Abstract
The 1,815,783-bp genome of a serotype M49 strain of Streptococcus pyogenes (group A streptococcus [GAS]), strain NZ131, has been determined. This GAS strain (FCT type 3; emm pattern E), originally isolated from a case of acute post-streptococcal glomerulonephritis, is unusually competent for electrotransformation and has been used extensively as a model organism for both basic genetic and pathogenesis investigations. As with the previously sequenced S. pyogenes genomes, three unique prophages are a major source of genetic diversity. Two clustered regularly interspaced short palindromic repeat (CRISPR) regions were present in the genome, providing genetic information on previous prophage encounters. A unique cluster of genes was found in the pathogenicity island-like emm region that included a novel Nudix hydrolase, and, further, this cluster appears to be specific for serotype M49 and M82 strains. Nudix hydrolases eliminate potentially hazardous materials or prevent the unbalanced accumulation of normal metabolites; in bacteria, these enzymes may play a role in host cell invasion. Since M49 S. pyogenes strains have been known to be associated with skin infections, the Nudix hydrolase and its associated genes may have a role in facilitating survival in an environment that is more variable and unpredictable than the uniform warmth and moisture of the throat. The genome of NZ131 continues to shed light upon the evolutionary history of this human pathogen. Apparent horizontal transfer of genetic material has led to the existence of highly variable virulence-associated regions that are marked by multiple rearrangements and genetic diversification while other regions, even those associated with virulence, vary little between genomes. The genome regions that encode surface gene products that will interact with host targets or aid in immune avoidance are the ones that display the most sequence diversity. Thus, while natural selection favors stability in much of the genome, it favors diversity in these regions.
Group A streptococcus ([GAS] Streptococcus pyogenes) causes a wide range of human diseases ranging from uncomplicated pharyngitis to life-threatening invasive disease. Acute post-streptococcal glomerulonephritis (APSGN) is one of the nonsuppurative sequelae that can occur following a GAS infection; the other common postinfection sequelae are rheumatic heart disease. Worldwide, it is estimated that approximately 470,000 cases of APSGN occur annually (23). Children and young adults are affected most commonly, with males having twice the incidence as females (74). By the 1940s, evidence was found that streptococcal skin infections were associated with APSGN, and these infections usually did not cause rheumatic fever, leading to the hypothesis that certain GAS strains were “rheumatogenic” while others were “nephritogenic” (41, 72). Further, divergent seasonal patterns of peak incidence exist separating nephritogenic and rheumatogenic GAS, with APSGN cases peaking in the late summer simultaneously with skin infections while rheumatogenic and throat infections reached the highest incidence in October (18). The study by Bisno and coworkers further demonstrated that during the summer peak of APSGN outbreaks, cases of rheumatic fever were virtually absent. Thus, clinical evidence strongly suggested the existence of a subpopulation of GAS that was adapted for colonization and infection of the skin and whose arsenal of virulence factors led to the onset of glomerulonephritis following infection.
Serological classification based upon the major surface M protein has been used to categorize GAS for well over half a century, and although strains causing glomerulonephritis are usually associated with certain M types, not all strains of a particular M type will have the potential to cause APSGN. Still, a correlation between M type, anatomical site of initial infection, and the appearance of APSGN has been observed. Nephritogenic strains of GAS associated with pyoderma and other skin infections tend to be associated most commonly with M antigen serotypes 2, 42, 49, 56, 57, and 60. By contrast, strains associated with rheumatic fever and throat infections are typically associated with M types 1, 4, 12, and 25 (17, 74, 78). These differences led to a classification of GAS into class I (throat isolates) or class II (skin isolates) types. Further work has demonstrated that these classifications can be refined to include specific genetic differences that include variations in the emm region, the FCT region associated with the T antigen or streptococcal pilus, the presence or absence of serum opacity factor (SOF), infection types, and differences in immunoglobulin G binding proteins (7, 10, 14, 33, 36, 49, 80, 86).
GAS strain NZ131 is a serotype M49 strain originally isolated from a clinical case of APSGN in New Zealand; NZ131 originally attracted attention because of its ability to undergo transformation at frequencies of up to 107 per microgram of plasmid DNA (75). Such frequencies are in contrast to many GAS strains that are often difficult to manipulate genetically. The ability of strain NZ131 to cause nephritis symptoms similar to those observed in humans in an animal model system has also been demonstrated (65, 66). Thus, its pathogenic potential and usefulness for genetic manipulation have led to the widespread use of strain NZ131 as a model organism in research investigations by a number of laboratories (2, 5, 22, 24-31, 34, 35, 37, 40, 42, 49, 53, 56-59, 62-66, 68, 70, 77, 84). Thus, the presented nucleotide sequence of the NZ131 genome will facilitate these ongoing as well as future studies.
To date, the genome sequences of 12 strains of GAS have been determined, representing strains of serotypes M1, M2, M3, M4, M5, M6, M12, M18, and M28; the genomes of two separate strain isolates have been determined for serotypes M1, M3, and M12 (Table 1). Genetic variation is essential to survival for all organisms, and each of these GAS genomes reveals that the endogenous prophages are major sources of diversity for this bacterium. However, in addition to prophages, additional unique genetic material can be found in GAS genomes that could promote fitness or survival of this organism. An illustration of this point is a cluster of genes found in the strain NZ131 genome (nudABC) as part of the emm49 pathogenic region that is apparently unique to M49 and M82 strains. Thus, the sequencing of this M49 serotype strain has provided not only complementary information to the many previous studies using NZ131 that allows them to be placed in a broader context but also new information that will open up additional investigations to shed light upon the genetic basis for the human disease caused by this and related GAS strains.
TABLE 1.
Summary of sequenced S. pyogenes genomes to date
| Strain | Size (bp) | M type | No. of prophages | Accession no. | Reference or source |
|---|---|---|---|---|---|
| SF370 | 1,852,441 | 1 | 4 | AE004092 | 41 |
| MGAS5005 | 1,838,554 | 1 | 3 | CP000017 | 71 |
| MGAS10270 | 1,928,252 | 2 | 5 | CP000260 | 11 |
| MGAS315 | 1,900,521 | 3 | 6 | AE14074 | 12 |
| SSI-1 | 1,894,275 | 3 | 6 | BA000034 | 66 |
| MGAS10750 | 1,937,111 | 4 | 4 | CP000262 | 11 |
| Manfredo | 1,841,271 | 5 | 5 | AM295007 | 51 |
| MGAS10394 | 1,899,877 | 6 | 8 | CP000003 | 4 |
| MGAS2096 | 1,860,355 | 12 | 2 | CP000261 | 11 |
| MGAS9429 | 1,836,467 | 12 | 3 | CP000259 | 11 |
| MGAS8232 | 1,895,017 | 18 | 5 | AE009949 | 80 |
| MGAS6180 | 1,897,573 | 28 | 4 | CP000056 | 47 |
| NZ131 | 1,815,783 | 49 | 3 | CP000829 | This work |
MATERIALS AND METHODS
Bacterial strains.
GAS strain NZ131 was originally isolated from a case of APSGN and was provided by Diana Martin, New Zealand Communicable Diseases Center, Porirua, New Zealand (75). For screening the nudABC gene cluster, 60 GAS strains from the collection of the University of Oklahoma Health Sciences Center covering a range of serotypes were selected (serotypes M1, M2, M3, M4, M6, M9, M11, M12, M22, M25, M28, M29, M49, M53, M58, M60, M82, M87, M103, M107, M111, and M118). These M49 strains were isolated from cases of APSGN, scarlet fever, or pharyngitis. In addition to these strains, a second group of 22 different M49 GAS strains was kindly provided by Bernard Beall from the CDC collection and were invasive strains collected in the United States during a six-year period from 2000 to 2006 (see Table S1 in the supplemental material). For strains that had not been previously typed by serology in the collection, M protein typing was done by PCR amplification of the variable nucleotide regions of the emm genes as previously described (6), sequencing of the PCR products, and comparison to known emm alleles. Twenty-two relatively conserved signal sequence residues and the first 83 residues of the mature M proteins were used for phylogenetic analysis. The emm sequence data for known strains were obtained from the S. pyogenes emm sequence database (Division of Bacterial and Mycotic Diseases, Centers for Disease Control and Prevention; http://www.cdc.gov/).
Genome sequencing and annotation.
DNA isolation, library construction, DNA sequencing, assembly, and final editing were done as previously reported (1, 39). The complete genome sequence has been deposited in the GenBank database under accession number CP000829. The GAS strain is available through the American Type Culture Collection (ATCC BAA-1633). The initial annotation was done with the assistance of Ross Overbeek, National Microbial Pathogen Data Resource. The software package BASys (83) and in-house perl scripts were used for construction of the circular genome map.
