Abstract
θ-Defensins are macrocyclic antimicrobial peptides that were previously isolated from leukocytes of a single species, the rhesus macaque. We now report the characterization of baboon θ-defensins (BTDs) expressed in bone marrow and peripheral blood leukocytes. Four cDNAs encoding θ-defensin precursors were characterized, allowing for the prediction of 10 theoretical θ-defensins (BTD-1 to BTD-10) produced by binary, head-to-tail splicing of nonapeptides excised from paired precursors. Five of the predicted θ-defensins were purified from baboon leukocytes, and synthetic versions of each were prepared. Anti-θ-defensin antibody localized the peptides in circulating neutrophils and monocytes and in immature and mature myeloid elements in bone marrow. Each of the BTDs possessed antimicrobial activity against bacterial and fungal test organisms in vitro. Peptide activities varied markedly despite a high degree of sequence conservation among the θ-defensins tested. Thus, baboons express numerous θ-defensins which appear to differentially contribute to host defense against diverse pathogens.
Antimicrobial peptides (AMPs) are effectors of the innate immune system. AMPs are expressed in cells (epithelia, neutrophils, and macrophages) that come into contact with potentially invasive microorganisms (17). In mammals, the two major classes of AMPs are defensins and cathelicidins. Cathelicidins are characterized by a conserved cathelin prodomain which lies N terminal to highly variable mature peptides that are released by activating proteases (27). Defensins are small, cationic peptides that are composed of three structural subclasses, α-, β-, and θ-defensins, differentiated by the spacing and pairing of their six disulfide-bonded cysteines (7, 9, 18). θ-Defensins are further distinguished by their macrocyclic backbone and as such represent the only known cyclic protein motif expressed in animals (16).
The biosynthesis of θ-defensins requires head-to-tail splicing of two 9-amino-acid sequences derived from θ-defensin precursors (16). θ-Defensins were first identified in neutrophils and monocytes of the rhesus monkey (21). Subsequently, Nguyen et al. (15) conducted a phylogenetic survey that revealed the existence of θ-defensin genes in other Old World monkeys and two apes (the siamang and orangutan), but none in New World monkeys or prosimians. Humans, chimpanzees, bonobos, and gorillas express θ-defensin pseudogenes in which the precursor mRNA contains a mutation producing a stop codon in the signal sequence, thus preventing translation of the θ-defensin precursor (15).
Rhesus θ-defensin-1 (RTD-1) is produced from the heterodimeric splicing of two θ-defensin precursors, proRTD1a and proRTD1b. Homodimeric excision/ligation reactions involving proRTD1a and proRTD1b were revealed by the isolation of RTD-2 and RTD-3 (12, 23). RTD-1, -2, and -3 have potent microbicidal activities against bacteria and fungi (23) and have antiviral activities against human immunodeficiency virus type 1 (3, 24) and herpes simplex virus (26). A synthetic θ-defensin designed based on the sequence of a human θ-defensin pseudogene was shown to possess antibacterial and antiviral activities (3, 26). In addition, θ-defensins are reported to bind and inactivate lethal toxin from Bacillus anthracis (25).
θ-Defensins are microbicidal in the presence of physiological concentrations of salt, divalent cations, and serum (21, 22). In contrast, the antimicrobial activities of α- and β-defensins are markedly reduced in the presence of salt and divalent cations (2, 11, 19). Acyclic RTD-1 was inactive against Staphylococcus aureus in physiologic saline, whereas the natural cyclic form of the peptide retained potent killing activity under these conditions (21). These data indicate that the cyclic backbone structure of θ-defensins confers salt insensitivity.
Here we report on the characterization of four θ-defensin precursor mRNAs expressed in baboon bone marrow and peripheral blood and on the isolation, synthesis, and activities of five of the corresponding θ-defensin peptides.
MATERIALS AND METHODS
BTD cDNA analysis.
Total RNA was prepared from bone marrow samples from healthy olive baboons (Papio anubis) (Southwest Foundation for Biomedical Research, San Antonio, TX; University of Oklahoma Health Sciences Center, Oklahoma City, OK) using RNA STAT-60 reagent (Tel-Test, Friendswood, TX) following the manufacturer's protocol. Reverse transcription-PCR (RT-PCR) was carried out by using standard methods with primers designed from published RTD sequences. The forward primer used for all baboon θ-defensin (BTD) cDNAs was 5′-GACGGCTGCTCTTGCTACAGG-3′, the reverse primer for BTD-a was 5′-CAAACGGCAGAATCCTCGTGTGC-3′, the reverse primer for BTD-b and -d was 5′-GACCCCAAACGCCTTTATAACAGTTG-3′, and the reverse primer for BTD-c was 5′-CCGCAAACGCCGTTATAACAGAC-3′. PCR products of the predicted size were visualized as 327- to 340-base-pair (bp) bands on a 1.5% agarose gel, subcloned into the pCR-2.1 TOPO vector (Invitrogen, Carlsbad, CA), and sequenced (Laguna Scientific, Laguna Nigel, CA).
Purification of BTDs from baboon leukocytes.
EDTA- or heparin-anticoagulated blood samples were obtained from olive baboons (Southwest Foundation for Biomedical Research, San Antonio, TX; University of Oklahoma Health Sciences Center, Oklahoma City, OK). Leukocytes were enriched by sedimentation of erythrocytes with 6% dextran or by centrifugation for 20 min at 270 × g. Residual erythrocytes were removed by hypotonic lysis with ice-cold water, and the remaining leukocytes were suspended in HEPES-buffered saline to a final concentration of 0.9% NaCl. Leukocyte preparations contained 75 to 85% neutrophils, 10 to 20% lymphocytes, and 3 to 6% monocytes.
