Abstract
Growth of Moraxella catarrhalis in a biofilm resulted in marked upregulation of two open reading frames (ORFs), aniA and norB, predicted to encode a nitrite reductase and a nitric oxide reductase, respectively (W. Wang, L. Reitzer, D. A. Rasko, M. M. Pearson, R. J. Blick, C. Laurence, and E. J. Hansen, Infect. Immun. 75:4959-4971, 2007). An ORF designated nsrR, which was located between aniA and norB, was shown to encode a predicted transcriptional regulator. Inactivation of nsrR resulted in increased expression of aniA and norB in three different M. catarrhalis strains, as measured by both DNA microarray analysis and quantitative reverse transcriptase PCR. Provision of a wild-type nsrR gene in trans in an nsrR mutant resulted in decreased expression of the AniA protein. DNA microarray analysis revealed that two other ORFs (MC ORF 683 and MC ORF 1550) were also consistently upregulated in an nsrR mutant. Consumption of both nitrite and nitric oxide occurred more rapidly with cells of an nsrR mutant than with wild-type cells. However, growth of nsrR mutants was completely inhibited by a low level of sodium nitrite. This inhibition of growth by nitrite was significantly reversed by introduction of an aniA mutation into the nsrR mutant and was completely reversed by the presence of a wild-type nsrR gene in trans. NsrR regulation of the expression of aniA was sensitive to nitrite, whereas NsrR regulation of norB was sensitive to nitric oxide.
Moraxella catarrhalis was once thought to be a harmless commensal inhabitant of the human upper respiratory tract, but it is now acknowledged to be an organism that is capable of causing disease in both the upper and lower respiratory tracts of humans (36). In infants and very young children, M. catarrhalis is a common cause of acute otitis media (27, 35, 56). In adults, the disease caused by M. catarrhalis can take a much different and more serious form. This organism has been shown to be an important cause of infectious exacerbations of chronic obstructive pulmonary disease (COPD) (37, 38, 50). It was recently estimated that as many as 2 million to 4 million exacerbations of COPD in the United States can be attributed to M. catarrhalis each year (38). Worldwide, COPD is reported to be the fourth leading cause of death in developed countries, and, according to recent estimates, COPD is projected to become the third leading cause of death worldwide by 2020 (for a review, see reference 8).
The ability of M. catarrhalis to colonize the mucosal surface of the nasopharynx is crucial to its ability to cause disease in other anatomic regions because such colonization provides a foothold for M. catarrhalis in its human host. In fact, colonization of the nasopharynx by M. catarrhalis is common during infancy, and a high rate of colonization with this organism is associated with an increased risk of otitis media (10). The mechanisms essential for colonization of the nasopharyngeal mucosa by M. catarrhalis have not been elucidated to date, although a number of M. catarrhalis gene products that may be involved in this process have been identified in the past few years (12, 21, 22, 29, 30, 34, 43, 44); essentially all of these gene products are surface-exposed proteins with demonstrated adhesive activity. In the human nasopharynx, it is likely that M. catarrhalis exists in a biofilm together with commensal bacteria on the mucosa, and there was a recent report describing M. catarrhalis biofilms detected on the mucosa of the middle ear in children with otitis media (16).
There have been only a few studies of biofilm formation by M. catarrhalis in vitro (6, 34, 41, 62), one of which identified genes which are upregulated when this organism grows in this manner (62). Among the genes whose expression was most highly upregulated during growth in a biofilm were several genes predicted to encode proteins involved in reducing nitrate (NO3−) to nitrous oxide (N2O) (Fig. 1A). These upregulated genes included the narGHJI gene cluster encoding a nitrate reductase complex, aniA encoding a nitrite reductase (also described as the major anaerobically induced outer membrane protein [20]), and norB encoding a nitric oxide reductase (Fig. 1A). While the ability to reduce NO3− to nitrite (NO2−) is a well-established characteristic of M. catarrhalis that can be useful for identification of this organism (7), the presence of the other two enzymes mentioned above indicated that M. catarrhalis possesses the potential to express a truncated denitrification pathway, lacking only the ability to convert N2O to gaseous nitrogen (N2). Moreover, the ability to produce nitric oxide (NO) from NO2− and the ability to reduce NO to N2O could provide M. catarrhalis with the ability to respire more efficiently under oxygen-limited conditions and to protect itself against NO generated by host defense mechanisms (42). Recent studies with Neisseria meningitidis, another pathogen that initially colonizes the mucosa of the nasopharynx, have shown that its nitric oxide reductase can affect NO-based signaling processes in human cells (51, 55).
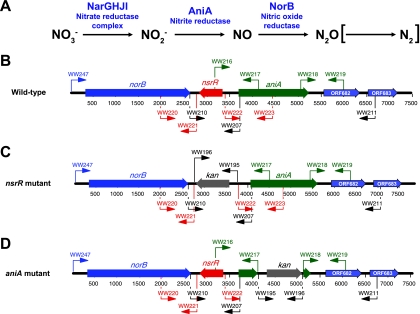
FIG. 1.
Schematic diagrams of M. catarrhalis gene products involved in the truncated denitrification pathway and construction of relevant mutants. (A) The truncated denitrification pathway in M. catarrhalis involves the first three enzymatic steps shown. M. catarrhalis apparently lacks the ability to reduce N2O to N2 (indicated by brackets). (B to D) Schematic diagrams of the M. catarrhalis chromosomal locus containing the norB, nsrR, and aniA genes and flanking regions in (B) wild-type O35E strain, (C) the O35E nsrR mutant, and (D) the O35E aniA mutant. The relative positions of the different primers used for PCR are indicated by arrows.
In this paper, we show that M. catarrhalis can reduce both NO2− and NO by virtue of its expression of the AniA and NorB proteins, respectively. We also show that an open reading frame (ORF) located between the aniA and norB genes encodes NsrR, a regulatory protein which controls the level of expression of both AniA and NorB. Inactivation of the M. catarrhalis nsrR gene resulted in overexpression of both AniA and NorB, which could be correlated with inhibition of growth by exogenously added NO2−. We also show that several other M. catarrhalis genes are regulated, directly or indirectly, by NsrR.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. catarrhalis strains used in this study are listed in Table 1. The base medium employed in this study was brain heart infusion (BHI) (Difco, Detroit, MI) medium, and broth cultures were incubated at 37°C with aeration. BHI medium was supplemented with kanamycin (15 μg/ml) or spectinomycin (15 μg/ml) when appropriate. All BHI agar plates were incubated at 37°C in an atmosphere containing 95% air and 5% CO2.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description or genotype | Reference or source |
|---|---|---|
| Strains | ||
| 7169 | Wild-type strain | 33 |
| 7169 nsrR | nsrR::Kanr | This study |
| 7169 aniA | aniA::Kanr | This study |
| ATCC 43617 | Wild-type strain | ATCC |
| ETSU-9 | Wild-type strain | Steven Berk |
| ETSU-9 aniA | aniA::Kanr | This study |
| ETSU-9 nsrR | nsrR::Kanr | This study |
| ETSU-9 nsrR(pWW115) | ETSU-9 nsrR containing pWW115 vector | This study |
| ETSU-9 nsrR(pWW150) | ETSU-9 nsrR containing a cloned wild-type nsrR gene from ATCC 43617 | This study |
| O35E | Wild-type strain | 18 |
| O35E aniA | aniA::Kanr | This study |
| O35E nsrR | nsrR::Kanr | This study |
| O35EΔnsrR | nsrR deletion mutant | This study |
| O35EΔnsrR aniA | nsrR aniA double mutant | This study |
| O35E nsrR(pWW115) | O35E nsrR containing pWW115 vector | This study |
| O35E nsrR(pWW150) | O35E nsrR containing a cloned wild-type nsrR gene from ATCC 43617 | This study |
| Plasmids | ||
| pWW115 | Specr, cloning vector for M. catarrhalis | 61 |
| pWW150 | pWW115 containing the cloned wild-type ATCC 43617 nsrR gene | This study |
To measure bacterial growth, bacteria were grown overnight on BHI agar and then suspended in BHI broth to a density of 260 Klett units. A 1-ml portion of the suspension was used to inoculate 20 ml of BHI broth in a 500-ml flask, which was agitated (200 rpm) at 37°C. Growth was measured turbidimetrically every hour.
