Abstract
Pentachlorophenol (PCP) is a toxic pollutant. Its biodegradation has been extensively studied in Sphingobium chlorophenolicum ATCC 39723. All enzymes required to convert PCP to a common metabolic intermediate before entering the tricarboxylic acid cycle have been characterized. One of the enzymes is tetrachloro-p-hydroquinone (TeCH) reductive dehalogenase (PcpC), which is a glutathione (GSH) S-transferase (GST). PcpC catalyzes the GSH-dependent conversion of TeCH to trichloro-p-hydroquinone (TriCH) and then to dichloro-p-hydroquinone (DiCH) in the PCP degradation pathway. PcpC is susceptible to oxidative damage, and the damaged PcpC produces glutathionyl (GS) conjugates, GS-TriCH and GS-DiCH, which cannot be further metabolized by PcpC. The fate and effect of GS-hydroquinone conjugates were unknown. A putative GST gene (pcpF) is located next to pcpC on the bacterial chromosome. The pcpF gene was cloned, and the recombinant PcpF was purified. The purified PcpF was able to convert GS-TriCH and GS-DiCH conjugates to TriCH and DiCH, respectively. The GS-hydroquinone lyase reactions catalyzed by PcpF are rather unusual for a GST. The disruption of pcpF in S. chlorophenolicum made the mutant lose the GS-hydroquinone lyase activities in the cell extracts. The mutant became more sensitive to PCP toxicity and had a significantly decreased PCP degradation rate, likely due to the accumulation of the GS-hydroquinone conjugates inside the cell. Thus, PcpF played a maintenance role in PCP degradation and converted the GS-hydroquinone conjugates back to the intermediates of the PCP degradation pathway.
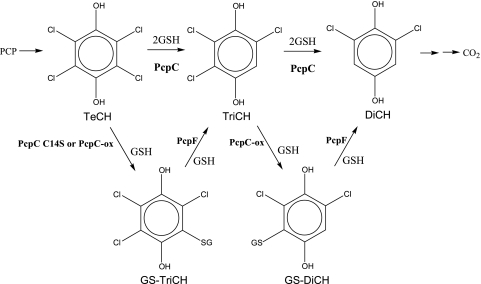
Pentachlorophenol (PCP) has been used mainly as a wood preservative and is a major environmental pollutant due to its high toxicity (4). Sphingobium chlorophenolicum ATCC 39723 mineralizes PCP to CO2, H2O, and HCl (15), and the complete metabolic pathway has been characterized (Fig. 1). PCP 4-monooxygenase (PcpB) oxidizes PCP to tetrachloro-p-benzoquinone, which can be reduced chemically by cellular reducing power or enzymatically by PcpD (quinone reductase) to tetrachloro-p-hydroquinone (TeCH) (5, 14, 18). TeCH reductive dehalogenase (PcpC) converts TeCH to trichloro-p-hydroquinone (TriCH) and then to 2,6-dichloro-p-hydroquinone (DiCH) (13, 19). DiCH 1,2-dioxygenase (PcpA) oxidizes DiCH to 2-chloromaleylacetate (12, 17), which is reduced by chloromaleylacetate reductase (PcpE) to maleylacetate and then to 3-oxoadipate (3). The latter is a common metabolic intermediate of aromatic compounds and can be channeled into the tricarboxylic acid cycle for complete mineralization. If all the enzymes function properly, no dead-end intermediates accumulate. However, PcpC, a glutathione (GSH) S-transferase (GST), is sensitive to oxygen (8). Its Cys-14 residue is readily oxidized by reactive oxygen species, and the oxidatively damaged PcpC (PcpC-ox) produces glutathionyl (GS) conjugates, GS-TriCH and GS-DiCH, which cannot be further metabolized by PcpC. Like PcpC-ox, the PcpC C14S mutant also converts TeCH to GS-TriCH and GS-DiCH (8). PcpC is easily oxidized during protein purification from S. chlorophenolicum, and the purified protein converts TeCH to GS-TriCH, GS-DiCH, and DiCH (19). Apparently, the purified protein from S. chlorophenolicum contains both PcpC and PcpC-ox. PcpC-ox converts TeCH to GS-TriCH and GS-DiCH conjugates, while PcpC converts TeCH to TriCH and then to DiCH. Since PcpC cannot further metabolize GS-TriCH and GS-DiCH, the conjugates accumulate in the reaction mixture (8). The production of the conjugates could also occur inside S. chlorophenolicum cells because oxidative stress is common to bacteria during aerobic growth. Thus, PcpC could be oxidatively damaged during aerobic degradation of PCP, and PcpC-ox would generate GS-TriCH and GS-DiCH, which could accumulate inside cells unless they were channeled back into the PCP degradation pathway. GS-TriCH and GS-DiCH could increase oxidative stress because they are substituted quinones, which undergo oxidation and reduction to generate superoxide and H2O2 (2). The genes physically associated with pcpC have been sequenced, and there is a putative GST gene (orf19) about 200 bp upstream of pcpC (3). In this report, we report the orf19 gene product as PcpF that converts GS-TriCH to TriCH and GS-DiCH to DiCH, respectively, channeling the conjugates back to the PCP degradation pathway.
