Abstract
There is an urgent need to search for biomarkers that are indicative of neurodegenerative diseases, as the clinical diagnosis of which remains unsatisfactory. Mass spectrometry (MS) has been playing an important role in studying peptide and protein identities, structures, modifications and interactions that collectively drive their biological functions. MS-based proteomics technology is thus well suited for the biomarker discovery. This article reviews the overall strategies and workflows employed for biomarker discovery and recent applications of MS-based proteomics in neurodegenerative diseases. Special emphasis is placed on the studies of protein post-translational modification pattern changes and differential peptidomics under these pathological conditions.
Keywords: Biomarker, neurodegenerative diseases, proteomics, peptidomics, post-translational modifications, mass spectrometry
Introduction
Neurodegenerative diseases, such as Alzheimer's disease (AD), Parkinson's disease (PD), amyotrophic lateral sclerosis (ALS) and prion diseases, are disorders caused by the deterioration of certain nerve cells. There are no cures for these disorders because the neurons of the central nervous system cannot regenerate on their own after cell death or damage. Diagnosis of neurodegenerative diseases is made primarily on clinical grounds including neuropsychological testing, limited laboratory tests, and brain imaging. However, there is considerable overlap in the clinical symptoms of these diseases, which complicates effective and accurate diagnosis, especially in their early stages. Therefore, there is an urgent need to develop a reliable approach/assay to diagnose neurodegenerative disorders early in their course, and to monitor responses of the patients to new therapies.
Biomarkers, a consensus definition of which is “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention” [1, 2], have become the interest of research in potential solution for disease diagnosis. The fact that proteins, rather than genes, are functionally responsible for almost all biological processes makes proteins attractive candidates in biomarker discovery.
Biomarkers find many applications in modern biology and medicine, from pregnancy tests to monitoring cholesterol levels. In recent years, there has been a growing interest in applying proteomics technology to research on clinical diagnostics of neurodegenerative diseases. Here we will review the use of mass spectrometry (MS) in the field of biomarker discovery in neurodegenerative diseases. While several early reviews presented detailed accounts on protein-based biomarker identification and discovery in various neurological disorders [3–7], this review will focus on the various post-translational modification (PTM) pattern changes and largely overlooked peptidomics involved in several major neurological diseases.
Workflow of Proteomics/Peptidomics
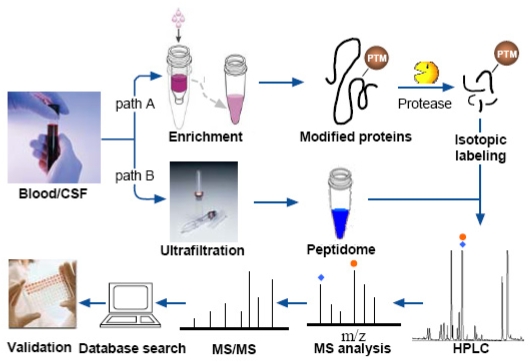
Generally, MS-based proteomic/peptidomic workflow involves the following five steps: (i) sample preparation; (ii) fractionation of protein/peptide samples; (iii) protein/peptide identification by MS or tandem MS; (iv) data processing by bioinformatics tools; and (v) validation of the results by alternative techniques [8]. Figure 1 depicts the major steps involved in MS-based proteomics /peptidomics for biomarker discovery.
Figure 1.
Overview of the workflow of MS-based proteomics/peptidomics for biomarker discovery. Path A illustrates a typical procedure for a proteomic study of post-translational modification (PTM), with an additional step of enriching the proteins with a particular PTM. Path B reveals the workflow of a peptidomics study, in which the peptidome is separated from the samples by ultrafiltration with low molecular weight filtrate preserved for subsequent analyses. After sample preparation steps the peptides are fractionated by HPLC and characterized by MS and MS/MS. The data processing is followed by a validation step, which usually involves immuno-based assays.
Step 1: Sample Preparation
There are two major sources for the search of protein biomarkers: one is body fluids such as cerebrospinal fluid (CSF) and serum, and the other is diseased tissue, which is post-mortem brain in the case of neurodegenerative diseases.
Body fluids represent an attractive medium for biomarker discovery attributed to their easy accessibility and protein-rich content. Being the only body fluid in direct contact with the brain, CSF is close to the site of pathology and instantaneously reflects the metabolic state of the brain under varying conditions [3, 9]. For this reason, comparison of proteomic profiles of human CSFs collected from diseased and normal individuals is particularly informative as the differentially expressed proteins may serve as putative biomarkers and may help to identify functional pathways that could shed more light on the pathogenesis of the disease of interest. Over the past several years, a number of groups have made efforts to characterize human CSF proteome and to search for biomarkers related to several major neurodegenerative diseases such as AD, PD and multiple sclerosis [10–15]. These studies have generated a list of CSF biomarker candidates associated with each disease that change in relative abundance. However, discrepancies were noted among different labs and the candidates can hardly be used for diagnosis.
The poor reproducibility of CSF biomarker candidates can be partially attributed to various challenges in sample preparation. The protein concentration of CSF is only 1/200 that of blood, with 70% of the mass being albumin and immunoglobins [16]. In addition, the CSF protein profile significantly overlaps with plasma. As a result, even a minute contamination with blood can have tremendous effect on the concentration of CSF proteins. The seemingly trivial sample storage process can sometimes introduce artifacts and give confounding results. For example, cystatin-C, a putative marker found to be increased in AD patients, was later proven by the same lab to be a mere processing artifact which could be eliminated if the samples were stored at −80°C [9, 17].
Although the blood is not in direct contact with the brain, it is a good alternative source for biomarker discovery because about 500ml of CSF are absorbed into the blood every day. Furthermore, damage to the blood-brain barrier, which occurs in neurodegenerative diseases, can enhance the exchange of proteins between brain and blood [18]. In addition, sampling blood is not as invasive as lumbar puncture for CSF collection. However, the dynamic range of proteins within plasma is known to exceed 1010, while only 106 protein concentration range can be assessed by the current most powerful MS technology when coupled with extensive separation [19]. It is therefore desirable to remove the most abundant proteins by immunodepletion methods [20–22] prior to subsequent fractionation and detection.
Although not applicable for pre-mortem biomarker discovery, post-mortem brain tissue is advantageous in allowing direct analysis of proteins from specific regions of interest and diseased loci. Instant freezing of the samples is necessary to avoid degradation of the proteins and their modifications [23]. The methods of protein extraction and solubilization differ with tissues of interest. A major challenge involved in brain proteomics is the isolation and separation of hydrophobic proteins. It should be kept in mind that some buffer ingredients, especially detergents, will hamper the subsequent separation and may not be compatible with MS analysis. The research groups of Caprioli and Andrén have circumvented the challenges of brain protein extraction by MS imaging of brain tissue sections directly placed on a metal sample plate in a molecular profiling study of animal model of PD [24]. Lubec et al published a thorough review of protocols used in brain proteomic research that offered detailed discussions about the above issues [25].
Step 2: Protein/Peptide Fractionation
Separation techniques used in proteomics research are either gel based fractionation methods such as two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), or non-gel based, such as one- or multidimensional liquid chromatography (LC) and capillary electrophoresis (CE). It is generally agreed that no single separation and detection technique can give a full account of the protein profile of a complex mixture such as blood. Although 2D-PAGE provides unparalleled resolving power and capability to visualize abundance changes, it suffers from several limitations such as poor performance on hydrophobic, highly acidic and basic proteins, difficulty in automation and limited dynamic range. LC techniques are complementary to gel-based approach and have become increasingly popular in proteomic research because they are reproducible, highly automated, and flexible in choosing different combinations of stationary and mobile phases. Popular fractionation methods used in proteomics studies include Multidimensional Protein Identification Technology (MudPIT) [26, 27], gel-based liquid chromatography (GeLC) [28, 29], affinity/reversed phase (RP) LC [30, 31] along with many others. If the low molecular weight region of the sample is the desired target for analysis, an ultrafiltration step is performed to isolate the peptidome from the interference of the proteins [32]. Filter devices with a cutoff of 10 kDa are generally used. More details of fractionation methods can be found in many extensive and thorough reviews [33–35].
One challenge with the fractionation and subsequent characterization of body fluid samples is the wide concentration range or proteins. The most abundant component in human body fluids, human serum albumin (HSA), contributes approximately 50% of the total protein content in plasma and CSF. If this component could be selectively removed, the chances of finding biomarkers in the lower-abundance range would be greatly improved. As part of the fractionation, several approaches with varying specificities are employed to deplete a number of most abundant proteins [20–22, 36]. One popular choice of abundant protein removal involves the use of immunodepletion based on IgG and IgY antibodies. Depleted serum from the multiple affinity removal chromatography has shown increased resolution in 2-DE and enhanced sensitivity of low-abundant proteins in a reproducible fashion [37].
