Abstract
The BipA protein of Escherichia coli has intriguing similarities to the elongation factor subfamily of GTPases, including EF-Tu, EF-G, and LepA. In addition, phenotypes of a bipA deletion mutant suggest that BipA is involved in regulation of a variety of pathways. These two points have led to speculation that BipA may be a novel regulatory protein that affects efficient translation of target genes through direct interaction with the ribosome. We isolated and characterized suppressors of the cold-sensitive growth phenotype exhibited by ΔbipA strains and identified insertion mutations in rluC. The rluC gene encodes a pseudouridine synthase responsible for pseudouridine modification of 23S rRNA at three sites, all located near the peptidyl transferase center. Deletion of rluC not only suppressed cold sensitivity but also alleviated the decrease in capsule synthesis exhibited by bipA mutants, suggesting that the phenotypic effects of BipA are manifested through an effect on the ribosome. The suppressor effect is specific to rluC, as deletion of other rlu genes did not relieve cold sensitivity, and further, more than a single pseudouridine residue is involved, as alteration of single residues did not produce suppressors. These results are consistent with a role for BipA in either the structure or the function of the ribosome and imply that wild-type ribosomes are dependent on BipA for efficient expression of target mRNAs and that the lack of pseudouridylation at these three sites renders the ribosomes BipA independent.
The bipA gene (previously also known as typA or yihK) is conserved in a large variety of bacterial species, including plant and insect symbionts; plant, animal, and human pathogens; and others (41; BlastN search results). Homologs of bipA can be identified either by using BipA as the query sequence or by searching for translational GTPase motifs; results show that homologs are present in bacterial extremophiles such as Psychrobacter cryohalolentis and Thiomicrospira crunogena and in some of the smallest genomes sequenced, such as that of Buchnera aphidicola (416 kb). Very recently, a bipA homolog was identified in the chloroplast of the halophytic plant Suaeda salsa (44), further emphasizing the high degree of conservation of this gene. Indeed, an analysis of 191 fully sequenced bacterial genomes identified a bipA homolog in all but 26 of the organisms examined (27). In addition to the extensive conservation of bipA, it was demonstrated previously that the bipA gene from Sinorhizobium meliloti is able to complement the cold-sensitive defect conferred by deletion of bipA in Escherichia coli K-12 and vice versa (23), suggesting conservation of function and indicating that BipA has a fundamental physiological role that has been retained throughout most of the microbial world.
Despite the prevalence of the gene, bipA has yet to be assigned a definitive function and, surprisingly for a highly conserved gene, is not essential (23, 34). However, clues to the function of BipA can be gleaned through consideration of the physiological conditions under which bipA has been identified and through examination of protein domain similarities. The bipA gene has been identified in several bacterial species in response to diverse genetic screens. As a result, BipA has been implicated as a regulator of a variety of cell processes, including resistance to bactericidal permeability-increasing protein (3, 35), pathogenicity (17), motility (14), capsule formation (40), establishment of symbiosis (23), and growth at low temperatures (34). The variety of cellular processes affected by bipA deletion led to the suggestion from several groups that BipA has a role both in housekeeping functions (23, 34) and in specialized activities related to stress and/or to host-microbe interactions (14, 23).
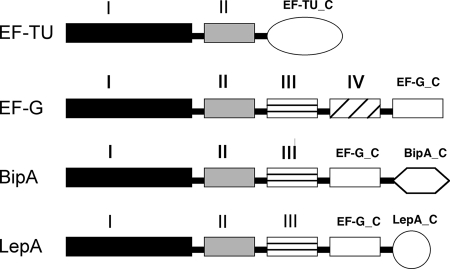
BipA is a 67.4-kDa protein with homology to the elongation factor family of GTPases, which includes EF-G, EF-Tu, and LepA (16, 35). All of these proteins exhibit ribosome-stimulated GTPase activity and share highly conserved domains (Fig. 1) (23). Each of these proteins contains domains I and II, while EF-G, LepA, and BipA all contain domain V of EF-G as well. In addition, LepA and BipA each have unique C-terminal domains. Thus, BipA and LepA have the GTP binding and translation factor motifs, as well as the C-terminal region of EF-G, but lack the anticodon stem-loop mimic of EF-Tu.
FIG. 1.
Domain architecture of elongation factor family members. The domain structures shown are predicted based on protein family analysis using the Pfam database (15). Except for EF-Tu, all family members have five distinct domains. All share the conservation of domains I and II. Domain I is the GTP binding region, and domain II is the signature translation factor β-barrel motif. The function of domain III is not clear, nor is this domain as highly conserved among family members. EF-G has a unique domain IV absent in other members, whereas domain V (the carboxy terminus of EF-G [EF-G_C]) is present in LepA and BipA. Both LepA and BipA have unique C-terminal regions (LepA_C and BipA_C). EF-Tu_C, C-terminal region of EF-Tu.
Like bipA, lepA is highly conserved but is dispensable, and a lepA deletion yields no observable phenotype (12). However, an exciting recent publication described a unique function for LepA (36). Qin and coworkers found that LepA induces a back-translocation reaction in elongating ribosomes, shifting the occupancy of the tRNAs from the E and P sites of the ribosomes back to the P and A sites, presumably allowing the ribosome to correct tRNA binding errors during translation. The authors proposed that LepA be defined as a third elongation factor in bacteria and be renamed EF4.
Based on protein similarities, these results suggest that BipA also plays a role in translation. Supportive of this hypothesis, an interaction between BipA and 70S ribosomes was observed previously by gel filtration analysis (33). In addition, the GTPase activity of BipA was stimulated by the addition of intact ribosomes, mRNA, and tRNA. EF-G was able to displace BipA from the ribosomes, suggesting that their binding sites overlap, as do the LepA and EF-G binding sites.
We hypothesize that the role of BipA is to regulate expression of target mRNAs through altered efficiency of translation, and we propose that the cold-sensitive phenotype of a bipA deletion strain reflects a deficiency in expression of a factor(s) required for robust growth at low temperatures. We envision a scenario in which ribosomes are dependent on BipA for efficient expression of these targets at low temperatures. Therefore, the isolation of suppressors of this phenotype should yield genes that allow bypassing of this deficiency, either by alternative expression of the missing gene product(s) or by relieving ribosomes of their dependency on BipA. We isolated and characterized such suppressors, and our findings demonstrate that dependence on BipA can be alleviated by deletion of rluC, resulting in ribosomes that lack pseudouridine modification at three sites on the 23S rRNA. This suppression is specific to rluC and requires abolishment of modification at all three sites.