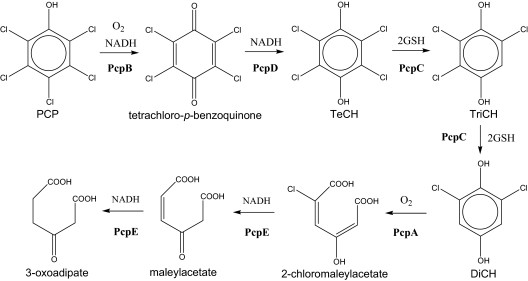
FIG. 1.
Pentachlorophenol degradation pathway of S. chlorophenolicum ATCC 39723. PcpB, PCP 4-monooxygenase; PcpD, tetrachloro-p-benzoquinone reductase; PcpC, TeCH reductive dehalogenase; PcpA, DiCH 1,2-dioxygenase; PcpE, chloromaleylacetate reductase; TeCH, tetrachloro-p-hydroquinone; TriCH, trichloro-p-hydroquinone; DiCH, 2,6-dichloro-p-hydroquinone.
MATERIALS AND METHODS
Chemicals and enzymes.
All chemicals were obtained from Aldrich Chemical Co. (Milwaukee, WI), Fisher Scientific Co. (Pittsburgh, PA), and Sigma Chemical Co. (St. Louis, MO). Catalase of bovine liver and GSH reductase of baker's yeast were obtained from Sigma Chemical Co. Restriction enzymes were obtained from New England Biolabs (Beverly, MA). PCR was performed with Taq DNA polymerase, and primers were purchased from Invitrogen (Carlsbad, CA).
Bacterial strains and culture conditions.
S. chlorophenolicum ATCC 39723 was cultured at 30°C in a mineral medium, with glutamate as the carbon and energy source (15). Escherichia coli strains TOP10 and BL21(DE3) were grown in Luria-Bertani (LB) medium or on LB agar plates at 37°C or as specified. Kanamycin (30 μg/ml) was added to the LB medium when required.
Gene cloning and protein expression.
Genomic DNA of S. chlorophenolicum was isolated with the PureGene DNA isolation kit (Gentra, Minneapolis, MN). The pcpF (orf19) gene and pcpC were amplified from the genomic DNA with primers GST2F/GST2R and PcpCF/PcpCR, respectively (Table 1). The pcpF PCR product was digested by NdeI and HindIII, and the pcpC PCR product was digested by NdeI and BamHI. The digested PCR products were ligated into the pET30-LIC vector (Novagen, Madison, WI). The ligation products were electroporated into E. coli BL21(DE3). Clones were confirmed by colony PCR and sequencing. The correct clones were directly used for protein production.
TABLE 1.
Primers used for gene cloning and mutation
| Primer | Sequencea |
|---|---|
| PcpCF | 5′-GAG GAT ATG CAT ATG CCT GAA GTC |
| PcpCR | 5′-CAT CTT CCG GAT CCC TAT TAA TCG |
| GST2F | 5′-AGT CAC CGA CAT ATG GGT CTT TTG |
| GST2R | 5′-TTC GAT GGG GAA GCT TTT TTG GTC |
| GST2Fi | 5′-AGG AAT CGC AAT ATC GCG |
| GST2Ri | 5′-ATG ACC GTG ATA GAT GGC |
| C14SF | 5′-ATT ACA CCA TGT CGA TCA GTT CGA TGA AGA CCC GT |
| C14SR | 5′-ACG GGT CTT CAT CGA ACT GAT CGA CAT GGT GTA AT |
| C53AF | 5′-TGC CGG TTT CGC CGC TCC CTG GGC GCA T |
| C53AR | 5′-ATG CGC CCA GGG AGC GGC GAA ACC GGC A |
| C248AF | 5′-TCT ATC ACG GTC ATT TCA AAG CCA ATC TGC GCC GGA TCG |
| C248AR | 5′-CGA TCC GGC GCA GAT TGG CTT TGA AAT GAC CGT GAT AGA |
Restriction sites are underlined.
Site-directed mutagenesis.
Site-directed mutagenesis was performed by using the Quick-Exchange kit from Stratagene (La Jolla, CA). The primers used for the conversions of C14S of PcpC as well as C53A and C248A of PcpF are listed in Table 1. The mutations were confirmed by sequencing, and the correct clones were transformed into E. coli BL21(DE3) for protein production.
Protein purification.