A freshly prepared solution of 2.5 M NaNO2 (in water) was filter sterilized and was added (when needed) to BHI broth to a final concentration of 5 mM before the medium was used. To study the effect of NO on the expression of certain M. catarrhalis genes, spermine NONOate {N-4-[1-(3-aminopropyl)-2-hydroxy-2-nitrosohydrazino]butyl-1,3-propanediamine} was added to bacterial cultures to generate NO (46). Briefly, a freshly prepared solution of 25 mM spermine NONOate (in 0.1 M NaOH) was filter sterilized and was added to a logarithmic-phase culture to a final concentration of 50 μM. Growth was allowed to continue for 45 min, and then spermine NONOate was added again to the same final concentration. After an additional 45-min growth period, the cells were harvested for extraction of total RNA. Control cultures received equivalent portions of 0.1 M NaOH.
Whole-cell lysate preparation and Western blot analysis.
Whole-cell lysates were prepared from BHI agar-grown cells as described previously (40). Western blot analysis was performed as described previously (60).
Construction of M. catarrhalis mutants. (i) nsrR mutants.
DNA fragments containing nucleotide sequences located immediately 5′ and 3′ of the nsrR ORF were amplified by PCR with oligonucleotide primer pairs WW220/WW221 and WW222/WW223 (Fig. 1B and Table 2) using M. catarrhalis ATCC 43617 genomic DNA as the template. The two DNA fragments were used as templates for overlapping extension PCR (24) with primers WW220 and WW223. The resultant amplicon, designated ΔNSRR and containing an internal SmaI site, was digested with both BamHI and SacI and then ligated into the M. catarrhalis plasmid cloning vector pWW115 (61) that had been digested with the same restriction enzymes. The ligation reaction mixture was used to electroporate M. catarrhalis ETSU-9. The plasmid isolated from one of the spectinomycin-resistant clones was designated pWW123. The kan cartridge from pAC7 was amplified by PCR as described previously (62) and then cloned into the SmaI site in the DNA insert in pWW123. The resultant recombinant plasmid, designated pWW124 and having the kan cartridge inserted in the same direction as the deleted nsrR gene, was used as the template for PCR amplification using primers WW220 and WW223. The resultant PCR product, designated ΔNSRR-KAN, was used to transform M. catarrhalis wild-type strains O35E, 7169, and ETSU-9. Kanamycin-resistant transformants were confirmed to be nsrR mutants (Fig. 1C) by performing anchored PCR using oligonucleotide primers WW210 and WW211 (Fig. 1B and Table 2), followed by nucleotide sequence analysis. A kanamycin-sensitive nsrR mutant was constructed by using the ΔNSRR DNA fragment described above to transform the kanamycin-resistant O35E nsrR mutant. One of the resultant kanamycin-sensitive transformants was confirmed to be an nsrR deletion mutant, designated O35EΔnsrR, by nucleotide sequence analysis.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| WW207 | TATCCCGGGTTCTCCTTATAGAAAGGT |
| WW210 | GACACGCCCATCACAGACTAA |
| WW211 | GCAACTCAACATGCTCATGAT |
| WW216 | TTGGATCCGCGCACTGTCAACGACTTTa |
| WW217 | GACAATCCATTTGCCCGGGTGGTTTGAGAACTGCACTb |
| WW218 | GGCAAATGGATTGTCTGGTAA |
| WW219 | CGACGAGCTCCCATTCTGCAATCTTAGGTc |
| WW220 | GAGGATCCACCACTTATATTTCACAa |
| WW221 | GTTTTAGATTGCCCGGGTCAATGATGGCTGGATTTAb |
| WW222 | CGGGCAATCTAAAACACTCAATCTCA |
| WW223 | TGGCGAGCTCCATATCAAAGGGCTGTAGAc |
| WW247 | TAGGATCCAATCACACTTAGGATTATCA |
| 683Fw | GGACATGATGGGCATGGTC |
| 683Rv | TAAGGCAGGTGCTGGCG |
| 1550Fw | AGACACCATCAGCGTTGAGCT |
| 1550Rv | AAGGTGACCTCTTCGGTGACAT |
The BamHI site is underlined.
The SmaI site is underlined.
The SacI site is underlined.
(ii) aniA mutants.
DNA fragments containing nucleotide sequences located immediately 5′ and 3′ of the aniA ORF were amplified by PCR with primer pairs WW216/WW217 and WW218/WW219 (Fig. 1B and Table 2) using M. catarrhalis ATCC 43617 genomic DNA as the template. The two DNA fragments were used as templates for overlapping extension PCR (24) with primers WW216 and WW219. The resultant amplicon, designated ΔANIA and containing an internal SmaI site, was digested with both BamHI and SacI and then ligated into pWW115 that had been digested with the same restriction enzymes. The ligation reaction mixture was used to electroporate M. catarrhalis ETSU-9. The plasmid isolated from one of the spectinomycin-resistant clones was designated pWW121, and the kan cartridge described above was cloned into the SmaI site in the DNA insert in pWW121. The resultant recombinant plasmid, designated pWW122 and having the kan cartridge inserted in the same direction as the deleted aniA gene, was used as the template for PCR amplification using primers WW216 and WW219. The resultant PCR product, designated ΔANIA-KAN, was used to transform M. catarrhalis wild-type strains O35E and ETSU-9. Kanamycin-resistant transformants were confirmed to be aniA mutants (Fig. 1D) by performing anchored PCR using oligonucleotide primers WW216 and WW211 (Fig. 1B and Table 2), followed by nucleotide sequence analysis.
(iii) aniA nsrR mutant.
The PCR amplicon ΔANIA-KAN described above was used to transform the M. catarrhalis O35EΔnsrR mutant. One of the kanamycin-resistant transformants was confirmed to be an nsrR aniA double mutant by performing anchored PCR followed by nucleotide sequence analysis.
Cloning of the M. catarrhalis nsrR gene for use in complementation analysis.
A DNA fragment containing the wild-type nsrR gene was PCR amplified by using oligonucleotide primers WW207 and WW210 (Fig. 1B) with M. catarrhalis ATCC 43617 genomic DNA as the template. The amplicon was ligated into the SmaI site in pWW115 and used to transform ETSU-9. A plasmid designated pWW150 containing the wild-type ATCC 43617 nsrR gene was isolated from one of the spectinomycin-resistant transformants.
RNA isolation and real-time qRT-PCR analysis.
Total RNA was isolated from broth-grown and biofilm-grown cells as described previously (62). Quantitative reverse transcriptase PCR (qRT-PCR) was performed as described previously (62). The oligonucleotide primer pairs used for qRT-PCR analysis involving norB (MC ORF 679), aniA (MC ORF 681), and the endogenous control MC ORF 1234 have been described previously (62). Other oligonucleotide primers used in this study are listed in Table 2.
Identification of NsrR-regulated genes by DNA microarray analysis.
Total RNA was isolated from M. catarrhalis wild-type strain O35E, 7169, and ETSU-9 cells and the nsrR mutants of these strains grown in BHI broth. DNA microarray analysis was performed as described previously (62) to identify genes whose expression was affected by NsrR. Independent experiments were performed to obtain a total of six sets of RNA samples (two samples per set) from three M. catarrhalis strain pairs (i.e., wild-type cells and nsrR mutant cells grown at the same time). These samples included one set from the 7169 strain pair, two sets from the ETSU-9 strain pair, and three sets from the O35E strain pair. “Dye swap” experiments were performed twice, once with ETSU-9 samples and once with O35E samples. Statistical analysis of the data was performed as described previously (62). NsrR-regulated gene expression in M. catarrhalis O35E was confirmed by qRT-PCR analysis. Two additional independent experiments were performed to isolate two sets of RNA samples from the M. catarrhalis O35E wild-type strain and nsrR mutant for qRT-PCR, and these analyses were performed twice for each set of RNA preparations as described previously (62).
Generation of polyclonal antibody.