Although rRNA modification is maintained in all organisms, the purpose has remained elusive. A recent finding showed that RluD-directed pseudouridylation promotes effective termination of translation (13), demonstrating that at least some functions of rRNA modification are functional rather than strictly structural. Our findings reported here support our hypothesis that BipA interacts with the ribosome and further suggest that RluC-mediated modification of 23S rRNA determines the dependence of ribosomes on BipA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All bacterial strains used were derivatives of E. coli K-12 and are listed in Table 1. The plasmids used for this study are described in Table 2. Bacteria were grown in modified Luria-Bertani (LB) medium (42) with 1% NaCl, with the addition of kanamycin (50 mg/liter), trimethoprim (150 mg/liter), ampicillin (125 mg/liter), tetracycline (25 mg/liter), or spectinomycin (40 mg/liter) when appropriate. SOC medium (29) was used for outgrowth during random transposon-insertion mutagenesis and site-directed mutagenesis. M9 minimal medium (42) was used to select for the parental strain after conjugation with the E. coli S17-1λpir donor strain during res-npt-res homologous recombineering (28, 30, 31). Strains containing pBAD18 or pBAD24 plasmid derivatives (20) were grown in the presence of glucose (0.4%) or arabinose (0.4%) to repress or induce, respectively, expression of cloned gene products. This level of glucose was chosen because our experience has been that 0.4% glucose provides more reproducible repression of the PBAD promoter than the usual 0.2%.
TABLE 1.
E. coli strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| MG1655 | rph-1 ilvG rfb-50 | 19 |
| AF600 | MG1655 ΔbipA::kan | 34 |
| KM32 | argE3 his-4 leuB6 proA2 thr-1 ara-14 galK2 lacY1 mtl-1 xyl-5 thi-1 rpsL31 tsx-33 supE44 Δ(recC ptr recB recD)::Plac-bet exo cmr | 30 |
| S17-1λpir | Tpr SmrrecA thi hsdRM+ RP4::2-Tc::Mu::Km Tn7 λpir | 24 |
| KK21 | KM32 ΔbipA::res-npt-res | This study |
| KK22 | KM32 ΔrluA::res-npt-res | This study |
| KK23 | KM32 ΔrluC::res-npt-res | This study |
| KK24 | KM32 ΔrluF::res-npt-res | This study |
| TB28 | MG1655 ΔlacZYA<frt> | 4 |
| JW1261 | Δ(araD-araB)567 ΔlacZ4787 rrnB-3 λ− ΔrluB::kan rph-1 Δ(rhaD-rhaB)568 hsdR514 | 2 |
| JW1121 | Δ(araD-araB)567 ΔlacZ4787 rrnB-3 λ− ΔrluE::kan rph-1 Δ(rhaD-rhaB)568 hsdR514 | 2 |
| KK28 | TB28 ΔbipA::res-npt-res | This study |
| KK30 | TB28 ΔbipA<res> | This study |
| KK31 | TB28 ΔbipA<res> ΔrluA::res-npt-res | This study |
| KK32 | TB28 ΔbipA<res> ΔrluB::kan | This study |
| KK33 | TB28 ΔbipA<res> ΔrluC::res-npt-res | This study |
| KK35 | TB28 ΔbipA<res> ΔrluE::kan | This study |
| KK36 | TB28 ΔbipA<res> ΔrluF::res-npt-res | This study |
| SQ110 | MG1655 ΔrrnABCDGH (ptRNA67) | C. L. Squires |
| KKQ113 | SQ110 ΔbipA::kan | This study |
| GEB495 | MC4100 ara+cpsB-lacZ Mud1 | 7 |
| SURA3 | W3110 ΔsurA3::Km | R. Kolter |
| KKC105 | GEB495 ΔbipA::tet | This study |
| KKC107 | GEB495 ΔsurA3::kan | This study |
| KKC114 | KKC105 ΔsurA3::kan | This study |
| KKC201 | KKC107 rluC::EZ-Tn5 <DHFR-1> | This study |
| KKC202 | KKC114 rluC::EZ-Tn5 <DHFR-1> | This study |
The symbols <res> and <frt> indicate the replacement of chromosomal DNA with the scar sequence after recombination with the ParA resolvase and the FLP recombinase, respectively.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pJMSB8 | Apr R6KoriV RP4 oriT | 24 |
| pKK1 | rluC+ (pBAD18) | This study |
| pKK4 | pKK1 carrying D144T substitution | This study |
| pRrnB | rrnB+ (pBAD24) | C. L. Squires |
| pRrnB-U955A | pRrnB(U955A) | C. L. Squires |
| pRrnB-U955G | pRrnB(U955G) | C. L. Squires |
| pRrnB-U2504A | pRrnB(U2504A) | C. L. Squires |
| pRrnB-U2580A | pRrnB(U2580A) | C. L. Squires |
| pRrnB-U2580C | pRrnB(U2580C) | C. L. Squires |
| pRrnB-U2580G | pRrnB(U2580G) | C. L. Squires |
| pKK24 | pRrnB(U955A U2580C) | This study |
| pKK25 | pRrnB(U955A U2504C) | This study |
| pKK26 | pRrnB(U2580C U2504G) | This study |
| pKK27 | pRrnB(U955A U2504C U2580A) | This study |
| pKK28 | pRrnB(U955A U2504C 2580C) | This study |
| pBMM-1 | res-npt-res (pBCSK+) | 28 |
Plasmid pKK1 was constructed by PCR amplification of rluC from the chromosomal DNA of strain MG1655 by using primers with restriction sites (EcoR1 and Xma1) near the termini (primer sequences are available upon request). The digested chromosomal fragment was ligated with pBAD18 digested with the same enzymes. Plasmid pKK4 was constructed by site-directed mutagenesis of pKK1, replacing the aspartate 144 codon (GAC) with a threonine codon (ACC). Plasmid pRrnB (pBAD24 backbone) and its derivatives pRrnB-U955A, pRrnB-U955G, pRrnB-U2504A, pRrnB-U2580A, pRrnB-U2580C, and pRrnB-U2580G were kindly provided by Catherine Squires, Aaron New, and Selwyn Quan (Tufts University and Stanford University). Plasmids pKK24 (carrying U955A and U2580C) and pKK25 (carrying U955A and U2504C) were constructed by site-directed mutagenesis of pRrnB-U955A. pKK26 (carrying U2580C and U2504G) was constructed by site-directed mutagenesis of pRrnB-U2580C; pKK27 (carrying U955A, U2504C, and U2580A) and pKK28 (carrying U955A, U2504C, and U2580C) were constructed by site-directed mutagenesis of pKK25.
