Abstract
Several Borrelia burgdorferi genes induced under mammalian host conditions have been purported to be important in Lyme disease pathogenesis based on their binding to host structures. These genes include the dbpBA locus, whose products bind host decorin and glycosoaminoglycans. Recently, the dbpBA genes were reported to be involved in borrelial infectivity. Here we extended the previous observations by using culture and quantitative PCR to evaluate low- and high-dose murine infection by a ΔdbpBA::Gentr derivative of B. burgdorferi strain B31. The results indicate that the ΔdbpBA::Gentr mutant is attenuated in the ability to initially colonize and then persist in multiple tissues. The mutant exhibited a colonization defect as early as 3 days postinfection, before the development of an adaptive immune response, and after low-dose infection of SCID mice, which are deficient in adaptive immunity. These findings suggest that the inability to adhere to host decorin may promote clearance of B. burgdorferi, presumably via innate immune mechanisms. In a high-dose infection, the mutant disseminated to several tissues, particularly joint tissue, but it was generally cleared from these tissues by 3 weeks postinfection. Finally, following high-dose infection of SCID mice, the dbpBA mutant exhibited only a mild colonization defect, suggesting that the adaptive response is involved in the clearance of the mutant in immunocompetent mice. Taken together, these results suggest that the DbpBA proteins facilitate the colonization of multiple tissues by B. burgdorferi and are required for optimal resistance to both innate and adaptive immune mechanisms following needle inoculation.
Borrelia burgdorferi is the etiologic agent of Lyme disease and traffics within an enzootic cycle that involves an arthropod vector and rodent mammalian reservoirs, but it can also infect other mammalian species, including humans. In humans, the bite of an infected Ixodes tick usually results in a red skin lesion, designated erythema migrans, and the illness is accompanied by general malaise and, in some cases, cardiac and neurologic sequela (for reviews, see references 39 and 58). Individuals who do not seek antibiotic therapy at this stage of the infection are at risk for developing manifestations associated with late Lyme disease, which in the United States are usually arthritis. In Europe, a neurologic pathology and an inflammatory skin condition known as acrodermatitis chronica atrophicans can occur in chronically infected individuals. As such, in areas where it is endemic, Lyme disease contributes to significant morbidity.
By virtue of its ability to transition between ticks and mammals, B. burgdorferi must modify gene expression quickly to adapt to such disparate environments. Previous studies using transcriptional profiling demonstrated that B. burgdorferi gene expression changes in response to pH, temperature, redox status, exposure to blood, and as-yet-uncharacterized mammal-specific factors (1, 2, 5, 10, 11, 15, 17, 26, 36, 40, 48, 51, 52, 59, 60, 63). One set of genes transcribed under conditions that mimic host-adapted conditions (either increased temperature, lower pH, redox status, or blood exposure) includes the dbpA and dbpB genes (5, 26, 40, 48, 52, 60), which are apparently cotranscribed from dbpB to dbpA (i.e., dbpBA) (22) and encode adhesins that bind to mammalian decorin (20, 21). Decorin is abundant in the extracellular matrix (ECM) of the dermal skin layer, as well as in joint tissue (7, 34, 43). The induction of dbpBA under simulated host conditions is consistent with the hypothesis that these genes have a role in borrelial pathogenesis, and the DbpBA proteins, along with other adhesins, are presumed to assist in cell and ECM attachment of B. burgdorferi to mediate colonization and initiate dissemination within the mammalian host (7, 20, 21, 34). To investigate this hypothesis, the dbpBA genes were deleted, and the resulting mutant was tested using BALB/c mice (56). In their initial study, Shi et al. concluded that DbpA and DbpB were not essential for mouse infection, although only 58% of the tissues were culture positive when a single large inoculum (105 B. burgdorferi cells) was used and no genetic complementation of intact dbpBA was evaluated (56). Another study indicated that the constitutive overexpression of dbpBA makes B. burgdorferi more infectious but impairs its ability to disseminate, suggesting that DbpBA has an important role in experimental infections (62). A more recent study by Shi et al. indicated that the dbpBA genes are required for virulence in mice following needle inoculation (55), a finding that was corroborated recently by Blevins et al. (4). Due to the differences in the studies mentioned above, the role of DbpBA in borrelial pathogenesis was independently evaluated and extended in the current study.
In this report, we demonstrate that deletion of dbpBA results in marked attenuation of B. burgdorferi in immunocompetent C3H mice following needle inoculation and we suggest that binding to host decorin early in the infectious process may be important in preventing clearance of B. burgdorferi via the innate immune response. The ability of the dbpBA mutant to colonize immunodeficient mice at a high-dose inoculum with which immunocompetent mice are effectively not infected suggests that, in addition to the early defect in survival observed, adaptive immunity further reduces the numbers of dbpBA mutant cells. These results suggest that binding to decorin within the ECM represents an important early step in the colonization of B. burgdorferi that promotes the establishment and persistence of this spirochete during experimental infection.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. All B. burgdorferi strains were grown in complete BSK-II medium as described previously (57). For selective pressure, B. burgdorferi was grown in BSK-II medium with antibiotics, where appropriate, at the following concentrations: kanamycin, 300 μg/ml; streptomycin, 50 μg/ml; and gentamicin, 50 μg/ml. All Escherichia coli strains were grown with aeration in LB media at 37°C. For experiments involving E. coli, antibiotics were used at the following concentrations: carbenicillin, 100 μg/ml; ampicillin, 100 μg/ml; spectinomycin, 100 μg/ml; gentamicin, 5 μg/ml; chloramphenicol, 15 μg/ml; and kanamycin, 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristics | Reference(s) or source |
|---|---|---|
| B. burgdorferi strains | ||
| ML23 | Missing lp25 | 30, 31 |
| ML23/pBBE22 | Missing lp25, complemented with BBE22 | 54 |
| JF105 | ML23 ΔdbpBA::Gentr | This study |
| JF105/pBBE22 | ML23 ΔdbpBA::Gentr, complemented with BBE22 | This study |
| JF105/pJBF17 | ML23 ΔdbpBA::Gentr, complemented with BBE22 and dbpBA | This study |
| E. coli strains | ||
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| ccdB survival T1R | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG tonA::Ptrc-ccdA | Invitrogen |
| Plasmids | ||
| pCR2.1-TOPO | Ampr Kanr; PCR cloning vector | Invitrogen |
| pCR-XL-TOPO | Kanr; PCR cloning vector | Invitrogen |
| pCR8/GW/TOPO | Spcr; Gateway PCR cloning/entry vector | Invitrogen |
| pDEST17 | Camr Ampr; Gateway destination vector | Invitrogen |
| pBSV2G | Gentr; borrelial shuttle vector | 16 |
| pBBE22 | Kanr; borrelial shuttle vector carrying BBE22 | 46 |
| pBBE22gate | Camr Kanr; pBBE22 modified to be a Gateway destination vector containing attR sitesa | This study |
| pNP3 | Gentr Kanr; ΔdbpBA::Gentr construct containing region of lp54 that contains 1.5 kb upstream and downstream of the end of dbpA (BBA24) and the start of dbpB (BBA25) | This study |
| pJBF12 | Spcr; intact dbpBA cloned into pCR8/GW/TOPO flanked by attL sites | This study |
| pJBF17 | Kanr; intact dbpBA recombined via Gateway system into pBBE22gate | This study |
See Materials and Methods.
PCR.
A PCR was conducted using either Herculase polymerase (Stratagene, La Jolla, CA) or Taq polymerase (Supermix; Invitrogen, Carlsbad, CA) as previously described (26, 52, 54). The oligonucleotide primers used in this study are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′) | Description |
|---|---|---|
| BBA21F | GATAAATAAAAGAAATTTATCTGAAA | Primer pair that amplifies a 1.7-kb fragment containing bba21 to sequences downstream of bba24 (dbpA) |
| dbpAsalmut | ACGCGTCGACATCCTTCTTTTACTGATGA | |
| BBA31com | CGCATTTGCAAGAGAATCAAGCGCATCGTCTT | Primer pair that amplifies a 2.8-kb fragment containing bba31 to sequences upstream of bba25 (dbpB) |
| dbpBsalmut | ACGCGTCGACCAAGACCATAACTATTGAATT | |
| 5pflgGentSal | ACGCGTCGACGAACGAATTGTTAGGTGGCGGTACT | Primer pair that amplifies a 0.9-kb fragment containing the aacC1 allele (Gentr) fused to the borrelial flgB promoter (PflgB-Gentr) in the ΔdbpBA::Gentr mutant only |
| 3pflgGentSal | ACGCGTCGACCCCGAGCTTCAAGGAAGATTTCCT | |
| 5ABMut | TTTATGTCTTGATTATCGGGCGAAGAGTTTA | When used with 3ABMut, amplifies 1.6- and 1.4-kb bands in the parent and ΔdbpBA::Gentr mutant, respectively; when used with 5pflgGentSal, amplifies a 1.1-kb fragment in the ΔdbpBA::Gentr mutant only |
| 3ABMut | AAGCCAGATTGCATAGCAAGCTTGAATTCCAA | When used with 3pflgGentSal, amplifies a 1.2-kb fragment in the ΔdbpBA::Gentr mutant only |
| ATTR1F | GCATGCCCTCGAATCAACAAGTTTG | Primer pair used to amplify the attR1-Camr-ccdB-attR2 region from pDEST17 for cloning into borrelial shuttle vectors to convert them to Gateway amenable vectors |
| ATTR1R | GCATGCCGCAGCCTCGAATCAACCAC | |
| DbpBA-F-XmaNco | CCCGGGCCATGGCTTGATTATCGGGCGAAGAG | Primer pair that amplifies a 1.7-kb fragment containing the intact dbpBA locus with 289 bp upstream and 131 bp downstream for cloning into pCR8/GW/TOPO |
| DbpBA-R-XmaNde | CCCGGGCATATGGCAAACTGGAAACAAGTC | |
| nTM17FrecA | GTGGATCTATTGTATTAGATGAGGCTCTCG | Primer pair used for real-time quantitative PCR detection of B. burgdorferi recA genea |
| nTM17RrecA | GCCAAAGTTCTGCAACATTAACACCTAAAG | |
| bactinF | CAAGTCATCACTATTGGCAACGA | Primer pair used for real-time quantitative PCR detection of the murine β-actin geneb |
| bactinR | CCAAGAAGGAAGGCTGGAAAA |
Construction of the ΔdbpBA::Gentr plasmid.
