Abstract
Clinical isolates of Chlamydia trachomatis that lack IncA on their inclusion membrane form nonfusogenic inclusions and have been associated with milder, subclinical infections in patients. The molecular events associated with the generation of IncA-negative strains and their roles in chlamydial sexually transmitted infections are not clear. We explored the biology of the IncA-negative strains by analyzing their genomic structure, transcription, and growth characteristics in vitro and in vivo in comparison with IncA-positive C. trachomatis strains. Three clinical samples were identified that contained a mixture of IncA-positive and -negative same-serovar C. trachomatis populations, and two more such pairs were found in serial isolates from persistently infected individuals. Genomic sequence analysis of individual strains from each of two serovar-matched pairs showed that these pairs were very similar genetically. In contrast, the genome sequence of an unmatched IncA-negative strain contained over 5,000 nucleotide polymorphisms relative to the genome sequence of a serovar-matched but otherwise unlinked strain. Transcriptional analysis, in vitro culture kinetics, and animal modeling demonstrated that IncA-negative strains isolated in the presence of a serovar-matched wild-type strain are phenotypically more similar to the wild-type strain than are IncA-negative strains isolated in the absence of a serovar-matched wild-type strain. These studies support a model suggesting that a change from an IncA-positive strain to the previously described IncA-negative phenotype may involve multiple steps, the first of which involves a translational inactivation of incA, associated with subsequent unidentified steps that lead to the observed decrease in transcript level, differences in growth rate, and differences in mouse infectivity.
Chlamydia trachomatis is the most common bacterial sexually transmitted infections (STI) in the United States and the leading cause of preventable blindness worldwide (12). Techniques for its genetic analysis are severely limited, restricting understanding of the molecular biology of this widespread pathogen. We previously described C. trachomatis isolates that form multiple nonfusogenic inclusions within single cells infected with multiple elementary bodies (EBs) (18). Inclusions formed by these isolates uniformly lacked the IncA protein in the inclusion membrane (18). These IncA-negative variant strains were found in approximately 2% of the chlamydial genital isolates in patients seen at STI clinics in the Seattle area of Washington. IncA-negative representatives of all routinely isolated serovars, except serovar G, were cultured from patients seen at these clinics (15). Nucleotide sequence analysis of the IncA-negative strains suggested that inactivating mutations occurred randomly across the gene. The genesis and persistence in vivo of these variant strains remain uncharacterized.
Because of the diversity and relative abundance of these strains in patient populations, we hypothesize that IncA-negative strains arise via mutation from wild-type strains and that variant strains have a unique niche in the patient population. If these hypotheses are true, we would predict that individual patients that carry both variant and wild-type strains can be identified and that in some cases the mixed populations should be nearly isogenic, with the exception of an inactivating mutation in incA. To test these hypotheses, we screened 55 low-passage IncA-negative isolates with anti-IncA monoclonal antibody (MAb) to identify possible minority IncA-positive populations. In three strains, serovar-matched minority populations that expressed and localized IncA to the inclusion membrane were identified. These “matched pairs” were separated from one another through limiting dilution cloning and compared with IncA-positive and -negative same-serovar strains isolated from independent patients. Analyses of genome sequence, incA transcription, and growth characteristics in vitro and in vivo demonstrated that the matched pair strains are genetically nearly isogenic and that the tested phenotypes of the IncA-positive and IncA-negative strains within a matched pair are more closely related than similar same-serovar pairs isolated from individual patients. The studies suggest that transition from an IncA-positive strain to an IncA-negative strain may represent an early stage in the phenotypic variation of C. trachomatis within patients and that other, as yet uncharacterized, changes may occur which further distinguish IncA-positive strains from IncA-negative strains.
MATERIALS AND METHODS
Chlamydiae and chlamydial culture.
Prototype strains D/UW-3/Cx and J/UW-36/Cx were used for reference sequence and PCR analysis. Clinical strains J/6276, J(s)/6276, J(s)/893, J(s)/1980, J/p225, Ia/165, Ia/p202, Ia/9878, Ia/1008, Ia(s)/1010, Ia/1011, Ia(s)/1025, Ia/1026, D/p248, D(s)/2923, D(s)/5058, D(s)/8039, E/9383, E(s)/1968, F/70, F(s)/70, F/381, F(s)/4022, F(s)/8068, and D(s)/561 were propagated from frozen samples in the University of Washington Chlamydia Repository. The “(s)” designation indicates that the strain demonstrates the nonfusogenic phenotype in culture (18). Specimen collection, culture isolation techniques, and serotyping were conducted as previously described (19). Infected HeLa or McCoy cells were incubated in minimal essential medium with 10% fetal bovine serum (Sigma-Aldrich) with or without cycloheximide (1 μg/ml) at 37°C in 5% CO2. Elementary bodies were partially purified by centrifugation of lysates of infected cells through a 30% Renografin pad (4).
Determination of infectious EB production.
McCoy cell monolayers in 12-mm shell vials were inoculated with chlamydiae suspended in SPG buffer (0.25 M sucrose, 10 mM sodium phosphate, 5 mM l-glutamic acid). All infections were conducted at a multiplicity of infection (MOI) of 0.5. Inocula were centrifuged onto the monolayers for 1 h at 1,200 × g. The inocula were then removed, and the cells were incubated in minimal essential medium with 10% fetal bovine serum. Infected monolayers were washed with phosphate-buffered saline and lysed for EB collection by incubation in distilled water for 2 min at 0, 24 h, and 48 h postinfection (p.i.). These lysates were plated in triplicate onto McCoy cell monolayers in 48-well plates using 10-fold dilutions for determination of inclusion-forming units (IFU). The average IFU output per well and the standard error of each average were determined for each of the triplicate cultures.
Antibodies and reagents for microscopy.
MAb directed at C. trachomatis IncA (3H7) was produced as described previously (1). MAb E6-H1, specific for chlamydial lipopolysaccharide (LPS), was a gift from Harlan Caldwell. Anti-serovar J major outer membrane protein (MOMP) (CC-1), anti-serovar I/Ia MOMP (PE-5), and anti-serovar F MOMP (FC-2) were acquired through the Washington Research Foundation (Seattle, WA). All secondary antibodies were purchased from Southern Biotechnology Associates, Inc. (Birmingham, AL). The DNA-specific fluorescent label 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; 2 μg/ml in mounting medium [Vectashield; Vector Laboratories]) was used to label host cell and bacterial DNA. Antibody labeling was performed on chlamydiae grown in HeLa cell monolayers on 12-mm coverslips as previously described (1).
