Abstract
The pathogenic mechanisms of Leptospira interrogans, the causal agent of leptospirosis, remain largely unknown. This is mainly due to the lack of tools for genetically manipulating pathogenic Leptospira species. Thus, homologous recombination between introduced DNA and the corresponding chromosomal locus has never been demonstrated for this pathogen. Leptospiral immunoglobulin-like repeat (Lig) proteins were previously identified as putative Leptospira virulence factors. In this study, a ligB mutant was constructed by allelic exchange in L. interrogans; in this mutant a spectinomycin resistance (Spcr) gene replaced a portion of the ligB coding sequence. Gene disruption was confirmed by PCR, immunoblot analysis, and immunofluorescence studies. The ligB mutant did not show decrease virulence compared to the wild-type strain in the hamster model of leptospirosis. In addition, inoculation of rats with the ligB mutant induced persistent colonization of the kidneys. Finally, LigB was not required to mediate bacterial adherence to cultured cells. Taken together, our data provide the first evidence of site-directed homologous recombination in pathogenic Leptospira species. Furthermore, our data suggest that LigB does not play a major role in dissemination of the pathogen in the host and in the development of acute disease manifestations or persistent renal colonization.
Leptospirosis is a widespread zoonosis that has emerged as a major public health problem in developing countries in Southeast Asia and South America (6, 22, 29). This increasingly common disease occurs in poor urban centers subject to frequent flooding (20). Rodents are the main reservoir of the disease, excreting the bacteria in their urine (14, 22). Humans are usually infected through contaminated water. More than 500,000 cases of severe leptospirosis are estimated to occur worldwide each year (46), and the fatality rate is 5 to 20% (29).
The control methods for leptospirosis implemented to date have been ineffective (29). A significant barrier to control and prevention of leptospirosis has been our limited understanding of the pathogenesis of the disease, due in part to the lack of genome sequences and tools to genetically manipulate the pathogens. Most of the barriers have now been overcome. The genomes of two pathogenic species and one saprophytic species have been sequenced (8, 32, 39, 40). Furthermore, we developed a transposon-mediated mutagenesis system for pathogenic Leptospira species (7). This advance allowed characterization of the first genetically defined virulence factor in pathogenic Leptospira spp. (41). However, the generation of targeted mutants of pathogenic species was not feasible until now.
High-molecular-weight leptospiral immunoglobulin-like repeat (Lig) proteins were previously identified as putative virulence factors in pathogenic Leptospira spp. (21, 26, 34). This family of three proteins, LigA, LigB, and LigC, belongs to the superfamily of bacterial immunoglobulin-like (Big) repeat domain proteins, which includes virulence determinants such as intimin from enteropathogenic Escherichia coli, invasin from Yersinia pseudotuberculosis, and BipA from Bordetella spp. (26). This superfamily appears to mediate pathogen-host cell interactions, such as invasion and host cell attachment, during infection. Choy et al. and Lin and Chang recently showed that recombinant Lig proteins can mediate in vitro interactions with host extracellular matrix proteins, including fibronectin, fibrinogen, collagen, and laminin (9, 23). In addition, lig genes are upregulated at physiological osmolarity (27) and encode surface-exposed proteins that are strongly recognized by sera from human leptospirosis patients (10, 26, 43). Finally, several studies have shown that Lig proteins are protective antigens in animal models of leptospirosis (21, 35, 42).
In this study, we produced a ligB mutant of L. interrogans by allelic exchange and evaluated the effect of the deletion in this mutant using both cell adhesion assays and animal models. The results provided the first demonstration of targeted mutagenesis of Leptospira pathogenic strains.
(This research was conducted by J. Croda in partial fulfillment of the requirements for a Ph.D. from the Department of Pathology, Medicine School, São Paulo University, and by C. P. Figueira and E. Wunder in partial fulfillment of the requirements for Ph.D. from Goncalo Moniz Research Center, Oswaldo Cruz Foundation, Brazil.)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Leptospires were cultivated in liquid Ellinghausen-McCullough-Johnson-Harris (EMJH) medium (13, 19) or on 1% agar plates at 30°C and were counted in a Petroff-Hausser counting chamber (Fisher Scientific). L. interrogans serovar Copenhageni strain Fiocruz L1-130, a virulent clinical isolate from Brazil (20, 32), was used in all experiments. E. coli was grown in Luria-Bertani medium. When necessary, spectinomycin or kanamycin was added to culture media at a concentration of 50 μg/ml.
Polyclonal and monoclonal antibodies.
We prepared immune sera against previously described recombinant fragments of LigA (LigANI; amino acid positions 625 to 1225) and LigB (LigBNI; amino acid positions 625 to 1257) (42). These fragments contain the 6th to 13th and 6th to 12th Big repeat domains of LigA and LigB, respectively, and do not include the portions of these molecules which have identical amino acid sequences (26). New Zealand White rabbits were immunized intravenously on days 0, 7, and 14 with three doses of 80 μg of recombinant protein fragments, using aluminum hydroxide as an adjuvant. Rabbits were bled on day 28 to obtain immune sera. For quality control, the reactivities of immune sera with recombinant and native Lig proteins were evaluated using enzyme-linked immunosorbent and immunoblot assays as previously described. Prebleed sera, as well as sera from rabbits immunized with phosphate-buffered saline (PBS) and alhydrogel, were used as control samples. Ascites fluid containing monoclonal antibodies (MAb) against a recombinant LigB protein fragment, LigBrep (26, 42), were provided by José Aleixo, Federal University of Pelotas. The LigBrep fragment corresponds to the six N-terminal Big repeat domains of the LigB molecule (amino acid positions 131 to 649).
Targeted mutagenesis.