Step 3: Protein/Peptide Identification and Quantitation by MS
Traditionally, proteins have been identified by de novo sequencing via Edman degradation, with subsequent detection of the released amino acid derivatives by UV absorbance spectroscopy. In the 1990s, the analysis of proteins had been revolutionized by the rapid development of MS ionization methods and instrumentation. Specifically, the emergence of two ionization methods in the late 1980s – electrospray ionization (ESI) [38] and matrix-assisted laser desorption/ionization (MALDI) [39] contributed to the predominant use of mass spectrometry for the analysis of large biomolecules including proteins and peptides. The selection of MS method is largely determined by the need of the biomarker research and the pertinent features that the various instrument types can provide. Instrument performance such as mass accuracy, resolving power, sensitivity and throughput should be taken into consideration. Domon and Aebersold have evaluated the characteristics of commonly used MS instruments in proteomics [40]. In addition to accurate mass measurement of molecular ions, the key to structural identification and characterization of peptides and proteins lies at the fragmentation capability offered by tandem MS techniques. Besides the more conventional collision-induced dissociation (CID) fragmentation techniques, two recently developed electron-based tandem MS fragmentation methods, electron capture dissociation (ECD) [41] and electron transfer dissociation (ETD) [42] have shown great promise for sequencing large peptides and small proteins, while preserving labile post-translational modifications.
MS-based quantitation methods have gained increased popularity and played more significant roles in biomarker discovery over the past several years, which can be complementary to the classical methods of differential protein gel or blot staining by dyes and fluorophores. Most of these MS-methods employ differential isotope labeling to create a specific mass tag that serves as the basis for relative quantitation. These mass tags can be introduced into proteins or peptides in a number of ways. One of the methods is metabolic labeling, which introduces a stable isotope signature into proteins during cell growth and division. A very popular approach called the stable isotope labeling by amino acids in cell culture (SILAC) [43] has been widely employed in quantitative proteomics studies. Chemical labeling is an alternative approach that performs specific chemical derivatization in vitro and introduces various tags to the peptide terminals and side chain functional groups. For example, Gygi et al [44] developed the isotope-coded affinity tag (ICAT) in which cysteine residues are specifically derivatized with a reagent containing either zero or eight deuterium atoms as well as a biotin group for affinity purification of cysteine-containing peptides. In chemical labeling methods relative quantitation is usually achieved by integration of MS signal over “heavy” and “light” labeled peptides in survey spectra. Isotope tags for relative and absolute quantification (iTRAQ) [45] employ a different concept by introducing tags that initially produce isobaric labeled peptides which precisely co-migrate in LC separations, and only upon fragmentation are the different tags distinguished by the mass spectrometer. Recently label-free approaches have been gaining more attention from the research community, which perform quantitation by either comparing the signal intensity of peptide precursor ions belonging to a particular protein [46], or counting the number of tandem MS fragmentation spectra identifying peptides of a given protein [47].
Step 4: Data Processing
The analysis of the thousands to millions of MS/MS spectra generated in proteomic studies can be a daunting task, which requires sophisticated algorithms. Over the last decade, many search engines/algorithms have been developed for handling such complex datasets, among which the most popular ones include SEQUEST [48], MASCOT [49], OMSSA [50], X!Tandem [51], MS-Tag [52] and many others. Databases for searching PTMs by MS data include UNIMOD [53], Deltamass [54], FindMod [55] and those that use MS/MS data for PTM characterization include SEQUEST, Modificomb [56] and MODi [57]. Although the emergence of automated database searching significantly increases the throughput of data analysis, it should be used with caution due to imperfect searching algorithms and possible errors existing in various databases. Validation of peptide and protein identification results has therefore become a necessary step. The use of randomized or reversed sequence databases has been introduced to evaluate false positive results [58].
Oftentimes bioinformatics tools beyond protein database search engines are needed to turn the enormous amount of data into useful information. For example, surface enhanced laser desorption ionization mass spectrometry (SELDI-MS) is one of the few methods that can be used to profile several hundred proteins from complex samples with a reasonable throughput. However, SELDI-MS profiles are characterized by complex spectra, high dimensionality, and significant noise, which all together make the discovery of biomarker peaks in clinical samples a challenging task. The need for computational methods is obvious in order to find peaks that correlate with phenotypes and to assess their statistical significance. Several classification techniques have been utilized for discriminating cancer samples from control samples using proteomic data. The two main components of these approaches are the feature selection method and a classification method to build a predictive model [59]. For example, Li et al used the signal-to-noise ratio for an initial feature selection and subsequently used “unified maximum separability analysis” repeatedly for classification in their breast cancer study [60].
Step 5: Validation
Once a panel of biomarker candidates has been identified in the discovery phase, a validation phase must be followed with a goal of selecting the ones with highest potential from the list for the clinical diagnosis [61]. While the biomarker candidates are typically identified based on mass spectrometry methods, it is often desirable to develop an independent analytical method to validate these putative markers. For example, immuno-based assays are often preferred for validation and development for clinical diagnosis due to its high sensitivity and throughput. Both Western blot and enzyme-linked immunosorbent assay (ELISA) are commonly used immuno-based techniques to confirm that the concentration of the candidate is significantly different between the control and the diseased state. Appropriate sets of blinded samples must be analyzed independently and the diagnostic sensitivity and specificity of the candidates must be determined [62, 63]. However, transferring MS-derived data to a working and validated immunological assay can be difficult due to the lack of commercially available antibodies or technical challenges and high cost associated with development of highly specific antibodies. Alternatively, several mass spectrometry methods have been employed for the characterization/validation phase of a clinical diagnostic test. For example, multiple reaction monitoring (MRM) MS using a triple quadrupole mass spectrometer has been employed for biomarker quantitation and validation. Targeted MS analysis using MRM enhances the lower detection limit for peptides by up to 100-fold by allowing rapid and continuous monitoring exclusively for the specific ions of interest [64]. Furthermore, MRM analysis coupled to stable isotope also offers multiplexing capability and increases the reliability of quantification by enhancing both the specificity and accuracy of analysis [64, 65].
General Status of Protein-based Biomarker Discovery in Neurodegenerative Diseases
In neurodegenerative disease research, proteomics technology has been mainly used in the analysis of the brain and CSF of both human and animal models with the goal of collecting information about gene products involved in such disorders, such as alterations in protein abundance and PTMs. Proteins with altered levels are potential drug targets or biomarkers. The biomarkers are useful for the disease diagnosis if they can be found in body fluids such as blood and urine.
Since the early application of proteomics in neuroscience, more than 300 unique proteins with changed levels or modifications have been reported to be associated with neurodegeneration and psychiatric disorders [4], most of which are involved in metabolism pathways, cytoskeleton formation, signal transduction, transport and detoxification. The proteins detected in these studies are mostly high-abundance CSF or brain proteins. A number of these proteins, for example, fructose-biphosphate aldolase and peroxiredoxins 1, were found to be altered in multiple disorders [4]. Admittedly, most of the changes have not been validated by other methods and many of them are irreproducible, statistically insignificant, or even contradictory to each other. Only after verification the resultant changes may be useful in identifying corresponding disorders.
Several reviews [3–7] have comprehensively covered the current status and progress of neurodegenerative disease-related proteomics studies in various major neurological disorders. In this review, we will focus our discussions on two sub-fields of MS-based proteomics investigation in neurodegenerative diseases, namely peptidomics and PTMs.
Peptidomics
The low molecular mass range (<10k Daltons) of both CSF and serum, although being a promising reservoir for biomarker discovery, remains largely uncharacterized. Analysis of endogenous peptides produced by aberrant cleavage of proteins in the diseased state can not only provide alternatives for disease diagnosis, but also shed light on the mechanisms and pathways involved in the neurodegenerative diseases. One significant advantage of using a MS-based approach is that it can unambiguously record protein fragment peaks in mining the low-mass proteome and peptidome. If a biomarker for a given disease state is a fragment of a larger protein, it may be extremely difficult to produce effective antibodies for conventional tests such as ELISA. Furthermore, coupling LC to MS overcomes the limit that 2-D gel electrophoresis has very low resolution for small proteins and peptides.
Due to the proximity to the brain, CSF has been the subject of several peptidomics studies recently. Many CSF peptides identified so far are biologically active. In an early study, Stark et al [66] developed organic phase extraction and MS-based profiling strategy and identified a number of peptide fragments of human CSF proteins. Later, Yuan and Desiderio [67] applied ultrafiltration with a limit of Mr<5 kDa and solid phase extraction (SPE), followed by LC-MS/MS to characterize human CSF peptidome. In this proof-of-principle study, 20 representative peptides derived from 12 proteins were identified. As outlined in Figure 1, by means of ultrafiltration, the CSF samples can be split into low molecular weight “peptidome” and higher molecular weight “proteome” fractions. While proteome fractions are subjected to tryptic digestions followed by LC-MS/MS identifications in conventional proteome mapping experiments, the endogenous CSF peptidome fractions are analyzed directly, by capillary LC coupled to tandem MS experiments. This approach has become the method of choice for peptidome profiling in CSF and other body fluid samples. By adopting similar strategy and using a hybrid LTQ-orbitrap mass spectrometer with high accuracy and high resolution, Zougman et al[32] were able to enhance the CSF peptidome profiling to confident identification of 563 peptides derived from 91 precursors.
