Abstract
Analysis of the complete sequence of the genome of Clostridium perfringens strain 13 resulted in identification of five genes, including pfoA (encoding theta toxin) and vrr (encoding VirR/VirS-regulated RNA), with consensus VirR-binding sequences upstream of the open reading frame (ORF), suggesting that expression of these genes may be regulated directly by the two-component VirR/VirS system. To test this possibility, we examined VirR/VirS system-mediated transcriptional regulation of three genes, virT, ccp (encoding alpha-clostripain), and virU, with the novel VirR-binding sequences. Northern analysis revealed that the steady-state levels (increases or decreases in the amounts of RNA expressed) of virT, ccp, and virU mRNAs were lower in a virR mutant strain than in the wild-type strain, as were the levels of the pfoA and vrr transcripts. The consensus VirR-binding sites were located similarly relative to the transcription start sites in the virT, ccp, and virU promoters. Mutation and overexpression analyses with virT and virU revealed that the virT gene product has a negative effect on expression of pfoA and ccp, whereas the virU gene product positively affects expression of pfoA, virT, ccp, and vrr. Nonsense and frameshift mutations in the virT or virU putative ORF did not affect the regulatory functions, suggesting that virT and virU may encode RNA regulators rather than proteins. These results suggest that a complex regulatory network, perhaps involving several regulatory RNA molecules, governs the expression of the VirR/VirS regulon in C. perfringens.
The gram-positive anaerobic bacterium Clostridium perfringens produces numerous extracellular toxins that are believed to play important roles in the pathogenicity of various diseases, including gas gangrene, which is also known as clostridial myonecrosis (9, 21). Because the toxins are thought to act synergistically in the development of gas gangrene (2), knowledge of the mechanisms that regulate expression of toxin genes is critical for understanding the pathogenesis of myonecrosis.
Bacterial two-component systems, consisting of a sensor histidine kinase and a response regulator, enable bacteria to respond to various environmental conditions through a phosphorelay between the sensor and the regulator. The two-component VirR/VirS system comprising the VirR response regulator and the VirS sensor protein is known to be involved in global regulation of the production of theta-toxin (also known as perfringolysin O), kappa-toxin (or collagenase), alpha-toxin (or phospholipase C), sialidase, protease, and hemagglutinin in C. perfringens (13, 24). The VirR/VirS system regulates the mRNA levels of plc (alpha-toxin), pfoA (theta-toxin), and colA (kappa-toxin) (4). Primer extension analysis revealed both VirR/VirS-dependent and independent promoters for pfoA and colA and a single VirR/VirS-dependent promoter for plc (4). The absence of a consensus binding site for phosphorylated VirR protein in the promoters of the colA and plc genes (4) suggests that complex regulatory networks might be involved in C. perfringens toxin production (26).
Four targets of the VirR/VirS system have been identified through differential display analyses. The VirR/VirS system was found to promote expression of ptp (encoding protein tyrosine phosphatase), cpd (encoding 2′,3′-cyclic nucleotide phosphodiesterase), and hyp7 (encoding a hypothetical 7-kDa protein) (3) and to inhibit expression of the ycgJ-metB-cysK-luxS (ygaG) operon (3, 20). It was suggested previously that hyp7 acts as a secondary regulator that positively regulates the levels of colA and plc mRNAs but not the level of pfoA mRNA (3). However, we reported previously that VirR/VirS-regulated RNA (VR-RNA) (encoded by vrr) transcribed from the Hyp7 coding region is a regulatory RNA that mediates the signal from the VirR/VirS system to control the expression of colA, plc, ptp, cpd, and ycgJ-metB-cysK-luxS, whereas pfoA is regulated directly by the VirR/VirS system (28). The VirR/VirS-VR-RNA cascade was also found to affect levels of plasmid-borne cpb2 (encoding beta2 toxin) and cna (encoding a possible collagen adhesin) mRNAs positively and negatively, respectively (19).
Two repeated sequences have been found upstream of the pfoA promoter (4), and it was reported previously that the VirR protein binds independently to these two repeats (CCCAGTTNTNCAC) (6). Interestingly, a monomeric repeat similar but not identical to the pfoA VirR-binding site has also been found in the promoter of vrr, the gene encoding VR-RNA (28). A CCAGTTNNNCAC core motif was highly conserved in both genes. These findings suggest that the VirR protein may bind to the vrr promoter, activating transcription of VR-RNA, which in turn activates colA and plc transcription, thus forming the basis for a regulatory cascade in the VirR/VirS regulon (28).
The complete genomic sequence of C. perfringens strain 13 has been reported (25). On the basis of sequence similarities with other known virulence genes, more than 20 candidate virulence genes were identified. By screening the genome for the previously identified VirR-binding consensus sequence, we identified genes potentially regulated by the VirR/VirS system. Five genes were found to have the consensus VirR-binding site in their putative promoter regions (25). In the present study, regulation of expression of these novel target genes was analyzed to improve our understanding of the VirR/VirS regulon in C. perfringens.