E. coli BL21(DE3) carrying pET30-pcpF was grown in 1 liter of LB medium at 37°C to a turbidity of 0.6 at 600 nm, induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside, and then incubated at room temperature for 5 h. All subsequent steps were performed at 4°C. The cells were harvested by centrifugation at 10,000 × g for 10 min and suspended in 10 ml of 20 mM potassium phosphate (KPi) buffer (pH 7.0). Freshly prepared phenylmethylsulfonyl fluoride in absolute ethanol was added to a concentration of 0.5 mM. The cells were disrupted by passing through a French pressure cell (model FA-030; Aminco, Urbana, IL) three times at 260 MPa. Cell debris was removed by centrifugation at 10,000 × g for 10 min, and membrane fraction was removed by ultracentrifugation at 183,960 × g (average) for 1 h. The supernatant was taken to 20% saturation of ammonium sulfate and centrifuged at 14,000 × g for 10 min. The supernatant was applied to a phenyl agarose (Sigma) column (12 by 1.5 cm) equilibrated with 20% saturation of ammonium sulfate in 20 mM KPi buffer (pH 7.0) containing 1 mM dithiothreitol (DTT). The column was washed with 30 ml of the equilibration buffer and then with a linear gradient of 20% to 0% saturation of ammonium sulfate in 200 ml of 20 mM KPi buffer containing 1 mM DTT. The protein was eluted around 10% saturation of ammonium sulfate. Fractions containing the targeted protein were pooled and precipitated with additional ammonium sulfate (to 70% saturation). The pellets were resuspended in 2 ml of 20 mM KPi buffer (pH 7.0) containing 1 mM DTT and dialyzed against the same buffer for 2 h. The sample was centrifuged to remove any precipitated proteins, and the supernatant was injected onto a 5-ml Econo-Pac High Q anion exchange column (Bio-Rad, Hercules, CA) equilibrated with the KPi buffer. Proteins were eluted by a gradient of 0 to 0.5 M NaCl (buffer A, 20 mM KPi [pH 7] with 1 mM DTT; buffer B, 20 mM KPi with 1 M NaCl and 1 mM DTT). Targeted protein was eluted off the column between 0.2 and 0.3 M NaCl, and fractions with the target protein were pooled and concentrated with Centriprep Ultracel YM-10 (Millipore, Bedford, MA). Glycerol was added to 10% before storage at −80°C.
The PcpC C14S mutant (PcpC C14S) was purified using the same procedures except that they were eluted off the agarose column between 20 and 15% saturation of ammonium sulfate and the High Q column between 0.15 and 0.25 M NaCl.
Enzyme assays.
PcpC and PcpC C14S activities were analyzed for the production of TriCH and GS-TriCH, respectively, in 40 μl of 70 mM KPi buffer (pH 6.5) containing 2 mM ascorbic acid, 200 μM TeCH, 5 mM GSH, and various amounts of PcpC or PcpC C14S. When small amounts of the enzymes were used and incubated for 1 or 2 min, TriCH and GS-TriCH were the primary products produced by the two enzymes, respectively. PcpF activity was assayed for the production of TriCH from GS-TriCH. PcpC C14S was used to convert TeCH to GS-TriCH and then heated at 65°C for 5 min to inactivate PcpC C14S. All assays were incubated at 30°C. The reactions were initiated by adding 1 μl of 200 mM GSH or 1 μl of an enzyme solution and stopped by adding 40 μl of a mixture of acetonitrile and acetic acid (9-to-1 ratio), centrifuged, and analyzed with high-performance liquid chromatography (HPLC) equipped with a C18 column and photodiode array detector, as previously described (19). The common GST activity was measured with 0.2 mM 1-chloro-2,4-dinitrobenzene as the substrate in 100 mM KPi buffer (pH 6.5) containing 0.2% Triton X-100, and continuous increase in absorbance at 340 nm was monitored (6).
GS-SG analysis.
The PcpF production of glutathione disulfide (GS-SG) from GS-TriCH and GSH was analyzed by using GSH reductase (Sigma) and NADPH. GS-TriCH (200 μM) produced by PcpC C14S was diluted to 50 μM with 20 mM KPi buffer containing 2 mM ascorbic acid and 5 mM GSH with a final volume of 1 ml, to which PcpF (30 μg) was added. GS-TriCH was completely converted to TriCH within 2 min as confirmed by HPLC analysis. Then, 300 μM NADPH and 6 μg of GSH reductase were added to determine the consumption of NADPH at 340 nm (6.22 mM−1 cm−1). One NADPH was used to reduce one GS-SG. GSH solutions contained some GS-SG, which was also estimated with the consumption of NADPH by GSH reductase and subtracted to obtain the GS-SG produced by PcpF.
Disruption of pcpF in S. chlorophenolicum.
