Abstract
Francisella tularensis is the causative agent of tularemia and is a category A select agent. Francisella novicida, considered by some to be one of four subspecies of F. tularensis, is used as a model in pathogenesis studies because it causes a disease similar to tularemia in rodents but is not harmful to humans. F. novicida exhibits a strong restriction barrier which reduces the transformation frequency of foreign DNA up to 106-fold. To identify the genetic basis of this barrier, we carried out a mutational analysis of restriction genes identified in the F. novicida genome. Strains carrying combinations of insertion mutations in eight candidate loci were created and assayed for reduced restriction of unmodified plasmid DNA introduced by transformation. Restriction was reduced by mutations in four genes, corresponding to two type I, one type II, and one type III restriction system. Restriction was almost fully eliminated in a strain in which all four genes were inactive. The strongest contributor to the restriction barrier, the type II gene, encodes an enzyme which specifically cleaves Dam-methylated DNA. Genome comparisons show that most restriction genes in the F. tularensis subspecies are pseudogenes, explaining the unusually strong restriction barrier in F. novicida and suggesting that restriction was lost during evolution of the human pathogenic subspecies. As part of this study, procedures were developed to introduce unmodified plasmid DNA into F. novicida efficiently, to generate defined multiple mutants, and to produce chromosomal deletions of multiple adjacent genes.
Restriction-modification (R-M) systems in bacteria limit the acquisition of foreign genes that enter the cell by infecting phage, conjugal transfer systems, or transformation. Incoming DNA is cleaved (restricted) if it has not been modified by methylation at specific sequences (24, 35). Almost all bacterial species carry R-M systems, with some species predicted to harbor dozens of R-M genes (11). There are four types of R-M systems (types I to IV), classified according to mechanism of action and the distribution of restriction, modification, and specificity functions among enzyme subunits (24, 27).
Francisella tularensis is an important human and zoonotic pathogen whose ecology and natural reservoirs are only partially understood. There are at least four subspecies which differ in their infectivity. Two of them, F. tularensis subsp. tularensis and F. tularensis subsp. holarctica, the causative agents of tularemia, are among the most infectious pathogens known and are classified as category A select agents due to their high infectivity, ease of dissemination, and severity of disease (5). Two others, “F. tularensis subsp. novicida” (Francisella novicida) and F. tularensis subsp. mediasiatica, do not generally cause disease in humans. The genomes of Francisella species are small (<2.0 Mbp), yet the bacteria are able to colonize hundreds of different animal and insect species, effectively evade host immune responses, and cause serious disease at even extremely small infective doses (e.g., fewer than 10 infecting cells of F. tularensis subsp. tularensis) (23). Genome comparisons have suggested that the highly virulent subspecies evolved from less virulent ancestors by processes that included significant gene loss (29, 31). Of the four commonly recognized species and subspecies, F. novicida is proposed to most closely represent the relatively avirulent ancestral strain (29, 31). F. novicida has emerged as an important model for studying virulence mechanisms in tularemia because it causes a tularemia-like disease in rodents but is not dangerous to humans (9, 13). F. novicida is also of inherent interest for its unique features among Francisella strains, which include rapid growth, relatively high virulence in mice, and the presence of one rather than two copies of the Francisella pathogenicity island (9, 18, 26).
An additional property which distinguishes F. novicida from the F. tularensis subspecies is the presence of a strong restriction barrier (16, 20). Plasmid DNA isolated from Escherichia coli or other Francisella subspecies transforms F. novicida as much as 106-fold less efficiently than plasmid DNA isolated directly from F. novicida (20). The unique presence of this barrier in F. novicida suggests that restriction may have been lost in the evolution of the more virulent lineages. The barrier is also of practical importance, for it has made genetic manipulations such as plasmid transformation and gene targeting in F. novicida challenging (7). In this study we have identified the principle determinants of restriction in F. novicida and have constructed a multiple mutant in which the restriction barrier is fully eliminated. The analysis substantiates the conjecture that restriction was lost in the evolution of highly virulent Francisella strains and provides useful tools for the genetic manipulation of F. novicida.
MATERIALS AND METHODS
Strains, growth media, culture conditions, plasmids, and molecular methods.
The F. novicida strains used were U112 (12) and transposon insertion derivatives from the near-saturation transposon mutant library (8). The E. coli strains used were CC118 [araD139 Δ(ara leu)7697 ΔlacX74 phoAΔ20 galE galK thi rpsE rpoB argEam recA1] (22), DH5α [fhuA2 Δ(argF-lacZ)U169 phoA glnV44 φ80Δ(lacZ)M15 gyrA96 recA1 endA1 thi-1 hsdR17] (36), and the dam-deficient strain CC160 [araD139 Δ(ara-leu)7697 ΔlaxX74 galE galK thi rpsL dam] (21). Nutrient growth medium for F. novicida was tryptic soy agar or broth (Difco) supplemented with 0.1% cysteine-HCl and 0.2% dextrose (TSBC or TSAC, respectively); for defined media, Chamberlain's defined medium (CDM) (4) and FnDM, a minimal medium described previously (8), were used. For E. coli, standard growth media were used (30). Antibiotics used for F. novicida were kanamycin (10 μg/ml), tetracycline (10 μg/ml), erythromycin (30 μg/ml), and hygromycin (225 μg/ml); for E. coli, tetracycline (15 μg/ml) and hygromycin (200 μg/ml) were used. Unless otherwise stated, strains were grown at 37°C. The plasmids used were F. novicida-E. coli shuttle plasmids pKK214gfp (1), which carries a tetracycline resistance marker, pMP633 (16), which carries a hygromycin resistance marker, the Flp recombinase-expressing plasmid pLG72 (8), and the pLG72 derivatives described below. Genomic DNA and plasmids were isolated using Qiagen kits (Valencia, CA).