Genome and sequence analysis.
Genome comparisons were done using MUMmer (54), and multiple DNA alignments were done using CLUSTAL W and Base-by-Base software (21, 82). Correspondence analysis of codon and amino acid usage to calculate the codon adaptation index (CAI) (73) of the NZ131 coding regions was performed using CodonW (http://codonw.sourceforge.net/). Gram-positive signal peptide prediction was performed using SignalP (8). Clustered regularly interspaced short palindromic repeat (CRISPR) region analysis of the NZ131 genome was done using the University of Paris-Sud 11 online CRISPRfinder program (http://crispr.u-psud.fr/Server/CRISPRfinder.php) (46). GAS multilocus sequence typing (MLST) was performed as previously described (36) and using the online S. pyogenes database query tool (http://spyogenes.mlst.net). FCT region and emm pattern analysis were done using previously established criteria (13, 51). Invasive and noninvasive M49 strains were used to survey for the presence of the NZ131 prophage integrases by PCR (see Table S2 in the supplemental material). PCR was performed using genomic DNA as templates under a condition of 25 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 60 s. PCR products were separated on a 2% agarose gel and visualized after ethidium bromide staining.
Analysis of nudABC in S. pyogenes strains.
To detect the nudA, nudB, and nudC genes in S. pyogenes strains, the primer pairs listed in Table S2 in the supplemental material were used to amplify internal regions of nudA, nudB, or nudC (303, 355, and 626 bp, respectively). PCR was performed using genomic DNA as templates as described above.
For transcript mapping of nudABC and their surrounding genes, PCR was performed using cDNA and primers that would amplify regions from adjoining genes (see Table S3 in the supplemental material). Total RNA (20 μg) was isolated from either SF370 (an nudABC mutant) or NZ131 (nudABC+) and reverse transcribed using the SuperScript II system (Invitrogen) for first-strand cDNA synthesis. For a reaction, 500 ng of random hexamers (Invitrogen) was mixed with the RNA in a total volume of 12 μl and heated to 70°C for 10 min. After the mixture was cooled to 25°C within 10 min, the reaction buffer was added according to the manufacturer's recommendations. After incubation at 25°C for 10 min, 1,800 U of SuperScript II was added to the reaction mixture and heated to 42°C within 10 min and incubated for 50 min. SuperScript II was heat inactivated at 70°C for 15 min, and the mixture was cooled to 4°C. RNA was removed using 2 U of RNase H (Invitrogen) and 5 μg of RNase A (Epicentre Technologies) for 10 min at 37°C in a 60-μl total volume. The cDNA was purified using the QiaQuick PCR purification kit (Qiagen). The concentration of cDNA was determined by the absorption at 260 nm. PCR was performed using primers that were positioned to amplify a region that included part of each adjoining open reading frame (ORF) and the separating intergenic sequence. Parallel reactions were employed using either chromosomal DNA (positive control) or purified RNA (negative control) for the reaction template. Amplified fragments were separated by agarose gel electrophoresis and visualized following ethidium bromide staining. The identities of the amplified sequences were confirmed by DNA sequencing.
RESULTS
Overview of the genome sequence of strain NZ131.
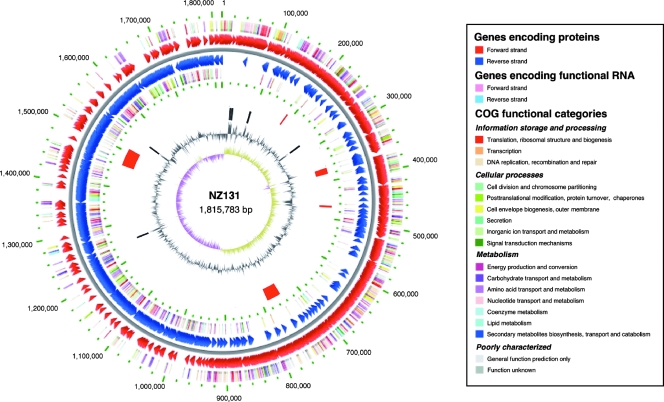
The genome of strain NZ131 is a single circular DNA molecule of 1,815,783 total bases (Fig. 1) and is the smallest S. pyogenes genome sequenced to date (Table 1). There are 1,699 probable protein-encoding ORFs that use 1,548,919 bases so that 85.3% of the genomic DNA is used as coding sequences. The base composition of the ORFs is 39.18% G+C while the composition of the total genome is 38.57%; both values are similar to the composition seen in the other completed GAS genomes. The multilocus sequence analysis indicates that the strain is MLST type 30 as are some other M49 strains found in the S. pyogenes MLST database (http://spyogenes.mlst.net). Six ribosomal gene clusters are present and occupy the same conserved locations noted in the other GAS genomes. A single identifiable riboswitch is positioned next to the gene for xanthine phosphoribosyltransferase (9); this element is universally conserved in all of the sequenced GAS genomes. The overall genome arrangement is colinear with the M1 genome strain and does not show the inversion around the origin of replication and terminus that is seen in genome strains Manfredo and SSI-1. As is the case with all sequenced strains, prophages and insertion (IS) elements make up much of the observed genetic diversity although strain NZ131 has unique genetic characteristics that may help define M49 strains. Predicted signal peptides were found in 195 of the encoded proteins.
FIG. 1.
Circular representation of the S. pyogenes strain NZ131 genome. Outer circle shows COG functional categories of coding regions in the clockwise direction. The lines in each concentric circle indicate the position of the represented feature; the color key is show to the right of the map. The second circle shows predicted coding regions transcribed on the forward (clockwise) DNA strand. The third circle shows predicted coding regions transcribed on the reverse (counterclockwise) DNA strand. The fourth circle shows COG functional categories of coding regions in the counterclockwise direction. The fifth circle shows mobile genetic elements and bacteriophage genomes (red). The sixth circle shows rRNA operons (dark gray). The seventh and eighth circles show the percent G+C content of the sequence and the percent G+C deviation by strand, respectively.
Codon usage analysis (see Fig. S1 in the supplemental material) showed that while codon triplets tended to be adapted to the overall percent G+C content of the genome (average codon G+C content, 38.7%), the third position tended to vary from this average and favored synonymous codons with either A or T in the third position (third position average G+C content, 29.2%). The CAI provides insight into how well adapted codon usage is for a given ORF. A high CAI value reflects the optimized use of the organism's preferred codons and is characteristic of highly expressed genes (73). Conversely, low CAI values may indicate genetic material that has been recently acquired and has not undergone codon optimization by natural selection. The calculation of the CAI for the NZ131 predicted ORFs showed that 78.2% of the ORFs had CAI values that fell within 1 standard deviation of the mean CAI of 0.4411 (see Fig. S1 in the supplemental material). As might be expected, genes encoding ribosomal proteins, glycolytic and other metabolic enzymes, and enzymes involved in transcription and translation mainly comprised the group with the highest CAI values (>2 standard deviations). Interestingly, this group of potentially highly expressed genes included several encoding hypothetical proteins, including Spy49_0729, a hypothetical protein that is conserved in many streptococcal species.
Genome prophages and other notable mobile genetic elements.
The role played by GAS prophages as vectors of virulence genes is well established, with examples provided by every sequenced genome. Three distinct prophage genomes were found in strain NZ131, occupying 101,578 bases of the bacterium's genome (5.6%) and encoding 112 predicted ORFs. Two of these prophages appear to have complete genomes while the third lacks many typical phage genes and is presumably a remnant that has undergone extensive deletion. The genetic arrangement of the two complete prophage genomes is typical and similar to the previously identified GAS prophages, with identifiable modules for integration and lysogeny, DNA replication, DNA packaging, structural proteins, host cell lysis, and virulence. However, as with all of these viral genomes, each contains unique genetic material. The absence of prophages at many of the commonly used bacterial attachment sites (attB) may reflect a certain genetic isolation or different selective pressures of these strains. For example, no prophage is integrated at the frequently used histone-like protein attachment site in contrast to the genomes of strains SF370, MGAS10750, SSI-1, MGAS8232, MGAS315, MGAS5005, Manfredo, and MGAS10394. The tmRNA gene that is the attachment site for bacteriophage T12 is also unoccupied in strain NZ131 although it is fully capable of participating in site-specific integration, as has been demonstrated using recombinant constructs (58, 60). Further, no prophage is integrated into the 5′ end of the DNA mismatch repair gene mutL ORF, which occurs in 6 of the 12 published genomes.
Prophage NZ131.1.
