Abstract
Although Mycobacterium tuberculosis and related species are considered to be typical endosomal pathogens, recent studies have suggested that mycobacteria can be present in the cytoplasm of infected cells and cause cytoskeleton rearrangements, the mechanisms of which remain unknown. Here, we used single-molecule force spectroscopy to demonstrate that the heparin-binding hemagglutinin (HBHA), a surface adhesin from Mycobacterium tuberculosis displaying sequence similarities with actin-binding proteins, is able to bind to actin. Force curves recorded between actin and the coiled-coil, N-terminal domain of HBHA showed a bimodal distribution of binding forces reflecting the detection of single and double HBHA-actin interactions. Force curves obtained between actin and the lysine-rich C-terminal domain of HBHA showed a broader distribution of binding events, suggesting they originate primarily from intermolecular electrostatic bridges between cationic HBHA domains and anionic actin residues. We also explored the dynamics of the HBHA-actin interaction, showing that the binding force and binding frequency increased with the pulling speed and contact time, respectively. Taken together, our data indicate that HBHA is able to specifically bind actin, via both its N-terminal and C-terminal domains, strongly suggesting a role of the HBHA-actin interaction in the pathogenesis of mycobacterial diseases.
More than a century after the discovery of their etiological agents, tuberculosis and leprosy remain major health threats for humans, but the molecular mechanisms that lead to the development of both diseases remain insufficiently well understood (2). Elucidation of these mechanisms, especially those allowing mycobacteria to adhere and disseminate, should facilitate the development of new prophylactic and/or therapeutic strategies.
One of the factors involved in mycobacterial adherence to nonprofessional phagocytes (17) and in extrapulmonary dissemination of Mycobacterium tuberculosis (23) is the heparin-binding hemagglutinin (HBHA) (for a recent review, see reference 15). This protein binds to sulfated glycoconjugates at the surfaces of epithelial cells via its C-terminal heparin-binding domain composed of several lysine-rich repeats (4, 22). It also promotes bacterial aggregation, presumably via specific coiled-coil interactions involving its N-terminal moiety (4). Using atomic force microscopy (AFM) in the single-molecule force spectroscopy (SMFS) mode (14), we recently provided quantitative insight into HBHA-heparin (6) and homophilic HBHA-HBHA interactions (31).
Interestingly, the expression of the HBHA-encoding gene is strongly upregulated in M. tuberculosis upon invasion of epithelial cells (5), suggesting that the adhesin may also play a role in host-cell interaction beyond the stage of binding. Latex beads coated with HBHA have been shown to be able to cross epithelial cell layers and to induce the reorganization of the actin filament network in confluent cell layers (19), the mechanism of which remains unknown.
Until recently, M. tuberculosis and related species were considered to be typical endosomal pathogens. However, recent reports have indicated that free M. tuberculosis, Mycobacterium marinum, and Mycobacterium leprae can be present in the cytoplasm (27, 28, 30) and demonstrated that intracellular M. marinum not only enters the cytoplasm of infected macrophages but also has the ability to be propelled by actin-based motility (27, 28), as observed for numerous intracellular pathogens, including Listeria monocytogenes, Shigella flexneri, and Rickettsia rickettsii (11, 27, 28, 29). Upon cell invasion, mycobacteria may thus be in contact with cytosolic components, including actin, which may be involved in epithelial cell trafficking. Here, we used SMFS to assess whether HBHA is capable of directly binding to actin, for which direct evidence has not yet been provided.
MATERIALS AND METHODS
Preparation of rHBHA derivatives.
Escherichia coli BL21(DE3) (pGD51) producing recombinant HBHA His-tagged at the N terminus (rHBHA N-His), was described elsewhere (4) and was kindly provided by G. Delogu. To obtain rHBHA C-His, pET-HBHAC was constructed as follows. The hbhA gene was amplified from genomic Mycobacterium bovis BCG DNA as a 371-bp fragment using the primers TCCGCTCGAGGCCGCGACTAGCCGG and ACCCAAGCTTTCAGTGGTGGTGGTGGTGGTGCTTCTGGGTGACCTTCTT, corresponding at the 3′ end of hbhA to a sequence encoding His6 residues. The amplicon was inserted into pCRIITopo (Invitrogen). A 406-bp XhoI fragment from this construct was then excised and used to replace the 495-bp XhoI fragment of pET-HBHA (22). The rHBHA N-His and rHBHA His-tagged at the C terminus (C-His) molecules were purified by heparin-Sepharose chromatography as described elsewhere (18).