Construction of temperature-sensitive plasmids pFFlp and pFFlp-hyg for expression of Flp recombinase in Francisella.
pLG72 (8) was digested with BglII (which cuts once within the plasmid at a site within the repA gene), end filled with T4 DNA polymerase, recircularized with T4 DNA ligase, and electroporated into U112. Screening of 48 transformants identified 2 with the ability to grow at 30°C but not 42°C. One of these was selected for further characterization. Sequence analysis of the entire repA gene within the corresponding plasmid (which we hereby name pFFlp) showed a single sequence difference from pLG72, an insertion of three base pairs (GAT) at codon 276 (the location of the aforementioned BglII site), resulting in the addition of an aspartate codon (D276DD). The plasmid was found to be maintained stably in F. novicida at 30°C but was lost from >80% of cells after overnight growth on solid medium at 37°C without selection. Its parent, pLG72, in contrast, was cured only with difficulty by screening hundreds of colonies after serial passage without selection (data not shown). We note that Maier and colleagues have also described a temperature-sensitive Francisella plasmid, pFNLTP9, generated by an amino acid substitution at a different location within the repA gene (M120I) and which could not be maintained at 42°C even in the presence of selection (20). We also constructed a derivative of pFFlp, pFFlp-hyg, in which the vector's tetracycline resistance marker was replaced with a hygromycin resistance marker. pFFlp-hyg will be useful in virulent Francisella sp. strains where tetracycline resistance is not an approved marker. To construct pFFlp-hyg, pMP633 (16) was digested with HpaI and StuI, releasing the hyg gene and its promoter region. pFFlp was digested with FspI, removing most of the tet gene. The two digestion reaction products were cleaned (Qiagen PCR cleanup kit), ligated together, and transformed into E. coli. Transformants selected on hygromycin were screened for the desired plasmid by restriction analysis. Candidate plasmids were tested to verify Flp recombinase activity and temperature-sensitive replication.
Transformation.
For electroporation of plasmid DNA into F. novicida, an overnight TSBC culture started from a fresh colony was subcultured 1:100 in 10 ml of TSBC and incubated at 37°C with aeration to an optical density at 600 nm (OD600) of 0.5 to 0.9. Cells were pelleted and resuspended twice in 0.5 M sucrose with 1 mM EDTA and twice in 0.5 M sucrose, then pelleted and resuspended in 0.5 M sucrose to a volume of ∼50 μl. Centrifugations were at approximately 3,000 × g for 5 min, and all steps were performed at room temperature. Concentrated cells (50 μl) were mixed with DNA, incubated at room temperature for 10 min, and electroporated (0.2-cm cuvette, 2.5 kV, 25 μF, 400 Ω; Gene Pulser; Bio-Rad, Hercules, CA). After adding 1 ml of TSBC, cells were incubated for 3 to 6 h at 37°C with aeration, and dilutions were plated on selective TSAC.
For transformation of chromosomal DNA into F. novicida, we used a variation of a previously described method (2, 32, 33). In brief, an overnight culture of the F. novicida recipient in CDM or FnDM was diluted 1:50 into CDM or FnDM, incubated at 37°C with aeration to an OD600 of 0.4 to 0.8, pelleted (∼3,000 × g at room temperature), and resuspended in a 1:10 volume of CDM or FnDM. DNA for transformation (e.g., 100 ng genomic DNA) and 0.1 ml concentrated cells were added to 1.0 ml transformation buffer (33) which was modified to exclude Mg2+ and Mn2+ (2). The mixture was incubated for 30 to 60 min at 37°C with gentle agitation (100 rpm shaker), then 2 ml of CDM was added and the mixture was incubated at 37°C with aeration for 3 to 6 h before plating on selective TSAC.
Flp-Frt recombination.
To carry out Flp-mediated recombination in strains bearing T20 insertions (the resistance marker in T20 is flanked by FRT sites), plasmid pFFlp was introduced by electroporation and transformants were selected by incubation at 30°C on TSAC with tetracycline. Transformants were restreaked for single colonies on TSAC with tetracycline and incubated at 30°C overnight. Isolated colonies from this streak were tested for kanamycin sensitivity (indicating Flp recombinase-mediated excision of the marker) by patching and incubation at 37°C. Usually 100% of colonies were Kans, whereas <0.1% of colonies from control transformations (using pKK214gfp, the parent of pLG72, and pFFlp, which does not bear the flp gene) were Kans. Kans patches were restreaked for single colonies on medium without selection and incubated at 37°C. Isolated colonies from this streak were tested for tetracycline sensitivity (indicating loss of pFFlp) by patching and incubation at 37°C. Usually >80% of colonies were Tets.
Construction of multiple mutants.
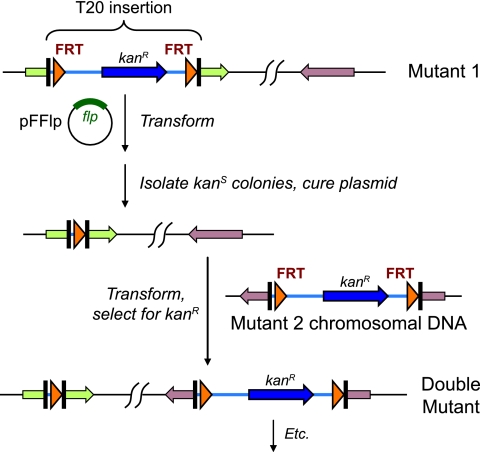
Multiple mutants were generated from T20 insertion alleles as follows and as shown in Fig. 1: (i) the resistance marker in a T20 insertion mutant was excised by Flp-mediated recombination and curing of the Flp-bearing plasmid (see above); (ii) a second allele was introduced by transformation of genomic DNA from the corresponding mutant (see above). The steps were repeated to generate double, triple, quadruple, and quintuple mutants representing combinations of the insertion alleles. Over 200 multiple mutants were generated; a complete list is provided in the supplemental material. Two key mutants, which are described in the Results section, were MFN192, a triple mutant bearing mutations in loci 2 (FTN_0710), 3 (FTN_1155), and 4 (FTN_1487), and MFN245, a quadruple mutant bearing mutations in loci 2, 3, 4, and 6 (FTN_1698).
