Abstract
Carotenoids are complex lipids that are known for acting against photodynamic injury and free radicals. We demonstrate here that σF is required for carotenoid pigment production in Mycobacterium smegmatis. We further show that a sigF mutant exhibits a transformation efficiency 104-fold higher than that of the parental strain, suggesting that σF regulates the production of components affecting cell wall permeability. In addition, a sigF mutant showed an increased sensitivity to hydrogen peroxide. An in silico search of the M. smegmatis genome identified a number of SigF consensus sites, including sites upstream of the carotenoid synthesis locus, which explains its SigF regulation.
Although Mycobacterium smegmatis was originally isolated from humans, this fast-growing mycobacterium species is mostly nonpathogenic and has been used as a model to investigate mycobacterial physiology (1). When grown on agar plates, the wild-type strain ATCC 607 is characterized by a whitish morphotype, which turns to yellow-orange upon several days of exposure to visible light. Many mycobacteria produce yellow, orange or, less frequently, salmon pink pigments either in the dark (scotochromogens) or upon exposure to light (photochromogens). These pigments have been characterized as carotenoids, a class of polyterpene lipids that occur widely throughout nature and whose functions are to act as free radical scavengers and to protect the cells against photodynamic injuries (6, 13).
Transposon insertion into sigF determines loss of pigmentation.
We constructed an M. smegmatis ATCC 607 insertional library using Tn611 (10, 24). The mutants were visually screened for a pigmentation defect. Only 1 out of 15,000 library clones displayed a phenotype readily differing from that of the wild-type strain. Indeed, while the ATCC 607 wild-type strain formed yellow-orange colonies, the colony of the mutant strain had a whitish color (Fig. 1A). This mutant was characterized by using ligation-mediated PCR (21), which revealed an insertion in the open reading frame encoding the alternative sigma factor σF, which is homologous to stress response and sporulation sigma factors in many bacteria (22). The σF protein sequences of M. smegmatis and Mycobacterium tuberculosis are highly similar (29), and in M. tuberculosis, σF is a general stress factor that appears to play a regulatory role in the structure and function of the mycobacterial cell wall (30).
FIG. 1.
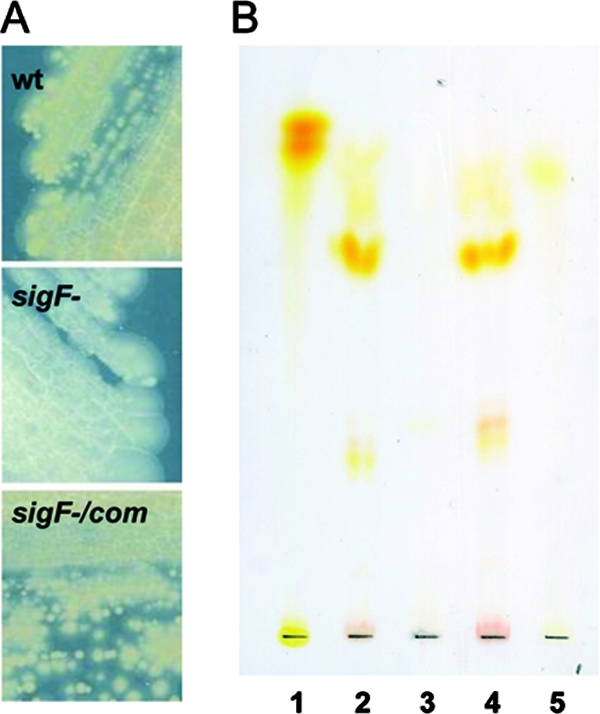
A) Pigment production by the wild-type strain (wt), the sigF mutant, and the complemented strain. B) TLC of lipid extracts from the wild-type, the sigF mutant, and the complemented strains. TLC was run using CHCl3-methanol (90:10), and no revelation has been performed. One mg of total lipid was loaded. Lanes: 1, β-carotene; 2, M. smegmatis ATCC 607 wild type; 3, M. smegmatis ATCC 607 sigF mutant; 4, M. smegmatis ATCC 607 sigF mutant (pNIP40B/sigF); 5, lycopene. The contrast of this image was improved by using processing software.