Prophage NZ131.1 appears to be a genetic remnant of a unique prophage that is not found in any of the other GAS genomes sequenced to date. It encompasses 16,182 bp with a 35.27% G+C content and encodes 23 predicted ORFs. The integration site is near the promoter of hypothetical protein Spy49_0371, but the attB duplication cannot be determined since the distal part of the phage genome and the attachment site apparently have been deleted. The surviving prophage DNA lacks genes for structural or lysis proteins, and no genes encoding recognizable virulence factors are observed. However, there are many genes encoding hypothetical proteins, and thus the possibility that one is associated with pathogenesis cannot be eliminated. Its presence in M49 strains seems to be sporadic since only 7 out of 38 M49 clinical isolates analyzed were positive for the prophage integrase gene (see Table S4 in the supplemental material). Therefore, while it is a prophage remnant, it does not seem to be a long-standing genetic element that is characteristic for M49 strains.
Prophage NZ131.2.
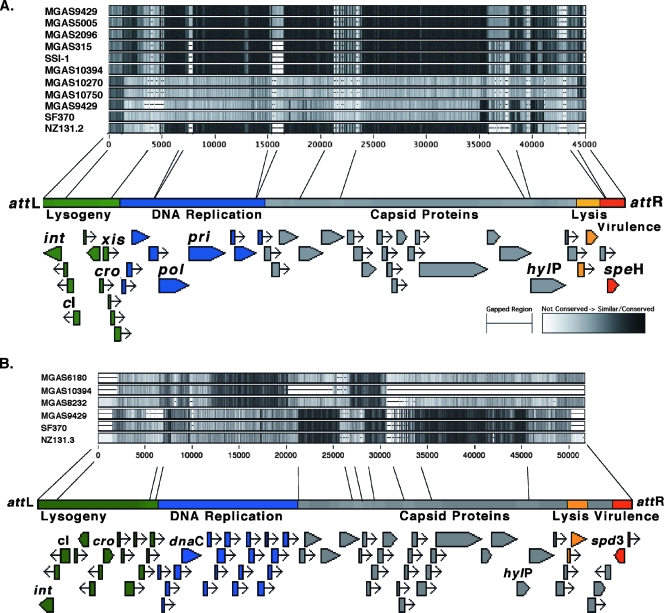
Prophage NZ131.2 has a 37,895-bp genome of 39.29% G+C content that encodes 45 ORFs. It is integrated into the 3′ end of the gene for dTDP-glucose 4,6-dehydratase such that normal coding of this gene is maintained. This attachment site is also used by a diverse group of prophages found in the Manfredo, SF370, MGAS10750, MGAS10270, and MGAS9429 genome strains. Accordingly, prophage NZ131.2 shares a nearly identical integrase gene with these prophages although the remainder of its genome is quite distinct and contains extended homologous regions with prophages from GAS strains MGAS9429, MGAS5005, MGAS2096, MGAS315, MGAS10394, and SSI-1 (Fig. 2). All of these prophages encode a phage DNA polymerase and at least one virulence-associated gene although no virulence gene is common to all. Prophage NZ131.2 encodes the virulence factor gene for streptococcal pyrogenic exotoxin H (speH). Prophages occupying this attachment site are possibly common in serotype M49 strains since 25 out of 38 clinical isolates were positive for the associated integrase gene (see Table S4 in the supplemental material).
FIG. 2.
Prophages NZ131.2 (A) and NZ131.3 (B). The genetic maps of the two complete prophage genomes found in strain NZ131 are shown. Above each is a multiple alignment of the NZ131 phage genome with the genome prophages that contain significant regions of homology. The predicted ORFs for each prophage are shown below the linear map and are color coded by probable functional region: lysogeny (green), DNA replication (blue), packaging and structural genes (gray), lysis (yellow), and virulence (red). Known genes are indicated next to their ORFs.
Prophage NZ131.3.
The third large prophage in the NZ131 genome, prophage NZ131.3 with 47,501 bp, is integrated into 5′ end of the conserved hypothetical protein Spy49_1532, separating it from its normal promoter. This particular attachment site, not previously observed in GAS strains, is identified by the sequence duplications flanking the prophage (5′-ATATGATGAATATGC-3′). The effect on host gene Spy49_1532 resulting from prophage integration into its promoter is unknown, but its expression may be altered or repressed, as in the case of the DNA mismatch repair gene mutL in GAS strain SF370 that is controlled by prophage excision in response to growth (71). The prophage genome has 37.96% G+C content and encodes 67 ORFs, including the virulence-associated streptodornase (spd3) and the paratox genes (3) as well as hyaluronidase. Prophage NZ131.3, while sharing DNA modules with previously discovered GAS genome phages, is less clearly related to any group and contains large regions of unique DNA sequence. Therefore, members of this prophage family may be genetically isolated from some other serotypes of GAS. Previous studies suggest that historic class II streptococci are associated with carriage of specific alleles of extoxins and not others (11); it may be that specific proteins, acting as surface receptors for phage attachment, limit the distribution of some phages. It is uncertain whether this prophage is present in the incomplete M49 S. pyogenes strain 591 whole-genome shotgun sequence (accession number AAFV01000000) since none of the phage-associated sequences or the flanking bacterial genome regions are present in the database. However, the presence of prophages at this attachment site may be sporadic in M49 strains since only 3 out of 38 were positive for the integrase gene (see Table S4 in the supplemental material).
IS elements.
A number of complete IS elements or fragments are found in the NZ131 genome, including several that appear to be common to most or all of the sequenced GAS genomes (for example, the tandem IS861 elements found in all genomes). Two copies of IS1161 are found, and one has inserted into the maltodextrose operon, deleting 7 of the 13 genes that compose this operon. One mobile element, encompassing genes Spy49_0166 to Spy49_0170, may be a transposon that encodes several proteins of unknown function including a potential transcription factor related to the mga global regulator of GAS. This particular mobile element has also been found in the SF370, MGAS6180, MGAS5005, MGAS9429, MGAS10270, and MGAS2096 genomes, and its frequent appearance suggests that it may confer selective value upon its hosts, perhaps even influencing virulence. The most interesting mobile element found in the genome is located in the cluster of virulence genes surrounding the emm gene encoding the major antiphagocytic M protein. As discussed below, a Nudix hydrolase may have been acquired as part of a complex transposon containing an IS861 element.
CRISPR elements.
CRISPR elements are repeat structures found in many prokaryotic genomes, including streptococci, and are composed of both tandem and interspaced repeats. A CRISPR locus is characterized by a succession of 21- to 47-bp direct repeat (DR) sequences separated by unique sequences of a similar length (spacers), giving a structure of the following type: DR1-spacer1-DR2-spacer2…DRn-1-spacern-1-DRn, where n is the number of repeats.
The spacers often are sequences derived from bacteriophage genomes, and the expression of CRISPR is thought to create anti-phage mRNA, interfering with the genetic program of the lytic cycle (4, 20, 61). Two predicted CRISPR regions are found in the NZ131 genome, one beginning at 827,277 bp and the other at 1,201,583 bp, and contain four and five spacers, respectively (Table 2). While several spacers have no known homologs, others contain prophage sequences that are associated with the lytic cycle and that have previously been identified by genome sequencing. These CRISPR elements provide a glimpse of previous encounters with bacteriophages in the evolutionary past by NZ131, and if indeed these elements represent an anti-phage mechanism in GAS, they suggest that certain prophage-associated genetic elements are restricted from contributing to future new phage infections in this strain.
TABLE 2.
CRISPR regions in the NZ131 genomea
| CRISPR region | Position in NZ131 (length [bp]) | Sequence | Homologous gene | Gene source (strain) | ORF |
|---|---|---|---|---|---|
| Element 1 | |||||
| DR | 827277-827577 (300) | GTTTTAGAGCTATGCTGTTTTGAATGGTCCCAAAAC | |||
| Spacer | 827313-827342 | AGCTGCATCGCTGTAGTATTTACCAATATA | Phage protein | MGAS10750 | Spy1302 |
| 827379-827409 | ACTGGGAAATTGATAAAATCGGCAATGCCCG | Phage endopeptidase | MGAS2096 | Spy0592 | |
| 827436-827475 | CAAGTTGTTTAATCGAAGAATTTCCCGTTG | Unannotated phage ORF | MGAS10270 | ||
| 827512-827541 | GTGCCTGTGGAGGAATTGATGAACATGCCT | Unknown | |||
| Element 2 | |||||
| DR | 1201583-1201947 (364) | ATTTCAATCCACTCACCCATGAAGGGTGAGAC | |||
| Spacer | 1201615-1201649 | AAGATGACACTGGTACTGCACATGTCGATTTAAAA | Unknown | ||
| 1201682-1201716 | AGATCTTAAAATGGATTCTTCTTCAGATATTTTTG | Unknown | |||
| 1201749-1201783 | GATAACAGTTGCTTTAGTCGATAAGTCGATTAGCG | Phage protein | SF370 | Spy1454 | |
| 1201816-1201850 | ATTATGTTTTGCCACATGAGAAAGTAAAAAATGGA | Phage protein | MGAS10394 | M6_Spy1540 | |
| 1201883-1201915 | TCCCTTATAATCGACAAAAAGCGCCGATTGATT | Hyaluronoglucosaminidase | MGAS10394 | M6_Spy1550 |
Two predicted CRISPR elements are found in the NZ131 genome, one beginning at 827,277 bp and the other at 1,201,583 bp. The DR consensus sequence of each CRISPR region and the spacer sequences for each are shown. Spacers whose sequences are part of known GAS genes have the homologous region identified by strain and ORF. In all cases, prophage genes are the source of these identifiable spacer sequences.