Clonal isolation of independent IncA-positive and -negative strains in cell culture.
Cloned isolates were initially produced from patients persistently infected with C. trachomatis expressing serovar Ia. These patients were cultured several times over the course of 2 to 5 years, and the chlamydiae recovered were originally used for different studies (6). A limiting dilution approach was used to generate microbiological clones from these isolates (17). Immunofluorescence was used to determine the IncA-positive or IncA-negative status of cloned strains from the clinical samples.
Screening IncA-negative clinical isolates for the presence of minority IncA-positive subpopulations.
Fifty-five clinical samples initially identified as containing IncA-negative C. trachomatis strains were examined for minority IncA-positive subpopulations. An aliquot of each isolate tested was inoculated onto McCoy cells at a high MOI and cultured for 40 h. Cells were then fixed with methanol and labeled with anti-IncA and anti-LPS MAbs. IncA-positive inclusions were identified in these populations by scanning the entire coverslip under a microscope at a low magnification and by using anti-IncA antibody.
Previous EB production experiments demonstrated that IncA-positive isolates have a slight to moderate growth rate advantage over IncA-negative strains in vitro, depending on the particular strain (21). This growth rate difference was exploited for the isolation of minority IncA-positive clones against a majority population of IncA-negative strains in selected patient samples. For this purpose, the original clinical specimens were passed in shell vials until a titer of >1,000 IFU/ml was attained. This culture was then lysed by sonic disruption and stored at −80°C. A set of vials was then inoculated (MOI of ∼1) using this lysate as a source of EBs. Single vials of the subsequent culture were sonicated every hour between 12 and 20 h p.i. Each sonicate was aliquoted and frozen at −80°C. After the samples were collected and frozen, one aliquot from each time point was thawed and inoculated onto McCoy cells in shell vials. These vials were cultured for 30 h p.i. and labeled with anti-LPS MAb. This experiment determined the earliest time point that EBs could be harvested from the frozen aliquots of infected-cell lysates. Because the IncA-positive strains tend to grow faster than the serovar-matched IncA-negative strains do, collection of EBs from the earliest time point possible enriched for IncA-positive C. trachomatis from the mixed clinical isolates. Lysates from the earliest cultures showing viable EB production were serially passed in the same manner until it was determined that the number of IFU of the IncA-positive strain was greater than that of the IncA-negative strain. This process required several serial passages for each strain. Once this was accomplished, a twofold limiting dilution method was used to microbiologically clone the IncA-positive chlamydial strain (17). Clonal isolation of the majority IncA-negative population from these isolates was accomplished directly via limiting dilution. Clones of these serovar-matched IncA-positive and -negative pairs from each isolate were amplified and stored at −80°C.
PCR amplification and nucleotide sequence analysis.
All references to gene number (locus tag) are based on the published C. trachomatis D/UW3 genome (16). To assess the genetic similarity of these candidate mutant and wild-type strain pairs recovered from the same patient specimen, we sequenced the entire open reading frames (ORFs) of four C. trachomatis polymorphic genes, namely, incA (CT119), omp1 (CT681, encoding MOMP), tarp (CT456) (5), and tox (CT166) (3). To sequence the entire ORF, we used specific PCR primers that are in the flanking regions of both >100 bp upstream of the 5′ ends and >100 bp downstream of the 3′ ends (Table 1). The complete ORF for each gene was amplified with PfuUltra (Stratagene), and both strands of the full-length gene were subjected to nucleotide sequence analysis. Consensus sequences were derived from assembled contigs with the Sequencher program (Gene Codes Corp., Ann Arbor, MI) and were compared with corresponding gene sequences in the reference D/UW-3 genome and in control strains of the same serovar.
TABLE 1.
Oligonucleotide primers or probes used in these studies
| Target | Primer or probe | Sequence (5′-3′) |
|---|---|---|
| incA | CT119F | ACATGTAAGCTGGTTTTTACATAG |
| CT119R | TGGCACCCTGTGTACAGTTAG | |
| omp1 | 681F | CTTTAACTAGGACGCAGTGCCGC |
| 681R | TGGACCCGACCGAAGCCGAGCC | |
| 81F2 | CTGAGATGTTTACAAATGCCGC | |
| U681R3 | AGACCATTTAACTCCAATGTA | |
| tarp | TPF1 | TTCTCCAGATACTTCAGAAAGC |
| TPF2 | CAAAGACTCTGACGGAGCTGGTG | |
| TPR1 | TGCACACATGTTTTCCAAGTC | |
| TPR2 | CAGGGTAAACGACGTCTAGG | |
| TPR3 | CCTCCAGGAATTCGTCCTCCG | |
| CT166 | CT166F | CTCCTCCAGAAGGAACGACAAC |
| CT166FA | CTTCAGAGAATAAGGTCACTAC | |
| CT166F2 | AAGAAAAACCTAAGACGACTCCG | |
| CT166F3 | AACGCGTGCAGAGTATTTAGAG | |
| CT166R | CCCAGCATACATGTCTCCATC | |
| incA RT-PCR | RT-PCR incA F | CACATTAGCAGGGAATGCTC |
| RT-PCR incA R | AGAGCCTTTAAGATCTGC | |
| incA | RCE1 | CTTTTTGTAGAGGGTGAT |
| incA Q-PCRa | Q-PCR incA F | AACTTTAAATGAGAAAGTGCCTATGACAA |
| Q-PCR incA R | ATCAGAAAGCCAACAAGATGT | |
| incA probeb | TATTTTCTTAATTTGTCCATCAAAGAA | |
| groEL Q-PCR | Q-PCR groEL F | TGTTACCGTTGCGAAAGAAGT |
| Q-PCR groEL R | TCCAGCTTTGTCAGCAGTTT | |
| groEL probeb | ACATGAAAATATGGGCGCTC |
Q-PCR, quantitative PCR.
Quantitative PCR probes are labeled with 6-carboxyfluorescein on the 5′ end and 6-carboxytetramethylrhodamine on the 3′ end.
Mouse infections.