To delete the dbpBA operon in B. burgdorferi, a 1.7-kb fragment that included the 3′ end of dbpA (BBA24) and sequences downstream, including part of BBA21, was amplified using the BBA21F and dbpAsalmut primers and borrelial genomic DNA as the template (Table 2). Next, a 2.8-kb fragment containing the start of dbpB (BBA25) and its upstream region, including a portion of BBA36, was amplified similarly using the BBA31com and dbpBsalmut primers (Table 2). Both fragments were cloned separately into the TA cloning vector pCR-XL-TOPO (Invitrogen), and transformants were selected for with kanamycin. After the sequences of the 1.7- and 2.8-kb fragments were confirmed, the 1.7-kb BBA21-dbpA-SalI fragment was purified following digestion with BamHI and SalI. The resulting product was then cloned into the SalI-BBA31-dbpB SalI fragment construct mentioned above that also was digested with BamHI and SalI. The resulting construct was missing most of the dbpB and dbpA sequences, and a single unique SalI site was generated in place of these sequences. The gentamicin resistance cassette was amplified using pBVS2G (generously provided by Patricia Rosa [16]) as the template, which contained the aacC1 gentamicin resistance (Gentr) gene linked to the strong borrelial flgB promoter, using 5flgGentsal and 3flgGentsal as the primers (Table 2). The resulting amplified product was TA cloned into the pCR-XL-TOPO vector, and E. coli transformants were selected based on coresistance to kanamycin and gentamicin. The resulting clone was sequenced, digested with SalI, and cloned into the unique SalI site engineered in the dbpBA deletion construct. E. coli transformants containing the Gentr dbpBA deletion construct were selected based on coresistance to kanamycin and gentamicin. The final construct containing the PflgB-Gentr cassette in place of dbpBA (ΔdbpBA::Gentr) was screened by digestion with SalI, confirmed by sequencing, and subsequently designated pNP3.
Gateway vector-borrelial shuttle vector construction.
To isolate a borrelial shuttle vector that could be easily modified using Invitrogen's Gateway recombination-based cloning system, the attR1-Camr-ccdB-attR2 region from pDEST17 (Invitrogen) that confers resistance to chloramphenicol was PCR amplified with SphI linkers using oligonucleotide primers ATTR1F and ATTR1R (Table 2). The amplified product was cloned into pCR2.1-TOPO (Invitrogen) and transformed into ccdB survival T1R cells. The resulting construct was digested with SphI, and the SphI-attR1-Camr-ccdB-attR2-SphI-containing fragment was cloned into pBBE22 (generously provided by Steve Norris [46]) cut with SphI, ligated, and transformed into ccdB survival T1R cells (Invitrogen). Transformants were selected with kanamycin and chloramphenicol, and the resulting plasmid construct was designated pBBE22gate. Subsequently, the dbpBA genes with 289 bp upstream and 131 bp downstream of the open reading frame was PCR amplified with oligonucleotide primers DbpBA-F-XmaNco and DbpBA-R-XmaNde (Table 2), and the resulting 1,689-bp band was cloned into the entry vector pCR8/GW/TOPO (Spcr; Invitrogen) that contains attL sites that flank the inserted fragment. Clones that conferred resistance to spectinomycin were screened, and the desired construct was designated pJBF12. An LR reaction, which involved the LR recombinase that promotes recombination with constructs containing attL and attR sites, was performed with pJBF12 and pBBE22gate, respectively, by using the manufacturer's specifications, except that both plasmids were incubated at 55°C for 10 min to relax supercoiling prior to addition of the LR recombinase. The LR reaction mixture was then incubated at 25°C for 1 h and then overnight at 15°C. Then the LR reaction mixture was transformed into DH5α cells, and transformants were selected with kanamycin and screened for sensitivity to chloramphenicol. The resulting plasmid with dbpBA recombined into pBBE22gate (resulting in concomitant loss of Camr and ccdB) was designated pJBF17.
Genetic manipulation of B. burgdorferi.
The B. burgdorferi strain B31 derivative ML23 (Table 1) was made electrocompetent as previously described (54) and transformed with pNP3 that contained the ΔdbpBA::Gentr construct. Transformants were selected by limiting dilution in liquid BSK-II containing gentamicin using 96-well plates as described previously (64) to obtain strain JF105 (ML23 ΔdbpBA::Gentr) (Table 1). Strain JF105 was then made electrocompetent and transformed with either pBBE22 (Kanr) to complement the infectivity defect inherent in the lp25-deficient ML23 strain background or with pJBF17 (Kanr) to complement the infectivity defect and provide intact dbpBA (Table 1).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analyses were performed as previously described (26, 52-54). Antiserum to DbpA was used in conjunction with appropriate substrates for chemiluminescent detection as described previously (26, 52-54).
Mouse infectivity: ID50 analysis.
The 50% infective doses (ID50) of ML23/pBBE22, JF105/pBBE22 (ΔdbpBA::Gentr), and JF105/pJBF17 were determined as previously described (31, 54). C3H/HeN mice were infected for 21 days, sacrificed, and processed to determine whether organs (skin at the site on inoculation, spleen, heart, bladder, lymph nodes, and tibiotarsal joint) were colonized by examining growth in BSK-II medium for 1 month after the animals were sacrificed. Each of the infectivity experiments was conducted two separate times over a 5-month period to assess reproducibility.
Mouse infectivity: kinetics-based analysis.
C3H/HeN mice were infected intradermally with either 103 or 105 cells of ML23/pBBE22, JF105/pBBE22 (ΔdbpBA::Gentr), or JF105/pJBF17, and 12 h later the mice were sacrificed and skin samples from the site of inoculation were cultured in BSK-II medium (31, 54). Additional kinetics-based infectivity analyses were conducted essentially as described above on days 3, 5, 7, 14, and 21 (30). Groups of mice were infected using inocula containing 103 and 105 cells for each B. burgdorferi strain tested (except ML23/pBBE22, for which only an inoculum containing 103 cells was used) in two separate infectivity experiments. Following sacrifice at the time points indicated above, the tissues described above for the ID50 analysis were removed and cultured in BSK-II medium.
Mouse infectivity: SCID mouse infectivity.
C3H-SCID mice (Harlan Laboratories, Philadelphia, PA) were infected with 103 and 105 cells of ML23/pBBE22, JF105/pBBE22 (ΔdbpBA::Gentr), and JF105/pJBF17 as previously described (30), except that the mice were sacrificed after 21 days.
Extraction of DNA from tissue for quantitative PCR analysis.
Joint and skin samples were aseptically removed and placed in 200 μl of cold phosphate-buffered saline. Total genomic DNA was extracted using a Roche High Pure PCR template preparation kit as described previously (37) except that a 2% collagenase solution (Sigma Aldrich) was added prior to the overnight incubation at 55°C.
Quantification of B. burgdorferi in infected tissues.
B. burgdorferi genomic equivalents in infected tissues were enumerated using an Applied Biosystems ABI 7500 fast real-time PCR system (Applied Biosystems Corp., Foster City, CA) in conjunction with Sybr green PCR Mastermix (Applied Biosystems). Approximately 100 ng of DNA was added to each reaction mixture. B. burgdorferi genome copies were detected using oligonucleotide primers nTM17FrecA and nTM17RrecA (Table 2) (35) at a final concentration of 0.3 μM to specifically detect the B. burgdorferi recA gene. The numbers of borrelial genomic copies were calculated by comparing the threshold cycle (CT) values with the values for serial dilutions of known amounts of B. burgdorferi genomic DNA, which were used as standards. Mouse genomic copies were detected using oligonucleotide primers bactinF and bactinR, which are specific for the mouse β-actin gene, at a final concentration of 0.3 μM. The numbers of β-actin copies were calculated by comparing the CT values with the values for serial dilutions of known amounts of the β-actin gene, which were used as standards (37). Triplicate measurements were obtained for all samples, and the values shown below are the numbers of B. burgdorferi recA copies per 106 mouse β-actin copies.