Virulence-associated hypervariable regions.
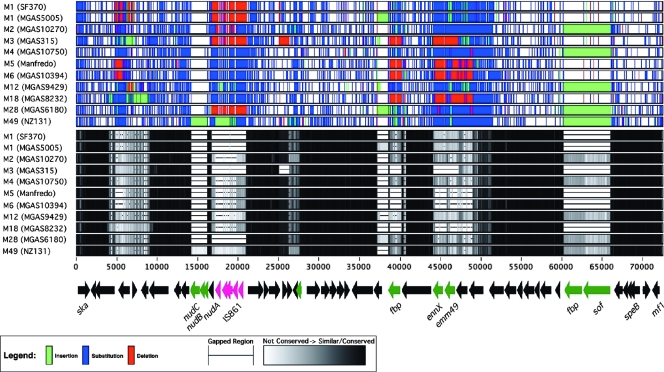
Outside of the prophage genomes and other mobile elements, much of each GAS genome is essentially colinear with the corresponding regions in the other genomes, taking into account the inversions near the origin of replication and the terminus in strains SSI-1 and Manfredo. However, two regions consistently show great variation and confer much of the individuality of a given strain. First, and central to the virulence of GAS, is the ∼73-kbp region of each genome containing the gene for the major antiphagocytic M protein (emm) as well as many other virulence-associated genes (vir regulon) (Fig. 3). It has been previously noted that significant variations exist between GAS serotypes in the emm region, with the presence of the SOF protein being one defining characteristic of historic class II GAS strains. Further, specific regions, particularly associated with emm and emm-like genes, show evidence of variation that may be related to horizontal transfer (85). When the different GAS genomes are compared over this region, the characteristic portions of the NZ131 genome are easily recognized (Fig. 3), which includes the novel group of genes containing a Nudix hydrolase, described below.
FIG. 3.
Comparison of the emm regions from the sequenced GAS genomes. The NZ131 genes from the ∼73-kbp region containing emm are shown at the bottom of the figure with the multiple alignment of this region with the corresponding regions from the other genome strains shown above. The alignment is presented by percentage similarity (grayscale) and by identifying insertions/deletions/substitutions (identified by color). For M types for which the genome has been sequenced more than once, only one example was selected unless significant differences existed. The genes associated with M49 streptococci are shown in green, and the genes associated with IS861 are shown in pink. Some genes are identified to provide orientation: ska (streptokinase), nudABC (Nudix hydrolase cluster [this work]), fbp (fibronectin binding protein), enn49, emm49 (serotype 49 M protein), sof, speB (streptococcal protease SpeB), and mf1 (mitogenic factor).
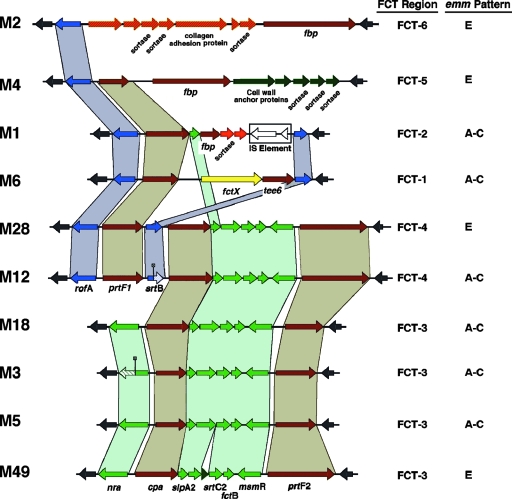
A second region demonstrating significant genome-to-genome variation is the FCT region containing many genes encoding extracellular matrix binding proteins such as protein F and the streptococcal pilus or T antigen (Fig. 4). Previous analysis of this region has identified genetic subgroups in this region (12, 51, 67), and the comparison of this region from the different sequenced GAS genomes confirms the degree of diversity in this region (Fig. 4). The strains can be subdivided into two groups based on whether they carry the gene for either the rofA or nra regulator. NZ131 is a member of the nra group that also includes the M3, M5, and M18 genome strains (FCT-3 group by the recently proposed classification scheme) (51). Although the M12 and M28 genomes have the rofA regulator, they share large regions of homology with the NZ131 group, suggesting that horizontal transfer of blocks of sequence has contributed to the current constellations. Further, Cpa, FctA, and PrtF2 are required components of the T-antigen complex, undoubtedly providing a selective pressure for maintaining this block (55). The diversity in this region and in the emm region argues that certain combinations of genes or clusters of genes represent successful combinations that promote colonization, virulence, or survival in infection of a human host. These combinations, along with the acquisition or loss of prophage-associated genes, are important in defining individual strains as well as groups of strains that are related by antigenicity.
FIG. 4.
FCT (pilus/T antigen) regions of GAS genome strains. The genes from the streptococcal FCT (pilus/T antigen) region from each genome strain were compared by CLUSTAL W alignment. The M3, M5, M18, and M49 genomes were closely related at the nucleotide level over this region, and all contained the nra regulator gene. By contrast, the remaining genomes (M1, M2, M4, M6, M12, and M28) substituted the rofA gene for nra and were more diverse as a group in general. Since the serotypes that have been analyzed by genome sequencing more than once (M1, M3, and M12) are essentially identical over this range, only one of each serotype is shown. The FCT region type (51) and the emm pattern (13) of each genome are indicated. Highly conserved regions shared by different genomes are indicated by color. The nra gene from the M3 genome and the srtB gene from the M12 genome have possible mutations in their ORFs that cause premature termination of transcription. The following genes are shown: cpa (collagen binding protein), prtF1 (fibronectin binding protein; also known as sfb1), prtF2 (fibronectin binding protein; also known as pflpI and fbaB), and fbp (fibronectin binding protein).
Identification and distribution of a unique gene cluster containing a Nudix hydrolase.
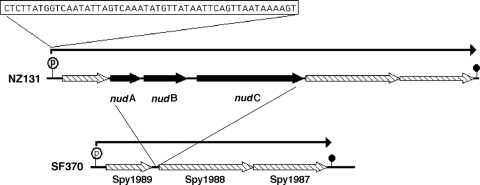
Among the unique genes found in the strain NZ131 genome were three ORFs positioned between genes for a ribosomal protein L11 methyltransferase (Spy49_1638) and a conserved hypothetical protein (Spy49_1642) (Fig. 5). On a regional scale, these genes are located in the cluster of virulence-associated genes centered on emm49, locally positioned in a cluster of genes between those encoding streptokinase and the C5a peptidase. The central gene of this group encodes a 146-amino-acid protein that contains the Nudix hydrolase motif: GX5EX7REUXEEXGU (where U is I, L, or V) (15) (Fig. 5). Nudix hydrolases are enzymes that cleave substrates that are nucleoside diphosphates linked to some other moiety, X. Members of the Nudix hydrolase family are widely distributed from viruses to human in nature (15), and they seem to hydrolyze potentially hazardous materials such as modified nucleotides or prevent the unbalanced accumulation of normal metabolites (15). These enzymatic activities have been suggested to play roles in cellular stress responses and, in bacteria, host cell invasion (15, 16, 32, 43, 44). The three genes appear to form a coordinated genetic group, and so the three ORFs were named nudA, nudB, and nudC, with nudB specifying the Nudix hydrolase and nudA and nudC encoding the two flanking Nudix-associated proteins.
FIG. 5.
The M49 streptococcal Nudix hydrolase region. The location of nudA, nudB, and nudC genes found in the M49 strain NZ131 are shown compared to the corresponding region from the M1 genome strain SF370 (39). Transcriptional analysis maps genes Spy1987, Spy1988, and Spy1989 onto a single polycistronic message in strain SF370. In strain NZ131, this operon has been expanded to five genes by the insertion of the three Nudix-associated genes (see Table S3 in the supplemental material). The predicted promoter is indicated by the encircled P, and the Rho-independent terminator is indicated by the filled circle.