A 600-bp internal fragment of pcpF was amplified by PCR using primers C14SFi and C14SRi (Table 1). The PCR product was cloned into pCR2.1-TOPO (Invitrogen). The plasmid was transformed into E. coli TOP10 competent cells and plated onto LB agar with kanamycin. Positive clones were identified by colony PCR, and plasmids were purified with the Qiagen plasmid purification kit (Qiagen, Germantown, MD). The plasmid (500 ng) was electroporated into 40 μl of competent S. chlorophenolicum ATCC 39723 cells as previously described (3). The integration of the plasmid into the chromosome of S. chlorophenolicum was selected by plating onto the mineral agar containing Na-glutamate (0.4%) and kanamycin (5 μg/ml). The pcpF disruption mutant was confirmed by colony PCR with primer pairs of GST2F/R and T7F/GST2R. T7F primed the sequence of the cloning vector pCR2.1-TOPO.
H2O2 treatment of PcpF.
Enzyme was treated by different concentrations of H2O2. One microliter of H2O2 at various concentrations was added to 8 μg of enzyme in 10 μl of KPi buffer, and the mixture was incubated at room temperature for 10 min. One microliter of catalase (26 μg/ml) was then added to terminate the reaction via incubation at room temperature for another 5 min. The treated enzyme was kept on ice and used within 1 h for activity measurements.
PCP degradation.
The S. chlorophenolicum wild type and pcpF disruption mutant were cultured in the mineral medium with glutamate to a turbidity of about 0.6 at 600 nm. The cultures were induced with 100 μM PCP and incubated with shaking until PCP was completely degraded. Cells were harvested and resuspended to a turbidity of 0.6 at 600 nm in the mineral medium with or without glutamate. Then, PCP was added to a target concentration of 100 μM PCP. The samples were incubated with shaking at 30°C, and the concentrations of PCP were detected on the basis of absorption at 320 nm with a UV-visible spectrophotometer.
PCP toxicity.
The S. chlorophenolicum wild type and pcpF disruption mutant were cultured in the mineral medium with glutamate to a turbidity of 1.0 at 600 nm. Serial 10-fold dilutions were done for each strain and then 10-μl portions of the dilutions were spotted onto the mineral agar plates containing Na-glutamate (0.4%) with various concentrations of PCP. Plates were incubated at 30°C for 1 week before taking pictures.
RESULTS
Overproduction and purification of PcpF and PcpC C14S.
The pcpF gene was cloned into the pET30-LIC vector and expressed in E. coli BL21(DE3). About 1.8 g of cells (wet weight) was obtained from 1-liter culture. A total of 88.9 mg of protein were recovered from cell extracts, and 27.3 mg of protein were obtained after phenyl agarose chromatography. PcpF was further purified by High Q chromatography, with a final yield of 17.2 mg. The purified protein generated a single band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown) with an apparent molecular mass of 35 kDa, agreeing with the calculated molecular mass of 35,383 Da. The pcpC gene was also cloned into the pET30-LIC vector, and a site-directed mutant pcpC C14S was generated. PcpC C14S was purified from E. coli BL21(DE3) cells to apparent homogeneity by ultracentrifugation, ammonium sulfate fractionation, phenyl agarose chromatography, and High Q chromatography.
Enzyme properties.
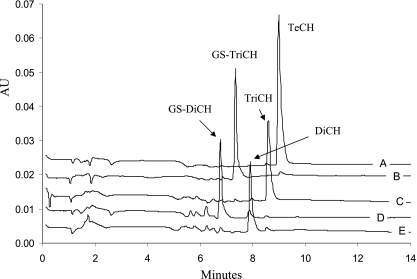
PcpC C14S was overproduced in E. coli. The cell extract after ultracentrifugation contained about 50% of the protein. We found that the partially purified PcpC C14S was sufficient to convert TeCH to GS-TriCH (Fig. 2A and B). After PcpC C14S reaction, the reaction mixture was heated at 65°C for 5 min to inactivate the enzyme. When purified PcpF was added, GS-TriCH was converted to TriCH (Fig. 2C). PcpF was inactivated at 65°C for 10 min, and the further addition of PcpC C14S produced GS-DiCH (Fig. 2D). After PcpC C14S inactivation by heating at 65°C for 5 min, the addition of PcpF converted GS-DiCH to DiCH (Fig. 2E). When PcpC C14S was coupled with PcpF, TeCH was converted to DiCH as was PcpC (data not shown). TeCH, GS-TriCH, TriCH, GS-DiCH, and DiCH were eluted off the HPLC column with retention times of 9.1, 7.3, 8.6, 6.8, and 8.0 min and characteristic absorption maxima of 306, 317, 302, 313, and 298 nm, respectively (Fig. 2). The specific activity of purified PcpF for TriCH production was estimated to be 3,829 ± 367 nmol min−1 mg−1 of protein (average and standard deviation of six samples). Since there was no apparent change in PcpF activities with GS-TriCH at concentrations ranging from 50 to 200 μM, the kinetic parameters were not determined, due to our HPLC assay requirement for relatively high substrate concentrations. The specific activity of purified PcpC C14S was 315 ± 19 nmol min−1 mg−1. The use of GSH by PcpF was confirmed by the formation of GS-SG. GS-SG was reduced by GSH reductase, and the consumption of NADPH was determined. For the conversion of 50 μM GS-TriCH to TriCH, 58 ± 4 μM GS-SG (average and standard deviation of three samples) was produced. PcpF did not transfer GSH onto 1-chloro-2,4-dinitrobenzene. The latter was not a total surprise, as not all GSTs use the compound as a substrate (16).