In addition to CSF peptidome, it is now recognized that the low molecular weight (LMW) fraction of the serum proteome may contain shed proteins and protein fragments emanating from physiologic and pathologic events taking place in all perfused tissues. Geho et al [68] proposed theoretical models, which predict that the vast majority of LMW biomarkers exist in association with circulating high molecular mass carrier proteins. Lopez et al [69] have recently examined the carrier-protein-bound fraction of serum to discover patterns of peptide ions that provide diagnostic signatures of AD. They pulled down the target peptides by affinity chromatography and analyzed them by MALDI-TOF. Although numerous LMW putative markers were detected, the amino acid sequence or identity of each marker, and how each one contributed to the classification in terms of specificity and sensitivity were not investigated.
An emerging exciting field in neuroscience-related peptidomics research is imaging mass spectrometry (IMS) (for review see [70]). In a typical procedure for IMS on tissues, a thin sliced tissue section is placed on a sample plate and MALDI matrix is deposited either as a thin layer or as a spot pattern. The sample is then introduced into the instrument for MS analysis and a laser is rastered across the tissue section collecting an array of mass spectra at each XY coordinates. This array of mass spectra can then be processed to produce individual molecular ion images, in which each pixel represents ion signals extracted from the corresponding spectrum. IMS offers several advantages over immunocytochemistry, an antibody-based alternative approach in determining the distribution of neuropeptides. These advantages include higher throughput, higher chemical specificity, and the ability to discover novel peptides. As an example, the distribution of five structurally related Aβ peptides in mouse brain sections has been determined by IMS with attomole sensitivity and 50-μm lateral resolution [71]. While most MALDI-based IMS studies rely on mass measurement alone to identify peptides and proteins, DeKeyser et al have taken advantage of the TOF/TOF mass analyzer that enables both post-source decay (PSD) and CID fragmentation in MS/MS, and provided increased confidence for assignment of a number of neuropeptide families [72]. The first attempt [24] to directly profile proteins and neuropeptides in the brain tissue of a rat model of PD demonstrated differential expression of numerous proteins as well as some changes in PTMs such as alterations of acetylation. In a later study [73], PEP-19, a 6.7-kDa polypeptide that belongs to a family of proteins involved in calcium transduction through their ability to interact with neuronal calmodulin, was found to be significantly decreased in model PD brain. Because IMS studies enabled the localization of specific peptide or protein molecules to the diseased region of the tissues, there is greater chance for this technology to discover potentially disease-relevant biomarkers. The limitation, however, is that the tissue is not as easily available as the body fluids. Also, the tissue-based disease biomarkers are more likely indicative of late-stage of disease progression. Nonetheless, once identified in diseased tissues via IMS technology, these putative (neuro)peptide or protein markers can serve as useful candidates to develop body fluid-based diagnostic assays.
Post-Translational Modifications (PTMs)
It has been extensively documented that a wide variety of PTMs can influence protein folding, modulate real-time dynamics, and regulate functions of most proteins. For example, phosphorylation and dephosphorylation of protein machinery play a critical role in intracellular signal transduction in the brain. Many other important PTMs such as glycosylation, methylation, acetylation, oxidation, nitrosylation and ubiquitination also regulate the functions, cellular targeting and degradation of proteins in the central nervous system (CNS). Thus, in addition to the change of protein concentrations, it is believed that aberrant PTM patterns of various proteins could be associated with the onset and progression of several neurodegenerative diseases.
Analysis of PTMs is challenging for many reasons. PTMs vary in sizes and physicochemical properties. Although PTMs are site specific, the diversity that they generate far exceeds the number of gene products. Furthermore, covalently attached PTMs are usually present at substoichiometric levels.
Western blot has been widely used to determine the presence of PTMs. However, this technique relies on the prior knowledge of the type and position of specific modifications and the availability of antibodies. It has low throughput and not ideal for studying highly complicated samples. In contrast, MS can be employed to discover novel modifications as well as monitor the known ones. The sites of PTMs can even be determined in MS/MS. Multiple proteins with the same modifications can be studied at once by MS coupled to specific isolation methods. Numerous MS-based proteomics strategies have been developed to study the PTM events. Specific chemical or affinity enrichment steps are usually incorporated into the sample preparation or fractionation stages of the general scheme of proteomic studies (Figure 1) [74, 75].
Phosphorylation
Phosphorylation, which occurs to the serine, threonine and tyrosine residues of proteins, is known as the most abundant and ubiquitous PTM involved in protein regulation and signal transduction. A few phosphorylation events have been found to be associated with the pathogenesis of neurodegenerative diseases. For example, microtubule-associated protein tau undergoes several PTMs, including hyperphosphorylation, and aggregates into paired helical filaments (PHFs), a component of neurofibrillary tangles (NFTs) characteristic of AD. The main known physiological functions of tau are stimulating microtubule assembly and stabilizing microtubule structure. It is well known that the phosphorylation of tau regulates its activity to bind to microtubules. The phosphorylation level of tau isolated from autopsied AD brains is 3- to 4- fold higher than the normal counterpart [76]. Upon dephosphorylation, the tau protein loses its toxicity. Gunnarsson et a have shown in vitro that memantine, a moderate affinity N-methylD-aspartate receptor antagonist approved for treatment of AD, reverse induced abnormal hyperphosphorylation of tau in hippocampal neurons of rats [77]. Consistent with the hyperphosphorylation of tau, several other neuronal proteins such as neurofilaments [78], β-tubulin [79] are also hyperphosphorylated in AD brain.
In the last decade, numerous studies have examined the total tau level in CSF with the hope to find a biomarker for diagnosis of AD. However, it is found that total tau level in CSF merely reflects nonspecific processes of neuronal degeneration and axonal damage [80]. In order to improve the specificity to discriminate AD from other neurodegenerative diseases, several groups have studied the phosphorylated tau (p-tau) level in CSF [81–84]. These studies used monoclonal antibodies specific to individual phosphorylated epitopes of tau. It was shown that p-tau level is more relevant than total tau level for diagnosis of AD.
MS-based approaches have been employed to identify phosphorylated proteins and quantitate the extent of phosphorylation. In the fragmentation process of the MS/MS, phosphopeptides in the positive-ion mode yield a neutral loss of H3PO4 at phosphoserine or phosphothreonine residues. Using this approach, Hanger et al discovered additional sites of phosphorylation on tau [85]. Several candidate kinases have been identified that can phosphorylate tau on sites found to be phosphorylated in PHF-tau. For example, Derkinderen et al identified phosphorylated Tyr-394 in PHF-tau from an AD brain and in human fetal brain tau using kinase c-Abl and mass spectrometry [86]. Reynolds et al identified by nanoelectrospray MS that three mitogen-activated protein kinases JNK, p38, ERK2, and glycogen synthase kinase 3β (GSK3β) are strong candidates as tau kinases that may be involved in the pathogenic hyperphosphorylation of tau in AD [87].
More recently, several methodologies have exploited the electrostatic properties of phosphate groups to enrich for phosphopeptides prior to MS analysis. Immobilized metal affinity chromatography (IMAC) was one of the first, which isolated the phosphopeptides by the highly selective affinity between phosphate groups and Ga(III) ions [88]. The phosphopeptides were then eluted from the metals by basic buffer, separated by RPLC and analyzed by MS/MS. More recently, other metal oxide species such as Fe(III)-IMAC [89], TiO2 [90] and ZrO2 [91] have been used for several phosphoproteomic analyses in model organisms and cellular organelles. Recently, comprehensive proteomic identification of phosphorylation sites in postsynaptic density preparations, a dense complex of proteins whose function is to detect and respond to neurotransmitters released from presynaptic axon terminals, has been achieved by Trinidad et al [92]. In this study, 723 unique phosphorylation sites were determined by coupling strong cation exchange (SCX) chromatography with IMAC. In another study, 331 phosphorylation sites representing 79 proteins were identified in the mouse synapse phosphoproteome [93]. However, no quantitation studies of these phosphoproteomes have been examined in the context of neurodegenerative diseases.
Glycosylation
The covalent attachment of oligosaccharides to asparagine residues (N-linked) or to serine and threonine residues (O-linked) is crucial for the protein activity, folding, stability, receptor-ligand recognition and cellular localization. However, there is only limited study of glycosylation changes in neurodegenerative diseases.
Several studies suggest that aberrant glycosylation changes occur in AD. Liu et al have shown that aberrant glycosylation may modulate tau protein at a substrate level so that it is easier to be phosphorylated and more difficult to be dephosphorylated at several phosphorylation sites in AD brain [94, 95]. Small and coworkers identified glycosylated isoforms of acetylcholinesterase and butyrylcholinesterase that are increased in AD CSF [96]. Further evidence of the importance of glycosylation in AD is provided by the findings that glycosylation regulates nicastrin [97], a presenilin complex component, and Asp-2 [98], a β-secretase protein, both of which play a role in cleavage of the amyloid precursor protein.
