kanamycin (100 μg ml−1), chloramphenicol (25 μg ml−1), and isopropyl β-d-1-thiogalactopyranoside (IPTG; 1 mM) where appropriate. A. pleuropneumoniae strains (Table 1) were grown at 37°C, with air plus 5% CO2, in brain heart infusion broth supplemented with NAD (10 μg ml−1; Sigma), kanamycin (100 μg ml−1), and chloramphenicol (2 μg ml−1) where appropriate.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| TOP10F′ | F′lacIqTn10(TetR) mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| BL21(DE3)pLysS | F−ompT hsdSB (rB− mB−) gal dcm (DE3) pLysS (CamR) | Invitrogen |
| JM109 | endA1 recA1 gyrA96 thi hsdR17 (rK− rK−) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB laqIqZΔM15] | Promega |
| S17 λ pir | [recA thi pro hsdR hsdM+ RP4::2-Tc::Mu::Km Tn7 lysogenized with λ pir phage] | 34 |
| A. pleuropneumoniae | ||
| 4074 | Serotype 1 | 29 |
| S1536 | Serotype 2 | 29 |
| S1421 | Serotype 3 | 29 |
| M62 | Serotype 4 | 29 |
| K17 | Serotype 5a | 29 |
| L20 | Serotype 5b | 29 |
| FEM⊘ | Serotype 6 | 29 |
| WF83 | Serotype 7 | 29 |
| 405 | Serotype 8 | 29 |
| 13261 | Serotype 9 | 29 |
| 13039 | Serotype 10 | 29 |
| 56153 | Serotype 11 | 29 |
| 8329 | Serotype 12 | 29 |
| N273 | Serotype 13 | 31 |
| 3606 | Serotype 14 | 31 |
| HS143 | Serotype 15 | 6 |
| 4074ΔaasP | aasP deletion and replacement with kanamycin cassette | This study |
| 4074ΔaasP (pNJO85) | 4074 aasP deletion mutant containing pNJO85 | This study |
| Plasmids | ||
| pCRT7/NT-TOPO | Cloning vector | Invitrogen |
| pRC7 | 4074 aasP gene cloned in pCRT7/NT-TOPO | This study |
| pUC4 | Cloning vector | Pharmacia |
| pRC3 | 4.96-kb fragment spanning the 4074 aasP region cloned in pUC4 | This study |
| pJMK30 | Source of kanamycin resistance cassette | 37 |
| pRC6 | pRC3 containing the kanamycin resistance cassette in the same orientation as the deleted aasP gene | This study |
| pJFF224-NX | A. pleuropneumoniae expression vector | 16 |
| pNJO85 | 4074 aasP gene cloned in pJFF224-NX | This study |
DNA manipulation.
Genomic DNA was extracted from A. pleuropneumoniae by using a DNeasy tissue kit (Qiagen). Plasmid DNA was prepared by using a QIAprep Spin kit (Qiagen). Restriction enzymes and T4 DNA ligase were purchased from Roche. All enzymatic reactions were carried out according to the manufacturer's instructions. DNA sequencing was carried out at the School of Biomedical Sciences (University of Nottingham) with an ABI 377 automated DNA sequencer.
Preparation of recombinant AasP.
The aasP gene was amplified from A. pleuropneumoniae 4074 by using oligonucleotide primers AASPF1 and AASPR1 (Table 2) with the Expand high-fidelity PCR system (Roche). The resulting amplicon was ligated into pCRT7/NT-TOPO (Invitrogen) according to the manufacturer's protocol. The resulting plasmid, pRC7, was used to transform E. coli BL21(DE3)pLysS; transformants were grown to log phase, induced with 1 mM IPTG for 3 h, and harvested by centrifugation. Recombinant six-histidine-tagged proteins were affinity purified under denaturing conditions following cell lysis in lysis buffer (50 mM potassium phosphate, pH 7.8, 400 mM NaCl, 100 mM KCl, 10% glycerol, 0.5% Triton X-100, 10 mM imidazole) with nickel-nitrilotriacetic acid spin columns (Qiagen), and eluted fractions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Recombinant AasP was excised from the gel and transferred to mini-D-tube dialyzers (Novagen), and the protein was electro-eluted according to the recommendations of the manufacturer. Proteins were then concentrated using YM-30 centrifugal filter units (Microcon).
TABLE 2.
