Abstract
A two-component regulatory system of Lactobacillus plantarum, encoded by genes designated lamK and lamR (hpk10 and rrp10), was studied. The lamK and lamR genes encode proteins which are highly homologous to the quorum-sensing histidine kinase LamC and the response regulator LamA, respectively. Transcription analysis of the lamKR operon and the lamBDCA operon and liquid chromatography-mass spectrometry analysis of production of the LamD558 autoinducing peptide were performed for ΔlamA, ΔlamR, ΔlamA ΔlamR deletion mutants and a wild-type strain. The results suggested that lamA and lamR are cooperating genes. In addition, typical phenotypes of the ΔlamA mutant, such as reduced adherence to glass surfaces and filamentous cell morphology, were enhanced in the ΔlamA ΔlamR mutant. Microarray analysis suggested that the same cell wall polysaccharide synthesis genes, stress response-related genes, and cell wall protein-encoding genes were affected in the ΔlamA and ΔlamA ΔlamR mutants. However, the regulation ratio was more significant for the ΔlamA ΔlamR mutant, indicating the cooperative effect of LamA and LamR.
Quorum-sensing systems are known to affect many intriguing bacterial phenotypes, such as virulence, competence, antibiotic production, and biofilm formation (10). In gram-positive bacteria quorum-sensing systems are mediated mainly by two-component systems (2CSs) consisting of a membrane-bound histidine protein kinase (HPK) and a response regulator (RR), which are regulated by autoinducing peptides (AIPs). Genes encoding these 2CSs are generally arranged in an operon consisting of the HPK-, RR-, and AIP-encoding genes (21). In a quorum-sensing 2CS, the HPK normally belongs to the subfamily designated HPK10 (11), and the RR belongs to the LytTR family (25). The 2CS transmits the AIP signal through the cell membrane and eventually triggers expression of the operon encoding the 2CS and AIP itself (autoinduction), as well as different target genes involved in, for example, virulence, competence, bacteriocin production, and biofilm formation.
The accessory gene regulator system (agr system) is one of the best-studied quorum-sensing systems in gram-positive bacteria. In pathogenic staphylococci, the agr system has a critical role in biofilm formation and probably in the invasion of host cells (5, 31), and more than 100 genes are under the control of this system (7, 26, 43-45). The agr gene clusters are conserved throughout the genus Staphylococcus and are divided into more than four groups, which are defined by the mutual inhibition by the peptides of the agr response in heterologous pairs. Recent genome sequencing projects have revealed that the genomes of several pathogenic bacteria, such as Listeria monocytogenes (3) and Enterococcus faecalis (12, 24), contain agr homologues and that these genes are important for virulence. In addition, systems homologous to the Agr system are present in some nonpathogenic commensal organsims, such as Lactobacillus plantarum (19), Bacillus subtilis (8), and Roseburia inulinivorans (30). However, the role of systems homologous to the agr system in nonpathogenic bacteria has not been clarified yet.
L. plantarum is a found in fermented food products, on plant material (6), and as a natural inhabitant of the human gastrointestinal tract (2). We previously studied the function and mechanism of the agr-like operon (lam) in Lactobacillus using molecular analysis based on the genome sequence of L. plantarum WCFS1, a single-colony isolate obtained from L. plantarum NCIMB8826, which was originally isolated from human saliva (National Collection of Industrial and Marine Bacteria, Aberdeen, United Kingdom) (13, 38). The genome sequence revealed that the lamBDCA operon (lp_3582 to lp_3580) contains an AIP-encoding gene (lamD), an HPK-encoding gene (lamC), an RR-encoding gene (lamA), and an AIP export/modification protein-encoding gene (lamB). A peptide was purified from the cultured media of a LamBD-overexpressing strain, and the most abundant peptide, which was designated LamD558, was found to be a cyclopentapeptide thiolactone (19, 37).
Global gene expression analysis using a microarray of a lamA knockout mutant (ΔlamA) showed that the cps2 operon (lp_1197 to lp_1211), which is one of the exopolysaccharide-related operons, was upregulated more than 10-fold in this mutant. Phenotypic analysis suggested that the ΔlamA cells showed reduced adherence to glass surfaces. These results suggested that the lamBDCA operon was related to the cell surface functionality of L. plantarum.
Interestingly, the ΔlamA mutant was able to produce 50% of the LamD558 peptide produced by the wild type and 30% of the transcripts of the lamBDCA operon produced by the wild type, suggesting that there was cross-regulation with other genes (4). The genome information revealed that 13 putative 2CSs were present in this strain, and 5 of them were predicted to be involved in quorum sensing, as deduced from their sequences, as previously reported (19, 22, 35, 36). In addition, the genes encoding one of these 2CSs, lp_3088 and lp_3087 (hpk10 and rrp10), are highly homologous to lamC and lamA, respectively. We speculated that this operon is also related to the lam system and designated it lamKR. In this study, we constructed a lamR knockout mutant (ΔlamR mutant) and a lamA lamR double-knockout mutant (ΔlamA ΔlamR mutant). The global gene expression profiles and phenotypes of these Δlam mutants were studied, and the results suggested that the lamKR operon encodes a novel quorum-sensing 2CS that controls the expression of cell surface-related genes of L. plantarum in concert with the lamBDCA operon.
MATERIALS AND METHODS
Strains, plasmids, and media.
The bacterial strains and plasmids used in this study are shown in Table 1. Escherichia coli JM109 was cultivated aerobically at 37°C in Luria-Bertani broth or in brain heart infusion broth when there was selection for erythromycin resistance (Difco). L. plantarum WCFS1 and its derivatives were cultivated in Mann-Rogosa-Shape broth (MRS) (Difco) at 30 or 37°C without agitation. Solid media contained 1.5% (wt/vol) agar. Where appropriate, erythromycin was added as follows: 150 μg ml−1 for E. coli or 5 μg ml−1 for L. plantarum WCFS1.
TABLE 1.
Bacterial strains and plasmids used in this study and relevant characteristics
| Bacterial strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| Escherichia coli JM109 | Cloning host for pUC19ery and its derivatives | Promega |
| Lactobacillus plantarum WCFS1 | Wild type, single-colony isolate from human saliva isolate NCIMB8826 | Haywardb |
| Lactobacillus plantarum MS ΔlamA | Double-crossover mutant of L. plantarum WCFS1 with lamA replaced by lamA::tag gene replacement cassette | Sturmec |
| Lactobacillus plantarum TFΔlamR | Double-crossover mutant of L. plantarum WCFS1 with lamR replaced by ΔlamR::tag gene replacement cassette | This study |
| Lactobacillus plantarum TFΔlamA/ΔlamR | Double-crossover mutant of L. plantarum WCFS1 with lamA and lamR replaced by lamA::tag and ΔlamR gene replacement cassettes, respectively | This study |
| pUC19ery | Ampr Emr, 3.8-kb derivative of pUC19 containing 1.1-kb HinPI fragment of pIL253 carrying the Eryr gene | van Kranenburgd |
| pDel-LamR-WCFS7 | pUC19ery vector carrying knockout ΔlamR gene replacement cassette | This study |
DNA isolation and construction of integration plasmids.
Genomic DNA was isolated from 5 ml of an L. plantarum WCFS1 culture as previously described and was used as a template for PCR. PCR was performed using proofreading Platinum Pfx DNA polymerase (Invitrogen).