A leukocyte preparation containing 3.3 × 109 cells was extracted with 60 ml of ice-cold 30% acetic acid and stirred at 8°C for 18 h. The extract was clarified by centrifugation for 20 min at 14,500 × g at 4°C, and supernatants were concentrated by centrifugal evaporation, pooled, and lyophilized. The lyophilate was dissolved in 20 ml of 5% acetic acid, clarified for 10 min at 14,500 × g, and loaded in 5-ml batches on a Bio-Gel P-60 column (2.5 by 60 cm) equilibrated in 5% acetic acid. Ten-milliliter fractions were collected at 42 ml/h with elution monitored at 280 nm. Fractions were analyzed by matrix-assisted laser desorption ionization-time-of-flight mass spectroscopy (MALDI-TOF MS) using a Voyager DE STR mass spectrometer. Putative θ-defensins were identified in fractions containing masses that matched those predicted from binary pairing of nonapeptide sequences in θ-defensin cDNAs (see above). Further evidence for the presence of θ-defensins in chromatographic fractions was obtained by observing a 348.36-Da mass change when putative θ-defensins were reduced with 1,4-dithiothreitol (DTT) and alkylated with iodoacetamide (23). In addition, 200-μl aliquots of P-60 column fractions were analyzed by 12.5% acid-urea (12.5% AU)-polyacrylamide gel electrophoresis (PAGE), and the migration of stained bands was compared to those of RTD-1, -2, and -3 used as standards (23).
θ-Defensins from Bio-Gel P-60 fractions were purified to homogeneity by sequential chromatography by semipreparative reverse-phase high-performance liquid chromatography (RP-HPLC) on a C18 column (10 by 250 mm) using 0.25%/min water-acetonitrile gradients containing 0.1% phosphoric acid, followed by RP-HPLC using 0.1% trifluoroacetic acid with water-acetonitrile gradient elution. Purified peptides were characterized by AU-PAGE, MALDI-TOF MS, and amino acid analysis. For amino acid analysis, BTDs were hydrolyzed in 6 N HCl for 2 h at 150°C and derivatized using the AccQ-Fluor reagent kit (Waters, Milford, MA). Amino acids were chromatographed on an AccQ-Tag column (3.9 by 150 mm) and analyzed using Millennium software.
α-Chymotrypsin digestion and peptide sequencing.
Five micrograms of natural BTD-7 was reduced with DTT, alkylated with 4-vinylpyridine, and purified by RP-HPLC. Lyophilized peptide was dissolved to 100 μg/ml in 1% ammonium bicarbonate (pH 8.0), digested with 50 μg of α-chymotrypsin-agarose beads (Sigma-Aldrich, St. Louis, MO) for 1 h at 30°C, and then acidified with 5% acetic acid. Peptide fragments were purified by analytical C18 RP-HPLC using a 2.1- by 100-mm column, and N-terminal Edman sequencing of the fragments was performed (Bio-Synthesis, Inc., Lewisville, TX).
BTD synthesis.
BTD-1, -2, -3, -4, and -7 were assembled at 0.2-mmol scale by solid-phase synthesis with 9-fluorenylmethoxy carbonyl (Fmoc) chemistry and O-(7-azabenzotriazol-1-yl)-1,13,3-tetramethyluronium hexafluorophosphate (HATU) activation on a Milligen 9050 automated synthesizer as previously described (23). Peptides were cleaved from the resin by treatment with reagent K for 4 h with agitation at room temperature. Crude synthetic products were reduced with DTT in 6 M guanidine-HCl, 0.2 M Tris-HCl, and 2 mM EDTA (pH 8.2) for 4 h at 50°C and purified by preparative RP-HPLC on a DeltaPak C18 cartridge (25 × 100 mm) (Waters, Milford, MA) with water-acetonitrile gradients containing 0.1% trifluoroacetic acid. Fractions were analyzed by MALDI-TOF MS, and those containing reduced BTD were concentrated by centrifugal evaporation, pooled, and diluted to 0.1 to 0.2 mg/ml with 1% acetic acid. The pH of the solution was adjusted to 7.8 to 8.0 using ammonium hydroxide and stirred in an open container for 18 h to form disulfide bonds. The oxidized product was purified by C18 RP-HPLC, and the fully oxidized state of the acyclic product was confirmed by MALDI-TOF MS. Acyclic peptides were lyophilized and converted to their hydrochloride salts by dissolution in 60 mM hydrochloric acid, lyophilized, and relyophilized three times from deioninized water. Peptide cyclization was carried out at 0.8 to 1.5 mg/ml in dimethyl sulfoxide containing 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide (EDC·HCl) and 1-hydroxybenzotriazole (HOBt). The pH was adjusted to 7.8 using N,N-diisopropylethylamine (DIEA), and cyclization was performed under nitrogen with stirring in the dark for 18 h at room temperature. The extent of cyclization was determined by RP-HPLC followed by MALDI-TOF MS. Cyclic peptides were purified to homogeneity using a C18 column (22 by 250 mm) and characterized by analytical C18 RP-HPLC, AU-PAGE, MALDI-TOF MS, and amino acid analysis.
Antibody production.
Anti-RTD-1 antibody was produced by immunizing a goat with a mixture of polymerized cyclic RTD-1 and polymerized acyclic RTD-1. Antigen was prepared by separately reducing 2 mg of synthetic cyclic and acyclic RTD-1 with DTT in 1 ml of 6 M guanidine-HCl, 0.2 M Tris-HCl, and 2 mM EDTA (pH 8.2) at 50°C for 4 h. Reduced peptides were acidified with 5% acetic acid and purified by RP-HPLC, and peptide-containing fractions were concentrated to 0.1 ml by centrifugal rotoevaporation. The pH of the peptide solutions was adjusted to 8.0 with ammonium hydroxide and stirred at room temperature in air for 24 h to induce disulfide cross-linking, and then the mixture was acidified to 5% acetic acid and lyophilized. For both RTD-1 and acyclic RTD-1, the oxidized products were shown to be polymerized by MS and PAGE. A 1:1 mixture of the cyclic and acyclic RTD-1 polymers was used as the immunogen (Elmira Biologicals Inc., Iowa City, IA). Immunoglobulin G (IgG) was purified from goat antiserum by ammonium sulfate precipitation followed by chromatography on a DEAE Econo-Pac column (Bio-Rad, Hercules, CA).