Strain constructions.
res-npt-res recombineering was performed as described previously (28). Briefly, the res-npt-res fragment from pBMM-1 was amplified by PCR using primers complementary to sequences at the termini of target genes (bipA, rluA, rluC, and rluF). The amplified fragments were introduced by electroporation into E. coli KM32, which expresses the phage lambda recombination system (30, 31). Kanamycin-resistant colonies were selected, and the correct insertions were verified by PCR. The npt cassette was removed from recombinant strains by conjugation with E. coli S17-1λpir(pJMSB8) (24). The deletion of bipA using a Tetr cassette was performed similarly except that no res sites were provided. Additional strains shown in Table 1 were constructed using standard genetic techniques (29, 42).
General molecular techniques.
DNeasy tissue and QIAprep spin miniprep kits (Qiagen Corp., Valencia, CA) were used for isolation of genomic DNA and plasmid DNA, respectively. PCR amplification was performed using the Failsafe PCR kit (Epicenter, Madison, WI). Plasmids and PCR fragments were purified from agarose gels using QIAquick gel extraction kits (Qiagen Corp.). Restriction enzymes were obtained from New England Biolabs (Beverly, MA) or Stratagene (La Jolla, CA). Site-directed mutagenesis of pKK1 was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Site-directed mutagenesis of pRrnB and its derivatives was performed with the Phusion site-directed mutagenesis kit (Finnzymes, MA). All kits were used according to the manufacturers' recommendations. Primers used for PCR amplification were obtained from Midland Certified Reagent Company (Midland, TX); sequences of primers will be supplied upon request.
Isolation of suppressors of cold sensitivity.
Electroporation-competent cells of AF600 (MG1655 ΔbipA) were transformed with 1 μl of the EZ-Tn5 <DHFR-1> transposome (Epicentre, Madison, WI). After outgrowth for 1 h in SOC medium at 37°C, the cells were pelleted by centrifugation and plated onto LB agar containing trimethoprim (150 mg/liter) for overnight incubation at 37°C. Five separate electroporations, yielding 5,000 colonies, were performed to create the insertion library. Approximately 100,000 cells from the library were plated onto LB agar containing trimethoprim (150 mg/liter), and the plates were incubated at 20°C and monitored regularly for fast-growing colonies. The fast-growing colonies were isolated and rechecked for a fast-growth phenotype by comparison with the parental strain. Potential suppressors were used as donors for P1 transduction into AF600, with selection for trimethoprim resistance, to ensure that suppression of cold sensitivity was linked to the Tn5 insertion.
Inverse PCR.
The sites of transposon insertion were identified by inverse PCR (26). Genomic DNA was digested with Nhe1 or BglII, neither of which cut within the transposon, and the digestion product was self-ligated in a dilute reaction mix. PCR was performed using primers oriented such that the genomic DNA flanking the transposon was amplified. The PCR product was gel purified and used as the template for DNA sequencing analysis (MWG Biotech Inc., High Point, NC).
Growth analysis.
Growth curve analyses were performed with a Bioscreen C microbiology reader from Labsystems (Helsinki, Finland). Overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.02, and 300 μl of diluted culture was added to each well of the honeycomb plate. Each strain was analyzed in triplicate. In the case of strains harboring pBAD18 or its derivatives, 3 μl of overnight culture was added to 300 μl of LB broth containing ampicillin and either glucose or arabinose as appropriate.
The pRrnB-derivative plasmids were introduced into KKQ113 by electroporation, and cells were plated onto LB agar plates containing ampicillin (125 mg/liter) and glucose (0.4%) for overnight incubation at 37°C. Three colonies from each transformation were chosen randomly and inoculated into LB broth containing ampicillin (125 mg/liter) and glucose (0.4%), and the cultures were incubated overnight at 37°C. These cultures were used the next day for growth curve analyses; all cultures were diluted to an OD600 of ∼0.02 in LB broth containing ampicillin (125 mg/liter) and arabinose (0.4%), and 300 μl of the diluted culture was added to the honeycomb plates.
Growth analyses were performed at 20°C with continuous shaking and absorbance readings taken every 30 min. We note that the values obtained are not directly comparable to those from a standard spectrophotometer due to the shorter path length of the honeycomb wells. In the case of strains containing pBAD18 derivatives, the analyses were performed with intermittent shaking, with the cultures shaken vigorously prior to each reading. We find that strains containing plasmids perform more reproducibly when grown by this technique.
β-Galactosidase assays.
β-Galactosidase activity assays were performed as described previously (29). Overnight cultures of strains harboring the cpsB-lacZ fusions (GEB495 strains) were diluted 1:100 in modified LB medium and grown at 30°C until the culture reached an OD of ∼0.6, at which time the assays were performed.
RESULTS
We demonstrated previously that deletion of bipA causes a temperature-specific defect manifested as slow growth at low temperatures (34). To gain insight into the physiological role of BipA, we isolated suppressor mutations that overcame the cold sensitivity with the goal of identifying mutations that bypassed the need for BipA at low temperatures. As shown in Fig. 2A, bipA mutants were delayed in their emergence from stationary phase and also exhibited increased doubling times in exponential phase. This slow growth was observed whether cells were diluted from stationary phase or from exponentially growing cultures (data not shown). Since the defect was not a complete cessation of growth, conventional isolation of spontaneous mutations was difficult because lawns of bacteria plated at 20°C would eventually grow and identification of fast-growing mutants within the lawn was problematic. We therefore chose to create random insertion mutations and identify fast-growing mutants from this collection. Because the mutation existed before the cells were plated at 20°C, fast growers arose quickly enough to be isolated from the background lawn of bacteria.
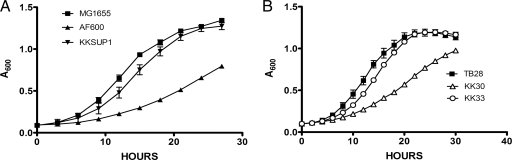
FIG. 2.
Deletion of rluC suppresses the cold sensitivity of the ΔbipA mutant. (A) Six fast-growing suppressor colonies were chosen for growth rate analysis. Each was grown in broth culture and analyzed at 20°C using a Bioscreen C reader. Each of the strains was analyzed in triplicate, and error bars represent the standard error of mean for each strain. AF600 grew slowly compared to the wild type MG1655, whereas all six suppressors had increased growth rates compared to that of AF600. The growth curve for only a single representative suppressor strain (KKSUP1) is shown. (B) Wild-type (TB28 rluC+ bipA+), ΔbipA (KK30), and ΔbipA ΔrluC (KK33) strains were analyzed to assess the ability of a precise rluC deletion to suppress cold sensitivity. As noted in the legend to panel A, each strain was analyzed in triplicate. All curves are shown with error bars; some bars are too small to be distinguishable.