Statistical analysis.
Quantitative PCR data were compared by transforming the number of B. burgdorferi genome copies (Nc) (normalized using 106 copies of mouse β-actin) to log10(Nc + 0.1) prior to one-factor (see Fig. 2) or two-factor (see Fig. 3A and 3B) analysis of variance. Parent, mutant, and complement copy numbers were compared by constructing appropriate orthogonal contrasts using the analysis of variance model coefficients. Calculations were performed using S-PLUS 7 for Windows (Insightful Corporation, Seattle, WA).
FIG. 2.
Quantitative real-time PCR analysis of the B. burgdorferi parental (ML23/pBBE22), ΔdbpBA::Gentr (JF105/pBBE22), and genetically complemented (JF105/pJBF17) strains in joint tissue following 21 days of infection in C3H mice. The absolute numbers of B. burgdorferi cells obtained from joint tissue were determined using mice infected with all three strains at a dose of 103 organisms. To normalize for mouse tissue in each sample tested, the number of β-actin copies was also determined. The results are expressed as the number of B. burgdorferi genome copies per 106 mouse β-actin copies. P values are indicated above the data.
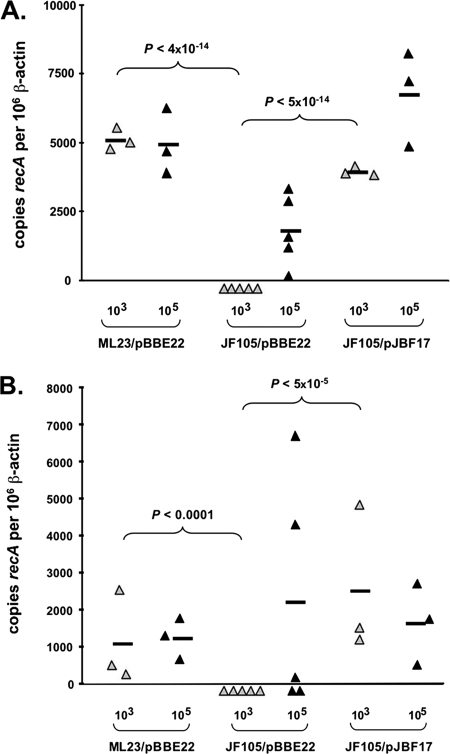
FIG. 3.
Quantitative PCR to determine the absolute numbers of spirochetes in joint tissue (A) and skin tissue (B) after 21 days of infection of C3H-SCID mice by the B. burgdorferi parental, ΔdbpBA::Gentr, and genetically complemented strains at the inocula indicated. The results are expressed as the number of B. burgdorferi genome copies per 106 mouse β-actin copies. P values are indicated above the data sets that are being compared.
RESULTS
Isolation of ΔdbpBA::Gentr mutant of B. burgdorferi.
To isolate a dbpBA deletion in B. burgdorferi, the region of the 54-kb linear plasmid (lp54) that encodes dbpBA (BBA24 [dbpA] and BBA25 [dbpB]) was amplified by PCR such that the genes were eliminated and a unique restriction site was generated simultaneously. Subsequently, a borrelial promoter-gentamicin resistance cassette was cloned into the engineered site to generate the ΔdbpBA::Gentr construct pNP3 (Fig. 1A). In order to isolate a ΔdbpBA::Gentr strain, pNP3 was transformed into strain ML23, which lacks the 25-kb linear plasmid (lp25) and, due to the absence of a restriction/modification system, is more readily transformable (54). Gentamicin-resistant isolates were obtained and analyzed by PCR (Fig. 1B) and Southern blotting (data not shown) to confirm the desired deletion. PCR analysis indicated that a 1.4-kb product was amplified from the ΔdbpBA::Gentr strain and the genetically complemented strain when primers 5ABmut and 3ABmut were used, whereas a 1.6-kb fragment was obtained when the same primers were used to amplify the native B. burgdorferi dbpBA region of lp54 (Fig. 1A and 1B). A smaller faint band was observed when the parent strain was used as the template, but it was slightly below the 1.4-kb band obtained for the ΔdbpBA::Gentr strain. For the genetically complemented strain both the 1.4- and 1.6-kb fragments were amplified, consistent with the presence of ΔdbpBA::Gentr and wild-type dbpBA, respectively (Fig. 1A and 1B). As expected, primers 3pflgGentSal and 5pflgGentSal amplified a product only from the mutant and genetically complemented strain, as did other primer combinations that amplified sequences only from strains carrying the ΔdbpBA::Gentr allele (i.e., either primers 5ABmut and 5pflgGentSal or primers 3ABmut and 3pflgGentSal), and the parent was negative for all of the PCRs when these three primer pairs were used (Fig. 1A and 1B). Western blot analysis confirmed that DbpA was not produced in the isolates screened (Fig. 1C, compare lane 1 [parent] with lane 2 [ΔdbpBA::Gentr]). Subsequent plasmid profile analysis indicated that all of the ΔdbpBA::Gentr clones obtained did not contain the 9-kb circular plasmid (cp9) and lp25, like the parent strain ML23 (31; data not shown). One clone, designated JF105, was used for all subsequent analyses.
FIG. 1.
Isolation and confirmation of dbpBA deletion mutants of infectious B. burgdorferi. (A) Strategy for deleting dbpBA. Plasmid pNP3, in which the BBA24 (dbpA) and BBA25 (dbpB) genes from lp54 were replaced by the gentamicin resistance gene aacC1, was used to transform B. burgdorferi strain B31 isolate ML23. Transformants were screened by PCR using primers shown to confirm the dbpBA deletion. Arrows 1 through 4 represent oligonucleotide primers 5ABMut, 3ABMut, 3pflgGentSal, and 5pflgGentSal, respectively (Table 2). One isolate that retained all plasmids (except cp9 and lp25) was obtained and designated JF105. (B) PCR analysis of the dbpBA deletion mutant and complemented strain. Lane 1, ML23/pBBE22 (parental strain); lane 2, JF105/pBBE22 (ΔdbpBA::Gentr); lane 3, JF105/pJBF17 (ΔdbpBA::Gentr with intact dbpBA). The primers used are indicated on the right (see panel A). (C) Western blot analysis of the ΔdbpBA::Gentr mutant and the genetically complemented strain. Samples were immunoblotted using antiserum specific for DbpA. Lane 1, ML23/pBBE22 (parental strain); lane 2, JF105/pBBE22 (ΔdbpBA::Gentr); lanes 3 and 4, JF105/pJBF17 (ΔdbpBA::Gentr with intact dbpBA). Lanes 1 and 2 contained protein from 1 × 107 borrelial cells, while lanes 3 and 4 contained whole-cell equivalents from 1.25 × 106 and 2.5 × 106 organisms, respectively. The arrow indicates the location of DbpA.
Genetic complementation of the ΔdbpBA::Gentr mutant.
The genetic background used for isolation of ΔdbpBA::Gentr strain JF105 was ML23, which is a strain B31 derivative that is missing lp25 (54). The lp25 plasmid is absolutely required for survival in mammals due to the presence of the BBE22 gene, which encodes a nicotinamidase essential for growth in these hosts (46). In order to use this genetic background to determine how the loss of a specific gene affects B. burgdorferi pathogenesis, the BBE22-containing region must be provided in trans, as was done previously for the BBK32::Strr strain JS315 (54). In addition, the genes that are inactivated or deleted need to be provided back for genetic complementation. To facilitate this, the borrelial shuttle vector pBBE22, which carries BBE22 and BBE23 (46), was modified with Invitrogen's Gateway system, which resulted in a plasmid backbone that allowed efficient recombination of target genes in conjunction with commercially available Gateway entry vectors. The resulting construct, designated pBBE22gate, was used to construct plasmids carrying dbpBA with its native promoter region, which yielded plasmid pJBF17. Plasmids pBBE22 and pJBF17 were then separately transformed into JF105. Following selection, the transformants were screened by PCR, Southern blotting, and plasmid profiling to ensure that the desired constructs were obtained and that no additional loss of plasmids had occurred (Fig. 1B and data not shown). The strains obtained had the same plasmid profiles as their genetic parent (data not shown). Furthermore, all the strains tested had similar growth kinetics in BSK-II medium (data not shown). Subsequently, immunoblot analysis was conducted to assess DbpA production (Fig. 1C). Protein lysates were prepared for ML23/pBBE22 (parental strain) (Fig. 1C, lane 1), JF105/pBBE22 (ΔdbpBA::Gentr) (lane 2), and JF105/pJBF17 (ΔdbpBA::Gentr with intact dbpBA) (lanes 3 and 4). Lanes 1 and 2 contained protein from 1 × 107 borrelial cells, while lanes 3 and 4 contained whole-cell equivalents from 1.25 × 106 and 2.5 × 106 organisms, respectively, and were immunoblotted and probed with antiserum specific for DbpA (Fig. 1C). The intensities of the bands in lanes 1 and 3 are approximately the same, indicating that genetic complementation of dbpBA on a borrelial shuttle vector resulted in an approximately eightfold increase in DbpA production. Serial dilutions of the dbpBA-complemented samples indicated that the amounts of protein in these samples were between four- and eightfold greater than the amount of protein obtained with genomically encoded dbpA (not shown), perhaps due to the increased copy number of the shuttle vectors. Alternatively, the high copy number may have altered the regulation of dbpBA by titrating out regulatory proteins, resulting in the overproduction of DbpA. The latter explanation seems unlikely given that the dbpBA genes are regulated in a positive manner by borrelial RpoS (9, 25).