Preparation of HBHA-modified surfaces and tips.
rHBHA N-His and rHBHA C-His were immobilized onto gold-coated supports and AFM tips, using the specific binding between the His tags and nitrilotriacetic acid (NTA)-terminated self-assembled monolayers (SAMs). AFM cantilevers (Microlevers, Veeco Metrology Group, Santa Barbara, CA) and silicon wafers (Siltronix, France) were coated using electron beam thermal evaporation with a 5-nm-thick Cr layer, followed by a 30-nm-thick Au layer. Before use, the gold-coated cantilevers and supports were rinsed with ethanol, dried with a gentle nitrogen flow and cleaned for 5 min by ultraviolet/ozone treatment (Jelight Co., Irvine, CA). They were immersed overnight in ethanol solutions containing 0.05 mM of NTA-terminated (20%) and tri(ethylene glycol[EG])-terminated (80%) alkanethiols (kindly supplied by N. L. Abbott; see reference 16 for details on the synthesis of these molecules) and rinsed with ethanol. Sonication was briefly applied to remove alkanethiol aggregates that may have been adsorbed. The SAM-coated surfaces were then immersed in 40 mM NiSO4 (pH 7.2) for 1 h and rinsed with phosphate-buffered saline (PBS). Finally, the samples were incubated in PBS containing 2 μg/ml His-tagged proteins for 2 h and further rinsed several times with PBS.
Preparation of actin-modified surfaces.
Actin was covalently immobilized onto SAMs of carboxyl-terminated alkanethiols. Cleaned gold surfaces were immersed overnight in ethanol solutions containing 1 mM of HS(CH2)10COOH (used as received) (Aldrich) and then rinsed with ethanol. Sonication was briefly applied to remove alkanethiol aggregates that may have been adsorbed. The SAMs were immersed for 30 min in a solution containing 20 mg/ml N-hydroxysuccinimide (NHS) (Sigma) and 50 mg/ml 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) (Sigma) and rinsed with water. The activated surfaces were then incubated with 0.1 mg/ml actin from bovine muscle (Sigma) in PBS for 2 h, rinsed with PBS, and kept in PBS solutions.
Validation of the modified surfaces.
The quality of the surface modifications described above was assessed by using X-ray photoelectron spectroscopy (XPS). Supports were rinsed with water and dried by flushing with a gentle nitrogen flow and then immediately introduced into the XPS vacuum chamber. The analyses were performed on a Kratos Axis Ultraspectrometer (Kratos Analytical, United Kingdom) equipped with a monochromatized aluminum X-ray source. The samples were fixed on a stainless steel multispecimen holder by using double-sided conductive tape. The angle between the normal axis of the sample surface and the electrostatic lens axis was 0°. The analyzed area was ∼700 μm by 300 μm. The constant pass energy of the hemispherical analyzer was set at 40 eV. The following sequence of spectra was recorded: survey spectrum, C1s, N1s, O1s, Au4f, S2p, and C1s, again to check the stability of charge compensation over time and the absence of degradation of the sample during the analyses. The binding energies were calculated with respect to the C-(C,H) component of the C1s peak of adventitious carbon fixed at 284.8 eV. Following subtraction of a linear baseline, molar fractions were calculated (CasaXPS program, Casa Software Ltd., United Kingdom) using peak areas normalized on the basis of acquisition parameters, sensitivity factors and the transmission function provided by the manufacturer.
AFM measurements.