MATERIALS AND METHODS
Strains, plasmids, medium, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. All C. perfringens strains were cultured in Gifu anaerobic medium (Nissui, Japan) at 37°C under anaerobic conditions as described previously (24). A single-crossover mutation was introduced into the virT gene of C. perfringens strain 13 with a pUC19-based suicide vector containing a 334-bp internal PCR fragment of virT and the ermB gene from pJIR418 (29). Escherichia coli DH5α was cultured as described previously (22). Plasmids pUC19 and pUC118 were used for cloning in E. coli, and pJIR418 (29) was used as an E. coli-C. perfringens shuttle vector.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| C. perfringens strains | ||
| 13 | Wild-type strain (type A) | 14 |
| TS133 | Strain 13 virR::Tetr | 24 |
| TS140 | Strain 13 Δvrr Emr | 28 |
| TS190 | Strain 13 virT::Emr | This study |
| E. coli DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Takara Bio Inc. |
| Plasmids | ||
| pBT405 | pJIR418 Ω(PstI 4.3-kb strain 13 genomic library) (virR+virS+ complementation vector), Ampr | Shimizu, unpublished data |
| pJIR418 | E. coli-C. perfringens shuttle vector, Cmr Emr | 29 |
| pSB1031 | pJIR418 Ω(PCR-amplified 637-bp fragment) (vrr+ complementation vector) | 4 |
| pTS930 | pJIR418 Ω(PCR-amplified 1,400-bp fragment) (virT+ complementation vector) | This study |
| pTS931 | pJIR418 Ω(PCR-amplified 539-bp fragment) (virU+ complementation vector) | This study |
| pTS932 | pTS930 (585T→A) | This study |
| pTS933 | pTS931 (182A→T) | This study |
| pTS934 | pUC118 Ω(PCR-amplified 334-bp fragment) (virT+ suicide vector), Emr | This study |
| pTS935 | pTS930 (1-bp frameshift at position 33) | This study |
| pTS936 | pTS931 (1-bp frameshift at position 15) | This study |
| pUC19 | Cloning vector, AmprlacZ′ pMB 1 ori | Takara Bio Inc. |
| pUC118 | Cloning vector, AmprlacZ′ pMB 1 ori | Takara Bio Inc. |
Tetr, resistance to tetracycline; Emr, resistance to erythromycin, Cmr, resistance to chloramphenicol; Ampr, resistance to ampicillin.
DNA manipulation.
Recombinant DNA was manipulated as described previously (22), unless otherwise noted. C. perfringens strains were transformed by electroporation as previously described (24).
Northern hybridization.
Total RNA from C. perfringens was extracted and Northern blotting was performed as previously described (1) with an AlkPhos-direct kit and CDP-star chemiluminescence (GE Healthcare). DNA probes were prepared from genomic DNA of C. perfringens strain 13 by performing PCR with the appropriate primer sets (Table 2). In some situations, the signal densities of the mRNA bands were measured with a densitometer. All Northern hybridization experiments were performed at least three times, and the reproducibility was confirmed. The results described below are representative results from the repeated experiments.
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′to 3′) | Use |
|---|---|---|
| virT-F | GAAGGTAAGTACCAAGATGA | Northern blotting |
| virT-R | TGATATTGCCACCCCAACTT | Northern blotting |
| ccp-F | GAGGCCGAAAAGACTGAAGG | Northern blotting |
| ccp-R | GCCCAATGTGGTATTGCTTG | Northern blotting |
| virU-F | CGATTCGTTTTTGATAGAAATGG | Northern blotting |
| virU-R | TTTATTTCTAGTTTTTCCTTTGATGA | Northern blotting |
| vrr-F | GACCAGTTACGCACAAAC | Northern blotting |
| vrr-R | GGACAGTTCTATTTCTAGG | Northern blotting |
| pfoA-F | GCAAGTATTGCAATGGCTTT | Northern blotting |
| pfoA-R | GTAAGTAATACTAGATCCAGGGT | Northern blotting |
| colA-F | GGTCTAGAGGATAGTGGAAGA | Northern blotting |
| colA-R | GTTCCTCTCATATCGTAAGT | Northern blotting |
| 0845-PE | TTTATCAATGGGTAACGTAAAGAAAGTACCAGATA | Primer extension |
| 0846-PE | GTTGAAACTCCTCCTAATAGAACGCTACCAATGGT | Primer extension |
| 0920-PE | TCGGTATAATAAAAATGATATTGTTACTAATATG | Primer extension |
| virT-NM | GAAAGTACCAGATTAAATAGAAATAG | Site-directed mutagenesis |
| virU-NM-1 | TTGGTATTATATATTAATATTTTCGTC | Site-directed mutagenesis |
| virU-NM-2 | GACGAAAATATTAATATATAATACCAA | Site-directed mutagenesis |
| virT-FM-F | ATCGCTTTAAGTTTATCATTTCTATTTTATCTGGT | Site-directed mutagenesis |
| virT-FM-R | ACCAGATAAAATAGAAATGATAAACTTAAAGCGAT | Site-directed mutagenesis |
| virU-FM-F | GATATGAAAGACGAAAAATTAATATAAAATACCAA | Site-directed mutagenesis |
| virU-FM-R | TTGGTATTTTATATTAATTTTTCGTCTTTCATATC | Site-directed mutagenesis |
Primer extension analysis.
Primer extension was carried out as described previously (17) by using an Amersham 5′ oligolabeling fluorescence kit and the Promega primer extension system. Oligonucleotide primers 0845-PE, 0846-PE, and 0920-PE used to determine the transcription start sites of CPE0845 (virT), CPE0846 (ccp), and CPE0920 (virU), respectively, are shown in Table 2. Signals were detected with a FluorImager analyzer (GE Healthcare).
Assays for perfringolysin O and alpha-clostripain.