An oligopeptide (YTKGKYGEQGLQPFDMEKAIRED) containing amino acids 236 to 258 of the predicted M. catarrhalis ATCC 43617 AniA protein was synthesized by the Protein Technology Center at the University of Texas Southwestern Medical Center and was used to immunize mice for production of polyclonal AniA antibody.
Determination of NO, N2O, and NO2− production and consumption.
To determine NO production in the presence of NO2−, wild-type, nsrR mutant, and aniA mutant M. catarrhalis O35E strains were grown in BHI broth to an optical density at 600 nm (OD600) of 2.0. The cells were then washed and resuspended in fresh BHI broth to an OD600 of 1.0, which was followed by addition of 5 mM (final concentration) NaNO2. NO production was monitored using an ISO-NOPMC Mark II electrode (WPI Instruments, Sarasota, FL) run through an MLT1122 analog adapter system (AD Instruments, Colorado Springs, CO) with standard curves generated according to the manufacturer's instructions. To measure NO2− consumption, wild-type, aniA, nsrR, and nsrR aniA M. catarrhalis O35E strains were grown in BHI broth to an OD600 of 2.0, washed, and resuspended in BHI broth to an OD600 of 1.0. After addition of 5 mM (final concentration) NaNO2, the concentration of NO2− remaining was determined using a standard Griess procedure. Briefly, culture supernatant fluid was prepared by pelleting cells after 5 min of incubation at 70°C. A 100-μl portion of this culture supernatant fluid was mixed with a 100-μl portion of a 1:1 mixture of 1% sulfanilamide (in 2.5% H3PO4) and 0.1% N-1-naphthylethylenediamine hydrochloride (in 2.5% H3PO4) and incubated at room temperature for 15 min. The NO2− concentration was determined by measuring the absorbance at 550 nm of the mixture and comparing the data to a standard curve. The NO-consuming activity of M. catarrhalis wild-type strain O35E and the isogenic O35E nsrR mutant was measured by growing cells to an OD600 of 2.0 in BHI medium and then washing and resuspending the cells in BHI broth to an OD600 of 1.0. Addition of 10 mM ProliNO (half-life, 1.8 s; AG Scientific, San Diego, CA) was used to generate 20 μM NO. The concentration of dissolved NO remaining at different times was monitored using an ISO-NOPMC Mark II electrode (WPI Instruments). The production of N2O by wild-type, aniA, nsrR, and nsrR aniA M. catarrhalis O35E strains was monitored using an oxygen-insensitive, N2O-specific probe (N2O-50-3112; Unisense AS, Aarhus, Denmark) connected to a picoammeter (PA2000; Unisense AS) and run through an A/D converter (ADC 216; Unisense AS). Cells were grown in BHI broth to an OD600 of 2.0 and resuspended in BHI medium to a final OD600 of 1.0, and this was followed by addition of 5 mM (final concentration) NaNO2.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the nucleotide sequences of the norB, nsrR, and aniA genes included in this study are as follows: for M. catarrhalis O35E, EU861986; for M. catarrhalis 7169, EU861987; and for M. catarrhalis ETSU-9, EU861988. The nucleotide sequences of the norB, nsrR, and aniA genes from M. catarrhalis ATCC 43617 have been deposited under accession number AX067454.
RESULTS
DNA sequence analysis of the locus containing the aniA and norB genes.
It was previously shown that growth of M. catarrhalis in the biofilm state resulted in upregulation of numerous different genes, including several genes predicted to encode proteins involved in a dissimilatory nitrate reduction pathway (62). In addition to genes encoding components of a nitrate reductase complex, genes encoding a predicted nitrite reductase (aniA) and a predicted nitric oxide reductase (norB) were among these most highly upregulated genes (62). Examination of the M. catarrhalis ATCC 43617 genome (GenBank accession numbers AX067426 to AX067466) revealed that the aniA and norB genes are located close to each other and are transcribed in the same direction (Fig. 1B). The predicted M. catarrhalis AniA protein was 68% identical to the Neisseria gonorrhoeae AniA nitrite reductase (formerly designated the anaerobically induced major outer membrane protein) (20). The M. catarrhalis norB gene encoded a predicted protein that was similar (64 to 65% identity) to the NorB nitric oxide reductases of both N. meningitidis and N. gonorrhoeae.
A third ORF (MC ORF 680) was located between the norB and aniA genes and was transcribed in the opposite direction (Fig. 1B). The protein encoded by this ORF was designated NsrR (for reasons described below). The 182-amino-acid NsrR protein most closely resembled (51% identity) a predicted BadM/Rrf2 transcriptional regulator found in Psychrobacter sp. strain PRwf-1 (GenBank accession no. YP_001279665.1). The M. catarrhalis NsrR protein was also similar (35% identity) to the N. meningitidis MC58 NsrR repressor, a protein which controls expression of genes encoding enzymes involved in dissimilatory nitrite reduction in this pathogen (46). Further examination of the M. catarrhalis NsrR protein sequence revealed that it contains the conserved domain of RirA, a novel iron-responsive transcriptional regulator (54, 57) found in nitrogen-fixing bacteria which may have a function similar to that of the Fur protein (for a review, see reference 49).
In a preliminary effort to determine the degree of conservation of the aniA, nsrR, and norB genes among M. catarrhalis strains, oligonucleotide primer pairs WW247/WW207 and WW210/WW211 (Fig. 1B and Table 2) were used to amplify overlapping DNA fragments using genomic DNA isolated from M. catarrhalis strains O35E, 7169, and ETSU-9 as the templates. Nucleotide sequence analysis showed that the predicted amino acid sequences of the AniA and NsrR proteins from these three strains were identical, whereas there were a few amino acid differences in the NorB proteins among these strains.
Construction of M. catarrhalis nsrR mutants.
The DNA microarray-based study which showed that both norB and aniA were among the most upregulated genes in biofilm-grown M. catarrhalis cells also indicated that these two genes were among the most downregulated genes when this organism was grown under iron-limiting conditions (62). Although these two genes showed the same apparent pattern of regulation, it is apparent from their arrangement in the chromosome that they are not in an operon (Fig. 1B). To determine whether the NsrR protein might be involved in the regulation of expression of the norB and aniA genes, we constructed nsrR mutations, first in strain ETSU-9 and then later in strains O35E and 7169, as described in Materials and Methods.
qRT-PCR analysis of aniA expression in ETSU-9 wild-type and nsrR mutant cells.
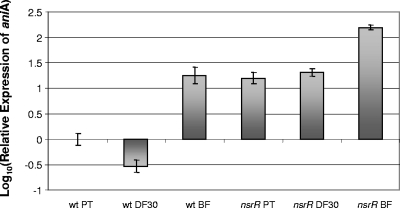
Total RNA samples obtained from M. catarrhalis ETSU-9 wild-type and nsrR mutant cells grown under planktonic conditions, under iron-limiting conditions, and in a biofilm as described previously (62) were used to measure expression of aniA by qRT-PCR. In the wild-type strain, expression of aniA was downregulated in iron-limiting media and was upregulated more than 10-fold in the biofilm (Fig. 2), in the same manner that was previously observed for M. catarrhalis ATCC 43617 (62). In contrast, expression of aniA was upregulated in the nsrR mutant under all three growth conditions, and the relative increase was greatest (more than 100-fold) in the biofilm-grown cells (Fig. 2). These results suggested that the nsrR gene might encode a regulatory protein that represses expression of aniA under aerobic growth conditions.
FIG. 2.
Expression of aniA by M. catarrhalis ETSU-9 wild-type and mutant strains under different growth conditions, as measured by qRT-PCR. Total RNA was isolated from wild-type ETSU-9 (wt) and ETSU-9 nsrR mutant (nsrR) cells grown in the planktonic state (PT) under iron limitation conditions in broth containing 30 μM Desferal (DF30) and in a continuous-flow biofilm system (BF) as described previously (62). qRT-PCR was performed as described in Materials and Methods.
Effect of NsrR on expression of AniA.