For allelic exchange, the gene fragment which encodes the LigB nonidentical region (nucleotides 1891 to 5450), also called the B2 region, was amplified from the genomic DNA of L. interrogans serovar Copenhageni strain Fiocruz L1-130 using primers LigBU1F-SmaI (5′ TCCCCCGGGGCTGAAATTAAAAATACCAGTGGAAG 3′) and P2R-SmaI (5′ TCCCCCGGGCCTATGTAGAGATAAGATCCACTTGC 3′). The PCR product was digested with SmaI and cloned into a PvuII-digested pGKm plasmid vector. The resulting plasmid was then digested with EcoRI, which removed a 699-bp sequence from the ligB B2 domain. The Spcr cassette was amplified from plasmid pGSpc DNA using primers Spc-EcorI3′ (5′ ACGGAATTCAACGTAAAGTAAG 3′) and Spc-EcorI5′ (5′ TCGGAATTCAACGCGTCCCGAGC 3′). The PCR product was digested with EcoRI and subsequently inserted into the plasmid from which the 699-bp ligB sequence had been removed to obtain plasmid pB2SK (Fig. 1B). For gene inactivation by plasmid insertion, an internal DNA fragment of ligB (nucleotides 1891 to 3771), also called the B1 region, was amplified from the genomic DNA of L. interrogans Copenhageni strain Fiocruz L1-130 using primers LigBU1F-SmaI (5′ TCCCCCGGGGCTGAAATTAAAAATACCAGTGGAAG 3′) and LigBU2R-SmaI (5′ TCCCCCGGGCACTTGGTTTAAGGAATTACAAACT 3′). The PCR product was digested with SmaI and cloned into a PvuII-digested pGSpc plasmid vector, resulting in plasmid pB1S. Plasmids pGKm and pGSpc (complete sequences are available on request) were derived from the cloning vector pGEM-7Zf(−) (Promega Corporation, Madison, WI) as previously described (15). We used the Enterococcus faecalis kanamycin and Staphylococcus aureus Spcr cassettes as previously described (5).
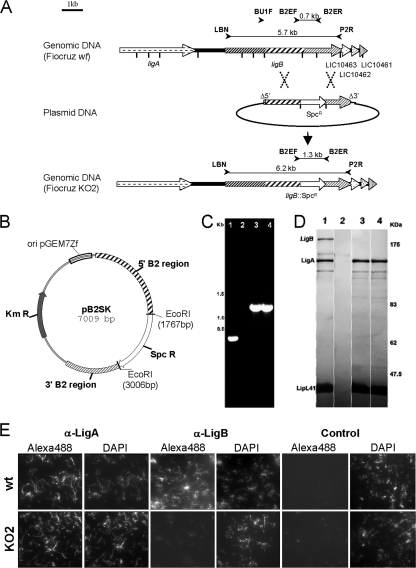
FIG. 1.
Disruption of ligB in L. interrogans strain Fiocruz L1-130. (A) Schematic representation of the genotype of the parental (Fiocruz wt) and ligB (Fiocruz KO2) mutant strains. The LigB protein has a tripartite structure which includes an N-terminal identical repeat region, a nonidentical repeat region, and the C-terminal region. The vertical bars indicate EcoRI restriction sites. The locations of primers used to check for allelic exchange, as well as the expected sizes of amplified products, are indicated. (B) Map of the pB2SK plasmid, in which the Spcr cassette was inserted between EcoRI sites in the B2 region of ligB. (C) PCR amplification of chromosomal DNA from the L. interrogans wild-type strain (lane 1), L. biflexa strain Patoc1 (lane 2), the L. interrogans ligB KO2 mutant (lane 3), and the L. interrogans ligB KO2 mutant reisolated from hamsters after infection (lane 4) with primers B2EF and B2ER, as shown in panel A. (D) Western blot of LigA and LigB expression in the L. interrogans wild-type strain (lane 1), L. biflexa strain Patoc1 (lane 2), the L. interrogans ligB mutant (lane 3), and the L. interrogans ligB mutant which was reisolated from hamsters (lane 4). Blots were also probed with LipL41 antiserum as a reference. (E) Immunofluorescence assays were performed with L. interrogans wild-type (wt) and ligB mutant (KO2) strains. Strains were labeled with antibodies against LigANI (α-LigA0), LigBNI (α-LigB), and a control. Alexa- and fluorescein isothiocyanate-conjugated secondary antibodies were used to detect surface-bound antibodies to LigANI and LigBNI, respectively. A DAPI counterstain was used to document the presence of leptospires. The photomicrograph show the results of one of three representative experiments.
The plasmid constructs, which are not replicative in Leptospira spp., were used to deliver the inactivated allele into L. interrogans by electroporation. Cells were grown to exponential phase (optical density at 420 nm, 0.10 to 0.20) and then centrifuged at 4,000 × g and concentrated to obtain 1010 bacteria/ml in sterile water. Suicide plasmids containing the inactivated allele were subjected to 5 to 30 s of UV treatment (254 nm, 400 μW/cm2) using a UV chamber (GS Gene linker; Bio-Rad). Two hundred microliters of cells was electroporated (1.8-kV, 200-Ω, 25-μF electric pulse in a prechilled 0.2-cm-diameter cuvette) in the presence of 100 to 500 ng of plasmid DNA and then transferred to 1 ml of EMJH liquid medium, in which the cells were incubated for 24 h at 30°C. The bacteria were then plated on EMJH medium supplemented with spectinomycin (50 μg/ml). After 4 to 6 weeks of incubation, Spcr colonies were picked and examined for allelic exchange in the target gene by PCR, Western blotting, and immunofluorescence analysis.
Genomic DNA analysis.
Genomic DNA was prepared from liquid cultures by use of a cell DNA purification kit (Maxwell, Promega, Madison, WI). To check for double homologous recombination, primers B2EF (5′ CATACACACTTGTAGTCAACAAACAAG 3′) and B2ER (5′ CGTAACGTAATTCGGAACCG 3′) and primers LBN (5′ GGGAATTCCATATGAAGAAAATATTTTGTATTTCG 3′) and P2R (5′ TATGTAGAGATAAGATCCACTTGC 3′) were used for amplification of the ligB locus (Fig. 1A). Gene inactivation by plasmid insertion was confirmed by using primers B1F (5′ ACCTGGAATTCCTCTAATACGGATATT 3′) and B1R (5′ GAATATAAAGGTTTGGAAAAAGAAACG 3′) for PCR amplification.
Immunoblotting.