The perfringolysin O activity in the C. perfringens culture supernatant was measured by the horse erythrocyte hemolysis method described previously (2). C. perfringens cells were cultured for 3 h to mid-log phase (see Fig. 2A) and collected by centrifugation. The supernatant was used for the hemolytic assay. Hemolytic activity was expressed as the reciprocal of the dilution that resulted in 50% hemolysis of 0.5% horse erythrocytes. The proteolytic activity of alpha-clostripain in the culture supernatant was determined with azocasein (Sigma Aldrich Japan) and the cysteine protease-specific inhibitors leupeptin and antipain (Wako Pure Chemicals) as previously described (11, 27). In brief, C. perfringens cells were cultured for 2 h to early log phase (see Fig. 2A) and collected by centrifugation, and 500 μl of the supernatant was mixed with an equal volume of an azocasein solution (5 mg/ml azocasein in 25 mM Tris-HCl [pH 7.5]-5 mM dithiothreitol) with or without 10 μM leupeptin or antipain. The mixture was incubated for 2 h at 37°C with gentle shaking, intact azocasein was removed by 3% trichloroacetic acid precipitation, and the absorbance at 450 nm of the supernatant was determined.
FIG. 2.

Growth curves and Northern blot analyses for the virR mutant (TS133) of C. perfringens. (A) Growth curves for C. perfringens strains with a 1% inoculum. All three C. perfringens strains grew with a doubling time of ∼20 min in Gifu anaerobic medium. ○, strain 13(pJIR418); □, strain TS133(pJIR418) (virR); ▵, strain TS133(pBT405) (virR+ virS+). OD 600 nm, optical density at 600 nm. (B) Total RNA was prepared from each culture at the indicated times (2 and 3 h). Either 10 μg (for virT and ccp) or 40 μg (for virU) of total RNA was resolved by agarose electrophoresis, blotted onto a nylon membrane, and hybridized with probes for virT (CPE0845), ccp (CPE0846), and virU (CPE0920). Lane 1, strain 13(pJIR418); lane 2, strain TS133(pJIR418); lane 3, strain TS133(pBT405).
Site-directed mutagenesis and frameshift mutagenesis.
Site-directed mutagenesis of the virT gene harbored by pTS930 was performed with an LA PCR in vitro mutagenesis kit (Takara Bio) with the mutagenic primer virT-NM (Table 2) to obtain pTS932. The virU gene on pTS931 was mutated by using a QuikChange site-directed mutagenesis kit (Stratagene) with primers virU-NM-F and virU-NM-R (Table 2) to obtain pTS933. Similarly, 1-base deletion frameshift mutations at positions 33 and 15 in the virT and virU coding regions, respectively, in the complemented plasmid vectors pTS930 and pTS931 (Table 1), respectively, were obtained by using a QuikChange site-directed mutagenesis kit (Stratagene) with primers virT-FM-F and virT-FM-R for virT and primers virU-FM-F and virU-FM-R for virU to construct pTS935 and pTS936 (Table 1). All procedures were performed according to the manufacturers' instructions.
RESULTS
Screening for VirR-binding sites in the C. perfringens genome.
We scanned the genomic sequence of C. perfringens (25) for VirR-binding sites (CCAGTTNNNCAC) located upstream of the open reading frame (ORF). Only five genes, pfoA, CPE0845 (virT), CPE0846 (ccp), CPE0920 (virU), and vrr, were found to have sequences similar to the VirR-binding site in their putative promoter regions (Fig. 1) (25). With the exception of virT and ccp, which are located back to back and are transcribed on opposite strands, these five genes are not clustered (Fig. 1). The deduced amino acid sequence of the putative protein encoded by ccp was highly similar to that of alpha-clostripain, a cysteine proteinase from Clostridium histolyticum (7). The expression of ccp was previously shown to be positively regulated by the VirR/VirS system (27). The putative proteins encoded by virT and virU showed no significant similarity to known proteins, and their functions remain unclear. Identification of the previously uncharacterized putative VirR-binding sites in virT, ccp, and virU led to an investigation of whether these three genes, like pfoA and vrr, are targets for direct regulation by the VirR/VirS system.
FIG. 1.
Schematic diagram of the locations of putative VirR-binding sites in five genes (pfoA, virT, ccp, virU, and vrr) on the chromosome of C. perfringens wild-type strain 13. The solid and cross-hatched arrows represent genes with VirR-binding sites and their flanking genes, respectively. Open circles indicate putative VirR-binding sites. The chromosomal locations of other genes mentioned in this paper are also indicated. The nucleotide numbers are the numbers for the chromosomal sequence of C. perfringens strain 13 (GenBank accession number BA000016).
Transcriptional regulation of virT, ccp, and virU by the VirR/VirS system.
We performed Northern analyses of RNA of C. perfringens strains with different mutant backgrounds at different growth stages to look for changes in the steady-state levels of virT, ccp, and virU that confirmed that that there is regulation by the VirR/VirS system. In C. perfringens wild-type strain 13, a 1.0-kb transcript of virT was clearly detected after 2 h of growth (early to mid-exponential phase) (Fig. 2B, upper left panel), and the level had decreased after 3 h of incubation (late exponential phase) (Fig. 2B, upper right panel). Similar patterns were observed for expression of the 1.6-kb alpha-clostripain (ccp) transcript (Fig. 2B, center panels) and the 0.5-kb virU transcript (Fig. 2B, lower panels) in the wild-type background. Even with 40 μg of total RNA blotted onto the membrane, the virU transcript signal was very low for strain 13, indicating that the level of expression of virU was significantly lower than the level of expression of virT or ccp. The time points of accumulation for virT, ccp, and virU mRNAs were similar to those for pfoA and vrr; all of the molecules were most abundant during the early exponential to mid-exponential growth phase (3, 4). Importantly, transcripts of virT, ccp, and virU either were undetectable or the bands were very weak for the virR mutant strain TS133 (Fig. 2B, lane 2). Expression of these genes was increased by transformation of intact virR and virS genes into TS133 (Fig. 2B, lane 3). These data clearly indicate that the steady-state RNA levels for virT, ccp, and virU were regulated positively by the VirR/VirS system in C. perfringens in a manner similar to the manner of regulation of pfoA and vrr (3, 4, 28). The higher levels of the virT, ccp, and virU transcripts in the complemented strains were likely due to the high copy number of the pBT405 complementation plasmid.