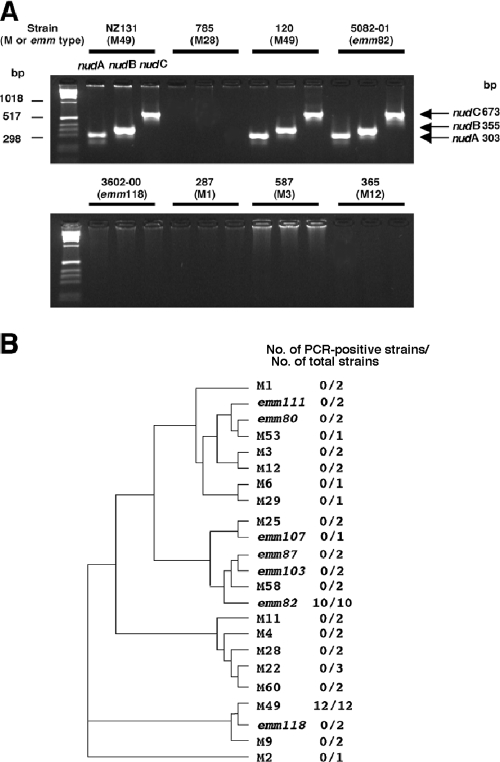
The occurrence of nudA, nudB, and nudC in GAS was determined by PCR analysis among a variety of M type strains (Fig. 6). The nudABC genes always occurred together in all strains that contained them and were found in all M49 and M82 strains tested (12 and 10, respectively) (Fig. 6B). By contrast, these genes were not found in any of the other strains analyzed. Consistent with these results, the published M1, M2, M3, M4, M5, M6, M12, M18, and M28 GAS genomes do not contain nudABC. Therefore, we concluded that the nudABC genes are strongly associated with the M49 and M82 strains, and it is noteworthy that M-type-specific genes have been rarely found except for SOF or emm-related genes. No associations were observed between the presence of nudABC and disease in this survey. The M49 S. pyogenes strain 591 whole-genome shotgun sequence did not contain this cluster of genes, but the absence of the cluster may simply reflect incomplete coverage of this genome.
FIG. 6.
M serotype association of nudABC in S. pyogenes. (A) A survey of S. pyogenes strains with various M serotypes or emm types found nudABC genes present in only emm49 and emm82 strains. Representative amplification products following specific PCR for the nudABC genes are shown. The nudABC genes were always present together in any PCR-positive strain. (B) Occurrence of nudA, nudB, and nudC in a variety of M types. Twenty-two relatively conserved signal sequence residues and the first 83 residues of the associated mature M protein genes were analyzed as recommended (http://www.cdc.gov/ncidod/biotech/strep/strepindex.htm). The phylogram based upon the M protein gene (emm) is presented below. M, typing by specific antiserums; emm, typing by nucleotide sequencing of the emm genes.
Comparison of the regions from strains SF370 and NZ131 shows that the nudABC cluster is an insertion into a predicted common operon containing the genes Spy1989, Spy1988, and Spy1987 (SF370 complement strand). Promoter analysis predicted a highly probable promoter (P = 1.0) positioned in front of Spy1989; secondary promoters were also predicted with lower probability within several of the ORFs, making their existence less likely. The analysis suggested that Spy1989 through Spy1987 would be transcribed on a polycistronic message, and thus it was probable that in NZ131 nudABC would be incorporated into this mRNA. To test this hypothesis, mRNA was isolated from strains SF370 and NZ131, converted into cDNA, and used as the template for PCR amplification with primers positioned to amplify the ends of adjacent ORFs along with the separating noncoding regions (see Table S3 in the supplemental material). These studies showed that the ORFs were transcribed in a polycistronic message, with the novel M49 genes existing as an insertion in this message in NZ131; the predicted mRNAs are indicated by lines with arrow above the ORFs in Fig. 5. The PCR amplification in these mapping studies was not due to chromosomal contamination since the isolated mRNA amplified no product until it was converted into cDNA by reverse transcription (not shown).
Codon usage analysis suggests that the addition of nudABC to this operon may have occurred recently. As discussed above, the CAI of a gene can provide evidence of recent acquisition by horizontal transfer. The CAI values for several genes, either of GAS or prophage origin, are shown in Fig. 7. The selected genes include a number of well-characterized virulence genes from strain NZ131 that are either common to all GAS strains (e.g., streptolysin O) or are bacteriophage encoded (speH). Additionally, the nudABC genes are shown with the surrounding genes common to all GAS strains. As shown in Fig. 7A, most of these genes show nearly average or better CAI values (≥0.4411). The nudABC genes, however, have lower CAI values than the surrounding universal GAS genes (Spy49_1637, Spy49 _1638, and Spy49 _1642) that are predicted to share the same polycistronic mRNA; in the case of nudA and nudC, this variance is more than 1 standard deviation below the mean. Such anomalous codon usage may result from the recent acquisition by serotype M49 and M82 GAS strains. Except for nudC, the difference in CAI is not reflected by a similar anomaly in percent G+C content, suggesting that recent horizontal transfer events may not result in noticeable base pair compositional variations from the surrounding DNA (Fig. 7B).
FIG. 7.
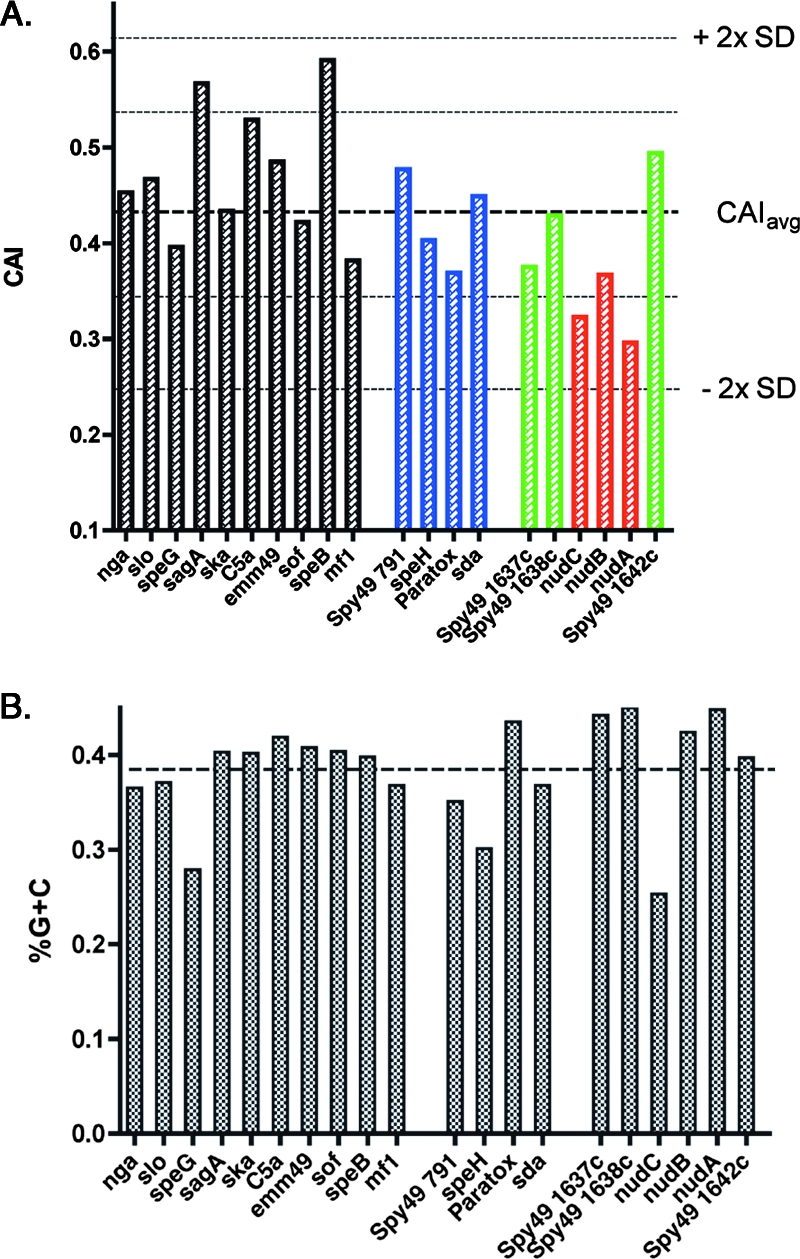
CAI analysis of nudABC and selected other genes. (A) The CAI for NADase (nga), streptolysin O (slo), streptolysin S (sagA), streptokinase (ska), C5a peptidase (C5a), M protein (emm49), SOF (sof), protease SpeB (speB), mitogenic factor (mf1), prophage protein Spy49_0791, exotoxin SpeH (speH), paratox, streptodornase (sda), hypothetical protein Spy49_1637c, methyltransferase Spy49_1638c, hypothetical protein encoded by nudC, Nudix hydrolase (nudB), hypothetical protein encoded by nudA, and hypothetical protein Spy49_1642c. Prophage-encoded genes are shown in blue, the members of the operon containing nudABC that are found universally in GAS strains are shown in green, and nudABC genes are shown in red. The lines indicate the mean CAI values and limits defined by 2 standard deviations (2× SD) above and below the mean. (B) The percent G+C content for each ORF listed above is shown. The dotted line indicates the average percent G+C content for the total genome.