Primers used in this study
| Primer function and name | DNA sequencea | Restriction site |
|---|---|---|
| Expression | ||
| AASPF1 | GGATCCTTGGAATCGCCTCATTCAGGC | BamHI |
| AASPR1 | GGATCCATTAGTTAGAAAGTTAAGCCG | BamHI |
| Mutagenesis | ||
| AASPF2 | CGCGGATCCCTTACCGCAATTCTACGAGC | BamHI |
| AASPR2 | CGCGGATCCCCGATTAATTGGCAATTACG | BamHI |
| AASPF3 | CGCGTCGACCGAATGTTGATTATGATCGC | SalI |
| AASPR3 | CGCGTCGACACAAGCGGTAGAGTCTGCGTAGGCTACGC | SalI |
| Complementation | ||
| AASPF4 | CGCGGATCCATTGCAAATCTGACCGCTTGG | BamHI |
| AASPR4 | CGCGGATCCATTAGTTAGAAAGTTAAGCCG | BamHI |
All primers were designed from the A. pleuropneumoniae 4074 genome sequence. Underlined sequences identify restriction enzyme sites; boldface indicates an A. pleuropneumoniae DNA uptake sequence.
Production of a rabbit antiserum against purified recombinant AasP.
New Zealand White female rabbits were immunized subcutaneously four times at 2-week intervals with 30 μg of recombinant AasP protein emulsified in Freund's complete (first immunization only) or incomplete adjuvant. After three injections, the animals were test bled, boosted once more, and sacrificed 10 days later.
SDS-PAGE and immunoblotting.
Proteins were electrophoretically separated using 10% polyacrylamide minigels (Mini-Protean III; Bio-Rad) and were stained using SimplyBlue Safestain (Invitrogen) or transferred to nitrocellulose membranes as previously described (27). Membranes were probed with mouse anti-pentahistidine antibody (Qiagen) or rabbit primary antibody diluted 1:1,000 in blocking buffer (5% [wt/vol] nonfat dry milk, 0.1% [vol/vol] Tween 20 in 1× phosphate-buffered saline [PBS]) and incubated for 2 h. After being washed three times in 1× PBS with 0.1% Tween 20 (PBST), membranes were incubated for 2 h with a goat anti-mouse (or anti-rabbit) immunoglobulin G-alkaline phosphatase conjugate (Sigma) at a dilution of 1:2,000 in blocking solution. After being washed with PBST, the blots were developed using BCIP (5-bromo-4-chloro-3-indolylphosphate)-Nitro Blue Tetrazolium liquid substrate (Sigma).
Construction of 4074ΔaasP.
A 4.96-kb fragment encompassing the aasP gene together with 1 kb of upstream and downstream sequence was amplified by PCR using primers AASPF2 and AASPR2 (Table 2). The amplified product was digested with BamHI and cloned into BamHI-digested pUC4 to yield pRC3 (Table 1). pRC3 was then subjected to inverse PCR using primers AASPF3 and AASPR3 (Table 2). These primers anneal at sites at the 3′ and 5′ ends of the aasP gene, respectively, and their use resulted in the amplification of a 5-kb amplicon in which the aasP gene was deleted. Use of these primers also led to the introduction into the amplicon of a unique SalI site and a 9-bp sequence (ACAAGCGGT), the latter of which was required for uptake of the final mutagenic construct into A. pleuropneumoniae. The SalI site was used to introduce the kanamycin resistance cassette from pJMK30 in place of the deleted aasP gene. One of the resulting plasmids, pRC6, containing the resistance cassette in the same orientation as the deleted aasP gene, was confirmed by restriction digestion and sequencing before being linearized and used to mutate 4074 by natural transformation and allelic exchange as described previously (9). The deletion in the resulting mutant (4074ΔaasP) was confirmed by PCR analysis and sequencing. Growth curve assays carried out using liquid cultures showed no significant differences between 4074ΔaasP and the wild-type strain (data not shown).
Complementation of AasP.
The 3-kb aasP gene was amplified from A. pleuropneumoniae 4074 by PCR using primers AASPF4 and AASPR4 (Table 2). The amplified product was digested with BamHI and cloned into BamHI-digested pJFF224-NX. The resulting plasmid (pNJO85) was confirmed by PCR analysis and sequencing before being mobilized into A. pleuropneumoniae 4074ΔaasP by conjugation according to a previously described method (34).
Preparation of secreted proteins.
A. pleuropneumoniae cells were grown to exponential phase in broth culture and harvested by centrifugation (15,000 × g for 10 min), and supernatants were filtered (0.2-μm Minisart syringe filter; Sartorius) and concentrated 1,000-fold (Vivaspin 2 protein concentrator with 30,000-Da molecular mass cutoff; Vivascience).
MS analysis of protein bands.
Protein bands excised from stained gels were destained and digested by trypsin as previously described (39). Peptide mixtures were subjected to peptide sequence analysis by nanoelectrospray ionization-quadruple time of flight (Q-TOF) tandem mass spectrometry (MS-MS) (Waters/Micromass), and the sequences obtained were used to search for matches by using the BLAST algorithm at http://www.expasy.org/tools/blast/.