Promoter analysis of the VirR/VirS-regulated genes.
To analyze the promoter regions of the three VirR/VirS-regulated genes, we identified transcription initiation sites for virT, ccp, and virU by performing a primer extension experiment with wild-type and virR mutant RNA templates. The virT-specific primer generated a single extension product with wild-type strain 13 RNA, whereas no product was obtained with the virR mutant strain TS133 (Fig. 3A, left panel). Similarly, both the ccp and virU gene-specific primers (Fig. 3A, middle and right panels, respectively) yielded single extension products with the wild-type RNA template, whereas no products were obtained with virR mutant strain RNA. These results indicate that transcription initiation from single sites in virT, ccp, and virU is dependent on the VirR/VirS system because little mRNA for these genes was present in the virR mutant strain.
FIG. 3.
Identification of the transcription initiation sites of the virT, ccp, and virU genes in C. perfringens. (A) Primer extension products derived using the oligonucleotide primers listed in Table 2 with template RNA prepared from 2-h cultures of C. perfringens strains (lane 1, wild-type strain 13; lane 2, virR strain TS133) were separated by electrophoresis on acrylamide gels. Sequencing reactions with the same primers and appropriate DNA templates were run on the same gel. The positions of extended products obtained with the virT, ccp, and virU primers are indicated by arrows, and the putative mRNA start sites are indicated by circles. (B) Deduced promoter sequences (−35 and −10) and consensus VirR-binding sites of the five VirR/VirS-regulated genes. The putative promoter sequences and mRNA start sites are indicated by boxes and circles, respectively. The deduced VirR-binding sequence of each gene is indicated by a dotted box, and conserved nucleotides are underlined. The promoter sequences of the theta-toxin (pfoA) and VR-RNA (vrr) genes are aligned, and the VB1 and VB2 regions are shown.
The length of each primer extension product was used to assign the position 1 site of transcription initiation for virT, ccp, and virU and to identify conserved elements (−35 and −10) in the promoter of each gene (Fig. 3B). We compared the locations of the VirR-binding sequences (CCAGTTWTNCA), the consensus promoter sequences (−35 and −10), and the mRNA start sites with those determined for pfoA and vrr in previous studies (4, 28) (Fig. 3B) and found that the relative distances between these elements were highly conserved in the five promoter regions. In particular, all five genes have a 42-bp interval between the VirR-binding sequences and the mRNA start site (Fig. 3B). The locations of the consensus promoter sequences (−35 and −10) were almost identical in all five genes.
In previous studies, two repeated sequences (VB1 and VB2) in the promoter region of pfoA (Fig. 3B) were identified as independent binding sites for the VirR protein (5, 6). The promoter regions of vrr, virT, ccp, and virU contained sequences similar to the VB2 sequence of pfoA (Fig. 3B). Although more divergent than the similarities between these four genes and pfoA in the VB2 consensus sequence, sequences similar to VB1 were found in these four genes (Fig. 3B), suggesting that the VB1 region may also be important for regulation of transcript by the VirR protein. The promoter structures of these five VirR-regulated genes are highly conserved and are distinct from the promoter structures of previously analyzed VirR/VirS-regulated genes (4, 19). The ability of the VirR protein to bind to the conserved sequences upstream of virT, ccp, and virU was confirmed in a previous study (5). Moreover, VirR has been reported to bind to some VirR boxes found in genes of two other strains of C. perfringens, ATCC13124 and SM101 (16), which suggests that the VirR-dependent regulatory system is present in various types of C. perfringens strains. Binding of a glutathione S-transferase-VirR fusion protein to the VirR-binding sequence was also examined using gel mobility shift assays, and this analysis confirmed that the VirR protein bound specifically to the conserved sequences in the promoter regions of pfoA, vrr, virT, ccp, and virU (data not shown).
Functional analysis of the virT and virU genes.
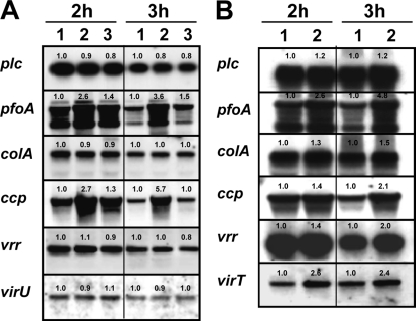
We were unable to predict the putative function of either virT or virU using the results of computer-based searches for sequence similarities. To explore the functional roles of these genes, we constructed virT isogenic mutants of strain 13 (see Materials and Methods). The resulting virT mutation in strain TS190, which was confirmed by Southern hybridization with a virT gene probe (data not shown), was used to examine expression of VirR/VirS-regulated genes. Compared to wild-type strain 13, in mutant strain TS190 there were at least 2.5-fold increases in the levels of both pfoA and ccp mRNAs during the logarithmic growth phase (2 and 3 h) (Fig. 4A). When mutant strain TS190 was complemented with the virT+ pTS930 plasmid [resulting in TS190(pTS930)], the level of each of the transcripts was reduced to the wild-type level, although complementation was not complete until the 3-h time point for unknown reasons (Fig. 4A). These data suggest that the virT gene product acts as a negative regulator of pfoA and ccp in the wild-type strain. However, no significant change in plc, colA, vrr, or virU expression was observed in TS190 (Fig. 4A), indicating that the negative effect of the virT gene product was specific to pfoA and ccp.
FIG. 4.