The aniA gene was inactivated in M. catarrhalis strains O35E and ETSU-9 as described in Materials and Methods (Fig. 1D). The aniA mutants were used initially as negative controls in a Western blot analysis in which AniA expression was measured in two different strain backgrounds. In addition, we cloned the wild-type nsrR gene from M. catarrhalis ATCC 43617 into pWW115. Recombinant plasmid pWW150 containing this nsrR gene was used to transform both the O35E and ETSU-9 nsrR mutants. Both of these mutants were also independently transformed with the plasmid vector pWW115.
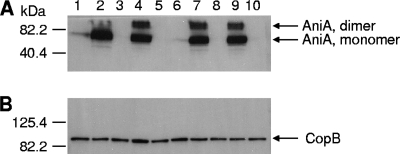
The wild-type O35E cells (Fig. 3A, lane 1) and wild-type ETSU-9 cells (Fig. 3A, lane 6) expressed very low levels of AniA. In contrast, the O35E nsrR mutant (Fig. 3A, lane 2) and the ETSU-9 nsrR mutant (Fig. 3A, lane 7) expressed readily detectable amounts of AniA. As expected, the O35E aniA mutant (Fig. 3A, lane 3) and the ETSU-9 aniA mutant (Fig. 3A, lane 8) did not express AniA protein. A plasmid-borne wild-type nsrR gene eliminated detectable expression of AniA in O35E nsrR(pWW150) (Fig. 3A, lane 5) and in ETSU-9 nsrR(pWW150) (Fig. 3A, lane 10). The presence of the plasmid vector alone in O35E nsrR(pWW115) (Fig. 3A, lane 4) and in ETSU-9 nsrR(pWW115) (Fig. 3A, lane 9) had no apparent effect on the expression of AniA by these two mutants.
FIG. 3.
Effect of NsrR on AniA protein expression in wild-type, mutant, and complemented mutant strains of M. catarrhalis. Whole-cell lysates of the strains were probed in a Western blot analysis with polyclonal AniA antiserum (A) or with the CopB-specific monoclonal antibody 10F3 (18) (B) as the primary antibody. Lane 1, wild-type strain O35E; lane 2, O35E nsrR mutant; lane 3, O35E aniA mutant; lane 4, O35E nsrR(pWW115); lane 5, O35E nsrR(pWW150); lane 6, wild-type ETSU-9; lane 7, ETSU-9 nsrR mutant; lane 8, ETSU-9 aniA mutant; lane 9, ETSU-9 nsrR(pWW115); lane 10, ETSU-9 nsrR(pWW150). The positions of the putative AniA monomers and dimers are indicated by arrows on the right in panel A. The CopB outer membrane protein was used as a loading control, and its position is indicated by an arrow on the right in panel B. The positions of molecular mass markers are indicated on the left in each panel.
Effect of NO2− on the growth of wild-type, mutant, and complemented mutant strains of M. catarrhalis.
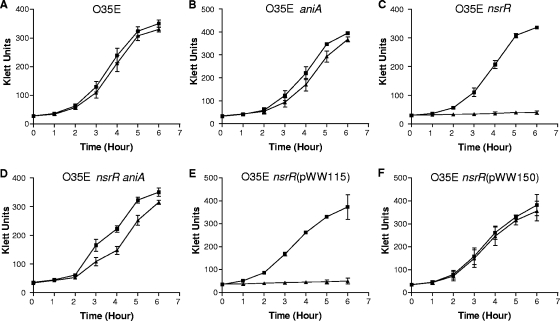
To begin to explore the biological significance of the M. catarrhalis NsrR protein, the effects of different nitrogen compounds on the growth of M. catarrhalis were investigated. Inclusion of 5 or 10 mM NaNO3 in the broth growth medium did not have any detectable effect on the growth of the wild-type O35E strain or its nsrR mutant (data not shown). The presence of 5 mM NaNO2 had little or no effect on growth of wild-type O35E (Fig. 4A). Similarly, NO2− had at most a very modest effect on the growth of the O35E aniA mutant (Fig. 4B). In contrast, growth of the O35E nsrR mutant was completely inhibited by 5 mM NO2− (Fig. 4C). The same inhibitory effect of NO2− was observed with nsrR mutants of the 7169 and ETSU-9 strains (data not shown). This inhibition of growth by NO2− was significantly reversed by the presence of an aniA mutation in the O35E nsrR mutant background (Fig. 4D) and was completely reversed by the presence of a wild-type nsrR gene in trans (Fig. 4F). As expected, the presence of the vector pWW115 alone in the O35E nsrR mutant had no effect on its growth in the presence of NO2− (Fig. 4E).
FIG. 4.
Effect of NO2− on the growth of wild-type, mutant, and complemented mutant strains of M. catarrhalis. Cells of wild-type strain O35E (A), the O35E aniA mutant (B), the O35E nsrR mutant (C), the O35E ΔnsrR aniA mutant (D), O35E nsrR(pWW115) (E), and O35E nsrR(pWW150) (F) were grown in BHI medium (▪) or in BHI medium containing 5 mM NaNO2 (▴). Most of the data are means for three independent growth experiments; the exception is the data in panel B, which are the means for two independent experiments.
Identification of NsrR-regulated M. catarrhalis genes.
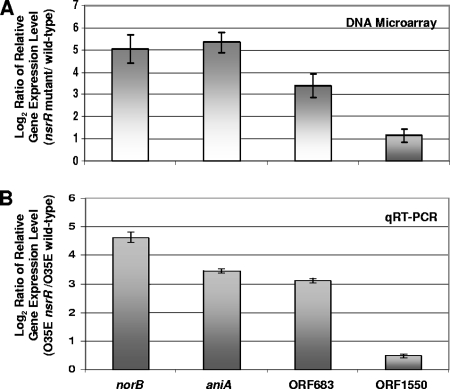
DNA microarray analysis was used to identify genes whose relative expression was upregulated at least twofold in all three nsrR mutants compared with the level of expression in the corresponding wild-type parental strains. The relative expression levels of four ORFs were shown to be increased at least twofold (P < 0.002) in all three nsrR mutants, as measured by DNA microarray analysis (Fig. 5A). These ORFs were norB (expression increased ∼30-fold), aniA (expression increased ∼30-fold), MC ORF 683 (expression increased ∼10-fold), and MC ORF 1550 (expression increased twofold). MC ORF 683 encodes a protein that has 39% identity with a hypothetical protein from Pseudoalteromonas haloplanktis (GENE ID 3708074 PSHAa1482). MC ORF 1550 encodes a protein with 67% identity to a photosynthetic reaction center (PRC)-barrel domain protein (GENE ID 5206683 PsycPRwf_2012) from a Psychrobacter species. The PRC-barrel domain is a conserved domain found in PRC subunits (2) and in some proteins involved in RNA metabolism (2, 32). qRT-PCR analysis was performed to confirm that expression of these four ORFs was negatively regulated by NsrR. In the M. catarrhalis O35E nsrR mutant, the expression of all four ORFs was upregulated compared to the expression in wild-type parental strain O35E (Fig. 5B). Finally, a putative NsrR-binding site, as defined by Rodionov et al. (47), was present upstream from all four of these ORFs (data not shown).
FIG. 5.
Identification of M. catarrhalis genes regulated by NsrR. (A) A DNA microarray analysis of total RNA isolated from wild-type strains O35E, 7169, and ETSU-9 and nsrR mutants of these strains was performed as described in Materials and Methods. Data for the genes consistently upregulated at least twofold (P < 0.002) in all three nsrR mutants are shown. The error bars indicate standard deviations. (B) qRT-PCR analysis of the relative levels of expression of norB, aniA, MC ORF 683, and MC ORF 1550 in nsrR mutant and wild-type cells of strain O35E. The endogenous control for the qRT-PCR measurements was expression of MC ORF 1234 (62). The maximum and minimum relative levels of expression are indicated by the error bars.
Effects of different nitrogenous compounds on expression of M. catarrhalis NsrR-regulated genes.