Western blot analysis.
One microgram of each synthetic peptide was resolved on a 12.5% AU-polyacrylamide gel and electroblotted onto 0.22-μm nitrocellulose by semidry transfer as previously described (23). Peptides were fixed to the membrane by incubation of the blot in 0.2% glutaraldehyde in phosphate-buffered saline for 1 h followed by quenching with 1 M ammonium chloride for 15 min. The blot was washed with TTBS (100 mM Tris buffer [pH 7.5], 0.9% sodium chloride, 0.1% Tween 20) and incubated with a 1:100 dilution of goat anti-RTD-1 IgG in phosphate-buffered saline for 18 h at 8°C. The secondary antibody (1:400,000 horseradish peroxidase-labeled horse anti-goat IgG) was incubated for 30 min at room temperature. Immunopositive bands were visualized with SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
Immunohistochemistry.
Cytospins of baboon peripheral blood leukocytes and bone marrow cells were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) and incubated in Fc receptor blocking solution, avidin, biotin, and 1.5% normal rabbit serum. Slides were incubated with 1:4 diluted anti-RTD-1 IgG or goat preimmune IgG (1:4) for 2 h at room temperature. Secondary antibody was biotinylated rabbit anti-goat IgG (1:200 dilution; 1 h at room temperature). Immunostaining was visualized using the avidin-biotin-glucose oxidase system, and cells were counterstained with nuclear fast red (Vector Laboratories, Burlingame, CA).
Antimicrobial assays.
Synthetic BTDs were evaluated for antimicrobial activities against bacteria (Staphylococcus aureus 502a and Escherichia coli ML35) and a fungus (Candida albicans 16820) using radial diffusion and liquid suspension assays as previously described (23). For the former, 5 μl of peptide was introduced into wells of seeded agarose and allowed to diffuse for 2 h at 37°C. Plates were then overlaid with agarose containing Trypticase soy broth or Sabouraud dextrose broth. After incubation at 37°C for 18 to 24 h, peptide activity was determined by measuring the diameter of clearing around each well. Microbicidal suspension assays were performed in 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer or 10 mM Tris buffer (pH 7.4), each containing 5 mM glucose as described previously (23).
E. coli ML35 membrane permeabilization.
Synthetic BTDs were evaluated for their ability to permeabilize E. coli ML35 cells as described previously (22). Approximately 2 × 106 CFU of E. coli ML35 per ml was added to various concentrations of peptide in buffer, and the hydrolysis of o-nitrophenyl-β-d-galactopyranoside (ONPG) was monitored at 405 nm for 60 min in a 96-well Spectra-Max 190 plate spectrophotometer using SOFTmaxPRO 3.1 software (Molecular Devices, Sunnyvale, CA). Inhibition of β-galactosidase activity was analyzed by incubating each peptide with 10 nM purified enzyme in 10 mM PIPES (pH 7.4). Dose-dependent inhibition of β-galactosidase was used to calculate an adjusted rate (R′) of ONPG hydrolysis in whole bacteria as follows: R′ = R + (−log10 C) × m + Δb, where R is the measured rate of hydrolysis, C is the peptide concentration, m is the slope obtained from a log-linear plot of the dose-dependent β-galactosidase inhibition by each peptide, and Δb is the difference of the y-axis intercepts obtained from plots of ONPG hydrolysis rates by purified β-galactosidase in 10 mM PIPES buffer and identical mixtures containing 1 μg/ml of each peptide.
Nucleotide sequence accession numbers.
The cDNA sequences for the following BTDs have been submitted to GenBank and assigned accession numbers FJ030939 for BTD-a, FJ030940 for BTD-b, FJ030941 for BTD-c, and FJ030942 for BTD-d.
Amino acid sequence accession numbers.
The θ-defensin peptide sequence data were submitted to UniProt Knowledgebase under accession numbers P86031 for BTD-a, P86032 for BTD-b, P86033 for BTD-c, and P86034 for BTD-d.
RESULTS
BTD cDNAs.
Studies of defensin genetics in nonhuman primates identified several full-length genes that encode cyclic defensins in Old World monkeys (15, 21). In contrast to an earlier report indicating that baboons lack θ-defensin genes (15), our preliminary studies suggested that θ-defensin mRNAs are expressed in the bone marrow of olive baboons (Papio anubis) and gelada baboons. RT-PCR analysis of bone marrow RNA from a single olive baboon disclosed three θ-defensin mRNAs (BTD-a, -b, and -c), each having 93 to 97% identity to orthologous genes (RTD1a, -1b, and -1c) in the rhesus monkey. Analysis of bone marrow RNA from a second baboon disclosed expression of a fourth θ-defensin precursor, BTD-d.
Figure 1A shows the deduced amino acid sequences of the BTD precursors aligned with those of rhesus macaques and that encoded by a human θ-defensin pseudogene wherein the open reading frame is interrupted by a stop codon (at residue 17). The θ-defensin genes encode polypeptide precursors with highly conserved signal and prosegment sequences followed by C-terminal dodecapeptide sequences that are more variable. Each of the BTD cDNA sequences shown in Fig. 1 was also identified following RT-PCR of RNA from baboon peripheral blood leukocytes. The differential (homodimeric and heterodimeric) pairing of BTD-a to BTD-d-derived nonamers would theoretically produce 10 cyclic peptides, BTD-1 to BTD-10, shown schematically in Fig. 1B (sequences and characteristics of the 10 BTDs shown in Table 1).