FIG. 2.
HPLC chromatograms of sequential reactions of TeCH transformation by PcpC C14S and PcpF. Reactions were carried out in 100 μl of 70 mM KPi buffer (pH 6.5) containing 2 mM ascorbic acid, 10 mM GSH, and 200 μM TeCH. (A) Reaction mixture only. A total of 20 μl of sample was removed, mixed with 20 μl of acetonitrile-acetic acid (9:1), centrifuged, and analyzed by HPLC. (B) Reaction mixture after the first PcpC C14S catalysis. Two microliters of PcpC C14S (ultracentrifuged cell extracts, 27.6 mg/ml) was added to the remaining 80 μl of reaction mixture, which was incubated at 30°C for 2 min and then heated at 65°C for 5 min before HPLC analysis of 20 μl of sample as described above. (C) Reaction mixture after the first PcpF catalysis and heat inactivation. One microliter of PcpF (2 mg/ml) was added to the remaining sample of reaction mixture B, which was incubated at 30°C for 2 min and then heated at 65°C for 10 min before HPLC analysis of 20 μl of sample as described above. (D) Reaction mixture after the second PcpC C14S (1 μl) catalysis and heat inactivation. (E) Reaction mixture after the second PcpF (0.5 μl) catalysis. HPLC data were extracted at 300 nm and exported in ASCII format and analyzed by Microsoft Excel. The chromatograms were separated by adding a constant of 0.005, 0.010, 0.015, 0.020, or 0.025 to the corresponding set of absorption unit (AU) data.
PcpC and PcpF activities did not change in S. chlorophenolicum wild-type cells after PCP induction of the pcp genes. PcpC activity was 2.5 ± 0.3 nmol min−1 mg−1 of protein in induced cell extracts and 2.2 ± 0.2 nmol min−1 mg−1 in uninduced cell extracts. PcpF activity was 0.41 ± 0.08 nmol min−1 mg−1 in induced cell extracts and 0.44 ± 0.07 nmol min−1 mg−1 in uninduced cell extracts. Hence, both pcpC and pcpF were constitutively expressed in S. chlorophenolicum, agreeing with previous reports about pcpC expression (3, 19). In the pcpF disruption mutant, the cell extracts did not show apparent PcpF activity after several minutes of incubation, while PcpC activity was unchanged at 2.6 ± 0.2 nmol min−1 mg−1. The results indicated that PcpF was the major, if not the only, enzyme responsible for channeling the conjugates back to the PCP degradation pathway in S. chlorophenolicum ATCC 39723. The pcpC and pcpF genes were not in a single operon, since there was no polar effect on PcpC activity in the pcpF disruption mutant. Furthermore, pcpC and pcpF were separated by 204 bp.
The Cys-14 residue of PcpC is readily oxidized during protein purification, and the damaged protein generates GS-TriCH and GS-DiCH conjugates (8). PcpF was also sensitive to oxidative damage. A 10-min exposure to H2O2 at 6.4 mM resulted in a 63% loss of PcpF activity. Cys residues are known to be oxidized by H2O2, and PcpF contained two Cys residues at positions 53 and 248. When site-directed mutagenesis was used to convert them to Ala residues, the PcpF C53A mutant completely lost its activity toward the conjugates, and PcpF C248A did not show any apparent change in its activity. The structure of PcpF was predicted on the basis of homologous protein structures at the Phyre website (http://www.sbg.bio.ic.ac.uk/phyre/) with GST iii from maize as the model (10). Cys-53 was located between the first β-sheet and first α-helix with the thiol in the substrate-binding pocket. The location indicates that it is likely to be involved in catalysis and its mutation should not affect the three-dimensional structure. Thus, Cys-53 of PcpF appeared to be involved in removing the GS moiety from the conjugates.
Effects of pcpF disruption in S. chlorophenolicum on PCP degradation and toxicity.