Primers lamR-XbaI and lamR-SmaI1 and primers lamR-SmaI2 and lamR-BamHI were used to amplify the 5′ and 3′ ends of lamR (Table 2) and the regions flanking lamR (approximately 1.2 kb on each side). PCR products were cloned into the nonreplicating integration vector pUC19ery (41) after digestion of the PCR products and vector with the appropriate restriction enzymes (Gibco BRL). Plasmids were transformed into competent cells of E. coli JM109 by a calcium procedure as recommended by the manufacturer (Promega). This resulted in plasmid pDel-LamR-WCFS7 containing the complete gene replacement cassette with the mutated RR gene.
TABLE 2.
Primers used for construction of deletion plasmids and Q-PCR analysis
| Primer | Sequence (5′-3′) | Remarks |
|---|---|---|
| lamR-XbaI | TCTAGAATTGGGACTGGTGGTGCTGTTTAC | Deletion plasmid construction |
| lamR-SmaI1 | CCCGGGAGACGGCTTGAGTTGGTTGCTGAC | Deletion plasmid construction |
| lamR-SmaI2 | CCCGGGATTTAGATGATGGTGAACTTGA | Deletion plasmid construction |
| lamR-BamHI | GAGCTCAAATAATCTTGTAATGCCTCTTG | Deletion plasmid construction |
| lamR-F | CAAGTGCTTAATGGGGTAA | Deletion plasmid construction |
| lp_930-F | CACGGGTGGCGGATAAGGTT | Alkaline shock protein |
| lp_930-R | TGTCATTTTGTTGCCGCCACTC | |
| lp_1197-2F | GACAAGCACGACCAACTCACA | Cps2A |
| lp_1197-2R | TAAGCCGACAACTAGCCCAATCAA | |
| lp_1204-F | ACGTCATCCATTTCGCTTTTT | Cps2H |
| lp_1204-R | GTCTCATTCCACGCATCTCTGT | |
| lp_3087-F | TATTAGCGCATGTCAAAAGTCAGC | LamR |
| lp_3087-R | CTCCGTATGCGTCGTCACAAAGAT | |
| lp_3088-F | TGAGCGCCATTTTTGTTACG | LamK |
| lp_3088-R | GCCCCGCTCTTGTCCCTTAG | |
| lp_3414-F | TGGGACGCATACGCAACCTAA | Extracellular protein |
| lp_3414-R | GCCAGCCAGCACCAGTCC | |
| lp_3575-F | CGCGTTTCAGATTGCCTTTTT | Integral membrane protein |
| lp_3575-R | CGTGCCAATCTTCCCAACAATA | |
| lp_3578-F | AAGCCGGTTGTTCCAGTTCAT | Catalase |
| lp_3578-R | GGCATCCGGCACCTCTTCT | |
| lp_3579-F | GCATGCCAAGATCCCCACGAA | Negative regulator of proteolysis |
| lp_3579-R | TCCGGCCCATGTCACGAA | |
| lp_3581-F | GATTGGCCTATTTGTCATT | LamC |
| lp_3581-R | GAGCTTCGATTTCATTCA | |
| lp_3582-F | CACCGGCTGTTACAAGAAAGAA | LamB |
| lp_3582-R | AAACTAATAACAGCGATACAACAA | |
| lp_3583-F | GATGGGCGCTTAACGGATGGAC | ATP-dependent protease subunit ClpL |
| lp_3583-R | TCTGGGCGGAAAAATGGTGCTAA | |
| LDH-F | TGATCCTCGTTCCGTYGATG | Lactate dehydrogenase |
| LDH-R | CCGATGGTTGCAGTTGAGTAAG |
Replacement of the L. plantarum WCFS1 lamR gene.
L. plantarum WCFS1 (13, 19) was transformed by electroporation, essentially as previously described (16), using electrocompetent cells of L. plantarum that were grown in MRS supplemented with 1% glycine and prepared in 30% polyethylene glycol 1450. L. plantarum cells were transformed with 2 μg of integration plasmid pDel-LamR-WCFS7, and integrants were selected by plating the resulting bacteria on MRS agar supplemented with erythromycin (5 μg ml−1) and incubating the plates in an anaerobic jar at 30°C for 2 to 4 days. Single-crossover integration was confirmed by colony PCR using primers lamR-F and lamR-BamHI (Table 2). For this purpose, single colonies were suspended in 20 μl Tris-EDTA and cells were disrupted by microwave treatment for 3 min at 750 W and subsequent incubation for 10 min at 95°C, after which PCR analysis was performed. The single-crossover mutant was subsequently propagated for 200 generations in MRS without erythromycin to obtain the anticipated erythromycin-sensitive (Ems) lamR replacement mutant after a second homologous recombination event. The anticipated gene organization of the lamR locus in the gene replacement mutant obtained was verified by enumeration of smaller fragments by colony PCR using primers lamR-F and lamR-BamHI, as well as by Southern blot analysis using standard procedures (29).
LC-MS analysis of culture supernatant.
L. plantarum strains (the wild-type strain and the ΔlamA, ΔlamR, and ΔlamA ΔlamR mutanta) were grown to an optical density at 600 nm (OD600) of 4.0 in a chemically defined medium supplemented with 1% glucose (18). The culture was diluted 1:20 to obtain an OD600 of 0.15 in 10 ml fresh chemically defined medium and incubated for 18 h at 30°C. Culture supernatant was partially purified with a Sep-Pak C18 cartridge column and was then analyzed by liquid chromatography (LC) (Agilent HP1100 column; Agilent Zorbax Eclipse XDB-C18; 2.1 by 50 mm) and mass spectrometry (MS) (JEOL Accutof T100LC; JEOL, Tokyo, Japan) as previously described (38). After scanning for molecular ions derived from column elutes, extracted ion chromatograms were plotted with detector counts at m/z 559.
Adherence assays.
A quantitative adherence assay was performed as previously described (34), with slight modifications. Briefly, cells were grown in 1 ml MRS in 24-well glass bottom plates (TPP, Switzerland) at 37°C for 48 h. The wells were washed twice with phosphate-buffered saline to remove loosely attached cells, and the remaining adherent cells were air dried for about 10 min. For staining, 0.5 ml crystal violet (0.1%) was added, and the preparations were incubated for 30 min and subsequently washed twice with water. The attached and stained cells were removed with 33% acetic acid, and the absorbance at 595 nm was determined. All the assays were performed in triplicate.
RNA extraction.
Cultures of the wild-type strain and the ΔlamA, ΔlamR, and ΔlamA ΔlamR mutants were grown in 50 ml MRS at 37°C using a starting OD600 of 0.2. Samples (25 ml) were harvested at each growth phase and immediately quenched and mixed with 4 volumes of quenching buffer (69% methanol, 66.7 mM HEPES; pH 6.5) at −80°C to stop further transcription, as described previously (27). Subsequently, samples were pelleted by centrifugation at −20°C in a prechilled centrifuge, and cells were resuspended in 0.5 ml cold Tris-EDTA buffer. RNA was isolated using the Macaloid method, essentially as described previously (20), and purified further by on-column DNase I treatment with a High Pure RNA isolation kit (Roche). For DNA microarray analysis, 20-μg RNA aliquots were prepared. All experiments were performed in duplicate.
cDNA preparation, fluorescent labeling, and hybridization for microarray analysis.