Mutant and wild-type L. interrogans Fiocruz L1-130 strains were grown in EMJH medium until the optical density at 420 nm was 0.2. Leptospira biflexa was also used as a control in these experiments. Bacteria were washed in PBS. After the concentration was adjusted to 2 × 108 bacteria/per well (20 μl), the cells were solubilized in 62.5 mM Tris hydrochloride (pH 6.8)-10% glycerol-5% 2-mercaptoethanol-2% sodium dodecyl sulfate. Crude protein extracts were resolved by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis using a discontinuous buffer system. After transfer to nitrocellulose membranes, immunoblots were blocked in 0.05 M Tris-buffered saline (pH 7.4)-0.05% (vol/vol) Tween 20 with 5% (wt/vol) nonfat dry milk. The blots were washed, incubated for 1 h at room temperature with a 1,000-fold dilution of mouse ascites containing MAb to the LigB identical repeat region (LigA/B) or with a 10,000-fold dilution of hyperimmune rabbit antisera to LipL41, and probed with goat anti-mouse and anti-rabbit immunoglobulin G (IgG) antibodies conjugated to alkaline phosphatase (Sigma). Immunoblots were developed in a nitroblue tetrazolium—5-bromo-4-chloro-3-indolylphosphate (BCIP) solution (Bio-Rad).
Immunofluorescence assays.
Immunofluorescence labeling was performed using a modified protocol of Cullen et al. (12). Suspensions containing 107 live leptospires in 10 μl of PBS were placed on poly-l-lysine-coated (Sigma) slides and incubated for 1 h in a humidified chamber. The slides were washed twice with PBS, blocked with PBS containing 1% bovine serum albumin (BSA) (Sigma) (PBS-BSA), and incubated for 1 h with hyperimmune rabbit antisera to the LigB nonidentical region (LigBNI) and the LigA nonidentical region (LigANI) (diluted 1:100 in PBS-BSA) and with control rabbit antisera to a leishmanial antigen. The slides were washed gently with PBS-BSA and incubated with donkey anti-mouse IgG antibodies conjugated to Alexa dye (Molecular Probes) or with goat anti-rat IgG antibodies conjugated to fluorescein isothiocyanate (Jackson ImmunoResearch Laboratories) for 1 h at 37°C. The slides were washed twice with PBS-BSA and incubated with 1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI) (Molecular Probes) for 1 h at room temperature. The slides were washed and then mounted in antifading solution (Prolong; Molecular Probes) and visualized by fluorescence microscopy (Olympus BX51).
Hamster model of acute infection.
Groups of four Golden Syrian male hamsters that were 5 to 8 weeks old were inoculated intraperitoneally with 10, 102, 104, and 106 cells of the wild-type and ligB mutant L. interrogans Fiocruz L1-130 strains. Negative control animals were inoculated intraperitoneally with 1 ml of EMJH medium. Animals were monitored daily for clinical signs of leptospirosis (prostration and jaundice) and survival. Surviving animals were killed after a 21-day postchallenge follow-up period. The 50% lethal dose (LD50) of L. interrogans strain Fiocruz L1-130 in 5- to 8-week-old hamsters was approximately 101 leptospires. Culture isolation and immunofluorescence studies were performed using kidney and liver samples (42) to determine whether surviving animals had a persistent infection. The protocols used for animal experiments followed the guidelines of the Animal Care and Use Committee of Fundação Oswaldo Cruz.
Rat model of chronic infection.
Groups of four or eight Wistar rats (Fiocruz, Rio de Janeiro, Brazil) that were 4 to 5 weeks old were inoculated intraperitoneally with 108 cells of the wild-type and ligB mutant strains in 1 ml EMJH medium. Control animals were inoculated intraperitoneally with 1 ml of sterile EMJH medium. Animals were sacrificed 15 days after infection. Necropsies were performed immediately after sacrifice. Kidney and liver samples were fixed in 4% formalin, embedded in paraffin, and cut into 4- to 5-μm sections for conventional histology analysis. Renal tissue samples were homogenized in 5 ml of EMJH liquid medium for 10 min. After separation of the supernatant from the tissues, 500 μl of the supernatant was used to inoculate 5 ml EMJH liquid medium, which was subsequently incubated at 29°C. The cultures were examined weekly for growth by dark-field microscopy for up to 6 weeks.
Histopathology studies.
Groups of three hamsters were inoculated with 106 cells of the wild-type and ligB mutant L. interrogans Fiocruz L1-130 strains and euthanized on day 9 postchallenge. Tissues (liver, kidneys, and lungs) were fixed in 10% buffered formaldehyde, embedded in paraffin, and sectioned using routine histological procedures to obtain 4- to 5-μm sections that were then stained with hematoxylin and eosin. For immunohistochemistry analysis, the paraffin was removed from the sections with xylene and ethanol. The tissues were blocked by incubation of sections with 1.0% BSA at room temperature for 20 min. The tissues were incubated with a 1,000-fold dilution of antiserum to LipL32 (17) at room temperature for 1 h. Samples were treated with 0.3% hydrogen peroxide for 15 min at room temperature and then incubated at room temperature for 30 min with goat anti-mouse or anti-rabbit antibodies conjugated to peroxidase (Histostain-Plus kit; Invitrogen). Enzyme reactions were developed using 3,3′-diaminobenzidine (Sigma).
Host cell adhesion assay.
Madin-Darby canine kidney (MDCK) cells were harvested by treating cell cultures with 0.05% trypsin and 0.02% EDTA in PBS and then plated on 24-well plates in Dulbecco's modified Eagle's medium (Cultilab) without antibiotics. Cell viability was determined by trypan blue exclusion, and 500-μl portions of cell suspensions containing 2 × 105 cells per ml were layered on round glass coverslips in 24-well tissue culture plates. The plates were incubated for 24 h and washed twice with PBS to remove nonadherent cells. Bacteria were suspended in warm (37°C) cell culture medium at a concentration of 2 × 107 cells per ml. Wild-type and ligB mutant strains were used at the same time in each experiment. A 500-μl aliquot of each bacterial suspension was then added to the wells at a bacterium/cell ratio of 100:1. The plates were incubated under static conditions for 1 h at 37°C (30). Experiments were performed in triplicate. Coverslips were washed three times in PBS to remove nonadherent bacteria. An immunofluorescence analysis was performed as described above. The first antibody was anti-LipL32 MAb or anti-Omp Salmonella MAb, and the second antibody was anti-mouse antibody conjugated with Alexa 488 (Molecular Probes). DAPI and Alcian Blue were used to stain the nucleus and the cytoplasm, respectively. The numbers of leptospires and MDCK cells were determined by examining 10 high-power fields during fluorescence microscopy. Student's t test was used to evaluate the significance of differences between the numbers of associated leptospires per host cell in incubations with wild-type and ligB mutant strains.