AFM images and force-distance curves were obtained in PBS solution (10 mM PBS, 150 mM NaCl, pH 7.4) at room temperature, using a Nanoscope IV Multimode AFM (Veeco Metrology Group, Santa Barbara, CA). The bottom side of hydrated supports was quickly dried using precision wipes (Kimwipes; Kimberly-Clark), and the supports were then immobilized on a steel sample puck using a small piece of adhesive tape. The mounted samples were immediately transferred into the AFM liquid cell, while avoiding dewetting. SMFS measurements were performed with triangular-shaped silicon nitride cantilevers functionalized as described above. All curves were recorded with a maximum applied force of ∼400 pN. To estimate the spring constants of the cantilevers, we measured their geometrical dimensions using scanning electron microscopy, as well as their free resonance frequency. The cantilever mechanical properties were then adjusted in order to match the calculated frequencies with the measured frequencies. The determined mechanical properties and the measured geometrical dimensions were then used to calculate the spring constants, which were typically ∼0.01 N/m.
RESULTS
Preparation of AFM tips and supports.
To address HBHA-actin interactions at the level of single molecules, genetically engineered His-tagged HBHA derivatives were attached, via their C-terminal or N-terminal end, to gold-coated AFM tips modified with Ni2+-NTA- and tri(EG)-terminated alkanethiols, a method that allows proteins to be uniformly oriented at low density. Actin was covalently bound to model surfaces modified with COOH-terminated alkanethiols using NHS/EDC.
The quality of the functionalized surfaces was assessed by using XPS and AFM imaging. Consistent with previous work (31), XPS revealed that gold surfaces treated with NTA/EG-terminated alkanethiols and with COOH-alkanethiols showed significant oxygen and sulfur concentrations, reflecting the presence of alkanethiol monolayers. Incubation of the NTA/EG surface with Ni2+ and rHBHA C-His or rHBHA N-His leads to an increase in the nitrogen concentration, reflecting the presence of substantial amounts of protein on the surface (31). Treatment of COOH surfaces with NHS/EDC and actin also leads to the same conclusion.
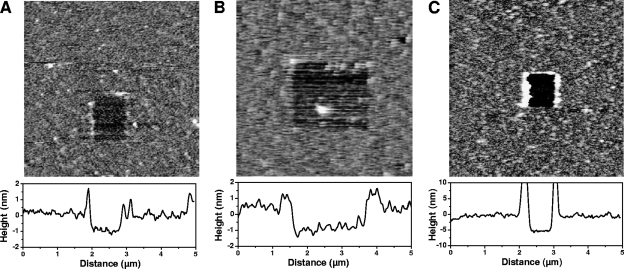
The protein-coated supports were further characterized using AFM topographic imaging in aqueous solution. Figures 1A and B show that surfaces modified with HBHA having either the N-terminal or the C-terminal end exposed were rather smooth and stable upon repeated scanning. To confirm the presence of protein layers, a small area was first recorded at large forces (>10 nN) for short periods of time, followed by imaging a larger portion of the same area under normal load. Imaging at large forces resulted in pushing the grafted material aside, thereby revealing the underlying support. The thickness of the removed films was found to be 1.0 ± 0.2 nm, confirming the presence of HBHA on the surface. Similar experiments were performed on actin-coated surfaces (Fig. 1C), demonstrating the presence of a film (5 ± 1 nm thick) on the surface. We also noted that performing scratching experiments on NTA/EG and COOH supports prior to peptide modification did not cause any morphological change, indicating that they resisted the large imaging forces.
FIG. 1.
Functionalization of surfaces with HBHA and actin. AFM topographic images (5 μm by 5 μm) recorded in PBS for NTA/EG-terminated supports after functionalization with HBHA having their N-terminal (A) or C-terminal (B) ends exposed, and for NHS/EDC-terminated supports after functionalization with actin (C). A small area was first scanned at large forces and high rates, followed by imaging of a larger image of the same region under small forces.
Force spectroscopy of the HBHA-actin interaction.