FIG. 1.
Alignment of prepro-θ-defensins. (A) BTD-a to -d amino acid sequences predicted from cDNA are aligned manually with RTD1a to -c, and human θ-defensin pseudogene (HTDp). Dots in aligned sequences denote amino acids identical to those in BTD-a, the asterisk symbol denotes the position of termination codon, and the # symbol denotes a stop codon that prematurely terminates translation. (B) Cyclic structures of 10 deduced BTD peptides derived from cDNA sequences.
TABLE 1.
Characterization of baboon θ-defensin sequences
| BTD | Deduced peptide sequence [precursor cDNA(s)]a | Charge | Native peptide mass
|
S-CAM peptide massd
|
||
|---|---|---|---|---|---|---|
| Theor.b | Exptlc | Theor. | Exptl | |||
| 1 | RCVCTRGFCRCVCRRGVC(a+b) | +5 | 2,054.58 | 2,055.69 | 2,402.94 | 2,404.21 |
| 2 | RCVCRRGVCRCVCRRGVC(b+b) | +6 | 2,061.61 | 2,062.72 | 2,409.97 | 2,411.73 |
| 3 | RCVCTRGFCRCVCTRGFC(a+a) | +4 | 2,047.54 | 2,048.38 | 2,395.90 | 2,396.98 |
| 4 | RCVCTRGFCRCICLLGIC(a+c) | +3 | 1,996.57 | 1,996.57 | 2,344.93 | 2,344.45 |
| 5 | RCVCRRGVCRCICLLGIC(b+c) | +4 | 2,003.61 | ND | ND | ND |
| 6 | RCICLLGICRCICLLGIC(c+c) | +2 | 1,945.61 | ND | ND | ND |
| 7 | RCVCTRGFCRCFCRRGVC(a+d) | +5 | 2,102.62 | 2,103.70 | 2,450.98 | 2,452.29 |
| 8 | RCVCRRGVCRCFCRRGVC(b+d) | +6 | 2,109.66 | ND | ND | ND |
| 9 | RCICLLGICRCFCRRGVC(c+d) | +4 | 2,051.66 | ND | ND | ND |
| 10 | RCFCRRGVCRCFCRRGVC(d+d) | +6 | 2,157.70 | ND | ND | ND |
The letters in parentheses designate the two precursor cDNA(s) from which the mature peptide sequence was derived (Fig. 1). For example, the two precursor cDNAs for BTD-1 were BTD-a and BTD-b, which is shown as (a+b).
Theor., theoretical. The calculated mass (Daltons) of the native or S-carboxamidomethylated (S-CAM) cyclic peptide is shown. ND, not determined.
Determined by MALDI-TOF MS. ND, not determined.
Isolated peptides were reduced with DTT and S-carboxamidomethylated (CAM) with iodoacetamide.
Purification of BTDs.
Acid extracts of baboon leukocytes were subjected to sequential gel filtration (Fig. 2A, inset) and HPLC purification (Fig. 2) steps as described in Materials and Methods. Putative θ-defensins were identified by MALDI-TOF MS before and after reduction and alkylation, allowing for the identification of tridisulfide peptides with masses matching one or more of those peptides shown in Fig. 1B. Each of the putative θ-defensins was found to comigrate with a synthetic version of one of the previously characterized rhesus θ-defensins. Homogeneous preparations of five baboon θ-defensins (Fig. 2B) were obtained by sequential HPLC steps (see Materials and Methods). Amino acid analysis was performed on samples of each peptide, and the amino acid compositions obtained (Table 2) allowed for the identification of five θ-defensins (BTD-1 to -4 and BTD-7) shown in Fig. 1B.
FIG. 2.
Purification of BTDs from leukocytes. (A) Acid extracts of peripheral blood leukocytes were chromatographed using a Bio-Gel P-60 gel filtration column (inset) equilibrated in 5% acetic acid. Fractions denoted by the bracket contain θ-defensin molecular masses. These fractions were pooled and rechromatographed by semipreparative RP-HPLC on a Vydac C18 column (10 by 250 mm). Numbers above the peaks indicate baboon θ-defensins. (B) BTD-1, -2, -3, -4, and -7 were purified to homogeneity, and 0.5 μg of each BTD was observed by analytical C18 RP-HPLC. BTD-2 is synthetic.
TABLE 2.
Amino acid compositions of purified BTDs
| Amino acid | Amino acid composition in:
|
||||
|---|---|---|---|---|---|
| BTD-1 | BTD-2 | BTD-3 | BTD-4 | BTD-7 | |
| Arg | 4.79 (5) | 6.00 (6) | 4.36 (4) | 3.26 (3) | 4.96 (5) |
| Cysa | 6.00 (6) | 6.00 (6) | 6.00 (6) | 6.00 (6) | 6.00 (6) |
| Gly | 1.98 (2) | 2.98 (2)b | 2.74 (2)b | 2.33 (2) | 2.60 (2)b |
| Ile | 0 (0) | 0 (0) | 0 (0) | 1.85 (2) | 0 (0) |
| Leu | 0 (0) | 0 (0) | 0 (0) | 2.20 (2) | 0 (0) |
| Phe | 1.00 (1) | 0 (0) | 2.00 (2) | 1.15 (1) | 1.89 (2) |
| Thr | 1.02 (1) | 0 (0) | 1.95 (2) | 1.14 (1) | 1.00 (1) |
| Val | 2.54 (3) | 3.67 (4) | 2.06 (2) | 1.00 (1) | 1.65 (2) |
Cysteine content was determined by reduction and alkylation, followed by MALDI-TOF MS.
Values are greater than expected due to glycine contamination.