FIG. 1.
Construction of multiple mutants in F. novicida. A strain with a T20 transposon insertion is transformed with pFFlp, a temperature-sensitive plasmid which expresses Flp recombinase. Recombinants which lose the kanamycin resistance marker within T20 due to Flp-mediated site-specific recombination between the FRT sites are identified by screening. The plasmid is cured. A T20 insertion in another locus is introduced by transformation of chromosomal DNA from a second mutant strain, generating a double mutant. The steps are repeated to add additional mutations.
A potential complication of expressing Flp recombinase in strains bearing FRT sites at multiple genomic locations (such as the multiple mutants described in this work) is genomic rearrangement. For FRT sites in reverse orientation to one another in the genome, recombination between them would produce an inversion of the intervening DNA. For cooriented sites, recombination would result in a deletion of the intervening DNA. For sites at locations distant from one another in the genome (such as the eight candidate R-M loci studied in this work), such a deletion would not be recovered due to loss of essential functions resident in the deleted fragment. To help reduce recovery of rearrangements for the multiple mutants constructed in this work, insertions were chosen whenever possible to be cooriented with one another. However, not all insertions in the triple, quadruple, and quintuple mutants were cooriented. While some inversions may therefore have occurred, such inversions would not be predicted to affect the phenotypes under study. To help ensure that genomic rearrangement was not a complicating factor, we independently constructed and tested multiple mutants for the various mutant combinations studied. Independently constructed mutants always showed consistent phenotypes (data not shown).
Generation of a large deletion.
Flp-mediated recombination between two cooriented FRT sites for which the intervening genomic sequence is not essential for growth offers a means for intentionally generating genomic deletions. We generated and confirmed a deletion of this kind as follows: several strains were created that carried insertions in the genes FTN_0287 (the locus 1 putative restriction gene) and FTN_0283, a pseudogene four genes distant from FTN_0287 (Fig. 2). After expressing Flp recombinase in these strains (by using pFFlp), we used PCR to test the resulting kanamycin-sensitive isolates for deletion of the genomic DNA between the two insertion sites. In one of two independent isolates examined, an amplification product corresponding to the deletion was observed, whereas in its parent (the same strain before Flp expression), the PCR test showed the presence of each individual insertion allele and not the deletion (data not shown).
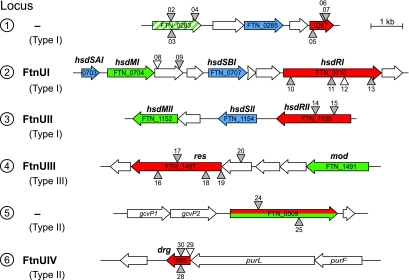
FIG. 2.
Candidate restriction loci in F. novicida. Restriction-modification genes predicted by the genome annotation (www.francisella.org) and by REBASE (rebase.neb.com) include three type I restriction genes, a type III restriction gene, a fused type II restriction and modification gene, and a type II restriction gene annotated as a dam-replacing family protein. Three additional possible restriction functions (FTN_0418, FTN_0449, and FTN_1378) are not shown but are described in the text. Putative functions are color coded: restriction (red), modification (green), specificity (blue). Gene names assigned in this work are shown in bold text above the genes. The four restriction-modification system names assigned are shown to the left. FTN_0283 (locus 1) is a modification methyltransferase pseudogene carrying a frameshift approximately two-thirds of the way through the gene (confirmed by sequencing). Numbered triangles represent transposon insertion alleles used in this study (see text and the supplemental material). Gray triangles, insertions bearing FRT site-specific recombination sites (T20 insertions); unfilled triangles, insertions without FRT sites. The orientations of the insertion within the genome are indicated by whether the triangles are shown above or below the lines representing the chromosome.
Assay of DNA restriction by plasmid transformation.
To assay transformation efficiency of unmodified plasmid relative to modified plasmid (relative transformation efficiency), we electroporated strains using defined mixtures of pKK214fgp isolated from U112 (the modified plasmid) and pMP633 isolated from E. coli CC118 (the unmodified plasmid). After outgrowth, the electroporation mix was plated separately on TSAC supplemented with tetracycline or hygromycin, and the relative efficiencies of transformation of the two plasmids were calculated by determining the number of transformants per ng DNA for each plasmid type. The quantities of the two plasmids in the defined mixtures (typically between 2 and 6 ng of the modified plasmid and, depending on the strain to be assayed, between 2 and 1,000 ng of the unmodified plasmid) were within the linear range for transformation of each plasmid type (data not shown). We found that when both plasmids were fully modified, the inherent transformation frequency of pMP633 was approximately eightfold higher than for pKK214gfp (n = 2; standard deviation = 3.4). Therefore, for the results presented in this report, we adjusted the calculated relative transformation efficiency ratios by this correction factor.
RESULTS
Multiple systems contribute to the restriction barrier in Francisella novicida.
The genome of F. novicida is predicted to encode multiple DNA restriction and modification functions (www.francisella.org). There are four restriction enzymes predicted with high confidence: three type I HsdR proteins and one probable type III restriction endonuclease (Fig. 2, loci 1 to 4). For most of these genes, putative specificity and methyltransferase functions are encoded by nearby genes (Fig. 2).