RESULTS
Allelic exchange mutagenesis of L. interrogans ligB.
Pathogenic Leptospira species possess between one and three lig genes. L. interrogans serovar Copenhageni strain Fiocruz L1-130 contains two lig genes, ligA (3,675 bp) and ligB (5,673 bp), which encode polypeptides with molecular masses of 128 and 201 kDa, respectively. The third gene, ligC, was identified as a pseudogene in this L. interrogans strain (26). The ligB locus was used as a target for mutagenesis by allelic exchange in L. interrogans strain Fiocruz L1-130. A gene replacement construct was generated by cloning ligB into a suicide vector, which deleted a portion of the ligB open reading frame, which was replaced by an Spcr cassette. The amounts of homologous L. interrogans DNA present on the two sides of the spectinomycin marker were 1.8 and 1 kb (Fig. 1A).
The origin of replication used in the plasmid construct was that from pGEM7Zf, which is nonfunctional in Leptospira spp. Thus, any Spcr colonies arising after electroporation of this plasmid into L. interrogans should have resulted from recombination of the plasmid with the host genome. Strain Fiocruz L1-130 was electroporated with the UV-irradiated plasmid construct as previously described (38) and plated on solid medium containing spectinomycin. A total of six transformation experiments were performed, one of which yielded two transformants. The two mutant clones were obtained on the same plate and may have been mutants of siblings.
To confirm that homologous recombination events occurred at the origin of the Spcr phenotype, the ligB locus was analyzed by PCR mapping of genomic DNA obtained from L. interrogans transformants. PCR amplification with primers B2EF and B2ER, which normally produce a 0.7-kb product with wild-type L. interrogans, generated a 1.3-kb DNA fragment with the two Spcr recombinants analyzed (Fig. 1B). The amplified product was the expected size if a ligB mutant resulted from integration of the Spcr cassette by double-crossover recombination in the ligB chromosomal locus (Fig. 1A and B). Furthermore, PCR amplification with primers B2N and P2R yielded a product which had a size that was consistent with a gene replacement event in ligB loci. Immunoblotting with an anti-LigA/B MAb confirmed that LigB-reactive polypeptides were not present in the mutant (Fig. 1C). Moreover, by evaluating the reactivity with antibodies raised against LigA, we were able to detect LigA, which is encoded by the lig gene located upstream of ligB (26). This finding indicates that ligB inactivation does not modify ligA expression. Immunofluorescence studies also demonstrated that the ligB mutant did not express LigB, whereas the wild-type strain did express this protein (Fig. 1D). In contrast, antisera to LigA labeled live ligB mutant and wild-type leptospiral strains similarly.
By using another approach, we cloned an internal fragment of ligB lacking the 5′ and 3′ ends of the open reading frame in a suicide vector. Five transformants were recovered after electroporation in L. interrogans strain Fiocruz L1-130 in one of four transformation experiments. With all clones tested we obtained integration of the plasmid via a single crossover event, which generated two copies of the targeted gene, one with a deletion at the 5′ end of the gene and the other with a deletion at the 3′ end, thereby rendering it inactive, as confirmed by immunoblotting (data not shown).
In addition to the differences in genotype mentioned above, the transformants resulting from allelic exchange and plasmid insertion did not produce LigB; hence, the term ligB mutant refers to the double-crossover recombinant KO2 mutant below unless indicated otherwise.
Loss of the ligB gene does not affect virulence and persistence in animal models.
The ligB mutant and wild-type strains had similar cell growth kinetics in liquid EMJH medium (the generation time for the parent and mutant strains was approximately 20 h). Inactivation of ligB did not affect cell morphology and motility.
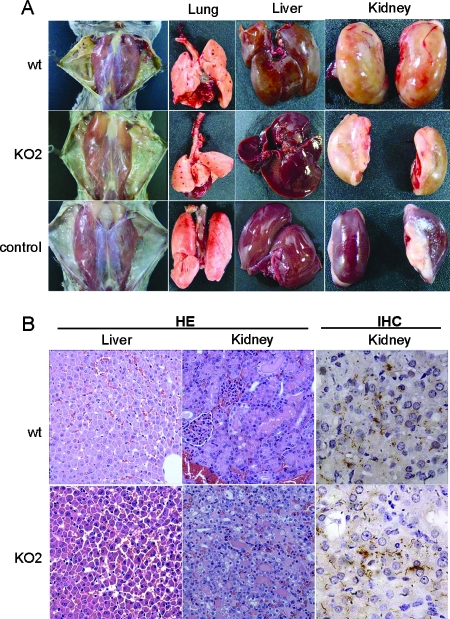
In order to determine whether LigB may have a role in virulence in vivo, we evaluated ligB mutants and the parental wild-type strain using the standard hamster model for acute leptospirosis. Different numbers of organisms (log increases in the challenge dose) were inoculated intraperitoneally to produce infection. The proportion of hamsters which died and the proportion which survived for each bacterial concentration were used to calculate the LD50. Three independent experiments in which groups of four animals were infected with each challenge dose were performed (Table 1). The LD50 was less than 100 bacteria for both the wild-type and mutant strains. Thus, the lack of LigB expression did not result in loss of virulence as measured by the LD50. In addition, no significant differences in the time to death were observed with the ligB and wild-type strains (data not shown). The general health status of the hamsters infected with the ligB and wild-type strains was also assessed. Infections with the ligB and wild-type strains produced similar pathological findings (jaundice, pulmonary hemorrhage, dissociation of hepatic trabecula, and acute damage of renal tubular epithelia with cell swelling in proximal segments). The immunohistochemistry results showed the same distribution of Leptospira in the renal parenchyma (Fig. 2).