Deletion of dbpBA attenuates the infectivity potential of B. burgdorferi.
To determine how the dbpBA deletion affected B. burgdorferi pathogenesis, immunocompetent C3H mice were infected with 10-fold serial dilutions of JF105/pBBE22 (with the ΔdbpBA::Gentr deletion) and JF105/pJBF17 (the dbpBA-complemented isolate), and the results were compared with the results for mice infected with ML23/pBBE22 (the infectious parent) in order to estimate the ID50 following 21 days of infection (Table 3). The data indicated that the infectious parent, ML23/pBBE22, which had an estimated ID50 of 150 organisms, exhibited infectivity similar to that of wild-type infectious B. burgdorferi (31, 47). The ID50 for ΔdbpBA::Gentr strain JF105/pBBE22 was more than 106 organisms, since no tissues were culture positive for any of the mice infected with 102, 103, or 104 cells of the mutant and only 10 and 28% of the tissues were culture positive for mice infected with 105 and 106 ΔdbpBA::Gentr B. burgdorferi cells, respectively (Table 3). In contrast, complementation with dbpBA restored the infectivity, and the calculated ID50 was 316 organisms (Table 3). This value is approximately twofold greater than the value for wild-type infectious B. burgdorferi (Table 3). Interestingly, whereas the positive-culture values for nearly all tissues were similar for the parental and complemented strains, the complemented strain exhibited an apparent defect in colonization of the spleen (Table 3). Nevertheless, the results obtained clearly indicate that the dramatic infectivity defect observed for the ΔdbpBA::Gentr B. burgdorferi strain is due to the lack of DbpBA and not the result of polar effects, a secondary mutation, or loss of an infectivity-associated plasmid. The infectivity defect of the ΔdbpBA::Gentr mutant observed in this study following needle inoculation is in contrast to the results of the initial study of Shi et al. (56), but it is in good agreement with the results reported more recently by Shi et al. (55) and Blevins et al. (4). Taken together, these results provide independent corroboration that dbpBA is required for maximal infectivity in the mouse model of experimental Lyme borreliosis following needle inoculation.
TABLE 3.
ID50 of the ΔdbpBA::Gentr strain, JF105/pBBE22, in immunocompetent C3H mice
| Isolate | Inoculum (cells) | No. of cultures positive/total no.
|
No. of mice positive/total no. | Calculated ID50 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lymph node | Skind | Heart | Spleen | Bladder | Joint | All sites | ||||
| ML23/pBBE22a | 104 | 2/2 | 2/2 | 2/2 | 2/2 | 2/2 | 2/2 | 12/12 | 2/2 | ∼150e |
| 103 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 24/24 | 4/4 | ||
| 102 | 1/3 | 1/3 | 1/3 | 1/3 | 1/3 | 1/3 | 6/18 | 1/3 | ||
| JF105/pBBE22b | 106 | 3/3 | 0/3 | 0/3 | 0/3 | 1/3 | 1/3 | 5/18 | 3/3 | >106 |
| 105 | 2/5 | 1/5 | 0/5 | 0/5 | 0/5 | 0/5 | 3/30 | 2/5 | ||
| 104 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||
| 103 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/36 | 0/6 | ||
| 102 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/18 | 0/3 | ||
| JF105/pJBF17c | 105 | 2/3 | 3/3 | 3/3 | 2/3 | 3/3 | 3/3 | 16/18 | 3/3 | 316 |
| 104 | 3/3 | 2/3 | 3/3 | 0/3 | 3/3 | 3/3 | 14/18 | 3/3 | ||
| 103 | 4/5 | 4/5 | 4/5 | 0/5 | 4/5 | 4/5 | 20/30 | 4/5 | ||
| 102 | 2/3 | 0/3 | 1/3 | 1/3 | 1/3 | 1/3 | 6/18 | 2/3 | ||
Parental strain B31 derivative which provides BBE22 in trans and is infectious (46).
ΔdbpBA::Gentr derivative of ML23/pBBE22.
ΔdbpBA::Gentr derivative of ML23/pBBE22 complemented with intact and expressed dbpBA.
The skin tested was skin from the site of inoculation.
The ID50 of ML23/pBBE22 is based on the results of numerous reproducible infectivity studies (31).
To obtain a quantitative assessment of colonization, real-time quantitative PCR was conducted. The results obtained support the ID50 findings since the ability of B. burgdorferi lacking dbpBA to colonize joint tissue following 21 days of infection was significantly impaired compared to the abilities of the infectious parent and complemented strain (P < 0.0001 and P < 0.006, respectively) (Fig. 2). Specifically, all of the joint samples from mice infected with 103 ΔdbpBA::Gentr B. burgdorferi cells were culture negative, and the bacteria were not detected by quantitative PCR (Fig. 2). The number of B. burgdorferi genomes obtained for the genetically complemented strain in joint tissue was less than the number obtained for the ML23/pBBE22 parent, and this may have reflected incomplete complementation of the phenotype in this assay (Table 3). Nevertheless, the quantitative PCR data support the notion that the infectious potential of the ΔdbpBA::Gentr mutant is significantly reduced following 21 days of infection.
Infection of C3H-SCID mice with ΔdbpBA::Gentr B. burgdorferi.
A possible explanation for the clearance of the ΔdbpBA::Gentr mutant observed is that the adaptive immune response results in rapid sterilization of the tissues coinciding with the appearance of the immunoglobulin M response to B. burgdorferi infection, as has been observed previously for a mutant missing lp28-1 (30). Previously, Shi et al. reported that SCID mice infected with 105 cells of B. burgdorferi having a dbpBA deletion were completely infectious, supporting the hypothesis that adaptive immunity is required for clearance of this mutant (56). To determine how the absence of an adaptive immune response impacted infectivity, we infected C3H-SCID mice with either 103 or 105 cells of the infectious parent, the ΔdbpBA::Gentr mutant, or the dbpBA-complemented strain. After 21 days the animals were sacrificed, and the tissues were scored for the ability of B. burgdorferi to grow in BSK-II medium. The results showed that after inoculation of 105 organisms, there was effectively no difference in infectivity among mice infected with the three strains, with the possible notable exception of the results for the skin, which appeared to be somewhat less efficiently colonized by the ΔdbpBA::Gentr mutant (Table 4). In contrast, when C3H-SCID mice were infected with 103 spirochetes, the ΔdbpBA::Gentr mutant could not colonize the C3H-SCID mice in any tissue tested, whereas the parent and genetically complemented strain colonized every tissue (Table 4). These observations suggest that following low-level infection (i.e., 103 spirochetes), the ability of the ΔdbpBA::Gentr mutant to survive in SCID mice is impaired in a manner similar to that observed with immunocompetent mice (Table 3) due to an inability to bind to host decorin and/or to evade innate killing (compare the data for the 103-cell inocula in Tables 3 and 4). Furthermore, adaptive immunity is needed to effectively clear the ΔdbpBA::Gentr mutant following inoculation of large doses, as shown by the differential infectivity observed for immunocompetent and SCID mice (Tables 3 and 4, respectively).
TABLE 4.
Infectivity of the ΔdbpBA::Gentr strain in C3H-SCID mice
| Isolate | Inoculum (cells) | No. of cultures positive/total no.
|
No. of mice positive/total no. | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Lymph nodes | Skina | Heart | Spleen | Bladder | Joint | All sites | |||
| ML23/pBBE22 | 105 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 18/18 | 3/3 |
| 103 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 18/18 | 3/3 | |
| JF105/pBBE22 | 105 | 5/5 | 3/5 | 5/5 | 5/5 | 5/5 | 5/5 | 28/30 | 5/5 |
| 103 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/30 | 0/5 | |
| JF105/pJBF17 | 105 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 18/18 | 3/3 |
| 103 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 18/18 | 3/3 | |
The skin tested was skin from the site of inoculation.