In addition to AD, glycosylation patterns have been found to be altered in other neurodegenerative diseases. For example, Reelin, a glycoprotein that is essential for the correct cytoarchitectonic organization of the developing CNS, is up-regulated in the brain and CSF in several neurodegenerative disorders, including frontotemporal dementia, progressive supranuclear palsy, PD as well as AD [99]. Furthermore, glycosylation pattern of Reelin differed in plasma and CSF, and the CSFs of control and diseased samples also exhibited different glycosylation patterns. These results support that glycoprotein Reelin is involved in the pathogenesis of a number of neurodegenerative diseases.
As with other protein PTM, glycopeptides derived from digesting glycoproteins are often suppressed by their non-glycosylated counterparts in the MS analysis. For this reason, enrichment strategies have been developed for glycopeptides. A general approach employs the natural affinity between glycans and a class of proteins known as lectins. Kaji et al developed a technique named isotope-coded glycosylation-site-specific tagging (IGOT) based on lectin column-mediated capture of glycopeptides produced by tryptic digestion of protein mixtures, followed by peptide-N-glycosidase-mediated incorporation of a stable isotope tag, 18O, specifically into the N-glycosylation site [100]. The identities and glycosylation sites are then characterized by multidimensional LC-MS. Another approach involves the use of serial lectin affinity chromatography (SLAC), in which two or more lectin columns with varying binding specificities are combined to select for subclasses of proteins and peptides containing various glycan structures [101]. SLAC can be coupled with isotopic labeling to compare the relative abundance of glycoproteins in human serum [102]. An alternative approach to lectin affinity chromatography exploits the chemical properties of glycans, in which the vicinal diols of mannose residues are oxidized to aldehydes, and then captured by forming stable Schiff-base bonds with solid-phase supported hydrazide [103].
Recently, lectin weak affinity chromatography (LWAC) and MS has been used to study in vivo O-GlcNAc, an O-linked glycosylation analogous to phosphorylation, from a postsynaptic density preparation [104]. Because relatively poor fragmentation in traditional CID is usually observed for O-GlcNAc modified peptides due to the favorable dissociation of labile O-GlcNAc, alternative fragmentation method ECD on a hybrid ion trap-Fourier transform ion cyclotron resonance (IT-FTICR) mass spectrometer was used for its ability to preserve labile PTMs [105]. The effectiveness of this strategy on complex peptide mixtures has been demonstrated through enrichment of 145 unique O-GlcNac-modified peptides, 65 of which are sequenced and belong to proteins with diverse functions in synaptic transmission. The combination of this work and an accompanying report [92] on the phosphoproteome of postsynaptic density preparations enables the construction of proposed models of complex protein regulation at the synapse through the potential interplay of these PTMs. It will be of interest to examine O-GlcNAc and its potential relationship with phosphorylation in proteomic studies of AD and other neurodegenerative disorders, but this has not been explored yet.
Oxidation
Increasing evidence indicates that oxidative stress plays a crucial role in some of the most important neurodegenerative diseases such as AD, mild cognitive impairment (MCI), PD, ALS and Huntington's disease (HD) [106].
The most widely used marker for oxidation damage to proteins is carbonylation – the conversion of amino acid hydroxyl side chains to the ketone or aldehyde derivative. Elevation of total carbonyl level of proteins has been reported for both AD and PD [107]. Initial attempts in the characterization of carbonylated proteins have involved oxyblot [108, 109], in which the carbonyl groups are derivatized with 2,4-dinitrophenylhydrazine (DNPH), followed by Western blotting of DNPH on 2-D gel separated proteins. The reactive regions of the gel are then excised for protein identification. The first use of MS-based proteomics identified several specifically oxidized proteins in AD brain: creatine kinase BB (CK), glutamine synthase (GS), ubiquitin carboxy-terminal hydrolase L-1 (UCH L-1), α-enolase and dihydropyrimidinase-related protein 2 (DRP2) [110, 111]. A potential caveat of this approach is that no direct evidence of oxidation from MS data is shown; the potential false positives may be produced as the correlation between Western blot signals and specific protein identities in gels may not be matched.
An alternative methodology employs biotin hydrazide Michael Addition to derivatize carbonyls, which then allows streptavidin capture of the oxidized proteins/peptides [112]. LC-MS/MS was used in this approach to overcome the problems of 2-D gel electrophoresis, in which certain types of proteins are often excluded or underrepresented. By combining this approach to bioinformatic tools, Soreghan et al successfully detected 117 oxidatively modified proteins, 59 of which were specifically associated with AD transgenic (PS1 + A beta PP) mice at 6 months of age [113].
A very valuable application of MS-based proteomics is to study the responsiveness to treatments of the diseases. For example, it is reported that decreased production of amyloid β-peptide (Aβ), the major constituent of senile plaques and a pathological hallmark of AD, by intracerebroventricular injection of an antisense oligonucleotide (AO) could reduce lipid peroxidation and protein oxidation and improve cognitive deficits in an aged senescence-accelerated mice model. Poon et al [114] positively identified decreased carbonylation on several proteins and increased expression of profiling 2 (Pro-2) in response to the lowering of Aβ levels by MALDITOF MS analysis and MASCOT database search.
Nitration
In addition to carbonylation, other forms of oxidative damage to proteins in neurodegenerative diseases are examined. For example, nitrotyrosine modifications of protein have been documented in oxidative stress-related pathologies in a variety of tissue types and can affect enzyme catalytic rates, protein interactions and phosphotyrosine signaling pathways. Nitration of protein tyrosine residues appears to be an early event in the lesions of ALS, PD, and AD. In particular, site-specific nitration can affect the function of several neurodegeneration-related proteins, such as Mn superoxide dismutase and neurofilament light subunit in ALS, α-synuclein and tyrosine hydroxylase in PD, and tau in AD [115, 116]. Although several highly specific immunological detection methods have been used to reveal protein nitration in vivo, MS-based proteomics approach offers the opportunity to discover novel protein candidates with nitrotyrosine modifications. Sacksteder et al [117] performed a comprehensive proteomic analysis of a whole mouse brain using LC/LCMS/MS and identified 29 proteins with 31 nitrotyrosine modifications among a total of 7792 proteins. Despite the low percentage of occurrence, more than half of these in vivo nitrated proteins have been shown to have functional links with PD or other neurodegenerative diseases. More recently, Zhang et al [118] developed a novel method for selective enrichment and detection of nitrotyrosine-modified peptides from complex samples, by chemically converting nitrotyrosine into free sulfhydryl groups followed by thiopropyl sepharose resin capture. An increased effectiveness of this method was demonstrated by the identifications of 150 unique nitrated peptides resulting from 102 proteins of mouse brain homogenate. With these method developments, the disease-related nitroproteome is a whole new territory that is yet to be explored.
Other PTMs
Ubiquitination normally labels misfolded or damaged proteins for ATP-dependent degradation through the well-known ubiquitinproteosome system. The PHFs in tangles of AD brains are ubiquitinated and the level of ubiquitin is increased several-fold in the cerebral cortex [119]. Although PHF-tau is highly ubiquitinated, it is not readily degraded. Instead, it is deposited as NFTs in AD brain. A possible cause proposed is a defective proteosome system in AD brain [120]. The majority of mass spectrometric methods for ubiquitination site mapping exploit the presence of a Gly-Gly motif on ubiquitinated lysine residues, resultant from tryptic digestion of the ubiquitination substrate, as a signature peptide [121, 122].
New variants for ubiquitination, such as sumoylation and neddylation, have been implicated in various neurodegenerative diseases. Sumoylation is a PTM by which small ubiquitin-like modifiers (SUMOs) are covalently conjugated to lysine residues on target proteins. Similar to ubiquitin, SUMOs colocalize with the neuronal inclusions associated with several neurodegenerative diseases, including multiple system atrophy and HD. The identification of huntingtin, ataxin-1, tau and α-synuclein as SUMO substrates provides further evidence of the involvement of this modification in the pathogenesis of the neurological diseases [123]. General approaches to the identification of novel cellular SUMO substrates rely upon purification of sumoylated proteins from cell lysates via affinity tags, followed by MS analysis [124]. A recent effort used activated-ion electron capture dissociation (AI-ECD) or infrared multiphoton dissociation (IRMPD) on an FTICR mass spectrometer to detect sumoylated proteins, via a dominant fragmentation with the loss of the SUMO modification through the N-Cɛ; bond cleavage within the modified lysine side chain [125]. Neddylation, another ubiquitin-like PTM, has also been studied by affinity purification and tandem MS [126]. In spite of the progress of proteomics studies, many details of the sumoylation and neddylation pathways, including the modification sites, remain unsolved.
Glycation, a non-enzymatic linkage of glucose or other reducing sugars to the amino groups of proteins, has also been associated with neurodegenerative diseases. Glycation often leads to subsequent oxidation, dehydration, condensation and formation of heterogeneous products called advanced glycation end products (AGEs). It has been demonstrated that AGEs can be identified immune-histochemically in both senile plaques and NFTs from patients with AD [127]. Glycation of Aβ significantly enhances its aggregation in vitro [128], and the glycation of tau, in addition to hyperphosphorylation, appears to facilitate the formation of PHFs [129]. It is therefore suggested that serum or CSF levels of toxic AGEs (TAGEs) can be used as a biomarker for early detection of AD [130]. Recently, MS has also been used for the study of glycated proteins [131]. For example, a reversed-phase liquid chromatography method followed by a neutral loss scan mass spectrometric method was developed for the screening of glycation in proteins [132]. The neutral loss scan was based on a unique sugar moiety neutral loss (-162 Da) in the fragmentation spectra of glycated peptides. Although proteomics approach has been used to study glycation in diabetes [131], its application in profiling glycated proteins involved in neuro-degenerative diseases is still rare.