Deletion of rluC suppresses ΔbipA cold sensitivity.
A library of random insertion mutations in a ΔbipA strain (AF600) was created using the EZ-Tn5 <DHFR-1> transposome. Approximately 100,000 mutants were screened to identify those that formed colonies on solid medium more quickly than the parent at 20°C. Of 20 promising colonies, 6 were chosen for further study. Growth curve analysis of the presumed suppressor mutants confirmed that all of these mutants grew more rapidly than AF600 and similarly to wild type MG1655 (Fig. 2A). To confirm that the improved growth rate was due to the Tn5 mutation, the insertions were transferred into naive AF600 by P1 transduction and growth was monitored again. In all cases, the cold-resistant growth phenotype was linked to the trimethoprim resistance (data not shown), confirming that insertion of Tn5 led to the suppressor phenotype.
To determine the location of the Tn5 insertions, inverse PCR was performed as described in Materials and Methods. In all six suppressors, Tn5 had been inserted within rluC, a gene encoding one of six pseudouridine synthases responsible for modification of 23S rRNA (9, 21). Two of the insertions mapped between nucleotides 448 and 449, two between nucleotides 613 and 614, and two between nucleotides 882 and 883 of rluC. Hence, we obtained three independent insertions in rluC, all of which alleviated the cold sensitivity of the ΔbipA strain. Diagnostic PCR with rluC-flanking primers confirmed that the remaining 14 suppressors also had disruptions of rluC, although the positions of these insertions were not mapped precisely.
To demonstrate conclusively that disruption of rluC suppresses the cold-sensitive phenotype of a ΔbipA strain, we constructed a complete, precise deletion of rluC. The genomic copy of rluC was replaced with a res-npt-res cassette in strain KM32, and the deletion-insertion was transferred into KK30 (ΔbipA) by P1 transduction. Again, growth curve analysis demonstrated that deletion of rluC alleviated the cold-sensitive phenotype of the ΔbipA mutant (Fig. 2B). Finally, the ΔrluC allele from the Keio collection (2) was introduced into a ΔbipA strain, and again, cold sensitivity was suppressed (data not shown).
Wild-type rluC complements a ΔrluC ΔbipA strain.
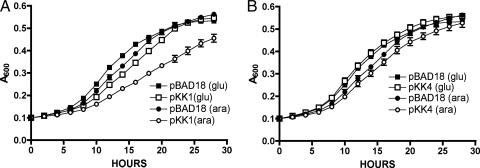
We next performed complementation analysis to rule out a polar effect on downstream genes due to insertion of the antibiotic cassette or the transposon. Wild-type rluC was cloned into pBAD18, placing the gene under the control of the arabinose-inducible PBAD promoter. The resultant plasmid, pKK1, was introduced into KK33 (ΔrluC ΔbipA). Expression of rluC from the plasmid was confirmed by analysis of crude cell extracts by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). KK33 containing the parental pBAD18 plasmid exhibited similar growth characteristics in glucose and arabinose (Fig. 3A). The strain containing pKK1 (rluC+) displayed growth characteristics similar to those of KK33/pBAD18 when glucose was added to repress expression of rluC. However, when rluC was induced by addition of arabinose, the growth rate decreased to that of the ΔbipA strain AF600 (Fig. 3A). Thus, the deletion of rluC suppressed the cold sensitivity induced by ΔbipA, while the expression of rluC+ from a plasmid complemented ΔrluC and reestablished the cold-sensitive growth defect.
FIG. 3.
Plasmid-borne rluC complements the ΔrluC suppression. rluC was cloned into pBAD18 and introduced into KK33 (ΔbipA ΔrluC), and analysis of the growth rates at 20°C was performed. (A) When rluC expression was repressed by addition of glucose (glu), growth was similar whether cells contained the parental plasmid pBAD18 or the rluC-carrying plasmid pKK1. Some leaky expression in glucose was apparent, demonstrated by a slight decrease in the growth rate of pKK1-containing cells. Addition of arabinose (ara) to induce rluC expression had no effect on cells carrying the pBAD18 control, but the pKK1-containing bacteria suffered a decrease in growth rate. All strains were assayed in triplicate. (B) Expression of rluC(D144A), containing an active-site mutation (carried by pKK4), failed to complement the deletion of rluC, as growth was similar to that of cells growing in the presence of glucose or that of cells containing pBAD18. All strains were assayed in triplicate. The altered growth conditions for this experiment resulted in strains' entering stationary phase sooner than the strains in experiments without plasmids.
The pseudouridine synthase activity of RluC is required for complementation.
These results demonstrated that production of the RluC protein is required for manifestation of the cold-sensitive phenotype of a ΔbipA mutant. Although the only known function of RluC is pseudouridylation of 23S rRNA, we felt it to be important to demonstrate that the contribution of RluC to cold sensitivity was indeed due to this activity. We replaced the conserved aspartate (D144) of RluC with threonine by site-directed mutagenesis; this aspartate-to-threonine change has been shown previously to disrupt the pseudouridylation activity of RluC (11). If the pseudouridylation activity is important for cold sensitivity, we would expect that the RluC D144T mutant protein [RluC(D144T)] would not complement the ΔbipA ΔrluC strain. Expression of RluC(D144T) was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis to demonstrate that the mutant protein was produced and that the mutation did not cause excessive degradation of the protein; no difference in the levels of wild-type and mutant protein expression was detected (data not shown).
Growth curve analyses showed that RluC(D144T) was unable to complement the ΔbipA ΔrluC double mutant, resulting in growth rates comparable to that of the control strain (Fig. 3B). These findings suggest that the cold-sensitive phenotype of a ΔbipA strain is dependent on modification of the rRNA by RluC, as either deletion of rluC or mutation of the active site abolished cold sensitivity.
Disruption of rluC suppresses the capsule synthesis defect of a ΔbipA strain.