We first sought to determine whether loss of any one of these four predicted restriction genes strongly compromised the observed restriction barrier. Individual mutants from the F. novicida transposon mutant library (8) were tested for increased efficiency of transformation of unmodified plasmid DNA. To obtain reproducible, quantitative results, we developed a “restriction assay” in which we transformed mixtures of modified and unmodified plasmids which could be genetically distinguished. The restriction activity of different mutants was then assayed by the relative transformation efficiency of the two plasmids (see Materials and Methods). Single mutants of the four putative restriction genes (Fig. 2, loci 1 to 4) each exhibited less-than-10-fold increases in transformation efficiency of unmodified relative to modified plasmid, suggesting that none of the four systems was predominantly responsible for the strong restriction barrier. These results did not rule out the possibility that the four systems play overlapping roles in restriction, so that elimination of any one of them alone would lead to little change in overall restriction.
Mutation of multiple systems substantially reduces restriction.
To assess whether more than one of the predicted restriction systems contributed to restriction, we constructed multiple mutants. We developed a technique for efficiently building multiple mutants using insertion alleles from the F. novicida transposon mutant library (8). The technique, depicted in Fig. 1, employs chromosomal DNA transformation to introduce marked transposon insertion alleles into recipient strains carrying unmarked insertion alleles created by Flp-FRT recombination. In brief, (i) the resistance marker within a T20 transposon insertion mutant is excised by introduction of a plasmid expressing Flp recombinase, resulting in an unmarked insertion; (ii) after curing the plasmid, a second insertion mutation is introduced by transformation of chromosomal DNA from a second transposon mutant; (iii) the marker excision and transformation steps are repeated to add additional mutations. To facilitate the plasmid curing step, we created a Flp expression plasmid, pFFlp, with temperature-sensitive replication in F. novicida (see Materials and Methods).
We constructed double, triple, and quadruple mutants carrying combinations of mutant alleles of the four strongly predicted restriction loci. Multiple insertion alleles were examined for each gene (see the supplemental material). Strains were then tested using the restriction assay described above. Figure 3A summarizes the data, showing all possible combinations of multiple mutations in these four strongly predicted restriction genes. Using alternative alleles at each locus produced comparable results (data not shown). The quadruple mutants and one of the triple mutant combinations displayed an approximately 1,000-fold decrease in restriction activity (Fig. 3A), representing a substantial attenuation of the restriction barrier. Most of the double and triple mutant combinations displayed intermediate levels of restriction.
FIG. 3.
Restriction activities of mutant strains of F. novicida. Transformation efficiencies of unmodified plasmid relative to modified plasmid (see Materials and Methods) are depicted for strains of various genotypes. The genotypes for the candidate restriction loci (Fig. 2 and text) are indicated at the bottom: +, intact; −, mutant. Points represent results from individual assays, with horizontal lines corresponding to the geometric means for the individual assays of strains of each genotype. (A) Mutant combinations of the four loci initially investigated. (B) Testing of additional candidate restriction genes (see text).
The results suggest that three of the four genes make significant contributions to the restriction barrier and that the effects of inactivation of each of the three loci are multiplicative. For example, mutations in locus 2 (FTN_0710) or locus 4 (FTN_1487) decreased restriction levels 5- to 10-fold, while strains carrying mutations in both loci showed an approximately 100-fold decrease. Mutations in locus 3 (FTN_1155), which individually (or when combined with locus 4 mutations) did not detectably reduce restriction, decreased restriction approximately 10-fold when combined with locus 2 mutations. Locus 1 (FTN_0287) appeared to have no effect on restriction, since all triple and quadruple mutant combinations in which locus 1 was mutated displayed the same levels of restriction as the corresponding multiple mutants in which locus 1 was intact (Fig. 3A). We conclude that loci 2, 3, and 4, but not locus 1, make significant contributions to restriction under our assay conditions.
A fourth restriction function.
Although the restriction barrier was substantially reduced in the multiple mutants examined, even the most severely affected still exhibited significant restriction, corresponding to a greater-than-100-fold decrease in the efficiency of transformation of unmodified plasmid (Fig. 3A). Thus, there must exist one or more additional restriction systems. Accordingly, we examined the genome for additional candidate loci. Several genes with homology to endonuclease functions were identified: FTN_0418, a predicted endonuclease with a URI domain; FTN_0449, a gene of unknown function with weak similarity (e = 0.009) to the type I HsdR COG group; FTN_0509, a protein of unknown function with similarity (32% identity) to an F. tularensis gene annotated as a restriction enzyme (Fig. 2, locus 5); FTN_1378, a hypothetical protein with similarity (32% identity) to a type I HsdR protein from Prosthecochloris aestuarii; FTN_1698, a Dam-replacing family protein with significant similarity (53% identity) to the type II restriction enzyme DpnI (Fig. 2, locus 6). Two of these genes, FTN_0509 and FTN_1698 (Fig. 2, loci 5 and 6), were also identified as potential restriction genes by REBASE, a database of known and postulated R-M genes in bacterial genomes (28).
To test whether any of these additional genes contributed to the restriction barrier, we introduced corresponding mutant alleles for four of them (FTN_0418 was not tested because no mutant alleles were available from the transposon insertion library [8]) into the strongest restriction-deficient triple and quadruple mutants described above. We then tested these strains for diminished restriction. We found that mutations in one of the candidate genes, the Dam-replacing family protein (FTN_1698; Fig. 2, locus 6), led to an approximately 500-fold reduction in plasmid restriction (Fig. 3B). Single mutants of FTN_1698 exhibited a 10- to 100-fold decrease in restriction relative to wild type (Fig. 3B). Mutations in the other three additional candidate loci examined did not significantly reduce restriction (Fig. 3B and data not shown). Thus, inactivation of the Dam-replacing type II function had the strongest effect on restriction of any of the individual genes tested.