Seven- to 8-week-old female BALB/c mice were purchased from Charles River Laboratory (Wilmington, MA). Animals were injected with 1.25 mg of Depo-Provera (progesterone) 10 and 3 days prior to infection. The mice were inoculated intravaginally directly into the cervix with 5.0 × 105 IFU/mouse of appropriate C. trachomatis strains in a final volume of 20 μl using a positive-displacement pipette. Swabs were collected on days 7 and 10. Swabbing was conducted with a Dacron swab that was inserted in the vagina and rotated 25 times. Swabs were placed in 1 ml of SPG buffer and immediately frozen at −80°C. Recovered IFU were enumerated by inoculating serial 10-fold dilutions of swab sample into the wells of 48-well plates which were centrifuged as described above and incubated for chlamydial growth for 48 h. Cells were then fixed with methanol and labeled with anti-LPS MAb. Fluorescence microscopy was used to count chlamydial inclusions from which the number of shed IFU/mouse was calculated. Five mice were inoculated per strain. Shedding of infectious chlamydiae in IncA-positive and -negative strains was compared by using a Student's t test. Repeated-measures analysis of variance was also used due to the correlated nature of the measurements.
Analysis of transcription in IncA-positive and -negative strains.
Total RNA was harvested by treating mock-infected or C. trachomatis-infected cells (MOI of 1) with the TRIzol reagent (Invitrogen, Carlsbad, CA). Amplification-grade RQ1 DNase (Promega, Madison WI) was added to remove residual genomic DNA. The RNA minikit (Qiagen, Valencia, CA) protocol was used to remove residual DNase and contaminants. RNA integrity was verified electrophoretically and by UV spectroscopy.
The NorthernMax-Gly kit (Ambion, Austin, TX) was used for Northern blot analysis of incA transcripts in cells infected with IncA-positive and -negative strains. DNA probes (a 316-bp incA and a 320-bp 16S rRNA PCR product) were labeled with digoxigenin-labeled deoxynucleoside triphosphates (Roche Diagnostics Corporation, Indianapolis, IN) and incubated with blots at 52°C for 12 to 16 h. Blots were washed and incubated with antidigoxigenin antibody prior to chemiluminescence detection.
Mapping of the transcriptional start for incA was conducted using a 5′ rapid amplification of cDNA end (5′-RACE; Invitrogen) procedure. cDNAs were generated via reverse transcription of 5 μg of total RNA using Superscript II polymerase (Invitrogen) and a gene-specific primer, RCE1 (Table 1). Sequencing of products was performed at the Center for Genome Research and Biocomputing at Oregon State University.
Quantitative reverse transcription-PCR (RT-PCR) was performed using the Superscript II first-strand synthesis system (Invitrogen). cDNA from total RNA (5 μg) was produced by using a protocol supplied by the manufacturer. Separate PCR amplification mixtures were used for target cDNAs from experimental and control samples. Control samples to normalize DNA content were cDNAs amplified with 16S rRNA and/or groEL primers. Threshold cycle values and the corresponding copy numbers for the controls were obtained from a standard curve generated by PCR amplification of several dilutions of amplicons with known quantity and compared to copy numbers obtained from PCRs of the target experimental samples. All assays were performed in triplicate.
Genomic DNA preparation and sequence analysis.
EBs of cloned chlamydial isolates D(s)/2923, F/70, F(s)/70, J/6276, and J(s)/6276 were purified as previously described (4). Purified EBs were incubated with RQ1 DNase I (Promega) for 60 min, and the DNase was inactivated with 2 mM EGTA. Chlamydial DNA was then extracted using a small-scale genomic preparation kit (Qiagen) after suspending the EBs in 5 mM dithiothreitol. Isolate D(s)/2923 genomic DNA was sequenced using classical Sanger sequencing methods at the Joint Genome Institute (Walnut Creek, CA). DNA from isolates F/70, F(s)/70, J/6276, and J(s)/6276 was further processed for Illumina-based sequence analysis using commercial DNA preparation kits (Illumina Inc., San Diego, CA) following the manufacturer's instructions.
Illumina-derived draft genomes were first assembled using the reference-guided assembly program Maq (http://maq.sourceforge.net/). Regions in the reference-guided assembled genome where Maq could not resolve sequence were then compared to contiguous sequences assembled through the use of de novo assembly software (VCAKE [10]), and a single contiguous draft sequence was produced. Single nucleotide polymorphisms (SNPs) between matched pairs were located using the Diffseq program from the Emboss software suite (14). Isolate genomes were also compared with the published C. trachomatis D/UW3 genome sequence (GenBank accession number AE001273; 14) using Diffseq to locate SNPs. The locations and effects of individual polymorphisms were first determined using an in-house SNP parsing program (http://web.engr.oregonstate.edu/∼davidsjo/). Any necessary manual gene variation analysis was performed using MacVector sequence analysis software (MacVector, Cary, NC). PCR analysis was used to verify 4 SNPs found between the serovar F-matched pair, as well as 5 of the 51 SNPs found in the serovar J-matched pair isolates.
Polymorphisms were then analyzed to determine which SNP in each serovar J pair was parental and which carried the product of the change. Each polymorphic nucleotide was compared to the corresponding nucleotide of two published C. trachomatis strains (D/UW3 [GenBank accession number AE001273] and A/HAR-13 [GenBank accession number CP000051]) and an additional four unpublished C. trachomatis genomes in our database (not shown). In cases where an individual SNP within one strain matched the corresponding nucleotide in all reference genomes, that strain was considered parental. The choice of parental strain for a particular SNP was recorded as unknown if there was not complete agreement between the sequence in one of the examined matched pair sequences and all of the reference strains used for comparison.
RESULTS
Analysis of sequential same-serovar patient isolates.