To prove that the loss of chromogenicity was specifically caused by the inactivation of sigF, we carried out complementation studies by expressing a wild-type copy of sigF at an ectopic locus within the mutant. In M. smegmatis, sigF seems to be part of the same transcription unit with rsbW, the gene encoding its putative anti-sigma factor, as in M. tuberculosis (3), since the sigF GTG start codon overlaps with the TGA stop codon of rsbW. Therefore, a fragment of 753 bp, containing the putative ribosome binding site upstream of the rsbW stop codon and the entire coding sequence of sigF, was amplified by PCR and cloned into the integrative expression vector pNIP40b between the XbaI and SpeI (indicated by lowercase letters) sites by using the primers RP283 (tctagaAAATTCCAGGAGGTCAGGTGACGTCGGAATACGCAGACGTTC) and RP271 (actagtGGCATTCCGAAGCGAGTTCC) (8, 28). The former primer was designed to contain an ribosome binding site (underlined). From previous experience, we know that cloning a gene at this site in pNIP40b leads to a transcriptional fusion with an upstream promoter and leads to expression of the transgene (7, 8, 28). The recombinant plasmid, pNIP40b/sigF, was introduced into the sigF mutant by electroporation as previously described (20). After the pulse, the cells were diluted in 0.5 ml of liquid medium and incubated for 14 h at 37°C. The resulting recombinant strains were isolated on Middlebrook 7H10 solid medium containing 0.05% Tween 80 in the presence of both kanamycin (25 μg/ml) and hygromycin (50 μg/ml). Quantitative reverse transcription-PCR (RT-PCR) experiments performed with a sigF-specific primer (RP799, GTCGATGGACAGCGTGTTGT) to start reverse transcription demonstrated that the level of sigF expression in the complemented strain was similar to that exhibited by the wild-type parental strain (data not shown). Primers were designed to be able to detect transcripts containing the entire sigF gene and not those containing a truncated form present in mutant and complemented strains. For PCR amplification, primers RP481 (CTGGTCGGTGAAGGTGCC) and RP482 (GTACGAACTGCCCGCGAT) were used (the underlined base indicates the transposon insertion site in the sigF mutant). The recombinant strains were also examined for complementation of the nonpigmented phenotype. As shown in Fig. 1A, the introduction of sigF into the mutant fully restored pigment production. We also carried out complementation using the sigF gene from M. tuberculosis. Although less efficient, the sigF gene of M. tuberculosis was able to complement the pigment deficiency of an M. smegmatis sigF mutant, demonstrating that these genes are true orthologs (data not shown), as already suggested by in silico studies (29).
σF controls the expression of a gene cluster involved in carotenoid biosynthesis.
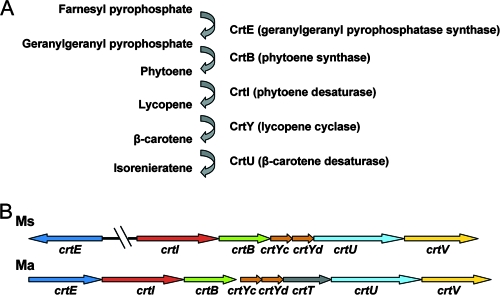
Thin-layer chromatography (TLC) analysis of the carotenoid fraction extracted from both wild-type and sigF mutant cells grown on 7H11 medium revealed four pale yellow pigment spots with different Rf values in the wild-type extract (lane 2) that were all missing in the extract of the mutant (lane 3) (Fig. 1B). These four yellow pigments were all present in the sigF-complemented strain (lane 4) (Fig. 1B). The more-apolar pigment, with an Rf value of 0.45, similar to that of the lycopene used as a control (lane 5), was purified by preparative TLC. It displayed a UV absorption spectrum similar to that of mycobacterial isorenieratene (formerly designated leprotene), with a λmax at 432, 457, and 483 nm (data not shown) (22). The other spots (including the major one) likely correspond to intermediary metabolites of the carotene family. The biosynthesis of the aromatic carotene isorenieratene is restricted to green photosynthetic bacteria and a few actinomycetes. Isorenieratene appears to be a characteristic pigment of almost all orange-pigmented mycobacteria (4), including M. phlei (12), M. aurum (18), M. avium, and M. intracellulare (26). A number of biochemical studies have documented the pathway leading to isorenieratene production (2, 16, 27). This metabolic pathway uses farnesyl pyrophosphate as a precursor, which leads to isorenieratene in five metabolic steps involving, subsequently, CrtE, CrtB, CrtI, CrtY, and CrtU as illustrated in Fig. 2A.