We further confirmed that the baboon peptides were indeed θ-defensins by determining the sequence of BTD-7. An α-chymotrypsin digestion product of the peptide was fractionated by RP-HPLC, and purified peptide fragments were characterized by Edman degradation. The sequences of two nonoverlapping peptide fragments (CRCFCRRGVCR and CVCTRGF) accounted for all residues identified by amino acid analysis. When the two sequences were laid head to tail, the entire sequence matched the primary structure and mass of BTD-7 (Table 1 and Fig. 1B). Analysis of the BTD-7 sequence indicates that it is composed of nonapeptides derived from the BTD-a and BTD-d precursors (Fig. 1B). Similarly, BTD-1 to -4 are produced by homodimeric or heterodimeric splicing reactions. BTD-1 and BTD-4 are the products of heterodimeric splicing of the BTD-a nonapeptide with BTD-b and BTD-c nonapeptides, respectively. BTD-2 and BTD-3 are produced by the homodimeric splicing of BTD-b- and BTD-a-derived nonapeptides, respectively (Table 1 and Fig. 1B). The relative abundance of the five θ-defensins in leukocyte preparations (based on recovery during purification) was approximately 150:110:55:15:1 (BTD-1/ BTD-3/ BTD-7/ BTD-4/ BTD-2), indicating a wide range of expression of individual θ-defensins. We failed to detect peptides corresponding to putative BTD-5, -6, -8, -9, or -10 (Fig. 1B) in leukocytes.
Characterization of synthetic and natural BTDs.
BTD-1, -2, -3, -4, and -7 were synthesized and purified for further studies. Purified linear peptides were oxidized in room air and cyclized as described above (Materials and Methods). Synthetic BTD preparations were 95 to 100% pure and equivalent to the corresponding natural peptides in RP-HPLC, AU-PAGE, and antimicrobial assays. As shown in Fig. 3, natural and synthetic BTD-1 behaved identically in each analysis. Characterization of the remaining synthetic peptides and comparison with the corresponding natural peptides gave equivalent results (data not shown).
FIG. 3.
Comparison of natural and synthetic BTD-1. (A) Analytical RP-HPLC of 0.5 μg of natural BTD-1 (N), 0.5 μg of synthetic BTD-1 (S), and 0.5 μg of a 1:1 mixture of natural and synthetic BTD-1 (M). (B) Silver-stained AU-PAGE of 4 × 106 cell equivalent leukocyte extract (E) or 0.5 μg each of natural BTD-1 (N), synthetic BTD-1 (S), and a 1:1 mixture of natural and synthetic BTD-1 (M). (C) Agar diffusion assays of synthetic and natural BTD-1. Results shown are representative of duplicate experiments.
Immunolocalization of θ-defensin peptides in baboon leukocytes and bone marrow cells.
Antibody produced against a polymerized form of RTD-1 was evaluated for its immunoreactivity with each of the baboon θ-defensins. Peptides were separated by AU-PAGE and analyzed by Western blotting with goat anti-RTD-1 antibody. As shown in Fig. 4, the antibody recognized all five synthetic BTDs (lanes 2 to 6) as well as RTD-1 to -3 (lane 7). Immunoreactivity was also observed in an extract of baboon leukocytes (lane 1), detecting a band that comigrated with BTD-1 and -7.
FIG. 4.
Western blotting of baboon and rhesus θ-defensins. Acid extracts of 4 × 106 baboon leukocytes (lane 1), 1 μg each of synthetic BTD-1, -2, -3, -4, and -7 (lanes 2 to 6, respectively), and a mixture of RTD-1, -2, and -3 (lane 7) were resolved on a 12.5% AU-polyacrylamide gel, transferred to a nitrocellulose membrane, and immunoblotted with goat anti-RTD-1 IgG.
BTD cellular expression and localization were evaluated by immunostaining cytocentrifuge preparations of baboon blood leukocytes with anti-RTD-1 IgG. Cytoplasmic staining of granules in neutrophils and monocytes, but not eosinophils, was observed (Fig. 5A). This pattern closely resembles the staining of α- and θ-defensins in rhesus macaque leukocytes (20, 21) and of α-defensins in human neutrophils (14). Immunostaining of baboon bone marrow cells showed BTD expression in myeloblasts as well as band and segmented neutrophils (Fig. 5C). These data indicate that θ-defensins are produced early in myelopoiesis, similar to θ-defensins in rhesus monkeys (21).
FIG. 5.
Immunohistochemical staining of BTDs. (A) Peripheral blood leukocytes stained with anti-RTD-1 IgG localized BTDs to the cytoplasmic granules of neutrophils (arrowheads) and monocytes (hollow arrows), but not eosinophils (yellow arrows). (B) Preimmune IgG staining of peripheral blood leukocytes. (C) Bone marrow cells show cytoplasmic staining of BTDs in neutrophilic precursors (hollow arrows) and neutrophils (arrowheads), but not eosinophils (yellow arrows). (D) Preimmune IgG staining of bone marrow cells.
Antimicrobial activities of BTDs.
Synthetic BTDs were evaluated for microbiostatic and microbicidal activities against gram-positive (S. aureus 502a) and gram-negative (E. coli ML35) bacteria and a fungus (C. albicans 16820) (Fig. 6 and 7). Data shown are representative of duplicate experiments. Like rhesus θ-defensins, baboon θ-defensins inhibited microbial growth in a dose-dependent manner, and both RTD-1 and BTD-1 had identical bacteriostatic activities against S. aureus and E. coli (data not shown). The five BTD peptides tested here produced nearly identical zones of clearing against S. aureus (Fig. 6, left). Somewhat greater activity variation was observed in assays against E. coli (Fig. 6, middle), with BTD-2 and BTD-3 being somewhat more effective than the other θ-defensins. Even greater differences in BTD activities were seen in diffusion assays against C. albicans (Fig. 6, right). The most potent peptide was BTD-2, which was approximately twice as active as BTD-4, the least active θ-defensin.
FIG. 6.