Clone-based DNA microarrays were used to determine the global gene transcription levels. Total-RNA samples (20 μg) were extracted from log-phase cultures, and Cy3- and Cy5-labeled cDNAs were prepared using a Cyscribe postlabeling kit (GE Healthcare, United Kingdom). Unincorporated dyes were removed from labeled fragments by using Autoseq G50 columns (GE Healthcare, United Kingdom). Slides were prehybridized for 45 min at 42°C in 20 ml prehybridization solution (1% bovine serum albumin, 5× SSC, 0.1% sodium dodecyl sulfate; filtered) (1× SSC is 0.15 NaCl plus 0.015 M sodium citrate), washed in filtered deionized water, and dried. Cohybridization with Cy5- and Cy3-labeled cDNA probes was performed overnight at 42°C for 16 h in Slidehyb#1 (Ambion, Austin, TX). The slides were then washed twice in 1× SSC-0.1% sodium dodecyl sulfate and twice in 1× SSC before they were scanned. The experiments were performed in duplicate with Cy5/Cy3 dye swaps.
Microarray scanning and data analysis.
Slides were scanned with a ScanArray Express 4000 scanner (Perkin Elmer, Wellesley, MA), and image analysis and processing were performed using the ImaGene version 4.2 software package (BioDiscovery, El Segundo, CA). The following criteria were used for flagging spots: (i) empty spot threshold, 2.0; (ii) poor spot threshold, 0.4; and (iii) negative spots. Routinely, more than 80% of all spots met these quality criteria. Raw data were stored in BASE (28). Flagged data were discarded, and the remaining high-quality spot data were normalized using a LOWESS fit with M-A transformed data. To calculate a regulatory ratio for each gene for the genes represented by clones on the microarray, a weighted average of the M values of all clones that overlapped with the gene of interest was calculated. The weight used for each clone was equal to the square of the overlap between the gene and clone divided by the total length of the gene. Consequently, this method weighted small overlapping fragments less than proportionally compared to larger overlapping fragments. Statistical analysis was performed with the statistical software program R (14), using the Maanova analysis of variance (ANOVA) model-fitting package (17). Significant effects due to mutation (two levels) and experiment (two levels) and their interactions were observed at different levels of significance (P < 0.05 and P < 0.01). Clones displaying these effects were selected and analyzed with the Eisen CLUSTER program (http://rana.lbl.gov/EisenSoftware.htm).
Q-PCR analysis.
cDNA was synthesized from purified RNA using random hexamers as a primers and Superscript III (Invitrogen). All the procedures were performed according to the manufacturer's instructions. The quantitative PCR (Q-PCR) primers which were used are shown in Table 2. The products were quantified using an iQ5 i-Cycler (Bio-Rad), and amplification was performed with a iQ5 Sybr green Supermix kit (Bio-Rad). All results were normalized using the level of the L. plantarum lactate dehydrogenase gene, and assays were performed in triplicate.
Cell morphology analysis.
For analysis of cell morphology the strains (wild-type strain and the ΔlamR, ΔlamA, and ΔlamA ΔlamR mutants) were grown under a wide range of conditions. Liquid cultures were grown anaerobically in MRS to an OD600 of 0.6 (mid-log phase) or >3 (late log phase). Solid-phase cultures were grown for 30 h on MRS agar or on strips of the porous ceramic Anopore (36 by 8 mm; Whatman, United Kingdom) placed on MRS agar. Cell morphology was examined immediately. Cells were visualized by staining with the fluorogenic dye Syto 9, followed by imaging with an Olympus BX-41 fluorescence microscope (15). Data were captured using an 8-bit Kappa charge-coupled device camera, and cell lengths were determined using TIFF images and ImageJ software (http://rsb.info.nih.gov/ij/). The lengths of at least 300 randomly selected cells were determined for each strain for each growth or storage condition, and assays were repeated at least twice. Calculations were performed using Microsoft Excel and custom scripts written for the R software package for multivariate (ANOVA) and posthoc (Tukey honestly significant difference) statistical tests (14). For the ANOVA the logarithmically transformed cell lengths were fitted to a linear model with lamA and lamR effects (two levels each; wild type and a mutant) and an incubation effect (five levels; mid-exponential growth phase, late exponential growth phase, incubation in the refrigerator for up to 21 days, incubation in the refrigerator for up to 21 days on Anopore, and incubation at room temperature for up to 7 days). The model included two- and three-way interaction effects.
Colony phenotype analysis.
A phenotypic analysis of colonies was performed by staining the colonies with a mixture of two fluorogenic dyes, followed by imaging entire colonies from above by using low-power fluorescence microscopy. Colonies of the wild-type strain and the ΔlamA, ΔlamR, and ΔlamA ΔlamR mutants were grown for 30 h on strips of Anopore placed on MRS agar and were then stained by transfer of the strips to microscope slides with a 1-mm-thick film of 1% (wt/vol) low-melting-point agarose containing the fluorogenic dyes Syto 9 and propidium iodide. This stained the colonies from beneath, essentially as previously described (15). Imaging and data capture were performed as described above using a ×4 Fluorotar objective lens (Olympus, The Netherlands), and images were merged using Photoshop software (Adobe). All images were taken with equivalent camera exposures.
RESULTS
Similarity of the lamKR and lamBDCA operons of L. plantarum WCFS1.
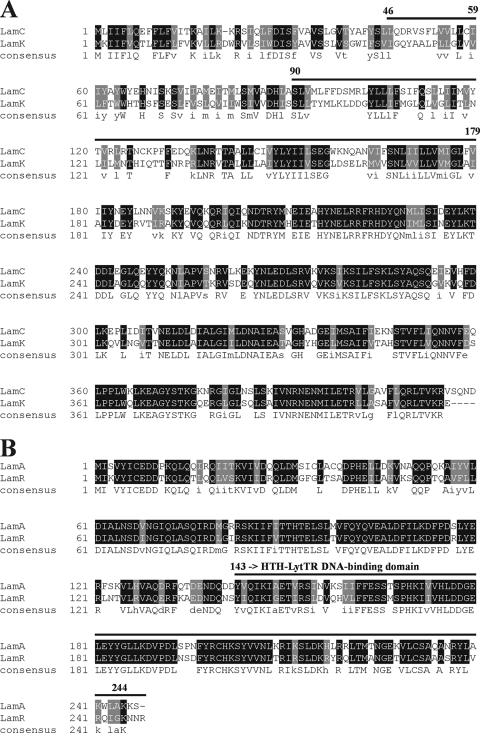
In the first part of this study, we compared the amino acid sequences of the HPKs and RRs which are encoded by the lamBDCA operon and the lamKR operon (Fig. 1). The results suggested that overall level of amino acid identity of the histidine kinase LamK and LamC was 55%, and the C-terminal ATPase domains were highly homologous. LamR and LamA have a helix-turn-helix Lyt-TR motif, which is a structure typical of quorum-sensing RRs (25) The overall level of amino acid identity of LamR and LamA was 70%.
FIG. 1.
Alignments of the sequences of (A) LamC and LamK and (B) LamA and LamR. Alignment was performed using ClustalW (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html) and BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html). Highly conserved residues are indicated by a black (identical) or gray (similar physicochemical characteristics) background. Bold lines with residue numbers indicate regions that define AIP specificity for the staphylococcal AgrC protein (A) or the HTH-LytTR DNA-binding domain of AgrA-like proteins (B).