FIG. 2.
A) Metabolic pathway leading to the production of isorenieratene in mycobacteria. Geranylgeranyl pyrophosphate (GGPP) is synthesized by CrtE (GGPP synthase) from farnesyl pyrophosphate. GGPP is converted to phytoene by CrtB (phytoene synthase). Phytoene is desaturated to lycopene by CrtI and transformed into β-carotene by CrtY (lycopene cyclase), an enzyme formed from the crtYc and crtYd gene products. The β-carotene is finally desaturated into isorenieratene by CrtU. B) Comparative genomic analysis of the carotenoid locus in M. smegmatis (Ms) and M. aurum (Ma).
In M. aurum the biosynthesis of isorenieratene is directed by a carotenogenic gene cluster containing eight open reading frames, each transcribed in the same direction (27). Exploring the M. smegmatis genome, we found a gene cluster containing the orthologs of six out of the eight open reading frames involved in the biosynthesis of isorenieratene in M. aurum (crtIBYcYdUV). The only gene missing in M. smegmatis was crtT, encoding a methyltransferase shown to be dispensable for isorenieratene production in Streptomyces griseus (16). Surprisingly, the crtE gene, encoding the GGPP synthase, was not part of the locus and was found more than 1.8 Mb away (Fig. 2B).
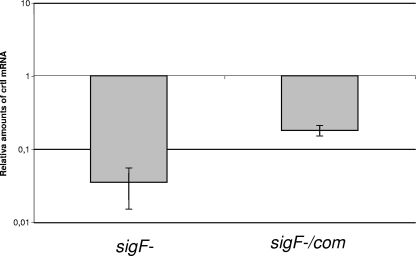
To check whether σF is required for regulation of expression of the crt genes, we measured the mRNA levels of crtI and crtE in the sigF mutant, the wild-type parental strain, and the complemented strain by quantitative real-time RT-PCR, using sigA mRNA as an internal invariant control (19). RNA was extracted from cells grown to mid-exponential phase at 37°C with shaking in Middlebrook 7H9 with 0.05% Tween 80. While crtE mRNA levels were the same in the three strains (data not shown), we found that the crtI mRNA level was reduced about 29-fold in the sigF mutant compared to the wild-type strain. This reduction was almost completely restored (80%) in the complemented strain (Fig. 3), suggesting that transcription of the crtIBYcYdUV cluster in M. smegmatis is under the transcriptional control of σF. In this regard, an lsr2 mutant has been recently reported to be nonpigmented in M. smegmatis (15); a transcriptomic analysis has shown that the sigF gene is expressed at a low level in this mutant (5) and likely explains this lack of pigmentation.
FIG. 3.
Quantitative RT-PCR of the crtI gene in the sigF mutant and the complemented strain, showing relative amounts of mRNA with respect to the wild type.
Interestingly, carotenoid biosynthesis in S. griseus and Streptomyces setonii is also regulated by a sigma factor (CrtS) that belongs to the same family of stress response sigma factors as mycobacterial σF and Bacillus subtilis σB (14, 17), reinforcing the hypothesis that in M. smegmatis the ctr genes are under σF control.
Since the M. tuberculosis SigF consensus sequence is known (3), we searched the crtI upstream region for similar sequences. Interestingly, we found a perfect match at position bp −104 that suggested that the σF effect on crt gene transcription is direct and not mediated by another effector. To further support this hypothesis, we mapped the crtI transcription start site by 5′-rapid amplification of cDNA ends (Roche) and were able to show that it is indeed localized 9 bp downstream of the −10 region of the putative SigF-dependent promoter consensus sequence (Fig. 4).
FIG. 4.
crtI upstream region. The −10 and −35 regions of the putative SigF-dependent promoter are underlined. The transcription start site and the translation start codon are shown in bold.
Mutation of σF induces a dramatic susceptibility to hydrogen peroxide.