Northern blot analyses of the virT mutant and wild-type strains transformed with the virU+ expression plasmid (pTS931). Total RNA was prepared from each culture at the times indicated (2 and 3 h), and 10 μg of each RNA preparation (40 μg for hybridization with virU probe) was resolved by agarose electrophoresis, blotted onto nylon membranes, and hybridized with plc, pfoA, colA, ccp, vrr, virT, and virU gene probes as indicated. The band densities relative to those for wild-type strain 13 are indicated above the bands. (A) Lane 1, wild-type strain 13(pJIR418); lane 2, strain TS190(pJIR418) (ΔvirT); lane 3, strain TS190(pTS930) (ΔvirT virT+). (B) Lane 1, wild-type strain 13(pJIR418); lane 2, wild-type strain 13(pTS931) (virU+).
For unknown reasons, repeated attempts to construct a strain 13 virU mutant using single-crossover or double-crossover recombination methods failed. As an alternative approach to test the function of virU, we introduced a virU overexpression plasmid, pTS931, into wild-type strain 13 to measure the effect of virU on the steady-state levels of putative VirR/VirS target genes. In 3-h cultures, the pfoA, ccp, vrr, and virT mRNA levels were all increased in response to the higher number of virU copies (Fig. 4B), suggesting that the virU gene encodes a positive regulator of pfoA, ccp, vrr, and virT expression. Although the abundance of plc and colA transcripts also increased slightly following transformation of pTS931 into strain 13 (Fig. 4B), we believe that this may have been a secondary effect of increased VR-RNA (Fig. 4B, vrr panels), which is known to enhance expression of plc and colA.
In addition to measuring changes in the levels of the pfoA and ccp mRNAs, we also measured the activities of the secreted gene products, perfringolysin O and alpha-clostripain, respectively, in supernatants from cultures of wild-type and mutant C. perfringens strains. Perfringolysin O activity was measured by determining the hemolytic activities of the culture supernatants with horse erythrocytes. The hemolytic activities of virT mutant strain TS190, TS190 with the plasmid expressing virT (pTS930), and wild-type strain 13 with the virU overexpression plasmid (pTS931) were 3.9-, 1.2- and 4.3-fold higher, respectively, than the hemolytic activity of wild-type strain 13 (Table 3). These results, which correspond well with the results of the Northern analyses, indicate that the activity encoded by wild-type virU stimulates the production of perfringolysin O, whereas the wild-type virT gene product inhibits the production of perfringolysin O.
TABLE 3.
Perfringolysin O activities of various C. perfringens strains
| Strain | Genotype | Perfringolysin O titer (log2)a | Difference (fold) compared to strain 13 |
|---|---|---|---|
| 13 | Wild type | 7.4 ± 0.5 | 1 |
| TS133 | Strain 13 virR::Tetr | 2.0 ± 0.1 | <0.1 |
| TS190 | Strain 13 ΔvirT | 9.4 ± 0.5 | 3.9 |
| TS190(pTS930) | Strain 13 ΔvirT (virT+) | 7.7 ± 0.4 | 1.2 |
| 13(pTS931) | Strain 13 (virU+) | 9.5 ± 0.7 | 4.3 |
Each value was calculated by using the results of triplicate independent experiments, and the values are means ± standard deviations.
Azocasein, a colorimetric substrate of alpha-clostripain and other cysteine proteinases, was used to measure alpha-clostripain activity specifically in culture supernatants containing two inhibitors of cysteine proteinases, leupeptin and antipain. In the presence of these inhibitors, the proteolytic activity of each strain, as measured by absorbance at 450 nm, decreased between 25 and 60% (Fig. 5A). The difference in activity represents alpha-clostripain-specific proteolysis of azocasein (Fig. 5B). The alpha-clostripain activity was increased in the virT mutant strain TS190, and when the mutation was complemented with pTS930 (virT+), the activity decreased to a level similar to the level in the wild-type strain (Fig. 5B), indicating that expression of alpha-clostripain is negatively regulated by virT. Similarly, the alpha-clostripain activity was higher in the virU-overexpressing strain than in the wild-type strain (Fig. 5B), suggesting that the virU gene product stimulates alpha-clostripain production. The similarities between the changes in perfringolysin O and alpha-clostripain activities and the changes in the levels of mRNAs for these genes in different genetic backgrounds (wild-type strain versus virT mutant or virU-overexpressing strain) suggest that the virT gene product negatively controls expression of pfoA and ccp and that the virU gene product enhances expression of pfoA and ccp in C. perfringens.
FIG. 5.

Alpha-clostripain activities of various C. perfringens strains. (A) The alpha-clostripain activity of each C. perfringens strain (indicated at the bottom) was determined with azocasein as a substrate under conditions with no inhibitor (−), with leupeptin (LP), or with antipain (AP). For each strain and treatment combination, the mean absorbance and standard deviation (error bar) calculated from three independent experiments are shown. (B) The difference in mean proteolytic activity between assays without inhibitors and assays with the leupeptin or antipain inhibitor (shown in panel A), which represents alpha-clostripain-specific proteolytic activity, was plotted for each C. perfringens strain. WT, wild type; OD 450 nm, optical density at 450 nm.
Mutational analyses of virT and virU.