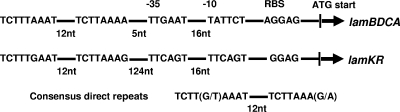
A comparison of flanking DNA sequences of the two operons suggested that a direct repeat with a high level of similarity was located in the 5′ flanking region of both operons (Fig. 2). These results suggested that lamKR might be a novel quorum-sensing operon that is similar to lamBDCA in terms of both gene expression and function.
FIG. 2.
Alignment of the promoter sequences of the lamBDCA operon and the lamKR operon. nt, nucleotides.
Cooperation of LamA and LamR in transcription regulation and LamD558 peptide production.
To confirm that LamA and LamR are functionally related, the ΔlamR and ΔlamA ΔlamR mutants were constructed. The growth of the ΔlamA, ΔlamR, ΔlamA ΔlamR mutants in MRS media was not significantly different from that of the wild-type strain (data not shown).
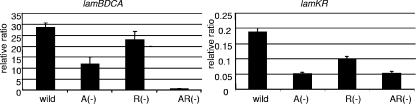
The transcription of the lamBDCA and lamKR operons and the production of the autoinducing peptide LamD558 were studied, and the results confirmed that the level of transcription of lamKR was much lower than that of lamBDCA, while the expression kinetics of lamKR were similar to those of lamBDCA in the wild-type strain (Fig. 3).
FIG. 3.
Gene expression ratios for the lamBDCA operon and the lamKR operon during cell growth for the wild-type strain and the ΔlamA, ΔlamR, and ΔlamA ΔlamR mutants. Cell cultures grown in MRS at 37°C without agitation were collected in the logarithmic growth phase. Total RNA was isolated and cDNA synthesis was performed as described in Materials and Methods. The vertical line indicates the relative level of gene expression compared to expression of the control lactate dehydrogenase gene. All Q-PCR analyses were performed in triplicate. A(−), ΔlamA mutant; R(−), ΔlamR mutant; AR(−), ΔlamA ΔlamR mutant.
The transcription of the lamBDCA operon in the ΔlamA mutant was repressed and was one-third of the transcription of the lamBDCA operon in wild-type strain, as previously reported (37). In the ΔlamR mutant, the transcription of this operon was affected only slightly and was 80% of that in the wild-type strain. A dramatic reduction was observed in ΔlamA ΔlamR mutant, in which the transcription of the lamBDCA operon was less than 5% of that in the wild-type strain (Fig. 3). The transcription of the lamKR operon in the ΔlamA and ΔlamA ΔlamR mutants was repressed and was one-third of that in the wild-type strain, and it was slightly repressed in the ΔlamR mutant.
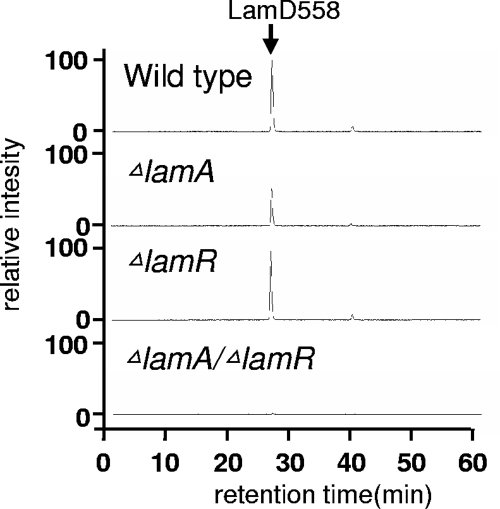
LC-MS analysis of culture supernatant suggested that the LamD558 production in the ΔlamR mutant was not significantly different from that in the wild-type strain. However, the lamR disruption significantly affected AIP production in the ΔlamA mutant. In the ΔlamA mutant the level of LamD558 was about 50% of the level in the wild-type strain, but no peptide production was observed in the ΔlamA ΔlamR mutant (Fig. 4). Taken together, the results show that LamR is functionally similar to LamA, and these two RR proteins appear to cooperate to regulate this quorum-sensing system.
FIG. 4.
LC-MS analysis of LamD558 production in the culture supernatants of wild-type and mutant strains of L. plantarum. The overnight culture supernatant of each strain was partially purified by using a Sep-Pak C18 cartridge column and then analyzed by LC-MS as described in Materials and Methods. Extracted ion chromatograms were plotted with detector counts at m/z 559, corresponding to the protonated molecular ion of LamD558.
Cooperative effect of lamA and lamR on global gene expression.
In order to study the effect of the lamR gene on global gene expression, microarray analyses were performed. No gene showed a significant (more than twofold) difference in the level of expression in the ΔlamR mutant compared to the wild-type strain (data not shown). This result suggested that the lack of LamR might be functionally covered by LamA, since the level of expression of lamR was much lower than the level of expression of lamA.
The results of comparisons of the ΔlamA ΔlamR mutant with the wild-type strain and of the ΔlamA ΔlamR mutant with the ΔlamA mutant are shown in Tables 3 and 4, respectively. Compared with the wild-type strain, 14 genes were upregulated more than threefold and 11 genes were upregulated more than fivefold in the ΔlamA ΔlamR mutant, while 39 genes were downregulated more than twofold and 20 genes were downregulated more than fivefold (Table 3). Most of these genes were previously reported to be regulated in the ΔlamA mutant (38). Typical examples of the commonly regulated genes were the genes in the cps2 operon (from lp_1197 to lp_1205), membrane protein-encoding genes (lp_0926, lp_3575, and lp_3577), stress response genes (lp_0930 and lp_3578), and the lamBDCA operon itself (lp_3580 to lp_3583).
TABLE 3.
Genes whose levels of expression were significantly different between the ΔlamA ΔlamR mutant and the wild-type strain
| Open reading frame | Gene | Product | Change (fold)a |
|---|---|---|---|
| Upregulated genes | |||
| lp_0783 | Lipoprotein precursor, peptide-binding protein OppA homolog | 3.1 | |
| lp_1197 | cps2A | Exopolysaccharide biosynthesis protein | 27.8 |
| lp_1198 | cps2B | Exopolysaccharide biosynthesis protein; chain length determiner Wzz | 54.9 |
| lp_1200 | galE2 | UDP-glucose 4-epimerase | 11.9 |
| lp_1201 | cps2E | Priming glycosyltransferase | 16.0 |
| lp_1202 | cps2F | Glycosyltransferase | 15.8 |
| lp_1203 | cps2G | Glycosyltransferase | 11.5 |
| lp_1204 | cps2H | Polysaccharide polymerase | 5.9 |
| lp_1205 | cps2I | Repeat unit transporter | 6.4 |
| lp_1207 | No product defined | 4.4 | |
| lp_3411 | Extracellular protein | 4.3 | |
| lp_3412 | Extracellular protein | 8.1 | |
| lp_3413 | Cell surface protein precursor | 13.5 | |
| lp_3414 | Extracellular protein | 7.4 | |
| Downregulated genes | |||
| lp_0023 | glgA | Starch (bacterial glycogen) synthase | −5.1 |
| lp_0111 | Oxidoreductase | −5.9 | |
| lp_0525 | kup1 | Potassium uptake protein | −3.3 |
| lp_0683 | Prophage P1 protein 60 | −4.0 | |
| lp_0885 | bglG1 | Transcription antiterminator | −12.4 |
| lp_0926 | Integral membrane protein | −21.4 | |
| lp_0927 | Hypothetical protein | −16.9 | |
| lp_0930 | asp2 | Alkaline shock protein | −5.3 |
| lp_1703 | Hypothetical protein | −4.7 | |
| lp_2658 | Glycosyltransferase (putative) | −3.2 | |
| lp_3045 | Short-chain dehydrogenase/oxidoreductase | −6.1 | |
| lp_3082 | Transport protein | −5.5 | |
| lp_3084 | Cell surface protein, ErfK family | −7.1 | |
| lp_3085 | asnB2 | Asparagine synthase (glutamine-hydrolyzing) | −7.7 |
| lp_3087 | lamR | RR | −4.2 |
| lp_3128 | Stress-induced DNA binding protein | −4.0 | |
| lp_3267 | gshR4 | Glutathione reductase | −3.5 |
| lp_3420 | gadB | Glutamate decarboxylase | −2.6 |
| lp_3575 | Integral membrane protein | −14.4 | |
| lp_3577 | Integral membrane protein | −5.0 | |
| lp_3578 | kat | Catalase | −5.6 |
| lp_3579 | nrpR5 | Negative regulator of proteolysis | −117.8 |
| lp_3580 | lamA | RR; accessory gene regulator protein A | −28.7 |
| lp_3582 | lamB | Accessory gene regulator protein B | −14.9 |
| lp_3583 | clpL | ATP-dependent Clp protease, ATP-binding subunit ClpL | −10.8 |
| lp_3586 | lox | Lactate oxidase | −32.4 |
Change in gene expression in the ΔlamA ΔlamR mutant compared to the wild-type strain.