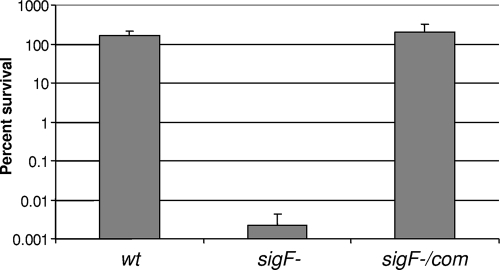
As σF regulates the transcription of the crtIBYcYdUV gene cluster, which ultimately leads to a loss of pigmentation, and carotenoids are notable in their biology as free radical scavengers, we hypothesized that the sigF mutant might be hypersensitive to hydrogen peroxide. To this end we incubated the wild-type, sigF mutant, and complemented strains in 5 mM H2O2 (0.017%) and monitored cell death by CFU counting. After 2 h of incubation, the sigF mutant inoculum decreased more than 104 times, whereas the wild-type and the sigF-complemented strains remained unaffected by this free radical treatment (Fig. 5). This experiment clearly demonstrates that SigF, as in other mycobacterial species, regulates pathways that are important for stress survival and particularly toward oxidative stress. Interestingly, a recent study independently reported this finding in the mc2155 strain and extended it to hypersensitivity to heat shock and acidic pH stresses (11). Whether these phenotypes are mediated in some way by carotenoid pigments remains to be tested. The pigment defect and hypertransformability were presumably not detected by Gebhard and colleagues, as the mc2155 strain is less pigmented than the parental ATCC 607 strain (unpublished observation) and already hypertransformable (15).
FIG. 5.
Quantitative assessment of the bacterial survival of the wild-type, sigF mutant, and sigF-complemented strains following exposure to hydrogen peroxide.
The sigF mutant exhibits higher transformation efficiency than the parental strain.
We noticed that the mutant appeared to be transformed with a higher efficiency than the parental strain. To check whether this difference was related to sigF, we characterized the wild-type, the sigF mutant, and the complemented strains for their abilities to be transformed by plasmid DNA. The replicative vector pLYGZeo (9) was electroporated as described above, and transformants were selected with 100 μg/ml of zeocin (Invitrogen). The transformation efficiency of the wild-type strain was ≤10 transformants/μg of DNA, whereas that of the sigF mutant was (9.5 ± 1.2) × 104 transformants/μg (mean ± standard error). This efficiency was reduced more than 100 times when a functional copy of the sigF gene was reintroduced into the mutant ([1.3 ± 0.1] × 102 transformants/μg). These results strongly suggest the presence of genes in the SigF regulon that inhibit the uptake of DNA during electroporation. Interestingly, the common M. smegmatis reference strain mc2155 is a mutant selected for its electroporation efficiency, which is 104 to 105 times greater than that of its parent strain (23). Whether or not this phenotype in mc2155 is due to a defect in sigF or in one of its downstream genes awaits clarification.
In silico identification of the SigF targets in M. smegmatis.
As the M. tuberculosis sigF complemented the M. smegmatis mutant and a perfect match with the sigF consensus sequence was found upstream of crtI, we probed the M. smegmatis genome for this sequence, using the string search tool Search Pattern (25). The word pattern search was GTTTCX14-18GGGTAT (allowing two mismatches per hexamer) and limited to intergenic regions located between nucleotides −10 and −200 upstream of coding regions. A number of genes were shown to contain a putative sigF consensus in their upstream untranslated region, suggesting that they may be part of the sigF regulon. These genes belong to several functional classes, such as cell wall processes, intermediary metabolism, regulation, or unknown function. Interestingly, the orthologs of some genes that have been shown to be regulated by σF in M. tuberculosis (30) were also potentially sigF regulated in M. smegmatis (see Table S1 in the supplemental material). However, few genes appear to be sigF regulated in both M. smegmatis and M. tuberculosis, suggesting that the σF regulons have highly diverged between the two species.
Conclusions.
Based on our data, we have shown that in M. smegmatis σF is involved in the regulation of genes required for carotenoid biosynthesis and probably other cell wall components. We also showed that the presence of a functional sigF gene decreases the M. smegmatis transformation efficiency, suggesting the involvement of some of the σF-regulated genes in the determination of cell wall permeability. The sigF gene is also involved in the resistance toward hydrogen peroxide. Finally, in silico analysis provided a list of potential σF targets in this species.