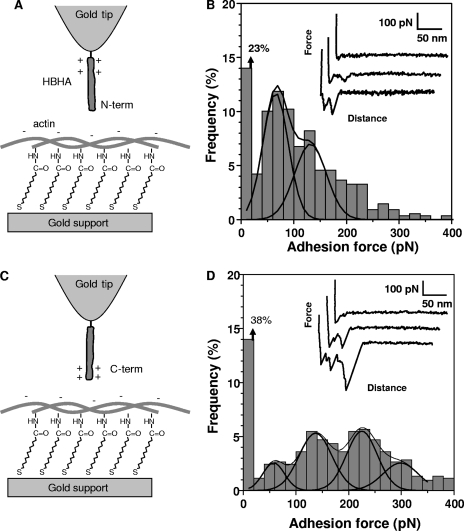
Having validated the functionalization strategies, force-distance curves were recorded between actin and the HBHA N-terminal region by using a pulling speed of 1,000 nm s−1 and an interaction time of 500 ms (Fig. 2A and B). A large fraction of the retraction curves (77%) displayed single adhesion peaks together with nonlinear elongation forces, yielding an adhesion force histogram with two maxima at 65 ± 3 pN and 131 ± 7 pN (as determined by Gaussian fits) (Fig. 2B). The specificity of the interaction was confirmed by showing a dramatic reduction of adhesion frequency (from 77% to 8%) when the measurements were performed with a BSA-coated tip instead of an HBHA-coated tip (data not shown).
FIG. 2.
Force spectroscopy of the HBHA-actin interaction. (A and C) Schematics of the surface chemistry used to functionalize model supports with actin and AFM tips with HBHA having their N-terminal (N-term) (A) or C-terminal (C-term) (C) ends exposed. (B and D) Representative force curves and adhesion force histograms (n = 512) recorded between actin and either the N-terminal domain (B) or the C-terminal domain (D) of HBHA. All curves were obtained using a retraction speed of 1,000 nm s−1 and an interaction time of 500 ms.
SMFS of the interaction between the lysine-rich, C-terminal domain of HBHA and actin (Fig. 2C) revealed a much broader distribution and a lower adhesion frequency (Fig. 2D). The adhesion histogram showed several maxima centered at 58 ± 6 pN, 137 ± 5 pN, 225 ± 8 pN, and 298 ± 25 pN. The observation of four maxima separated by 60 to 80 pN suggests that this value corresponds to the adhesion strength quantum between individual HBHA and actin molecules. To understand the measured forces, it has to be considered that the C-terminal region of HBHA has a cationic character, since it is very rich in cationic lysines (18). This property has been shown to mediate the interaction of the C-terminal domain with heparin (4, 6, 22). It is therefore tempting to attribute the 58-pN adhesion force to intermolecular electrostatic bridges between the cationic groups of the C-terminal domain and anionic residues of actin. Similar findings were reported for the homophilic interactions associated with the S-layer protein CbsA: the cationic C-terminal region showed larger adhesion forces than the N-terminal region, presumably reflecting intermolecular electrostatic bridges between the cationic lysines and anionic aspartates/glutamates from two interacting peptides (32).
Dynamics of the HBHA-actin interaction.