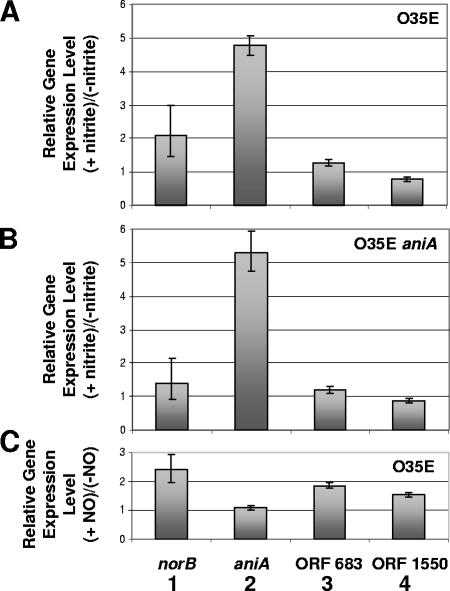
qRT-PCR analysis was performed to examine the effects of NO2− and NO on the relative expression levels of aniA, norB, MC ORF 683, and MC ORF 1550. It is important to note that the oligonucleotide primer pair used for detection of aniA transcripts (62) was located inside the 5′ end of the aniA ORF but was upstream of the region that was deleted in the aniA mutant (Fig. 1D). Therefore, use of this primer pair in qRT-PCR could detect expression of the truncated aniA transcript in the aniA mutant. Transcription of aniA was increased about fivefold in both the O35E wild-type strain (Fig. 6A) and the O35E aniA mutant (Fig. 6B) in the presence of NO2−. Addition of NO2− to cells of the wild-type O35E strain resulted in the production of a readily detectable level of NO, whereas the O35E aniA mutant produced little if any detectable NO under these same conditions (Fig. 7A). Taken together, these results suggested that NsrR-regulated expression of aniA was sensitive to NO2−.
FIG. 6.
Effects of nitrogenous compounds on expression of genes regulated by NsrR. (A and B) Total RNA extracted from cells of M. catarrhalis wild-type strain O35E (A) and the O35E aniA mutant (B) grown in the presence or absence of 5 mM NaNO2 were used for qRT-PCR. (C) Total RNA extracted from logarithmic-phase M. catarrhalis O35E cells grown for 90 min in the presence or absence of the NO-generating compound spermine NONOate was used for qRT-PCR. The relative levels of expression of norB (bars 1), aniA (bars 2), MC ORF 683 (bars 3), and MC ORF 1550 (bars 4) are shown. The endogenous control for the qRT-PCR measurements was the expression of MC ORF 1234 (62). The maximum and minimum relative levels of expression are indicated by the error bars.
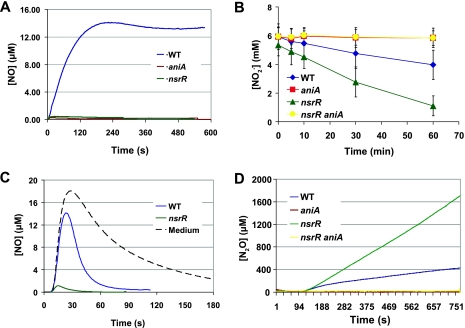
FIG. 7.
Metabolism of nitrogenous compounds by M. catarrhalis wild-type strain O35E and the strain O35E mutants. (A) Production of NO from NO2−. Cell suspensions of the wild-type strain (WT), the aniA mutant, and the nsrR mutant in BHI medium supplemented with 5 mM NaNO2 were monitored for 10 min for the presence of NO using an NO-specific electrode. The probe specificity was affirmed by the ability of an NO scavenger, Carboxy-PTIO, to quench the measurable signal (not shown). (B) Consumption of NO2−. Cell suspensions of the wild-type strain, the aniA mutant, the nsrR mutant, and the nsrR aniA mutant in BHI medium supplemented with 5 mM NaNO2 were monitored for the presence of NO2− using the Griess reaction. (C) Consumption of NO. Cell suspensions of the wild-type strain and the nsrR mutant were exposed to 20 μM NO produced by addition of 10 mM ProliNO. The dissolved NO concentration was monitored using an NO-specific electrode. For reference, the NO-consuming activity of BHI medium was determined (dotted line). (D) Production of N2O. Cell suspensions of the wild-type strain, the aniA mutant, the nsrR mutant, and the nsrR aniA mutant in BHI medium supplemented with 5 mM NaNO2 (administered 2 min into the assay) were monitored for the presence of N2O using an N2O-specific electrode.
Addition of NO2− resulted in elevated expression (i.e., twofold) of norB in the O35E wild-type strain (Fig. 6A) but did not increase expression of norB significantly in the O35E aniA mutant (Fig. 6B). Expression of the other two ORFs (MC ORF 683 and MC ORF 1550) in the putative NsrR regulon was not affected by the presence of NO2− (Fig. 6A and 6B). When the NO-generating agent spermine NONOate was added to a logarithmic-phase culture of M. catarrhalis wild-type strain O35E, expression of norB was increased approximately twofold after 90 min (Fig. 6C). In contrast, expression of aniA was not affected by NO (Fig. 6C). These results directly confirmed that the expression of aniA is not sensitive to NO, whereas the expression of norB is sensitive to NO.
Metabolism of nitrogenous compounds by M. catarrhalis.
The production of NO from NO2− by wild-type strain O35E but not by the O35E aniA mutant (Fig. 7A) confirmed that the M. catarrhalis AniA protein is indeed a nitrite reductase which can produce NO by reducing NO2−. In addition, it was shown that the O35E nsrR mutant consumed NO2− much faster than the wild-type parent strain O35E (Fig. 7B). This was most likely the result of the elevated expression of AniA in the O35E nsrR mutant (Fig. 3A, lane 2) because the O35E aniA mutant did not consume NO2− during the same period of time (Fig. 7B). More importantly, inactivation of the aniA gene in the O35E nsrR mutant completely eliminated consumption of NO2− (Fig. 7B).
Examination of the consumption of NO by M. catarrhalis strains showed that the O35E nsrR mutant consumed NO much faster than the O35E wild-type strain (Fig. 7C). This most likely resulted from the increased expression of norB in the O35E nsrR mutant (Fig. 5). This very rapid consumption of NO by the nsrR mutant provides an explanation for the very low level of detectable NO produced from NO2− by this nsrR mutant (Fig. 7A).
In a preliminary experiment, the M. catarrhalis O35E nsrR mutant generated a much higher level of N2O than its parent strain (Fig. 7D) following the addition of NO2−. This nsrR mutant produced 2.5 mM N2O from 5 mM NaNO2 in about 50 min, whereas the wild-type parent strain generated about 0.4 mM N2O in the same time period (data not shown). As expected, inactivation of the aniA gene resulted in no detectable level of N2O production from nitrite by either the O35E aniA mutant or the O35E ΔnsrR aniA mutant (Fig. 7D).
DISCUSSION
Interest in the reduction of nitrogenous compounds (i.e., denitrification) by bacterial pathogens has increased in the past few years (for a review, see reference 42). Denitrification is essentially the reduction of soluble anionic nitrogen oxides to volatile uncharged gases (NO, N2O, or N2), which results in a net loss of nitrogen from the local environment. Bacteria often perform denitrification as a substitute for aerobic respiration, where a nitrogen oxide rather than molecular oxygen serves as the electron acceptor. Accordingly, denitrification is employed to generate an electrochemical gradient across the cytoplasmic membrane via coupling sites that expel protons to the periplasmic space. In contrast to aerobic respiration, where molecular oxygen is associated with a terminal oxidase during its tetravalent reduction to water, denitrification involves the generation of many freely diffusible intermediates that interact with several independent reductases. The reaction generally results in the following sequential reduction: NO3− → NO2− → NO → N2O → N2. However, there is a great deal of diversity among denitrifying microbes with respect to the types of enzymes involved, as well as the completeness of the reaction (for a review, see reference 64).
While bacteria (e.g., some Pseudomonas species) that are able to live in several different environments (including a human host) have well-studied denitrification pathways that result in the conversion of NO3− to gaseous nitrogen (64), only recently has it been determined that that some obligate human pathogens (e.g., N. meningitidis) also can reduce some nitrogenous compounds to the level of N2O (3, 45). Indeed, under some conditions, nitrate reductase activity encoded by the narGHJI gene cluster of Mycobacterium bovis BCG can contribute to virulence in an animal model (63), and expression of a regulator (NnrA) of denitrification genes in Brucella melitensis is required for virulence in mice (15). Recent studies with N. meningitidis indicated that the nitric oxide reductase NorB is primarily responsible for the nitric oxide detoxification accomplished by this pathogen, as measured in both macrophages and a nasopharyngeal mucosa organ culture system (52).