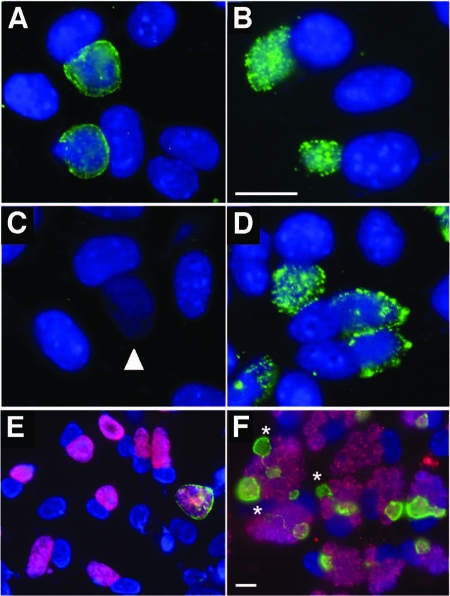
Table 2 shows a timeline of culture results from two patients identified in a previous report as having multiple sequential same-serovar infections (6). Selected samples from these experiments were serotyped and screened for the nonfusing phenotype. Each tested isolate was confirmed as serovar Ia, but the nature of inclusion formation in cells infected at a high MOI varied among isolates. Fluorescence microscopic analysis with anti-IncA antibody was used to determine the IncA phenotype in these isolates (Fig. 1A to D). On one occasion each for two patients, the nonfusing (IncA-negative) phenotype was recovered in cloned isolates [isolate Ia(s)/1010 and Ia(s)/1025]. Isolates recovered and cloned at other times from the same patients were serovar Ia and IncA positive (Fig. 1). These results supported the hypothesis that clonally related IncA-positive and IncA-negative C. trachomatis might be found in the same patients and vary in abundance within individuals. We then examined our collection of nonfusogenic clinical isolates with anti-IncA antibodies to determine the frequency of these IncA-negative isolates that might have a minority IncA-positive subpopulation. In these experiments, single IncA-positive inclusions were occasionally observed against a background of IncA-negative chlamydial inclusions (Fig. 1E and F). Five of 55 IncA-negative isolates contained a minority subpopulation of IncA-positive C. trachomatis inclusions, and three of these isolates had the same serotype that the IncA-negative majority did. The remaining isolates contained C. trachomatis that were not of the same serovar and most probably were collected from a patient that had acquired a mixed infection. The three matched pairs were serovar F, serovar J, and serovar D. The proportion of IncA-positive versus IncA-negative strains in these populations ranged from approximately 0.001 [J/J(s); Fig. 1E] to 0.3 [D/Ds, not shown; F/F(s); Fig. 1F]. Figure 1F shows the serovar F-matched pair inoculated onto cells at a high MOI. Several cells in this image were infected with both strains, with the developmental forms localized to individual IncA-positive or -negative vacuoles. This demonstrates the nonfusogenicity of the IncA-negative inclusions (red) with each other and with the smaller number of IncA-positive inclusions (green). Note that several of the IncA-positive inclusions are linked by IncA-laden fibers. A diluted sample of this preparation yields single inclusions of both types in the infected cells, resulting in an image similar to that shown in Fig. 1E.
TABLE 2.
Culture points in two individuals persistently infected with C. trachomatis serovar Ia
| Patienta and time point | Culture result
|
|
|---|---|---|
| Strain | IncA statusb | |
| Patient 1 | ||
| T = 0 | 1025 | − |
| T = 6 mo | 1026 | + |
| T = 12 mo | NDc | NT |
| Patient 2 | ||
| T = 0 | 1008 | + |
| T = 36 mo | 1010 | − |
| T = 48 mo | 1011 | + |
Patient 1 was culture/LCR positive for a total period of 3 years. Patient 2 was culture/LCR positive for a total period of 6 years.
−, negative; +, positive; NT, not tested (sample unavailable).
ND, no designated strain.
FIG. 1.
Fluorescence microscopic analysis of McCoy cells infected with different IncA-positive and -negative serovar-matched pairs. In all panels, DNA is labeled blue with DAPI. (A to D) IncA labeling (green in panels A and C) and anti-serovar Ia MOMP labeling (green in panels B and D) of serial isolates Ia/1026 (A and B) and Ia(s)/1025 (C and D). Note the unlabeled, IncA-negative inclusion in panel C (white arrowhead). The bar in panel B represents 10 μm, and panels A to D are shown at the same magnification. (E and F) Primary isolation cultures for the serovar J- and serovar F-matched pairs (panel E and F, respectively). In both images, labeling of IncA is green and serotype-specific labeling of MOMP is red. (E) A rare (1/10,000) IncA-positive inclusion against the background of a largely IncA-negative, serovar J C. trachomatis (isolate 6276). (F) A high-titer clinical sample that contains both IncA-positive and -negative C. trachomatis of serovar F (isolate 70). The IncA-positive strain within this pair represents approximately 30% of the population. Note secondary inclusion formation (indicated by asterisks) and intricate fiber production emanating from IncA-positive inclusions in some cells. The bar in panel F represents 10 μm, and panels E and F are shown at the same magnification.
DNA sequence analysis of different matched pairs.
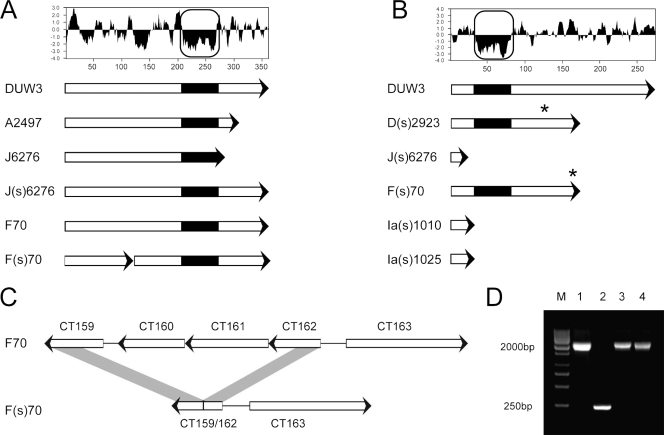
PCR-based and whole-genome sequencing was used to address the question of the clonal relatedness of individual matched pairs. Dean et al. (6) demonstrated that the serovar Ia MOMP sequences were identical or very nearly identical in the strains examined in their work. Our studies began with the nucleotide sequence analysis of four genes, incA (CT119), omp1 (CT681), tox (CT166), and tarp (CT456) from five selected serovar Ia strains isolated from a previous study of chlamydial persistence in patients. We found that the coding sequence for incA in the complete incA ORF in the Ia(s) strains exhibited a single base pair insertion at position 36, compared to the incA sequence from the wild-type serovar Ia strains (Fig. 2). Otherwise, all sequences from the Ia strains collected from single patients were identical.
FIG. 2.
ORF analysis of regions showing variation found in the matched pair isolates. (A) Kyte-Doolittle hydrophilicity plot of gene CT135, and gene representations of lab strain D/UW3 (DUW3), ocular isolate A2496 (11), and matched pair isolates. (B) Kyte-Doolittle hydrophilicity plot of the predicted CT119 (incA) protein sequence, and ORF representations of lab strain D/UW3 and clinical isolates showing the IncA-negative phenotype. The boxed regions in panels A and B highlight a putative Inc motif. Asterisks indicate locations of frameshift mutations leading to downstream truncation. (C) A region of the plasticity zone that is deleted in isolate F(s)/70 relative to strain F/70, resulting in putative CT159/CT162 fusion. (D) PCR analysis confirming the deletion found in isolate F(s)/70, with the length of each product indicated to the left. Lanes: M, molecular size markers (M); 1, strain F/70; 2, strain F(s)/70; 3, strain F(s)/4022; 4, strain F(s)/8068.