Protein and nucleic acid sequence analysis.
Public databases containing previously published protein and DNA sequences were searched using the BLAST and PSI-BLAST programs available at http://www.ncbi.nlm.nih.gov/blast/Blast.cgi. The genome database of A. pleuropneumoniae 4074 was interrogated at http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi.
RESULTS
Cloning, expression, and purification of recombinant AasP.
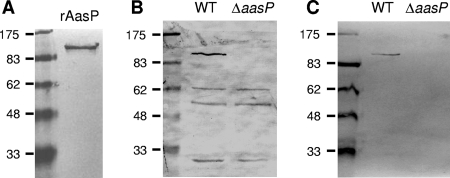
To examine the potential autocleavage of AasP, rabbit AasP-specific polyclonal antiserum (RαAasP) was generated. To facilitate this, the gene from A. pleuropneumoniae 4074 (corresponding to amino acids 30 to 932) was amplified and cloned into the expression vector pCRT7/NT-TOPO in order to express N-terminally six-histidine-tagged recombinant AasP protein. After induction of E. coli cells harboring the AasP expression plasmid, a recombinant protein of ca.100 kDa (consistent with the predicted molecular mass of AasP) was strongly expressed. Immunoblots probed with anti-pentahistidine antibodies confirmed the identity of this band as the recombinant protein through detection of the histidine tag (data not shown). The fusion protein was then affinity purified using Ni-nitrilotriacetic acid columns and further purified by electroelution from gels and used to generate RαAasP. Immunoblot analysis confirmed that RαAasP reacted to the purified recombinant AasP protein (Fig. 1A).
FIG. 1.
Immunoblot analysis confirms that RαAasP recognizes recombinant AasP (rAasP) (A) and the corresponding ca. 104-kDa AasP protein in whole-cell lysates (B) and secreted proteins (C) of wild-type (WT) A. pleuropneumoniae 4074 but not in the ΔaasP mutant derivative. No additional AasP-specific bands are apparent, suggesting that intact AasP is the active form of the protein.
Construction of an A. pleuropneumoniae aasP null-mutant strain.
To examine the role of AasP as a maturation protease, an aasP knockout derivative of A. pleuropneumoniae was generated. To achieve this, the aasP gene plus flanking DNA was amplified and cloned, and inverse PCR was employed to remove the aasP open reading frame and to introduce a 9-bp uptake sequence. The product was then ligated to a kanamycin resistance marker, and the resulting plasmid was linearized and used to transform A. pleuropneumoniae 4074. With this strategy, the aasP gene was successfully mutated to yield 4074ΔaasP. The genotype of this mutant was confirmed by PCR and sequencing (data not shown).
AasP is not proteolytically cleaved during export.
Many autotransporters, for example, N. meningitidis AspA/NalP, exhibit autoproteolytic activity (36). Such cell surface processing often modulates the functions of autotransporter proteins and may result in the release of the functional passenger domain of the protein. In addition to the strongly reactive AasP band, immunoblot analysis of A. pleuropneumoniae whole-cell lysates showed three cross-reactive bands at ca. 62, 55, and 30 kDa (Fig. 1B). However, these bands were also present in preparations of the ΔaasP mutant, demonstrating that these proteins were not AasP cleavage products. Similarly, no AasP cleavage products could be detected using similar immunoblot analysis of secreted-protein preparations (Fig. 1C). Taken together, these results demonstrate that AasP is not proteolytically cleaved in this strain under the conditions used and suggests that the intact form of the protein is the active form of AasP.
AasP acts as a surface maturation protease.