M. catarrhalis is an obligate aerobe and cannot grow under anaerobic conditions (1, 23). The ability of this organism to reduce NO3− to the level of NO2− and beyond has been documented previously (for a review, see reference 7). However, only recently were the genes that likely encode the relevant enzymes identified in a study of global gene regulation in M. catarrhalis (62). Genes encoding a nitrate reductase complex (NarGHJI), a predicted nitrite reductase (AniA), and a predicted nitric oxide reductase (NorB) were all found to be highly upregulated when M. catarrhalis was grown in a biofilm in vitro (62). In the present study, we identified an ORF (nsrR) which, when inactivated, allowed increased expression of both aniA and norB. This effect was not strain specific but occurred in the three different nsrR mutants tested in this study (Fig. 2 and 5A). DNA microarray analysis of the transcriptomes of the same nsrR mutants showed that at least two other ORFs (MC ORF 683 and MC ORF 1550) were upregulated in the absence of NsrR expression (Fig. 5). Analysis of the regions immediately upstream from these four ORFs revealed, in all four instances, the presence of a sequence with homology to the NsrR-binding site defined previously in studies of other bacteria (47). While this putative NsrR regulon may appear to be rather small, the NsrR regulon in N. meningitidis (19) was recently shown to contain only five genes, including aniA and norB. In contrast, the NsrR regulon in Escherichia coli is much larger and contains at least 20 genes (11).
While both aniA and norB appear to be members of an NsrR regulon in M. catarrhalis, expression of these two genes can be differentially regulated in response to certain nitrogenous compounds. Expression of aniA was upregulated about fivefold in both the wild-type O35E strain (Fig. 6A) and the O35E aniA mutant (Fig. 6B) when NO2− was present, even though this aniA mutant did not generate NO from NO2− (Fig. 7A). Nitrite was previously shown to at least partially relieve NsrR-dependent inhibition of aniA expression in N. gonorrhoeae (39) and in Nitrosomonas europaea (5). Moreover, similar to results obtained with the N. meningitidis aniA gene (46), chemically generated NO did not affect expression of the M. catarrhalis aniA gene (Fig. 6C).
The M. catarrhalis nsrR mutant expressed norB transcripts at a level that was more than an order of magnitude higher than the level of expression in the wild-type parental strain (Fig. 5). In the wild-type O35E strain, expression of norB was increased about twofold either when nitrite was added (Fig. 6A) or when NO was produced from spermine NONOate (Fig. 6C). The latter finding indicates that NO can relieve the NsrR-dependent repression of norB expression in M. catarrhalis. NO-sensitive NsrR repression of norB transcription was observed in both N. meningitidis (46) and N. gonorrhoeae (26). Because the M. catarrhalis aniA mutant did not generate NO from NO2− (Fig. 7A), addition of NO2− did not affect expression of norB in this mutant (Fig. 6B).
It has been established previously that growth of M. catarrhalis in the biofilm state results in a very high level of expression of the aniA gene (as well as the narGHJI and norB genes) (62), suggesting that there is probably a transcriptional regulator which upregulates expression of aniA in this growth environment, where oxygen is likely limiting. One candidate activator is the transcriptional regulator of fumarate and nitrate reduction (FNR), which functions in response to oxygen (for a review, see reference 28) and which has previously been shown to be the transcriptional activator of the aniA gene in N. gonorrhoeae (25, 31). BLAST-based searching of the available nucleotide sequences of the M. catarrhalis ATCC 43617 genome (GenBank accession numbers AX067426 to AX067466) did not reveal any candidates for an fnr gene encoding a protein with the [4Fe-4S] cluster. In addition, a search of nucleotide sequences comprising most, if not all, of the M. catarrhalis 7169 genome did not reveal any homology with fnr from N. meningitidis MC58 at either the nucleotide or encoded protein level (Anthony Campagnari, personal communication). However, examination of the nucleotide sequence upstream of the M. catarrhalis aniA gene revealed the presence of a putative FNR-binding site. Interestingly, MC ORF 921 (62) encodes a protein with 26% identity to DnrD (gene id 5096479 dnrD), a regulator which lacks the [4Fe-4S] center and which belongs to a new subgroup of the FNR-CRP regulator family (58, 59). Whether the MC ORF 921-encoded protein or another regulator is involved in the activation of the expression of aniA under oxygen-limiting growth conditions remains to be determined.
An unexpected finding in this study was the complete inhibition of growth of the O35E nsrR mutant in the presence of 5 mM NaNO2 (Fig. 4C); independently performed viability experiments indicated that nitrite was bacteriostatic for the nsrR mutant (data not shown). The growth of this nsrR mutant was not affected by addition of 10 mM NaNO3 (data not shown), likely because the narGHJI genes (encoding the nitrate reductase complex) are expressed at a very low level under well-aerated conditions (62), thereby limiting the rate at which NO2− could be produced. This nitrite-dependent growth inhibition of the nsrR mutant could be reversed by introducing a second mutation in the aniA gene (Fig. 4D) or by the presence of a wild-type M. catarrhalis nsrR gene in trans (Fig. 4F). The higher level of expression of the aniA and norB genes in the nsrR mutant resulted in enhanced consumption of both NO2− (Fig. 7B) and NO (Fig. 7C), together with elevated production of N2O (Fig. 7D). Additional experiments involving exposure of suspensions of M. catarrhalis cells to both NO and N2O independently revealed that NO had a modest bactericidal effect on both wild-type and nsrR mutant cells, whereas N2O had no apparent effect on the viability of these cells (data not shown). At this point, the mechanism for the observed inhibition of growth of the nsrR mutants in the presence of NO2− remains to be determined. However, expression of NsrR may provide some protection for M. catarrhalis in the presence of a low level of nitrite in the environment.
Some bacteria express an NO-inducible flavohemoglobin protein (Hmp), a nitric oxide dioxygenase that plays an important role in NO detoxification by converting NO to NO3− aerobically (13, 17) or by converting NO to N2O anaerobically (17). The expression of Salmonella enterica serovar Typhimurium Hmp was repressed by NsrR both aerobically and anaerobically (4, 14). In this enteric pathogen, Hmp is the enzyme primarily responsible for metabolism of NO under aerobic conditions and is required for virulence in a mouse model (4). In E. coli, expression of Hmp also was repressed by NsrR (11), and Hmp is required for NO consumption, for conferring resistance to nitrosative stress (17), and for protecting bacteria from killing within macrophages (53). The growth of wild-type S. enterica serovar Typhimurium and an nsrR mutant was not inhibited by the nitrosating agent S-nitrosoglutathione, whereas the growth of both an nsrR hmp double mutant and an hmp mutant was completely inhibited by S-nitrosoglutathione (14). The complete inhibition of growth of M. catarrhalis nsrR mutants by 5 mM NO2− (Fig. 4C) indicates that this bacterium does not have an Hmp protein for NO detoxification, and BLAST-based searching of the M. catarrhalis ATCC 43617 genome did not detect any genes encoding Hmp-like proteins. Instead, M. catarrhalis likely is dependent on the expression of NorB for NO detoxification.
N. meningitidis and M. catarrhalis appear to share some of the basic components of a truncated denitrification pathway (62), although only the former organism can grow anaerobically. The organization of the norB, nsrR, and aniA genes is different in these two pathogens; the norB and aniA ORFs are located adjacent to each other in the N. meningitidis MC58 genome (GenBank accession number NC_003112) but are transcribed divergently, and the nsrR gene is located elsewhere in the N. meningitidis genome. Interestingly, exogenously added nitrite appears to be toxic for an M. catarrhalis nsrR mutant under aerobic conditions, whereas an N. meningitidis nsrR mutant is able to grow more rapidly on nitrite under oxygen-limited conditions (46). In addition, in contrast to an M. catarrhalis nsrR mutant (Fig. 4A and 4C), an N. meningitidis nsrR mutant grew more slowly than its wild-type parent strain under aerobic conditions (46). These results reinforce the fact that there is a fundamental difference between these two pathogens in terms of respiratory capability.