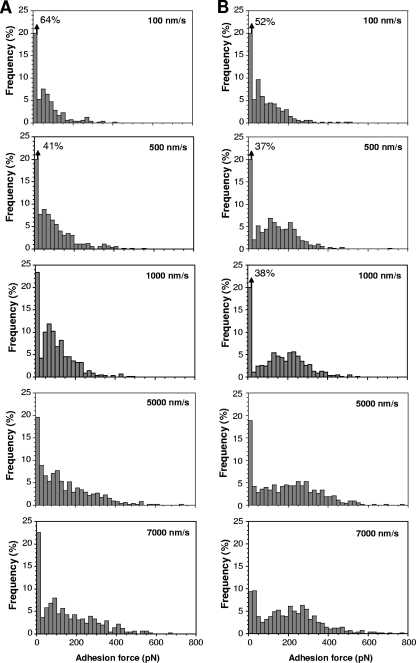
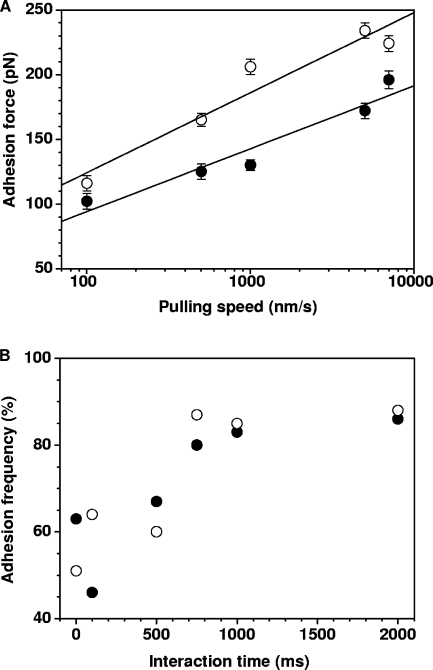
We next explored the dynamics of the HBHA-actin interaction by varying the pulling speed (Fig. 3). Consistent with results of earlier AFM molecular recognition studies (1, 3, 9, 10, 20, 31), we found that the HBHA-actin binding force increased linearly with the logarithm of the pulling speed, both for the N-terminal and C-terminal domains (Fig. 4A).
FIG. 3.
Dependence of the HBHA-actin interaction force on the pulling speed. Adhesion force histograms obtained in PBS between actin and HBHA having either their N-terminal (A) or C-terminal (B) ends exposed, using a pulling speed of 100 nm s−1, 500 nm s−1, 1,000 nm s−1, 5,000 nm s−1, and 7,000 nm s−1. Constant interaction time (500 ms) and approach speed (1,000 nm s−1) were used.
FIG. 4.
Dynamics of the HBHA-actin interaction. (A) Plot of the adhesion force measured between actin and HBHA having either their N-terminal (closed symbols) or C-terminal (open symbols) ends exposed, as a function of the logarithm of the pulling speed applied during retraction, while keeping the interaction time (500 ms) and the approach speed (1,000 nm s−1) constant. Data represent the mean of 512 measurements, as well as the standard error of the mean. (B) Plot of the binding frequency as a function of the interaction time measured at a constant approach and retraction speed of 1,000 nm s−1. Data represent the mean of 512 measurements.
The effect of the interaction time on the binding frequency (i.e., number of curves with adhesion events) was also examined, while keeping the pulling speed constant (6). The binding frequency of both regions reached a plateau corresponding to ∼90% binding probability after 1 s (Fig. 4B). Presumably, this fairly slow binding process reflects the prolonged interaction time required for conformational changes, allowing for optimal fitting between the filamentous HBHA and actin molecules.
DISCUSSION
Although many pathogenic mycobacteria have long been considered to be typical endosomal pathogens, evidence is now accumulating that at least some of them may escape the phagosome and gain access to the cytoplasmic milieu, where they are in contact with cytosolic components of the host cell (27, 28, 30). In some cases, actin rearrangement has been observed upon mycobacterial invasion of host cells, and certain mycobacteria, such as M. marinum, have been shown to be propelled by actin-based motility. However, the mechanism of direct or indirect interactions of intracellular mycobacteria with actin has not been elucidated yet. In particular, the mycobacterial components of this interaction have not yet been identified. We have previously shown that M. tuberculosis HBHA-coated beads are able to penetrate into epithelial cell layers and cause actin rearrangements (19), making this protein one of the prime candidates for mycobacterial interactions with actin. HBHA-like molecules are produced by all pathogenic mycobacteria, and their amino acid sequences and domain structures are well conserved (for a review, see reference 15).
In this study, we provide the first biophysical evidence for a direct binding of HBHA to actin. Strong single HBHA-actin interactions were demonstrated by using SMFS. The measured actin-binding forces were different for the N-terminal and the C-terminal HBHA domains, suggesting two different binding mechanisms, i.e., specific binding of the N-terminal coiled-coil structures and mere electrostatic interactions of the lysine-rich C-terminal domain of HBHA with actin. The binding force between either domain or actin increased with the pulling speed, and the binding frequencies increased with the contact time. These features strongly suggest that these forces reflect specific HBHA-actin interactions.