Supplementary Material
Acknowledgments
We acknowledge INSERM for funding this project under the Avenir program to J.M.R., Chargé de Recherches at INSERM and MIUR-COFIN 2006 grant no. 2006064583 (awarded to R.M.). D.K. is supported by the Chancelleries des Universités de Paris.
We thank referee number 2 for the hydrogen peroxide hypothesis. We acknowledge F. Laval (IPBS, Toulouse, France) for valuable advice on mass spectrometry, E. Rampazzo (University of Padova) for technical support and valuable discussion, and G. Palù (University of Padova) for continuous support.
Footnotes
Published ahead of print on 19 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alvarez, E., and E. Tavel. 1885. Recherches sur le bacille de Lustgarden. Arch. Physiol. Norm. Pathol. 6303-321. [Google Scholar]
- 2.Armstrong, G. A. 1997. Genetics of eubacterial carotenoid biosynthesis: a colorful tale. Annu. Rev. Microbiol. 51629-659. [DOI] [PubMed] [Google Scholar]
- 3.Beaucher, J., S. Rodrigue, P. E. Jacques, I. Smith, R. Brzezinski, and L. Gaudreau. 2002. Novel Mycobacterium tuberculosis anti-sigma factor antagonists control sigma F activity by distinct mechanisms. Mol. Microbiol. 451527-1540. [DOI] [PubMed] [Google Scholar]
- 4.Britton, G., T. W. Goodwin, D. J. Brown, and N. J. Patel. 1980. Carotenoid biosynthesis by cultures and cell-free preparations of Flavobacterium R1560. Methods Enzymol. 67264-270. [DOI] [PubMed] [Google Scholar]
- 5.Colangeli, R., D. Helb, C. Vilcheze, M. H. Hazbon, C. G. Lee, H. Safi, B. Sayers, I. Sardone, M. B. Jones, R. D. Fleischmann, S. N. Peterson, W. R. Jacobs, and D. Alland. 2007. Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis. PLoS Pathog. 3e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David, H. L. 1974. Carotenoid pigments of Mycobacterium kansasii. Appl. Microbiol. 28696-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Mendonca-Lima, L., M. Picardeau, C. Raynaud, J. Rauzier, Y. O. de la Salmoniere, L. Barker, F. Bigi, A. Cataldi, B. Gicquel, and J. M. Reyrat. 2001. Erp, an extracellular protein family specific to mycobacteria. Microbiology 1472315-2320. [DOI] [PubMed] [Google Scholar]
- 8.Deshayes, C., F. Laval, H. Montrozier, M. Daffe, G. Etienne, and J. M. Reyrat. 2005. A glycosyltransferase involved in biosynthesis of triglycosylated glycopeptidolipids in Mycobacterium smegmatis: impact on surface properties. J. Bacteriol. 1877283-7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao, L. Y., R. Groger, J. S. Cox, S. M. Beverley, E. H. Lawson, and E. J. Brown. 2003. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect. Immun. 71922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia, M. J., C. Guilhot, R. Lathigra, M. C. Menendez, P. Domenech, C. Moreno, B. Gicquel, and C. Martin. 1994. Insertion sequence IS1137, a new IS3 family element from Mycobacterium smegmatis. Microbiology 1402821-2828. [DOI] [PubMed] [Google Scholar]
- 11.Gebhard, S., A. Hümpel, A. D. McLellan, and G. M. Cook. 2008. The alternative sigma factor SigF of Mycobacterium smegmatis is required for survival of heat shock, acidic pH and oxidative stress. Microbiology 1542786-2795. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin, T. W., and M. Jamikorn. 1956. Studies in carotenogenesis. 17. The carotenoids produced by different strains of Mycobacterium phlei. Biochem. J. 62269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grange, J. 1996. Mycobacteria and human disease, 2nd ed., p. 11-40. Oxford University Press, New York, NY.