TABLE 1.
Virulence of the wild-type and ligB mutant L. interrogans Fiocruz L1-130 strains in the hamster model of leptospirosis
| Strain | No. deaths (% of total) with a challenge dose ofa:
|
|||
|---|---|---|---|---|
| 106 Bacteria | 104 Bacteria | 102 Bacteria | 101 Bacteria | |
| Expt 1 | ||||
| Wild type | 4 (100) | 4 (100) | 4 (100) | NDb |
| ligB KO2 mutant | 4 (100) | 4 (100) | 4 (100) | ND |
| Expt 2 | ||||
| Wild type | 4 (100) | 4 (100) | 4 (100) | ND |
| ligB KO2 mutant | 4 (100) | 4 (100) | 4 (100) | ND |
| Expt 3 | ||||
| Wild type | ND | 3 (75) | 4 (100) | 3 (75) |
| ligB KO2 mutant | ND | 4 (100) | 4 (100) | 4 (100) |
Groups of four hamsters were inoculated with each challenge dose.
ND, not determined.
FIG. 2.
Pathology in hamsters infected with the ligB mutant. (A) Gross appearance of hamsters infected with the wild-type (wt) and mutant ligB mutant (KO2) strains and a representative uninfected control hamster. (B) Livers and kidneys from hamsters infected with the wild-type and ligB mutant strains of L. interrogans. Tissues were stained with hematoxylin and eosin (HE) (magnification, ×400), and the immunohistochemistry analysis was performed with antiserum specific for LipL32 (IHC) (magnification, ×1,000).
The virulence of the ligB mutant was evaluated using the rat model for renal colonization. In three separate experiments, groups of four or eight rats were infected intraperitoneally with 108 cells of either the ligB mutant or wild-type strain as previously described (1). Rats were sacrificed 15 days after infection. The ligB mutant behaved like the wild type. High levels of both strains were recovered in rat kidneys (Table 2). In hamster and rat experiments, the double-crossover disruptant KO2 was recovered from animals at the time of sacrifice and 2 weeks postchallenge, respectively, and the genotype was confirmed by PCR and immunoblot analysis (Fig. 1). This suggests that the ligB disruption was stable in the absence of selection.
TABLE 2.
Renal colonization of Rattus norvegicus with the wild-type and ligB mutant strains of L. interrogans Fiocruz L1-130 after experimental challenge
| Strain | No. animals with evidence of Leptospira renal colonization (% of total) based ona:
|
|
|---|---|---|
| Culture isolation | Immunofluorescence | |
| Expt 1 | ||
| Wild type | 6 (75) | 8 (100) |
| ligB KO2 mutant | 8 (100) | 8 (100) |
| Expt 2 | ||
| Wild type | 4 (100) | 4 (100) |
| ligB KO2 mutant | 4 (100) | 4 (100) |
| Expt 3 | ||
| Wild type | 8 (100) | 8 (100) |
| ligB KO2 mutant | 5 (63) | 5 (63) |
Groups of eight rats and groups of four rats were inoculated with 108 leptospires in experiments 1 and 3 and in experiment 2, respectively.
In vitro adherence of the ligB mutant to MDCK cells.
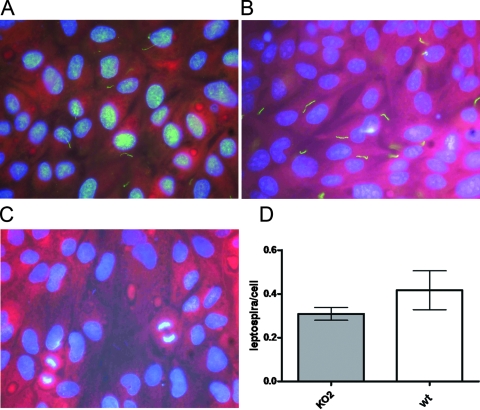
Interactions of L. interrogans wild-type and mutant strains with cultured epithelial cells were assayed by examining the adherence of leptospires to epithelial monolayers of MDCK cells. Cell monolayers were incubated using a multiplicity of infection of 100 bacteria per MDCK cell, and subsequent binding was quantified by microscopic analysis. There was not any statistically significant difference between the L. interrogans ligB mutant and the wild-type strain in the number of bacteria associated with MDCK cells (Fig. 3). These findings suggest that the ligB genotype does not influence in vitro host cell association.
FIG. 3.
Adherence of the L. interrogans ligB mutant to MDCK epithelial cells. The adherence of the L. interrogans wild-type (A) and ligB mutant KO2 (B) strains to MDCK epithelial cells was examined. Anti-Salmonella OmpA MAb was used as a control (C). Representative micrographs obtained by fluorescence microscopy are shown. (D) Attachment ratios (means ± standard deviations) determined using 10 random fields. wt, wild type.
DISCUSSION
Compared to other bacterial species, the genetic data for leptospires and determination of the molecular basis of the pathogenesis of these organisms are in their infancy. Analysis of the complete genome sequences of pathogenic Leptospira species revealed that more than 50% of the predicted open reading frames did not exhibit similarity to genes encoding proteins with known functions (8, 32, 39, 40). Until a method for constructing pathogenic leptospire mutants is developed, any function of leptospire proteins, including virulence factors, remains speculative. Previous attempts to inactivate genes in pathogenic Leptospira species have been unsuccessful. The putative role of LigB in virulence (21, 26, 34) prompted us to generate an L. interrogans ligB mutant.
We used approaches used previously for saprophytic Leptospira species (5, 16, 24, 25, 28, 37, 38, 45) to carry out gene targeting by homologous recombination in the pathogen L. interrogans. Although the efficiency of the transformations was low, our results show the feasibility of performing allelic exchange in pathogenic Leptospira spp. by homologous recombination. Our previous attempts to generate homologous recombination in L. interrogans were not successful (unpublished data), presumably due to the target gene chosen. The use of a large region of homologous DNA (more than 1 kb) may have increased the probability of homologous recombination. The ligAB locus appears to be the target of fragment rearrangements and recombination events. It has been suggested that ligA was created from ligB by gene duplication, since the fragments which encode the first six Big domains are identical in the two genes (26). Furthermore, sequence analysis of lig genes from collections of pathogenic Leptospira species resulted in evidence of recombination between Leptospira species at this locus (unpublished data). Finally, in addition to allelic exchange derived from a double homologous recombination event, we showed that targeted integration of a suicide plasmid which contains a 5′- and 3′-truncated fragment of the gene of interest can facilitate targeted mutagenesis in L. interrogans.