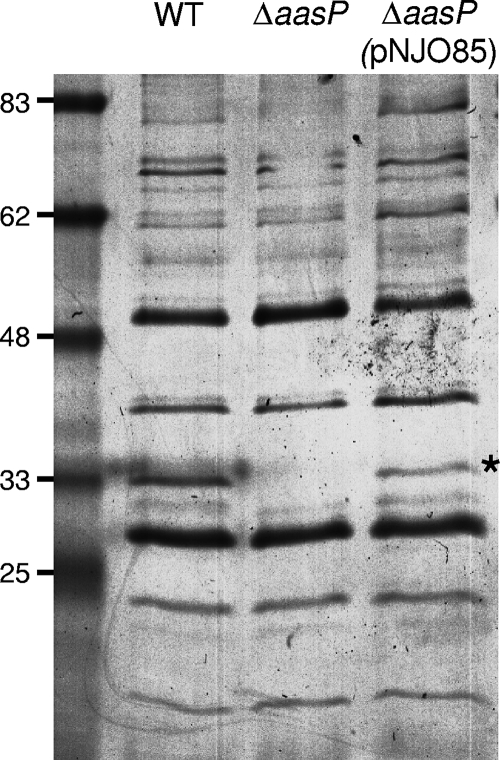
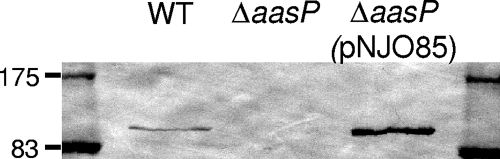
AasP is predicted to contain a proteolytic domain in the Gly349-to-Gly359 region, suggesting that AasP may act as a maturation protease cleaving other A. pleuropneumoniae proteins at the cell surface to facilitate their release, in a mature, functional form, into the extracellular environment. To address this question, secreted proteins from the 4074 wild type and its ΔaasP derivative were purified, concentrated, and separated by SDS-PAGE. Coomassie staining revealed a ca. 33-kDa band in secreted-protein preparations of the wild type which was absent from similar preparations of the mutant (Fig. 2, lanes 1 and 2). No other differences between the samples were apparent with this method. To confirm that this phenotype was not the result of an unwanted secondary mutation or a polar effect introduced as a result of the aasP mutation, a wild-type copy of the gene was introduced in trans into 4074ΔaasP by using the pJFF224-NX-based plasmid pNJO85. Introduction of pNJO85 led to high levels of AasP expression (Fig. 3) and to the production of the 33-kDa protein in the secreted protein fraction of this strain (Fig. 2, lane 3).
FIG. 2.
SDS-PAGE analysis of secreted proteins from the A. pleuropneumoniae 4074 wild type (WT), the ΔaasP mutant derivative, and a mutant harboring the complementation plasmid pNJO85 reveals the presence of a ca. 33-kDa protein (marked with a star) that is absent in the ΔaasP mutant preparation.
FIG. 3.
Immunoblot analysis of whole-cell lysates confirms the high-level expression of AasP in ΔaasP(pNJO85). Equivalent amounts of protein were loaded in each well. WT, wild type.
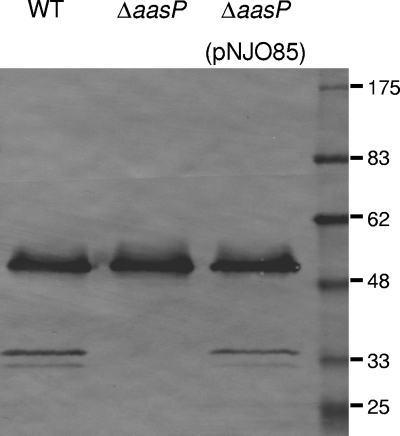
The ca. 33-kDa band was excised from the stained gel, analyzed by MS, and identified as a fragment of outer membrane lipoprotein A (OmlA). This identification was subsequently confirmed by immunoblot analysis, in which secreted protein fractions were probed with rabbit anti-OmlA, and a ca. 33-kDa reactive band was detected in the secreted-protein preparations of the wild-type and complemented strains but not in the 4074ΔaasP preparation (Fig. 4). The additional, ca. 50-kDa reactive band observed in all preparations represents full-length OmlA, which, despite having a predicted molecular mass of 40-kDa, migrates aberrantly on SDS-PAGE gels (18). A second, weakly reactive band at ca. 30 kDa was also detected in the secreted-protein preparations of the wild-type and complemented strains but not in the 4074ΔaasP preparation (Fig. 4) and may represent an additional OmlA cleavage product which was not apparent in Coomassie-stained preparations.
FIG. 4.
Immunoblot detection of OmlA fragments in the secreted protein fractions of the A. pleuropneumoniae 4074 wild type (WT), the ΔaasP mutant derivative, and ΔaasP(pNJO85).
Mapping of the trypsin-derived OmlA peptides identified by Q-TOF MS revealed that all fragments detected from the ca. 33-kDa reactive band corresponded to the central and C-terminal parts of the OmlA protein (Fig. 5). The ca. 30-kDa band was also excised and analyzed by MS; however, the small amount of protein present prevented identification or mapping of the trypsin-derived peptides. Overall, this suggests that while the majority of OmlA remains intact and anchored in the outer membrane via an N-terminal lipid domain, a proportion of OmlA molecules are proteolytically processed, and at least one fragment corresponding to the central and C-terminal parts of OmlA is released into the extracellular environment.
FIG. 5.
Amino acid sequence of the A. pleuropneumoniae 4074 OmlA protein (GenBank accession number AAD00608). Underlined regions correspond to peptides identified by nanoelectrospray ionization-Q-TOF MS-MS from the ca. 33-kDa OmlA fragment found in the secreted proteins of the A. pleuropneumoniae 4074 wild-type and complemented strains but not the ΔaasP mutant derivative.