Microbiostatic activities of BTDs. Synthetic BTD-1, -2, -3, -4, and -7 were tested for their antimicrobial activities against the indicated organisms in an agar diffusion assay. Zones of clearing were measured after incubation for 18 to 24 h. Results shown are representative of duplicate experiments.
FIG. 7.
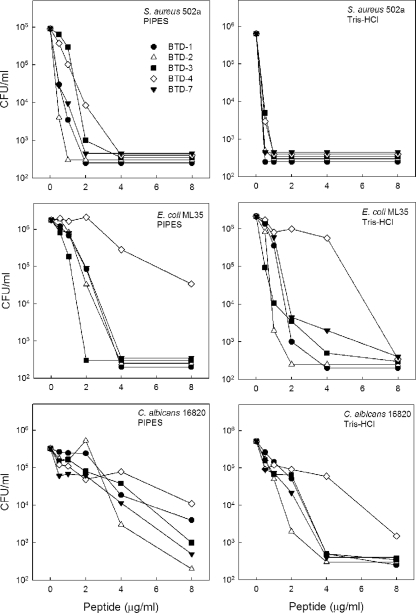
Microbicial activities of BTDs. Synthetic BTDs were incubated with 2 × 106 CFU of each organism for 2 h at 37°C using two buffering conditions, 10 mM PIPES (pH 7.4) and 5 mM glucose (left panels) or 10 mM Tris-HCl (pH 7.4) and 5 mM glucose (right panels). The numbers of CFU were determined after the plates were incubated at 37°C for 18 to 24 h. The limit of detection (1 colony/plate) was equal to 1 × 103 CFU/ml in the incubation mixture.
We previously showed that the microbicidal activities of rhesus monkey θ-defensins were markedly affected by the ionic milieu of the incubation medium (22). Therefore, we evaluated BTD microbicidal activities using two buffering solutions, 10 mM PIPES (pH 7.4) containing 5 mM glucose (Fig. 7, left panels) or 10 mM Tris-HCl (pH 7.4) containing 5 mM glucose (Fig. 7, right panels). In both buffer systems, all five BTDs were able to kill >90% of all microorganisms tested at 8 μg/ml, the highest concentration tested. Overall, the greatest microbicidal effect was against S. aureus where >3 log units of killing was obtained with 4 μg/ml in PIPES buffer and with 1 μg/ml in Tris buffer (Fig. 7, top two panels). BTD killing of E. coli was similar in PIPES and Tris buffers (Fig. 7, middle panels); BTD-4 was the least active BTD against E. coli, but it still showed 90 to 100% killing at 8 μg/ml. Each of the baboon θ-defensins except BTD-4 killed >3 log units of C. albicans at 4 to 8 μg/ml in Tris buffer, but only BTD-2 and BTD-7 (at 8 μg/ml) killed to this degree in PIPES buffer (Fig. 7, bottom panels). Overall, BTDs exhibited somewhat greater microbicial activity in Tris buffer than in PIPES buffer against C. albicans, especially at 4 to 8 μg/ml. These data are consistent with results seen from diffusion assays and demonstrate that baboon θ-defensin activities vary depending on the target microorganism and the buffer system employed.
BTD permeabilization of E. coli ML35.
BTDs were analyzed for their mechanism of E. coli killing by determining the extent of bacterial permeabilization produced in the ONPG hydrolysis assay described in Materials and Methods. As indicated in Fig. 8A, each peptide elicited dose-dependent efflux of ONP from E. coli. Previously we demonstrated that rhesus macaque θ-defensins enter the cytoplasm of E. coli ML35 where they inhibit β-galactosidase to various degrees. To correct for BTD-mediated inhibition of β-galactosidase, purified enzyme was incubated with each peptide, and ONPG conversion was determined (Fig. 8B). A corrected rate of ONPG hydrolysis was then calculated as described in Materials and Methods and plotted as a function of peptide concentration (Fig. 8C). At low peptide concentrations (<1 μg/ml), the rate of ONPG hydrolysis was similar for BTD-1, -3, -4, and -7, whereas BTD-2 was 1.5- to 3-fold more active than the other BTDs. In addition, the concentration at which the rate of ONPG hydrolysis plateaued varied substantially among the five peptides (Fig. 8C), indicating that the interaction with the cell envelope differs among the BTDs.
FIG. 8.
E. coli ML35 permeabilization by BTDs. (A) Synthetic BTDs were incubated with 2 × 106 CFU E. coli in 10 mM PIPES (pH 7.4) and 3 mM ONPG, and ONP efflux was monitored spectrophotometrically at 405 nm. (B) BTD inhibition of purified β-galactosidase (β-gal) was determined by monitoring the rates of ONPG hydrolysis in a cell-free system. (C) The adjusted rates of ONPG hydrolysis were calculated using the inhibition coefficients determined from panel B to correct the values in panel A. Results shown are representative of duplicate experiments.
DISCUSSION
Investigations of rhesus macaque leukocytes led to the discovery of the macrocyclic θ-defensin peptide family and revealed a unique biosynthetic pathway that produces the mature octadecapeptide (12, 21, 23). Subsequently, genetic studies by Nguyen and colleagues (15) demonstrated the presence of θ-defensin precursor genes in a number of Old World Monkeys (rhesus and pigtail macaques and colobus monkeys), siamangs (a lesser ape), and orangutans (a great ape), whereas New World Monkeys and prosimians lacked θ-defensin genes. Of note was the finding that humans, gorillas, bonobos, and chimpanzees possess only pseudogenes in which a conserved premature stop codon interrupts the open reading frame in the θ-defensin precursor signal peptide. In the same report, data on two Old World Monkeys, the gelada baboon and the silver leaf langur, appeared to lack either intact genes or pseudogenes encoding θ-defensins. This finding was at variance with our studies indicating that θ-defensin precursor mRNAs were expressed in bone marrow of olive and gelada baboons. We therefore undertook studies to evaluate the expression of θ-defensin mRNAs and peptides in these Old World species.