Preliminary sequence analysis was also conducted on the serovar J- and F-matched pairs. The nucleotide sequence analysis of incA from the IncA-negative strain J(s)/6276 had a single base pair deletion at position 18 that resulted in a reading frameshift and inactivation of the gene (3). This deletion was not represented in the IncA-positive strain isolated from the same patient. incA was also sequenced from the strains F/70 and F(s)/70, a matched pair of serovar F. The coding sequence of incA was inactivated in strain F(s)/70 by a single deletion at position 515. Therefore, the sequences at incA were identical within each of the serovar Ia-, J-, and F-matched pairs, with the exception of the single insertion or deletion that interrupted the gene (Fig. 2).
Genomic sequence analysis.
Five chlamydial genomes were sequenced to investigate the possibility that matched pairs were more closely isogenic than a serotype-matched, but otherwise unrelated, pair of IncA-positive and IncA-negative strains. The Illumina sequencing technology was used to generate genome sequence from cloned strains for each strain within the serovar J- and F-matched pairs. The genome sequence of the serovar F and F(s)-matched pair differed within incA as described in the previous paragraph and differed at four other loci within the genome (Table 3). The most significant difference was a 1,941-bp deletion within the plasticity zone (13) of the F(s) isolate, leading to the deletion of ORFs CT160 and CT161 and the fusion of ORFs CT159 and CT162 (Fig. 2). This deletion was not found in any other examined strain, including two additional serovar F(s) isolates (Fig. 2D). ORFs CT160 and CT161 encode hypothetical proteins with no known homologs in other organisms. There are three other SNPs that varied in these genomes, two within a truncated CT135 in the IncA-negative strain and one in an intergenic region.
TABLE 3.
Summary of nucleotide differences among the sequenced chlamydial strains
| Strain comparison | Total no. of SNPs | No. of indels | No. of genes affected by indels | No. of amino acid substitutions in gene | No. of SNPs in intergenic region |
|---|---|---|---|---|---|
| F/70 vs F(s)/70 | 5 | 3a | 6b | 0 | 1 |
| J/6276 vs J(s)/6276 | 51 | 2 | 2 | 36 | 13 |
| D/UW3 vs D(s)/2923 | 5,764 | 168 | 54 | 2,376 | 1,018 |
Two of these indels consist of a single base pair. The other indel is the 1,941-bp deletion found in strain F(s)70.
The genes affected by these indels include two from genes affected by single base pair indels (CT119 and CT135) and four genes affected by the 1,941-bp deletion (CT159, CT160, CT161, and CT162).
The two matched serovar J strains differed at 51 nucleotide positions within the genome, one of which was the change in incA as described above. Surprisingly, the only other ORF that is affected by a deletion in this matched pair is CT135, which, in this case, is truncated in the wild-type strain but intact in the IncA-negative strain (Fig. 2). Outside of the differences in incA and CT135, the SNPs in the serovar J-matched pair include 13 within intergenic regions and 36 within coding regions (Table 3). Amino acids are changed in 24 of these genes. In order to determine the probable parental sequence within an individual SNP, the nucleotide at each position was compared with genomic data from published (16) and unpublished genome sequences (Table 4). These comparisons showed that 24 of the SNPs most probably changed from parental sequences in the IncA-negative strain, while 25 most probably changed from parental sequences in the IncA-positive strain. This suggested that both strains accumulated mutations at approximately the same rate. Similarly, there were no apparent differences in any category of genetic change between the fusogenic and nonfusogenic strain, including the number of transitions, transversions, deletions, or choice of mutated base. There was, however, a bias in the movement toward A+T in the cumulative mutations, with 32 of the total changes being C or G to A or T. In contrast, 13 total changes went from A or T to G or C.
TABLE 4.
Characterization of genomic changes in the serovar J-matched pair
| Change | Total no. of changes | No. of transitions | No. of transversions | No. of deletions | No. of the following changes:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A to G | C to T | G to A | T to C | C to A | T to A | C to G | C/G to A/T | A/T to C/G | |||||
| From IncA positive to IncA negative | 25 | 22 | 2 | 1 | 3 | 8 | 8 | 4 | 0 | 0 | 2 | 16 | 7 |
| From IncA negative to IncA positive | 24 | 19 | 4 | 1 | 6 | 10 | 3 | 0 | 3 | 1 | 0 | 16 | 6 |
| Unknown | 2 | ||||||||||||
Conventional shotgun sequencing was used to determine the genome sequence of strain D(s)/2923, an IncA-negative C. trachomatis isolate that was isolated in the absence of an IncA-positive match (18). Genome sequence analysis of strain D(s)/2923 identified a single nucleotide deletion in incA that resulted in a 99-amino-acid C-terminal truncation. The complete genome differed from the prototype D/UW3 sequence by 5,764 bp, including 47 cases where indels (insertions or deletions) were differently present in genes of the two strains. The amino acid sequence of MOMP in D(s)/2923 was 99.0% identical to the D/UW3 MOMP sequence.
The above results demonstrate that the genome sequence of strains within a same-patient serovar-matched pair were very similar and, in one case, nearly isogenic. In contrast, an IncA-negative strain isolated in the absence of a serovar-matched IncA-positive strain had many more SNPs relative to the genome sequence of a serovar-matched C. trachomatis strain.
Transcriptional analysis of IncA-positive and -negative strains.