To quantify the B. burgdorferi present in C3H-SCID mouse tissues, samples of the skin and joint tissue were analyzed using quantitative PCR. The results indicate that the spirochetal loads in the joints of C3H-SCID mice infected with infectious B. burgdorferi cells were greater (approximately 10-fold greater) (Fig. 3) than the spirochetal loads in the joints of immunocompetent mice (Fig. 2), consistent with the idea that an adaptive immune response is able to keep the infection in check but does not clear B. burgdorferi from the tissues (Fig. 2). Furthermore, the values obtained for the ΔdbpBA::Gentr mutant are consistent with the culture data obtained, because all mice infected with 103 ΔdbpBA::Gentr spirochetes were cultivation negative (Table 4) and based on quantitative PCR the numbers of organisms were significantly less than the numbers of cells of the parent and complemented strain (P < 4 × 10−14 and P < 5 × 10−14, respectively) (Fig. 3A). In contrast, C3H-SCID mice infected with 105 ΔdbpBA::Gentr B. burgdorferi cells were culture positive (Table 4), and spirochete genomes were detectable in all five joint samples tested, although the levels were lower than those of the infectious parent (however, the difference was not statistically significant [P < 0.4]) (Fig. 3A). As predicted, the colonization of the joint tissue of C3H-SCID mice by the complemented strain was indistinguishable from the colonization of the joint tissue of C3H-SCID mice by the parent strain after inoculation of either a low or high dose (P ∼ 0.65 and P ∼ 0.6, respectively) (Fig. 3A).
The efficiencies of colonization of the skin of C3H-SCID mice by the parent and complemented strains were statistically indistinguishable both when inocula containing 103 cells were used and when inocula containing 105 cells were used (P ∼ 0.65 and P ∼ 0.95, respectively) (Fig. 3B). As predicted by our inability to culture ΔdbpBA::Gentr B. burgdorferi from the skin (or any other tissue) after injection of 103 organisms, quantitative PCR of skin samples detected no bacteria, and the differences were significant when the data were compared to data for both the parent and complemented strains (P < 1 × 10−4 and P < 5 × 10−5, respectively) (Fig. 3B). After inoculation of 105 mutant bacteria, spirochete genomes were detectable in the skin of three of five mice (Fig. 3B), which is in good agreement with the culture data obtained (Table 4). Two of the three positive skin samples contained inordinately high numbers of B. burgdorferi cells, but the reason for the variability is not clear. Nevertheless, with the exception of these two samples, all quantitative PCR values obtained for the ΔdbpBA::Gentr B. burgdorferi strain in joint tissue or skin, with low or high doses, were lower than the values for the parent and complemented strains, providing compelling evidence that the ability of the ΔdbpBA::Gentr strain to establish an infection is impaired even in immunodeficient mice.
Kinetics of infection for ΔdbpBA::Gentr B. burgdorferi.
Since the loss of dbpBA compromised the ability of B. burgdorferi to colonize mice following 21 days of infection of immunocompetent mice, we wanted to determine both the temporal basis and spatial basis of clearance. Also, since we determined that adaptive immunity played a part in the clearance of the ΔdbpBA::Gentr mutant (Table 4), we were interested in assessing colonization at earlier time points, when the innate immune response may play a role in reducing the colonization of the mutant. To this end, we performed a kinetics-based infectivity experiment with either 103 or 105 cells of the parent, the ΔdbpBA::Gentr mutant, and the genetically complemented strain using immunocompetent C3H mice. Following 12 h of infection of immunocompetent C3H mice, lymph node tissue was positive for one-half of the animals tested that were infected with 103 B. burgdorferi cells irrespective of the strain used; all other tissue sites tested (skin and spleen) were culture negative for the 103-cell dose independent of the dbpBA status of the samples tested (Table 5). For mice infected with 105 spirochetes, all strains tested colonized the site of intradermal inoculation in all animals tested, and a disseminated infection was also observed in most mice (50 to 70% of distant sites depending on the strain tested [Table 5]). Thus, when culture assays are used, the loss of dbpBA does not completely eliminate the ability of B. burgdorferi to survive in mouse skin, as well as other tissues, early in infection. However, at this point we cannot exclude the possibility that the absolute numbers of ΔdbpBA::Gentr strain cells are much less than the absolute numbers of cells of the infectious parent and the genetically complemented strain.
TABLE 5.
Infectivity of the B. burgdorferi parental, ΔdbpBA::Gentr, and genetically complemented strains following 12 h of infection in immunocompetent C3H mice
| Isolate | Inoculum (cells) | No. of cultures positive/total no.
|
No. of mice positive/total no. | |||
|---|---|---|---|---|---|---|
| Lymph nodes | Skina | Spleen | All sites | |||
| ML23/pBBE22 | 105 | 4/5 | 5/5 | 3/5 | 12/15 | 5/5 |
| 103 | 3/5 | 0/5 | 0/5 | 3/15 | 3/5 | |
| JF105/pBBE22 | 105 | 4/5 | 5/5 | 3/5 | 12/15 | 5/5 |
| 103 | 2/5 | 0/5 | 0/5 | 2/15 | 2/5 | |
| JF105/pJBF17 | 105 | 3/5 | 5/5 | 2/5 | 10/15 | 5/5 |
| 103 | 2/4 | 0/4 | 0/4 | 2/12 | 2/4 | |
The skin tested was skin from the site of inoculation.
When assays were performed over 21 days after inoculation of 103 or 105 cells, the parent strain was capable of disseminating and persisting in all tissues tested. In contrast, at the low dose (103 spirochetes), the ΔdbpBA::Gentr mutant colonized only one of five mice (in only three tissues) after 14 days. At the higher dose (105 spirochetes), the mutant was capable of dissemination, albeit impaired, in some of the mice, as shown by the ability to culture borrelial cells lacking dbpBA from a small minority of the tissues tested throughout the duration of the kinetics experiment (Fig. 4). Specifically, the heart and bladder were negative for all infections and at all time points tested with the ΔdbpBA::Gentr B. burgdorferi strain. Moderate dissemination was observed in the spleen (1/5 sites positive only at day 3), in the skin at the inoculation site (1/5 sites positive only at day 21), and in the lymph nodes (3/5 and 2/5 sites positive at days 14 and 21) (Fig. 4). Interestingly, the joint tissue was the tissue colonized best by the mutant, with 1/3, 1/3, and 4/5 sites positive at days 5, 7, and 14, respectively. Although the joint tissue of four of five mice was infected at day 14, none of these mice was still infected at day 21, indicating that the ΔdbpBA::Gentr B. burgdorferi strain that could colonize this tissue was not able to persist, so no tissues were culture positive by day 21 after low-dose infection and only a small percentage of tissues were infected at this time point following high-dose infection. Since our study ended at 21 days, we do not know if the organism in the sole skin sample that became culture positive between days 14 and 21 could persist or whether additional sites might become culture positive at a later time.
FIG. 4.
Kinetics of B. burgdorferi dissemination in C3H mice. C3H mice were infected with the B. burgdorferi parental, ΔdbpBA::Gentr, and genetically complemented strains using 103 spirochetes, and the ΔdbpBA::Gentr and genetically complemented strains were also infected using 105 organisms. Mice were sacrificed on days 3, 5, 7, 14, and 21, and the tissues were cultured to determine B. burgdorferi growth. The strains and doses tested are indicated below the x axis. The percentages of culture-positive samples are indicated on the y axis. The results are expressed as percentages of culture-positive samples for the strains and concentrations of spirochetes tested. The values represent data for three to six mice depending on the strain and time point evaluated. An asterisk indicates that all samples tested were culture negative. LN, lymph nodes.
The colonization defects revealed in this kinetic study were due to the dbpBA mutation, because the genetically complemented strain (JF105/pJBF17) was, with few exceptions, capable of wild-type levels of colonization. In mice infected with 105 cells of the complemented strain there were dissemination and nearly wild-type levels of persistence (Fig. 4). After administration of a dose containing 103 cells, there was an apparent decrease in colonization by the dbpBA-complemented strain (JF105/pJBF17) at day 21, but this was due mostly to the complete lack of infectivity in one of the five mice tested (Fig. 4). In addition, this kinetics experiment revealed that this strain was cleared from the spleen between days 7 and 14, consistent with our previous analysis of splenic colonization at day 21 (Table 3). It is possible that the increased amount of DbpBA produced by the genetically complemented strain resulted in clearance due to enhanced binding of antiserum against DbpA and DbpB, as proposed by Xu et al. (62). The slight differences between the parental and complemented strains aside, taken together, the results indicate that a B. burgdorferi strain lacking DbpBA was able to disseminate in some of the mice infected, but to a markedly reduced extent, and, more importantly, that the ability of this strain to persistently infect immunocompetent mice was greatly impaired.