TABLE 4.
Genes whose levels of expression were significantly different between the ΔlamA ΔlamR and ΔlamA mutants
| Open reading frame | Gene | Product | Change (fold)a |
|---|---|---|---|
| Upregulated genes | |||
| lp_0152 | Transcription regulator | 2.8 | |
| lp_0177 | malG | Maltose/maltodextrin ABC transporter, permease protein | 2.2 |
| lp_0179 | amy2 | α-Amylase | 2.3 |
| lp_0180 | msmK1 | Multiple-sugar ABC transporter, ATP-binding protein | 3.2 |
| lp_0302 | Extracellular protein | 2.4 | |
| lp_0547 | ftsH | Cell division protein FtsH, ATP-dependent zinc metallopeptidase | 2.5 |
| lp_0578 | npsA | Nonribosomal peptide synthetase NpsA | 2.1 |
| lp_1197 | cps2A | Exopolysaccharide biosynthesis protein | 2.1 |
| lp_1198 | cps2B | Exopolysaccharide biosynthesis protein; chain length determiner Wzz | 2.6 |
| lp_1200 | galE2 | UDP-glucose 4-epimerase | 2.4 |
| lp_1201 | cps2E | Priming glycosyltransferase | 2.7 |
| lp_1203 | cps2G | Glycosyltransferase | 2.3 |
| lp_1730 | map3 | Maltose phosphorylase | 2.1 |
| lp_2151 | pdhD | Pyruvate dehydrogenase complex, E3 component; dihydrolipoamide dehydrogenase | 2.1 |
| lp_2352 | Amino acid ABC transporter, ATP-binding protein | 2.6 | |
| lp_3411 | Extracellular protein | 2.1 | |
| lp_3412 | Extracellular protein | 7.6 | |
| lp_3413 | Cell surface protein precursor | 9.8 | |
| lp_3414 | Extracellular protein | 7.2 | |
| Downregulated genes | |||
| lp_0254 | cysE | Serine O-acetyltransferase | −2.2 |
| lp_0255 | metC1 | Cystathionine beta-lyase | −2.2 |
| lp_2833 | Transport protein | −2.4 | |
| lp_3084 | Cell surface protein, ErfK family | −2.3 | |
| lp_3085 | asnB2 | Asparagine synthase (glutamine hydrolyzing) | −2.4 |
| lp_3087 | lamK | RR | −2.4 |
| lp_3575 | Integral membrane protein | −3.0 | |
| lp_3579 | nrpR5 | Negative regulator of proteolysis | −10.7 |
| lp_3580 | lamA | RR; accessory gene regulator protein A | −16.3 |
| lp_3582 | lamB | Accessory gene regulator protein B | −7.2 |
Change in gene expression in the ΔlamA ΔlamR mutant compared to the ΔlamA mutant.
In addition, we observed that these genes were more significantly up- or downregulated in the ΔlamA ΔlamR mutant than in the ΔlamA mutant. For instance, the first three genes in the cps2 operon, lp_1197, lp_1198, and lp_1199, were upregulated 8.3-, 9.1-, and 4.4-fold, respectively, in the ΔlamA mutant compared to the wild-type strain (data not shown), while they were upregulated 19.0-, 15.7-, and 15.3-fold in the ΔlamA ΔlamR mutant compared to the wild-type strain (Table 3).
To more clearly determine the effect of knocking out both lam operons, a direct comparison of the ΔlamA ΔlamR and ΔlamA mutants was performed. Nineteen genes were upregulated significantly (more than twofold), while 10 genes were downregulated more than twofold in the ΔlamA ΔlamR mutant compared to the ΔlamA mutant (Table 4). As expected, the cps2 operon was activated nearly twofold in the ΔlamA ΔlamR mutant compared with the ΔlamA mutant. An additive effect was also observed for the downregulated genes, such as genes in the lamKR operon (lp_3087), lp_3084, lp_3085, lp_3575, and lp_3579.
To our surprise, the operon comprising lp_3412 to lp_3414 was synergistically regulated in the ΔlamA ΔlamR mutant. The levels of expression of this operon in the ΔlamA and ΔlamR mutants were not significantly different, but this operon was uniquely upregulated more than fivefold in the ΔlamA ΔlamR mutant compared the wild-type strain and the ΔlamR and ΔlamA mutants (Tables 3 and 4).
Q-PCR analysis.
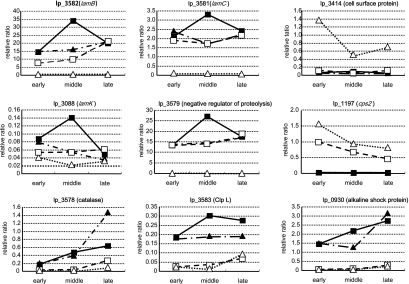
RNA samples extracted from the mutants in the early, middle, and late log phases were analyzed by Q-PCR to confirm the observations obtained in the microarray analysis (Fig. 5). The kinetics data suggested that the lamBDCA operon (lp_3580 and lp_3581) was completely repressed in the ΔlamA ΔlamR mutant and partially repressed in the ΔlamA and ΔlamR mutants throughout the growth phases. The difference was most significant in the mid-log phase. Similar regulation was observed for the lamKR operon (lp_3088) and the stress-related protein-encoding genes, such as lp_3579 (negative regulator of proteolysis), lp_0929 and lp_0930 (alkaline shock protein), and lp_3578 (catalase). Most of these genes are localized in the loci which were designated “life style adaptation regions” (22).
FIG. 5.