AasP is naturally expressed in A. pleuropneumoniae.
Due to their accessible outer membrane location and roles in virulence, autotransporters have been suggested as possible candidate vaccines against several infections caused by gram-negative bacteria (38). One important prerequisite for a vaccine candidate is that a large proportion of target strains should possess and express the relevant antigen. Baltes et al. reported that the aasP gene could be detected in all A. pleuropneumoniae reference strains (1). We confirmed these results independently using PCR (data not shown) and extended this analysis to demonstrate that the genes are expressed in vitro in the same panel of reference strains by using immunoblot analysis of whole-cell extracts. The ca. 104-kDa AasP protein was detected in all strains tested, with the exception of 4074ΔaasP, demonstrating that the gene is expressed in diverse lineages and serotypes of A. pleuropneumoniae (Fig. 1B and 6).
FIG. 6.
AasP is expressed across divergent lineages and serogroups of A. pleuropneumoniae. Immunoblot analysis using RαAasP confirmed that AasP is detected in whole-cell lysates of all strains tested. Lane numbers correspond to serogroups; the strains tested were A. pleuropneumoniae S1536 (serogroup 2), S1421 (serogroup 3), M62 (serogroup 4), K17 (serogroup 5a), L20 (serogroup 5b), FEMØ (serogroup 6), WF83 (serogroup 7), 405 (serogroup 8), 13261 (serogroup 9), 13039 (serogroup 10), 56153 (serogroup 11), 8329 (serogroup 12), N273 (serogroup 13), 3606 (serogroup 14), and HS143 (serogroup 15).
DISCUSSION
There is currently a need for a better understanding of the pathogenic mechanisms underlying infection and persistence of A. pleuropneumoniae in the porcine host. One class of bacterial proteins with a likely role in these processes is the autotransporter proteins. AasP is the only subtilisin-like autotransporter of A. pleuropneumoniae reported to date (1). AasP was found to be expressed in porcine lung tissue during chronic infection with the serotype 7 strain AP76 (1). Under anaerobic conditions, AasP expression requires the global anaerobic regulator HlyX (1), and a ca. 100-kDa protein, presumed to be AasP, was visualized in silver-stained outer membrane preparations of A. pleuropneumoniae AP76 (1).
We independently identified AasP in a serotype 1 strain due to its sequence similarity to well-characterized autotransporter proteins with proteolytic activities. On the basis of this, we hypothesized that AasP may undergo autocleavage resulting in release of the passenger domain. However, our results confirm that AasP remains intact as a 104-kDa outer membrane protein. The small amount of AasP detected in the secreted-protein preparation of wild-type A. pleuropneumoniae (Fig. 1C) is likely to represent AasP found in outer membrane blebs which have been copurified with secreted proteins.
AasP shares significant identity with subtilisin-like serine protease autotransporters, including AspA/NalP of N. meningitidis (36) and SphB1 of B. pertussis (13), and, like them, acts as a surface maturation protease. SphB1 is involved in the maturation of the preprotein FhaB at the cell surface, producing filamentous hemagglutinin (12). Secretion of FhaB across the outer membrane occurs through an outer membrane channel-forming protein, designated FhaC (21). While a prediction of AasP protease activity can be hypothesized by analogy with other proteins, we are the first to confirm this experimentally. Confirmation was achieved by analyzing the secreted proteins of the wild-type and ΔaasP derivative strains of A. pleuropneumoniae and showing differences in protein content. Significantly, this phenotype was complemented by reintroduction of a wild-type copy of aasP in trans by use of a pJFF224-NX-based shuttle vector. Higher levels of expression of AasP in the complemented strain were presumably due to gene dosage effects, as the plasmid vector has a copy number of approximately 10 to 12 copies per cell. Unexpectedly, ca. 33-kDa, and possibly ca. 30-kDa, fragments of OmlA were found to be released into the extracellular milieu by wild-type A. pleuropneumoniae and the complemented strain but not by the aasP mutant derivative. To our knowledge, this is the first report of OmlA processing and release by A. pleuropneumoniae. Interestingly, the amount of fragmented OmlA present in the secreted protein fraction of the complemented strain was similar to that in the wild type (Fig. 4), even though expression levels of AasP were higher in the complemented strain (Fig. 3). This would suggest that the amount of OmlA processed is not dependent on the concentration of AasP but may be dependent on the concentration of OmlA, the proportion of OmlA in a protease-accessible conformation, or the concentration of any unidentified cofactor(s). OmlA is a 40-kDa outer membrane protein found to be expressed independently of the availability of iron (18) but is upregulated in the presence of bronchoalveolar lavage fluid (25). OmlA is expressed by isolates of all A. pleuropneumoniae serotypes, but sequence variation between alleles results in three antigenically distinct forms. OmlA1 is present in serotypes 1, 2, 8, 9, 11, and 12 (18); OmlA5 in serotypes 5a, 5b, and 10 (11, 23); and OmlA7 in serotypes 3, 4, 6, and 7 (22). The anti-OmlA antiserum used in this study was raised against OmlA1 and does not recognize the OmlA5 and OmlA7 variants in immunoblots (18), preventing us from determining conclusively whether AasP-mediated OmlA processing occurs in strains expressing these variants. Both OmlA1 and OmlA5 can individually protect pigs from death following challenge with the homologous serotype (11, 18), and both are included in the subunit vaccine Pleurostar APP (Novartis). Although OmlA1 has a predicted molecular mass of 40-kDa, it migrates at approximately 50 kDa on SDS-PAGE gels. This difference has been suggested to be a result of the high number of proline residues in the protein (18). The aberrant mobility makes an accurate prediction of the site of AasP-mediated cleavage difficult, although our results suggest that one cleavage site is before amino acid Asp133 since this is the first amino acid contained within the ca. 33-kDa OmlA peptides identified by Q-TOF MS. N-terminal sequencing would confirm the site of cleavage. The function of OmlA in A. pleuropneumoniae is not known, and thus, the significance of AasP-mediated cleavage is not clear.