DISCUSSION
As the first step in successful infection of a host, a pathogen must bind to host structures and either replicate and disseminate from the binding site or subsequently invade host cells. For either extracellular or intracellular pathogens, successful colonization requires an ability to replicate and disseminate to peripheral sites or to infect adjacent cells or tissues, after which a secondary colonization step can occur. In the case of B. burgdorferi, deposition in the dermis of a mammal resulting from the bite of an infected tick introduces the pathogen to a number of potential binding substrates in this locale, most notably ECM proteoglycan ligands.
Previous biochemical studies demonstrated that B. burgdorferi can bind to several host structures found in the ECM, including decorin and fibronectin, and on host cells, including integrin receptors, glycosoaminoglycans, and surface-associated factor H, via the activity of DbpA, DbpB, BBK32, P66, Bgp, and factor H binding proteins (6, 12, 14, 19-21, 23, 27, 28, 33, 38, 41, 42, 44, 45, 49, 54). Accordingly, it has long been argued that adherence to these sites is a key step in borrelial pathogenesis (for reviews, see references 8 and 13). However, until recently, it was difficult to assess the role of a particular gene or its product in borrelial pathogenesis due to an inability to genetically manipulate B. burgdorferi. Recently, many different investigators have independently developed methodologies to isolate and characterize isogenic mutants of infectious B. burgdorferi strains, and, with the advent of these advances (for a review, see reference 50), several borrelial adhesins have been inactivated and their effects on the resulting virulence have been examined. Specifically, isogenic dbpBA, BBK32, and bgp mutants of infectious B. burgdorferi strains have been isolated (42, 54-56). A p66 mutant has been obtained only in a noninfectious background (12), so its role in borrelial pathogenesis is not known yet. Knockout of bgp had no effect on B. burgdorferi virulence when the organism was assayed using immunodeficient mice (42), whereas the BBK32 mutant exhibited only a slight infectivity defect (54). The lack of a dramatic phenotype for either the bgp or BBK32 mutant implies that other adhesins may compensate for the known binding properties of the bgp and BBK32 proteins or that these molecules play a subtle role in the pathogenesis of B. burgdorferi. With regard to compensatory function, the ability to bind to glycosoaminoglycans has been reported for Dbp proteins, Bgp, and BBK32 (8, 13, 18, 19, 41, 42). Thus, if the binding to a given adhesin is directed primarily against a glycosoaminoglycan structure, then inactivation of such a gene (e.g., bgp) may have a more subtle effect on borrelial pathogenesis.
For dbpBA, recent publications from one laboratory suggested (i) that DbpA and DbpB were not essential for infection in immunocompetent mice, (ii) that overexpression of dbpA increased the infectivity of B. burgdorferi, and (iii) that DbpB and DbpBA were required for full virulence of B. burgdorferi following needle inoculation (55, 56, 62). To reconcile these apparently disparate observations, we examined how the loss of DbpBA affects the ability of B. burgdorferi to colonize C3H mice by calculating an ID50 and performing a kinetics-based infectivity analysis.
Our results clearly showed that deletion of dbpBA attenuates the pathogenic potential of B. burgdorferi based on ID50 analyses after 21 days of infection following needle inoculation (Table 3), which independently corroborated the results most recently reported by Shi et al. (55) and Blevins et al. (4). Specifically, the ΔdbpBA::Gentr strain was isolated from only 10 and 28% of the sites when inocula containing 105 and 106 spirochetes, respectively, were used and from no tissue sample when inocula containing fewer spirochetes were used (Table 3). In addition, genetic complementation of the ΔdbpBA::Gentr strain (JF105/pBBE22) with intact dbpBA (JF105/pJBF17) restored infectivity in mice in most instances, indicating that the infectivity defect observed in the ΔdbpBA::Gentr strain was due to the deletion of dbpBA and not the result of polar effects, an unlinked mutation, or loss of an infectivity-associated plasmid from B. burgdorferi (Table 3). Finally, the quantitative real-time PCR analysis corroborated the infectivity data for immunocompetent mice as the ΔdbpBA::Gentr B. burgdorferi strain was not detected in joints when an inoculum containing 103 organisms was used. Conversely, analysis of the parent and dbpBA-complemented strain samples showed that there was ample B. burgdorferi in the joint tissues tested, although the levels for the complemented strain samples were lower than the levels for the parent strain samples (Fig. 2). Taken together, these results indicate that DbpA and/or DbpB is required for maximal infectivity in the mouse model following 21 days of infection.
Because the ΔdbpBA::Gentr strain has such a dramatic phenotype, we were interested in determining the temporal and spatial basis of clearance of this strain. The kinetics-based analysis indicated that, particularly after a high-dose intradermal inoculum was used, the ΔdbpBA::Gentr strain was able to disseminate to several tissues, including the joint tissue, in four of five of the mice infected after 14 days but was unable to maintain a persistent infection (Fig. 4). The inability of the ΔdbpBA::Gentr strain to maintain an infection might be explained by two possible mechanisms that are not mutually exclusive. First, the inability of the ΔdbpBA::Gentr strain to bind to host decorin could result in more efficient clearance of the mutant from the host. Although unproven, it is conceivable that the lack of binding to host decorin results in free-floating ΔdbpBA::Gentr cells in interstitial fluid in the skin that are more readily phagocytosed by innate immune cells. Previously, Liang et al. reported that the interaction of B. burgdorferi with host decorin protected the spirochete from clearance, presumably due to the inability of antibodies directed against the borrelial Dbp proteins to bind to and eliminate the spirochete (34). This may be an important factor in the inability of the ΔdbpBA::Gentr mutant to persist, but it does not explain the rapid clearance of the mutant from the infected mice before adaptive immune mechanisms clear the organism. Thus, the second possibility is that the Dbp proteins provide a mechanism to override the initial innate immune response, which allows the spirochete to disseminate and remain infective. The exact mechanism used by the Dbp proteins is not known, but it may involve interactions with other host proteins that alter the immunological response to favor borrelial colonization and persistence.
Previously, Brown et al. reported that decorin-deficient mice (Dcn−/−) exhibited a reduction in B. burgdorferi colonization, particularly at low doses (7). In the subsequent study of Liang et al., when the size of the B. burgdorferi inoculum was increased, few differences between wild-type and decorin-deficient mice were observed except for the joint and skin, where in the decorin-deficient mice there was reduced colonization following 2 weeks of infection (34). Interestingly, the numbers for the skin and joint tissue were significantly reduced following 2 months of infection compared to the data for the Dcn+/+ mice, suggesting that the organisms might localize to different sites in wild-type and Dcn−/− mice and/or that the adaptive immune response is able to process B. burgdorferi more efficiently in the Dcn−/− mice (34).
Although the infectivity of B. burgdorferi was reduced in the Dcn−/− mice, B. burgdorferi was able to colonize the decorin-deficient mice with most of the inocula tested (7). The striking difference in infectivity observed for the ΔdbpBA::Gentr strain reported here compared to the results for the decorin-deficient mice implies that DbpA and DbpB may have additional roles in borrelial pathogenesis over and above their adherence activity with host decorin. If adherence to decorin were the only function attributable to DbpA and DbpB, then the infectivity phenotype for decorin-deficient mice and wild-type B. burgdorferi and the infectivity phenotype for wild-type mice and the ΔdbpBA::Gentr B. burgdorferi strain should be comparable. The difference observed suggests that Dbp proteins may affect the host response to B. burgdorferi infection. Alternatively, as indicated above, it is possible that the inability of the ΔdbpBA::Gentr strain to bind decorin puts the B. burgdorferi cells at greater risk of being phagocytosed by innate immune cells. Studies to examine these possibilities are under way.
It is highly likely that the innate and adaptive immune responses are both involved in the clearance of the ΔdbpBA::Gentr isolate, as shown by the SCID mouse infectivity data (Table 4 and Fig. 3). Specifically, whereas the low-dose inoculum of the ΔdbpBA::Gentr strain tested (103 organisms) was cleared in SCID mice, infection of SCID mice by the parent strain and by the genetically complemented strain was indistinguishable from infection of immunocompetent mice by these strains based on the culture-positive phenotype (Table 4). This is in stark contrast to the results for complete infection of SCID mice with 105 organisms independent of the status of the dbpBA genes, suggesting that clearance of the ΔdbpBA::Gentr strain in immunocompetent mice is also dependent on the adaptive immune response (Table 4). Previously, Shi et al. showed that SCID mice infected by the dbpBA mutant were indistinguishable from SCID mice infected by the infectious parent, but these workers evaluated only infections with 105 B. burgdorferi cells (56). With only data for this high dose, one might erroneously conclude that one function of DbpBA is to avoid clearance by adaptive immunity. If adaptive immunity were the sole immune clearance mechanism, then the ΔdbpBA::Gentr mutant would disseminate to all tissues, with clearance initiated when borrelia-specific antibodies are generated, as was observed for B. burgdorferi cells lacking lp28-1 (30). The enhanced clearance of the ΔdbpBA::Gentr strain suggests that decorin binding and/or the binding to other host factors via the Dbp proteins protects the spirochetes from rapid clearance via innate immune cells, presumably resident macrophages, langerhans cells, and/or neutrophils. Alternatively, the loss of Dbp proteins from the surface of B. burgdorferi may put the spirochetes at risk since high levels of DbpA and DbpB would likely be produced during infection. Thus, the loss of an abundant surface protein may compromise the overall integrity of the borrelial outer membrane and lead to enhanced innate immune clearance, as recently proposed by Xu et al. (61).