Analysis of RT-PCR products derived from olive baboon bone marrow disclosed the presence of four θ-defensin mRNAs (Fig. 1), and these data facilitated the isolation of five θ-defensins from peripheral blood leukocytes. On the basis of previous findings in rhesus monkeys, we predicted that baboon θ-defensins would be composed of 18 amino acids that would derive from binary combinations of the BTD-a to -d nonapeptides highlighted in Fig. 1. Of the 10 possible θ-defensin products thus generated, 5 were isolated. The identity of each peptide was confirmed by synthesizing the deduced structure and/or by direct sequencing of the natural peptide. This is only the second animal species from which naturally occurring cyclic peptides have been isolated, thus confirming the binary excision/ligation biosynthetic pathway previously characterized in rhesus monkeys.
The sequences of the θ-defensins expressed in baboons and rhesus monkeys are very similar, but no single peptide is expressed by both species. The net charges among the peptides (either isolated or predicted) range from +2 to +6 for both species, and there is a wide range of expression levels among the peptides. Five of the six predicted macaque θ-defensins have been purified from myeloid tissues (23; unpublished data), but only 5 of the 10 predicted baboon peptides have been isolated thus far. This may be a reflection of expression levels and/or efficiency of the steps involved in purifying individual peptides.
The relative abundance of the BTDs isolated in this study reflects the ratios resulting from combining the leukocytes from 10 baboons, and the relative BTD levels may differ in individual animals. As noted, RT-PCR analysis of bone marrow RNA from one olive baboon failed to amplify BTD-d cDNA, whereas it was present in others. Thus, it is likely that BTD gene copy number is polymorphic, similar to that observed with primate α- and β-defensins (1, 5, 6, 13). In addition, Leonova et al. identified the rhesus θ-defensin gene, RTD1a, in only two of four monkeys examined (12).
Like the θ-defensins in rhesus monkeys, baboon θ-defensins were immunolocalized to cytoplasmic granules in neutrophils and neutrophilic progenitors in the bone marrow. Like other granule components, they are mobilized for delivery to the phagosome following phagocytosis by granulocytes. In the phagolysosome, they interact with ingested organisms as components of a complex mixture of antimicrobial effector molecules. While the relative contribution of θ-defensins to vacuolar killing is not yet known, our in vitro experiments demonstrate that each of the peptides possesses antimicrobial activity and that much of this activity is microbicidal. Each of the peptides tested had similar antibacterial activities as measured in diffusion plate assays, whereas antifungal activities were more variable. Since the readout in diffusion assays does not differentiate between microbiostasis and killing per se, we tested for bactericidal and fungicidal activities of each peptide. Despite the finding that all θ-defensins killed >95% of each organism at concentrations of <8 μg/ml (∼4 μM), there were clear differences in peptide potency; of note was the finding that BTD-4, the least cationic (net charge of +3) of the θ-defensins tested was the least active peptide in each antimicrobial assay. However, previous studies have shown there to be only weak correlation between antimicrobial efficacy and cationic charge (4, 20). Therefore, differences in BTD antimicrobial activities are likely the result of other factors, such as the ability to traverse the cell envelope as discussed below. As observed previously (22), the selection of assay markedly affected the microbicidal activity, pointing to the importance of understanding the microenvironment of the phagolysosome and other microenvironments where defensins are believed to function in vivo.
The results of permeabilization assays provided additional insights into the interaction of the peptides with bacteria. Each baboon θ-defensin disrupted the E. coli cell envelope as evidenced by influx of ONPG and its hydrolysis by cytoplasmic β-galactosidase. A first approximation of the dose-dependent permeabilization (Fig. 8A) demonstrated large differences in the potency of the five θ-defensins. However, previous studies showed that rhesus θ-defensins directly inhibit β-galactosidase by entering the E. coli cytosol and inhibiting the enzyme in situ (22). It was therefore necessary to adjust for β-galactosidase inhibition by each θ-defensin (Fig. 8B) allowing for an estimate of the actual permeabilization mediated by each peptide (Fig. 8C). It should be noted that the corrected rates of ONPG hydrolysis must be considered estimates because the extent of θ-defensin uptake at each concentration is not known. However, the shape of the curve(Fig. 8C) for each peptide is consistent with previous permeabilization studies using E. coli ML35 as the target cell (10, 22). Moreover, there is generally a good correlation between the rate of ONPG hydrolysis (Fig. 8C) and the microbicidal activity (Fig. 7) of each peptide against E. coli.
The olive baboon is a primate model frequently used to study human diseases. However, it is well-known that baboons and humans respond very differently to microbial antigens (8). We hypothesize that the lack of θ-defensins in humans may underlie some of the differences in innate immune responses observed in humans and baboons.
Acknowledgments
This study was supported by NIH grants AI22931, AI58129, and DE15517 (M.E.S.); RR013986 (Southwest National Primate Research Center); and RR12317 and RR16556 (Oklahoma University Health Science Center).
We thank Dat Tran and Prasad Tongaonkar for critical review of the manuscript.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 13 October 2008.