Kinetics of gene expression: ratios of the wild-type strain to the ΔlamA, ΔlamR, and ΔlamA ΔlamR mutants. Cell cultures grown in MRS at 37°C without agitation were collected in the early, middle, and late logarithmic growth phases. Then total RNA was isolated and cDNA synthesis was performed as described in Materials and Methods. The vertical line indicates the relative gene expression level compared to expression of the control lactate dehydrogenase gene. All the Q-PCR analyses were performed in triplicate. ▪, wild-type strain; □, ΔlamA mutant; ▴, ΔlamR mutant; ▵, ΔlamA ΔlamR mutant.
It was also confirmed that the operon comprising lp_3012 to lp_3014 was strongly activated in all growth phases and that the highest level was observed in the early growth phase in the ΔlamA ΔlamR mutant. Neither upregulation nor growth-dependent gene expression of this operon was observed in the ΔlamA and ΔlamR mutants and the wild-type strain.
Effect of lamR on adherence properties of L. plantarum WCFS1.
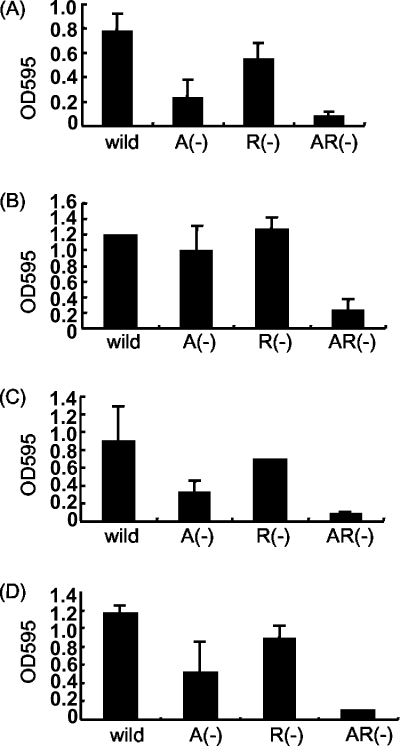
The glass adherence phenotype was determined for the ΔlamA, ΔlamR, and ΔlamA ΔlamR mutants and the wild-type strain. As previously reported (38), around fourfold fewer cells of the ΔlamA mutant than of the wild-type strain adhered to a glass surface. In contrast, the number of the ΔlamR mutant cells that adhered to a glass surface was almost 75% of the number of the wild-type cells that adhered. The ΔlamA ΔlamR mutant cells showed the least adherence to glass. The number of cells that adhered to a glass surface was less than one-eighth of the number of wild-type cells that adhered (Fig. 6A). When the results shown in Tables 3 and 4 and Fig. 5 were compared, there was clearly a negative correlation between the number of adherent cells and the level of expression of the cps2 operon; that is, as cps2 expression increased, the number of adherent cells decreased. This strongly suggested that the cps2 operon might be important primarily for the glass adherence property of L. plantarum cells.
FIG. 6.
Quantification of adherence of L. plantarum WCFS1 to a glass surface. Cell cultures were cultivated in MRS media containing (A) glucose, (B) maltose, (C) galactose, and (D) raffinose as a unique carbon source for 48 h at 37°C. The quantification methods used are described in Materials and Methods. A(−), ΔlamA mutant; R(−), ΔlamR mutant; AR(−), ΔlamA ΔlamR mutant.
In order to study the effect of the carbon source on glass adherence, the same experiments were performed using cell cultures grown in MRS in which glucose was replaced by maltose, galactose, or raffinose (Fig. 6B, 6C, and 6D). The level of glass adherence was slightly affected by the carbon source. In maltose-containing medium, the level of glass adherence was higher than that in glucose-containing medium. In all media, the lamA disruption was more effective in reducing the adherence than the lamR disruption, and the ΔlamA ΔlamR mutant showed the least adherence.
Effect of the lamA and lamR genes on cell morphology and colony structure.
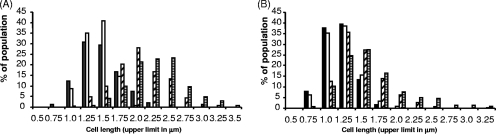
In the cell morphology experiments, we found that there were differences in cell length between the ΔlamA, ΔlamR, and ΔlamA ΔlamR mutants and the wild-type strain. In order to confirm this, the cell length was quantified using digital images. Multiway ANOVA, using cell length, was performed to assess the cell lengths of the ΔlamR, ΔlamA, and ΔlamA ΔlamR mutants compared with the wild-type strain under a range of different storage and growth conditions (Fig. 7). The results strongly suggested that the cell length depended on the expression of both lam operons. Both the ΔlamA and ΔlamR mutations resulted in increases in the mean cell length (ANOVA Pr>F values, <2 × 10−16 and 3.9 × 10−13, respectively), and the strongest effect was observed for the ΔlamA mutation. Again, the interaction of lamA and growth conditions was stronger than the interaction of lamR and growth conditions (Pr>F values, <2 × 10−16 and 0.01, respectively). The increase in the length of the ΔlamA mutant cells was most pronounced under high-cell-density conditions (Fig. 7A). The effect was observed with late-log-phase cells cultured in liquid MRS (Fig. 7B). A more limited effect was also observed in mid-log-phase cells (data not shown). The trend toward more elongated cells was generally stronger for the ΔlamA ΔlamR mutant and weaker for the ΔlamR mutant and wild-type strain (Fig. 7). ANOVA for the contribution that each Δlam mutation made to cell length supported the observation that the interaction between the two lam systems was significant (Pr>F, 3.5 × 10−6), meaning that an additive effect was observed when both mutations were present. Also, a significant three-way interaction was observed, meaning that the environmental conditions influenced the degree to which the Δlam mutations interacted (Pr>F, 1.2 × 10−4). In particular, the interaction was most pronounced when cells were in colony form. Although the significance of these interactions in a linear model does not provide direct evidence for a biochemical interaction between the lam systems, it does suggest that there is at least an indirect interaction between the two systems and that this is consistent with the gene regulation data obtained in this study.
FIG. 7.
Cell morphology of Δlam mutants: frequency plots of data used for ANOVA. Sample plots for the two most informative conditions tested, colonial growth (A) and late-log-phase liquid culture (B), are shown. The bars indicate the distributions of cell lengths for 300 cells (expressed as percentages) for the wild-type strain (filled bars), the ΔlamA mutant (bars with diagonal lines), the ΔlamR mutant (open bars), and the ΔlamA ΔlamR mutant (bars with horizontal lines).
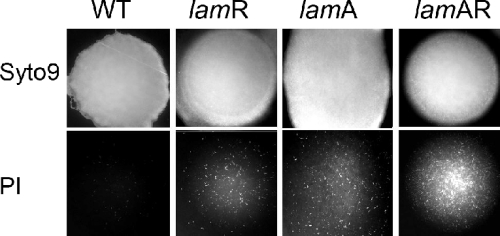
Colonies of the ΔlamA, ΔlamR, and ΔlamA ΔlamR mutants and the wild-type strain appeared to be similar to the eye in terms of size and morphology. However, when preparations were double stained using the fluorogenic dyes Syto 9 and propidium iodide, phenotypic differences became apparent (Fig. 8). For the wild-type strain, the colonies were stained almost completely with Syto 9. At a high magnification only rare (<0.1%) filamented cells (length, >20 μm) showed strong propidium iodide staining. All three Δlam mutants exhibited increased propidium iodide staining in the colony center compared to the wild-type strain. In some cases the difference was most extreme in the ΔlamA ΔlamR mutant, and in others the ΔlamA and ΔlamA ΔlamR mutants appeared to be similar (Fig. 8). Propidium iodide preferentially stains cells with compromised or damaged membranes, and it is likely that the colony structure or the integrity of cells in the colony center is affected by the Δlam mutations.