A variety of phenotypes have been associated with deletion of bipA, including an effect on capsule synthesis in E. coli K5 (40). To determine whether deletion of rluC also suppressed these additional phenotypes, we first needed to assess the effect of ΔbipA on capsule synthesis in K-12 strains. In E. coli K-12, the colanic acid capsule is synthesized by the products of the cps genes, which are regulated by a complex system that includes the Rcs pathway. Under usual laboratory conditions, the cps genes are expressed at very low levels. Insult to the cellular envelope and the subsequent activation of the Rcs pathway induce expression of the cps genes (7). Deletion of the surA gene, encoding a periplasmic peptidyl-prolyl isomerase, is one way to provide such activation due to the accumulation of unfolded proteins in the periplasm (25, 39).
To assay capsule expression in E. coli K-12, we utilized strain GEB495 (7), containing the cpsB-lacZ Mud1 fusion (5, 43). Although we hypothesize that BipA affects translation, we predict that its primary target is upstream of cpsB and that, therefore, an operon fusion should indirectly reflect alterations in BipA activity. As expected, in the absence of injury or stress to the cell, cpsB expression was very low (Fig. 4), such that any decrease in expression by deletion of bipA could not be ascertained. Therefore, to induce capsule genes including cpsB, we introduced a surA deletion; we observed a significant increase in cpsB-lacZ expression in the ΔsurA strain (Fig. 4). Deletion of bipA in this background resulted in a sevenfold decrease in expression of cpsB-lacZ. This result suggests that BipA is involved in positive regulation of colanic acid synthesis in K-12 strains of E. coli.
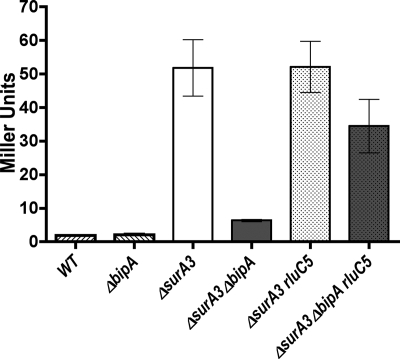
FIG. 4.
Deletion of rluC suppresses the capsule synthesis defect of a ΔbipA strain. Capsule synthesis was assayed using strain GEB495 containing the cpsB-lacZ Mud1 fusion. Strains were grown at 30°C to an OD600 of ∼0.6, and β-galactosidase assays were performed. The deletion of surA significantly increased capsule expression. When bipA was deleted in the ΔsurA strain, capsule expression was decreased sevenfold. The deletion of rluC partially suppressed the effect of ΔbipA and restored cpsB-lacZ expression. WT, wild type; rluC5, ΔrluC.
To determine whether the deletion of rluC suppresses the effect of ΔbipA on capsule expression, the rluC disruption mutation was introduced into the ΔsurA and ΔbipA ΔsurA strains (Fig. 4). The deletion of rluC in the ΔbipA ΔsurA strain resulted in an increase of approximately fourfold in the expression of the cpsB-lacZ fusion. Therefore, although suppression was not complete, rluC deletion affected two distinct phenotypes caused by bipA deletion, indicating that the function of BipA is no longer required when rRNA modification is altered.
Suppression is specific to rluC.
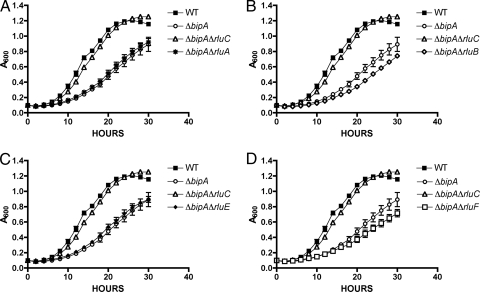
There are six different pseudouridine synthases which modify the 23S rRNA at different positions (9, 11, 21, 37, 38, 45). To determine whether the suppression of ΔbipA phenotypes is specific for ΔrluC, we constructed deletions of rluA, rluB, rluE, and rluF in KK30 (ΔbipA). We did not construct a deletion of rluD because of the known detrimental effect on growth and the resultant rapid growth of suppressors (37). None of these deletions suppressed the cold sensitivity of the ΔbipA strain (Fig. 5), suggesting that the suppressor function is specific to rluC and presumably to the specific sites modified by RluC.
FIG. 5.
The suppression of cold sensitivity is specific to rluC. Additional rlu genes were deleted independently in the ΔbipA strain, and growth at 20°C was analyzed. Results for the wild-type strain (WT; TB28) and the ΔbipA (KK30) and ΔbipA ΔrluC (KK33) strains are shown in each graph. Also shown are results for the ΔbipA ΔrluA (KK31) (A), ΔbipA ΔrluB (KK32) (B), ΔbipA ΔrluE (KK35) (C), and ΔbipA ΔrluF (KK36) (D) strains. Only the rluC deletion suppressed the cold-sensitive phenotype of the ΔbipA strain. All assays were performed in triplicate.
Because RluC modifies three sites in the 23S rRNA, while each of the other enzymes alters only one site (with the exception of RluD, which also introduces three pseudouridines), we thought it possible that deletion of multiple rlu genes might alter the rRNA in a manner analogous to deletion of rluC and thus suppress the effects of ΔbipA. Therefore, we constructed strains with deletions of two of the rlu genes (rluA, rluB, rluE, and rluF) in all possible pairs or with all combinations of three deletions; none of these was able to suppress the cold-sensitive growth defect (data not shown). Therefore, the suppression activity is specific to rluC and most likely to the specific sites modified by RluC.
All three pseudouridines contribute to the ΔbipA phenotype.
RluC modifies three separate uridine residues in 23S rRNA; these are located at nucleotide positions 955, 2504, and 2580 (9, 21). To identify the residue or residues required for the BipA-dependent phenotype, we utilized the six-rrn-operon deletion strain (SQ110) provided by C. L. Squires. This strain has deletions of six of the seven chromosomal rrn operons, leaving only rrnE intact. The rrnB operon can be supplied on a plasmid (pRrnB), allowing molecular manipulation to introduce mutations at specific sites in the rRNA. Although the cellular population of 23S rRNA will be a mixture of the plasmid-encoded mutant form and the single chromosome-encoded wild type, the effects of mutations in this strain can usually be detected. There is a strain with all seven chromosomal copies deleted, with the only 23S rRNA provided from the plasmid, but this strain was exceedingly unstable in our hands, making interpretation of results ambiguous.
We deleted bipA from the six-rrn deletion strain (SQ110) by P1 transduction and subsequently introduced either pRrnB (carrying the wild-type rrnB operon) or derivative mutant plasmids and assayed growth of the resultant strains. The mutant plasmids encoded 23S rRNA with mutations at each of the three RluC target sites, either alone, in pairs, or at all three positions.