Recent structural observations have indicated that the M. tuberculosis HBHA has a homodimeric structure, which involves the N-terminal coiled-coil moiety of the protein (7). It is not known if in the cytosol of infected cells the HBHA dimers are dissociated to interact with actin. The single-molecule interactions shown here suggest that dissociated HBHA monomers interact with actin. Homotypic interaction forces of the N-terminal coiled-coil domain of HBHA (31) are similar to the forces of the HBHA-actin interaction measured here, suggesting that in the presence of actin, dissociation of HBHA dimers may indeed occur and that HBHA monomers then bind to actin. This hypothesis would deserve further investigation, e.g., by studying whether the actin-binding activity is changed when adding C-terminal, N-terminal, or whole HBHA monomers in the solution.
Interestingly, multiple sequence alignments of HBHA show significant sequence similarities with a number of different actin-binding proteins (Fig. 5). These proteins include tropomyosin 1, ezrin 1, and the heavy chain of myosin 9. Like HBHA, tropomyosin 1 is an elongated coiled-coil dimer present in both muscle and nonmuscle cells. However, the interaction of tropomyosin with actin has mostly been characterized in muscle cells (13). Ezrin 1 is an actin-binding protein that links the actin filaments to the cell membrane and is a member of the ERM (ezrin, radixin, and moesin) family. The actin-binding domain of ezrin 1 has been identified. It is a region rich in alpha helices (21) and indeed corresponds to the portion of the protein that is homologous to HBHA. Of note, ERM proteins have been implicated in the entry and cellular signaling cascades of many invasive bacterial pathogens (8, 12, 24, 25). Finally, myosin 9 also binds actin via its alpha helix-rich coiled-coil domain (26). The region of myosin 9 that is homologous to HBHA corresponds to the C-terminal domain known to bind to actin.
FIG. 5.
Multiple alignment of HBHA with different actin-binding proteins. Only the domains homologous to HBHA are shown. Residues identical between at least two aligned proteins are indicated in white in either black or gray boxes. Negative (−) and positive (+) charges conserved among at least three aligned proteins are indicated. Accession numbers for tropomyosin 1 (TPM1), ezrin 1 (EZR1), and myosin 9 (MYO9) are NP_001018007.1, P15311, and P35579, respectively.
These observations strongly suggest that HBHA-actin interactions play a role in the modulation of the cell cytoskeleton via direct binding to actin, and therefore perhaps in epithelial trafficking of Mycobacterium tuberculosis. This may provide a reason for the upregulation of the hbhA gene expression once M. tuberculosis has invaded the epithelial cell (5). Increased production of HBHA within the invaded cell may help the mycobacteria to more efficiently interact with actin, which may increase its capacity to move within the cell and be transcytosed from the apical to the basal side of epithelial cell layers. This indicates a bifunctional role of HBHA, as an epithelial cell adhesin and an intracellular actin-binding factor.
Acknowledgments
We dedicate this article to the memory of Franco D. Menozzi.
This work was supported by the National Foundation for Scientific Research (FNRS), the Foundation for Training in Industrial and Agricultural Research (FRIA), the Université Catholique de Louvain (Fonds Spéciaux de Recherche), the Federal Office for Scientific, Technical, and Cultural Affairs (Interuniversity Poles of Attraction Programme), the Région Wallonne, and the Research Department of the Communauté Française de Belgique (Concerted Research Action).
Y.F.D. is a Research Associate of the FNRS.
Footnotes
Published ahead of print on 3 October 2008.