- 14.Kato, F., T. Hino, A. Nakaji, M. Tanaka, and Y. Koyama. 1995. Carotenoid synthesis in Streptomyces setonii ISP5395 is induced by the gene crtS, whose product is similar to a sigma factor. Mol. Gen. Genet. 247387-390. [DOI] [PubMed] [Google Scholar]
- 15.Kocincova, D., A. K. Singh, J. L. Beretti, H. Ren, D. Euphrasie, J. Liu, M. Daffe, G. Etienne, and J. M. Reyrat. 2008. Spontaneous transposition of IS1096 or ISMsm3 leads to glycopeptidolipid overproduction and affects surface properties in Mycobacterium smegmatis. Tuberculosis (Edinburgh) 88390-398. [DOI] [PubMed] [Google Scholar]
- 16.Krugel, H., P. Krubasik, K. Weber, H. P. Saluz, and G. Sandmann. 1999. Functional analysis of genes from Streptomyces griseus involved in the synthesis of isorenieratene, a carotenoid with aromatic end groups, revealed a novel type of carotenoid desaturase. Biochim. Biophys. Acta 143957-64. [DOI] [PubMed] [Google Scholar]
- 17.Lee, H. S., Y. Ohnishi, and S. Horinouchi. 2001. A sigma B-like factor responsible for carotenoid biosynthesis in Streptomyces griseus. J. Mol. Microbiol. Biotechnol. 395-101. [PubMed] [Google Scholar]
- 18.Levy-Frebault, V., and H. L. David. 1979. Mutations affecting pigment synthesis in Mycobacterium aurum. J. Gen. Microbiol. 115317-323. [DOI] [PubMed] [Google Scholar]
- 19.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31715-724. [DOI] [PubMed] [Google Scholar]
- 20.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor sigma E: role in global gene expression and survival in macrophages. Mol. Microbiol. 41423-437. [DOI] [PubMed] [Google Scholar]
- 21.Prod'hom, G., B. Lagier, V. Pelicic, A. J. Hance, B. Gicquel, and C. Guilhot. 1998. A reliable amplification technique for the characterization of genomic DNA sequences flanking insertion sequences. FEMS Microbiol. Lett. 15875-81. [DOI] [PubMed] [Google Scholar]
- 22.Rodrigue, S., R. Provvedi, P. E. Jacques, L. Gaudreau, and R. Manganelli. 2006. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 30926-941. [DOI] [PubMed] [Google Scholar]
- 23.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 41911-1919. [DOI] [PubMed] [Google Scholar]
- 24.Sonden, B., D. Kocincova, C. Deshayes, D. Euphrasie, L. Rhayat, F. Laval, C. Frehel, M. Daffe, G. Etienne, and J. M. Reyrat. 2005. Gap, a mycobacterial specific integral membrane protein, is required for glycolipid transport to the cell surface. Mol. Microbiol. 58426-440. [DOI] [PubMed] [Google Scholar]
- 25.Staden, R. 1988. Methods to define and locate patterns of motifs in sequences. Comput. Appl. Biosci. 453-60. [DOI] [PubMed] [Google Scholar]
- 26.Tarnok, I., and Z. Tarnok. 1970. Carotene and xanthophylls in mycobacteria. I. Technical procedures; thin-layer chromatographic patterns of mycobacterial pigments. Tubercle 51305-312. [DOI] [PubMed] [Google Scholar]
- 27.Viveiros, M., P. Krubasik, G. Sandmann, and M. Houssaini-Iraqui. 2000. Structural and functional analysis of the gene cluster encoding carotenoid biosynthesis in Mycobacterium aurum A+. FEMS Microbiol. Lett. 18795-101. [DOI] [PubMed] [Google Scholar]
- 28.Vultos, T. D., I. Mederle, V. Abadie, M. Pimentel, J. Moniz-Pereira, B. Gicquel, J. M. Reyrat, and N. Winter. 2006. Modification of the mycobacteriophage Ms6 attP core allows the integration of multiple vectors into different tRNAala T-loops in slow- and fast-growing mycobacteria. BMC Mol. Biol. 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waagmeester, A., J. Thompson, and J. M. Reyrat. 2005. Identifying sigma factors in Mycobacterium smegmatis by comparative genomic analysis. Trends Microbiol. 13505-509. [DOI] [PubMed] [Google Scholar]
- 30.Williams, E. P., J. H. Lee, W. R. Bishai, C. Colantuoni, and P. C. Karakousis. 2007. Mycobacterium tuberculosis SigF regulates genes encoding cell wall-associated proteins and directly regulates the transcriptional regulatory gene phoY1. J. Bacteriol. 1894234-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.