At least one of the M. catarrhalis gene products involved in this truncated denitrification pathway is expressed in vivo when this bacterium grows in its human host. The AniA protein, designated Msp78 (48), was recently shown to be present when M. catarrhalis grew in the respiratory tracts of patients with COPD who had acquired and then subsequently cleared an infection with this pathogen (48). Similarly, the AniA protein of N. gonorrhoeae was also shown to be expressed in vivo in humans (9). Ongoing studies in our laboratory are focused on further elucidation of the mechanisms controlling expression of the proteins involved in this truncated denitrification pathway and on the relevance of these enzymes to host-pathogen interactions.
Acknowledgments
This study was supported by Public Health Service grant AI36344 to E.J.H. and by Public Health Service grant AI39557 to F.C.F. A.R.R. received support from Public Health Service training grant T32 AI55396. W.M.H. and D.A.S. were supported by NSF Microbial Interactions and Processes grant MCB-0604448 to D.A.S.
We thank John Nelson, Anthony Campagnari, and Steven Berk for providing the clinical isolates of M. catarrhalis used in this study and Anthony Campagnari for searching the M. catarrhalis 7169 genome for fnr.
Footnotes
Published ahead of print on 26 September 2008.
REFERENCES
- 1.Ahmad, F., H. Young, D. T. McLeod, M. J. Croughan, and M. A. Calder. 1987. Characterisation of Branhamella catarrhalis and differentiation from Neisseria species in a diagnostic laboratory. J. Clin. Pathol. 401369-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anantharaman, V., and L. Aravind. 2002. The PRC-barrel: a widespread, conserved domain shared by photosynthetic reaction center subunits and proteins of RNA metabolism. Genome Biol. 3:RESEARCH0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anjum, M. F., T. M. Stevanin, R. C. Read, and J. W. Moir. 2002. Nitric oxide metabolism in Neisseria meningitidis. J. Bacteriol. 1842987-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bang, I. S., L. Liu, A. Vazquez-Torres, M. L. Crouch, J. S. Stamler, and F. C. Fang. 2006. Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin hmp. J. Biol. Chem. 28128039-28047. [DOI] [PubMed] [Google Scholar]
- 5.Beaumont, H. J., S. I. Lens, W. N. Reijnders, H. V. Westerhoff, and R. J. van Spanning. 2004. Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol. Microbiol. 54148-158. [DOI] [PubMed] [Google Scholar]
- 6.Budhani, R. K., and J. K. Struthers. 1998. Interaction of Streptococcus pneumoniae and Moraxella catarrhalis: investigation of the indirect pathogenic role of beta-lactamase-producing moraxellae by use of a continuous-culture biofilm system. Antimicrob. Agents Chemother. 422521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catlin, B. W. 1990. Branhamella catarrhalis: an organism gaining respect as a pathogen. Clin. Microbiol. Rev. 3293-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman, K. R., D. M. Mannino, J. B. Soriano, P. A. Vermeire, A. S. Buist, M. J. Thun, C. Connell, A. Jemal, T. A. Lee, M. Miravitlles, S. Aldington, and R. Beasley. 2006. Epidemiology and costs of chronic obstructive pulmonary disease. Eur. Respir. J. 27188-207. [DOI] [PubMed] [Google Scholar]
- 9.Clark, V. L., J. S. Knapp, S. Thompson, and K. W. Klimpel. 1988. Presence of antibodies to the major anaerobically induced gonococcal outer membrane protein in sera from patients with gonococcal infections. Microb. Pathog. 5381-390. [DOI] [PubMed] [Google Scholar]
- 10.Faden, H. S., Y. Harabuchi, J. J. Hong, and Tonawanda/Williamsburg Pediatrics. 1994. Epidemiology of Moraxella catarrhalis in children during the first 2 years of life: relationship to otitis media. J. Infect. Dis. 169:1312-1317. [DOI] [PubMed] [Google Scholar]
- 11.Filenko, N., S. Spiro, D. F. Browning, D. Squire, T. W. Overton, J. Cole, and C. Constantinidou. 2007. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J. Bacteriol. 1894410-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsgren, A., M. Brant, M. Karamehmedovic, and K. Riesbeck. 2003. The immunoglobulin D-binding protein MID from Moraxella catarrhalis is also an adhesin. Infect. Immun. 713302-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner, P. R., A. M. Gardner, L. A. Martin, and A. L. Salzman. 1998. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. USA 9510378-10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilberthorpe, N. J., M. E. Lee, T. M. Stevanin, R. C. Read, and R. K. Poole. 2007. NsrR: a key regulator circumventing Salmonella enterica serovar Typhimurium oxidative and nitrosative stress in vitro and in IFN-gamma-stimulated J774.2 macrophages. Microbiology 1531756-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haine, V., M. Dozot, J. Dornand, J. J. Letesson, and X. De Bolle. 2006. NnrA is required for full virulence and regulates several Brucella melitensis denitrification genes. J. Bacteriol. 188:1615-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall-Stoodley, L., F. Z. Hu, A. Gieseke, L. Nistico, D. Nguyen, J. Hayes, M. Forbes, D. P. Greenberg, B. Dice, A. Burrows, P. A. Wackym, P. Stoodley, J. C. Post, G. D. Ehrlich, and J. E. Kerschner. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hausladen, A., A. J. Gow, and J. S. Stamler. 1998. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc. Natl. Acad. Sci. USA 9514100-14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helminen, M. E., I. Maciver, J. L. Latimer, L. D. Cope, G. H. McCracken, Jr., and E. J. Hansen. 1993. A major outer membrane protein of Moraxella catarrhalis is a target for antibodies that enhance pulmonary clearance of the pathogen in an animal model. Infect. Immun. 61:2003-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heurlier, K., M. J. Thomson, N. Aziz, and J. W. Moir. 2008. The nitric oxide (NO)-sensing repressor NsrR of Neisseria meningitidis has a compact regulon of genes involved in NO synthesis and detoxification. J. Bacteriol. 1902488-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoehn, G. T., and V. L. Clark. 1999. Isolation and nucleotide sequence of the gene (aniA) encoding the major anaerobically induced outer membrane protein of Neisseria gonorrhoeae. Infect. Immun. 60. 114695-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holm, M. M., S. L. Vanlerberg, I. M. Foley, D. D. Sledjeski, and E. R. Lafontaine. 2004. The Moraxella catarrhalis porin-like outer membrane protein CD is an adhesin for human lung cells. Infect. Immun. 721906-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm, M. M., S. L. Vanlerberg, D. D. Sledjeski, and E. R. Lafontaine. 2003. The Hag protein of Moraxella catarrhalis strain O35E is associated with adherence to human lung and middle ear cells. Infect. Immun. 714977-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams. 1994. Bergey's manual of determinative bacteriology. Williams and Wilkins, Baltimore, MD.