REFERENCES
- 1.Aldred, P. M., E. J. Hollox, and J. A. Armour. 2005. Copy number polymorphism and expression level variation of the human alpha-defensin genes DEFA1 and DEFA3. Hum. Mol. Genet. 142045-2052. [DOI] [PubMed] [Google Scholar]
- 2.Bals, R., X. Wang, Z. Wu, T. Freeman, V. Bafna, M. Zasloff, and J. M. Wilson. 1998. Human beta-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J. Clin. Investig. 102874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole, A. M., T. Hong, L. M. Boo, T. Nguyen, C. Zhao, G. Bristol, J. A. Zack, A. J. Waring, O. O. Yang, and R. I. Lehrer. 2002. Retrocyclin: a primate peptide that protects cells from infection by T- and M-tropic strains of HIV-1. Proc. Natl. Acad. Sci. USA 991813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullor, J. S., S. Wood, W. Smith, L. Panico, and M. E. Selsted. 1991. Bactericidal potency and mechanistic specificity of neutrophil defensins against bovine mastitis pathogens. Vet. Microbiol. 2949-58. [DOI] [PubMed] [Google Scholar]
- 5.Dork, T., and M. Stuhrmann. 1998. Polymorphisms of the human beta-defensin-1 gene. Mol. Cell Probes 12171-173. [DOI] [PubMed] [Google Scholar]
- 6.Eisenhauer, P., S. S. Harwig, D. Szklarek, T. Ganz, and R. I. Lehrer. 1990. Polymorphic expression of defensins in neutrophils from outbred rats. Infect. Immun. 583899-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3710-720. [DOI] [PubMed] [Google Scholar]
- 8.Haudek, S. B., B. E. Natmessnig, W. Furst, S. Bahrami, G. Schlag, and H. Redl. 2003. Lipopolysaccharide dose response in baboons. Shock 20431-436. [DOI] [PubMed] [Google Scholar]
- 9.Lehrer, R. I. 2004. Primate defensins. Nat. Rev. Microbiol. 2727-738. [DOI] [PubMed] [Google Scholar]
- 10.Lehrer, R. I., A. Barton, K. A. Daher, S. S. Harwig, T. Ganz, and M. E. Selsted. 1989. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Investig. 84553-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehrer, R. I., T. Ganz, D. Szklarek, and M. E. Selsted. 1988. Modulation of the in vitro candidacidal activity of human neutrophil defensins by target cell metabolism and divalent cations. J. Clin. Investig. 811829-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leonova, L., V. N. Kokryakov, G. Aleshina, T. Hong, T. Nguyen, C. Zhao, A. J. Waring, and R. I. Lehrer. 2001. Circular minidefensins and posttranslational generation of molecular diversity. J. Leukoc. Biol. 70461-464. [PubMed] [Google Scholar]
- 13.Linzmeier, R. M., and T. Ganz. 2005. Human defensin gene copy number polymorphisms: comprehensive analysis of independent variation in alpha- and beta-defensin regions at 8p22-p23. Genomics 86423-430. [DOI] [PubMed] [Google Scholar]
- 14.Mackewicz, C. E., J. Yuan, P. Tran, L. Diaz, E. Mack, M. E. Selsted, and J. A. Levy. 2003. Alpha-defensins can have anti-HIV activity but are not CD8 cell anti-HIV factors. AIDS 17F23-F32. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen, T. X., A. M. Cole, and R. I. Lehrer. 2003. Evolution of primate theta-defensins: a serpentine path to a sweet tooth. Peptides 241647-1654. [DOI] [PubMed] [Google Scholar]
- 16.Selsted, M. E. 2004. Theta-defensins: cyclic antimicrobial peptides produced by binary ligation of truncated alpha-defensins. Curr. Protein Pept. Sci. 5365-371. [DOI] [PubMed] [Google Scholar]
- 17.Selsted, M. E., and A. J. Ouellette. 1995. Defensins in granules of phagocytic and non-phagocytic cells. Trends Cell Biol. 5114-119. [DOI] [PubMed] [Google Scholar]
- 18.Selsted, M. E., and A. J. Ouellette. 2005. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6551-557. [DOI] [PubMed] [Google Scholar]
- 19.Selsted, M. E., D. Szklarek, and R. I. Lehrer. 1984. Purification and antibacterial activity of antimicrobial peptides of rabbit granulocytes. Infect. Immun. 45150-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang, Y.-Q., J. Yuan, C. J. Miller, and M. E. Selsted. 1999. Isolation, characterization, cDNA cloning, and antimicrobial properties of two distinct subfamilies of α-defensins from rhesus macaque leukocytes. Infect. Immun. 676139-6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang, Y. Q., J. Yuan, G. Osapay, K. Osapay, D. Tran, C. J. Miller, A. J. Ouellette, and M. E. Selsted. 1999. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science 286498-502. [DOI] [PubMed] [Google Scholar]
- 22.Tran, D., P. Tran, K. Roberts, G. Ösapay, J. Schaal, A. Ouellette, and M. E. Selsted. 2008. Microbicidal properties and cytocidal selectivity of rhesus macaque theta defensins. Antimicrob. Agents Chemother. 52944-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tran, D., P. A. Tran, Y. Q. Tang, J. Yuan, T. Cole, and M. E. Selsted. 2002. Homodimeric theta-defensins from rhesus macaque leukocytes: isolation, synthesis, antimicrobial activities, and bacterial binding properties of the cyclic peptides. J. Biol. Chem. 2773079-3084. [DOI] [PubMed] [Google Scholar]
- 24.Wang, W., A. M. Cole, T. Hong, A. J. Waring, and R. I. Lehrer. 2003. Retrocyclin, an antiretroviral theta-defensin, is a lectin. J. Immunol. 1704708-4716. [DOI] [PubMed] [Google Scholar]
- 25.Wang, W., C. Mulakala, S. C. Ward, G. Jung, H. Luong, D. Pham, A. J. Waring, Y. Kaznessis, W. Lu, K. A. Bradley, and R. I. Lehrer. 2006. Retrocyclins kill bacilli and germinating spores of Bacillus anthracis and inactivate anthrax lethal toxin. J. Biol. Chem. 28132755-32764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasin, B., W. Wang, M. Pang, N. Cheshenko, T. Hong, A. J. Waring, B. C. Herold, E. A. Wagar, and R. I. Lehrer. 2004. θ defensins protect cells from infection by herpes simplex virus by inhibiting viral adhesion and entry. J. Virol. 785147-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zanetti, M. 2004. Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 7539-48. [DOI] [PubMed] [Google Scholar]