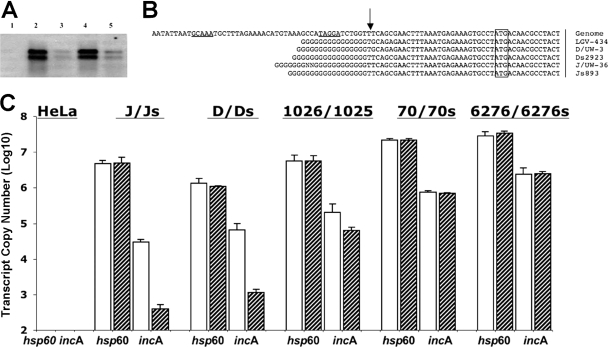
Northern blot analysis and quantitative RT-PCR were used to investigate transcript levels in cloned isolates of IncA-positive and -negative strains. Examination of a set of unmatched IncA-positive and -negative strains demonstrated that incA transcripts are found in both IncA-positive and -negative strains. However, transcript abundance was reduced approximately 10-fold in tested IncA-negative strains (Fig. 3). Two transcripts were also found in each strain, one that measured approximately full length for incA (∼850 bp) and one slightly smaller species. Quantitative PCR was then used to examine cloned strains isolated from patients infected singly with either an IncA-positive or -negative strain, and each cloned isolate from the serovar F- and serovar J-matched pairs. IncA-negative strains collected independently of a matched pair had transcript levels that were consistent with the Northern blotting results (Fig. 3C). However, both the IncA-positive and -negative members of each matched pair had incA transcript levels that were very similar. These data demonstrated that two groups of IncA-negative strains exist with respect to incA transcription, one group had transcript levels similar to those of a serotype-matched strain, and another type that had reduced levels of incA transcript.
FIG. 3.
Transcription of incA in prototype and clinical C. trachomatis strains. (A) Northern blot analysis of wild-type and IncA-negative C. trachomatis strains, probed with full-length incA. Lanes: 1, uninfected HeLa cells; 2, J/UW36; 3, J(s)/893; 4, D/UW3; 5, D(s)/2923. (B) Results of 5′-RACE analysis of the incA transcriptional start site in wild-type and variant strains. The strains analyzed are indicated on the far right, with the D/UW3 genome sequence shown in the top line. The arrow indicates the identified transcriptional start site. The boxed nucleotides shows the predicted translational start site. Underlined sequences on the top line indicate predicted −10 and −35 regions of the serovar D genome sequence. (C) Quantitative RT-PCR assays were performed for different C. trachomatis strains to determine the abundance of incA and hsp60 transcripts. Cells were infected at an MOI of 3. In each case, the IncA-positive strain is indicated by the white bar, while the IncA-negative strain is indicated by the hatched bar. To normalize transcript levels in each total RNA sample used for incA quantitation, the abundance of hsp60 transcripts was measured and used as a control.
The transcriptional start site of incA, as mapped with 5′-RACE analysis, was identical in all tested IncA -positive and -negative strains (Fig. 3B). Additionally, there was no evidence of changes in the sequences upstream of incA that might correlate to the reduced level of transcript.
In vitro growth.
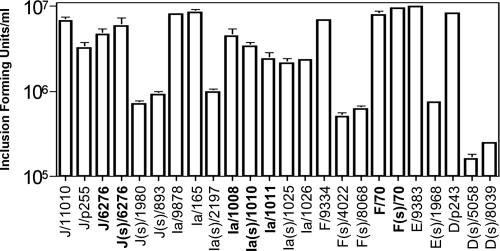
Each of the C. trachomatis isolates used in the mouse experiments, plus each of the strains from the serovar Ia- and F-matched pairs, were used in EB output experiments following culture in McCoy cells (Fig. 4). Each strain was inoculated at an MOI of approximately 0.5. Between 1.3 × 104 and 3.1 × 104 EBs were harvested in cultures conducted immediately after inoculation, and variation in this recovery was not a function of IncA positivity or negativity (not shown). Each strain infected and grew within the cells, but there were clear differences in the numbers of EBs produced in the infections. Consistent with the work of Xia et al. (21), the IncA-negative strains isolated outside a matched pair accumulated at least 10-fold-fewer EBs at both 24 h (Fig. 4) and 48 h p.i. (not shown). In contrast, IncA-positive and -negative members of each matched pair produced very similar numbers of infectious EBs.
FIG. 4.
Production of infectious chlamydia during growth in murine epithelial cells in vitro. Chlamydiae were collected for quantitative culture 24 h postinfection of the primary monolayer. The y axis is logarithmic in scale. Each value represents an average of three wells, and standard errors are indicated with error bars. An (s) designation in a strain name indicates a strain that is IncA negative. Members of a matched pair are shown in bold type.
Mouse infections.
We examined the ability of IncA-positive and -negative strains to colonize the murine genital tract. BALB/c mice were infected and then cultured on days 7 and 10 p.i. (Fig. 5). One matched pair (serovar J) was used in these studies, and several other clinical isolates were tested, none of which were part of a matched pair. Wild-type strains of serovars J, D, and E were compared to serotype-matched IncA-negative strains.
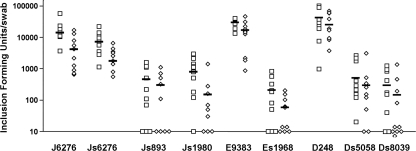
FIG. 5.
Chlamydial shedding from mice infected vaginally with closely related and unrelated same-serovar strains of IncA-positive isolates compared to corresponding IncA-negative variants. The y axis is logarithmic and represents harvested inclusion-forming units per mouse at the particular time point. The open squares show culture data from samples collected 7 days postchallenge. The diamonds show culture data from samples collected 10 days postchallenge. The bar in each data set shows the mean value of the isolates for a particular isolate on a particular day. Data points on the x axis represent culture values below our limits of detection.
The levels of EBs produced by the serovar J and J(s) strains in the matched pair were similar and not statistically different, while the EB level produced by each IncA-negative strain outside a matched pair was significantly different from the level produced by each serotype-matched wild-type strain (P < 0.001, Student's t test). Analyses of variance also demonstrated that there was a significant difference (P < 0.001) between chlamydial production in all IncA-positive versus unmatched IncA-negative strains. These data support the hypothesis that IncA-negative strains that are part of a matched pair grow at a rate similar to those of IncA-positive strains in vivo. In contrast, IncA-negative strains that were not part of a matched pair had an in vivo colonization deficiency relative to both the wild-type strains and the single tested IncA-negative strain that was part of a matched pair.