The strain bearing mutations in loci 2, 3, 4, and 6 exhibited a 105- to 106-fold decrease in restriction compared to wild type. We designated this strain MFN245 (its parent, the locus 2, 3, 4 triple mutant, was named MFN192; see the supplemental material for additional strain names). The average transformation frequency of unmodified plasmid relative to modified plasmid for MFN245 was approximately 1 (Fig. 3B), indicating a complete or nearly complete absence of restriction acting on the test plasmid.
We have named the four restriction systems showing measurable activity (loci 2, 3, 4, and 6) FtnUI, FtnUII, FtnUIII, and FtnUIV, respectively. In consideration of precedent and recommended naming conventions for the different types of restriction systems (3, 27), we have named the individual genes comprising each system as follows (and as shown in Fig. 2): for FtnUI (locus 2; a type I R-M system), hsdSAI (FTN_0703), hsdMI (FTN_0704), hsdSBI (FTN_0707), and hsdRI (FTN_0710); for FtnUII (locus 3; type I), hsdMII (FTN_1152), hsdSII (FTN_1154), and hsdRII (FTN_1155); for FtnUIII (locus 4; type III), res (FTN_1487) and mod (FTN_1491); for FtnUIV (locus 6; type II), drg (FTN_1698).
FTN_1698 (drg) restricts adenine-methylated DNA.
Dam-replacing proteins are an unusual class of type II restriction enzymes which generally restrict DNA that is adenine-methylated at GATC sites by Dam (15). Unmethylated GATC sites are not cleaved (3). Dam-replacing restriction enzymes, which include DpnI, are encoded by a gene family (drg) found in place of Dam methylase genes (3). The Dam-replacing proteins are “orphan” restriction enzymes in that linked (unnecessary) modification genes are absent (Fig. 2, locus 6). To test directly whether FTN_1698 (drg) performs a function similar to these drg genes, we tested the transformation efficiency of plasmid isolated from either dam+ or dam-deficient E. coli (Fig. 4). For transformation into strains in which drg was intact (wild-type U112 or the locus 2, 3, 4 triple mutant MFN192), DNA isolated from dam-deficient E. coli displayed a transformation efficiency that was 10- to 1,000-fold higher than DNA isolated from dam+ E. coli. In addition, the efficiency of transformation of DNA from dam-deficient E. coli into these strains was equivalent to that of DNA isolated from dam+ E. coli into isogenic drg mutant strains (the locus 6 single mutant or the locus 2, 3, 4, 6 quadruple mutant MFN245, respectively) (Fig. 4). When drg was mutated, the Dam methylation state of the transforming DNA had no effect on transformation efficiency (Fig. 4). These data indicate that the drg (locus 6) gene product specifically restricts DNA which has been methylated by Dam.
FIG. 4.
Transformation of Dam-methylated and Dam-unmethylated DNA into F. novicida. Transformation efficiencies of plasmid isolated from dam+ or dam-deficient E. coli relative to plasmid isolated from F. novicida are depicted for strains of various genotypes. Strain genotypes for the four putatively active restriction loci are indicated at the bottom (+, intact; −, mutant). Points represent results from individual assays; horizontal lines represent geometric means.
The restriction-deficient strains are modification proficient.
As shown above, plasmid from dam+ E. coli can be introduced into MFN245 at up to 106-greater efficiency than into U112. If MFN245 remains modification proficient, then plasmids isolated from MFN245 should transform U112 with substantially greater efficiency than unmodified plasmids or plasmid isolated from dam-deficient E. coli. To determine the modification capability of MFN245, we tested the transformation efficiency into U112 of a plasmid isolated from dam+ E. coli, dam-deficient E. coli, U112, MFN192, or MFN245 (Table 1). We found that plasmid isolated from MFN245 transformed U112 nearly as efficiently as plasmid isolated from U112. The absence of Dam methylation by F. novicida MFN245 accounts for a 10- to 100-fold increase in the plasmid's transformability into U112 compared to DNA from dam+ E. coli (for DNA from dam-deficient E. coli, we observed a 10-fold increase in this instance [Table 1]). The approximately 103-fold additional increase in transformation efficiency observed implies that MFN245 remains almost completely modification proficient for the other three R-M systems, FtnUI, FtnUII, and FtnUIII (loci 2, 3, and 4). MFN192 (the loci 2, 3, 4 triple mutant) and several other partially restriction-deficient mutants were also tested and found to be equally modification proficient (Table 1 and data not shown).
TABLE 1.
Modification capabilities of the restriction-deficient mutantsa
| Plasmid source | Transformation efficiency relative to fully modified plasmid |
|---|---|
| E. coli (dam+) | 1.5 × 10−4 |
| E. coli (dam deficient) | 1.0 × 10−3 |
| U112 | 1.0 |
| MFN192 (hsdRI hsdRII res) | 0.46 |
| MFN245 (hsdRI hsdRII res drg) | 0.35 |
A Hygr-marked plasmid isolated from the strains listed was cotransformed with a Tetr-marked plasmid isolated from U112 to determine the transformation frequency of the Hygr-marked plasmid relative to the fully modified plasmid. Values represent geometric means of two separate assays and are normalized to the geometric mean value for plasmid isolated from U112. Variance was within 0.5 logs under each condition.
DISCUSSION
We report a comprehensive genetic analysis of the functions underlying the strong restriction barrier in F. novicida. Four of eight different potential restriction systems were found to contribute to the restriction of unmodified plasmid DNA introduced by transformation. The active functions corresponded to two type I restriction systems, one type II restriction system, and one type III restriction system. A strain carrying mutations inactivating all four systems appeared to be fully restriction-negative and displayed a 105 to 106 greater transformation efficiency than the wild type.
The type II system (drg) (Fig. 2, locus 6) was quantitatively the most important of the four restriction functions and corresponds to an unusual class of enzyme which specifically restricts Dam-methylated DNA and is found in bacterial genomes in place of dam (3). The finding suggests that exposure to DNA derived from Dam-bearing bacteria may have been significant in the history of the F. novicida species. Dam methylase genes are particularly widespread in enteric bacteria but are also present in other genera (15, 17). Given the cleavage specificity of drg for Dam-methylated DNA, it is not surprising that no dam gene is found in the F. novicida genome.