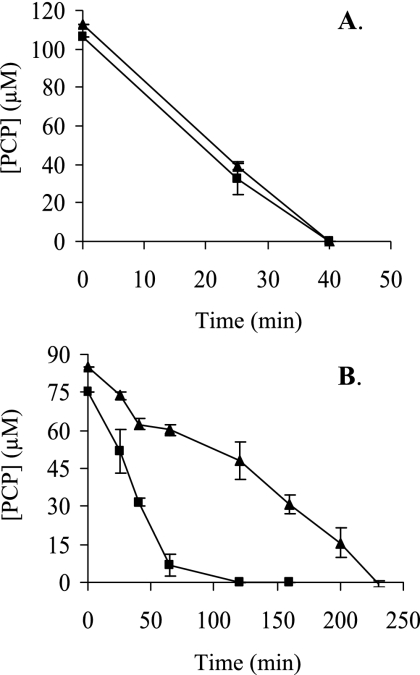
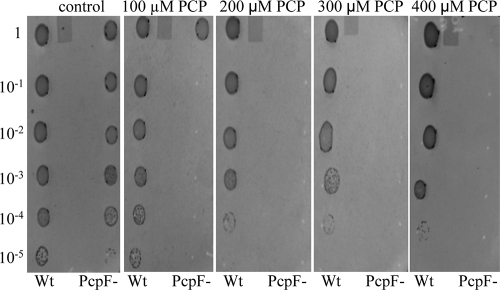
The pcpF gene was inactivated in S. chlorophenolicum. The disruption mutant was confirmed by colony PCR (data not shown). The effects of pcpF disruption on PCP degradation by S. chlorophenolicum were tested. Both the wild type and its pcpF disruption mutant degraded 100 μM PCP without any apparent difference when growing on glutamate. When the PCP-induced cells (with the pcp genes expressed) were harvested and resuspended in fresh mineral medium, the presence of glutamate made a difference for PCP degradation. The wild-type and mutant cells completely degraded 100 μM PCP within 40 min in the presence of glutamate (Fig. 3A). However, when the induced cells were suspended in the mineral medium without glutamate, the wild-type cells degraded 100 μM PCP in 1 h, while it took the mutant 4 h to complete the degradation (Fig. 3B). The possible toxicity of the unremoved GS-TriCH and GS-DiCH on S. chlorophenolicum was further tested by growing on agar plates containing different concentrations of PCP. The wild-type cells grew on glutamate plates with 400 μM PCP, while the disruption mutant cells displayed significant inhibition even with 100 μM PCP (Fig. 4).
FIG. 3.
PCP degradation by the S. chlorophenolicum wild type and pcpF mutant. Cells were growing on glutamate, induced with PCP, harvested, washed, and resuspended to an optical density at 600 nm of 0.6 in mineral medium with glutamate (A) and without glutamate (B). Immediately, PCP was added to a target concentration of 100 μM, and its degradations by the wild type (▪) and the pcpF mutant (▴) were measured.
FIG. 4.
PCP toxicity to the S. chlorophenolicum wild type and pcpF mutant. The wild type (Wt) and pcpF mutant (PcpF−) were cultured to an optical density at 600 nm of 1 and plated in serial 10-fold dilutions onto mineral agar plates containing glutamate and various PCP concentrations. The plates were incubated at 30°C for 7 days.
DISCUSSION
The PCP degradation pathway in S. chlorophenolicum has been extensively studied with the enzymes and genes identified and characterized (3). One of the enzymes, PcpC, as a TeCH reductive dehalogenase, is easily damaged by H2O2 (8). PcpC-ox converts TeCH into GS-TriCH and GS-DiCH conjugates, and those conjugates cannot be further metabolized by PcpC. The pcpF gene, next to pcpC on the chromosome of S. chlorophenolicum (3), codes for a putative GST. A BLASTP search reveals that PcpF is highly conserved in the Bacteria domain, including enterobacteria, proteobacteria, cyanobacteria, and firmicutes (gram positive). The top 100 hits have a high degree of homology to PcpF, with the first on the list, YqjG of Bradyrhizobium sp. strain BTAi1, displaying 63.7% identity and the last on the list, SPO3222 of Silicibacter pomeroyi DSS-3, showing 52.6% identity by global alignments (9). Since the homologous proteins are all hypothetical GSTs, the role of PcpF in PCP degradation is not apparent from sequence analysis. However, our biochemical and genetic analyses demonstrate that PcpF channels the GS-TriCH and GS-DiCH generated by PcpC-ox back to the PCP degradation pathway in S. chlorophenolicum.
The role of PcpF-catalyzed conversion of GS-TriCH and GS-DiCH to TriCH and DiCH was demonstrated by biochemical assays with purified PcpF. Although PcpC converts TeCH to TriCH and then to DiCH without the production of the conjugates, PcpC-ox and PcpC C14S produced GS-TriCH and GS-DiCH conjugates (8). When GS-TriCH and GS-DiCH were generated by PcpC C14S from TeCH and TriCH, PcpF converted them to TriCH and DiCH, respectively (Fig. 2). The reactions catalyzed by these enzymes were proposed as shown in Fig. 5. When PcpF was coupled with PcpC C14S, TeCH was quantitatively converted to DiCH. Since PcpF did not use TeCH or TriCH directly, the reactions were concerted actions of PcpC C14S and PcpF.