Recently, Blevins et al. reported that a dbpBA deletion mutant of strain 297 was severely defective for infectivity following needle inoculation of immunocompetent mice (4), similar to the infectivity results presented here. Interestingly, the strain 297 mutant did not exhibit as dramatic a phenotype when the infection was initiated using infected ticks. While one possible interpretation of these findings is that the DbpA and DbpB proteins do not play an important role during mammalian infection (contrary to conclusions based on needle inoculation studies), it is also possible that some defect in the infectivity of a dbpBA mutant after tick inoculation might be revealed upon further investigation. For example, whereas 100% (two of two) of mice that were infected using 5 ticks containing the parental strain became infected, 50% (one of two) and 67% (four of six) of mice that were infected using 5 and 10 ticks containing the dbpBA mutant, respectively, became infected. It is also not clear whether the 297 dbpBA mutant was capable of disseminated infection following tick infection since the infectivity of tick-infected mice was evaluated only by testing ear skin at a site presumably adjacent to where the tick feeding occurred. Finally, if in fact DbpA and DbpB play a less critical role after tick inoculation than after needle inoculation, the results may reflect the ability of tick saliva to locally immunosuppress the host immune response (3, 24, 29, 32) such that the B. burgdorferi dbpBA deletion strain deposited in the skin can migrate to immunoprotected niches in the skin, which are then protected against subsequent clearance by the adaptive immune response. This explanation is consistent with the data that we obtained with SCID mice, which showed that a dbpBA deletion mutant can colonize immunodeficient mice when higher doses are used (Table 4).
In summary, we showed that loss of the dbpBA genes results in a dramatic loss of infectivity in the mouse model of experimental Lyme disease. Furthermore, kinetics analyses indicated that B. burgdorferi cells that do not synthesize DbpBA exhibit muted dissemination when high-dose inocula are used and are unable to persist. The ability of SCID mice to clear the B. burgdorferi ΔdbpBA::Gentr strain when low doses were used suggests that the DbpBA proteins are required for early survival due to binding to host decorin, which may provide a microenvironment that prevents innate immune clearance in a manner reminiscent of tick-based infectivity, where the tick saliva may locally immunosuppress the host response (3, 24, 29, 32). As indicated above, this may explain the difference between the infectivity potential observed for ticks and the infectivity potential observed for needle inoculation, where the dbpBA mutant was able to colonize mice following a blood meal (4). Nevertheless, the results presented here extend previously reported data by demonstrating that ΔdbpBA::Gentr cells are cleared early during the infectious process in immunocompetent and SCID mice. The number of cells that are capable of surviving in immunocompetent mice is then greatly reduced when the host mounts a borrelia-specific adaptive immune response. In addition, these studies provided independent corroboration of the conclusion that the dbpBA genes are important for the maximum pathogenic potential of B. burgdorferi following needle inoculation (55, 56). Finally, the large difference between the infection potential of wild-type B. burgdorferi in decorin-deficient mice (7) and the infection potential of the ΔdbpBA::Gentr deletion strain in wild-type mice shown here suggests that, in addition to their role in binding host decorin, the Dbp proteins may modulate the host immune response through interaction with additional host structures in a manner that allows B. burgdorferi to establish and maintain an infection.
Acknowledgments
We are grateful to Steve Norris for the generous gift of plasmid pBBE22 and Patti Rosa for providing the borrelial shuttle vector pBSV2G. We thank Jennifer Faske for outstanding technical assistance. We also thank Vernon Tesh for his critical evaluation of the manuscript.
This work was supported by a Microbial Pathogenesis Training Grant from the Texas A&M Health Science Center and Texas A&M University (to E.H.W.) and by Public Health Service grants R03-AR049383 (to N.P.), R01-AI037601 (to J.M.L.), and R01-AI058086 (to J.T.S. and M.H.) from the National Institutes of Health.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 22 September 2008.
REFERENCES
- 1.Akins, D. R., K. W. Bourell, M. J. Caimano, M. V. Norgard, and J. D. Radolf. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Investig. 1012240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins, D. R., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol. Microbiol. 18507-520. [DOI] [PubMed] [Google Scholar]
- 3.Anguita, J., N. Ramamoorthi, J. W. Hovius, S. Das, V. Thomas, R. Persinski, D. Conze, P. W. Askenase, M. Rincon, F. S. Kantor, and E. Fikrig. 2002. Salp15, an Ixodes scapularis salivary protein, inhibits CD4+ T cell activation. Immunity 16849-859. [DOI] [PubMed] [Google Scholar]
- 4.Blevins, J. S., K. E. Hagman, and M. V. Norgard. 2008. Assessment of decorin-binding protein A to the infectivity of Borrelia burgdorferi in the murine models of needle and tick infection. BMC Microbiol. 882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks, C. S., P. S. Hefty, S. E. Jolliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 713371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks, C. S., S. R. Vuppala, A. M. Jett, A. Alitalo, S. Meri, and D. R. Akins. 2005. Complement regulator-acquiring surface protein 1 imparts resistance to human serum in Borrelia burgdorferi. J. Immunol. 1753299-3308. [DOI] [PubMed] [Google Scholar]
- 7.Brown, E. L., R. M. Wooten, B. J. Johnson, R. V. Iozzo, A. Smith, M. C. Dolan, B. P. Guo, J. J. Weis, and M. Hook. 2001. Resistance to Lyme disease in decorin-deficient mice. J. Clin. Investig. 107845-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabello, F. C., H. P. Godfrey, and S. A. Newman. 2007. Hidden in plain sight: Borrelia burgdorferi and the extracellular matrix. Trends Microbiol. 15350-354. [DOI] [PubMed] [Google Scholar]
- 9.Caimano, M. J., C. H. Eggers, K. R. Hazlett, and J. D. Radolf. 2004. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 726433-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll, J. A., C. F. Garon, and T. G. Schwan. 1999. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 673181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Champion, C. I., D. R. Blanco, J. T. Skare, D. A. Haake, M. Giladi, D. Foley, J. N. Miller, and M. A. Lovett. 1994. A 9.0-kilobase-pair circular plasmid of Borrelia burgdorferi encodes an exported protein: evidence for expression only during infection. Infect. Immun. 622653-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coburn, J., and C. Cugini. 2003. Targeted mutation of the outer membrane protein P66 disrupts attachment of the Lyme disease agent, Borrelia burgdorferi, to integrin αvβ3. Proc. Natl. Acad. Sci. USA 1007301-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coburn, J., J. R. Fischer, and J. M. Leong. 2005. Solving a sticky problem: new genetic approaches to host cell adhesion by the Lyme disease spirochete. Mol. Microbiol. 571182-1195. [DOI] [PubMed] [Google Scholar]
- 14.Coburn, J., L. Magoun, S. C. Bodary, and J. M. Leong. 1998. Integrins αvβ3 and α5β1 mediate attachment of Lyme disease spirochetes to human cells. Infect. Immun. 661946-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das, S., S. W. Barthold, S. S. Giles, R. R. Montgomery, S. R. Telford III, and E. Fikrig. 1997. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and the mammalian host. J. Clin. Investig. 99987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elias, A. F., J. L. Bono, J. J. Kupko III, P. E. Stewart, J. G. Krum, and P. A. Rosa. 2003. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J. Mol. Microbiol. Biotechnol. 629-40. [DOI] [PubMed] [Google Scholar]
- 17.Fikrig, E., S. W. Barthold, W. Sun, W. Feng, S. R. Telford III, and R. A. Flavell. 1997. Borrelia burgdorferi P35 and P37 proteins, expressed in vivo, elicit protective immunity. Immunity 6531-539. [DOI] [PubMed] [Google Scholar]
- 18.Fischer, J. R., K. T. LeBlanc, and J. M. Leong. 2006. Fibronectin binding protein BBK32 of the Lyme disease spirochete promotes bacterial attachment to glycosaminoglycans. Infect. Immun. 74435-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer, J. R., N. Parveen, L. Magoun, and J. M. Leong. 2003. Decorin-binding proteins A and B confer distinct mammalian cell type-specific attachment by Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 1007307-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, B. P., E. L. Brown, D. W. Dorward, L. C. Rosenberg, and M. Hook. 1998. Decorin-binding adhesins from Borrelia burgdorferi. Mol. Microbiol. 30711-723. [DOI] [PubMed] [Google Scholar]