DISCUSSION
The sequencing and analysis of the strain NZ131 genome have led to several important insights into serotype M49 strains of GAS and will provide a platform to expand the previous research studies using this strain. First, a new group of M49-type-specific genes has been identified that along with the previously identified sof- and emm-related genes better refine the identity of these strains. This cluster of specific genes (nudABC) was identified in the emm49 pathogenic region of the S. pyogenes NZ131 genome and may be unique to M49 and M82 strains although a more exhaustive survey would be required for a definitive answer. The nudABC genes are an insertion into a three-gene operon of unknown function (Spy49_1637, Spy49_1638, and Spy49_1642) that is common to all GAS strains, resulting in its expansion to six genes while maintaining the common promoter. Gene nudB was found to be a member of the Nudix hydrolase family while the two associated genes (nudA and nudC) remain functionally unidentified. However, both of these associated genes are intriguing: nudA is related to proteins found in bacteria associated with extreme environments, while nudC is a predicted transmembrane protein.
Gene nudA encodes a member of a group of conserved hypothetical proteins of unknown function defined by COG4043 in the Clusters of Orthologous Groups database (81). Remarkably, the other members of this protein group are found in the extremophiles Bacillus halodurans, Methanopyrus kandleri, Pyrococcus abyssi, and Pyrococcus horikoshii (50, 76, 79), suggesting a possible role for these proteins in withstanding some environmental stress. The third ORF, nudC, is 1,083 nucleotides in length and encodes a 360-amino-acid protein with no matches to homologous proteins in the current databases. However, the predicted NudC peptide contains seven transmembrane domains (48, 52), suggesting that this peptide is positioned in the GAS cell membrane. Considered as a group, the three genes may be involved in some stress response pathway that includes a membrane-associated element that may function in environmental sensing or transport. Since nephritogenic strains have been associated with skin infections (19, 41, 72), these genes may facilitate survival in an environment that is more variable and unpredictable than the uniform warmth and moisture of the throat. Further, the insertion of these genes into an operon common to all GAS strains suggests that the other encoded proteins in this group, which are universal to GAS, may be also associated with survival against challenges in the host environment. It is unclear, however, whether nudABC contributes to the symptomology of APSGN since other serotypes associated with this disease (M types 2 and 60) lack this group.
Other historic class II GAS genomes (SOF+) have been analyzed: an emm2 strain (MGAS10270), an emm4 strain (MGAS10750), and an emm28 strain (MGAS6180) that are representative of strains associated with puerperal sepsis (childbirth fever) (45). A comparison of these strains reveals that while all share defining characteristics of historic class II strains such as being SOF+, they differ in many ways that probably reflect divergent adaptations and evolutionary histories. None of these strains, for example, has the nudABC gene cluster found in NZ131. No prophage attachment sites are common between these other SOF+ strains and NZ131. The CRISPR elements found between the strains have DRs identical to the DR of CRISPR element 1 from NZ131 (Table 2); however, the strains often have unrelated spacers of phage-derived sequences found between these repeats (data not shown), again suggesting a different evolutionary history of previous phage encounters. An allele of streptokinase is found in the M49 strain NZ131 that is distinct from the other strains, and previous studies employing a rabbit infection model have shown that the NZ131 allele is associated with increased nephritogenecity (65). Significantly, a number of striking differences exist when the emm and pilus/T-antigen regions are compared (Fig. 4 and 5). Thus, while it may be useful to define a class II group of M type strains based upon specific variants in the emm region, the presence of SOF, and differences in immunoglobulin G binding proteins (7, 10, 14, 33, 36, 49, 80, 86), the associated disease and site of bacterial colonization may be the most useful characteristics in GAS classification.
The utility of strain NZ131 for electrotransformation was the feature that initially attracted attention to its use (75), and it was subsequently found to be competent for transformation by both circular and linear DNA (69). The ability to genetically manipulate strain NZ131 has contributed to its widespread use in research; however, the cellular basis for this enhanced ability to take up genetic material is unclear and is certainly distinct from the natural ability for transformation seen in other streptococcal species like Streptococcus pneumoniae or Streptococcus mutans. As with the other sequenced GAS genomes, NZ131 has genes associated with competence in other species of Streptococcus such as the alternate sigma factor comX. However, genes that have been demonstrated to be important or essential for competence and transformation in related streptococci (such as the comABCDE system of S. pneumoniae and the comEDC-cslBA-bsmH cluster from S. mutans) are missing from NZ131 and the other S. pyogenes genome strains. Hildago-Grass et al. described a locus (sil) that is associated with enhanced virulence and interstrain DNA transfer (47). Two genome strains, MGAS8232 (M18) and MGAS10750 (M4), have a virtually complete sil locus, while the remaining genome strains, including NZ131, have only the blpM-homolog gene. Indeed, the actual mechanism of genetic transfer mediated by sil is unclear since the possibility of transduction by endogenous prophages was not ruled out by the described experiments (47). Thus, the electrocompetence seen in NZ131 may be due to some characteristic that is unrelated to natural transformation systems and whose primary role in the cell may even be unrelated to DNA uptake or metabolism. There is no obvious defect in DNA repair in NZ131, so electrocompetence is probably not the result of a heightened tolerance for mismatches that normally would inhibit DNA recombination (71). Some alteration to the cell surface leading to better binding of DNA or increased stability of membrane pores following electrical shock may allow an increased level of recombination to occur. Whatever the source of this unique feature, it is not something common to all M49 strains since many are poorly transformable (75). It is possible that little capsule is produced during logarithmic growth on synthetic medium (A. Suvorov, unpublished observation), and this trait may facilitate DNA uptake. However, the strain is fully capable of capsule production, having a canonical capsule operon and overexpressing capsule upon the inactivation of regulatory gene covR (38) (unpublished observation), and so the role of capsule expression in electrocompetence remains unclear. Understanding the cellular basis for the variation in electrocompetence between different GAS strains will be an important advance in the ability to genetically manipulate any strain of interest. Analysis of global gene expression patterns in NZ131 and comparison to other strains that are poorly transformable may help identify the determinants influencing this process.
The examination of 13 examples of GAS genomes continues to shed light upon the evolutionary history of this human pathogen. The contribution of endogenous prophages to the biology of GAS is reaffirmed in every genome. However, as the nudABC region shows, novel genetic elements that are not obviously associated with mobile genetic elements can also help share and define particular GAS strains. Additionally, the data strongly suggest that horizontal transfer of genetic material has apparently led to the existence of highly variable virulence-associated regions that are marked by multiple rearrangements and genetic diversification. By contrast, other regions, even those associated with virulence like the one containing streptolysin O, vary little between genomes. The genome regions that encode surface gene products that will interact with host targets or aid in immune avoidance are the ones that display the most sequence diversity, and while natural selection favors stability in much of the genome, it favors diversity in these regions. The characteristics of the strain NZ131 that have been explored in many previous studies of its genetics and pathogenicity can now be complemented with the genome data presented here, and the combination should fuel future research that will increase our understanding of this important human pathogen.
Supplementary Material
Acknowledgments
For this work, W.M.M. was supported in part by NIH grant P20 RR016478 from the INBRE Program of the National Center for Research Resources (NCRR), by grant P20 RR015564 from the NCRR, and by NIH grant R15A1072718 . We acknowledge the use of the S. pyogenes MLST database that is located at Imperial College London and is funded by the Wellcome Trust.
We are grateful to Bernard Beall, CDC, Atlanta, GA, for providing the collection of serotype M49 invasive strains and to Ross Overbeek, National Microbial Pathogen Data Resource, for assistance with the annotation. We thank Gorana Savic and Mona Balkis for expert technical help.