FIG. 8.
Colony phenotypes of Δlam mutants and wild-type L. plantarum: merged images of typical colonies of L. plantarum WCFS1 and lam mutants (all approximately 2 mm in diameter) after double staining from beneath with Syto 9 and propidium iodide and imaging by low-power fluorescence microscopy. WT, wild type.
Taken together, these data suggested qualitatively that the structure of the colonial growth of the Δlam mutants was affected and that functional lam operons play a role in the integrity of cells for colony growth and survival.
DISCUSSION
In this study, we thoroughly investigated the function of the lamKR operon in terms of gene regulation, AIP production, and phenotypes. All of the results strongly suggested that the two lam 2CSs function cooperatively. Due to the high levels of homology of the encoded proteins, it is likely that the HPKs and RRs encoded by lamKR and lamCA cross talk with each other, as is the case in E. coli (48). It is also possible that LamA and LamR bind to the promoter regions of both operons and thereby regulate the expression of each other's genes at the transcriptional level, since similar direct repeats were observed in the upstream regions of lamBDCA and lamKR. PCR experiments with 22 L. plantarum strains using primers designed from the consensus sequence of the amino acid regions between LamA and LamR and between LamC and LamK showed that 20 strains contained lam genes (data not shown). This may suggest that the lam system is widespread among L. plantarum strains. However, more direct in vitro analyses, such as cross-phosphorylation assays with LamA and LamR and gel shift assays with LamA and LamR and with lamKR and lamBDCA promoter regions, should be performed to prove these hypotheses. Such analyses are under way in our lab.
Our data also suggested that disruption of lamR affected the global gene expression and phenotypes, such as glass adherence and cell length. As expected, the phenotypic differences between the ΔlamR mutant and the wild-type strain were similar to those between the ΔlamA mutant and the wild-type strain and were most obvious in the ΔlamA ΔlamR mutant.
Biofilm formation is one of the typical quorum-sensing-dependent phenotypes. Several reports have suggested that bacterial biofilm formation is under the control of quorum-sensing systems. In the genus Staphylococcus, agr mutants have been reported to show increased biofilm formation and primary attachment on polystyrene (43, 45). Further, studies by Tannnock et al. (39), Wen et al. (47), and Yoshida et al. (49) suggested that LuxS-based signaling affects biofilm formation. Our results provided another example of initiation of biofilm formation or adherence that was regulated by a quorum-sensing system. However, the genes that have a direct effect on these phenotypes are still unknown. Our microarray and Q-PCR analyses revealed that many gene expression differences observed for the ΔlamA mutant were quantitatively enhanced in the ΔlamA ΔlamR mutant. The most probable genes that are responsible for glass adherence are the cps2 genes, because the number of glass-adherent ΔlamA ΔlamR mutant cells and the level of expression of the cps2 genes showed a strong negative correlation. It is likely that polysaccharides produced by the enzymes encoded by the cps2 operon were responsible for the poor glass adherence phenotype.
Cell morphology was another phenotype that was affected by lam genes. To our knowledge, this report provides the first direct link between quorum-sensing systems and cell morphology in gram-positive bacteria, although such a link has been reported previously for gram-negative bacteria (33). Microarray analysis revealed that the level of expression of the cps2 operon was positively correlated to the cell length, suggesting that this operon may play a role in cell length, as well as biofilm formation. The alternative possibility is that filamentous phenotypes are the result of the stress response of L. plantarum. Some of the stress-related genes, such as lp_3575 (integral membrane protein), lp_3577 (integral membrane protein), lp_3578 (catalase), lp_3579 (negative regulator of proteolysis), lp_3583 (ATP-dependent Clp protease subunit ClpL), and lp_3586 (lactate oxidase), were downregulated nearly 10-fold in the ΔlamA or ΔlamA ΔlamR mutant. The results of Syto 9 and propidium iodide staining also supported the argument that a lack of lamA and lamR results in strongly stressed cells (Fig. 8). It is well known that bacterial cells have a filamentous morphology under stress conditions (1, 40, 46). A study which demonstrated that a B. subtilis mutant with a knockout of ClpP, a proteolytic enzyme with bacterial ATP-dependent proteases, had a filamentous cell morphology appears to be particularly relevant (23). Another study, which showed that ClpL also is involved in cold sensitivity and stress in Streptococcus thermophilus (42), was also intriguing since we observed that the differences in cell length of Δlam mutants were more pronounced after cold storage for 1 to 3 weeks (data not shown). It is likely that expression of the lam system is more important at a high cell density, since no difference was observed between the Δlam mutants and the wild-type strain in liquid culture.
Interestingly, an operon which encodes cell surface proteins (lp_3412, lp_3413, and lp_3414) was strongly upregulated throughout growth in the ΔlamA ΔlamR mutant (Tables 3 and 4 and Fig. 5). This operon was overexpressed in neither the ΔlamA mutant nor the ΔlamR mutant, suggesting that a complete lack of the lam system is necessary for the overexpression. Although the function of this operon has not been addressed yet, a recent bioinformatics study suggested that the lp_3413 protein is a membrane-anchored protein and that the lp_3412 and lp_3414 proteins may bind to the lp_3413 protein to form a functional complex (32). Another interesting study suggested that lp_3412, lp_3413, and lp_3414 are upregulated when the L. plantarum strain is in the gut environment (9).
In conclusion, lamA and lamR are important for L. plantarum to adapt to particular environmental niches where they can grow at high densities. Future studies on the survivability and persistence of Δlam mutants in L. plantarum-specific environments, such as the gastrointestinal tract, oral cavity, or fermented foods, may provide more information about the functional role of the lam systems in nature.
Footnotes
Published ahead of print on 19 September 2008.
REFERENCES
- 1.Ackerley, D. F., Y. Barak, S. V. Lynch, J. Curtin, and A. Matin. 2006. Effect of chromate stress on Escherichia coli K-12. J. Bacteriol. 1883371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahrne, S., S. Nobaek, B. Jeppsson, I. Adlerberth, A. E. Wold, and G. Molin. 1998. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J. Applied Microbiol. 8588-94. [DOI] [PubMed] [Google Scholar]
- 3.Autret, N., C. Raynaud, I. Dubail, P. Berche, and A. Charbit. 2003. Identification of the agr locus of Listeria monocytogenes: role in bacterial virulence. Infect. Immun. 714463-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bron, P. A., C. Grangette, A. Mercenier, W. M. de Vos, and M. Kleerebezem. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 1865721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buzzola, F. R., L. P. Alvarez, L. P. N. Tuchscherr, M. S. Barbagelata, S. M. Lattar, L. Calvinho, and D. O. Sordelli. 2007. Differential abilities of capsulated and noncapsulated Staphylococcus aureus isolates from diverse agr groups to invade mammary epithelial cells. Infect. Immun. 75886-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caplice, E., and G. F. Fitzgerald. 1999. Food fermentations: role of microorganisms in food production and preservation. Int. J. Food Microbiol. 50131-149. [DOI] [PubMed] [Google Scholar]
- 7.Chan, W. C., B. J. Coyle, and P. Williams. 2004. Virulence regulation and quorum sensing in staphylococcal infections: competitive AgrC antagonists as quorum sensing inhibitors. J. Med. Chem. 474633-4641. [DOI] [PubMed] [Google Scholar]
- 8.Comella, N., and A. D. Grossman. 2005. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol. Microbiol. 571159-1174. [DOI] [PubMed] [Google Scholar]
- 9.De Vries, M. C. 2006. Analyzing global gene expression of Lactobacillus plantarum in the human gastro-intestinal tract. Wageningen University, Wageningen, The Netherlands.