It has been reported that in C. perfringens regulatory RNA molecules control expression of several genes, including toxin genes (17, 28). Considering the relatively small size of the virT and virU ORFs (609 and 216 bp, respectively), it is possible that these genes, like vrr and virX, encode regulatory RNAs. To test this hypothesis, nonsense mutations were introduced into the protein-encoding regions of virT and virU with plasmid vectors (pTS932 and pTS933, respectively), and then differences between the ccp and pfoA steady-state mRNA levels in samples of wild-type and mutant cell total RNAs were determined (Fig. 6). When the virT nonsense mutant gene in pTS932 was introduced into the isogenic virT mutant strain TS190, the relative levels of ccp and pfoA transcripts in TS190(pTS932) were not different from the levels in TS190(pTS930) containing an intact virT gene (Fig. 6A, panel a, lanes 3 and 4). Similarly, a nonsense mutation was constructed in the coding region of virU in plasmid pTS933, which was transformed into wild-type strain 13 (Fig. 6B). The results indicated that there was a difference in the level of vrr, pfoA, or ccp mRNA between wild-type strain 13 overexpressing intact virU (pTS931) and wild-type strain 13 overexpressing mutated virU (pTS933) (Fig. 6B, panel a, lanes 2 and 3). Furthermore, one-base deletions at positions 33 and 15 in the virT and virU coding regions (Fig. 6) were introduced to generate frameshift mutations, resulting in plasmids pTS935 and pTS936, respectively. Plasmid pTS935 was transformed into the isogenic virT mutant strain TS190 and wild-type strain 13, and the mRNA levels for pfoA and ccp were determined by Northern hybridization. As shown in Fig. 6, the relative levels of ccp and pfoA transcripts in TS190(pTS935) with frameshifted virT were not different from the levels in TS190(pTS930) containing an intact virT gene. Similarly, the transcript levels of ccp and pfoA were not different in strain 13 harboring pTS936 (with frameshifted virU) and strain 13 harboring pTS931. Taken together, these results indicate that there was no difference in the regulatory functions of virT and virU whether the ORFs were intact or not intact. These data strongly suggest that both the virT and virU genes encode regulatory RNA molecules, not proteins, that regulate the VirR/VirS regulon. The promoters of pfoA, virT, ccp, vrr, and virU were screened, and no conserved sequence motifs (besides the VirR consensus sequence) were found, indicating that it is unlikely that there are other regulatory RNAs or proteins that are shared by these VirR/VirS-regulated genes.
FIG. 6.
Site-directed mutagenesis of the virT (A) and virU (B) genes and the effects of the mutations on steady-state levels of VirR-regulated gene mRNAs. The nonsense and frameshift codons engineered in the virT and virU coding regions (left panels) are indicated by large boxes. The positions of the nonsense mutation (NM) and frameshift mutation (FM) are indicated by TAA and an asterisk, respectively. Plasmids carrying mutated virT and virU genes (designated pTS932 and pTS935 for virT and pTS933 and pTS935 for virU) were transformed into different C. perfringens strains. The band densities relative to those for wild-type strain 13 are indicated above the bands. (A) (Panel a) Lane 1, wild-type strain 13(pJIR418); lane 2, strain TS190(pJIR418) (ΔvirT); lane 3, strain TS190(pTS930 (ΔvirT virT+); lane 4, strain TS190(pTS932) (ΔvirT virTNM). (Panel b) Lane 1, wild-type strain 13(pJIR418); lane 2, strain TS190(pJIR418) (ΔvirT); lane 3, strain TS190(pTS930) (ΔvirT virT+); lane 4, strain TS190(pTS935) (ΔvirT virTFM). (B) (Panel a) Lane 1, wild-type strain 13(pJIR418); lane 2, wild-type strain 13(pTS931) (virU+); lane 3, wild-type strain 13(pTS933) (virUNM). (Panel b) Lane 1, wild-type strain 13(pJIR418); lane 2, wild-type strain 13(pTS931) (virU+); lane 3, wild-type strain 13(pTS9360 (virUFM). Northern analyses were performed with the indicated probes (right panels).
DISCUSSION
A genome-wide search for promoter-proximal VirR-binding sites previously identified the theta-toxin-encoding gene pfoA (6) and the vrr gene, which encodes regulatory VR-RNA (28), and also identified three new genes, virT, ccp, and virU, as potential members of the VirR/VirS regulon (25). To test the veracity of this in silico identification, genetic and molecular analyses of virT, ccp, and virU functions and regulation by the VirR/VirS system were performed.
Comparative Northern analyses of wild-type and virR mutant strains of C. perfringens revealed that virT, ccp, and virU are positively regulated by the VirR/VirS system at the RNA level. The sequence and location of a putative VirR-binding consensus site (VB1/VB2) and its location (from position −40 to position −80 upstream of the initiation site) were conserved in the pfoA, vrr, virT, ccp, and virU promoters, suggesting that VirR may bind directly to these sites to activate transcription (5). Gel shift assays confirmed that the VirR protein binds specifically to the conserved promoter sequences. These data led us to conclude that the VirR/VirS regulon involves five genes regulated directly by the VirR protein in C. perfringens (Fig. 7).
FIG. 7.
Schematic diagram of the VirR/VirS regulon. The diagram was constructed by using the results of this and previous studies (3, 4, 12, 17, 18, 20, 24, 25, 28).
The deduced amino acid sequence encoded by ccp is highly similar to the amino acid sequence of alpha-clostripain from C. histolyticum (EC 3.4.22.8) (7), which is a heterodimeric cysteine endopeptidase with specificity for Arg-X peptidyl bonds (7, 8). The two polypeptide chains (termed light and heavy) of the native enzyme are encoded by a single 1,581-bp gene, and the junction between the two polypeptide DNA sequences encodes a linker nonapeptide (7). Alpha-clostripain has been implicated in damage of cells in fetal rat calvaria (10) and may contribute to the virulence of other clostridial infections (23). Detection of alpha-clostripain protein and the enzymatic activity in the supernatants of C. perfringens cultures (27) suggest that alpha-clostripain may be a virulence factor. The C. perfringens genome lacks many genes needed for amino acid biosynthesis, and alpha-clostripain may participate in degradation of host proteins that yields nutrients required for C. perfringens survival and growth (25). Further studies are needed to elucidate the roles of alpha-clostripain in C. perfringens nutrient uptake and pathogenicity.