FIG. 5.
Proposed role of PcpF in the PCP degradation pathway. PcpC catalyzes two consecutive steps in PCP degradation. Both PcpC C14S and PcpC-ox catalyze the formation of GS-TriCH and GS-DiCH conjugates. PcpF channels the conjugates back to the PCP degradation pathway. GS-TriCH, glutathionyl-TriCH; GS-DiCH, glutathionyl-DiCH; PcpC-ox, oxidatively damaged PcpC; PcpF, GS-quinol lyase. Other abbreviations are defined in the legend for Fig. 1.
Both PcpC and PcpF are sensitive to reactive oxygen species. It is known that the Cys-14 residue of PcpC is the target for oxidation, and PcpC-ox produced GS-TriCH and GS-DiCH (8). PcpF has two Cys residues at positions of 53 and 248. Our site-directed mutagenesis demonstrated that Cys-53 was critical for the enzyme activity toward the conjugates, while Cys-248 was not. The Cys residues can be damaged by H2O2. During PCP degradation by S. chlorophenolicum, it is possible that only very small fractions of PcpC and PcpF are oxidatively damaged. PcpC-ox will produce very small amounts of GS-quinol conjugates, and the undamaged PcpF can convert them to TriCH and DiCH, back to the PCP degradation pathway (Fig. 5). The small fraction of oxidatively damaged PcpF is not functional, which should not cause any detrimental effects to the cells.
Genetic analysis further supports the maintenance role of PcpF during PCP degradation. The presence of another carbon and energy source made a major difference for PCP degradation by the pcpF mutant. The S. chlorophenolicum wild type and its pcpF disruption mutant both degraded 100 μM PCP at similar rates with glutamate, while the mutant significantly slowed down PCP degradation in the absence of glutamate. Since the cells were growing on glutamate when induced with PCP, glutamate was the major carbon and energy source for the cells. After harvested and resuspended in fresh medium without glutamate, the cells lost the major carbon and energy source. It is known that carbon-starved cells are under significant oxidative stress (11). Thus, it is likely that glutamate-starved cells contain more PcpC-ox molecules and consequently more GS-TriCH and GS-DiCH conjugates. For the wild type, PcpF returns them back to the PCP degradation pathway. For the pcpF mutant, the conjugates may accumulate inside the cells. The quinol moieties in the conjugates can undergo oxidation and reduction to generate more reactive oxygen species, toxic to the cells (2). The toxic effects of conjugates to the cells were further tested with plate assays containing various concentrations of PCP. The PCP degradation assay in liquid culture was a short-time exposure to PCP (Fig. 3), while the plate assay was a long-time exposure to PCP. Only small amounts of cells were spotted onto the agar surface, and the cell numbers were significantly reduced in higher dilutions. The few cells on the agar plate grew on glutamate and PCP simultaneously, and PCP could not be rapidly consumed due to low cell density. Even if GS-TriCH and GS-DiCH conjugates were produced at a very slow rate by a small fraction of PcpC-ox, they might be accumulated in the pcpF mutant cells. The accumulation of the conjugates was suggested as the pcpF mutant degraded PCP more slowly (Fig. 3B) and was more sensitive to PCP (Fig. 4).
PcpF is a GST, as indicated by its ability to catalyze GSH-dependent GS-hydroquinone lyase activity and by its sequence similarity with other GSTs. Another GST, LigG, has been shown to catalyze a similar lyase reaction, removing the GS moiety from α-GS-β-hydroxypropiovanillone (7). PcpF and LigG share only 20.1% sequence identity by global alignment (9). The Cys-15-Pro-16 sequence at the N terminus of LigG is aligned with the Cys-53-Pro-54 sequence at the N terminus of PcpF. The same sequence has been reported to play a catalytic role in a human omega class GST, GSTO1-1, with Cys-32 forming a disulfide bond with GSH and Pro-33 stabilizing the Cys thiolate (1). LigG and GSTO1-1 share 22.9% sequence identity, and PcpF and GSTO1-1 have 20.1% sequence identity. Although the sequence homology is not great among the three GSTs, both PcpF and LigG are likely to form disulfide bonds with the GS moiety from the conjugate substrates before the Cys-SH is regenerated with a GSH by a thiol-disulfide exchange reaction.
Acknowledgments
We thank Man-I Kuan for help with some experiments and Sara Belchik for suggestions in preparing the manuscript.
This research was funded by the United States National Science Foundation grant MCB-0323167 and the National Institute of Health grant R21-DK070274.
Footnotes
Published ahead of print on 26 September 2008.