DISCUSSION
We previously showed that incA is inactivated by frameshifts, deletions, and nonsense mutations, leading to strains of the organism that form nonfusogenic inclusions (15). Although there was some ambiguity concerning this issue in previous publications (15, 18), complete sequence analysis on cloned strains demonstrate that all truly IncA-negative nonfusogenic strains have an interrupted incA coding sequence. Retrospective epidemiological studies demonstrated that the nonfusing strains produced lower numbers of elementary bodies, fewer inflammatory cells, and less severe symptomology in patients (8) and may have growth defects in vitro (21). However, in our analysis of C. trachomatis isolated in the Seattle/King County area in Washington, the incidence of IncA-negative strains approaches 2% of all non-LGV genital serovars except serovar G (18). Therefore, we hypothesize that IncA-negative strains either are generated very commonly in vivo or manifest a unique phenotype allowing them to compete successfully with IncA-positive strains in patients. The first evidence that variation within incA might occur in a single patient was identified in a pair of patients infected with genetically similar strains that were either IncA positive or negative at different culture points during infection (Table 2). These strains were microbiologically cloned for a previous study (6), and the original culture is not available. However, the ability to clone, by limiting dilution, sequential IncA-positive and -negative strains suggested that the mixed population of IncA-positive and -negative C. trachomatis strains persists in patients in vivo. The identification of these serial cultures led us to examine the possibility that individual nonfusogenic isolates in our collection had a minority IncA-positive subpopulation. It was fully possible that we missed such mixed populations in the original screen, since the screen for variant isolates involved the formation of multiple inclusions in cells, a property that would be conserved in a mixed IncA-negative and -positive population (15). Several such strains were identified, including some that were of mixed serovars and three that were of the same serovar. The identification of these same-serovar, IncA-negative and -positive “matched pairs” led to two hypotheses. First, deletions accumulate within the gene during culture in vivo, leading to a subpopulation that is IncA negative, and, second, this variation led to diversity that allows IncA-negative strains to compete with wild-type strains in patients.
If the matched pairs represent very recent mutation events, possibly in the patient being cultured, there should be very little variation between the genomes in a matched pair. A rapid genome sequencing approach was used to demonstrate that this was the case. The serovar F-matched pair, in which the IncA-positive strains represented approximately 20% of the population, had differences at four positions in the genome. The serovar J-matched pair, in which the IncA-positive subpopulation was represented at less than 0.1%, had differences at 51 positions. These data support the conclusion that the matched pairs had recently diverged from one another. In contrast, two serovar D strains that vary at incA but that were isolated independently, differed at over 5,000 bp. We hypothesize that, at least in the case of the serovar F-matched pair, this divergence may have occurred in the patient from which the sample was collected.
We hypothesize that the shift was from the IncA-positive phenotype to the IncA-negative phenotype for two reasons. First, the difference between the IncA-positive and IncA-negative matched pairs are either a deletion (in the serovar F or J pairs) or an insertion in the sequence (the serovar Ia pairs), which lead to different truncated protein products. There are no changes in the reading frame that suggest that a suppressing mutation is present in the IncA-positive strains relative to the IncA-negative strains. Thus, in order to mutate from an IncA-negative strain to a IncA-positive strain, a progeny IncA-positive strain would need to have a correcting mutation at the exact same position in the gene. Second, one of the few differences between the serovar F and F(s) matched pair is a 1,941-bp deletion in the F(s) strain. The presence of this deletion can more easily be explained by the IncA-positive strain being the parent of the IncA-negative strain.
Genome sequencing identified four inactivated genes in the serovar F IncA-negative strain, relative to the matched IncA-positive strain. The inactivated genes included incA, CT135, and a large deletion that removed CT160 and CT161 (Fig. 2). While there were 51 nucleotide differences in the serovar J-matched pair, only two involved indels in coding sequences—the change inactivating incA and a change involving CT135. In this case, however, the IncA-positive, wild-type strain had an inactivated CT135 ORF, while the IncA-negative variant had an intact ORF. This ORF was also shown to be one of approximately 20 variable genes among trachoma biovar strains (11). CT135 is an uncharacterized gene encoding a hypothetical protein that is conserved among Chlamydia trachomatis, Chlamydia muridarum, and Chlamydia suis. Distant homologs are present in the other chlamydiae, but they are not in a syntenous region of the chromosome. Within C. trachomatis, the predicted CT135 protein contains a hydrophobicity motif suggestive of possible localization to the inclusion membrane (Fig. 2) (1). There is no reason to believe that direct inactivation of CT135 is associated with any aspect of the IncA-negative phenotype, as the gene is intact in strain D(s)/2923 and in the IncA-negative member of the serovar J-matched pair. However, the results of collected analysis of this gene suggest that it is commonly variable in otherwise closely related clinical strains, perhaps varying the infectious process by differential localization of the encoded protein to the inclusion membrane.
The absence of IncA on the inclusion membrane is directly associated with lack of fusogenicity in clinical isolates, because IncA molecules on adjacent inclusions participate in homotypic vesicle fusion processes (7, 9, 18). However, we have previously demonstrated that there are growth rate differences, host gene expression differences, and patient phenotype differences between IncA-positive and IncA-negative strains. While these additional traits were associated with the absence of IncA, it may be that lacking IncA is a marker for these differences and not the actual cause of at least some of these phenotypes (21). It is possible that inactivation of IncA is a stage in a randomly selected process that leads to the clinical phenotypes observed in our previous work. The results in the present report provide four lines of evidence for this model. First, the results of genome sequencing suggest that the strains isolated as matched pairs have very little sequence variation relative to strains isolated independently. Second, quantitative PCR data demonstrated no difference in incA transcript production in the serovar J- and F-matched pairs, while unmatched IncA-negative strains had markedly lower levels of transcript. Third, the level of infectious chlamydiae produced was different in IncA-negative strains within a match versus strains isolated independently of a corresponding match. Each strain in the serovar Ia, F, and J pairs had nearly equivalent numbers of EBs produced following infection, while the unmatched IncA-negative strains had at least 10-fold-lower EB counts at both 24 and 48 h postinoculation. Finally, the strains in the serovar J-matched pair had similar infectivities in the mouse model, while all other IncA-negative strains were significantly less infectious in mice. The in vivo and in vitro culture data suggest that these results can be explained by a reduction in growth rate by the unmatched IncA-negative strains. However, the reduction in incA transcription in the unmatched strains, under conditions where a constitutive gene (hsp60) was standardized, emphasizes that a reduction in growth rate is likely associated with other changes in the genesis of distinct IncA-negative strains.
While we were unable to demonstrate a measurable growth rate difference in the in vitro culture of the members of the matched pairs, our cloning strategy for isolation of the IncA-positive minority members of a matched pair demonstrated that these strains could be educed from their IncA-negative partners by harvesting EBs produced at early time points following infection. This suggests that the IncA-positive strains are able to produce EBs more quickly in culture. It is possible that examining EB production earlier, both in the in vitro and the in vivo culture experiments, might identify a measurable growth rate difference between IncA-positive and -negative strains in a matched pair.