Although we were unable to predict the functions of the molecules encoded by virT and virU based on homology with other genes (25), the activities of these molecules clearly influence VirR/VirS gene regulation; the virT product acts as a negative regulator of expression, and the virU product acts as a positive regulator of expression (Fig. 4). The inability to alter these effects by nonsense mutations in virT and virU suggests that these genes, like vrr (28) and virX (17), encode regulatory RNA molecules rather than proteins (Fig. 6). A possible secondary structure was examined for the predicted virT and virU RNA molecules. The predicted secondary structures of the whole virT and virU RNAs were tight and compact overall, similar to the structure predicted for VR-RNA (data not shown) (28). The transcriptional terminator downstream of the virT and virU regions was also searched, and only virU was found to have inverted repeat sequences. Furthermore, Northern analyses were performed with 50-mer synthetic sense and antisense oligonucleotide probes to look for changes in the steady-state levels of virT and virU mRNAs. Both virT and virU transcripts were detected in wild-type strain 13 with antisense probes, whereas no signals were obtained with the sense virT and virU probes (data not shown).
Unexpectedly, the number of regulatory RNA molecules found to be involved in regulation of virulence (and other) genes in C. perfringens is increasing. Based on recent reports of the importance of small RNA molecules in regulation of transcription and/or translation in both prokaryotes and eukaryotes (12, 15), many RNA molecules involved in other aspects of C. perfringens gene regulation may still be unknown. In the case of the virT and virU RNAs, the absence of a putative consensus sequence for direct annealing of these RNAs to the promoters of the virT- and virU-regulated genes pfoA, virT, ccp, virU, and vrr suggests that these regulatory RNAs may affect the activity of other proteins or RNA regulators for these five genes. Because the effects of virT and virU mutations on transcription were much more subtle than those of the VirR/VirS system or VR-RNA, virT and virU may fine-tune transcription of VirR/VirS-regulated genes to maintain balanced gene expression. Future studies of the effects of virT and virU on gene regulation, such as DNA microarray analyses of C. perfringens cultured under changing environmental conditions, may provide a more detailed view of the overall effects of these regulatory genes.
The conclusion that the VirR/VirS system directly regulates only five genes (pfoA, vrr, virT, ccp, and virU) in C. perfringens via VirR binding is somewhat surprising. It has been reported that the VirR/VirS system influences expression of many other genes, including plc (encoding alpha-toxin), colA (encoding kappa-toxin), cpd (encoding 2′,3′-cyclic nucleotide phosphodiesterase), ptp (encoding protein tyrosine phosphatase), ycgJ (encoding a hypothetical protein), metB (encoding cystathionine gamma-lyase), cysK (encoding cysteine synthase), and luxS (encoding the autoinducer 2 production protein) (3, 4, 18, 20). However, for plc, colA, cpd, ptp, and ycgJ-metB-cysK-luxS, VR-RNA has been shown to be a secondary RNA regulator (3, 28) (Fig. 7). Another RNA regulator, virX, controls the levels of pfoA, plc, and colA mRNAs independent of the VirR/VirS regulatory cascade (17) (Fig. 7). Furthermore, cell-cell signaling by autoinducer 2 synthesized by the luxS gene product, which may be mediated through an unidentified two-component system (18), also plays an important role in the regulation of toxin production.
It is clear that the VirR/VirS regulon consists of two classes of genes, the genes that are regulated directly by the VirR/VirS system and the genes that are regulated indirectly (Fig. 7). The mechanism of regulation of the VirR/VirS regulon, including the actions of the virT and virU regulatory RNA molecules, should be clarified by comprehensive DNA microarray-based analyses of gene expression.
Acknowledgments
We thank Harumi Yaguchi for supplying the vrr mutant strain and Kaori Honjo, Yukari Tajima, and Hameem I. Kawsar for helpful discussions.
This work was supported by the Research for the Future Program of the Japan Society for the Promotion of Science, by Grants-in Aid for Scientific Research (B) from the Japan Society for the Promotion of Science, and by KAKENHI (Grant-in-Aid for Scientific Research on Priority Areas “Applied Genomics”) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 12 September 2008.