Footnotes
Published ahead of print on 26 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ajdic, D., W. McShan, R. McLaughlin, G. Savic, J. Chang, M. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. Roe, and J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 9914434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajdic, D., W. M. McShan, D. J. Savic, D. Gerlach, and J. J. Ferretti. 2000. The NAD-glycohydrolase gene of Streptococcus pyogenes. FEMS Microbiol. Lett. 191235-241. [DOI] [PubMed] [Google Scholar]
- 3.Aziz, R. K., R. A. Edwards, W. W. Taylor, D. E. Low, A. McGeer, and M. Kotb. 2005. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J. Bacteriol. 1873311-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 3151709-1712. [DOI] [PubMed] [Google Scholar]
- 5.Bates, C. S., C. Toukoki, M. N. Neely, and Z. Eichenbaum. 2005. Characterization of MtsR, a new metal regulator in group A streptococcus, involved in iron acquisition and virulence. Infect. Immun. 735743-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beall, B., R. Facklam, and T. Thompson. 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beall, B., G. Gherardi, M. Lovgren, R. R. Facklam, B. A. Forwick, and G. J. Tyrrell. 2000. emm and sof gene sequence variation in relation to serological typing of opacity-factor-positive group A streptococci. Microbiology 1461195-1209. [DOI] [PubMed] [Google Scholar]
- 8.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 9.Bengert, P., and T. Dandekar. 2004. Riboswitch finder—a tool for identification of riboswitch RNAs. Nucleic Acids Res. 32W154-W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bessen, D., and V. A. Fischetti. 1990. A human IgG receptor of group A streptococci is associated with tissue site of infection and streptococcal class. J. Infect. Dis. 161747-754. [DOI] [PubMed] [Google Scholar]
- 11.Bessen, D. E., M. W. Izzo, T. R. Fiorentino, R. M. Caringal, S. K. Hollingshead, and B. Beall. 1999. Genetic linkage of exotoxin alleles and emm gene markers for tissue tropism in group A streptococci. J. Infect. Dis. 179627-636. [DOI] [PubMed] [Google Scholar]
- 12.Bessen, D. E., and A. Kalia. 2002. Genomic localization of a T serotype locus to a recombinatorial zone encoding extracellular matrix-binding proteins in Streptococcus pyogenes. Infect. Immun. 701159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bessen, D. E., A. Manoharan, F. Luo, J. E. Wertz, and D. A. Robinson. 2005. Evolution of transcription regulatory genes is linked to niche specialization in the bacterial pathogen Streptococcus pyogenes. J. Bacteriol. 1874163-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bessen, D. E., L. G. Veasy, H. R. Hill, N. H. Augustine, and V. A. Fischetti. 1995. Serologic evidence for a class I group A streptococcal infection among rheumatic fever patients. J. Infect. Dis. 1721608-1611. [DOI] [PubMed] [Google Scholar]
- 15.Bessman, M. J., D. N. Frick, and S. F. O'Handley. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 27125059-25062. [DOI] [PubMed] [Google Scholar]
- 16.Bessman, M. J., J. D. Walsh, C. A. Dunn, J. Swaminathan, J. E. Weldon, and J. Shen. 2001. The gene ygdP, associated with the invasiveness of Escherichia coli K1, designates a Nudix hydrolase, Orf176, active on adenosine (5′)-pentaphospho-(5′)-adenosine (Ap5A). J. Biol. Chem. 27637834-37838. [DOI] [PubMed] [Google Scholar]
- 17.Bisno, A. L. 1995. Non-suppurative poststreptococcal sequelae: rheumatic fever and glomerulonephritis, p. 1799-1810. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, vol. 2. Churchill Livingstone, New York, NY. [Google Scholar]
- 18.Bisno, A. L., I. A. Pearce, H. P. Wall, M. D. Moody, and G. H. Stollerman. 1970. Contrasting epidemiology of acute rheumatic fever and acute glomerulonephritis. N. Engl. J. Med. 283561-565. [DOI] [PubMed] [Google Scholar]
- 19.Bisno, A. L., M. Svartman, and G. H. Stollerman. 1971. Cross-reacting and monotypic T antigens of nephritogenic pyoderma streptococci: the type 56 system. J. Immunol. 1061493-1498. [PubMed] [Google Scholar]
- 20.Bolotin, A., B. Quinquis, A. Sorokin, and S. D. Ehrlich. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 1512551-2561. [DOI] [PubMed] [Google Scholar]
- 21.Brodie, R., A. J. Smith, R. L. Roper, V. Tcherepanov, and C. Upton. 2004. Base-By-Base: single nucleotide-level analysis of whole viral genome alignments. BMC Bioinformatics 596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchanan, J. T., A. J. Simpson, R. K. Aziz, G. Y. Liu, S. A. Kristian, M. Kotb, J. Feramisco, and V. Nizet. 2006. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 16396-400. [DOI] [PubMed] [Google Scholar]
- 23.Carapetis, J. R., A. C. Steer, E. K. Mulholland, and M. Weber. 2005. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5685-694. [DOI] [PubMed] [Google Scholar]
- 24.Chaussee, M., D. Ajdic, and J. Ferretti. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect. Immun. 671715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaussee, M. A., E. A. Callegari, and M. S. Chaussee. 2004. Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. J. Bacteriol. 1867091-7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaussee, M. A., E. J. McDowell, L. D. Rieck, E. A. Callegari, and M. S. Chaussee. 2006. Proteomic analysis of a penicillin-tolerant rgg mutant strain of Streptococcus pyogenes. J. Antimicrob. Chemother. 58752-759. [DOI] [PubMed] [Google Scholar]
- 27.Chaussee, M. S., R. L. Cole, and J. P. van Putten. 2000. Streptococcal erythrogenic toxin B abrogates fibronectin-dependent internalization of Streptococcus pyogenes by cultured mammalian cells. Infect. Immun. 683226-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaussee, M. S., D. Gerlach, C. E. Yu, and J. J. Ferretti. 1993. Inactivation of the streptococcal erythrogenic toxin B gene (speB) in Streptococcus pyogenes. Infect. Immun. 613719-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaussee, M. S., G. A. Somerville, L. Reitzer, and J. M. Musser. 2003. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J. Bacteriol. 1856016-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 69822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conyers, G. B., and M. J. Bessman. 1999. The gene, ialA, associated with the invasion of human erythrocytes by Bartonella bacilliformis, designates a Nudix hydrolase active on dinucleoside 5′-polyphosphates. J. Biol. Chem. 2741203-1206. [DOI] [PubMed] [Google Scholar]
- 33.Courtney, H. S., D. L. Hasty, Y. Li, H. C. Chiang, J. L. Thacker, and J. B. Dale. 1999. Serum opacity factor is a major fibronectin-binding protein and a virulence determinant of M type 2 Streptococcus pyogenes. Mol. Microbiol. 3289-98. [DOI] [PubMed] [Google Scholar]
- 34.Dmitriev, A. V., E. J. McDowell, K. V. Kappeler, M. A. Chaussee, L. D. Rieck, and M. S. Chaussee. 2006. The Rgg regulator of Streptococcus pyogenes influences utilization of nonglucose carbohydrates, prophage induction, and expression of the NAD-glycohydrolase virulence operon. J. Bacteriol. 1887230-7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorschner, R. A., V. K. Pestonjamasp, S. Tamakuwala, T. Ohtake, J. Rudisill, V. Nizet, B. Agerberth, G. H. Gudmundsson, and R. L. Gallo. 2001. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J. Investig. Dermatol. 11791-97. [DOI] [PubMed] [Google Scholar]
- 36.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 692416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eriksson, A., and M. Norgren. 2003. Cleavage of antigen-bound immunoglobulin G by SpeB contributes to streptococcal persistence in opsonizing blood. Infect. Immun. 71211-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Federle, M., K. McIver, and J. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 1813649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. Lai, S. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 984658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frank, C., K. Steiner, and H. Malke. 1995. Conservation of the organization of the streptokinase gene region among pathogenic streptococci. Med. Microbiol. Immunol. 184139-146. [DOI] [PubMed] [Google Scholar]
- 41.Futcher, P. H. 1940. Glomerulonephritis following infections of the skin. Arch. Intern. Med. 651192-1210. [Google Scholar]
- 42.Gase, K., J. J. Ferretti, C. Primeaux, and W. M. McShan. 1999. Identification, cloning, and expression of the CAMP factor gene (cfa) of group A streptococci. Infect. Immun. 674725-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaywee, J., S. Radulovic, J. A. Higgins, and A. F. Azad. 2002. Transcriptional analysis of Rickettsia prowazekii invasion gene homolog (invA) during host cell infection. Infect. Immun. 706346-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaywee, J., W. Xu, S. Radulovic, M. J. Bessman, and A. F. Azad. 2002. The Rickettsia prowazekii invasion gene homolog (invA) encodes a Nudix hydrolase active on adenosine (5′)-pentaphospho-(5′)-adenosine. Mol. Cell Proteomics 1179-185. [DOI] [PubMed] [Google Scholar]