Conclusions and Outlook
Due to the sample complexity of the body fluids and brain tissue, a thorough analysis will require a combination of multiple enrichment and separation steps, and efficient detection methods. Despite its current limitations, MS-based proteomics has established itself as the leading technology for a high-throughput qualitative and quantitative analysis of protein mixtures. The rapid development of mass analyzers with higher mass resolution and superior sequencing capabilities, separation systems with unsurpassed performance will facilitate the biomarker discovery in all kinds of pathological conditions, including neuro-degenerative diseases. However, the analysis of difficult types of proteins that are present at low abundance, hydrophobic, or extensively modified, remains to be challenging and will require further development of improved analytical tools and methodologies.
Being the most commonly acquired neurodegenerative diseases, AD and PD have been the “stars” for most of the proteomics research efforts, which can be reflected from the dominating number of publications on these two diseases. However, it is equally important to study the less common types of diseases such as Pick's disease, Alexander's disease, and Creutzfeldt-Jakob disease (CJD). General changes in metabolism may be linked to altered neuronal function that is common to a variety of neurodegenerative disorders, which is probably why some protein changes are common in multiple diseases. Therefore, it is essential to identify biomarkers that not only can differentiate diseased from control samples, but also be disease-specific.
Currently, biomarker discovery efforts have focused more on discovering novel candidates than on further validating these putative biomarkers and testing their diagnostic values. This is largely due to a lack of efficient strategies for determining which candidates are worth the investment of time and research resources required for assay development and optimization. The targeted quantitative MS-based methods can serve as a crucial bridge between discovery and validation by focusing on those high-ranking candidate proteins.
It is believed that multiple candidate biomarkers can improve the accuracy of diagnosing neurodegenerative diseases, especially when they are used in a panel of diagnostic assays in the context of combining the use of neuroimaging and clinical data. Furthermore, deeper understanding of the metabolic pathways of the candidate biomarkers will greatly facilitate the interpretation of assays of individual as well as a panel of analytes. The emerging methodologies that combine affinity-enrichment, quantitative techniques and PTM-specific proteomics have begun to reveal the molecular features of complex cellular networks and will contribute to our better understanding of the detailed molecular mechanisms of neurological diseases. With the current technologies, however, the field still has many challenges to overcome to achieve the ultimate goals.
Acknowledgments
This work was supported in part by National Institutes of Health through grant AI0272588 and the Wisconsin Alumni Research Foundation at the University of Wisconsin-Madison. L.L. acknowledges an Alfred P. Sloan Research Fellowship.
References
- 1.Atkinson AJ, Colburn WA, Degruttola VG, Demets DL, Downing GJ, Hoth DF, Oates JA, Peck CC, Schooley RT, Spilker BA. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 2.Wagner JA, Williams SA, Webster CJ. Biomarkers and surrogate end points for fit-for-purpose development and regulatory evaluation of new rugs. Clin Pharmacol Ther. 2007;81:104–107. doi: 10.1038/sj.clpt.6100017. [DOI] [PubMed] [Google Scholar]
- 3.Davidsson P, Sjogren M. The use of proteomics in biomarker discovery in neurodegenerative diseases. Dis Markers. 2005;21:81–92. doi: 10.1155/2005/848676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fountoulakis M, Kossida S. Proteomics-driven progress in neurodegeneration research. Electrophoresis. 2006;27:1556–1573. doi: 10.1002/elps.200500738. [DOI] [PubMed] [Google Scholar]
- 5.Andrade EC, Krueger DD, Nairn AC. Recent advances in neuroproteomics. Curr Opin Mol Ther. 2007;9:270–281. [PMC free article] [PubMed] [Google Scholar]
- 6.Schulenborg T, Schmidt O, Van Hall A, Meyer HE, Hamacher M, Marcus K. Proteomics in neurodegeneration – disease driven approaches – Review. J Neural Transm. 2006;113:1055–1073. doi: 10.1007/s00702-006-0512-8. [DOI] [PubMed] [Google Scholar]
- 7.Drabik A, Bierczynska-Krzysik A, Bodzonkulakowska A, Suder P, Kotlinska J, Silberring J. Proteomics in neurosciences. Mass Spectrom Rev. 2007;26:432–450. doi: 10.1002/mas.20131. [DOI] [PubMed] [Google Scholar]
- 8.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 9.Carrette O, Demalte I, Scherl A, Yalkinoglu O, Corthals G, Burkhard P. Hochstrasser DF and Sanchez JC. A panel of cerebrospinal fluid potential biomarkers for the diagnosis of Alzheimer's disease. Proteomics. 2003;3:1486–1494. doi: 10.1002/pmic.200300470. [DOI] [PubMed] [Google Scholar]
- 10.Pan S, Wang Y, Quinn JF, Peskind ER, Waichunas D, Wimberger JT, Jin JH, Li JG, Zhu D, Pan C, Zhang J. Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J Proteome Res. 2006;5:2769–2779. doi: 10.1021/pr060251s. [DOI] [PubMed] [Google Scholar]
- 11.Abdi F, Quinn JF, Jankovic J, McIntosh M, Leverenz JB, Peskind E, Nixon R, Nutt J, Chung K, Zabetian C. Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J Alzheimers Dis. 2006;9:293–348. doi: 10.3233/jad-2006-9309. [DOI] [PubMed] [Google Scholar]
- 12.Puchades M, Hansson SF, Nilsson CL, Andreasen N, Blennow K, Davidsson P. Proteomic studies of potential cerebrospinal fluid protein markers for Alzheimer's disease. Mol Brain Res. 2003;118:140–146. doi: 10.1016/j.molbrainres.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Castano EM, Roher AE, Esh CL, Kokjohn TA, Beach T. Comparative proteomics of cerebrospinal fluid in neuropathologically-confirmed Alzheimer's disease and non-demented elderly subjects. Neurol Res. 2006;28:155–163. doi: 10.1179/016164106X98035. [DOI] [PubMed] [Google Scholar]
- 14.Pan S, Rush J, Peskind ER, Galasko D, Chung K, Quinn J, Jankovic J, Leverenz JB, Zabetian C, Pan C. Application of Targeted Quantitative Proteomics Analysis in Human Cerebrospinal Fluid Using a Liquid Chromatography Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Tandem Mass Spectrometer (LC MALDI TOF/TOF) Platform. J Proteome Res. 2008;7:720–730. doi: 10.1021/pr700630x. [DOI] [PubMed] [Google Scholar]
- 15.Finehout EJ, Franck Z, Choe LH, Relkin N, Lee KH. Cerebrospinal fluid proteomic biomarkers for Alzheimer's disease. Ann Neurol. 2007;61:120–129. doi: 10.1002/ana.21038. [DOI] [PubMed] [Google Scholar]
- 16.Yuan X, Desiderio DM. Proteomics analysis of human cerebrospinal fluid. J Chromatogr B. 2005;815:179–189. doi: 10.1016/j.jchromb.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 17.Carrette O, Burkhard PR, Hughes S, Hochstrasser DF, Sanchez JC. Truncated cystatin C in cerebrospiral fluid: Technical artefact or biological process? Proteomics. 2005;5:3060–3065. doi: 10.1002/pmic.200402039. [DOI] [PubMed] [Google Scholar]
- 18.Zipser BD, Johanson CE, Gonzalez L, Berzin TM, Tavares R, Hulette CM, Vitek MP. Hovanesian V and Stopa EG. Microvascular injury and blood-brain barrier leakage in Alzheimer's disease. Neurobiol Aging. 2007;28:977–986. doi: 10.1016/j.neurobiolaging.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs JM, Adkins JN, Qian WJ, Liu T, Shen YF, Camp DG, Smith RD. Utilizing human blood plasma for proteomic biomarker discovery. J Proteome Res. 2005;4:1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 20.Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics. 2006;6:6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Echan LA, Tang HY, Ali-Khan N, Lee K, Speicher DW. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005;5:3292–3303. doi: 10.1002/pmic.200401228. [DOI] [PubMed] [Google Scholar]
- 22.Lei Huang Harvie G, Feitelson JS, Gramatikoff K, Herold DA, Allen DL, Amunngama R, Hagler RA, Pisano MR, Zhang WW, Fang XM. Immunoaffinity separation of plasma proteins by IgY microbeads: Meeting the needs of proteomic sample preparation and analysis. Proteomics. 2005;5:3314–3328. doi: 10.1002/pmic.200401277. [DOI] [PubMed] [Google Scholar]