The two type I restriction genes which contribute to the restriction barrier (hsdRI and hsdRII; loci 2 and 3 in Fig. 2) may have partially overlapping specificities. This was suggested by the finding that hsdRII (locus 3) mutations by themselves or in combination with locus 1 or locus 4 mutations did not significantly decrease restriction but did cause a decrease when combined with hsdRI (locus 2) mutations (Fig. 3A). On the other hand, hsdRI mutations always diminished restriction, even in the absence of hsdRII mutations, suggesting that if there is a functional redundancy between these two genes, it is only partial. There appears to be partial redundancy in the contributions to the restriction barrier of some of the other genes as well. Specifically, the effect of loss of restriction by the drg (locus 6) gene product, whether due to gene inactivation or the absence of Dam methylation of incoming DNA, was significantly greater (>100-fold) when hsdRI, hsdRII, and res (loci 2, 3, and 4) were inactivated than when they were intact (∼10-fold) (Fig. 4). Since the drg gene product apparently restricts with different specificity than the other systems, this observation can likely be explained by overlapping effects on the integrity of the incoming plasmid molecules.
Two of the genes which were annotated as restriction genes—one by the F. novicida genome annotation (FTN_0287) and both by REBASE (rebase.neb.com)—appeared to make no contribution to restriction (FTN_0287 and FTN_0509; loci 1 and 5 in Fig. 2). FTN_0287 is annotated as a type I restriction subunit but encodes a much smaller gene product (255 codons) than the two type I enzymes which were shown to be active (1,036 and 782 codons). Hence, FTN_0287 may be a gene remnant that is no longer active. This possibility is reinforced by the finding that the modification subunit gene neighboring it, FTN_0283, is a pseudogene. The second apparently inactive gene, FTN_0509, is annotated by rebase.neb.com as a fused type II restriction and modification gene, suggesting an unusual structure which also may have lost restriction capability. Hence, these two genes may both be inactive as restriction genes. It is possible, however, that one or both of them encode active restriction enzymes but that the enzyme recognition sites were not present on the plasmids used to assay restriction in this study.
Several new tools and techniques were developed in this work which should be of practical value to the Francisella research community. The full restriction-negative strain, MFN245, is modification-positive (Table 1). Therefore, the traditionally difficult task of transforming E. coli-derived plasmids into F. novicida can easily be surmounted by first transforming into MFN245 and then isolating the plasmid from MFN245 and transforming into the wild-type strain. MFN245 should even be suitable as a direct recipient of ligation reactions, facilitating the construction of plasmids which replicate exclusively in Francisella spp. Furthermore, the absence of the restriction barrier in MFN245 may facilitate targeted mutagenesis by transformation of linear PCR products using procedures analogous to those developed for other Francisella species (14, 19).
A simple alternative procedure to increase the efficiency of F. novicida transformation with plasmid DNA is to isolate the plasmid directly from dam-deficient E. coli strains. Although such plasmid preparations will not transform as efficiently as plasmid isolated from F. novicida MFN245, this approach should be adequate for many applications.
This report also presents a general technique for making multiple mutants in F. novicida. The technique utilizes chromosomal transformation to introduce specific transposon insertion alleles and transient expression of Flp recombinase to recycle the resistance marker, allowing the construction of multiple mutants through iteration of these steps (Fig. 1). Quintuple mutants were generated without difficulty in this work, and in principle many additional mutations could also be introduced. The technique can also be applied to intentionally produce deletions between chromosomal loci, allowing, for example, the deletion of nonessential chromosomal islands or operons. We generated one such deletion (of approximately 4.8 kb) in the course of this work (see Materials and Methods).
Our results help explain why the strong restriction barrier exhibited by F. novicida is not found in fully virulent Francisella tularensis subspecies. Analysis of the available sequenced genomes shows that most of the restriction genes present in these subspecies are pseudogenes (Table 2). The only intact annotated restriction genes are the following: in F. tularensis subsp. tularensis, a type III gene and (in strain WY96-3418 but not in Schu4 or FSC198) a type I gene; in F. tularensis subsp. holarctica, a type III gene (in strain OSU18 only; none in strains LVS or FTA); in F. tularensis subsp. mediasiatica, a type II gene and a type III gene. The high number of restriction genes present as pseudogenes in these genomes suggests that there has been significant loss of restriction capability in all three subspecies but not in F. novicida. Experimental results support this conclusion, as transformation of plasmid DNA isolated from E. coli is orders of magnitude more efficient in tested strains of F. tularensis subsp. holarctica and F. tularensis subsp. tularensis than F. novicida (20). Since F. novicida is proposed to most closely resemble the ancestral F. tularensis strain (29, 31), these findings suggest that restriction may have been more important in the ancestral strain than in the more virulent lineages.
TABLE 2.