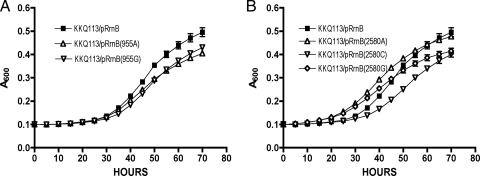
When strains carrying the rrn mutant plasmids were plated onto LB agar with glucose (which should inhibit transcription of the mutant rRNA from the plasmid, although glucose is known to allow leaky expression [20]), immediate differences in growth characteristics were apparent. Strains containing the wild-type plasmid or plasmids with single mutations at either nucleotide 955 or 2580 (pRrnB-U955A, pRrnB-U955G, pRrnB-U2580A, pRrnB-U2580C, or pRrnB-U2580G) all grew well, while the strain with pRrnB-U2504A, carrying an alteration at nucleotide 2504, produced very small colonies. Upon restreaking of the latter strain, suppressors grew rapidly, as demonstrated by fast-growing “pop-up” colonies. The U2504 residue has been shown previously to be required for in vitro assembly of ribosomes (18) and translational accuracy (32); therefore, this severe growth defect was not surprising. Our observation supports the previous suggestion that U2504 is critical and demonstrates that changes at position 2504 are not well-tolerated. However, we found that individual changes at positions 955 and 2580 did not significantly affect growth.
Strains with plasmids carrying two mutations all produced small colonies, and subsequent rapid growth of suppressors was observed, similar to the pRrnB-U2504A strain. The U955A-U2580G (pKK24) combination was particularly detrimental, even though the individual changes at position 955 or 2580 did not significantly affect growth. Because of the poor growth characteristics and rapid suppressor development, the double mutants and the U2504 mutants were not studied further.
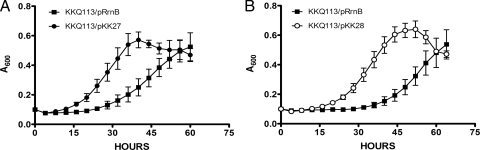
Surprisingly, strains with plasmids harboring changes at all three sites (pKK27 [U955A, U2504C, and U2580A] and pKK28 [U955A, U2504C, and U2580C]) were relatively stable, with uniform colony morphologies and no apparent pop-up suppressors. These strains had colony phenotypes similar to those of strains with plasmids carrying individual 955 or 2580 changes. Growth curve analyses were performed with the single mutants and the triple mutants to determine whether the lack of pseudouridylation at only one site was sufficient to suppress cold sensitivity or if a combination of sites was involved. None of the single mutations was able to suppress the cold-sensitive growth phenotype of KKQ113 (Fig. 6), suggesting that more than one pseudouridylation must be abolished to overcome the effect of bipA deletion. However, mutation of all three pseudouridylation sites did suppress the ΔbipA phenotype, demonstrating that replacement of uridine with an alternate nucleotide did not interfere with the suppressor effect (Fig. 7). These results indicate that suppression of a bipA deletion phenotype requires the absence of modification at more than a single pseudouridine, and perhaps at all three.
FIG. 6.
Mutations at a single pseudouridylation site are not sufficient to relieve the cold sensitivity of the ΔbipA strain. KKQ113 (ΔbipA in the six-rrn deletion strain) was transformed with single-site mutant plasmids, and analysis of growth rates at 20°C was performed. pRrnB was the parent plasmid carrying wild-type rrnB and promoted cold-sensitive growth. All assays were performed in triplicate. (A) The change of residue U955 to adenine (in pRrnB-U955A) or guanine (in pRrnB-U955G) did not abolish cold sensitivity. (B) The alteration of nucleotide U2580 to adenine (in pRrnB-U2580A), cytosine (in pRrnB-U2580C), or guanine (in pRrnB-U2580G) also did not abolish the cold sensitivity.
FIG. 7.
The alteration of all three RluC-dependent pseudouridylation sites relieves the cold sensitivity of the ΔbipA strain. KKQ113 (ΔbipA in the six-rrn deletion strain) was transformed with plasmids containing mutations at all three sites modified by RluC, and analysis of growth rates at 20°C was performed. pRrnB was the parent plasmid carrying wild-type rrnB and promoted cold-sensitive growth. All assays were performed in triplicate. Two different combinations of triple-site mutations eliminated the cold-sensitive growth defect of a bipA mutant. (A) Plasmid pKK27 contains U955A, U2504C, and U2580A. (B) Plasmid pKK28 has U955A, U2504C, and U2580C. The decrease in growth at the ends of the curves is ascribed to the consumption of the arabinose and, therefore, decreased expression of the plasmid-borne rrnB operon.
DISCUSSION
Although BipA has been identified previously in several different bacterial species through a variety of screens, little has been resolved regarding its physiological role or mechanism of action. Two features of BipA have guided speculation as to its function. First, the clear similarity of BipA to members of the EF-Tu family of GTPases and the presence of the same domain structure found in these proteins led to predictions that it binds to the ribosome (17, 23). Indeed, subsequent studies demonstrated that BipA does interact with the ribosome and that it is displaced by EF-G, implying that BipA and EF-G bind to overlapping sites. Further, the GTPase activity of BipA is dramatically enhanced by 70S ribosomes, mRNA, and tRNA, suggesting that a complete translation reaction is required for optimal hydrolytic activity (33). Second, deletions in bipA affect a variety of cellular functions, including host-pathogen interactions, host-symbiont relationships, motility, capsule formation, and growth at low temperatures. These pleiotropic phenotypes suggest that BipA affects multiple pathways, either directly or indirectly. The combination of these observations led to the prevailing hypothesis that BipA regulates translation by an unprecedented mechanism, presumably involving binding to the ribosome and GTP hydrolysis.
One model for BipA activity was proposed following the observation that BipA regulates the DNA binding protein Fis (33). Analysis of the fis sequence led to the suggestion that mRNAs dependent on BipA for efficient expression contain longer than average ribosome binding regions, resulting in the formation of a very stable mRNA-16S rRNA duplex that inhibits initiation of translation. According to this idea, binding of BipA would relieve this constraint by destabilizing the duplex and promoting initiation. However, further experiments failed to support the involvement of BipA in fis regulation, resulting in the withdrawal of the pertinent publication (33; K. Krishnan, unpublished data). Subsequently, as no direct target of BipA is now known, it is difficult to postulate models based on sequences or structures of mRNAs. While it may be true that BipA enhances translation initiation, there is no evidence to either support or refute the specific model proposed.
The data presented here are consistent with a model in which BipA is directly associated with ribosomal activity and in which abolishing pseudouridylation at three sites renders ribosomes independent of BipA. We can envision several possible mechanisms of BipA action; two are discussed here: a role in ribosome assembly and a direct role in the translation process.