REFERENCES
- 1.Board, P. G., M. Coggan, G. Chelvanayagam, S. Easteal, L. S. Jermiin, G. K. Schulte, D. E. Danley, L. R. Hoth, M. C. Griffor, A. V. Kamath, M. H. Rosner, B. A. Chrunyk, D. E. Perregaux, C. A. Gabel, K. F. Geoghegan, and J. Pandit. 2000. Identification, characterization, and crystal structure of the omega class glutathione transferases. J. Biol. Chem. 27524798-24806. [DOI] [PubMed] [Google Scholar]
- 2.Bolton, J. L., M. A. Trush, T. M. Penning, G. Dryhurst, and T. J. Monks. 2000. Role of quinones in toxicology. Chem. Res. Toxicol. 13135-160. [DOI] [PubMed] [Google Scholar]
- 3.Cai, M., and L. Xun. 2002. Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J. Bacteriol. 1844672-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crosby, D. G. 1981. Environmental chemistry of pentachlorophenol. Pure Appl. Chem. 531052-1080. [Google Scholar]
- 5.Dai, M., J. B. Rogers, J. R. Warner, and S. D. Copley. 2003. A previously unrecognized step in pentachlorophenol degradation in Sphingobium chlorophenolicum is catalyzed by tetrachlorobenzoquinone reductase (PcpD). J. Bacteriol. 185302-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Habig, W. H., M. J. Pabst, and W. B. Jakoby. 1974. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 2497130-7139. [PubMed] [Google Scholar]
- 7.Masai, E., A. Ichimura, Y. Sato, K. Miyauchi, Y. Katayama, and M. Fukuda. 2003. Roles of the enantioselective glutathione S-transferases in cleavage of β-aryl ether. J. Bacteriol. 1851768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarthy, D. L., S. Navarrete, W. S. Willett, P. C. Babbitt, and S. D. Copley. 1996. Exploration of the relationship between tetrachlorohydroquinone dehalogenase and the glutathione S-transferase superfamily. Biochemistry 3514634-14642. [DOI] [PubMed] [Google Scholar]
- 9.Myers, E. W., and W. Miller. 1988. Optimal alignments in linear space. Comput. Appl. Biosci. 411-17. [DOI] [PubMed] [Google Scholar]
- 10.Neuefeind, T., R. Huber, P. Reinemer, and J. Knaeblein. 1997. Cloning, sequencing, crystallization and X-ray structure of glutathione S-transferase-III from Zea mays var. mutin: a leading enzyme in detoxification of maize herbicides. J. Mol. Biol. 274577-587. [DOI] [PubMed] [Google Scholar]
- 11.Nyström, T. 2006. Oxidation of bacterial proteome in response to starvation. Methods Biochem. Anal. 4989-95. [DOI] [PubMed] [Google Scholar]
- 12.Ohtsubo, Y., K. Miyauchi, K. Kanda, T. Hatta, H. Kiyohara, T. Senda, Y. Nagata, Y. Mitsui, and M. Takagi. 1999. PcpA, which is involved in the degradation of pentachlorophenol in Sphingomonas chlorophenolica ATCC 39723, is a novel type of ring-cleavage dioxygenase. FEBS Lett. 459395-398. [DOI] [PubMed] [Google Scholar]
- 13.Orser, C. S., J. Dutton, C. Lange, P. Jablonski, L. Xun, and M. Hargis. 1993. Characterization of a Flavobacterium glutathione S-transferase gene involved reductive dechlorination. J. Bacteriol. 1752640-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orser, C. S., C. C. Lange, L. Xun, T. C. Zahrt, and B. J. Schneider. 1993. Cloning, sequence analysis, and expression of the Flavobacterium pentachlorophenol 4-monooxygenase gene in Escherichia coli. J. Bacteriol. 175411-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saber, D. L., and R. L. Crawford. 1985. Isolation and characterization of Flavobacterium strains that degrade pentachlorophenol. Appl. Environ. Microbiol. 501512-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheehan, D., G. Meade, V. M. Foley, and C. A. Dowd. 2001. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 3601-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xun, L., J. Bohuslavek, and M. Cai. 1999. Characterization of 2,6-dichloro-p-hydroquinone 1,2-dioxygenase (PcpA) of Sphingomonas chlorophenolica ATCC 39723. Biochem. Biophys. Res. Commun. 266322-325. [DOI] [PubMed] [Google Scholar]
- 18.Xun, L., and C. S. Orser. 1991. Purification and properties of pentachlorophenol hydroxylase: a flavoprotein from Flavobacterium sp. strain ATCC 39723. J. Bacteriol. 1734447-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xun, L., E. Topp, and C. S. Orser. 1992. Purification and characterization of a tetrachloro-p-hydroquinone reductive dehalogenase from a Flavobacterium sp. J. Bacteriol. 1748003-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]