While OmlA was the only AasP-processed protein identified under the experimental conditions used in this study, given the identity between AasP and SphB1, it would be interesting to determine whether A. pleuropneumoniae expresses FhaB- and FhaC-like proteins and whether the former is a substrate for AasP-mediated proteolysis. Genes encoding FhaB- and FhaC-like proteins are present in the A. pleuropneumoniae 4074 genome sequence (GenBank accession no. ZP_00204494 and ZP_00135051, respectively), although the FhaC-encoding gene contains frameshift mutations and may not be functional. It should be noted, however, that although AasP and SphB1 share sequence similarity, significant differences are apparent. For example, in contrast to SphB1 (and AspA/NalP), AasP is not predicted to be a lipoprotein. These differences may explain why AasP appears to cleave a different subset of protein substrates. To confirm the proteolytic activity of AasP and to identify additional substrates, we have attempted to use recombinant AasP protein in in vitro proteolysis assays. However, recombinant AasP has no detectable proteolytic activity; presumably, this has been abolished by the denaturing conditions used to purify the protein.
Due to their accessible outer membrane location and roles in virulence, autotransporters have been suggested as possible candidate vaccines against several infections caused by gram-negative bacteria (38). Indeed, some commercially available vaccines already contain autotransporters; for example, pertactin is included in some acellular vaccines against whooping cough, caused by B. pertussis (20). Given our observation that AasP is universally expressed by A. pleuropneumoniae, this suggests that AasP is worthy of future study as a possible candidate vaccine against this devastating porcine pathogen.
In conclusion, we have functionally characterized the AasP autotransporter protein, which is universally expressed by A. pleuropneumoniae strains in vitro and is responsible for cleavage and release of fragments of OmlA from the cell surface. This is the first demonstration of a function for any autotransporter protein in Actinobacillus pleuropneumoniae.
Acknowledgments
This work was partly supported by the Biotechnology and Biological Sciences Research Council.
We acknowledge the practical help of Rusheng Chew and Ahmad Kamal Azri bin Abi Musa Asa'ari. We thank G. Gerlach for the gift of rabbit polyclonal anti-OmlA1 antisera, G. Frey for the gift of pJFF224-NX, and K. Bailey for useful discussions.
Editor: A. Camilli
Footnotes
Published ahead of print on 13 October 2008.