For members of the superfamily containing the Big proteins, previous studies have demonstrated that compared to the wild-type strain, an intimin-deficient enteropathogenic E. coli strain is defective for adherence to cultured cells and for intestinal colonization (33). Similarly, analysis of a Y. enterocolitica inv mutant suggested that invasin is necessary for efficient translocation of the bacteria across the intestinal epithelium (36). ligB is conserved among all pathogenic Leptospira species and is upregulated when bacteria confront the host environment (26, 34). Furthermore, expression of ligB is correlated with the virulence status of Leptospira strains (26). Therefore, the working hypothesis has been that LigB is essential for the bacteria to survive, disseminate, and/or colonize in the host.
Yet we found that loss of ligB was not associated with a loss of virulence phenotypes. Inoculation of ligB mutants produced the same acute disease manifestations and lethal outcomes that were observed in hamsters infected with wild-type strains. In addition, inoculation of ligB mutants resulted in efficient renal colonization in experimental rats similar to that observed with wild-type strains.
Moreover, we found that the L. interrogans ligB mutant was able to adhere to epithelial cells in vitro. The interaction of L. interrogans with host cells is critical for dissemination in the host (4). Although the LigB protein has been shown to bind in vitro to host extracellular matrix moieties (9, 23), our findings suggest that there may be other modes of leptospiral attachment to host epithelial cells. As a caveat, we did not examine whether ligB mutants bind to extracellular matrix components, including fibronectin, and further studies are required to evaluate this possibility. Furthermore, the mechanism of association of the pathogen with the host cell in vivo may be quite different than what is observed in vitro; therefore, we cannot exclude the possibility that Lig proteins mediate host cell interactions based on observations made with in vitro assays alone.
Because of the location of the Spcr cassette in the 3′ end of ligB, a truncated LigB protein could have been expressed. However, immunoblot analysis using polyclonal and monoclonal antibodies against recombinant fragments located upstream of the disruption site did not allow identification of any fragments in the mutant strains. Furthermore, whereas these antibodies stained strongly with the wild-type strain in immunofluorescence studies, no signal was associated with the mutant strains. It is therefore unlikely that a truncated ligB fragment was expressed in the mutants.
In the present study, the data obtained with the ligB mutant suggest that an absence of LigB does not lead to a loss of virulence and a loss of colonization in the acutely and chronically infected animal models, respectively. In our challenge experiments, hamsters and rats were infected by intraperitoneal inoculation of leptospires. We cannot exclude the possibility that LigB may play a role in penetration of the host or other early events during infection. Alternatively, the fact that the ligB mutant remained virulent may have been due to functional redundancy in the bacteria. The numerous lipoproteins which are present in leptospires (11) in addition to the LigB protein may compensate for the loss of LigB expression. Several surface-associated Leptospira proteins, including LigA, have been shown to interact in vitro with extracellular matrix components (2, 3, 18, 31, 44). Thus, the function of LigB may be replaced to various extents by other lipoproteins which may play a role in host-cell interactions. LigA and LigB proteins contain Big domains that may have redundant functions (9, 27, 34). Choy et al. demonstrated that domains within LigA and LigB proteins bind specifically to fibronectin in vitro (9). A phenotype distinct from that of the parental strain may occur only when both genes are disrupted. Therefore, further studies should include generation of ligA and ligAB mutants.
In conclusion, we demonstrated for the first time that site-directed homologous recombination can be successfully achieved in pathogenic Leptospira. The approaches used in this study, therefore, make it feasible to produce knockout mutations in putative virulence-associated genes in Leptospira and evaluate the roles that these genes may play in leptospiral pathogenesis.
Acknowledgments
This work was supported by a Fiocruz-Pasteur Scientific Cooperation Agreement, by the Brazilian National Research Council (Instituto Milênio 420067/2005), by the French Ministry of Research ANR Jeunes Chercheurs (no. 05-JCJC-0105-01), and by the National Institutes of Health (grants 5 R01 AI052473, 2 R01 AI034431, and 2 D43 TW00919). J. Croda received scholarship 2007/00083-2 from the Research Support Foundation of the State of São Paulo.
Editor: A. Camilli
Footnotes
Published ahead of print on 22 September 2008.