- 23.Fountoulakis M, Hardmeier R, Hoger H, Lubec G. Postmortem changes in the level of brain proteins. Exp Neurol. 2001;167:86–94. doi: 10.1006/exnr.2000.7529. [DOI] [PubMed] [Google Scholar]
- 24.Pierson J, Norris JL, Aerni HR, Svenningsson P, Caprioli RM, Andren PE. Molecular Profiling of Experimental Parkinson's Disease: Direct Analysis of Peptides and Proteins on Brain Tissue Sections by MALDI Mass Spectrometry. J Proteome Res. 2004;3:289–295. doi: 10.1021/pr0499747. [DOI] [PubMed] [Google Scholar]
- 25.Lubec G, Krapfenbauer K, Fountoulakis M. Proteomics in brain research: potentials and limitations. Prog Neurobiol. 2003;69:193–211. doi: 10.1016/s0301-0082(03)00036-4. [DOI] [PubMed] [Google Scholar]
- 26.Wolters DA, Washburn MP, Yates JR. An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 27.Delahunty CM, Yates JR. MudPIT: multidimensional protein identification technology. Biotechniques. 2007;43:563, 565, 567. [PubMed] [Google Scholar]
- 28.Graham RLJ, Sharma MK, Ternan NG, Weatherly DB, Tarleton RL, McMullan G. A semi-quantitative GeLC-MS analysis of temporal proteome expression in the emerging nosocomial pathogen. Ochrobactrum anthropi. Genome Biol. 2007;8:R110. doi: 10.1186/gb-2007-8-6-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolff S, Otto A, Albrecht D, Zeng JS, Buttner K, Gluckmann M, Hecker M, Becher R. Gel-free and gel-based proteomics in Bacillus subtilis – A comparative study. Mol Cell Proteomics. 2006;5:1183–1192. doi: 10.1074/mcp.M600069-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotech. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 31.Xiong L, Andrews D, Regnier F. Comparative proteomics of glycoproteins based on lectin selection and isotope coding. J Proteome Res. 2003;2:618–625. doi: 10.1021/pr0340274. [DOI] [PubMed] [Google Scholar]
- 32.Zougman A, Pilch B, Podtelejnikov A, Kiehntopf M, Schnabel C, Kumar C, Mann M. Integrated analysis of the cerebrospinal fluid peptidome and proteome. J Proteome Res. 2008;7:386–399. doi: 10.1021/pr070501k. [DOI] [PubMed] [Google Scholar]
- 33.Fournier ML, Gilmore JM, Martin-Brown SA, Washburn MP. Multidimensional separations-based shotgun proteomics. Chem Rev. 2007;107:3654–3686. doi: 10.1021/cr068279a. [DOI] [PubMed] [Google Scholar]
- 34.Stasyk T, Huber LA. Zooming in: Fractionation strategies in proteomics. Proteomics. 2004;4:3704–3716. doi: 10.1002/pmic.200401048. [DOI] [PubMed] [Google Scholar]
- 35.Roe MR, Griffin TJ. Gel-free mass spectrometry-based high throughput proteomics: Tools for studying biological response of proteins and proteomes. Proteomics. 2006;6:4678–4687. doi: 10.1002/pmic.200500876. [DOI] [PubMed] [Google Scholar]
- 36.Shores KS, Knapp DR. Assessment approach for evaluating high abundance protein depletion methods for cerebrospinal fluid (CSF) proteomic analysis. J Proteome Res. 2007;6:3739–3751. doi: 10.1021/pr070293w. [DOI] [PubMed] [Google Scholar]
- 37.Björhall K, Miliotis T, Davidsson P. Comparison of different depletion strategies for improved resolution in proteomic analysis of human serum samples. Proteomics. 2005;5:307–317. doi: 10.1002/pmic.200400900. [DOI] [PubMed] [Google Scholar]
- 38.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 39.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 40.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 41.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Amer Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 42.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 44.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotech. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 45.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Bondarenko PV, Chelius D, Shaler TA. Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal Chem. 2002;74:4741–4749. doi: 10.1021/ac0256991. [DOI] [PubMed] [Google Scholar]
- 47.Liu H, Sadygov RG, Yates JR. A Model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 48.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Amer Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 49.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 50.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 51.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 52.Clauser KR, Baker P, Burlingame AL. Role of accurate mass measurement (+/−10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal Chem. 1999;71:2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- 53.Creasy DM, Cottrell JS. Unimod: Protein modifications for mass spectrometry. Proteomics. 2004;4:1534–1536. doi: 10.1002/pmic.200300744. [DOI] [PubMed] [Google Scholar]
- 54.Lehmann WD, Bohne A, Von Der Lieth CW. The information encrypted in accurate peptide masses – improved protein identification and assistance in glycopeptide identification and characterization. J Mass Spectrom. 2000;35:1335–1341. doi: 10.1002/1096-9888(200011)35:11<1335::AID-JMS70>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 55.Wilkins MR, Gasteiger E, Gooley AA, Herbert BR, Molloy MP, Binz P-A, Ou K, Sanchez J-C, Bairoch A, Williams KL, Hochstrasser DF. High-throughput mass spectrometric discovery of protein post-translational modifications. J Mol Biol. 1999;289:645–657. doi: 10.1006/jmbi.1999.2794. [DOI] [PubMed] [Google Scholar]
- 56.Savitski MM, Nielsen ML, Zubarev RA. ModifiComb, a new proteomic tool for mapping substoichiometric post-translational modifications, finding novel types of modifications, and fingerprinting complex protein mixtures. Mol Cell Proteomics. 2006;5:935–948. doi: 10.1074/mcp.T500034-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Kim S, Na S, Sim JW, Park H, Jeong J, Kim H, Seo Y, Seo J, Lee KJ, Paek E. MODi : a powerful and convenient web server for identifying multiple post-translational peptide modifications from tandem mass spectra. Nucleic Acids Res. 2006;34:W258–W263. doi: 10.1093/nar/gkl245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Campii DG, Smith RD. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: The human proteome. J Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 59.Wagner M, Naik D, Pothen A, Kasukurti S, Devineni R, Adam B-L, Semmes OJ, Wright G. Computational protein biomarker prediction: a case study for prostate cancer. BMC Bioinformatics. 2004;5:26. doi: 10.1186/1471-2105-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48:1296–1304. [PubMed] [Google Scholar]
- 61.Labaer J. So, you want to look for biomarkers (introduction to the special biomarkers issue) J Proteome Res. 2005;4:1053–1059. doi: 10.1021/pr0501259. [DOI] [PubMed] [Google Scholar]
- 62.Decramer S, Wittke S, Mischak H, Zurbig P, Walden M, Bouissou F, Bascands J-L, Schanstra JP. Predicting the clinical outcome of congenital unilateral ureteropelvic junction obstruction in newborn by urinary proteome analysis. Nat Med. 2006;12:398–400. doi: 10.1038/nm1384. [DOI] [PubMed] [Google Scholar]
- 63.Thal LJ, Kantarci K, Reiman EM, Klunk WE, Weiner MW, Zetterberg H, Galasko D, Pratico D, Griffin S, Schenk D, Siemers E. The role of biomarkers in clinical trials for Alzheimer disease. Alzheimer Dis Assoc Dis. 2006;20:6–15. doi: 10.1097/01.wad.0000191420.61260.a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotech. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 65.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Stark M, Danielsson O, Griffiths WJ, Jornvall H, Johansson J. Peptide repertoire of human cerebrospinal fluid: novel proteolytic fragments of neuroendocrine proteins. J Chromatogr B. 2001;754:357–367. doi: 10.1016/s0378-4347(00)00628-9. [DOI] [PubMed] [Google Scholar]
- 67.Yuan X, Desiderio DM. Human cerebrospinal fluid peptidomics. J Mass Spectrom. 2005;40:176–181. doi: 10.1002/jms.737. [DOI] [PubMed] [Google Scholar]
- 68.Geho DH, Liotta LA, Petricoin EF, Zhao W, Araujo RP. The amplified peptidome: the new treasure chest of candidate biomarkers. Curr Opin Chem Biol. 2006;10:50–55. doi: 10.1016/j.cbpa.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 69.Lopez MF, Mikulskis A, Kuzdzal S, Bennett DA, Kelly J, Golenko E, Dicesare J, Denoyer E, Patton WF, Ediger R. High-resolution serum proteomic profiling of Alzheimer disease samples reveals disease-specific, carrier-protein-bound mass signatures. Clin Chem. 2005;51:1946–1954. doi: 10.1373/clinchem.2005.053090. [DOI] [PubMed] [Google Scholar]