- 24.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8528-535. [PubMed] [Google Scholar]
- 25.Householder, T. C., W. A. Belli, S. Lissenden, J. A. Cole, and V. L. Clark. 1999. cis- and trans-acting elements involved in regulation of aniA, the gene encoding the major anaerobically induced outer membrane protein in Neisseria gonorrhoeae. J. Bacteriol. 181541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Householder, T. C., E. M. Fozo, J. A. Cardinale, and V. L. Clark. 2000. Gonococcal nitric oxide reductase is encoded by a single gene, norB, which is required for anaerobic growth and is induced by nitric oxide. Infect. Immun. 685241-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2547-559. [DOI] [PubMed] [Google Scholar]
- 28.Korner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27559-592. [DOI] [PubMed] [Google Scholar]
- 29.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 1821364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipski, S. L., C. Akimana, J. M. Timpe, R. M. Wooten, and E. R. Lafontaine. 2007. The Moraxella catarrhalis autotransporter McaP is a conserved surface protein that mediates adherence to human epithelial cells through its N-terminal passenger domain. Infect. Immun. 75314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lissenden, S., S. Mohan, T. Overton, T. Regan, H. Crooke, J. A. Cardinale, T. C. Householder, P. Adams, C. D. O'Conner, V. L. Clark, H. Smith, and J. A. Cole. 2000. Identification of transcription activators that regulate gonococcal adaptation from aerobic to anaerobic or oxygen-limited growth. Mol. Microbiol. 37839-855. [DOI] [PubMed] [Google Scholar]
- 32.Lovgren, J. M., G. O. Bylund, M. K. Srivastava, L. A. Lundberg, O. P. Persson, G. Wingsle, and P. M. Wikstrom. 2004. The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits. RNA 101798-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luke, N. R., and A. A. Campagnari. 1999. Construction and characterization of Moraxella catarrhalis mutants defective in expression of transferrin receptors. Infect. Immun. 675815-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luke, N. R., J. A. Jurcisek, L. O. Bakaletz, and A. A. Campagnari. 2007. Contribution of Moraxella catarrhalis type IV pili to nasopharyngeal colonization and biofilm formation. Infect. Immun. 755559-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy, T. F. 2000. Bacterial otitis media: pathogenetic considerations. Pediatr. Infect. Dis. J. 19S9-S15. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, T. F. 2005. Moraxella (Branhamella) catarrhalis and other gram-negative cocci, p. 2529. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Mandell, Douglas, and Bennett's principles and practice of infectious disease. Elsevier Inc., Philadelphia, PA.
- 37.Murphy, T. F. 1996. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol. Rev. 60267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy, T. F., A. L. Brauer, B. J. Grant, and S. Sethi. 2005. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am. J. Respir. Crit. Care Med. 172195-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Overton, T. W., R. Whitehead, Y. Li, L. A. Snyder, N. J. Saunders, H. Smith, and J. A. Cole. 2006. Coordinated regulation of the Neisseria gonorrhoeae-truncated denitrification pathway by the nitric oxide-sensitive repressor, NsrR, and nitrite-insensitive NarQ-NarP. J. Biol. Chem. 28133115-33126. [DOI] [PubMed] [Google Scholar]
- 40.Patrick, C. C., A. Kimura, M. A. Jackson, L. Hermanstorfer, A. Hood, G. H. McCracken, Jr., and E. J. Hansen. 1987. Antigenic characterization of the oligosaccharide portion of the lipooligosaccharide of nontypeable Haemophilus influenzae. Infect. Immun. 552902-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearson, M. M., C. A. Laurence, S. E. Guinn, and E. J. Hansen. 2006. Biofilm formation by Moraxella catarrhalis in vitro: roles of the UspA1 adhesin and the Hag hemagglutinin. Infect. Immun. 741588-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philippot, L. 2005. Denitrification in pathogenic bacteria: for better or worst? Trends Microbiol. 13191-192. [DOI] [PubMed] [Google Scholar]
- 43.Plamondon, P., N. R. Luke, and A. A. Campagnari. 2007. Identification of a novel two-partner secretion locus in Moraxella catarrhalis. Infect. Immun. 752929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy, M. S., T. F. Murphy, H. S. Faden, and J. M. Bernstein. 1997. Middle ear mucin glycoprotein; purification and interaction with nontypeable Haemophilus influenzae and Moraxella catarrhalis. Otolaryngol. Head Neck Surg. 116175-180. [DOI] [PubMed] [Google Scholar]
- 45.Rock, J. D., M. R. Mahnane, M. F. Anjum, J. G. Shaw, R. C. Read, and J. W. Moir. 2005. The pathogen Neisseria meningitidis requires oxygen, but supplements growth by denitrification. Nitrite, nitric oxide and oxygen control respiratory flux at genetic and metabolic levels. Mol. Microbiol. 58800-809. [DOI] [PubMed] [Google Scholar]
- 46.Rock, J. D., M. J. Thomson, R. C. Read, and J. W. Moir. 2007. Regulation of denitrification genes in Neisseria meningitidis by nitric oxide and the repressor NsrR. J. Bacteriol. 1891138-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodionov, D. A., I. L. Dubchak, A. P. Arkin, E. J. Alm, and M. S. Gelfand. 2005. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol. 1e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruckdeschel, E. A., C. Kirkham, A. J. Lesse, Z. Hu, and T. F. Murphy. 2008. Mining the Moraxella catarrhalis genome: identification of potential vaccine antigens expressed during human infection. Infect. Immun. 761599-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudolph, G., H. Hennecke, and H. M. Fischer. 2006. Beyond the Fur paradigm: iron-controlled gene expression in rhizobia. FEMS Microbiol. Rev. 30631-648. [DOI] [PubMed] [Google Scholar]
- 50.Sethi, S., N. Evans, B. J. Grant, and T. F. Murphy. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347465-471. [DOI] [PubMed] [Google Scholar]
- 51.Stevanin, T. M., J. R. Laver, R. K. Poole, J. W. Moir, and R. C. Read. 2007. Metabolism of nitric oxide by Neisseria meningitidis modifies release of NO-regulated cytokines and chemokines by human macrophages. Microbes Infect. 9981-987. [DOI] [PubMed] [Google Scholar]
- 52.Stevanin, T. M., J. W. Moir, and R. C. Read. 2005. Nitric oxide detoxification systems enhance survival of Neisseria meningitidis in human macrophages and in nasopharyngeal mucosa. Infect. Immun. 733322-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stevanin, T. M., R. C. Read, and R. K. Poole. 2007. The hmp gene encoding the NO-inducible flavohaemoglobin in Escherichia coli confers a protective advantage in resisting killing within macrophages, but not in vitro: links with swarming motility. Gene 39862-68. [DOI] [PubMed] [Google Scholar]
- 54.Todd, J. D., M. Wexler, G. Sawers, K. H. Yeoman, P. S. Poole, and A. W. Johnston. 2002. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology 1484059-4071. [DOI] [PubMed] [Google Scholar]
- 55.Tunbridge, A. J., T. M. Stevanin, M. Lee, H. M. Marriott, J. W. Moir, R. C. Read, and D. H. Dockrell. 2006. Inhibition of macrophage apoptosis by Neisseria meningitidis requires nitric oxide detoxification mechanisms. Infect. Immun. 74729-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verduin, C. M., C. Hol, A. Fleer, H. van Dijk, and A. Van Belkum. 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin. Microbiol. Rev. 15:125-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viguier, C., O. Cuiv, P. Clarke, and M. O'Connell. 2005. RirA is the iron response regulator of the rhizobactin 1021 biosynthesis and transport genes in Sinorhizobium meliloti 2011. FEMS Microbiol. Lett. 246:235-242. [DOI] [PubMed] [Google Scholar]
- 58.Vollack, K. U., E. Hartig, H. Korner, and W. G. Zumft. 1999. Multiple transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses and cognate metabolic processes. Mol. Microbiol. 311681-1694. [DOI] [PubMed] [Google Scholar]
- 59.Vollack, K. U., and W. G. Zumft. 2001. Nitric oxide signaling and transcriptional control of denitrification genes in Pseudomonas stutzeri. J. Bacteriol. 1832516-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, W., A. S. Attia, L. Liu, T. Rosche, N. J. Wagner, and E. J. Hansen. 2006. Development of a shuttle vector for Moraxella catarrhalis. Plasmid 5550-57. [DOI] [PubMed] [Google Scholar]
- 61.Wang, W., and E. J. Hansen. 2006. Plasmid pWW115, a cloning vector for use with Moraxella catarrhalis. Plasmid 56133-137. [DOI] [PubMed] [Google Scholar]
- 62.Wang, W., L. Reitzer, D. A. Rasko, M. M. Pearson, R. J. Blick, C. Laurence, and E. J. Hansen. 2007. Metabolic analysis of Moraxella catarrhalis and the effect of selected in vitro growth conditions on global gene expression. Infect. Immun. 754959-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weber, I., C. Fritz, S. Ruttkowski, A. Kreft, and F. C. Bange. 2000. Anaerobic nitrate reductase (narGHJI) activity of Mycobacterium bovis BCG in vitro and its contribution to virulence in immunodeficient mice. Mol. Microbiol. 351017-1025. [DOI] [PubMed] [Google Scholar]
- 64.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]