REFERENCES
- 1.Athanazio, D. A., E. F. Silva, C. S. Santos, G. M. Rocha, M. A. Vannier-Santos, A. J. McBride, A. I. Ko, and M. G. Reis. 2008. Rattus norvegicus as a model for persistent renal colonization by pathogenic Leptospira interrogans. Acta Trop. 105176-180. [DOI] [PubMed] [Google Scholar]
- 2.Atzingen, M. V., A. S. Barbosa, T. De Brito, S. A. Vasconcellos, Z. M. de Morais, D. M. Lima, P. A. Abreu, and A. L. Nascimento. 2008. Lsa21, a novel leptospiral protein binding adhesive matrix molecules and present during human infection. BMC Microbiol. 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbosa, A. S., P. A. Abreu, F. O. Neves, M. V. Atzingen, M. M. Watanabe, M. L. Vieira, Z. M. Morais, S. A. Vasconcellos, and A. L. Nascimento. 2006. A newly identified leptospiral adhesin mediates attachment to laminin. Infect. Immun. 746356-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barocchi, M. A., A. I. Ko, M. G. Reis, K. L. McDonald, and L. W. Riley. 2002. Rapid translocation of polarized MDCK cell monolayers by Leptospira interrogans, an invasive but nonintracellular pathogen. Infect. Immun. 706926-6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauby, H., I. Saint Girons, and M. Picardeau. 2003. Construction and complementation of the first auxotrophic mutant in the spirochaete Leptospira meyeri. Microbiology 149689-693. [DOI] [PubMed] [Google Scholar]
- 6.Bharti, A. R., J. E. Nally, J. N. Ricaldi, M. A. Matthias, M. M. Diaz, M. A. Lovett, P. N. Levett, R. H. Gilman, M. R. Willig, E. Gotuzzo, and J. M. Vinetz. 2003. Leptospirosis: a zoonotic disease of global importance. Lancet Infect. Dis. 3757-771. [DOI] [PubMed] [Google Scholar]
- 7.Bourhy, P., H. Louvel, I. Saint Girons, and M. Picardeau. 2005. Random insertional mutagenesis of Leptospira interrogans, the agent of leptospirosis, using a mariner transposon. J. Bacteriol. 1873255-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulach, D. M., R. L. Zuerner, P. Wilson, T. Seemann, A. McGrath, P. A. Cullen, J. Davis, M. Johnson, E. Kuczek, D. P. Alt, B. Peterson-Burch, R. L. Coppel, J. I. Rood, J. K. Davies, and B. Adler. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. USA 10314560-14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choy, H. A., M. M. Kelley, T. L. Chen, A. K. Moller, J. Matsunaga, and D. A. Haake. 2007. Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect. Immun. 752441-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croda, J., J. G. Ramos, J. Matsunaga, A. Queiroz, A. Homma, L. W. Riley, D. A. Haake, M. G. Reis, and A. I. Ko. 2007. Leptospira immunoglobulin-like proteins as a serodiagnostic marker for acute leptospirosis. J. Clin. Microbiol. 451528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen, P. A., D. A. Haake, and B. Adler. 2004. Outer membrane proteins of pathogenic spirochetes. FEMS Microbiol. Rev. 28291-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullen, P. A., X. Xu, J. Matsunaga, Y. Sanchez, A. I. Ko, D. A. Haake, and B. Adler. 2005. Surfaceome of Leptospira spp. Infect. Immun. 734853-4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellinghausen, H. C., and W. G. McCullough. 1965. Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am. J. Vet. Res. 2645-51. [PubMed] [Google Scholar]
- 14.Faine, S. A., B. Bolin, C., and P. Perolat. 1999. Leptospira and leptospirosis, 2nd ed. MediSci, Melbourne, Australia.
- 15.Girons, I. S., P. Bourhy, C. Ottone, M. Picardeau, D. Yelton, R. W. Hendrix, P. Glaser, and N. Charon. 2000. The LE1 bacteriophage replicates as a plasmid within Leptospira biflexa: construction of an L. biflexa-Escherichia coli shuttle vector. J. Bacteriol. 1825700-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guegan, R., J. M. Camadro, I. Saint Girons, and M. Picardeau. 2003. Leptospira spp. possess a complete haem biosynthetic pathway and are able to use exogenous haem sources. Mol. Microbiol. 49745-754. [DOI] [PubMed] [Google Scholar]
- 17.Haake, D. A., G. Chao, R. L. Zuerner, J. K. Barnett, D. Barnett, M. Mazel, J. Matsunaga, P. N. Levett, and C. A. Bolin. 2000. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 682276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauk, P., F. Macedo, E. C. Romero, S. A. Vasconcellos, Z. M. de Morais, A. S. Barbosa, and P. L. Ho. 2008. In LipL32, the major leptospiral lipoprotein, the C terminus is the primary immunogenic domain and mediates interaction with collagen IV and plasma fibronectin. Infect. Immun. 762642-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, R. C., and V. G. Harris. 1967. Differentiation of pathogenic and saprophytic leptospires. J. Bacteriol. 9427-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko, A. I., M. Galvão Reis, C. M. Ribeiro Dourado, W. D. J. Johnson, and L. W. Riley. 1999. Urban epidemic of severe leptospirosis in Brazil. Salvador Leptospirosis Study Group. Lancet 354820-825. [DOI] [PubMed] [Google Scholar]
- 21.Koizumi, N., and H. Watanabe. 2004. Leptospiral immunoglobulin-like proteins elicit protective immunity. Vaccine 221545-1552. [DOI] [PubMed] [Google Scholar]
- 22.Levett, P. N. 2001. Leptospirosis. Clin. Microbiol. Rev. 14296-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, Y. P., and Y. F. Chang. 2007. A domain of the Leptospira LigB contributes to high affinity binding of fibronectin. Biochem. Biophys. Res. Commun. 362443-448. [DOI] [PubMed] [Google Scholar]