- 21.Guo, B. P., S. J. Norris, L. C. Rosenberg, and M. Hook. 1995. Adherence of Borrelia burgdorferi to the proteoglycan decorin. Infect. Immun. 633467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagman, K. E., P. Lahdenne, T. G. Popova, S. F. Porcella, D. R. Akins, J. D. Radolf, and M. V. Norgard. 1998. Decorin-binding protein of Borrelia burgdorferi is encoded within a two-gene operon and is protective in the murine model of Lyme borreliosis. Infect. Immun. 662674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartmann, K., C. Corvey, C. Skerka, M. Kirschfink, M. Karas, V. Brade, J. C. Miller, B. Stevenson, R. Wallich, P. F. Zipfel, and P. Kraiczy. 2006. Functional characterization of BbCRASP-2, a distinct outer membrane protein of Borrelia burgdorferi that binds host complement regulators factor H and FHL-1. Mol. Microbiol. 611220-1236. [DOI] [PubMed] [Google Scholar]
- 24.Hovius, J. W., M. Levi, and E. Fikrig. 2008. Salivating for knowledge: potential pharmacological agents in tick saliva. PLoS Med. 5e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubner, A., X. Yang, D. M. Nolen, T. G. Popova, F. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 9812724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyde, J. A., J. P. Trzeciakowski, and J. T. Skare. 2007. Borrelia burgdorferi alters its gene expression and antigenic profile in response to CO2 levels. J. Bacteriol. 189437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jozsi, M., M. Oppermann, J. D. Lambris, and P. F. Zipfel. 2007. The C-terminus of complement factor H is essential for host cell protection. Mol. Immunol. 442697-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, J. H., J. Singvall, U. Schwarz-Linek, B. J. Johnson, J. R. Potts, and M. Höök. 2004. BBK32, a fibronectin binding MSCRAMM from Borrelia burgdorferi, contains a disordered region that undergoes a conformational change on ligand binding. J. Biol. Chem. 27941706-41714. [DOI] [PubMed] [Google Scholar]
- 29.Kotsyfakis, M., A. Sa-Nunes, I. M. Francischetti, T. N. Mather, J. F. Andersen, and J. M. Ribeiro. 2006. Antiinflammatory and immunosuppressive activity of sialostatin L, a salivary cystatin from the tick Ixodes scapularis. J. Biol. Chem. 28126298-26307. [DOI] [PubMed] [Google Scholar]
- 30.Labandeira-Rey, M., J. Seshu, and J. T. Skare. 2003. The absence of linear plasmid 25 or 28-1 of Borrelia burgdorferi dramatically alters the kinetics of experimental infection via distinct mechanisms. Infect. Immun. 714608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labandeira-Rey, M., and J. T. Skare. 2001. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect. Immun. 69446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leboulle, G., M. Crippa, Y. Decrem, N. Mejri, M. Brossard, A. Bollen, and E. Godfroid. 2002. Characterization of a novel salivary immunosuppressive protein from Ixodes ricinus ticks. J. Biol. Chem. 27710083-10089. [DOI] [PubMed] [Google Scholar]
- 33.Li, X., X. Liu, D. S. Beck, F. S. Kantor, and E. Fikrig. 2006. Borrelia burgdorferi lacking BBK32, a fibronectin-binding protein, retains full pathogenicity. Infect. Immun. 743305-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang, F. T., E. L. Brown, T. Wang, R. V. Iozzo, and E. Fikrig. 2004. Protective niche for Borrelia burgdorferi to evade humoral immunity. Am. J. Pathol. 165977-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liveris, D., G. Wang, G. Girao, D. W. Byrne, J. Nowakowski, D. McKenna, R. Nadelman, G. P. Wormser, and I. Schwartz. 2002. Quantitative detection of Borrelia burgdorferi in 2-millimeter skin samples of erythema migrans lesions: correlation of results with clinical and laboratory findings. J. Clin. Microbiol. 401249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lybecker, M. C., and D. S. Samuels. 2007. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol. Microbiol. 641075-1089. [DOI] [PubMed] [Google Scholar]
- 37.Maruskova, M., M. D. Esteve-Gassent, V. L. Sexton, and J. Seshu. 2008. Role of the BBA64 locus of Borrelia burgdorferi in early stages of infectivity in a murine model of Lyme disease. Infect. Immun. 76391-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDowell, J. V., J. Wolfgang, E. Tran, M. S. Metts, D. Hamilton, and R. T. Marconi. 2003. Comprehensive analysis of the factor H binding capabilities of Borrelia species associated with Lyme disease: delineation of two distinct classes of factor H binding proteins. Infect. Immun. 713597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nadelman, R. B., and G. P. Wormser. 1998. Lyme borreliosis. Lancet 352557-565. [DOI] [PubMed] [Google Scholar]
- 40.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 711689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parveen, N., M. Caimano, J. D. Radolf, and J. M. Leong. 2003. Adaptation of the Lyme disease spirochaete to the mammalian host environment results in enhanced glycosaminoglycan and host cell binding. Mol. Microbiol. 471433-1444. [DOI] [PubMed] [Google Scholar]
- 42.Parveen, N., K. A. Cornell, J. L. Bono, C. Chamberland, P. Rosa, and J. M. Leong. 2006. Bgp, a secreted glycosaminoglycan-binding protein of Borrelia burgdorferi strain N40, displays nucleosidase activity and is not essential for infection of immunodeficient mice. Infect. Immun. 743016-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Höök. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48585-617. [DOI] [PubMed] [Google Scholar]
- 44.Probert, W. S., and B. J. Johnson. 1998. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol. Microbiol. 301003-1015. [DOI] [PubMed] [Google Scholar]
- 45.Probert, W. S., J. H. Kim, M. Hook, and B. J. Johnson. 2001. Mapping the ligand-binding region of Borrelia burgdorferi fibronectin-binding protein BBK32. Infect. Immun. 694129-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purser, J. E., M. B. Lawrenz, M. J. Caimano, J. K. Howell, J. D. Radolf, and S. J. Norris. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48753-764. [DOI] [PubMed] [Google Scholar]
- 47.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 9713865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 991562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogers, E. A., and R. T. Marconi. 2007. Delineation of species-specific binding properties of the CspZ protein (BBH06) of Lyme disease spirochetes: evidence for new contributions to the pathogenesis of Borrelia spp. Infect. Immun. 755272-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosa, P. A., K. Tilly, and P. E. Stewart. 2005. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat. Rev. Microbiol. 3129-143. [DOI] [PubMed] [Google Scholar]
- 51.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 922909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seshu, J., J. A. Boylan, F. C. Gherardini, and J. T. Skare. 2004. Dissolved oxygen levels alter gene expression and antigen profiles in Borrelia burgdorferi. Infect. Immun. 721580-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seshu, J., J. A. Boylan, J. A. Hyde, K. L. Swingle, F. C. Gherardini, and J. T. Skare. 2004. A conservative amino acid change alters the function of BosR, the redox regulator of Borrelia burgdorferi. Mol. Microbiol. 541352-1363. [DOI] [PubMed] [Google Scholar]
- 54.Seshu, J., M. D. Esteve-Gassent, M. Labandeira-Rey, J. H. Kim, J. P. Trzeciakowski, M. Hook, and J. T. Skare. 2006. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol. Microbiol. 591591-1601. [DOI] [PubMed] [Google Scholar]
- 55.Shi, Y., Q. Xu, K. McShan, and F. T. Liang. 2008. Both decorin-binding proteins A and B are critical for the overall virulence of Borrelia burgdorferi. Infect. Immun. 761239-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi, Y., Q. Xu, S. V. Seemanapalli, K. McShan, and F. T. Liang. 2006. The dbpBA locus of Borrelia burgdorferi is not essential for infection of mice. Infect. Immun. 746509-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skare, J. T., E. S. Shang, D. M. Foley, D. R. Blanco, C. I. Champion, T. Mirzabekov, Y. Sokolov, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1995. Virulent strain associated outer membrane proteins of Borrelia burgdorferi. J. Clin. Investig. 962380-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steere, A. C., J. Coburn, and L. Glickstein. 2004. The emergence of Lyme disease. J. Clin. Investig. 1131093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stevenson, B., T. G. Schwan, and P. A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 634535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tokarz, R., J. M. Anderton, L. I. Katona, and J. L. Benach. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole-genome DNA array. Infect. Immun. 725419-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu, Q., K. McShan, and F. T. Liang. 2008. Essential protective role attributed to the surface lipoproteins of Borrelia burgdorferi against innate defences. Mol. Microbiol. 6915-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, Q., S. V. Seemanaplli, K. McShan, and F. T. Liang. 2007. Increasing the interaction of Borrelia burgdorferi with decorin significantly reduces the 50 percent infectious dose and severely impairs dissemination. Infect. Immun. 754272-4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang, X., M. S. Goldberg, T. G. Popova, G. B. Schoeler, S. K. Wikel, K. E. Hagman, and M. V. Norgard. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 371470-1479. [DOI] [PubMed] [Google Scholar]
- 64.Yang, X. F., U. Pal, S. M. Alani, E. Fikrig, and M. V. Norgard. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 199641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]