REFERENCES
- 1.Baumgartner, W., P. Hinterdorfer, W. Ness, A. Raab, D. Vestweber, H. Schindler, and D. Drenckhahn. 2000. Cadherin interaction probed by atomic force microscopy. Proc. Natl. Acad. Sci. USA 974005-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bermudez, L. E., and F. J. Sangari. 2001. Cellular and molecular mechanisms of internalization of mycobacteria by host cells. Microb. Infect. 337-42. [DOI] [PubMed] [Google Scholar]
- 3.Bustanji, Y., C. R. Arciola, M. Conti, E. Mandello, L. Montanaro, and B. Samori. 2003. Dynamics of the interaction between a fibronectin molecule and a living bacterium under mechanical force. Proc. Natl. Acad. Sci. USA 10013292-13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delogu, G., and M. J. Brennan. 1999. Functional domains present in the mycobacterial hemagglutinin, HBHA. J. Bacteriol. 1817464-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delogu, G., M. Sanguinetti, B. Posteraro, S. Rocca, S. Zanetti, and G. Fadda. 2006. The hbhA gene of Mycobacterium tuberculosis is specifically upregulated in the lungs but not in the spleens of aerogenically infected mice. Infect. Immun. 743006-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupres, V., F. D. Menozzi, C. Locht, B. H. Clare, N. L. Abbott, S. Cuenot, C. Bompard, D. Raze, and Y. F. Dufrêne. 2005. Nanoscale mapping and functional analysis of individual adhesins on living bacteria. Nat. Methods 2515-520. [DOI] [PubMed] [Google Scholar]
- 7.Esposito, C., M. V. Pethoukov, D. I. Svergun, A. Ruggiero, C. Pedone, E. Pedone, and R. Berisio. 2008. Evidence for an elongated dimeric structure of heparin-binding hemagglutinin from M. tuberculosis. J. Bacteriol. 1904749-4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eugène, E., I. Hoffmann, C. Pujol, P. O. Couraud, S. Bourdoulous, and X. Nassif. 2002. Microvilli-like structures are associated with the internalization of virulent capsulated Neisseria meningitidis into vascular endothelial cells. J. Cell Sci. 1151231-1241. [DOI] [PubMed] [Google Scholar]
- 9.Evans, E., and K. Richie. 1997. Dynamic strength of molecular adhesion bonds. Biophys. J. 721541-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert, Y., M. Deghorain, L. Wang, B. Xu, P. D. Pollheimer, H. J. Gruber, J. Errington, B. Hallet, X. Haulot, C. Verbelen, P. Hols, and Y. F. Dufrêne. 2007. Single-molecule force spectroscopy and imaging of the vancomycin/d-Ala-d-Ala interaction. Nano Lett. 7796-801. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg, M. B. 2001. Actin-based motility of intracellular microbial pathogens. Microbiol. Mol. Biol. Rev. 65595-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goosney, D. L., R. De Vinney, and B. B. Finlay. 2001. Recruitment of cytoskeletal and signalling proteins to enteropathogenic and enterohemorrhagic Escherichia coli pedestals. Infect. Immun. 693315-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunning, P., G. O'Neill, and E. Hardeman. 2008. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol. Rev. 881-35. [DOI] [PubMed] [Google Scholar]
- 14.Hinterdorfer, P., and Y. F. Dufrêne. 2006. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods 3347-355. [DOI] [PubMed] [Google Scholar]
- 15.Locht, C., D. Raze, C. Rouanet, C. Genisset, J. Segers, and F. Mascart. 2008. The mycobacterial heparin-binding hemagglutinin, virulence factor and antigen useful for diagnostics and vaccine development, p. 305-322. In M. Daffé and J.-M. Reyrat (ed.), The mycobacterial cell wall. ASM Press, Washington, DC.