Putative restriction genes in bacterial strains
| Strain | Annotated restriction genesa (no. of intact genes/no. of pseudogenes)
|
||||
|---|---|---|---|---|---|
| Type I | Type II | Type III | Type IV | Total | |
| F. tularensis subsp. novicida U112 | 3/0 | 2/0 | 1/0 | 0/0 | 6/0 |
| F. tularensis subsp. tularensis strains | |||||
| Schu4 | 0/2 | 0/1 | 1/1 | 0/0 | 1/4 |
| FSC198 | 0/2 | 0/1 | 1/1 | 0/0 | 1/4 |
| WY96-3418 | 1/1 | 1/0 | 0/2 | 0/0 | 2/3 |
| F. tularensis subsp. holarctica strains | |||||
| LVS | 0/3 | 0/1 | 0/0 | 0/0 | 0/4 |
| FTA | 0/2 | 0/1 | 0/0 | 0/0 | 0/3 |
| OSU18 | 0/2 | 0/1 | 1/0 | 0/0 | 1/3 |
| F. tularensis subsp. mediasiatica FSC147 | 0/2 | 1/0 | 1/0 | 0/0 | 2/2 |
| Yersinia pseudotuberculosis strains | |||||
| IP32953 | 2/0 | 0/0 | 0/0 | 2/0 | 4/0 |
| IP31758b | 1/0 | 0/0 | 0/0 | 0/0 | 1/0 |
| Yersinia pestis strains | |||||
| CO92 | 0/0 | 0/0 | 0/0 | 2/0 | 2/0 |
| KIM | 0/0 | 0/0 | 0/0 | 2/0 | 2/0 |
| Burkholderia thailandensis E264 | 1/0 | 4/0 | 1/0 | 0/0 | 6/0 |
| Burkholderia pseudomallei strains | |||||
| K96243 | 1/0 | 1/0 | 1/0 | 0/0 | 3/0 |
| 1710b | 0/0 | 0/0 | 2/0 | 0/0 | 2/0 |
| Burkholderia mallei strains | |||||
| NCTC10247 | 0/0 | 0/0 | 1/0 | 0/0 | 1/0 |
| ATCC 23344 | 0/0 | 0/0 | 1/0 | 0/0 | 1/0 |
Based on information in REBASE (rebase.neb.com).
IP31758 is significantly more pathogenic to humans than IP32953 (6).
Interestingly, we identified two other genera (Yersinia and Burkholderia) where a reduction in restriction functions seems to correlate with evolution toward virulence, suggesting a common evolutionary theme (Table 2). In Y. pestis, a serious human pathogen, there are fewer restriction genes than in the less pathogenic Y. pseudotuberculosis, from which Y. pestis recently diverged (34). An unusually virulent strain of Y. pseudotuberculosis, IP31758 (6), also carries fewer restriction genes than the less virulent type strain of the same species, IP32953. In Burkholderia, the human pathogenic species B. pseudomallei and B. mallei have significantly fewer restriction genes than the related nonpathogenic species B. thailandensis (10, 25). In the strains examined in both of these genera (Yersinia and Burkholderia), no restriction genes present as pseudogenes were identified (Table 2).
In the case of Francisella tularensis, since the virulent strains are characterized by a high number of pseudogenes across many functional categories (29), it may be that restriction was simply one of many capacities not strongly selected for in the relatively short evolutionary histories of the virulent subspecies. Alternatively, the maintenance of a strong restriction barrier in F. novicida (and presumably the ancestral F. tularensis strain) may indicate a greater exposure to incoming foreign DNA in this strain, whether due to environmental niche or genetic factors modulating such exposure. It is also possible, however, that the loss of restriction in some Francisella strains (or in strains of other bacterial species) may have enabled more rapid evolution toward virulence by facilitating the acquisition of virulence genes by horizontal transfer.
Supplementary Material
Acknowledgments
We thank Francis Nano and Mitchell Brittnacher for helpful discussions.
This work was supported by National Institutes of Health grants AI074609 and AI057141.
Footnotes
Published ahead of print on 3 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abd, H., T. Johansson, I. Golovliov, G. Sandstrom, and M. Forsman. 2003. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, L. S., M. Z. Gu, S. C. Cowley, W. W. Leung, and F. E. Nano. 1991. Transformation and allelic replacement in Francisella spp. J. Gen. Microbiol. 1372697-2703. [DOI] [PubMed] [Google Scholar]
- 3.Cantalupo, G., C. Bucci, P. Salvatore, C. Pagliarulo, V. Roberti, A. Lavitola, C. B. Bruni, and P. Alifano. 2001. Evolution and function of the neisserial dam-replacing gene. FEBS Lett. 495178-183. [DOI] [PubMed] [Google Scholar]
- 4.Chamberlain, R. E. 1965. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13232-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darling, R. G., C. L. Catlett, K. D. Huebner, and D. G. Jarrett. 2002. Threats in bioterrorism. I: CDC category A agents. Emerg. Med. Clin. North Am. 20273-309. [DOI] [PubMed] [Google Scholar]
- 6.Eppinger, M., M. J. Rosovitz, W. F. Fricke, D. A. Rasko, G. Kokorina, C. Fayolle, L. E. Lindler, E. Carniel, and J. Ravel. 2007. The complete genome sequence of Yersinia pseudotuberculosis IP31758, the causative agent of Far East scarlet-like fever. PLoS Genet. 3e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank, D. W., and T. C. Zahrt. 2007. Genetics and genetic manipulation in Francisella tularensis. Ann. N. Y. Acad. Sci. 110567-97. [DOI] [PubMed] [Google Scholar]
- 8.Gallagher, L. A., E. Ramage, M. A. Jacobs, R. Kaul, M. Brittnacher, and C. Manoil. 2007. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc. Natl. Acad. Sci. USA 1041009-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kieffer, T. L., S. Cowley, F. E. Nano, and K. L. Elkins. 2003. Francisella novicida LPS has greater immunobiological activity in mice than F. tularensis LPS, and contributes to F. novicida murine pathogenesis. Microbes Infect. 5397-403. [DOI] [PubMed] [Google Scholar]
- 10.Kim, H. S., M. A. Schell, Y. Yu, R. L. Ulrich, S. H. Sarria, W. C. Nierman, and D. DeShazer. 2005. Bacterial genome adaptation to niches: divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC Genomics 6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi, I., A. Nobusato, N. Kobayashi-Takahashi, and I. Uchiyama. 1999. Shaping the genome-restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev. 9649-656. [DOI] [PubMed] [Google Scholar]