REFERENCES
- 1.Baltes, N., F. F. R. Buettner, and G. F. Gerlach. 2007. Selective capture of transcribed sequences (SCOTS) of Actinobacillus pleuropneumoniae in the chronic stage of disease reveals an HlyX-regulated autotransporter protein. Vet. Microbiol. 123110-121. [DOI] [PubMed] [Google Scholar]
- 2.Baltes, N., I. Hennig-Pauka, and G. F. Gerlach. 2002. Both transferrin binding proteins are virulence factors in Actinobacillus pleuropneumoniae serotype 7 infection. FEMS Microbiol. Lett. 209283-287. [DOI] [PubMed] [Google Scholar]
- 3.Baltes, N., M. N′diaye, I. D. Jacobsen, A. Maas, F. F. R. Buettner, and G. F. Gerlach. 2005. Deletion of the anaerobic regulator HlyX causes reduced colonization and persistence of Actinobacillus pleuropneumoniae in the porcine respiratory tract. Infect. Immun. 734614-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandara, A. B., M. L. Lawrence, H. P. Veit, and T. J. Inzana. 2003. Association of Actinobacillus pleuropneumoniae capsular polysaccharide with virulence in pigs. Infect. Immun. 713320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bélanger, M., D. Dubreuil, J. Harel, C. Girard, and M. Jacques. 1990. Role of lipopolysaccharides in adherence of Actinobacillus pleuropneumoniae to porcine tracheal rings. Infect. Immun. 583523-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackall, P. J., H. L. Klaasen, H. van den Bosch, P. Kuhnert, and J. Frey. 2002. Proposal of a new serovar of Actinobacillus pleuropneumoniae: serovar 15. Vet. Microbiol. 8447-52. [DOI] [PubMed] [Google Scholar]
- 7.Boekema, B. K. H. L., J. P. M. van Putten, N. Stockhofe-Zurwieden, and H. E. Smith. 2004. Host cell contact-induced transcription of the type IV fimbria gene cluster of Actinobacillus pleuropneumoniae. Infect. Immun. 72691-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bossé, J. T., H. Janson, B. J. Sheehan, A. J. Beddek, A. N. Rycroft, J. S. Kroll, and P. R. Langford. 2002. Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect. 4225-235. [DOI] [PubMed] [Google Scholar]
- 9.Bossé, J. T., J. H. E. Nash, S. J. Kroll, and P. R. Langford. 2004. Harnessing natural transformation in Actinobacillus pleuropneumoniae: a simple method for allelic replacements. FEMS Microbiol. Lett. 233277-281. [DOI] [PubMed] [Google Scholar]
- 10.Buettner, F. F. R., A. Maas, and G. F. Gerlach. 2008. An Actinobacillus pleuropneumoniae arcA deletion mutant is attenuated and deficient in biofilm formation. Vet. Microbiol. 127106-115. [DOI] [PubMed] [Google Scholar]
- 11.Bunka, S., C. Christensen, A. A. Potter, P. J. Willson, and G. F. Gerlach. 1995. Cloning and characterization of a protective outer membrane lipoprotein of Actinobacillus pleuropneumoniae serotype 5. Infect. Immun. 632797-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coutte, L., S. Alonso, N. Reveneau, E. Willery, B. Quatannens, C. Locht, and F. Jacob-Dubuisson. 2003. Role of adhesin release for mucosal colonization by a bacterial pathogen. J. Exp. Med. 197735-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coutte, L., R. Antoine, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2001. Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. EMBO J. 205040-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deslandes, V., J. Nash, J. Harel, J. Coulton, and M. Jacques. 2007. Transcriptional profiling of Actinobacillus pleuropneumoniae under iron-restricted conditions. BMC Genomics 872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenwick, B., and S. Henry. 1994. Porcine pleuropneumonia. J. Am. Vet. Med. Assoc. 2041334-1340. [PubMed] [Google Scholar]
- 16.Frey, J. 1992. Construction of a broad host range shuttle vector for gene cloning and expression in Actinobacillus pleuropneumoniae and other Pasteurellaceae. Res. Microbiol. 143263-269. [DOI] [PubMed] [Google Scholar]
- 17.Frey, J. 1995. Virulence in Actinobacillus pleuropneumoniae and RTX toxins. Trends Microbiol. 3257-261. [DOI] [PubMed] [Google Scholar]
- 18.Gerlach, G. F., C. Anderson, S. Klashinsky, A. Rossi-Campos, A. A. Potter, and P. J. Willson. 1993. Molecular characterization of a protective outer membrane lipoprotein (OmlA) from Actinobacillus pleuropneumoniae serotype 1. Infect. Immun. 61565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. A. A. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hijnen, M., Q. He, R. Schepp, P. Van Gageldonk, J. Mertsola, F. R. Mooi, and G. A. M. Berbers. 2008. Antibody responses to defined regions of the Bordetella pertussis virulence factor pertactin. Scand. J. Infect. Dis. 4094-104. [DOI] [PubMed] [Google Scholar]
- 21.Hodak, H., B. Clantin, E. Willery, V. Villeret, C. Locht, and F. Jacob-Dubuisson. 2006. Secretion signal of the filamentous haemagglutinin, a model two-partner secretion substrate. Mol. Microbiol. 61368-382. [DOI] [PubMed] [Google Scholar]