Several GTPases are important for correct ribosome assembly (22). Additionally, although the role of rRNA modifications remains unclear, it has been suggested previously that they provide a structural determinant for ribosome assembly or function (8, 10). Therefore, it is possible that BipA contributes to the correct assembly of normally modified ribosomes. BipA and RluC may function in the same assembly pathway; once modified by RluC, ribosomes would require BipA for correct assembly. The suppression of ΔbipA phenotypes by ΔrluC would then suggest that unmodified ribosomes are able to assemble via an alternate, BipA-independent pathway. Although not impossible, it seems unlikely that the cell would have two alternate assembly pathways for ribosomes, one for modified ribosomes and one for unmodified ribosomes. Additionally, the assembly GTPases belong to different families of GTPases than does BipA, with little to no sequence similarity (6); BipA is a member of the translational GTPase family.
Given the extensive similarity of BipA to LepA and EF-G and the overlapping ribosomal binding sites (6, 33), we think it more probable that BipA acts on ribosomes during the elongation phase of translation. Displacement of BipA by EF-G may indicate a competition in which BipA is present only transiently or may reflect a specialized binding state for BipA, for example, when ribosomes are stalled. Similar to this idea, LepA has a specialized role in promoting a back-translocation reaction that allows the ribosome to correct tRNA binding errors (36). Surprisingly, however, given the seeming importance of this function, lepA is not essential, and deletion of this gene produces no defined phenotype (12). We speculated that perhaps lepA and bipA perform similar roles and thus can substitute for each other. If this was true, we would predict that a lepA bipA double mutation would be lethal or that the two mutations would at least have additive effects on cell growth. We therefore constructed a strain with this double mutation and found that the phenotype mimicked that of the bipA mutant and that growth was no more impaired than that of a single-deletion mutant (data not shown). Therefore, we surmise that BipA function is independent of LepA and that the role of BipA is distinct.
Recent evidence has shown that, contrary to conventional wisdom, at least some rRNA modifications have a functional role (13). RluD also modifies the 23S rRNA at three sites, inserting pseudouridines at positions 1911, 1915, and 1917, all of which are located in helix 69. Unlike deletion of rluC, however, deletion of rluD confers a severe growth defect. Isolation of suppressors demonstrated that RluD (and pseudouridylation at those three sites) is involved in translation termination by release factor 2 and that the primary reason for slow growth is attributable to the termination defect. Although a ribosome assembly defect is also observed in an rluD mutant, it is speculated to be a secondary effect due to synthesis of abnormal proteins (13). Thus, pseudouridylation may be important for specific steps of the translation cycle, and the contribution of sites other than those modified by RluD is still to be determined.
We do not think that BipA or RluC pseudouridylation is involved in termination, as is RluD, because mutations in bipA or in rluC do not inhibit growth under normal conditions (9, 34). Additionally, the locations of the pseudouridines catalyzed by RluD and RluC activities are notably different. The RluD-inserted modifications are located in helix 69, which contacts both the A- and P-site tRNAs, as well as the bridge B2a that connects the two subunits of the ribosome and is known to be involved in termination (1). The sites of modification directed by RluC, however, are all located near or within the peptidyl transferase center and thus are more likely to affect peptide bond formation (46).
Our data suggest that under certain conditions (such as low temperatures), ribosomes are dependent on BipA. Whether this dependence reflects ribosome assembly or translational accuracy or efficiency, we do not know, although we favor the model in which BipA affects translational efficiency. The loss of rRNA modification at three specific sites renders the ribosomes independent of BipA, implying a functional alteration in the ribosome that not only permits BipA-independent translation of target mRNAs but also does not interfere with translation of other mRNAs. This scenario raises an intriguing question as to why the cell retains both BipA and pseudouridylation at these sites when dispensing with both incurs no detrimental effect. We presume that both contribute to the efficiency of translation and that growth conditions under which the double deletion produces a growth disadvantage could be found.
Acknowledgments
We are extremely grateful to Catherine Squires, Aaron New, Selwyn Quan, and Janine Maddock for their generous gifts of bacterial strains and plasmids, the construction of some mutants, and thoughtful discussion.
This work was funded by an ND-EPSCoR dissertation grant to K.K. and a UND faculty seed grant to A.M.F.
Footnotes
Published ahead of print on 26 September 2008.
REFERENCES
- 1.Ali, I. K., L. Lancaster, J. Feinberg, S. Joseph, and H. F. Noller. 2006. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol. Cell 23865-874. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 22006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker, H. C., N. Kinsella, A. Jaspe, T. Friedrich, and C. D. O'Connor. 2000. Formate protects stationary-phase Escherichia coli and Salmonella cells from killing by a cationic antimicrobial peptide. Mol. Microbiol. 351518-1529. [DOI] [PubMed] [Google Scholar]
- 4.Bernhardt, T. G., and P. A. de Boer. 2004. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol. Microbiol. 521255-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brill, J. A., C. Quinlan-Walshe, and S. Gottesman. 1988. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 1702599-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldon, C. E., P. Yoong, and P. E. March. 2001. Evolution of a molecular switch: universal bacterial GTPases regulate ribosome function. Mol. Microbiol. 41289-297. [DOI] [PubMed] [Google Scholar]
- 7.Castanié-Cornet, M. P., K. Cam, and A. Jacq. 2006. RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli. J. Bacteriol. 1884264-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow, C. S., T. N. Lamichhane, and S. K. Mahto. 2007. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conrad, J., D. Sun, N. Englund, and J. Ofengand. 1998. The rluC gene of Escherichia coli codes for a pseudouridine synthase that is solely responsible for synthesis of pseudouridine at positions 955, 2504, and 2580 in 23S ribosomal RNA. J. Biol. Chem. 27318562-18566. [DOI] [PubMed] [Google Scholar]