- 45.Green, N. M., S. Zhang, S. F. Porcella, M. J. Nagiec, K. D. Barbian, S. B. Beres, R. B. Lefebvre, and J. M. Musser. 2005. Genome sequence of a serotype M28 strain of group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J. Infect. Dis. 192760-770. [DOI] [PubMed] [Google Scholar]
- 46.Grissa, I., G. Vergnaud, and C. Pourcel. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35W52-W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hidalgo-Grass, C., M. Ravins, M. Dan-Goor, J. Jaffe, A. E. Moses, and E. Hanski. 2002. A locus of group A Streptococcus involved in invasive disease and DNA transfer. Mol. Microbiol. 4687-99. [DOI] [PubMed] [Google Scholar]
- 48.Hofmann, K., and W. Stoffel. 1993. TMBASE-A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374166. [Google Scholar]
- 49.Jeng, A., V. Sakota, Z. Li, V. Datta, B. Beall, and V. Nizet. 2003. Molecular genetic analysis of a group A Streptococcus operon encoding serum opacity factor and a novel fibronectin-binding protein, SfbX. J. Bacteriol. 1851208-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, M. Sekine, S. Baba, H. Kosugi, A. Hosoyama, Y. Nagai, M. Sakai, K. Ogura, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Ohfuku, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, and H. Kikuchi. 1998. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 555-76. [DOI] [PubMed] [Google Scholar]
- 51.Kratovac, Z., A. Manoharan, F. Luo, S. Lizano, and D. E. Bessen. 2007. Population genetics and linkage analysis of loci within the FCT region of Streptococcus pyogenes. J. Bacteriol. 1891299-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305567-580. [DOI] [PubMed] [Google Scholar]
- 53.Kuo, C. F., Y. H. Luo, H. Y. Lin, K. J. Huang, J. J. Wu, H. Y. Lei, M. T. Lin, W. J. Chuang, C. C. Liu, Y. T. Jin, and Y. S. Lin. 2004. Histopathologic changes in kidney and liver correlate with streptococcal pyrogenic exotoxin B production in the mouse model of group A streptococcal infection. Microb. Pathog. 36273-285. [DOI] [PubMed] [Google Scholar]
- 54.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lizano, S., F. Luo, and D. E. Bessen. 2007. Role of streptococcal T antigens in superficial skin infection. J. Bacteriol. 1891426-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malke, H., and K. Steiner. 2004. Control of streptokinase gene expression in group A and C streptococci by two-component regulators. Indian J. Med. Res. 119(Suppl.)48-56. [PubMed] [Google Scholar]
- 57.Malke, H., K. Steiner, W. M. McShan, and J. J. Ferretti. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. 296259-275. [DOI] [PubMed] [Google Scholar]
- 58.McShan, W. M., R. E. McLaughlin, A. Nordstrand, and J. J. Ferretti. 1998. Vectors containing streptococcal bacteriophage integrases for site-specific gene insertion. Methods Cell Sci. 2051-57. [Google Scholar]
- 59.McShan, W. M., and D. J. Savic. 2006. The timing of streptolysin O release is controlled by the sloR operon. Int. Congr. Ser. 1289199-202. [Google Scholar]
- 60.McShan, W. M., Y.-F. Tang, and J. J. Ferretti. 1997. Bacteriophage T12 of Streptococcus pyogenes integrates into the gene for a serine tRNA. Mol. Microbiol. 23719-728. [DOI] [PubMed] [Google Scholar]
- 61.Mojica, F. J., C. Diez-Villasenor, J. Garcia-Martinez, and E. Soria. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60174-182. [DOI] [PubMed] [Google Scholar]
- 62.Montanez, G. E., M. N. Neely, and Z. Eichenbaum. 2005. The streptococcal iron uptake (Siu) transporter is required for iron uptake and virulence in a zebrafish infection model. Microbiology 1513749-3757. [DOI] [PubMed] [Google Scholar]
- 63.Nizet, V., B. Beall, D. Bast, V. Datta, L. Kilburn, D. Low, and J. De Azavedo. 2000. Genetic locus for streptolysin S production by group A Streptococcus. Infect. Immun. 684245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nizet, V., T. Ohtake, X. Lauth, J. Trowbridge, J. Rudisill, R. A. Dorschner, V. Pestonjamasp, J. Piraino, K. Huttner, and R. L. Gallo. 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414454-457. [DOI] [PubMed] [Google Scholar]
- 65.Nordstrand, A., W. M. McShan, J. J. Ferretti, S. E. Holm, and M. Norgren. 2000. Allele substitution of the streptokinase gene reduces the nephritogenic capacity of group A streptococcal strain NZ131. Infect. Immun. 681019-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nordstrand, A., M. Norgren, J. J. Ferretti, and S. E. Holm. 1998. Streptokinase as a mediator of acute post-streptococcal glomerulonephritis in an experimental mouse model. Infect. Immun. 66315-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramachandran, V., J. D. McArthur, C. E. Behm, C. Gutzeit, M. Dowton, P. K. Fagan, R. Towers, B. Currie, K. S. Sriprakash, and M. J. Walker. 2004. Two distinct genotypes of prtF2, encoding a fibronectin binding protein, and evolution of the gene family in Streptococcus pyogenes. J. Bacteriol. 1867601-7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Savic, D., and J. Ferretti. 1997. Evidence for a site specific genomic rearrangement in the slo region of Streptococcus pyogenes. Adv. Exp. Med. Biol. 418983-985. [DOI] [PubMed] [Google Scholar]
- 69.Savic, D. J., and J. J. Ferretti. 2003. Novel genomic rearrangement that affects expression of the Streptococcus pyogenes streptolysin O (slo) gene. J. Bacteriol. 1851857-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Savic, D. J., W. M. McShan, and J. J. Ferretti. 2002. Autonomous expression of the slo gene of the bicistronic nga-slo operon of Streptococcus pyogenes. Infect. Immun. 702730-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scott, J. R., P. Thompson-Mayberry, S. Lahmamsi, C. J. King, and W. M. McShan. 2008. Phage-associated mutator phenotype in group A streptococcus. J. Bacteriol. 1906290-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seegal, D., and D. P. Earle. 1941. A consideration of certain biological differences between glomerulonephritis and rheumatic fever. Am. J. Med. Sci. 201528-529. [Google Scholar]
- 73.Sharp, P. M., and W. H. Li. 1987. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 151281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Silva, F. G. 1998. Acute postinfectious glomerulonephritis and glomerulonephritis complicating persistent bacterial infection, p. 389-453. In J. C. Jennette, J. L. Olson, M. M. Schwartz, and F. G. Silva (ed.), Hepinstall's pathology of the kidney, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 75.Simon, D., and J. J. Ferretti. 1991. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol. Lett. 82219-224. [DOI] [PubMed] [Google Scholar]
- 76.Slesarev, A. I., K. V. Mezhevaya, K. S. Makarova, N. N. Polushin, O. V. Shcherbinina, V. V. Shakhova, G. I. Belova, L. Aravind, D. A. Natale, I. B. Rogozin, R. L. Tatusov, Y. I. Wolf, K. O. Stetter, A. G. Malykh, E. V. Koonin, and S. A. Kozyavkin. 2002. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. USA 994644-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Steiner, K., and H. Malke. 2002. Dual control of streptokinase and streptolysin S production by the covRS and fasCAX two-component regulators in Streptococcus dysgalactiae subsp. equisimilis. Infect. Immun. 703627-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stollerman, G. H. 1975. Rheumatic fever and streptococcal infection, p. 1-303. In G. H. Stollerman (ed.), Clinical cardiology monographs. Grune and Stratton, New York, NY.
- 79.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 284317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Talkington, D. F., B. Schwartz, C. M. Black, J. K. Todd, J. Elliott, R. F. Breiman, and R. R. Facklam. 1993. Association of phenotypic and genotypic characteristics of invasive Streptococcus pyogenes isolates with clinical components of streptococcal toxic shock syndrome. Infect. Immun. 613369-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2833-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Van Domselaar, G. H., P. Stothard, S. Shrivastava, J. A. Cruz, A. Guo, X. Dong, P. Lu, D. Szafron, R. Greiner, and D. S. Wishart. 2005. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 33W455-W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Watanabe, Y. 2001. Cloning of group A streptococcal pyrogenic exotoxin-B gene and its recombinant protein expression in culture supernatant. J. Nippon Med. Sch. 68222-232. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 85.Whatmore, A. M., V. Kapur, J. M. Musser, and M. A. Kehoe. 1995. Molecular population genetic analysis of the enn subdivision of group A streptococcal emm-like genes: horizontal gene transfer and restricted variation among enn genes. Mol. Microbiol. 151039-1048. [DOI] [PubMed] [Google Scholar]
- 86.Whatmore, A. M., V. Kapur, D. J. Sullivan, J. M. Musser, and M. A. Kehoe. 1994. Non-congruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol. Microbiol. 14619-631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.