- 22.Ito, H., M. Osaki, I. Uchida, T. Ohya, and T. Sekizaki. 1998. Demonstration of the third antigenically distinct outer membrane lipoprotein (OmlA) in Actinobacillus pleuropneumoniae serotype 7. FEMS Microbiol. Lett. 167303-308. [DOI] [PubMed] [Google Scholar]
- 23.Ito, H., I. Uchida, T. Sekizaki, E. Ooishi, T. Kawai, T. Okabe, A. Taneno, and N. Terakado. 1995. Molecular cloning of an Actinobacillus pleuropneumoniae outer membrane lipoprotein (OmlA) from serotype 5a. Microb. Pathog. 1829-36. [PubMed] [Google Scholar]
- 24.Jacobsen, I., I. Hennig-Pauka, N. Baltes, M. Trost, and G. F. Gerlach. 2005. Enzymes involved in anaerobic respiration appear to play a role in Actinobacillus pleuropneumoniae virulence. Infect. Immun. 73226-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobsen, I. D., J. Meens, N. Baltes, and G. F. Gerlach. 2005. Differential expression of non-cytoplasmic Actinobacillus pleuropneumoniae proteins induced by addition of bronchoalveolar lavage fluid. Vet. Microbiol. 109245-256. [DOI] [PubMed] [Google Scholar]
- 26.Jobert, J. E., C. Savoye, R. Cariolet, M. Kobisch, and F. Madec. 2000. Experimental aerosol transmission of Actinobacillus pleuropneumoniae to pigs. Can. J. Vet. Res. 6421-26. [PMC free article] [PubMed] [Google Scholar]
- 27.Kizil, G., I. Todd, M. Atta, S. P. Borriello, K. Ait-Tahar, and D. A. A. Ala'Aldeen. 1999. Identification and characterization of TspA, a major CD4+ T-cell- and B-cell-stimulating Neisseria-specific antigen. Infect. Immun. 673533-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo, R. Y., C. A. Strathdee, P. E. Shewen, and B. J. Cooney. 1991. Molecular studies of Ssa1, a serotype-specific antigen of Pasteurella haemolytica A1. Infect. Immun. 593398-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacDonald, J., and A. N. Rycroft. 1992. Molecular cloning and expression of ptxA, the gene encoding the 120-kilodalton cytotoxin of Actinobacillus pleuropneumoniae. Infect. Immun. 602726-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Negrete-Abascal, E., M. E. Reyes, R. M. García, S. Vaca, J. A. Girón, O. García, E. Zenteno, and M. de La Garza. 2003. Flagella and motility in Actinobacillus pleuropneumoniae. J. Bacteriol. 185664-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen, R., L. O. Andresen, T. Plambeck, J. P. Nielsen, L. T. Krarup, and S. E. Jorsal. 1997. Serological characterization of Actinobacillus pleuropneumoniae biotype 2 strains isolated from pigs in two Danish herds. Vet. Microbiol. 5435-46. [DOI] [PubMed] [Google Scholar]
- 32.Ramjeet, M., V. Deslandes, F. St. Michael, A. D. Cox, M. Kobisch, M. Gottschalk, and M. Jacques. 2005. Truncation of the lipopolysaccharide outer core affects susceptibility to antimicrobial peptides and virulence of Actinobacillus pleuropneumoniae serotype 1. J. Biol. Chem. 28039104-39114. [DOI] [PubMed] [Google Scholar]
- 33.Shakarji, L., L. G. Mikael, R. Srikumar, M. Kobisch, J. W. Coulton, and M. Jacques. 2006. FhuA and HgbA, outer membrane proteins of Actinobacillus pleuropneumoniae: their role as virulence determinants. Can. J. Microbiol. 52391-396. [DOI] [PubMed] [Google Scholar]
- 34.Sheehan, B. J., J. T. Bossé, A. J. Beddek, A. N. Rycroft, J. S. Kroll, and P. R. Langford. 2003. Identification of Actinobacillus pleuropneumoniae genes important for survival during infection in its natural host. Infect. Immun. 713960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor, D. J. 1999. Actinobacillus pleuropneumoniae, 7th ed. Blackwell Science, Oxford, United Kingdom.
- 36.Turner, D. P., K. G. Wooldridge, and D. A. A. Ala'Aldeen. 2002. Autotransported serine protease A of Neisseria meningitidis: an immunogenic, surface-exposed outer membrane, and secreted protein. Infect. Immun. 704447-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Vliet, A. H., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 1805291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells, T. J., J. J. Tree, G. C. Ulett, and M. A. Schembri. 2007. Autotransporter proteins: novel targets at the bacterial cell surface. FEMS Microbiol. Lett. 274163-172. [DOI] [PubMed] [Google Scholar]
- 39.Wooldridge, K. G., M. Kizil, D. B. Wells, and D. A. A. Ala'Aldeen. 2005. Unusual genetic organization of a functional type I protein secretion system in Neisseria meningitidis. Infect. Immun. 735554-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Y., J. M. Tennent, A. Ingham, G. Beddome, C. Prideaux, and W. P. Michalski. 2000. Identification of type 4 fimbriae in Actinobacillus pleuropneumoniae. FEMS Microbiol. Lett. 18915-18. [DOI] [PubMed] [Google Scholar]