We have never identified a spurious IncA-negative strain being generated in any IncA-positive culture in vitro, and thus, it is not likely that changing from an IncA-positive strain to an IncA-negative strain occur at an appreciable level in laboratory culture. IncA-negative strains are, however, found at a rate of approximately 2% in clinical isolates from patients seen at STI clinics in the Seattle area of Washington. The changes are not a function of an expanding clone in the patient population, because the mutations inactivating incA are randomly distributed across the coding sequence and are found in strains representing several different serovars. These traits suggest that there exists an unidentified phenotype associated with the variants that facilitates their selection and maintenance in vivo. Additionally, the identification of matched pairs from clinical samples demonstrates that IncA-negative strains can grow to high levels with IncA-positive strains in the human host.
A role for the IncA-negative strains in the biology of chlamydial infection and disease remains to be elucidated. Antibody against IncA is present in sera from infected patients (2), and a homolog of IncA is a CD8+ T-cell antigen in Chlamydophila pneumoniae (20). Thus, it is possible that IncA is a significant antigen involved in protection against infection. This might lead to selection of randomly generated IncA-negative strains that can avoid the host immune response. However, it is also possible that IncA-negative strains occupy an uncharacterized niche within the host, and their unique ability to occupy this niche allows for their survival within the larger population of IncA-positive strains. Studies on the biology and fitness of IncA-negative C. trachomatis continue in our laboratory.
Acknowledgments
Pacita Roberts provided important statistical analyses in this research. Sara Weeks is acknowledged for technical assistance.
This research is supported in part by grants from the National Institutes of Health (AI48769 and AI031448).
Editor: R. P. Morrison
Footnotes
Published ahead of print on 13 October 2008.
REFERENCES
- 1.Bannantine, J. P., R. S. Griffiths, W. Viratyosin, W. J. Brown, and D. D. Rockey. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell. Microbiol. 235-47. [DOI] [PubMed] [Google Scholar]
- 2.Bannantine, J. P., W. E. Stamm, R. J. Suchland, and D. D. Rockey. 1998. Chlamydia trachomatis IncA is localized to the inclusion membrane and is recognized by antisera from infected humans and primates. Infect. Immun. 666017-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belland, R. J., M. A. Scidmore, D. D. Crane, D. M. Hogan, W. Whitmire, G. McClarty, and H. D. Caldwell. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. USA 9813984-13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 311161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clifton, D. R., K. A. Fields, S. S. Grieshaber, C. A. Dooley, E. R. Fischer, D. J. Mead, R. A. Carabeo, and T. Hackstadt. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. USA 10110166-10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean, D., R. J. Suchland, and W. E. Stamm. 2000. Evidence for long-term cervical persistence of Chlamydia trachomatis by omp1 genotyping. J. Infect. Dis. 182909-916. [DOI] [PubMed] [Google Scholar]
- 7.Delevoye, C., M. Nilges, A. Dautry-Varsat, and A. Subtil. 2004. Conservation of the biochemical properties of IncA from Chlamydia trachomatis and Chlamydia caviae: oligomerization of IncA mediates interaction between facing membranes. J. Biol. Chem. 27946896-46906. [DOI] [PubMed] [Google Scholar]
- 8.Geisler, W. M., R. J. Suchland, D. D. Rockey, and W. E. Stamm. 2001. Epidemiology and clinical manifestations of unique Chlamydia trachomatis isolates that occupy nonfusogenic inclusions. J. Infect. Dis. 184879-884. [DOI] [PubMed] [Google Scholar]
- 9.Hackstadt, T., M. A. Scidmore-Carlson, E. I. Shaw, and E. R. Fischer. 1999. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell. Microbiol. 1119-130. [DOI] [PubMed] [Google Scholar]
- 10.Jeck, W. R., J. A. Reinhardt, D. A. Baltrus, M. T. Hickenbotham, V. Magrini, E. R. Mardis, J. L. Dangl, and C. D. Jones. 2007. Extending assembly of short DNA sequences to handle error. Bioinformatics 232942-2944. [DOI] [PubMed] [Google Scholar]
- 11.Kari, L., W. M. Whitmire, J. H. Carlson, D. D. Crane, N. Reveneau, D. E. Nelson, D. C. Mabey, R. L. Bailey, M. J. Holland, G. McClarty, and H. D. Caldwell. 2008. Pathogenic diversity among Chlamydia trachomatis ocular strains in nonhuman primates is affected by subtle genomic variations. J. Infect. Dis. 197449-456. [DOI] [PubMed] [Google Scholar]
- 12.Mabey, D. 2008. Trachoma: recent developments. Adv. Exp. Med. Biol. 60998-107. [DOI] [PubMed] [Google Scholar]
- 13.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 281397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16276-277. [DOI] [PubMed] [Google Scholar]
- 15.Rockey, D. D., W. Viratyosin, J. P. Bannantine, R. J. Suchland, and W. E. Stamm. 2002. Diversity within inc genes of clinical Chlamydia trachomatis variant isolates that occupy non-fusogenic inclusions. Microbiology 1482497-2505. [DOI] [PubMed] [Google Scholar]
- 16.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282754-759. [DOI] [PubMed] [Google Scholar]
- 17.Suchland, R. J., A. Bourillon, E. Denamur, W. E. Stamm, and D. M. Rothstein. 2005. Rifampin-resistant RNA polymerase mutants of Chlamydia trachomatis remain susceptible to the ansamycin rifalazil. Antimicrob. Agents Chemother. 491120-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suchland, R. J., D. D. Rockey, J. P. Bannantine, and W. E. Stamm. 2000. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect. Immun. 68360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suchland, R. J., and W. E. Stamm. 1991. Simplified microtiter cell culture method for rapid immunotyping of Chlamydia trachomatis. J. Clin. Microbiol. 291333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wizel, B., B. C. Starcher, B. Samten, Z. Chroneos, P. F. Barnes, J. Dzuris, Y. Higashimoto, E. Appella, and A. Sette. 2002. Multiple Chlamydia pneumoniae antigens prime CD8+ Tc1 responses that inhibit intracellular growth of this vacuolar pathogen. J. Immunol. 1692524-2535. [DOI] [PubMed] [Google Scholar]
- 21.Xia, M., R. J. Suchland, R. E. Bumgarner, T. Peng, D. D. Rockey, and W. E. Stamm. 2005. Chlamydia trachomatis variant with nonfusing inclusions: growth dynamic and host-cell transcriptional response. J. Infect. Dis. 1921229-1236. [DOI] [PubMed] [Google Scholar]