- 10.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41139-227. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, L. E., and M. Perego. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 1865629-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayward, A. C., and G. H. G. Davis. 1956. The isolation and classification of Lactobacillus strains from Italian saliva samples. Br. Dent. J. 1012733-2741. [Google Scholar]
- 14.Ihaka, R., and R. Gentleman. 1996. R: a language for data analysis and graphics J. Comput. Graph. Statist. 5299-314. [Google Scholar]
- 15.Ingham, C. J., M. van den Ende, D. Pijnenburg, P. C. Wever, and P. M. Schneeberger. 2005. Growth and multiplexed analysis of microorganisms on a subdivided, highly porous, inorganic chip manufactured from Anopore. Appl. Environ. Microbiol. 718978-8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Josson, K., T. Scheirlinck, F. Michiels, C. Platteeuw, P. Stanssens, H. Joos, P. Dhaese, M. Zabeau, and J. Mahillon. 1989. Characterization of a gram-positive broad-host-range plasmid isolated from Lactobacillus hilgardii. Plasmid 219-20. [DOI] [PubMed] [Google Scholar]
- 17.Kerr, M. K., M. Martin, and G. A. Churchill. 2000. Analysis of variance for gene expression microarray data. J. Comput. Biol. 7819-837. [DOI] [PubMed] [Google Scholar]
- 18.Kets, E. P. W., E. A. Galinski, and J. A. M. de Bont. 1994. Carnitine: a novel compatible solute in Lactobacillus plantarum. Arch. Microbiol. 162243-248. [Google Scholar]
- 19.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. E. J. Fiers, W. Stiekema, R. M. K. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 1001990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. De Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216281-291. [DOI] [PubMed] [Google Scholar]
- 21.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55165-199. [DOI] [PubMed] [Google Scholar]
- 22.Molenaar, D., F. Bringel, F. H. Schuren, W. M. de Vos, R. J. Siezen, and M. Kleerebezem. 2005. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 1876119-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Msadek, T., V. Dartois, F. Kunst, M.-L. Herbaud, F. Denizot, and G. Rapoport. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27899-914. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, A. D. L. Akkermans, W. M. de Vos, and H. Nagasawa. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41145-154. [DOI] [PubMed] [Google Scholar]
- 25.Nikolskaya, A. N., and M. Y. Galperin. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 302453-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 481429-1449. [DOI] [PubMed] [Google Scholar]
- 27.Pieterse, B., R. H. Jellema, and M. J. van der Werf. 2006. Quenching of microbial samples for increased reliability of microarray data. J. Microbiol. Methods 64207-216. [DOI] [PubMed] [Google Scholar]
- 28.Saal, L., C. Troein, J. Vallon-Christersson, S. Gruvberger, A. Borg, and C. Peterson. 2002. BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol. 3software0003.1-software0003.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Scott, K. P., J. C. Martin, G. Campbell, C.-D. Mayer, and H. J. Flint. 2006. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans.” J. Bacteriol. 1884340-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shompole, S., K. T. Henon, L. E. Liou, K. Dziewanowska, G. A. Bohach, and K. W. Bayles. 2003. Biphasic intracellular expression of Staphylococcus aureus virulence factors and evidence for Agr-mediated diffusion sensing. Mol. Microbiol. 49919-927. [DOI] [PubMed] [Google Scholar]
- 32.Siezen, R. J., J. Boekhorst, L. Muscariello, D. Molenaar, B. Renckens, and M. Kleerebezem. 2006. Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific gram-positive bacteria. BMC Genomics 7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 1835187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stepanovic, S., D. Vukovic, I. Dakic, B. Savic, and M. Svabic-Vlahovic. 2000. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40175-179. [DOI] [PubMed] [Google Scholar]
- 35.Sturme, M. H. J. 2005. Analysis of quorum sensing regulatory systems in the human isolate Lactobacillus plantarum. Wageningen University, Wageningen, The Netherlands.
- 36.Sturme, M. H. J., C. Francke, R. J. Siezen, W. M. de Vos, and M. Kleerebezem. 2007. Making sense of quorum sensing in lactobacilli: a special focus on Lactobacillus plantarum WCFS1. Microbiology 1533939-3947. [DOI] [PubMed] [Google Scholar]
- 37.Sturme, M. H. J., M. Kleerebezem, J. Nakayama, A. D. L. Akkermans, E. E. Vaughan, and W. M. de Vos. 2002. Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie van Leeuwenhoek 81233-243. [DOI] [PubMed] [Google Scholar]
- 38.Sturme, M. H. J., J. Nakayama, D. Molenaar, Y. Murakami, R. Kunugi, T. Fujii, E. E. Vaughan, M. Kleerebezem, and W. M. de Vos. 2005. An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J. Bacteriol. 1875224-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tannock, G. W., S. Ghazally, J. Walter, D. Loach, H. Brooks, G. Cook, M. Surette, C. Simmers, P. Bremer, F. Dal Bello, and C. Hertel. 2005. Ecological behavior of Lactobacillus reuteri 100-23 is affected by mutation of the luxS gene. Appl. Environ. Microbiol. 718419-8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomas, C. A., K. V. Alsaker, H. P. J. Bonarius, W. T. Hendriksen, H. Yang, J. A. Beamish, C. J. Paredes, and E. T. Papoutsakis. 2003. DNA array-based transcriptional analysis of asporogenous, nonsolventogenic Clostridium acetobutylicum strains SKO1 and M5. J. Bacteriol. 1854539-4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Kranenburg, R., J. D. Marugg, I. I. van Swam, N. J. Willem, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24387-397. [DOI] [PubMed] [Google Scholar]
- 42.Varcamonti, M., S. Arsenijevic, L. Martirani, D. Fusco, G. Naclerio, and M. De Felice. 2006. Expression of the heat shock gene clpL of Streptococcus thermophilus is induced by both heat and cold shock. Microb. Cell Factories 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vuong, C., C. Gerke, G. A. Somerville, E. R. Fischer, and M. Otto. 2003. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188706-718. [DOI] [PubMed] [Google Scholar]
- 44.Vuong, C., F. Gotz, and M. Otto. 2000. Construction and characterization of an agr deletion mutant of Staphylococcus epidermidis. Infect. Immun. 681048-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 1821688-1693. [DOI] [PubMed] [Google Scholar]
- 46.Wehrl, W., M. Niederweis, and W. Schumann. 2000. The FtsH protein accumulates at the septum of Bacillus subtilis during cell division and sporulation. J. Bacteriol. 1823870-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen, Z. T., and R. A. Burne. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 1862682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto, K., K. Hirao, T. Oshima, H. Aiba, R. Utsumi, and A. Ishihama. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 2801448-1456. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida, A., T. Ansai, T. Takehara, and H. K. Kuramitsu. 2005. LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl. Environ. Microbiol. 712372-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]