REFERENCES
- 1.Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 25611905-11910. [PubMed] [Google Scholar]
- 2.Awad, M. M., A. E. Bryant, D. L. Stevens, and J. I. Rood. 1995. Virulence studies on chromosomal alpha-toxin and theta-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of alpha-toxin in Clostridium perfringens-mediated gas gangrene. Mol. Microbiol. 15191-202. [DOI] [PubMed] [Google Scholar]
- 3.Banu, S., K. Ohtani, H. Yaguchi, T. Swe, S. T. Cole, H. Hayashi, and T. Shimizu. 2000. Identification of novel VirR/VirS-regulated genes in Clostridium perfringens. Mol. Microbiol. 35854-864. [DOI] [PubMed] [Google Scholar]
- 4.Ba-Thein, W., M. Lyristis, K. Ohtani, I. T. Nisbet, H. Hayashi, J. I. Rood, and T. Shimizu. 1996. The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J. Bacteriol. 1782514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung, J. K., B. Dupuy, D. S. Deveson, and J. I. Rood. 2004. The spatial organization of the VirR boxes is critical for VirR-mediated expression of the perfringolysin O gene, pfoA, from Clostridium perfringens. J. Bacteriol. 1863321-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, J. K., and J. I. Rood. 2000. The VirR response regulator from Clostridium perfringens binds independently to two imperfect direct repeats located upstream of the pfoA promoter. J. Bacteriol. 18257-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dargatz, H., T. Diefenthal, V. Witte, G. Reipen, and D. von Wettstein. 1993. The heterodimeric protease clostripain from Clostridium histolyticum is encoded by a single gene. Mol. Gen. Genet. 240140-145. [DOI] [PubMed] [Google Scholar]
- 8.Gilles, A. M., J. M. Imhoff, and B. Keil. 1979. Alpha-clostripain. Chemical characterization, activity, and thiol content of the highly active form of clostripain. J. Biol. Chem. 2541462-1468. [PubMed] [Google Scholar]
- 9.Hatheway, C. L. 1990. Toxigenic clostridia. Clin. Microbiol. Rev. 366-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hefley, T., J. Cushing, and J. S. Brand. 1981. Enzymatic isolation of cells from bone: cytotoxic enzymes of bacterial collagenase. Am. J. Physiol. 240C234-C238. [DOI] [PubMed] [Google Scholar]
- 11.Janoir, C., S. Péchiné, C. Grosdidier, and A. Collignon. 2007. Cwp84, a surface-associated protein of Clostridium difficile, is a cystein protease with degrading activity on extracellular matrix proteins. J. Bacteriol. 189:7174-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson, J., and P. Cossart. 2003. RNA-mediated control of virulence gene expression in bacterial pathogens. Trends Microbiol. 11280-285. [DOI] [PubMed] [Google Scholar]
- 13.Lyristis, M., A. E. Bryant, J. Sloan, M. M. Awad, I. T. Nisbet, D. L. Stevens, and J. I. Rood. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol. Microbiol. 12761-777. [DOI] [PubMed] [Google Scholar]
- 14.Mahony, D. E., and T. I. Moore. 1976. Stable L-forms of Clostridium perfringens and their growth on glass surfaces. Can. J. Microbiol. 22953-959. [DOI] [PubMed] [Google Scholar]
- 15.Masse, E., N. Majdalani, and S. Gottesman. 2003. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 6120-124. [DOI] [PubMed] [Google Scholar]
- 16.Myers, G. S. A., D. A. Rasko, J. K. Cheung, J. Ravel, R. Seshadri, R. T. DeBoy, Q. Ren, J. Varga, M. M. Awad, L. M. Brinkac, S. C. Daugherty, D. H. Haft, R. J. Dodson, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. A. Sullivan, H. Khouri, G. I. Dimitrov, K. L. Watkins, S. Mulligan, J. Benton, D. Radune, D. J. Fisher, H. S. Atkins, T. Hiscox, B. H. Jost, S. J. Billington, J. G. Songer, B. A. McClane, R. W. Titball, J. I. Rood, S. B. Melville, and I. T. Paulsen. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 161031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohtani, K., S. K. Bhowmik, H. Hayashi, and T. Shimizu. 2002. Identification of a novel locus that regulates expression of toxin genes in Clostridium perfringens. FEMS Microbiol. Lett. 209113-118. [DOI] [PubMed] [Google Scholar]
- 18.Ohtani, K., H. Hayashi, and T. Shimizu. 2002. The luxS gene is involved in cell-cell signaling for toxin production in Clostridium perfringens. Mol. Microbiol. 44171-179. [DOI] [PubMed] [Google Scholar]
- 19.Ohtani, K., H. I. Kawsar, K. Okumura, H. Hayashi, and T. Shimizu. 2003. The VirR/VirS regulatory cascade affects transcription of plasmid-encoded putative virulence genes in Clostridium perfringens. FEMS Microbiol. Lett. 222137-141. [DOI] [PubMed] [Google Scholar]
- 20.Ohtani, K., H. Takamura, H. Yaguchi, H. Hayashi, and T. Shimizu. 2000. Genetic analysis of the ycgJ-metB-cysK-ygaG operon negatively regulated by the VirR/VirS system in Clostridium perfringens. Microbiol. Immunol. 44525-528. [DOI] [PubMed] [Google Scholar]
- 21.Petit, L., M. Gibert, and M. R. Popoff. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7104-110. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 23.Seddon, S. V., and S. P. Borriello. 1992. Proteolytic activity of Clostridium difficile. J. Med. Microbiol. 36307-311. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu, T., W. Ba-Thein, M. Tamaki, and H. Hayashi. 1994. The virR gene, a member of a class of two-component response regulators, regulates the production of the perfringolysin O, collagenase, and hemagglutinin in Clostridium perfringens. J. Bacteriol. 1761616-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu, T., A. Okabe, and J. I. Rood. 1997. Regulation of toxin production in Clostridium perfringens, p. 451-470. In J. I. Rood, G. Songer, B. A. McClane, and R. W. Titball (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, London, United Kingdom.
- 27.Shimizu, T., K. Shima, K. Yoshino, K. Yonezawa, T. Shimizu, and H. Hayashi. 2002. Proteome and transcriptome analysis of the virulence genes regulated by the VirR/VirS system in Clostridium perfringens. J. Bacteriol. 1842587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu, T., H. Yaguchi, K. Ohtani, S. Banu, and H. Hayashi. 2002. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol. Microbiol. 43257-265. [DOI] [PubMed] [Google Scholar]
- 29.Sloan, J., T. A. Warner, P. T. Scott, T. L. Bannam, D. I. Berryman, and J. I. Rood. 1992. Construction of a sequenced Clostridium perfringens-Escherichia coli shuttle plasmid. Plasmid 27207-219. [DOI] [PubMed] [Google Scholar]