- 24.Louvel, H., J. M. Betton, and M. Picardeau. 2008. Heme rescues a two-component system Leptospira biflexa mutant. BMC Microbiol. 825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louvel, H., S. Bommezzadri, N. Zidane, C. Boursaux-Eude, S. Creno, A. Magnier, Z. Rouy, C. Medigue, I. Saint Girons, C. Bouchier, and M. Picardeau. 2006. Comparative and functional genomic analyses of iron transport and regulation in Leptospira spp. J. Bacteriol. 1887893-7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsunaga, J., M. A. Barocchi, J. Croda, T. A. Young, Y. Sanchez, I. Siqueira, C. A. Bolin, M. G. Reis, L. W. Riley, D. A. Haake, and A. I. Ko. 2003. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol. Microbiol. 49929-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsunaga, J., Y. Sanchez, X. Xu, and D. A. Haake. 2005. Osmolarity, a key environmental signal controlling expression of leptospiral proteins LigA and LigB and the extracellular release of LigA. Infect. Immun. 7370-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazouni, K., G. Pehau-Arnaudet, P. England, P. Bourhy, I. Saint Girons, and M. Picardeau. 2006. The Scc spirochetal coiled-coil protein forms helix-like filaments and binds to nucleic acids generating nucleoprotein structures. J. Bacteriol. 188469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBride, A. J., D. A. Athanazio, M. G. Reis, and A. I. Ko. 2005. Leptospirosis. Curr. Opin. Infect. Dis. 18376-386. [DOI] [PubMed] [Google Scholar]
- 30.Merien, F., G. Baranton, and P. Perolat. 1997. Invasion of Vero cells and induction of apoptosis in macrophages by pathogenic Leptospira interrogans are correlated with virulence. Infect. Immun. 65729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merien, F., J. Truccolo, G. Baranton, and P. Perolat. 2000. Identification of a 36-kDa fibronectin-binding protein expressed by a virulent variant of Leptospira interrogans serovar icterohaemorrhagiae. FEMS Microbiol. Lett. 18517-22. [DOI] [PubMed] [Google Scholar]
- 32.Nascimento, A. L., A. I. Ko, E. A. Martins, C. B. Monteiro-Vitorello, P. L. Ho, D. A. Haake, S. Verjovski-Almeida, R. A. Hartskeerl, M. V. Marques, M. C. Oliveira, C. F. Menck, L. C. Leite, H. Carrer, L. L. Coutinho, W. M. Degrave, O. A. Dellagostin, H. El-Dorry, E. S. Ferro, M. I. Ferro, L. R. Furlan, M. Gamberini, E. A. Giglioti, A. Goes-Neto, G. H. Goldman, M. H. Goldman, R. Harakava, S. M. Jeronimo, I. L. Junqueira-de-Azevedo, E. T. Kimura, E. E. Kuramae, E. G. Lemos, M. V. Lemos, C. L. Marino, L. R. Nunes, R. C. de Oliveira, G. G. Pereira, M. S. Reis, A. Schriefer, W. J. Siqueira, P. Sommer, S. M. Tsai, A. J. Simpson, J. A. Ferro, L. E. Camargo, J. P. Kitajima, J. C. Setubal, and M. A. Van Sluys. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 1862164-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nougayrède, J. P., P. J. Fernandes, and M. S. Donnenberg. 2003. Adhesion of enteropathogenic Escherichia coli to host cells. Cell. Microbiol. 5359-372. [DOI] [PubMed] [Google Scholar]
- 34.Palaniappan, R. U., Y. F. Chang, S. S. Jusuf, S. Artiushin, J. F. Timoney, S. P. McDonough, S. C. Barr, T. J. Divers, K. W. Simpson, P. L. McDonough, and H. O. Mohammed. 2002. Cloning and molecular characterization of an immunogenic LigA protein of Leptospira interrogans. Infect. Immun. 705924-5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palaniappan, R. U., S. P. McDonough, T. J. Divers, C. S. Chen, M. J. Pan, M. Matsumoto, and Y. F. Chang. 2006. Immunoprotection of recombinant leptospiral immunoglobulin-like protein A against Leptospira interrogans serovar Pomona infection. Infect. Immun. 741745-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pepe, J. C., and V. L. Miller. 1993. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc. Natl. Acad. Sci. USA 906473-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Picardeau, M., H. Bauby, and I. Saint Girons. 2003. Genetic evidence for the existence of two pathways for the biosynthesis of methionine in the Leptospira spp. FEMS Microbiol. Lett. 225257-262. [DOI] [PubMed] [Google Scholar]
- 38.Picardeau, M., A. Brenot, and I. Saint Girons. 2001. First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol. Microbiol. 40189-199. [DOI] [PubMed] [Google Scholar]
- 39.Picardeau, M., D. M. Bulach, C. Bouchier, R. L. Zuerner, N. Zidane, P. J. Wilson, S. Creno, E. S. Kuczek, S. Bommezzadri, J. C. Davis, A. McGrath, M. J. Johnson, C. Boursaux-Eude, T. Seemann, Z. Rouy, R. L. Coppel, J. I. Rood, A. Lajus, J. K. Davies, C. Medigue, and B. Adler. 2008. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS ONE 3e1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren, S. X., G. Fu, X. G. Jiang, R. Zeng, Y. G. Miao, H. Xu, Y. X. Zhang, H. Xiong, G. Lu, L. F. Lu, H. Q. Jiang, J. Jia, Y. F. Tu, J. X. Jiang, W. Y. Gu, Y. Q. Zhang, Z. Cai, H. H. Sheng, H. F. Yin, Y. Zhang, G. F. Zhu, M. Wan, H. L. Huang, Z. Qian, S. Y. Wang, W. Ma, Z. J. Yao, Y. Shen, B. Q. Qiang, Q. C. Xia, X. K. Guo, A. Danchin, I. Saint Girons, R. L. Somerville, Y. M. Wen, M. H. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422888-893. [DOI] [PubMed] [Google Scholar]
- 41.Ristow, P., P. Bourhy, F. W. da Cruz McBride, C. P. Figueira, M. Huerre, P. Ave, I. S. Girons, A. I. Ko, and M. Picardeau. 2007. The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 3e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva, E. F., M. A. Medeiros, A. J. McBride, J. Matsunaga, G. S. Esteves, J. G. Ramos, C. S. Santos, J. Croda, A. Homma, O. A. Dellagostin, D. A. Haake, M. G. Reis, and A. I. Ko. 2007. The terminal portion of leptospiral immunoglobulin-like protein LigA confers protective immunity against lethal infection in the hamster model of leptospirosis. Vaccine 256277-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srimanote, P., N. Wongdeethai, P. Jieanampunkul, S. Samonkiert, C. Leepiyasakulchai, T. Kalambaheti, and V. Prachayasittikul. 2008. Recombinant ligA for leptospirosis diagnosis and ligA among the Leptospira spp. clinical isolates. J. Microbiol. Methods 7273-81. [DOI] [PubMed] [Google Scholar]
- 44.Stevenson, B., H. A. Choy, M. Pinne, M. L. Rotondi, M. C. Miller, E. Demoll, P. Kraiczy, A. E. Cooley, T. P. Creamer, M. A. Suchard, C. A. Brissette, A. Verma, and D. A. Haake. 2007. Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS ONE 2e1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tchamedeu Kameni, A. P., E. Couture-Tosi, I. Saint-Girons, and M. Picardeau. 2002. Inactivation of the spirochete recA gene results in a mutant with low viability and irregular nucleoid morphology. J. Bacteriol. 184452-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.World Health Organization. 1999. Leptospirosis worldwide, 1999. Wkly. Epidemiol. Rec. 74237-242. [PubMed] [Google Scholar]