- 70.McDonnell LA, Heeren RMA. Imaging mass spectrometry. Mass Spectrom Rev. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 71.Stoeckli M, Staab D, Staufenbiel M, Wiederhold K-H, Signor L. Molecular imaging of amyloid [beta] peptides in mouse brain sections using mass spectrometry. Anal Biochem. 2002;311:33–39. doi: 10.1016/s0003-2697(02)00386-x. [DOI] [PubMed] [Google Scholar]
- 72.Dekeyser SS, Kutz-Naber KK, Schmidt JJ, Barrett-Wilt GA, Li L. Imaging mass spectrometry of neuropeptides in decapod crustacean neuronal tissues. J Proteome Res. 2007;6:1782–1791. doi: 10.1021/pr060603v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Skold K, Svensson M, Nilsson A, Zhang X, Nydahl K, Caprioli RM, Svenningsson P, Andren PE. Decreased striatal levels of PEP-19 following MPTP lesion in the mouse. J Proteome Res. 2006;5:262–269. doi: 10.1021/pr050281f. [DOI] [PubMed] [Google Scholar]
- 74.Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol. 2006;7:391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- 75.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotech. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 76.Kenessey A, Yen SH. The extent of phosphorylation of fetal-Tau is comparable to that ff Phf-Tau from Alzheimer paired helical filaments. Brain Res. 1993;629:40–46. doi: 10.1016/0006-8993(93)90478-6. [DOI] [PubMed] [Google Scholar]
- 77.Gunnarsson MD, Kilander L, Basun H, Lannfelt L. Reduction of phosphorylated tau during memantine treatment of Alzheimer's disease. Dement Geriatr Cogn. 2007;24:247–252. doi: 10.1159/000107099. [DOI] [PubMed] [Google Scholar]
- 78.Wang JZ, Tung YC, Wang YP, Li XT, Iqbal K, Grundke-Iqbal I. Hyperphosphorylation and accumulation of neurofilament proteins in Alzheimer disease brain and in okadaic acid-treated SY5Y cells. FEBS Lett. 2001;507:81–87. doi: 10.1016/s0014-5793(01)02944-1. [DOI] [PubMed] [Google Scholar]
- 79.Vijayan S, El-Akkad E, Grundke-Iqbal I, Iqbal K. A pool of beta-tubulin is hyperphosphorylated at serine residues in Alzheimer disease brain. FEBS Lett. 2001;509:375–381. doi: 10.1016/s0014-5793(01)03201-x. [DOI] [PubMed] [Google Scholar]
- 80.Hampel H, Mitchell A, Blennow K, Frank RA, Brettschneider S, Weller L, Moller HJ. Core biological marker candidates of Alzheimer's disease – perspectives for diagnosis, prediction of outcome and reflection of biological activity. J Neural Trans. 2004;111:247–272. doi: 10.1007/s00702-003-0065-z. [DOI] [PubMed] [Google Scholar]
- 81.Hampel H, Buerger K, Kohnken R, Teipel SJ, Zinkowski R, Moeller HJ, Rapoport SI, Davies P. Tracking of Alzheimer's disease progression with cerebrospinal fluid tau protein phosphorylated at threonine 231. Ann Neurol. 2001;49:545–546. [PubMed] [Google Scholar]
- 82.Hampel H, Buerger K, Zinkowski R, Teipel SJ, Goernitz A, Andreasen N, Sjoegren M, Debernardis J, Kerkman D, Ishiguro K, Ohno H, Vanmechelen E, Vanderstichele H, McCulloch C, Moller HJ, Davies P, Blennow K. Measurement of phosphorylated tau epitopes in the differential diagnosis of Alzheimer disease – A comparative cerebrospinal fluid study. Arch Gen Psychiat. 2004;61:95–102. doi: 10.1001/archpsyc.61.1.95. [DOI] [PubMed] [Google Scholar]
- 83.Hu YY, He SS, Wang XC, Duan QH, Grundkeiqbal I, Iqbal K, Wang JZ. Levels of nonphosphorylated and phosphorylated tau in cerebrospinal fluid of Alzheimer's disease patients – An ultrasensitive bienzyme-substrate-recycle enzyme-linked Immunosorbent assay. Am J Pathol. 2002;160:1269–1278. doi: 10.1016/S0002-9440(10)62554-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Buerger K, Zinkowski R, Teipel SJ, Tapiola T, Arai H, Blennow K, Andreasen N, Hofmannkiefer K, Debernardis J, Kerkman D, McCulloch C, Kohnken R, Padberg F, Pirttila T, Schapiro MB, Rapoport SI, Moller HI, Davies P, Hampel H. Differential diagnosis of Alzheimer disease with cerebrospinal fluid levels of tau protein phosphorylated at threonine 231. Arch Neurol. 2002;59:1267–1272. doi: 10.1001/archneur.59.8.1267. [DOI] [PubMed] [Google Scholar]
- 85.Hanger DP, Betts JC, Loviny TLF, Blackstock WP, Anderton BH. New phosphorylation sites identified in hyperphosphorylated tau (paired helical filament-tau) from Alzheimer's disease brain using nanoelectrospray mass spectrometry. J Neurochem. 1998;71:2465–2476. doi: 10.1046/j.1471-4159.1998.71062465.x. [DOI] [PubMed] [Google Scholar]
- 86.Derkinderen P, Scales TME, Hanger DP, Leung K-Y, Byers HL, Ward MA, Lenz C, Price C, Bird IN, Perera T, Kellie S, Williamson R, Noble W, Van Etten RA, Leroy K, Brion JP, Reynolds CH, Anderton BH. Tyrosine 394 is phosphorylated in Alzheimer's paired helical filament Tau and in fetal Tau with c-Abl as the candidate tyrosine kinase. J Neurosci. 2005;25:6584–6593. doi: 10.1523/JNEUROSCI.1487-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reynolds CH, Betts JC, Blackstock WP, Nebreda AR, Anderton BH. Phosphorylation sites on Tau identified by nanoelectrospray mass spectrometry. differences in vitro between the mitogen-activated protein kinases ERK2, c-Jun N-terminal kinase and P38, and glycogen synthase kinase-3 beta. J Neurochem. 2000;74:1587–1595. doi: 10.1046/j.1471-4159.2000.0741587.x. [DOI] [PubMed] [Google Scholar]
- 88.Posewitz MC, Tempst P. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal Chem. 1999;71:2883–2892. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 89.Barnouin KN, Hart SR, Thompson AJ, Okuyama M, Waterfield M, Cramer R. Enhanced phosphopeptide isolation by Fe(III)-IMAC using 1,1,1,3,3,3-hexafluoroisopropanol. Proteomics. 2005;5:4376–4388. doi: 10.1002/pmic.200401323. [DOI] [PubMed] [Google Scholar]
- 90.Pinkse MWH, Mohammed S, Gouw LW, van Breukelen B, Vos HR, Heck AJR. Highly robust, automated, and sensitive on line TiO2-based phosphoproteomics applied to study endogenous phosphorylation in Drosophila melanogaster. J Proteome Res. 2008;7:687–697. doi: 10.1021/pr700605z. [DOI] [PubMed] [Google Scholar]
- 91.Kweon HK, Hakansson K. Metal oxide-based enrichment combined with gas-phase ion-electron reactions for improved mass spectrometric characterization of protein phosphorylation. J Proteome Res. 2008;7:749–755. doi: 10.1021/pr070386d. [DOI] [PubMed] [Google Scholar]
- 92.Trinidad JC, Specht CG, Thalhammer A, Schoepfer R, Burlingame AL. Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol Cell Proteomics. 2006;5:914–922. doi: 10.1074/mcp.T500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 93.Collins MO, Yu L, Coba MP, Husi H, Campuzano L, Blackstock WP, Choudhary JS, Grant SGN. Proteomic analysis of in vivo phosphorylated synaptic proteins. J Biol Chem. 2005;280:5972–5982. doi: 10.1074/jbc.M411220200. [DOI] [PubMed] [Google Scholar]
- 94.Liu F, Zaidi T, Iqbal K, Grundke-Iqbal I, Gong CX. Aberrant glycosylation modulates phosphorylation of tau by protein kinase A and dephosphorylation of tau by protein phosphatase 2A and 5. Neuroscience. 2002;115:829–837. doi: 10.1016/s0306-4522(02)00510-9. [DOI] [PubMed] [Google Scholar]
- 95.Liu F, Iqbal K, Grundke-Iqbal I, Hart GW, Gong C-X. O-GlcNAcylation regulates phosphorylation of tau: A mechanism involved in Alzheimer's disease. Proc Natl Acad Sci USA. 2004;101:10804–10809. doi: 10.1073/pnas.0400348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saez-Valero J, Fodero LR, Sjogren M, Andreasen N, Amici S, Gallai V, Vanderstichele H, Vanmechelen E, Parnetti L, Blennow K, Small DH. Glycosylation of acetyl-cholinesterase and butyrylcholinesterase changes as a function of the duration of Alzheimer's disease. J Neurosci Res. 2003;72:520–526. doi: 10.1002/jnr.10599. [DOI] [PubMed] [Google Scholar]
- 97.Yang D-S, Tandon A, Chen F, Yu G, Yu H, Arawaka S, Hasegawa H, Duthie M, Schmidt SD, Ramabhadran TV. Mature glycosylation and trafficking of nicastrin modulate its binding to presenilins. J Biol Chem. 2002;277:28135–28142. doi: 10.1074/jbc.M110871200. [DOI] [PubMed] [Google Scholar]
- 98.Charlwood J, Dingwall C, Matico R, Hussain I, Johanson K, Moore S, Powell DJ, Skehel JM, Ratcliffe S, Clarke B. Characterization of the glycosylation profiles of Alzheimer's beta -secretase protein Asp-2 expressed in a variety of cell lines. J Biol Chem. 2001;276:16739–16748. doi: 10.1074/jbc.M009361200. [DOI] [PubMed] [Google Scholar]