- 12.Larson, C. L., W. Wicht, and W. L. Jellison. 1955. A new organism resembling P. tularensis isolated from water. Public Health Rep. 70253-258. [PMC free article] [PubMed] [Google Scholar]
- 13.Lauriano, C. M., J. R. Barker, S. S. Yoon, F. E. Nano, B. P. Arulanandam, D. J. Hassett, and K. E. Klose. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. USA 1014246-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, J., X. Zogaj, J. R. Barker, and K. E. Klose. 2007. Construction of targeted insertion mutations in Francisella tularensis subsp. novicida. BioTechniques 43487-490, 492. [DOI] [PubMed] [Google Scholar]
- 15.Lobner-Olesen, A., O. Skovgaard, and M. G. Marinus. 2005. Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 8154-160. [DOI] [PubMed] [Google Scholar]
- 16.LoVullo, E. D., L. A. Sherrill, L. L. Perez, and M. S. Pavelka, Jr. 2006. Genetic tools for highly pathogenic Francisella tularensis subsp. tularensis. Microbiology 1523425-3435. [DOI] [PubMed] [Google Scholar]
- 17.Low, D. A., N. J. Weyand, and M. J. Mahan. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 697197-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludu, J. S., O. M. de Bruin, B. N. Duplantis, C. L. Schmerk, A. Y. Chou, K. L. Elkins, and F. E. Nano. 2008. The Francisella pathogenicity island protein PdpD is required for full virulence and associates with homologues of the type VI secretion system. J. Bacteriol. 1904584-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludu, J. S., E. B. Nix, B. N. Duplantis, O. M. de Bruin, L. A. Gallagher, L. M. Hawley, and F. E. Nano. 2008. Genetic elements for selection, deletion mutagenesis and complementation in Francisella spp. FEMS Microbiol. Lett. 27886-93. [DOI] [PubMed] [Google Scholar]
- 20.Maier, T. M., A. Havig, M. Casey, F. E. Nano, D. W. Frank, and T. C. Zahrt. 2004. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl. Environ. Microbiol. 707511-7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manoil, C., and J. Bailey. 1997. A simple screen for permissive sites in proteins: analysis of Escherichia coli lac permease. J. Mol. Biol. 267250-263. [DOI] [PubMed] [Google Scholar]
- 22.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 828129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLendon, M. K., M. A. Apicella, and L. A. Allen. 2006. Francisella tularensis: taxonomy, genetics, and immunopathogenesis of a potential agent of biowarfare. Annu. Rev. Microbiol. 60167-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray, N. E. 2002. 2001 Fred Griffith review lecture. Immigration control of DNA in bacteria: self versus non-self. Microbiology 1483-20. [DOI] [PubMed] [Google Scholar]
- 25.Ong, C., C. H. Ooi, D. Wang, H. Chong, K. C. Ng, F. Rodrigues, M. A. Lee, and P. Tan. 2004. Patterns of large-scale genomic variation in virulent and avirulent Burkholderia species. Genome Res. 142295-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owen, C. R., E. O. Buker, W. L. Jellison, D. B. Lackman, and J. F. Bell. 1964. Comparative studies of Francisella tularensis and Francisella novicida. J. Bacteriol. 87676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts, R. J., M. Belfort, T. Bestor, A. S. Bhagwat, T. A. Bickle, J. Bitinaite, R. M. Blumenthal, S. Degtyarev, D. T. Dryden, K. Dybvig, K. Firman, E. S. Gromova, R. I. Gumport, S. E. Halford, S. Hattman, J. Heitman, D. P. Hornby, A. Janulaitis, A. Jeltsch, J. Josephsen, A. Kiss, T. R. Klaenhammer, I. Kobayashi, H. Kong, D. H. Kruger, S. Lacks, M. G. Marinus, M. Miyahara, R. D. Morgan, N. E. Murray, V. Nagaraja, A. Piekarowicz, A. Pingoud, E. Raleigh, D. N. Rao, N. Reich, V. E. Repin, E. U. Selker, P. C. Shaw, D. C. Stein, B. L. Stoddard, W. Szybalski, T. A. Trautner, J. L. Van Etten, J. M. Vitor, G. G. Wilson, and S. Y. Xu. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 311805-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts, R. J., T. Vincze, J. Posfai, and D. Macelis. 2007. REBASE—enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 35D269-D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohmer, L., C. Fong, S. Abmayr, M. Wasnick, T. J. Larson Freeman, M. Radey, T. Guina, K. Svensson, H. S. Hayden, M. Jacobs, L. A. Gallagher, C. Manoil, R. K. Ernst, B. Drees, D. Buckley, E. Haugen, D. Bovee, Y. Zhou, J. Chang, R. Levy, R. Lim, W. Gillett, D. Guenthener, A. Kang, S. A. Shaffer, G. Taylor, J. Chen, B. Gallis, D. A. D'Argenio, M. Forsman, M. V. Olson, D. R. Goodlett, R. Kaul, S. I. Miller, and M. J. Brittnacher. 2007. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 8R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Svensson, K., P. Larsson, D. Johansson, M. Bystrom, M. Forsman, and A. Johansson. 2005. Evolution of subspecies of Francisella tularensis. J. Bacteriol. 1873903-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyeryar, F. J., Jr., and W. D. Lawton. 1969. Transformation of Pasteurella novicida. J. Bacteriol. 1001112-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyeryar, F. J., and W. D. Lawton. 1970. Factors affecting transformation of Pasteurella novicida. J. Bacteriol. 1041312-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterfield, N. R., B. W. Wren, and R. H. Ffrench-Constant. 2004. Invertebrates as a source of emerging human pathogens. Nat. Rev. Microbiol. 2833-841. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, G. G., and N. E. Murray. 1991. Restriction and modification systems. Annu. Rev. Genet. 25585-627. [DOI] [PubMed] [Google Scholar]
- 36.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 173469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.