- 16.Luk, Y.-Y., M. L. Tingey, D. J. Hall, B. A. Israel, C. J. Murphy, P. J. Bertics, and N. L. Abbott. 2003. Using liquid crystals to amplify protein-receptor interactions: design of surfaces with nanometer-scale topography that present histidine-tagged protein receptors. Langmuir 191671-1680. [Google Scholar]
- 17.Menozzi, F. D., J. H. Rouse, M. Alavi, M. Laude-Sharp, J. Muller, R. Bischoff, M. J. Brennan, and C. Locht. 1996. Identification of a heparin-binding hemagglutinin present in mycobacteria. J. Exp. Med. 184993-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menozzi, F. D., R. Bischoff, E. Fort, M. J. Brennan, and C. Locht. 1998. Molecular characterization of the mycobacterial heparin-binding hemagglutinin, a mycobacterial adhesin. Proc. Natl. Acad. Sci. USA 9512625-12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menozzi, F. D., V. M. Reddy, D. Cayet, D. Raze, A.-S. Debrie, M.-P. Dehouck, R. Cecchelli, and C. Locht. 2006. Mycobacterium tuberculosis heparin-binding haemagglutinin adhesion (HBHA) triggers receptor-mediated trancytosis without altering the integrity of tight junctions. Microbes Infect. 81-9. [DOI] [PubMed] [Google Scholar]
- 20.Merkel, R., P. Nassoy, A. Leung, K. Ritchie, and E. Evans. 1999. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 39750-53. [DOI] [PubMed] [Google Scholar]
- 21.Niggli, V., and J. Rossy. 2008. Ezrin/radixin/moesin: versatile controllers of signaling molecules and of the cortical cytoskeleton. Int. J. Biochem. Cell. Biol. 40344-349. [DOI] [PubMed] [Google Scholar]
- 22.Pethe, K., M. Aumercier, E. Fort, C. Gatot, C. Locht, and F. D. Menozzi. 2000. Characterization of the heparin-binding site of the mycobacterial heparin-binding hemagglutinin adhesin. J. Biol. Chem. 27514273-14280. [DOI] [PubMed] [Google Scholar]
- 23.Pethe, K., S. Alonso, F. Biet, G. Delogu, M. J. Brennan, C. Locht, and F. D. Menozzi. 2001. The heparin-binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature 412190-194. [DOI] [PubMed] [Google Scholar]
- 24.Pistor, S., T. Chakraborty, U. Walter, and J. Wehland. 1995. The bacterial actin nucleator protein ActA of Listeria monocytogenes contains multiple binding sites for host microfilament proteins. Curr. Biol. 5517-525. [DOI] [PubMed] [Google Scholar]
- 25.Selbach, M., S. Moese, S. Backert, P. R. Jungblut, and T. F. Meyer. 2004. The Helicobacter pylori CagA protein induces tyrosine dephosphorylation of ezrin. Proteomics 42962-2968. [DOI] [PubMed] [Google Scholar]
- 26.Sellers, J. R. 2000. Myosins: a diverse superfamily. Biochim. Biophys. Acta 14963-22. [DOI] [PubMed] [Google Scholar]
- 27.Stamm, L. M., J. H. Morisaki, L.-Y. Gao, R. L. Jeng, K. L. McDonald, R. Roth, S. Takeshita, J. Heuser, M. D. Welch, and E. J. Brown. 2003. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J. Exp. Med. 1981361-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamm, L. M., M. A. Pak, J. H. Morisaki, S. B. Snapper, K. Rottner, S. Lommel, and E. J. Brown. 2005. Role of the WASP family proteins for Mycobacterium marinum actin tail formation. Proc. Natl. Acad. Sci. USA 10214837-14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens, J. M., E. E. Galyov, and M. P. Stevens. 2006. Actin-dependent movement of bacterial pathogens. Nat. Rev. Microbiol. 491-101. [DOI] [PubMed] [Google Scholar]
- 30.van der Wel, N., D. Hava, D. Houben, D. Fluitsma, M. van Zon, J. Pierson, M. Brenner, and P. J. Peters. 2007. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 1291287-1298. [DOI] [PubMed] [Google Scholar]
- 31.Verbelen, C., V. Dupres, D. Raze, F. Dewitte, C. Locht, and Y. F. Dufrêne. 2007. Single-molecule force spectroscopy of mycobacterial adhesin-adhesin interactions. J. Bacteriol. 1898801-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verbelen, C., J. Antikainen, T. K. Korhonen, and Y. F. Dufrêne. 2007. Exploring the molecular forces within and between CbsA S-layer proteins using single molecule force spectroscopy. Ultramicroscopy 1071004-1011. [DOI] [PubMed] [Google Scholar]