- 99.Botella-Lopez A, Burgaya F, Gavin R, Garcia-Ayllon MS, Gomez-Tortosa E, Pena-Casanova J, Urena JM, Del Rio JA, Blesa R, Soriano E, Saez-Valero J. Reelin expression and glycosylation patterns are altered in Alzheimer's disease. Proc Natl Acad Sci USA. 2006;103:5573–5578. doi: 10.1073/pnas.0601279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K-I, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotech. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 101.Cummings RD, Kornfeld S. Fractionation of asparagine-linked oligosaccharides by serial lectin-agarose affinity chromatography. A rapid, sensitive, and specific technique. J Biol Chem. 1982;257:11235–11240. [PubMed] [Google Scholar]
- 102.Qiu R, Regnier FE. Use of Multidimensional lectin affinity chromatography in differential glycoproteomics. Anal Chem. 2005;77:2802–2809. doi: 10.1021/ac048751x. [DOI] [PubMed] [Google Scholar]
- 103.Zhang H, Li X-J, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotech. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 104.Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 105.Zubarev RA. Electron-capture dissociation tandem mass spectrometry. Curr Opin Biotechnol. 2004;15:12–16. doi: 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 106.Mariani E, Polidori MC, Cherubini A, Mecocci P. Oxidative stress in brain aging, neurodegenerative and vascular diseases: An overview. J Chromatogr B. 2005;827:65–75. doi: 10.1016/j.jchromb.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 107.Giasson BI, Ischiropoulos H, Lee VMY, Trojanowski JQ. The relationship between oxidative/nitrative stress and pathological inclusions in Alzheimer's and Parkinson's diseases. Free Radical Bio Med. 2002;32:1264–1275. doi: 10.1016/s0891-5849(02)00804-3. [DOI] [PubMed] [Google Scholar]
- 108.Korolainen MA, Goldsteins G, Alafuzoff I, Koistinaho J, Pirttila T. Proteomic analysis of protein oxidation in Alzheimer's disease brain. Electrophoresis. 2002;23:3428–3433. doi: 10.1002/1522-2683(200210)23:19<3428::AID-ELPS3428>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 109.Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, Markesbery WR. Protein oxidation in the brain in Alzheimer's disease. Neuroscience. 2001;103:373–383. doi: 10.1016/s0306-4522(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 110.Castegna A, Aksenov M, Aksenova M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part 1: Creatine kinase bb, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radical Bio Med. 2002;33:562–571. doi: 10.1016/s0891-5849(02)00914-0. [DOI] [PubMed] [Google Scholar]
- 111.Castegna A, Aksenov M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part II: dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- 112.Soreghan BA, Yang F, Thomas SN, Hsu J, Yang AJ. High-throughput proteomic-based identification of oxidatively induced protein carbonylation in mouse brain. Pharm Res. 2003;20:1713–1720. doi: 10.1023/b:pham.0000003366.25263.78. [DOI] [PubMed] [Google Scholar]
- 113.Soreghan BA, Lu BW, Thomas SN, Duff K, Rakhmatulin EA, Nikolskaya T, Chen T, Yang AJ. Using proteomics and network analysis to elucidate the consequences of synaptic protein oxidation in a PS1+A beta PP mouse model of Alzheimer's disease. J Alzheimers Dis. 2005;8:227–241. doi: 10.3233/jad-2005-8302. [DOI] [PubMed] [Google Scholar]
- 114.Poon HF, Farr SA, Banks WA, Pierce WM, Klein JB, Morley JE, Butterfield DA. Proteomic identification of less oxidized brain proteins in aged senescence-accelerated mice following administration of antisense oligonucleotide directed at the A[beta] region of amyloid precursor protein. Mol Brain Res. 2005;138:8–16. doi: 10.1016/j.molbrainres.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 115.Reynolds MR, Berry RW, Binder LI. Nitration in neurodegeneration: Deciphering the “Hows” “nYs”. Biochemistry. 2007;46:7325–7336. doi: 10.1021/bi700430y. [DOI] [PubMed] [Google Scholar]
- 116.Danielson SR, Andersen JK. Oxidative and nitrative protein modifications in Parkinson's disease. Free Radical Bio Med. 2008;44:1787–1794. doi: 10.1016/j.freeradbiomed.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sacksteder CA, Qian WJ, Knyushko TV, Wang H, Chin MH, Lacan G, Melega WP, Camp DG, Smith RD, Smith DJ. Endogenously nitrated proteins in mouse brain: links to neurodegenerative disease. Biochemistry. 2006;45:8009–8022. doi: 10.1021/bi060474w. [DOI] [PubMed] [Google Scholar]
- 118.Zhang Q, Qian WJ, Knyushko TV, Clauss TRW, Purvine SO, Moore RJ, Sacksteder CA, Chin MH, Smith DJ, Camp DG. A method for selective enrichment and analysis of nitrotyrosine-containing peptides in complex proteome samples. J Proteome Res. 2007;6:2257–2268. doi: 10.1021/pr0606934. [DOI] [PubMed] [Google Scholar]
- 119.Wang GP, Khatoon S, Iqbal K, Grundkeiqbal I. Brain ubiquitin is markedly elevated in Alzheimer disease. Brain Res. 1991;566:146–151. doi: 10.1016/0006-8993(91)91692-t. [DOI] [PubMed] [Google Scholar]
- 120.Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer's disease. J Neurochem. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- 121.Wang D, Cotter RJ. Approach for determining protein ubiquitination sites by MALDI-TOF mass spectrometry. Anal Chem. 2005;77:1458–1466. doi: 10.1021/ac048834d. [DOI] [PubMed] [Google Scholar]
- 122.Kirkpatrick DS, Denison C, Gygi SP. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dorval V, Fraser PE. SUMO on the road to neurodegeneration. Biochimica et Biophysica Acta (BBA) – Mol Cell Res. 2007;1773:694–706. doi: 10.1016/j.bbamcr.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 124.Li T, Evdokimov E, Shen R-F, Chao C-C, Tekle E, Wang T, Stadtman ER, Yang DCH, Chock PB. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: A proteomic analysis. Proc Natl Acad Sci USA. 2004;101:8551–8556. doi: 10.1073/pnas.0402889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cooper HJ, Tatham MH, Jaffray E, Heath JK, Lam TT, Marshall AG, Hay RT. Fourier transform ion cyclotron resonance mass spectrometry for the analysis of small ubiquitin-like modifier (SUMO) modification: identification of lysines in RanBP2 and SUMO targeted for modification during the E3 autoSUMOylation reaction. Anal Chem. 2005;77:6310–6319. doi: 10.1021/ac058019d. [DOI] [PubMed] [Google Scholar]
- 126.Jones J, Wu K, Yang Y, Guerrero C, Nillegoda N, Pan Z-Q, Huang L. A targeted proteomic analysis of the ubiquitin-like modifier Nedd8 and associated proteins. J Proteome Res. 2008;7:1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smith MA, Taneda S, Richey PL, Miyata S, Yan SD, Stern D, Sayre LM, Monnier VM, Perry G. Advanced maillard reaction end-products are associated with Alzheimer-disease pathology. Proc Natl Acad Sci USA. 1994;91:5710–5714. doi: 10.1073/pnas.91.12.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vitek MP, Bhattacharya K, Glendening JM, Stopa E, Vlassara H, Bucala R, Manogue K, Cerami A. Advanced glycation end-products contribute to amyloidosis in Alzheimer-disease. Proc Natl Acad Sci USA. 1994;91:4766–4770. doi: 10.1073/pnas.91.11.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yan SD, Chen X, Schmidt AM, Brett J, Godman G, Zou YS, Scott CW, Caputo C, Frappier T, Smith MA. Glycated Tau-protein in Alzheimer-disease – A mechanism for induction of oxidant stress. Proc Natl Acad Sci USA. 1994;91:7787–7791. doi: 10.1073/pnas.91.16.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Takeuchi M, Sato T, Takino JI, Kobayashi Y, Furuno S, Kikuchi S, Yamagishi SI. Diagnostic utility of serum or cerebrospinal fluid levels of toxic advanced glycation end-products (TAGE) in early detection of Alzheimer's disease. Med Hypotheses. 2007;69:1358–1366. doi: 10.1016/j.mehy.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 131.Lapolla A, Fedele D, Seraglia R, Traldi P. The role of mass spectrometry in the study of non-enzymatic protein glycation in diabetes: An update. Mass Spectrom Rev. 2006;25:775–797. doi: 10.1002/mas.20090. [DOI] [PubMed] [Google Scholar]
- 132.Gadgil HS, Bondarenko PV, Treuheit MJ, Ren D. Screening and sequencing of glycated proteins by neutral loss scan LC/MS/MS method. Anal Chem. 2007;79:5991–5999. doi: 10.1021/ac070619k. [DOI] [PubMed] [Google Scholar]