- 10.Decatur, W. A., and M. J. Fournier. 2002. rRNA modifications and ribosome function. Trends Biochem. Sci. 27344-351. [DOI] [PubMed] [Google Scholar]
- 11.Del Campo, M., Y. Kaya, and J. Ofengand. 2001. Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli. RNA 71603-1615. [PMC free article] [PubMed] [Google Scholar]
- 12.Dibb, N. J., and P. B. Wolfe. 1986. lep operon proximal gene is not required for growth or secretion by Escherichia coli. J. Bacteriol. 16683-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ejby, M., M. A. Sørensen, and S. Pedersen. 2007. Pseudouridylation of helix 69 of 23S rRNA is necessary for an effective translation termination. Proc. Natl. Acad. Sci. USA 10419410-19415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farris, M., A. Grant, T. B. Richardson, and C. D. O'Connor. 1998. BipA: a tyrosine-phosphorylated GTPase that mediates interactions between enteropathogenic Escherichia coli (EPEC) and epithelial cells. Mol. Microbiol. 28265-279. [DOI] [PubMed] [Google Scholar]
- 15.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freestone, P., S. Grant, I. Toth, and V. Norris. 1995. Identification of phosphoproteins in Escherichia coli. Mol. Microbiol. 15573-580. [DOI] [PubMed] [Google Scholar]
- 17.Grant, A. J., M. Farris, P. Alefounder, P. H. Williams, M. J. Woodward, and C. D. O'Connor. 2003. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 48507-521. [DOI] [PubMed] [Google Scholar]
- 18.Green, R., and H. F. Noller. 1996. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA 21011-1021. [PMC free article] [PubMed] [Google Scholar]
- 19.Guyer, M. S., R. R. Reed, J. A. Steitz, and K. B. Low. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb. Symp. Quant. Biol. 45135-140. [DOI] [PubMed] [Google Scholar]
- 20.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, L., J. Ku, M. Pookanjanatavip, X. Gu, D. Wang, P. J. Greene, and D. V. Santi. 1998. Identification of two Escherichia coli pseudouridine synthases that show multisite specificity for 23S RNA. Biochemistry 3715951-15957. [DOI] [PubMed] [Google Scholar]
- 22.Karbstein, K. 2007. Role of GTPases in ribosome assembly. Biopolymers 871-11. [DOI] [PubMed] [Google Scholar]
- 23.Kiss, E., T. Huguet, V. Poinsot, and J. Batut. 2004. The typA gene is required for stress adaptation as well as for symbiosis of Sinorhizobium meliloti 1021 with certain Medicago truncatula lines. Mol. Plant-Microbe Interact. 17235-244. [DOI] [PubMed] [Google Scholar]
- 24.Kristensen, C. S., L. Eberl, J. M. Sanchez-Romero, M. Givskov, S. Molin, and V. De Lorenzo. 1995. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J. Bacteriol. 17752-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazar, S. W., and R. Kolter. 1996. SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 1781770-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyristis, M., A. E. Bryant, J. Sloan, M. M. Awad, I. T. Nisbet, D. L. Stevens, and J. I. Rood. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol. Microbiol. 12761-777. [DOI] [PubMed] [Google Scholar]
- 27.Margus, T., M. Remm, and T. Tenson. 2007. Phylogenetic distribution of translational GTPases in bacteria. BMC Genomics 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meberg, B. M., A. L. Paulson, R. Priyadarshini, and K. D. Young. 2004. Endopeptidase penicillin-binding proteins 4 and 7 play auxiliary roles in determining uniform morphology of Escherichia coli. J. Bacteriol. 1868326-8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Murphy, K. C. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 1802063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246321-330. [DOI] [PubMed] [Google Scholar]
- 32.O'Connor, M., W. M. Lee, A. Mankad, C. L. Squires, and A. E. Dahlberg. 2001. Mutagenesis of the peptidyltransferase center of 23S rRNA: the invariant U2449 is dispensable. Nucleic Acids Res. 29710-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owens, R. M., G. Pritchard, P. Skipp, M. Hodey, S. R. Connell, K. H. Nierhaus, and C. D. O'Connor. 2004. A dedicated translation factor controls the synthesis of the global regulator Fis. EMBO J. 233375-3385. (Retraction, 26:4607, 2007.) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Pfennig, P. L., and A. M. Flower. 2001. BipA is required for growth of Escherichia coli K12 at low temperature. Mol. Genet. Genomics 266313-317. [DOI] [PubMed] [Google Scholar]
- 35.Qi, S. Y., Y. Li, A. Szyroki, I. G. Giles, A. Moir, and C. D. O'Connor. 1995. Salmonella typhimurium responses to a bactericidal protein from human neutrophils. Mol. Microbiol. 17523-531. [DOI] [PubMed] [Google Scholar]
- 36.Qin, Y., N. Polacek, O. Vesper, E. Staub, E. Einfeldt, D. N. Wilson, and K. H. Nierhaus. 2006. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 127721-733. [DOI] [PubMed] [Google Scholar]
- 37.Raychaudhuri, S., J. Conrad, B. G. Hall, and J. Ofengand. 1998. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 41407-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raychaudhuri, S., L. Niu, J. Conrad, B. G. Lane, and J. Ofengand. 1999. Functional effect of deletion and mutation of the Escherichia coli ribosomal RNA and tRNA pseudouridine synthase RluA. J. Biol. Chem. 27418880-18886. [DOI] [PubMed] [Google Scholar]
- 39.Rouvière, P. E., and C. A. Gross. 1996. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 103170-3182. [DOI] [PubMed] [Google Scholar]
- 40.Rowe, S., N. Hodson, G. Griffiths, and I. S. Roberts. 2000. Regulation of the Escherichia coli K5 capsule gene cluster: evidence for the roles of H-NS, BipA, and integration host factor in regulation of group 2 capsule gene clusters in pathogenic E. coli. J. Bacteriol. 1822741-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott, K., M. A. Diggle, and S. C. Clarke. 2003. TypA is a virulence regulator and is present in many pathogenic bacteria. Br. J. Biomed. Sci. 60168-170. [DOI] [PubMed] [Google Scholar]
- 42.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Trisler, P., and S. Gottesman. 1984. lon transcriptional regulation of genes necessary for capsular polysaccharide synthesis in Escherichia coli K-12. J. Bacteriol. 160184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, F., N. Q. Zhong, P. Gao, G. L. Wang, H. Y. Wang, and G. X. Xia. 2008. SsTypA1, a chloroplast-specific TypA/BipA-type GTPase from the halophytic plant Suaeda salsa, plays a role in oxidative stress tolerance. Plant Cell Environ. 31982-994. [DOI] [PubMed] [Google Scholar]
- 45.Wrzesinski, J., K. Nurse, A. Bakin, B. G. Lane, and J. Ofengand. 1995. A dual-specificity pseudouridine synthase: an Escherichia coli synthase purified and cloned on the basis of its specificity for psi 746 in 23S RNA is also specific for psi 32 in tRNA(phe). RNA 1437-448. [PMC free article] [PubMed] [Google Scholar]
- 46.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292883-896. [DOI] [PubMed] [Google Scholar]